Abstract
We recently characterized the substrate scope of wild-type RebH and evolved variants of this enzyme with improved stability for biocatalysis. The substrate scopes of both RebH and the stabilized variants, however, are limited primarily to compounds similar in size to tryptophan. We have now used a substrate walking approach to further evolve RebH variants with expanded substrate scope. Two particularly notable variants were identified: 3-SS, which provides high conversion of tricyclic tryptoline derivatives; and 4-V, which accepts a broad range of large indoles and carbazoles. This constitutes the first reported use of directed evolution to enable functionalization of substrates not accepted by wild-type RebH and demonstrates the utility of RebH variants for site-selective halogenation of biologically active compounds.
Keywords: halogenase, directed evolution, RebH, substrate walking, biocatalysis
Halogenated aromatic compounds are essential building blocks in chemical synthesis[1] and are commonly found in pharmaceuticals[2] and agrochemicals;[3] an estimated one quarter of all such compounds are halogenated.[4] This is in part because halogenation can significantly impact molecular pharmacology, which is highlighted by the effects of chlorination or bromination on a diverse range of drugs spanning antibiotics,[5] anticancer agents,[6] and psychoactive compounds[7]. It has also been demonstrated that the presence of a halogen substituent can greatly alter drug metabolism. In one study of the microsomal clearance of over 220,000 compounds, chlorination was found to predictably increase or decrease the metabolic half-life of a drug, depending on the location of the substitution.[8]
Despite the importance of halogen substituents, conventional arene halogenation methods, such as those proceeding via electrophilic aromatic substitution (EAS), often suffer from poor regioselectivity and require harsh reaction conditions.[9] It is therefore notable that flavin-dependent halogenase (FDH) catalyzed arene halogenation, proposed to proceed via EAS,[10] provides high regioselectivity using air and halide salts as the terminal oxidant and halogen source, respectively. We recently reported that the tryptophan 7-halogenase RebH, a FDH from the rebeccamycin biosynthetic pathway,[11] can halogenate a range of indoles and naphthalenes on a preparative scale.[12] In contrast to previous reports for another FDH, PrnA,[13] RebH was found to halogenate many of these substrates at sites other than those most electronically activated, providing regioselectivity that would not be obtained from conventional EAS. The unnatural substrates halogenated by RebH in both our[12] and others’[14] work were, however, comparable in size to the native substrate, tryptophan (1, Scheme 1), and increasing structural differences led to lower turnover numbers and less favorable kinetic parameters.[12] Given a recent report in which cross-linked RebH aggregates were used for gram scale halogenation,[15] the narrow substrate scope and low activity of RebH on unnatural substrates stand as key limitations to its general utility for preparative halogenation.
Scheme 1.
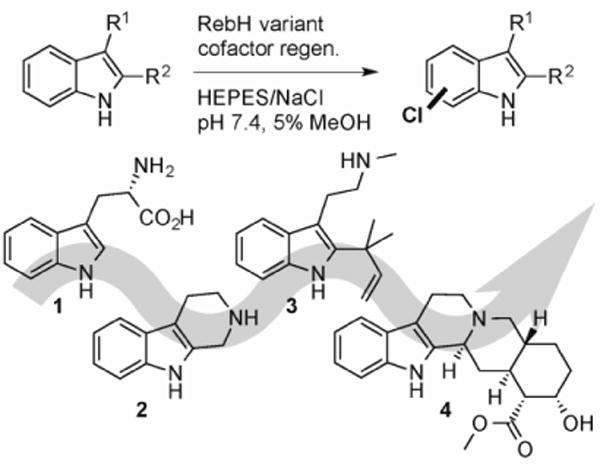
General scheme for RebH catalyzed halogenation and substrates used for directed evolution via substrate walking.
