Abstract
The CRISPR-Cas9 system has transformed genome engineering of model organisms from possible to practical. CRISPR-Cas9 can be readily programmed to generate sequence-specific double-strand breaks that disrupt targeted loci when repaired by error-prone non-homologous end joining or to catalyze precise genome modification through homology-directed repair (HDR). Here we describe a streamlined approach for rapid and highly efficient engineering of the Drosophila genome via CRISPR-Cas9-mediated HDR. In this approach, transgenic flies expressing Cas9 are injected with plasmids to express guide RNAs (gRNAs) and positively marked donor templates. We detail target site selection; gRNA plasmid generation; donor template design and construction; and the generation, identification and molecular confirmation of engineered lines. We also present alternative approaches and highlight key considerations for experimental design. The approach outlined here can be used to rapidly and reliably generate a variety of engineered modifications, including genomic deletions and replacements, precise sequence edits, and incorporation of protein tags.
Keywords: CRISPR, Cas9, homology directed repair, genome engineering, Drosophila
INTRODUCTION
The CRISPR-Cas9 system is significantly advancing the ability of researchers to engineer targeted genome modifications for functional studies of genes and genetic elements. In Drosophila, the CRISPR-Cas9 system has been used to disrupt, delete, replace, tag and edit multiple genes and genetic elements (Bassett et al., 2013; Gratz et al., 2013a; Gratz et al., 2014; Kondo and Ueda, 2013; Lee et al., 2014; Port et al., 2014; Ren et al., 2013; Sebo et al., 2014; Xue et al., 2014; Yu et al., 2014; Yu et al., 2013). The rapid and widespread adoption of CRISPR-Cas9 illustrates the utility of this novel genome engineering platform for generating a wide variety of modifications, and its power for addressing fundamental biological questions, understanding and treating disease, and engineering agriculturally relevant species and their pests.
Endogenous CRISPR-Cas9 systems have been adapted as simple and highly robust genome engineering tools that are being widely adopted by the research community. The most widely used Streptococcus pyogenes system was simplified to two components to facilitate genome engineering: a common endonuclease called Cas9 and a single chimeric RNA referred to as a guide RNA (gRNA) (Jinek et al., 2012). gRNAs interact with Cas9 and guide the nuclease to specific DNA sequences through an easily programmed 20-nt target sequence that directly base pairs with complementary DNA. Upon binding its target, Cas9 utilizes its two nuclease domains to generate a double-strand break (DSB). The only known requirement for a potential cleavage site is the presence of a 3-bp protospacer adjacent motif (PAM) of the form NGG immediately 3′ of the 20-nt target sequence. Thus, S. pyogenes CRISPR-Cas9 target sites occur an average of once in every eight basepairs of genomic sequence.
Induction of a DSB in genomic DNA triggers repair by one of two general cellular repair pathways, both of which can be co-opted for genome engineering. Non-homologous end-joining (NHEJ) is an error-prone process in which broken ends are simply ligated together. This repair pathway can yield small insertions and deletions (indels) that disrupt function at cleavage sites. In contrast, homology-directed repair (HDR) employs homologous DNA sequences as templates for precise repair. By supplying donor templates comprising exogenous sequence flanked by homology-containing stretches (commonly referred to as homology arms), the HDR pathway can be appropriated to make precise modifications including defined deletions, sequence substitutions, or insertions. Beyond the genome engineering applications of the CRISPR-Cas9 system, nuclease-dead Cas9 is being used as a sequence-specific repressor or activator of gene expression and is being developed as a tool for probing genome structure and function without causing mutations (Anton et al., 2014; Bikard et al., 2013; Chen et al., 2013; Cheng et al., 2013; Fujita and Fujii, 2013; Gilbert et al., 2013; Kearns et al., 2014; Maeder et al., 2013; Perez-Pinera et al., 2013; Qi et al., 2013)
Here we detail a rapid and efficient CRISPR-Cas9 method for HDR-mediated engineering of the Drosophila genome (Gratz et al., 2013a; Gratz et al., 2014). We have used this approach to generate numerous genome modifications, including gene replacements, in-frame protein tag insertions, and conditional alleles. The Basic Protocol covers target site selection; gRNA generation; donor design and construction; and the generation, identification and molecular confirmation of engineered lines. We begin with key considerations for experimental design and discuss alternative approaches.
STRATEGIC PLANNING
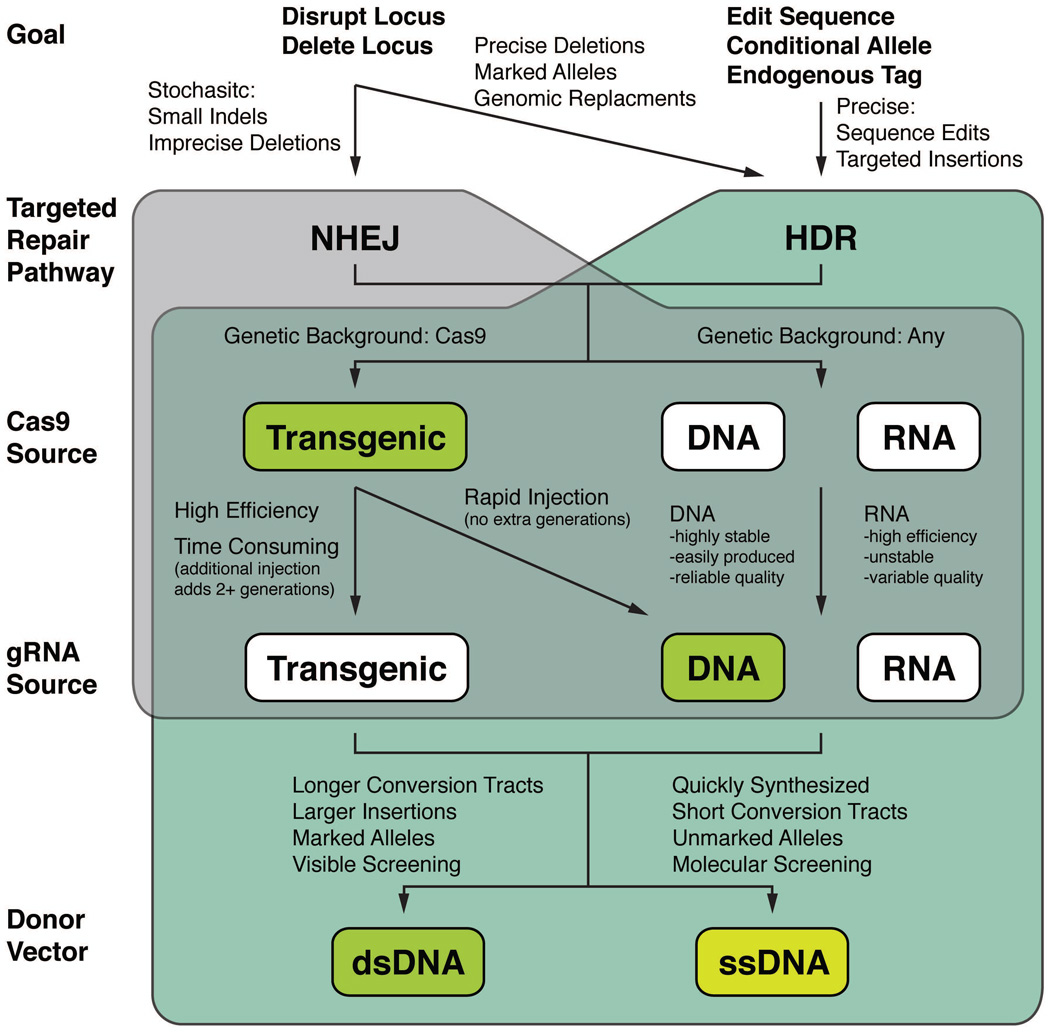
Figure 1 shows a decision tree that can be used as a guide in designing the appropriate strategy for different types of CRISPR-Cas9 genome engineering experiments. Here we discuss the key considerations for each decision point.
-
What do you want to achieve?
Loss-of-function (lof) allele: If your goal is to generate a lof allele, an approach that relies on either NHEJ or HDR can be employed to achieve your aim. The key difference in practice is whether a donor repair template is included. If you choose to go with NHEJ (no donor), you will have two options: (1) disrupt the locus by targeting a single cleavage event in a critical sequence and recovering disruptive indels or (2) delete the locus with two flanking gRNAs. You will need to screen candidate mutants using an appropriate molecular approach unless you can screen phenotypically for the desired mutant. The simplest way to identify relatively small indels is by using high-resolution melt analysis (HRMA), while PCR can be used to detect larger deletions (Bassett et al., 2013; Gratz et al., 2013a). Following sequence verification, you will have an unmarked lof allele.
Complex modifications: To engineer defined modifications, such as specific changes to a nucleotide sequence or insertion of a tag or other exogenous DNA (e.g. FRT sites for a conditional allele), you will need to employ the HDR pathway by supplying a donor repair template and screening for the desired repair event. This pathway can also be employed to generate defined deletions or to insert screenable marker genes that facilitate recovery of engineered flies. Subsequently, markers that have been flanked by LoxP or FRT sites can be readily removed to minimize alterations to the engineered locus or retained to provide a marked allele.
-
Irrespective of whether your goal is to employ the NHEJ or HDR pathway, you will need to decide how you will introduce Cas9.
Transgenic Cas9 source: Unless your experimental goals limit you to a specific genetic strain, we recommend using a transgenic Cas9 source for the highest efficiency and reliability. We have generated transgenic fly lines expressing Cas9 under the control of the germline vasa promoter. These and related fly lines are available at the Bloomington Drosophila Stock Center (Gratz et al., 2014; Port et al., 2014; Sebo et al., 2014).
Injection of Cas9 as DNA or RNA: If you wish to work in a particular genetic background (or non-melanogaster species), you can introduce Cas9 as either plasmid DNA or mRNA through injection. RNA-based approaches can generate targeted mutations at higher efficiency than the DNA constructs attempted to date, but yield somewhat more experiment-to-experiment variability, possibly due to complications encountered with the injection of RNA (Bassett et al., 2013; Yu et al., 2013). A disadvantage in using RNA is the relatively high cost compared to DNA, which is inexpensive to generate and easy to handle. The primary drawback of injected Cas9 DNA is that, while it appears to work quite reliably, it does so with lower efficiency than Cas9 mRNA (Gratz et al., 2013a; Ren et al., 2013). Optimized promoters are likely to mitigate this difference. In some cases, such as attempts to generate recessive lethal alleles, the lower efficiency observed with DNA constructs may provide an advantage by reducing the levels of biallelic targeting.
-
All CRISPR experiments require a gRNA to guide Cas9 to the targeted locus. As with Cas9, gRNAs can be supplied transgenically or injected as DNA or RNA.
Transgenic gRNA source: gRNAs can be integrated into the genome using phiC31-mediated transgenesis (Kondo and Ueda, 2013; Port et al., 2014; Xue et al., 2014). This approach has the advantage of catalyzing extremely efficient NHEJ – and likely HDR, though to date only one such experiment has been published (Port et al., 2014). The disadvantage of an integrated gRNA approach is the time and cost of generating a unique gRNA-expressing transgenic fly line for each targeting experiment. Integrating a gRNA more than doubles the timeline for obtaining an engineered fly line and costs several hundred dollars if injections are outsourced, as is commonly the case. However, given the advantage of increased efficacy, this approach is particularly useful if a gene or genomic sequence will be manipulated frequently.
Injection of gRNA as DNA or RNA: Injection of gRNAs, as either DNA or RNA, is the most rapid method for engineering flies and both are highly efficient, particularly when combined with transgenic sources of Cas9 (Gratz et al., 2014; Ren et al., 2013; Xue et al., 2014; Yu et al., 2014).
-
For HDR, but not NHEJ, you will need to supply a donor repair template. Both dsDNA and single-stranded DNA (ssDNA) donors have been used successfully in Drosophila.
dsDNA donor: dsDNA donors are highly versatile as they can incorporate large DNA sequences (Gratz et al., 2014; Port et al., 2014; Xue et al., 2014; Yu et al., 2014). We have generated vectors for rapidly cloning locus-specific homology arms into donors with visible markers. The pHD-DsRed-attP and pHD-DsRed vectors, described in detail below, are available from Addgene.
ssDNA donor: ssDNA donor templates can be used to incorporate small modifications (Gratz et al., 2013a; Port et al., 2014; Xue et al., 2014). The primary advantage of using ssDNA donors is that they can be rapidly synthesized, obviating the need for cloning. However, most synthesis companies have a size limit of ~200 nt for single-stranded DNA synthesis, so they cannot be used for larger modifications such as the incorporation of fluorescent tags or for the inclusion of a selection marker for identifying engineered flies. Thus, molecular screening is required, increasing the labor necessary to identify and recover the intended allele.
Figure 1.
Strategic planning flowchart. The options outlined in this protocol are indicated by green boxes. See text (Strategic Planning) for a detailed discussion of each choice point, including the advantages and disadvantages of each strategy.
In the Basic Protocol below, we detail our preferred method for efficient generation of engineered flies via HDR: injection of vasa-Cas9 flies with gRNA plasmids and a positively marked dsDNA donor template. This choice represents a favorable balance of time, cost, efficiency, and reliability. With this approach, we obtain engineered alleles within one month at a total reagent cost of approximately $150. Injections generally cost an additional $200 if outsourced. In our experience, an average of 25% (range = 7–42%) of fertile injected flies transmit the targeted event to their progeny. In Alternate Protocol 1, we detail HDR with ssDNA donor templates. Alternate Protocol 2 covers HDR in other genetic backgrounds using Cas9 supplied as DNA. In Alternate Protocol 3, we outline our approach for NHEJ using a transgenic source of Cas9 and gRNA supplied as DNA. This approach has also been used successfully by Ren et al. (2013). Together these protocols offer a versatile toolset amenable for generating a variety of genome modifications in Drosophila.
BASIC PROTOCOL
Target Site Selection
Selection of high-quality target sites is essential for the success of any CRISPR-based genome engineering experiment. It is important to identify target sites that will generate DSBs close to the location of the intended modification. In choosing a target site, location must be balanced with target-site specificity and, thus, the potential for off-target DSBs. While originally raised as a significant concern in the editing of transformed cell lines (Fu et al., 2013), with careful target site selection, off-target cleavage does not seem to be a significant problem for genome editing of organisms or human stem cells (Bassett et al., 2013; Chiu et al., 2013; Duan et al., 2014; Gratz et al., 2013a; Gratz et al., 2014; Kiskinis et al., 2014; Smith et al., 2014; Suzuki et al., 2014; Veres et al., 2014; Yang et al., 2013). Nonetheless, our current understanding of Cas9-induced cleavage is far from complete, so it is important to select the most specific sites possible to minimize the potential for off-target mutagenesis. To facilitate the rapid identification of high-quality target sites, we have developed a web-based tool, CRISPR Optimal Target Finder, that identifies gRNA cleavage sites and evaluates their specificity (Gratz et al., 2014).
It is essential that target sites be identified in sequence obtained from the fly strain that will be edited, not the reference genome. Polymorphisms between a given fly strain and the reference genome are frequent, especially in intergenic regions, and could eliminate or significantly decrease cleavage if they occur within your target sequence. Thus, CRISPR Optimal Target Finder identifies gRNA target sites in user-supplied DNA sequence rather than reference genome sequences. In the Basic Protocol, we use vasa-Cas9 flies. However, as described in Alternate Protocol 2 below, our approach can be readily adapted to engineer any fly strain.
Materials
- vasa-Cas9 fly stocks (Bloomington Drosophila Stock Center)
- y1 M{vas-Cas9.RFP-}ZH-2A w1118/FM7a, P{Tb1}FM7-A (stock number 55821)
- w1118; PBac{vas-Cas9}VK00037/CyO, P{Tb1}CprCyO-A (stock number 56552)
- w1118; PBac{vas-Cas9}VK00027 (stock number 51324)
Total DNA purification kit
PCR and sequencing primers
Phusion High-Fidelity DNA Polymerase
Gel extraction kit
Protocol steps
Isolate genomic DNA from the Cas9 (or other) fly strain in which the genome modifications will be made. Purify total DNA from about 50 adult flies. This large genomic DNA preparation can be used for many subsequent CRISPR experiments.
- Design PCR primers to amplify a region of about 500-1,000-bp centered around the target region.
- Only one gRNA is necessary to catalyze HDR. We often design our experiments to include two gRNAs targeting either end of the region to be modified (See Figure 3). However, the effect of one vs. two gRNAs, if any, on efficiency or choice of cellular repair pathway is not known. Two gRNAs increase the likelihood that at least one gRNA will efficiently induce cleavage. On the other hand, two gRNAs increase the potential of off-target cleavage.
Prepare a 50-µl reaction to amplify the target region.
Use gel electrophoresis to purify the PCR product.
Sequence the PCR product using Sanger sequencing.
Identify CRISPR target sites in the sequenced region. Submit the sequence returned from step 5 to the CRISPR Optimal Target Finder (tools.flycrispr.molbio.wisc.edu/targetFinder).