Several groups have used RebH and other FDHs in metabolic engineering efforts that involve typtophan halogenation and conversion of halogenated tryptophan into halogenated natural product derivatives.[16] A point mutant of RebH has been reported to preferentially halogenate tryptamine over tryptophan, and direct halogenation of this smaller alkaloid precursor has aided the aforementioned metabolic engineering efforts.[17] On the other hand, no examples in which FDHs are used for late stage halogenation of complex, biologically active compounds on a preparative scale have been reported, presumably due to substrate scope limitations. We recently used directed evolution to engineer thermostable RebH variants that provide increased conversion of several unnatural substrates.[18] Using techniques developed for this effort, and starting from one of the stable variants identified, we sought to evolve the substrate scope of RebH via substrate walking (Scheme 1). This approach involves improving enzyme activity on a known substrate with structural homology to a target substrate, identifying variants that fortuitously possess activity on the target substrate, and repeating this process to gradually evolve the substrate scope of the enzyme.[19]
We found that RebH variant 1-PVM from our thermostability lineage provided increased conversion of numerous substrates, including tryptamine and tryptoline (2, Scheme 1, Table 1). Initially, we intended to further improve 1-PVM activity on 2, as we hoped that variants with improved activity on this tricyclic molecule would also have activity on larger tryptophan-based natural products. Both RebH and 1-PVM, however, provided insufficient conversion of 2 for reliable UPLC analysis under the conditions used for high throughput screening (crude lysate in 96 well plates). Because 1-PVM is both more stable and has nearly twofold higher activity on 2 than RebH (Table 1), it was used as the parent for our first round of evolution. We hoped that improving 1-PVM activity on 1, which was similar to that of RebH and could be detected by UPLC, would provide variants with sufficient activity on 2 to enable further evolution using this substrate.
Table 1.
Representative UPLC conversions and (enzyme loadings) for substrates used in substrate walking based mutagenesis of RebH.
| Generation | Enzyme | Mutation[a] | Method (substrate)[b] | %Conv. 1 (%Loading)[c] |
%Conv. 2 (%Loading) |
%Conv. 3 (%Loading) |
%Conv. 4 (%Loading) |
|---|---|---|---|---|---|---|---|
| 0 | RebH | – | – | 53 (0.2) | 3 (0.5) | <1 (5) | 1 (5) |
| 1 | 1-PVM | S2P/M71V/K145M | epPCR[d] (1) | 43 (0.2) | 5 (0.5) | <1 (5) | 5 (5) |
| 2 | 2-T | N467T | epPCR (1) | 53 (0.2) | 7 (0.5) | <1 (5) | 9 (5) |
| 3 | 3-SS | G112S/N470S | epPCR (2) | 22 (0.2) | 64 (0.5) | 6 (5) | 9 (5) |
| 3 | 3-S | S112G | point mut.(3)[e] | 43/5[f] (0.2) | 50 (0.5) | 29 (5) | 39 (5) |
| 4 | 4-V | A442V | epPCR (3) | 39/3[f] (0.2) | 43 (0.5) | 48 (5) | 38 (5) |
Mutations relative to the parent on the previous line.
Method used to introduce mutations and (substrate used in the screening effort noted).
Conversions of substrates determined by UPLC and (mol% loadings of enzymes used).
Error-prone PCR.
Point mutation introduced via SOE PCR.[20]
Second number refers to conversion to dihalogenated product.
A library of 1-PVM variants, each containing an average of 1–2 amino acid mutations, was generated by error-prone PCR,[18] and 1080 colonies were picked and expressed in 96 well plates. Halogenation of 1 was assayed using UPLC, and any variants that provided increased conversion relative to 1-PVM included in each plate were submitted to a secondary screen for increased conversion of 2. All hits were verified by gene sequencing and large scale reactions conducted using purified enzyme. From this library, one variant possessing a single N467T mutation (and therefore called 2-T) gave a modest 1.2-fold increase in conversion of 1, and, when submitted to the secondary screen, showed a 1.3-fold increase in conversion of 2. Although also modest, this improvement elevated the conversion of 2 to nearly 10% under the conditions used for screening. We found that at this level of conversion, we could reliably detect 1.5-fold increases in conversion of 2, and thus decided to screen directly on 2 for our next round of mutagenesis.