- Select the genome release you wish to search. Target Finder is currently configured to search D. melanogaster release 5 by default.
- Release 6 is also available, as are genomes for D. simulans, D. yakuba, D. mauritiana, D. sechellia, D. ananassae, D. erecta, D. persimlis, D. pseudoobscura, D. virilis, D. mojavensis, D. willistoni, D. grimshawi, Anopheles gambiae (strains M and S), Aedes aegypti, Apis mellifera, Tribolium castaneum (release 2 and draft release 4), and C. elegans.
Select the length of the target sites you would like to identify. Target Finder will identify all target sequences of the selected length in the 500–1000 bp sequenced region from step 2. Full-length target sites of 20-nt offer the highest cleavage efficiency and are the most commonly used. Target sequences of a shorter length have been shown to increase the specificity of Cas9 cleavage while decreasing cleavage efficiency (Fu et al., 2014).
- Target Finder will identify all CRISPR target sequences in the input sequence. If you would like to limit identified target sites to those that start with a G (for efficient expression from the U6 promoter when supplying gRNA as plasmid DNA) or those that start with GG (for efficient expression from the T7 promoter using in vitro transcription of gRNAs), select the appropriate option.
- Alternatively, a G can simply be added to the 5′ end of any target sequence when cloning the gRNA plasmid (see below) for efficient U6 transcription.
-
Once CRISPR targets are identified, their specificity will be evaluated based on user-selected criteria. The final 12 nt of the CRISPR target sequence, often referred to as the ‘seed’ sequence, are more critical for specificity than the distal eight nucleotides. The CRISPR Optimal Target Finder algorithms consider both the number and location of mismatches in the evaluation of potential off-target cleavage sites.
High-stringency (default setting) defines potential off-target sites as those with (i) perfect matches (zero mismatches) to the seed sequence or (ii) one mismatch in the seed sequence and one or zero mismatches in the distal sequence.
Maximum stringency defines potential off-target as sites with (i) perfect matches (zero mismatches) to the seed sequence, (ii) one mismatch in the seed sequence and four or fewer mismatches in the distal sequence, or (iii) two mismatches in the seed sequence and a maximum of one mismatch in the distal sequence.
PAM: By default, the program will only consider sequences adjacent to a canonical NGG PAM in the evaluation of potential off-target cleavage sites. Putative off-target sites adjacent to a non-canonical PAM sequence of the form NAG can be considered by selecting the ‘NGG and NAG’ option. In transformed cell lines, target sites adjacent to an NAG PAM were cleaved at one-fifth the efficiency of those adjacent to a canonical NGG PAM sequence (Hsu et al., 2013).
We recommend use of the default settings for most applications in Drosophila where little off-target cleavage has been observed to date (Bassett et al., 2013; Gratz et al., 2013a; Gratz et al., 2014).
Select specific target site(s) for your genome engineering project by balancing proximity to the site you are editing and the potential for off-target cleavage. Target Finder returns all identified target sites in order of specificity. For each target site, the program also provides oligonucleotide sequences designed for gRNA plasmid cloning into a pU6-BbsI-gRNA vector.
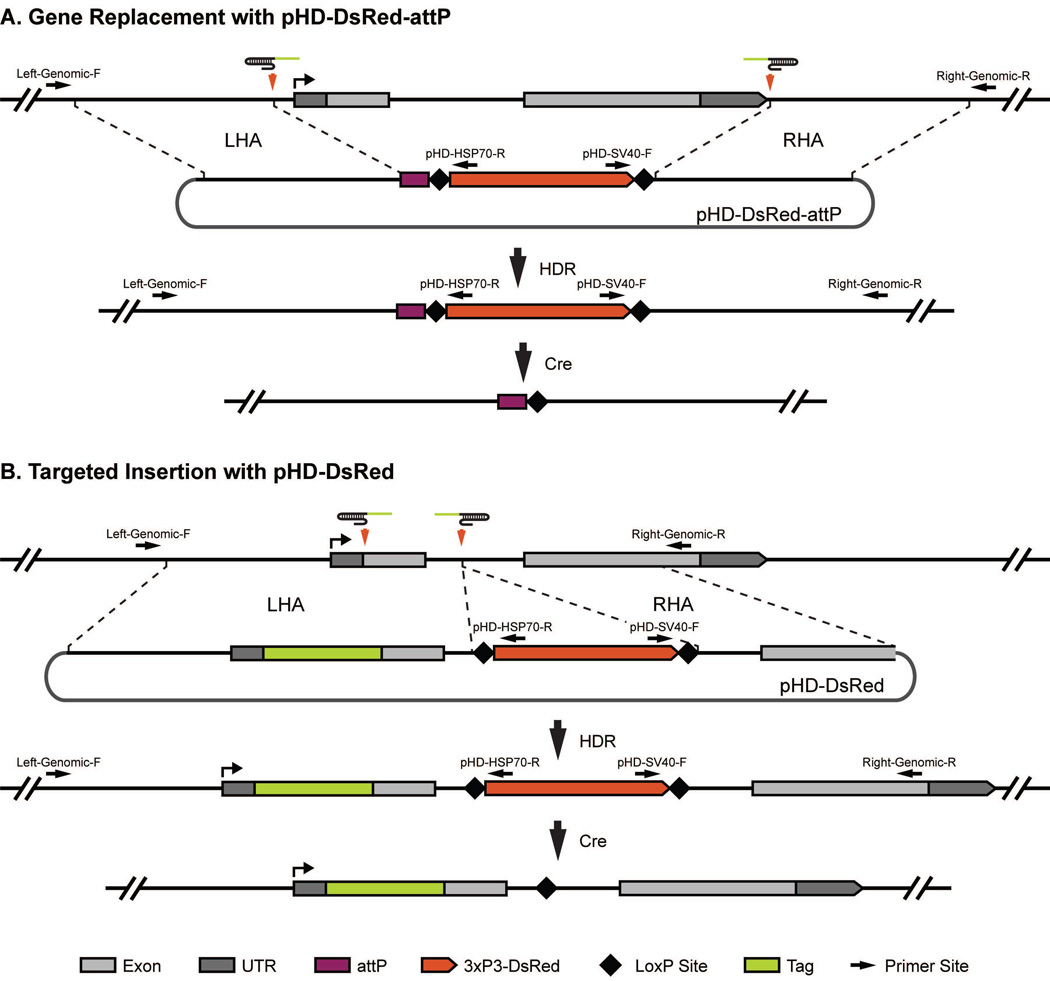
Figure 3.
Donor construct design. The (A) pHD-DsRed-attP vector and (B) pHD-DsRed donor vectors and their typical uses are depicted. Both vectors contain a removable 3xP3-dsRed marker flanked by LoxP sites and two multiple-cloning sites for insertion of the left (LHA) and right (RHA) homology arms. pHD-DsRed-attP also contains the recombination-based docking site attP. (A) In the case of replacing a locus using the pHD-DsRed-attP vector, two target sites flanking the region to be replaced are chosen. Homologous sequences immediately flanking the cleavage sites should be cloned into the MCSs. Upon Cas9-mediated cleavage and HDR, the region between the two gRNA cut sites is replaced with the attP site and removable DsRed marker. Using Cre recombinase, the DsRed marker can be removed leaving only the attP docking site and a single LoxP site. (B) In the case of tagging a gene using the pHD-DsRed vector, select a target site close to the tag insertion site and another target site in a nearby intron where the DsRed marker will be placed. Homology arms will include sequences immediately flanking the cleavage sites. In addition, one of the homology arms will contain the in-frame tag and sequences between the tag and the DsRed marker. Upon Cas9 cleavage and HDR, the untagged region is replaced with a tagged region and a visible 3xP3-DsRed marker. Using Cre recombinase, the DsRed marker can be removed leaving only the tagged coding sequence and a single LoxP site. Black arrows indicate the primer binding sites used for molecular characterization for candidate alleles. Note that the two locus specific primers are in the genomic region outside of the homology regions used in the donor vector.
gRNA Plasmid Preparation
To supply gRNAs containing the target-specific sequences from a plasmid DNA source, we have generated vectors for rapid cloning of target-specific sequences using short complementary oligonucleotides and a simple annealing and ligation process. The pU6-BbsI-gRNA vectors utilize the small RNA promoter of a Drosophila U6 gene to express the gRNA.
Materials
pU6-BbsI-gRNA (Addgene; Plasmid 45946)
BbsI endonuclease
Sense and antisense gRNA oligonucleotides
T4 Polynucleotide Kinase
T4 DNA Ligase
Gel extraction kit
E. coli DH5α or other suitable cloning strain
Plasmid miniprep kit
- Sequencing Primers
- T7 (5′-TAATACGACTCACTATAGGG-3′)
- T3 (5′- AATTAACCCTCACTAAAGGG-3′)
Endotoxin-Free Plasmid Midi Kit
-
12.Order a pair of short complementary oligonucleotides for each target site. You can design them yourself using the following guidelines. The oligonucleotide design incorporates the target sequence and cohesive ends for cloning into the pU6-BbsI-gRNA backbone. The top strand should be designed in the format of 5′-CTTCG(N)19-3′, where G(N)19 corresponds to your unique target site sequence beginning with a G for efficient transcription from the Drosophila U6 promoter (Figure 2). The bottom strand is designed in the format of 5′-AAAC(N)19C-3′, with (N)19C representing the reverse compliment of the targeting sequence. Alternatively, to aid in design process, the CRISPR Optimal Target Finder has a feature that will output the oligonucleotide sequences needed for cloning selected target sites.
- You can either use T4 Polynucleotide Kinase (PNK) to add the 5′ phosphates to standard oligonucleotides, as described below, or order 5′ phosphorylated oligonucleotides.
- As noted above, target sites without an endogenous 5′ G can be used by simply adding a G to the 5′ end of a 20-nt target site in the format of G(N)20 to achieve efficient transcription. In this case the top strand should be designed in the format of 5′-CTTCG(N)20-3′ and the bottom strand designed in the format of 5′-AAAC(N)20C-3′.
-
13.
Resuspend oligonucleotides at a concentration of 100 µM in nuclease-free H2O.
-
14.
Combine 1 µL of the top strand oligonucleotide (100 µM stock), 1 µL of the bottom strand oligonucleotide (100 µM stock), 1 µL of T4 DNA Ligase buffer (10X), 6 µL of H2O, and 1 µL of T4 Polynucleotide Kinase (10U/µL). Incubate at 37°C for 30 minutes, 95°C for 5 min, then ramp to 25°C at a rate of −5°C/min.
-
15.Digest 1 µg of pU6-BbsI-gRNA or pU6-BbsI-U63-gRNA with 10 Units BbsI for two hours at 37°C.
- The pU6-BbsI-gRNA plasmid expresses the gRNA under the control of the U6-2 promoter. The pU6-BbsI-U63-gRNA plasmid expresses the gRNA under the control of the U6-3 promoter, but is otherwise identical. Port et al. (2014) reported that U6-3 was more efficient than either U6-1 or U6-2. However, we have not observed consistent differences between the two promoters in our HDR experiments. This might be explained by low numbers in all cases (only 3 flies per promoter were analyzed in the published work and we have not made exhaustive comparisons); differences between integrated and injected gRNA or between NHEJ- and HDR-based experiments; and/or locus or gRNA-specific differences. We use the two promoters interchangeably.
-
16.
Use gel electrophoresis to purify the digested vector. Determine the DNA concentration using a spectrophotometer.
-
17.
Ligate the annealed insert into pU6-BbsI-gRNA. Combine 1 µL of annealed insert (Step 14), 50 ng of BbsI digested pU6-BbsI-gRNA, 1 µL of T4 DNA Ligase buffer, 1 µL of T4 DNA Ligase, and enough H2O to bring the reaction to 10 µL. Incubate at 25°C for 1 hour.
-
18.
Transform the ligation reaction into DH5α cells and select colonies on plates containing 100 µg/mL ampicillin. A control transformation of digested pU6-BbsI-gRNA vector alone can be performed to ensure no contaminating undigested plasmid was collected in Step 16.
-
19.Isolate plasmids from 2–4 individual colonies using a plasmid miniprep kit. Screen for plasmids with incorporated oligonucleotides by Sanger sequencing using T7 and/or T3 primers.
- Alternatively, positive colonies can be identified via colony PCR using a target site oligonucleotide in combination with a T3 or T7 primer. Use a pipette tip to scratch a visible amount of an individual colony into a PCR tube and use the tip to inoculate 4 mL of LB with ampicillin. Add 10 ul of PCR master mix to the PCR tube and use the appropriate cycling parameters to amplify the diagnostic product.
-
20.Prepare DNA for injection from a positive clone using an Endotoxin-Free Plasmid Midi Kit.
- As with all Drosophila injection-based techniques, preparing high-quality DNA is critical to successful CRISPR-mediated genome engineering.
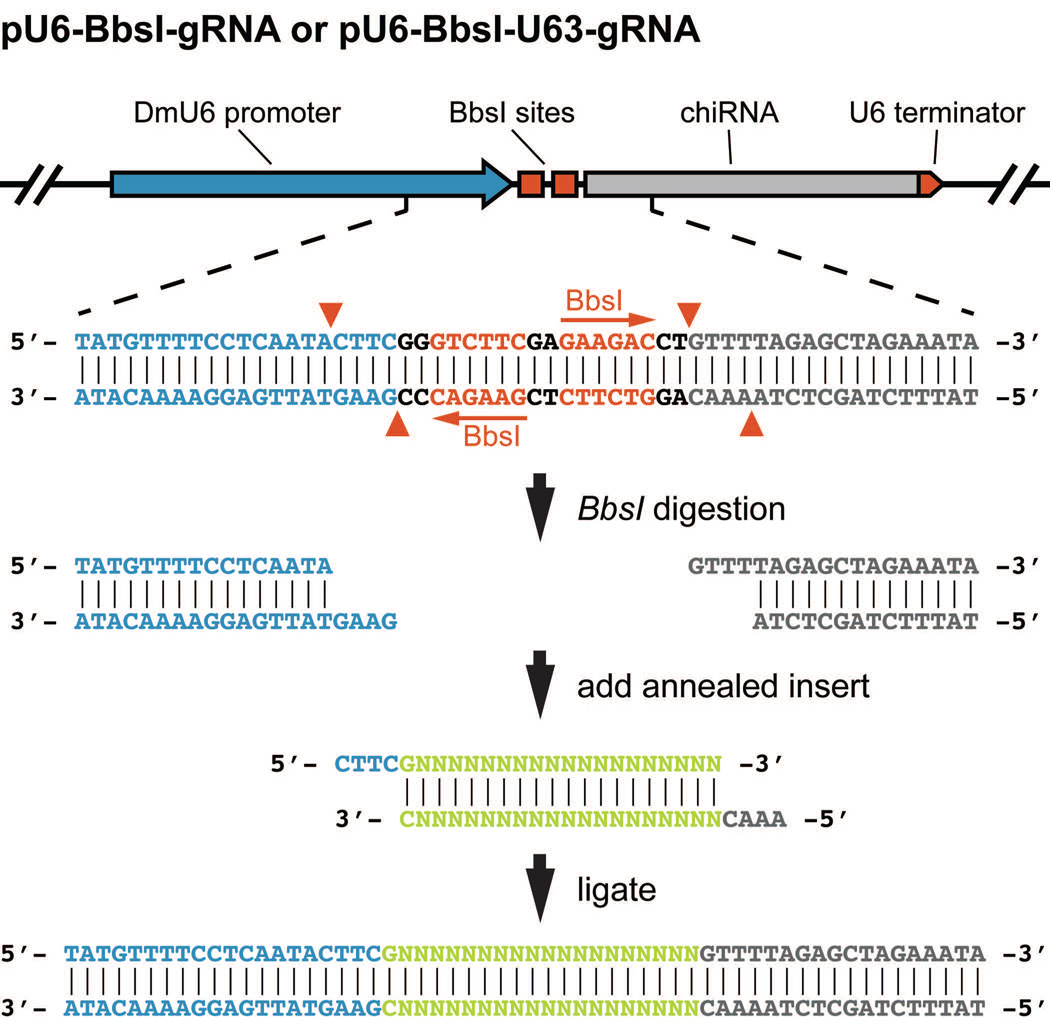
Figure 2.
gRNA plasmid cloning. The pU6-BbsI-gRNA vector contains two BbsI cut sites between the Drosophila U6-2 (snRNA:U6:96Ab) promoter and the common portion of the gRNA. Specific target site sequences are synthesized as complementary oligonucleotides designed to generate appropriate cohesive 5′ overhangs once annealed. The annealed oligonucleotides, once phosphorylated, are then ligated into the BbsI digested pU6-BbsI-gRNA vector. U6-2 sequence (blue), BbsI recognition sequences (red), target site specific sequence (green), and the common portion of the gRNA (grey) are indicated. Red arrowheads denote the breakpoints generated by BbsI cleavage.