The second round library was constructed as described above but using 2-T as a parent, and 1080 variants were again picked and expressed. Reactions were conducted using 2 as the substrate, and any variants that showed 1.5-fold or higher activity in lysate were grown and purified to confirm their activity. Several mutations giving activity above this threshold were found, and one of these, N470S, led to nearly 7-fold higher conversion of 2. This mutation and a second mutation identified in this round of screening, G112S, were combined to give variant 3-SS, which provided 9-fold higher conversion of 2 relative to 2-T.
Preparative chlorination of 2 using 1 mol% 3-SS afforded 93% conversion to 6-chlorotryptoline (Table 2). This represents a significant improvement over our previously reported chlorination of 2 using 10 mol% RebH, which gave a nearly 1:1 mixture of 6- and 7-chlorotryptoline at only 58% conversion[12]; only trace 7-chlorination was observed using 3-SS. Many of the mutations found in 3-SS are in the enzyme active site (N467T, N470S, and G112S; see Fig. S2) and could impact the binding of 2 and thus the halogenation selectivity on this substrate.[21] Steady-state kinetic analysis of these variants was conducted to probe the effects of these mutations on kinetic parameters (Table 3). We found that the catalytic efficiency scaled accordingly with the increased conversion observed for successive variants. While this effect appears to have been solely due to increased kcat for variants 1-PVM and 2-T, the final variant, 3-SS, possesses greatly increased kcat and greatly decreased Km, resulting in a catalytic efficiency nearly 70-fold greater than WT.
Table 2.
Yields for 10 mg RebH variant-catalyzed halogenation reactions[a]
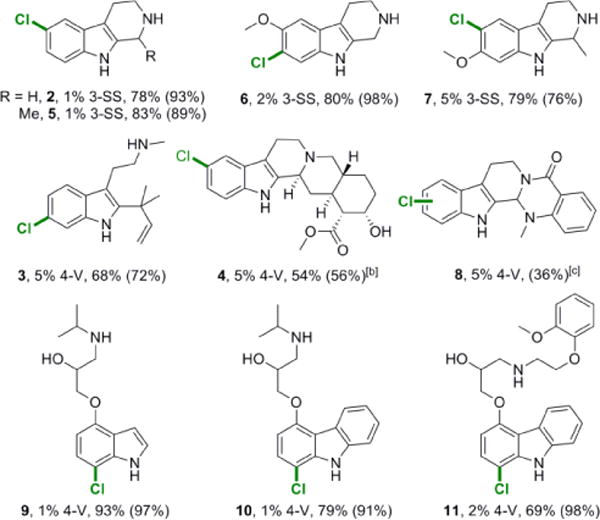
|
Substrate numbers, mol% enzyme loadings, isolated yields, and (UPLC conversions) are provided.
In addition to the major product of 10-chlorination shown, 11-chlorination was also observed to afford a minor product.
Multiple closely eluting products were observed which were not individually isolated, so only total UPLC conversion is shown.
Table 3.
Kinetic data for chlorination of tryptoline (2) by RebH variants.