Donor Vector Design
dsDNA donor vectors can be made in many configurations to facilitate the generation of an endless variety of genome modifications (Figure 3). The Basic Protocol focuses on the design of donor constructs using the pHD-DsRed-attP or pHD-DsRed vectors available through Addgene. The design of ssDNA donors is described in Alternate Protocol 1 below. The pHD-DsRed-attP vector is used for generating marked knock-out alleles that harbor an attP phage recombination site for serial manipulations of the target locus catalyzed by phiC31. pHD-DsRed is used for generating positively marked targeted insertions or sequence edits. Both vectors include a removable DsRed marker expressed strongly in the eye for visual identification of lines with targeted events and contain multiple cloning sites for inserting locus-specific homology arms.
-
21.Identify the two ~1-Kb sequences that flank the cleavage site and the genomic region that will be modified.
- Extensive analysis of zinc-finger nuclease-catalyzed HDR demonstrated that homology arms of 1 Kb mediate efficient HDR (Beumer et al., 2013), and we and others have found this to be the case with CRISPR-based HDR (Gratz et al., 2014; Port et al., 2014; Xue et al., 2014; Yu et al., 2014).
-
22.Ensure that neither AarI nor SapI sequences occur within your homology arms, as these restriction enzymes are used for cloning the donor vector.
- If your homology arms contain AarI or SapI sites, alter your design and/or experimental protocol accordingly. For example, if the 5′ homology arm contains an AarI cut site, it may be necessary to ‘invert’ the locations of the 5′ and 3′ homology arms in the donor such that the donor cassette is placed on the antisense strand. If the 3′ arm contains an AarI cut site, it will simply be necessary to clone the 5′ homology arm first. Alternatively, the multiple cloning sites can be used instead of AarI or SapI.
-
23.
Design and order primers to amplify both homology arms. Homology arms should contain sequence immediately adjacent to your cleavage sites (Cas9-mediated DSBs are generated 3-bp upstream of the PAM) for efficient HDR. When deleting or replacing a locus, the homology arms will simply comprise the sequence flanking the deleted region (Figure 3A). For inserting exogenous sequences or editing genomic sequences, design overlapping extension PCR oligonucleotides to construct a fragment that contains (i) a 1-Kb homology region, (ii) the insertion or edit you wish to make, and (iii) the genomic sequence between the edit and the marker insertion site, usually in an adjacent intron (Figure 3B). For detailed guidance on the design of overlapping extension PCR primers, we refer the reader to a previous Current Protocols publication (Miklos et al., 2012).
Left Homology Arm (AarI): The forward primer should follow the format 5′-NNNNCACCTGCNNNNTCGC(N)20-3′ where the AarI site is italicized, the cohesive end generated by AarI digestion in underlined, Ns represent spacer sequences required for efficient cleavage and accurate cohesive end generation, and (N)20 represents the hybridization sequence. The reverse primers vary slightly for the two vectors because only one includes the attP sequence in the AarI overhang. For the pHD-DsRed-attP vector, the reverse primer should follow the format 5′-NNNNCACCTGCNNNNCTAC(N)20-3′ (Figure 4A and B). For pHD-DsRed cloning, the reverse primer should follow the format 5′-NNNNCACCTGCNNNNTTAT(N)20-3′ (Figure 4A and C).
Right Homology Arm (SapI): The right homology arm primer design is the same for both pHD-DsRed-attP and pHD-DsRed. The forward primer should follow the format 5′-NNNNGCTCTTCNTAT(N)20-3′ where the SapI site is italicized, the cohesive end generated by SapI digestion is underlined, Ns represent spacer sequences required for efficient cleavage and accurate cohesive end generation, and (N)20 represents the hybridization sequence (Figure 4D). The reverse primer should follow the format 5′-NNNNGCTCTTCNGAC(N)20-3′ (Figure 4D).- Alternatives to overlapping extension PCR abound, including Gibson and Golden Gate cloning or DNA synthesis, which is becoming an increasingly cost-effective option.
-
24.
If your target sequences remain intact in your homology arms, incorporate silent mutations to disrupt the PAM or two seed sequence nucleotides into your design. This will ensure that neither your donor construct nor the successfully engineered locus are targets for cleavage.
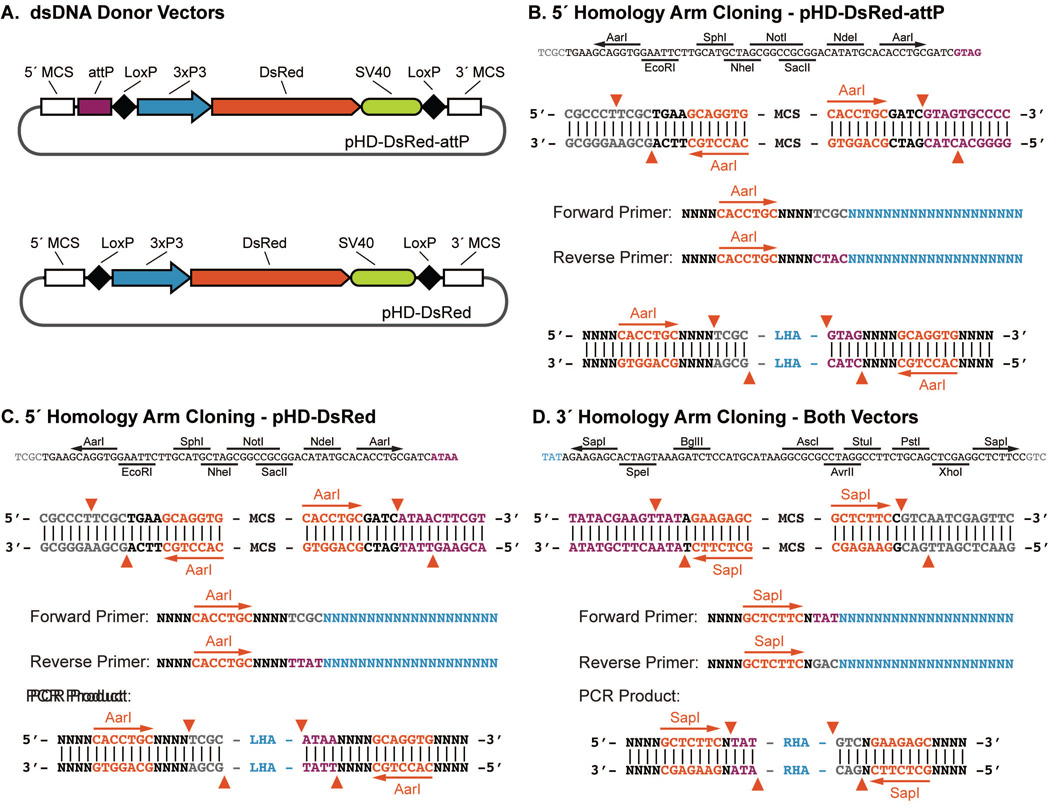
Figure 4.
Donor plasmid cloning. (A) Schematic of the pHD-DsRed-attP and pHD-DsRed donor vector including the MCSs. (B–D) Multiple cloning site sequences and primer design for type IIS restriction site (AarI or SapI) based cloning of homology arms for the LHA of pHD-DsRed-attP (B), the LHA of pHD-DsRed (C), and the RHA of both pHD-DsRed-attP and pHD-DsRed (D). Note that the LHA of pHD-DsRed-attP and pHD-DsRed requires slightly different primers due to the presence or absence of the attP site. Vector backbone sequences (grey), attP/LoxP sequence (purple), AarI/SapI recognition sites (red), and locus specific hybridization sequences (blue) are indicated. Red arrows indicate the breakpoints generated by AarI or SapI digestion.
Donor Vector Construction
Below we outline the steps for rapidly constructing dsDNA donor constructs using the type IIS restriction sites AarI and SapI in the pHD-DsRed-attP or pHD-DsRed vectors for seamless integration of homology arms. These vectors also contain multiple cloning sites for an alternative cloning method.
Materials
pHD-DsRed-attP (Addgene; Plasmid 51019, also called pDSRed-attP)
pHD-DsRed (Addgene; Plasmid 51434)
Phusion High-Fidelity DNA Polymerase
dNTPs
Gel extraction kit
AarI endonuclease
SapI endonuclease
T4 DNA Ligase
Gel extraction kit
E. coli DH5α or other suitable cloning strain
LB with ampicillin
Plasmid Mini Kit
- Primers
- pHD-BB-1 (5′-ACGAAAGGCTCAGTCGAAAG-3′)
- pHD-BB-2 (5′-TGATATCAAAATTATACATGTCAACG-3′)
- pHD-HSP70-R (5′-CGGTCGAGGGTTCGAAATCGATAAG-3′)
- pHD-SV40-F (5′-GGCCGCGACTCTAGATCATAATC-3′)
Endotoxin-Free Plasmid Midi Kit (Macherey-Nagel NucleoBond Xtra Midi EF or similar)
-
25.Prepare a 50-µl reaction to amplify each homology arm. Homology arms should be amplified from the targeted fly line as maximal homology increases the efficiency of HDR (Deng and Capecchi, 1992; Nassif and Engels, 1993).
- 0.5 µl of template DNA
- 2.5 µl of 10mM forward primer
- 2.5 µl of 10mM reverse primer
- 1.0 µl of 10mM each dNTPs
- 10 µl of 5X HF Phusion Buffer
- 0.2 µl of Phusion polymerase
- Water to 50 µl
-
26.Perform PCR with the following parameters.
1 cycle: 2 min 94°C (initial denaturation) 30 cycles: 10 sec 98°C (denaturation) 30 sec 60°C (annealing) 30 sec 72°C (extension) 1 cycle: 10 min 72°C (final extension) -
27.
Purify homology arms via gel electrophoresis. Determine the DNA concentration using a spectrophotometer.
-
28.
Simultaneously digest 1 µg of the pHD-DsRed-attP or pHD-DsRed vector and 1 µg of the left homology arm PCR product with 4 Units of AarI for four hours at 37°C.
-
29.
Use gel electrophoresis to purify both the AarI digested pHD-DsRed-attP or pHD-DsRed vector and left homology arm PCR product. Determine the DNA concentration using a spectrophotometer.
-
30.
Ligate the left homology arm into pHD-DsRed-attP or pHD-DsRed vector. Combine 50 ng of AarI digested pHD-DsRed-attP, 3:1 molar ratio of digested left homology arm PCR product, 1 µL of T4 DNA Ligase buffer, 1 µL of T4 DNA Ligase, and enough H2O to bring the reaction to 10 µL. Incubate at 25°C for 1 hour.
-
31.
Transform the ligation reaction into DH5α cells and select colonies on plates containing 100 µg/mL ampicillin. A control transformation of digested plasmid alone can be included to ensure the sample is not contaminated with undigested plasmid.
-
32.Select 2–4 individual colonies and culture in 4 mL of LB overnight. Isolate plasmid DNA using a plasmid miniprep kit. Screen for plasmids with incorporated homology arms by Sanger sequencing using pHD-BB-1 and pHD-HSP70-R.
- Alternatively, positive colonies can be identified via colony PCR as described above.
-
33.
Simultaneously digest 1 µg of positive constructs from Step 32 and 1 µg of the right homology arm PCR product with 5 Units of SapI for two to four hours at 37°C.
-
34.
Use gel electrophoresis to purify both the SapI digested construct and right homology arm PCR product. Determine the DNA concentration using a spectrophotometer.
-
35.
Ligate the right homology arm into pHD-DsRed-attP. Combine 50 ng of SapI digested pHD-DsRed-attP, 36.6 ng of digested left homology arm PCR product, 1 µL of T4 DNA Ligase buffer, 1 µL of T4 DNA Ligase, and enough H2O to bring the reaction to 10 µL. Incubate at 25°C for 1 hour.
-
36.
Transform the ligation reaction into DH5α cells and select colonies on plates containing 100 µg/mL ampicillin. A control transformation of digested plasmid alone can be included to ensure the sample is not contaminated with undigested plasmid.
-
37.Isolate plasmids from 2–4 individual colonies using a plasmid miniprep kit. Screen for plasmids with incorporated oligonucleotides by Sanger sequencing using pHD-BB-2 and pHD-SV40-F.
- Alternatively, positive colonies can be identified via colony PCR as described above.
-
38.
Prepare high-quality DNA for injection from a positive clone using an Endotoxin-Free Plasmid Midi Kit.
Injection of CRISPR Components
CRISPR components are injected using standard Drosophila injection techniques. Here we provide the injection mixture for HDR in vasa-Cas9 flies.
Materials
pU6-BbsI-gRNA(s)
pHD-DsRed-attP or pHD-DsRed donor vector
Sterile PCR-grade water
vasa-Cas9 fly stocks (Bloomington Drosophila Stock Center)
-
39.Prepare the injection mixture with 500 ng/µl plasmid donor vector and 100 ng/µl of each gRNA vector in sterile PCR-grade water.
- This parameter can be altered based on your experience. We have tested a range of concentrations and find that gRNA plasmid concentrations between 50 and 250 ng/µl and dsDNA donor concentrations between 250 and 500 ng/µl yield successful HDR.
-
40.
Inject 150–300 embryos using standard Drosophila techniques.
Identification and Molecular Confirmation of CRISPR Alleles
Following injection and an appropriate outcross of injected flies, candidate CRISPR alleles are easily identified by screening for flies with red fluorescent eyes in F1 progeny. Once these candidates have been crossed to a balancer line, they can be sacrificed for molecular characterization to verify recovery of the intended genome modification. Below we describe our strategy of performing three PCRs that, in combination with Sanger sequencing, confirm targeted and precise editing (See Figure 3).
Materials
Adult fly homogenization buffer (see Reagents and Solutions for recipe)
- PCR and sequencing primers
- pHD-HSP70-R (5′-CGGTCGAGGGTTCGAAATCGATAAG-3′)
- pHD-SV40-F (5′-GGCCGCGACTCTAGATCATAATC-3′)
- Left-Genomic-F (target locus specific)
- Right-Genomic-R (target locus specific)
Phusion High-Fidelity DNA Polymerase
dNTPs
Gel extraction kit
-
41.
Design primers (see Figure 3). First, we perform two flanking PCRs to amplify regions extending from within the DsRed marker cassette to the right and left flanking genomic regions. To confirm precise incorporation at the targeted locus, it is critical that the genomic primer-binding sites are outside the homology arms of the donor vector as depicted in Figure 3. Candidate alleles are then confirmed via a spanning PCR using the two genomic primers. This reaction will amplify the entire modified region and rule out undesirable crossover (“ends-in”) repair events that result in the incorporation of the entire donor vector including backbone (Yu et al., 2014).
-
42.
Isolate genomic DNA from the F1 candidate flies identified by DsRed expression in the eyes. Anesthetize a single fly and place it in a 0.2 mL PCR tube. Using a P200 pipette tip draw up 50 µL of freshly prepared adult fly homogenization buffer. Keeping the buffer in the pipette, use the tip to disrupt and homogenize the fly. Once the fly is homogenized, dispense the remaining buffer.
-
43.
Incubate at 37°C for 30–45 minutes followed by a 5 minute incubation at 95°C.
-
44.
Use 1 µL of genomic DNA per 50 µL PCR.
-
45.Prepare 3 separate 50 µL reactions for each candidate.
- Primers sets:
-
1- Left-Genomic-F to pHD-HSP70-R
-
2- pHD-SV40-F to Right-Genomic-R
-
3- Left-Genomic-F to Right-Genomic-R
-
1
- Recipe:
- 1 µl of the genomic DNA from step 42
- 2.5 µl of 10mM forward primer
- 2.5 µl of 10mM reverse primer
- 1.0 µl of 10mM each dNTPs
- 10 µl of 5X Phusion Buffer
- 0.2 µl of Phusion polymerase
- Water to 50 µl
-
46.
Use gel electrophoresis to purify positive PCR products.
-
47.
Sequence the PCR product using Sanger sequencing to confirm expected sequence.
ALTERNATE PROTOCOL 1
HDR with Single-Stranded DNA Donors
When engineering small modifications, it may be desirable to use ssDNA donors, which can be rapidly synthesized. However, ssDNA donors are generally limited to 200 nt and, thus, cannot be used for engineering large modifications (such as the integration of a fluorescent tag). They also cannot be designed to include a visible marker for screening, so molecular screening is required, which increases the time and labor required to recover engineered flies.
Protocol steps
Design an ssDNA donor with homology arms corresponding to sequences immediately adjacent to the targeted cleavage sites and the intended modification or insertion. Homology arms of ~40–60 nt have been shown to mediate efficient ssDNA-based HDR in Drosophila (Banga and Boyd, 1992; Beumer et al., 2013; Gratz et al., 2013a; Port et al., 2014; Xue et al., 2014). The orientation of the ssDNA is critical to the success of the experiment. During DNA repair, free 3′ ends created by rescission at the DSB invade homologous DNA. Therefore, to serve as a template for repair, it is essential for the ssDNA be complementary to the free 3′ end. Note that the PAM is not required for cleavage of single-stranded DNA so it is important that the ssDNA does not include an intact target site or it may be a target of cleavage (Jinek et al., 2012).