| Variant | Mol % | kcat [min−1] | Km [μM] | kcat/Km [min−1 μM−1] |
|---|---|---|---|---|
| RebH | 5 | 0.013 | 144 | 9.05 × 10−5 |
| 1-PVM | 5 | 0.040 | 215 | 1.86 × 10−4 |
| 2-T | 3 | 0.056 | 236 | 2.37 × 10−4 |
| 3-SS | 0.5 | 0.537 | 8.6 | 6.24 × 10−3 |
We further explored the substrate scope of 3-SS by examining its activity on a range of additional substrates. We found that 3-SS is particularly effective for halogenating tricyclic indoles similar to 2 (Table 2). Eleagnine (5), an alkaloid from Chrysophyllum albidum with potent analgesic effects,[22] has the structure of 2 with an additional methyl group at the 1-position. We found this added steric bulk distal from the indole moiety interesting, as we aimed to further expand the substrate scope of RebH to encompass compounds significantly larger in the area of this methyl group. Although RebH shows very low conversion of 5, 3-SS has high activity on this compound – over 60-fold higher than that observed with wild-type RebH (Table 4). With only 1 mol% enzyme loading of 3-SS, 89% conversion of 5 was seen on the 10 mg scale. We found that 3-SS also gave good conversion of two tryptoline derivatives with substituents on the benzene ring of the indole moiety: pinoline (6), or 6-methoxytryptoline, a metabolite with monoamine oxidase A inhibition activity,[23] which was chlorinated solely at the 7-position; and tetrahydroharmine (7), or 7-methoxyeleagnine, an alkaloid from Banisteriopsis caapi with antiviral and antifungal activity,[24] which was chlorinated solely at the 6-position. These two substrates show that these RebH variants are able to tolerate substituents at multiple positions on the indole moiety, which occur frequently in biologically active natural products.
Table 4.
Ratio of improvement of variant vs. RebH for each substrate in Table 2.
| Substrate | Variant | Mol % | Activity Ratio[a] |
|---|---|---|---|
| Tryptoline (2) | 3-SS | 0.5 | 65.5 |
| Eleagnine (5) | 3-SS | 0.5 | 67.1 |
| Pinoline (6) | 3-SS | 0.5 | 2.0 |
| Tetrahydroharmine (7) | 3-SS | 5 | 17.6 |
| Desbromo-dFBr (3) | 4-V | 5 | N/A[b] |
| Yohimbine (4) | 4-V | 5 | 38.0 |
| Evodiamine (8) | 4-V | 5 | 16.5 |
| Pindolol (9) | 4-V | 0.2 | 1.3 |
| Carazolol (10) | 4-V | 0.2 | 4.9 |
| Carvedilol (11) | 4-V | 0.5 | 8.2 |
Activity ratio is ratio of conversion seen with variant tested vs WT. Reaction conditions were those shown in Scheme 2.
WT showed no quantifiable activity, and thus a ratio cannot be determined.
The goal of our substrate walking effort was to expand the substrate scope of RebH to include biologically active compounds that are significantly larger than L-tryptophan or the tryptoline derivatives described above. One substrate in particular caught our interest, deformylflustrabromine (dFBr), a tryptamine metabolite first isolated from Flustra foliacea. dFBr is a potent inhibitor of bacterial biofilm formation, but only when it possesses the 6-bromo substituent on its indole moiety; no detectable biofilm inhibition is observed when this substituent is not present.[5] To the best of our knowledge, however, the biological activities of the 4-, 5-, and 7-bromo isomers and the chlorinated analogues have not been reported. The preparation of these compounds for their subsequent biological evaluation seemed like a prime target for RebH variants given the substrate scope outlined above. We synthesized 3,[25] the non-halogenated indole analogue of dFBr, and while RebH has no quantifiable activity on this compound, 3-SS did provide low conversion. Notably, we found that variant 3-S, which lacks the G112S mutation but still possesses N470S, gave 5-fold higher conversion of 3 than did 3-SS.
As the conversion of 3 afforded by 3-S was sufficient to observe low activity in crude lysate in 96 well plates, we screened for improved activity on 3 for our third round of evolution just as was done on 2 for our second round. After verifying the hits with purified enzyme, one variant was found with nearly 1.7-fold increased activity on 3. This variant, 4-V, possesses a single A442V mutation that is quite distant from the active site of the enzyme. It is well established that mutations distant from enzyme active sites can have profound effects on their activity,[26] and such mutations are especially difficult to find with structure-guided mutagenesis. We used 4-V for preparative chlorination of 3, and found that this enzyme provided exclusively 6-chlorination, providing a facile synthesis of the 6-chloro analogue of dFBr. The high activity of 4-V on 3, despite the bulk of the inverse prenyl group, encouraged us to explore the substrate scope of this variant. We first looked at two pentacyclic compounds with well studied biological activities: yohimbine (4), an alkaloid from Catharansus roseus that is also an α2 adrenergic receptor antagonist;[27] and evodiamine (8), an alkaloid from the Tetradium family of plants.[28] Both of these compounds are significantly larger than any others we had assayed to this point – 4 has over twice the molecular weight of 2 – but to our surprise, both showed significant conversion with 4-V (Table 2). While 4-V produces two distinct monochlorinated derivatives of 8, as well as one dichlorinated derivative, only monochlorinated products were observed with 4, and at a sufficient level of conversion for a preparative scale halogenation. After performing the preparative scale reaction, we observed that 4 is chlorinated to 56% conversion in a roughly 3:1 ratio at the 10- and 11-positions, respectively.