The injection mixture should contain 100 ng/µl of ssDNA.
Engineered flies will need to be identified by modification-specific PCR amplification or a modification-spanning PCR followed by sequencing. DNA can be obtained from F1 flies after outcrossing. Alternatively, a non-lethal method, such as isolating wing or leg DNA can be used prior to outcrossing of F1 candidates.
ALTERNATE PROTOCOL 2
HDR in Any Genetic Background
For many applications, it is necessary or desirable to engineer a specific fly strain. This is easily accomplished using an injectable source of Cas9 such as pBS-Hsp70-Cas9. Targeting efficiency is lower than with a transgenic Cas9 source, so it is advisable to inject a larger number of embryos.
Additional Materials
pBS-Hsp70-Cas9 plasmid (Addgene; plasmid 46294)
-
1.
Prepare the injection mixture with 250–500 ng/µl of pBS-Hsp70-Cas9, 500 ng/µl plasmid donor vector and 100 ng/µl of each gRNA vector in sterile PCR-grade water.
-
2.
Inject 250–500 embryos using standard Drosophila techniques.
ALTERNATE PROTOCOL 3
Generation of Disruptive Indels and Deletions via NHEJ
If your goal is to generate a disruptive allele, you can target the NHEJ repair pathway by introducing Cas9 and one or two gRNAs in the absence of a donor repair template. Using one gRNA to target a single cleavage event in critical sequence, you can recover disruptive indels. With two gRNAs, you can delete the intervening sequence.
Protocol steps
- Design and generate gRNAs. For introducing a disruptive indel, design one gRNA in a location where the insertion or deletion of a small number of nucleotides is expected to interfere with function. Indels are often less than 10 bp, but can be much longer (Koike-Yusa et al., 2014). For deleting a genomic region, design two gRNAs flanking the region you wish to delete. We have used this approach to delete regions as large as 14 Kb in Drosophila (Gratz et al., 2014). Note that the deletion will not be precise and you will likely observe the loss or gain of a small number of nucleotides at the repaired junction.
- The generation of deletions by NHEJ with two gRNAs can be conducted in lines with a marked element in the targeted locus (Gratz et al., 2014). This allows for the detection of deletions by loss of the marker in the element.
- The injection mixture should contain 100–250 ng/µl of each gRNA.
- When using a single gRNA, we have increased the concentration to 500 ng/µl.
Engineered flies will need to be identified molecularly. Indels can be identified by HRMA, while PCR can be used to detect larger deletions.
REAGENTS AND SOLUTIONS
Adult fly homogenization buffer:
10 mM Tris-HCl (pH 8.2)
25 mM NaCl
1 mM EDTA
Store at room temperature for up to 6 months
1 µl of 20 mg/mL proteinase K added to 99 µl of homogenization buffer just prior to use
COMMENTARY
Background Information
The CRISPR-Cas9 system is a highly accessible and effective tool for genome engineering in Drosophila (Bassett and Liu, 2014; Gratz et al., 2013b; Harrison et al., 2014; Kondo, 2014). The Basic Protocol outlined above details an optimized CRISPR-Cas9 approach that has several advantages. We use a transgenic source of Cas9, expressed in the germline under the control of the vasa promoter, to achieve highly efficient and reliable genome engineering. The introduction of gRNA using rapidly constructed plasmids is quick and inexpensive. Our donor vectors facilitate streamlined cloning of locus-specific donor templates, and the incorporation of a removable DsRed marker makes identification of candidate alleles markedly easier than identification through molecular characterization.
While not covered in our protocol, other groups have successfully applied the CRISPR-Cas9 system in Drosophila using a variety of methods for introducing gRNAs and Cas9. NHEJ has been successfully accomplished using transgenic Cas9 + gRNA supplied as RNA (Xue et al., 2014), transgenic Cas9 + transgenic gRNA (Kondo and Ueda, 2013; Port et al., 2014; Xue et al., 2014), Cas9 DNA + gRNA plasmid (Gratz et al., 2013a; Ren et al., 2013), and Cas9 mRNA + gRNA supplied as RNA (Bassett et al., 2013; Yu et al., 2013). Successful HDR has been reported using Cas9 DNA, gRNA plasmid and either a dsDNA or ssDNA donor (Gratz et al., 2013a; Gratz et al., 2014). However, efficiency is higher with a transgenic Cas9 source, and all other HDR experiments reported in Drosophila to date have been conducted in Cas9-expressing flies using either gRNA plasmid (Gratz et al., 2014), gRNA supplied as RNA (Xue et al., 2014; Yu et al., 2014), or a transgenic gRNA source (Port et al., 2014).
Critical Parameters
Sequencing of target sites: Due to the prevalence of polymorphisms between distinct genetic backgrounds in Drosophila, it is critical to sequence the intended target locus in the genetic background in which the genome engineering experiment will be performed. Even a single basepair change in a target site can be detrimental to the success of the experiment.
Donor template construction: To protect both the donor template and the modified locus from unintended cleavage, it is critical that your donor template not contain an intact gRNA target site
Molecular confirmation of engineered lines: Because unexpected events can always occur during DNA repair, it is important to thoroughly confirm all candidate alleles. To do this, we suggest three PCRs (See Figure 3B) that together will confirm that engineered DNA has been incorporated at the target locus and that the locus is free of additional modifications, including the integration of donor vector backbone sequences (Yu et al., 2014).
Finally, it is important to note that, while CRISPR-Cas9 is working quite well in Drosophila, the system is not yet fully understood. For example, locus- and sequence-specific effects on cleavage efficiency are poorly understood. The Perrimon group (Harvard Medical School) has developed a tool that uses data from high throughput experiments in S2 cells to predict cleavage efficiency based on gRNA target sequence (www.flyrnai.org/evaluateCrispr/). An understanding of how donor composition and other experimental design features may influence the efficiency of HDR or the DNA repair pathway utilized by the cell awaits further study.
Troubleshooting
Poor viability: Reduce the concentration of gRNAs and donor vector in the injection mixture. While this may reduce efficiency, we have found that reducing the overall concentration of the injection mixture can increase viability. If the poor viability is due to highly efficient generation of a lethal allele, use an injected DNA source of Cas9 (pBS-hsp70-Cas9) instead of vasa-Cas9. This will decrease cleavage efficiency, and thus the occurrence of biallelic events, facilitating the recovery of recessive lethal lesions.
Poor efficiency: In the event that a given targeting experiment fails to yield engineered alleles, the gRNAs should be tested for cleavage efficiency. For genome engineering strategies using a pair of gRNAs, a simple PCR spanning the two gRNA target sites can be performed on embryos 24 hours after injection of both gRNAs into vasa-Cas9. Amplicons indicating a deletion between the two targeted cleavage sites demonstrate that both gRNAs are capable of generating DSBs. For strategies using one gRNA, cleavage efficiency can be tested using HRMA or a mismatch-specific nuclease assay on embryos 24 hours post injection. This approach can also be used to assess gRNA efficiency prior to embarking on a CRISPR-based experiment.
Anticipated Results
Using the approach described above to replace genes with attP docking sites in multiple experiments, an average of 24% of injected flies produced correctly engineered progeny. The deleted genes ranged in size from 2 to 27 Kb. Interestingly, we have not observed a strong correlation between deletion size and efficiency, suggesting that locus- or gRNA-specific effects may play a larger role in determining differences in efficiency between targeting experiments. We have also used this approach to insert in-frame tags in a number of loci at a similar average efficiency of 26%.
The highest probability off-target cleavage sites can be identified by sequence similarity to the targeted site. In a subset of our experiments, we have assayed these sites and found no evidence of off-target cleavage (Gratz et al., 2014). Based on this and similar findings by others, we expect that with careful target site selection, engineered fly lines can be generated with few or no off-target mutations in most cases (Bassett et al., 2013; Gratz et al., 2013a; Gratz et al., 2014).
Time Considerations
Target site selection, preparation of gRNA plasmids, and construction of donor templates can be accomplished in 1 week with 1–4 hours hands-on time per day. Embryo injection can be accomplished in 1 day with 4 hours hands-on time. After 10 days, injected flies can be outcrossed in approximately 1 hour. After another 10 days, F1 progeny are screened and outcrossed (2 hours) before molecular confirmation, which can be completed in 2 days (5 hours hands-on time).