Given that we had demonstrated that RebH variants could accept greatly expanded bulk distal to the indole moiety of these unnatural substrates, as well as substitution at the 5- and 6-positions on the indole moiety, we decided to explore the impact of large substitutions at the 4-position of the indole ring. Pindolol (9), a nonselective beta blocker,[29] possesses a sizeable substituent with alcohol and amine functionalities via an ether linkage at the indole 4-position. We found that 4-V was able to fully convert this compound to the 7-chlorinated product, even at lower (<1%) enzyme loadings. A similar compound, carazolol (10),[30] possesses a carbazole rather than an indole moiety, and we found that this compound was selectively halogenated at the analogous position. This was the first time we had observed selective conversion of a carbazole in high yield by any RebH variant, and encouraged by this result we decided to test carvedilol (11), another carbazole with an even bulkier substituent at the 5-position (analogous to the indole 4-position) that is a nonselective beta blocker/alpha-1 blocker.[31] Carvedilol has a molecular weight of over 400 g/mole, twice the size of the native tryptophan and even larger than 4. Despite this added steric bulk, we found that 4-V was able to again give full conversion of 11 at only 2% enzyme loading, with exquisite selectivity to only a single chlorinated product. The confined nature of the RebH active site in crystal structures containing bound tryptophan substrate[32] makes rationalizing binding of the larger substrates explored in this work difficult, but it may be that some of these, particularly 11, are not fully contained in the active site.
In summary, beginning from a thermostablized variant of RebH, 1-PVM, RebH variants with significantly expanded substrate scope were identified using only three rounds of random mutagenesis and screening on progressively larger substrates. These variants catalyzed selective halogenation of several unnatural substrates displaying a range of sizes, substitutions, and biological activities. Given the established impact that halogenation has on the biological activities of compounds and the dearth of methods to perform selective halogenations such as those we have described, we believe that these variants could prove useful for producing high-value derivatives of biologically active compounds. While the mutations discovered offer great insight for future targeted libraries[33] the discovery of crucial mutations distant from the active site, such as A442V in variant 4-V, highlight the value of random mutagenesis in discovering important yet difficult to predict mutations[26]. We hope to gain insight into the role of this mutation and the others identified during our evolution efforts using protein crystallography with bound substrates, which we are currently exploring.
Supplementary Material
Scheme 2.

General scheme for RebH variant-catalyzed arene halogenations. [a] Cofactor regen system consisted of 0.5 mol % MBP-RebF and 50 U mL−1 glucose dehydrogenase.
Footnotes
This work was supported by an NIH Pathways to Independence Award (5R00GM087551) and a Searle Scholar Award (11-SSP-202) to J.C.L., and by a NIH Chemistry and Biology Interface training grant to J.T.P. (T32 GM008720),
Supporting information for this article, including complete experimental procedures, is available on the WWW under http://dx.doi.org/10.1002/anie.201xxxxxx.
Contributor Information
Mr. James T. Payne, Department of Chemistry, University of Chicago, 5735 S. Ellis Ave., Chicago, IL 60637 (USA)
Dr. Catherine B. Poor, OPX Biotechnologies, 2425 55th St., Suite 100, Boulder, CO 80301
Prof. Jared C. Lewis, Email: jaredlewis@uchicago.edu, Department of Chemistry, University of Chicago, 5735 S. Ellis Ave., Chicago, IL 60637 (USA).