ACKNOWLEDGEMENTS
We thank the members of the Harrison, Wildonger and O’Connor-Giles labs for invaluable input throughout this work. Plasmids and transgenic fly lines described here are available through the non-profit distributor Addgene and the Bloomington Drosophila Stock Center, respectively. Additional information and resources are available at flyCRISPR.molbio.wisc.edu and tools.flycrispr.molbio.wisc.edu/targetFinder. All software code is available upon request. This work was funded by startup funds from the University of Wisconsin, a grant from the McKnight Foundation to KOCG, grants from the National Institute of Neurological Disorders and Stroke, National Institutes of Health to KOCG (R01 NS078179 and R21 NS088830) and JW (R00 NS072252), and grants from the Wisconsin Partnership Program and March of Dimes to M.M.H.
Footnotes
INTERNET RESOURCES
Websites:
flyCRISPR
CRISPR fly design
FlyCas9
www.shigen.nig.ac.jp/fly/nigfly/cas9/
OXfCRISPR
oxfcrispr.org
Reagents:
Bloomington Drosophila Stock Center CRISPR stocks
flystocks.bio.indiana.edu/Browse/misc-browse/CRISPR.htm
Addgene CRISPR plasmids
Target finder tools:
CRISPR Optimal Target Finder
tools.flycrispr.molbio.wisc.edu/targetFinder/
DRSC Find CRISPRs
www.flyrnai.org/crispr/index.html
CRISPR Design
E-CRISP
Discussion groups
flyCRISPR Discussion group
LITERATURE CITED
- Anton T, Bultmann S, Leonhardt H, Markaki Y. Visualization of specific DNA sequences in living mouse embryonic stem cells with a programmable fluorescent CRISPR/Cas system. Nucleus. 2014:5. doi: 10.4161/nucl.28488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banga SS, Boyd JB. Oligonucleotide-directed site-specific mutagenesis in Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:1735–1739. doi: 10.1073/pnas.89.5.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AR, Liu JL. CRISPR/Cas9 and genome editing in Drosophila. Journal of genetics and genomics = Yi chuan xue bao. 2014;41:7–19. doi: 10.1016/j.jgg.2013.12.004. [DOI] [PubMed] [Google Scholar]
- Bassett AR, Tibbit C, Ponting CP, Liu JL. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell reports. 2013;4:220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer KJ, Trautman JK, Mukherjee K, Carroll D. Donor DNA Utilization during Gene Targeting with Zinc-finger Nucleases. G3. 2013 doi: 10.1534/g3.112.005439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic acids research. 2013;41:7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, Huang B. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, Rangarajan S, Shivalila CS, Dadon DB, Jaenisch R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell research. 2013;23:1163–1171. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu H, Schwartz HT, Antoshechkin I, Sternberg PW. Transgene-free genome editing in Caenorhabditis elegans using CRISPR-Cas. Genetics. 2013;195:1167–1171. doi: 10.1534/genetics.113.155879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C, Capecchi MR. Reexamination of gene targeting frequency as a function of the extent of homology between the targeting vector and the target locus. Molecular and cellular biology. 1992;12:3365–3371. doi: 10.1128/mcb.12.8.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Lu G, Xie Z, Lou M, Luo J, Guo L, Zhang Y. Genome-wide identification of CRISPR/Cas9 off-targets in human genome. Cell research. 2014 doi: 10.1038/cr.2014.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature biotechnology. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nature biotechnology. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T, Fujii H. Efficient isolation of specific genomic regions and identification of associated proteins by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) using CRISPR. Biochemical and biophysical research communications. 2013;439:132–136. doi: 10.1016/j.bbrc.2013.08.013. [DOI] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O’Connor-Giles KM. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013a;194:1029–1035. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, O’Connor-Giles KM. Highly Specific and Efficient CRISPR/Cas9-Catalyzed Homology-Directed Repair in Drosophila. Genetics. 2014;196:961–971. doi: 10.1534/genetics.113.160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Wildonger J, Harrison MM, O’Connor-Giles KM. CRISPR/Cas9-mediated genome engineering and the promise of designer flies on demand. Fly. 2013b;7:249–255. doi: 10.4161/fly.26566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, Jenkins BV, O’Connor-Giles KM, Wildonger J. A CRISPR view of development. Genes & development. 2014;28:1859–1872. doi: 10.1101/gad.248252.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature biotechnology. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns NA, Genga RM, Enuameh MS, Garber M, Wolfe SA, Maehr R. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development. 2014;141:219–223. doi: 10.1242/dev.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiskinis E, Sandoe J, Williams LA, Boulting GL, Moccia R, Wainger BJ, Han S, Peng T, Thams S, Mikkilineni S, Mellin C, Merkle FT, Davis-Dusenbery BN, Ziller M, Oakley D, Ichida J, Di Costanzo S, Atwater N, Maeder ML, Goodwin MJ, Nemesh J, Handsaker RE, Paull D, Noggle S, McCarroll SA, Joung JK, Woolf CJ, Brown RH, Eggan K. Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1. Cell Stem Cell. 2014;14:781–795. doi: 10.1016/j.stem.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera Mdel C, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nature biotechnology. 2014;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- Kondo S. New horizons in genome engineering of Drosophila melanogaster. Genes & genetic systems. 2014;89:3–8. doi: 10.1266/ggs.89.3. [DOI] [PubMed] [Google Scholar]
- Kondo S, Ueda R. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics. 2013;195:715–721. doi: 10.1534/genetics.113.156737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Kwak SJ, Kim J, Kim AK, Noh HM, Kim JS, Yu K. RNA-guided genome editing in Drosophila with the purified Cas9 protein. G3. 2014;4:1291–1295. doi: 10.1534/g3.114.012179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nature methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklos AE, Hughes RA, Ellington AD. Design and assembly of large synthetic DNA constructs. In: Ausubel Frederick M, et al., editors. Current protocols in molecular biology. Vol. 3. 2012. Chapter Unit3 23. [DOI] [PubMed] [Google Scholar]
- Nassif N, Engels W. DNA homology requirements for mitotic gap repair in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:1262–1266. doi: 10.1073/pnas.90.4.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, Guilak F, Crawford GE, Reddy TE, Gersbach CA. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nature methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F, Chen HM, Lee T, Bullock SL. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E2967–E2976. doi: 10.1073/pnas.1405500111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Sun J, Housden BE, Hu Y, Roesel C, Lin S, Liu LP, Yang Z, Mao D, Sun L, Wu Q, Ji JY, Xi J, Mohr SE, Xu J, Perrimon N, Ni JQ. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:19012–19017. doi: 10.1073/pnas.1318481110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebo ZL, Lee HB, Peng Y, Guo Y. A simplified and efficient germline-specific CRISPR/Cas9 system for Drosophila genomic engineering. Fly. 2014;8:52–57. doi: 10.4161/fly.26828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C, Gore A, Yan W, Abalde-Atristain L, Li Z, He C, Wang Y, Brodsky RA, Zhang K, Cheng L, Ye Z. Whole-Genome Sequencing Analysis Reveals High Specificity of CRISPR/Cas9 and TALEN-Based Genome Editing in Human iPSCs. Cell Stem Cell. 2014;15:12–13. doi: 10.1016/j.stem.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Yu C, Qu J, Li M, Yao X, Yuan T, Goebl A, Tang S, Ren R, Aizawa E, Zhang F, Xu X, Soligalla RD, Chen F, Kim J, Kim NY, Liao HK, Benner C, Esteban CR, Jin Y, Liu GH, Li Y, Izpisua Belmonte JC. Targeted gene correction minimally impacts whole-genome mutational load in human-disease-specific induced pluripotent stem cell clones. Cell Stem Cell. 2014;15:31–36. doi: 10.1016/j.stem.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veres A, Gosis BS, Ding Q, Collins R, Ragavendran A, Brand H, Erdin S, Talkowski ME, Musunuru K. Low Incidence of Off-Target Mutations in Individual CRISPR-Cas9 and TALEN Targeted Human Stem Cell Clones Detected by Whole-Genome Sequencing. Cell Stem Cell. 2014;15:27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z, Ren M, Wu M, Dai J, Rong YS, Gao G. Efficient gene knock-out and knock-in with transgenic Cas9 in Drosophila. G3. 2014;4:925–929. doi: 10.1534/g3.114.010496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Chen H, Liu J, Zhang H, Yan Y, Zhu N, Guo Y, Yang B, Chang Y, Dai F, Liang X, Chen Y, Shen Y, Deng WM, Chen J, Zhang B, Li C, Jiao R. Various applications of TALEN- and CRISPR/Cas9-mediated homologous recombination to modify the Drosophila genome. Biology open. 2014;3:271–280. doi: 10.1242/bio.20147682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Ren M, Wang Z, Zhang B, Rong YS, Jiao R, Gao G. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics. 2013;195:289–291. doi: 10.1534/genetics.113.153825. [DOI] [PMC free article] [PubMed] [Google Scholar]