References
- 1.Diederich F, Stang PJ, editors. Metal-Catalyzed Cross-Coupling Reactions. Wiley-VCH; New York: 1998. [Google Scholar]
- 2.a) Hernandes MZ, Cavalcanti SMT, Moreira DRM, de Azevedo Junior WF, Leite ACL. Curr Drug Targets. 2010;11:303–314. doi: 10.2174/138945010790711996. [DOI] [PubMed] [Google Scholar]; b) Naumann K. Pest Manage Sci. 2000;56:3–21. [Google Scholar]
- 3.Jeschke P. Pest Manage Sci. 2010;66:10–27. doi: 10.1002/ps.1829. [DOI] [PubMed] [Google Scholar]
- 4.Herrera-Rodriguez LN, Khan F, Robins KT, Meyer HP. Chem Today. 2011;29:31. [Google Scholar]
- 5.Bunders CA, Minvielle MJ, Worthington RJ, Ortiz M, Cavanagh J, Melander C. J Am Chem Soc. 2011;133:20160. doi: 10.1021/ja209836z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams PG, Buchanan GO, Feling RH, Kauffman CA, Jensen PR, Fenical W. J Org Chem. 2005;70:6196–6203. doi: 10.1021/jo050511+. [DOI] [PubMed] [Google Scholar]
- 7.Smith BM, Smith JM, Tsai JH, Schultz JA, Gilson CA, Estrada SA, Chen RR, Park DM, Prieto EB, Gallardo CS, Sengupta D, Dosa PI, Covel JA, Ren A, Webb RR, Beeley NRA, Martin M, Morgan M, Espitia S, Saldana HR, Bjenning C, Whelan KT, Grottick AJ, Menzaghi F, Thomsen WJ. J Med Chem. 2008;51:305. doi: 10.1021/jm0709034. [DOI] [PubMed] [Google Scholar]
- 8.Sun H, Keefer CE, Scott DO. Drug Metab Lett. 2011;5:232. doi: 10.2174/187231211798472575. [DOI] [PubMed] [Google Scholar]
- 9.Smith K, El-Hiti GA. Curr Org Synth. 2004;1:253–274. [Google Scholar]
- 10.Dong C, Flecks S, Unversucht S, Haupt C, van Pée KH, Naismith JH. Science. 2005;309:2216–2219. doi: 10.1126/science.1116510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeh E, Garneau S, Walsh CT. Proc Natl Acad Sci. 2005;102:3960–3965. doi: 10.1073/pnas.0500755102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Payne JT, Andorfer MC, Lewis JC. Angew Chem Int Ed. 2013;52:5271–5274. doi: 10.1002/anie.201300762. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem. 2013;125:5379–5382. Note: we previously referred to 6- and 7-halogenation of tryptoline as 5- and 6-halogenation, by analogy to the numbering used for L-tryptophan, but here we use the established numbering for carbazole derivatives. [Google Scholar]
- 13.Hölzer M, Burd W, Reißig H-U, van Pée K-H. Adv Synth Catal. 2001;343:591–595. [Google Scholar]
- 14.Frese M, Guzowska PH, Voß H, Sewald N. Chem Cat Chem. 2014;6:1270–1276. [Google Scholar]
- 15.Frese M, Sewald N. Angew Chem Int Ed. 2014 doi: 10.1002/anie.201408561. Advance online publication. [DOI] [Google Scholar]
- 16.a) Roy AB, Grüschow S, Cairns N, Goss RJM. J Am Chem Soc. 2010;132:12243. doi: 10.1021/ja1060406. [DOI] [PubMed] [Google Scholar]; b) Runguphan W, Qu X, O’Connor SE. Nature. 2010;468:461. doi: 10.1038/nature09524. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Sánchez C, Zhu L, Braña AF, Salas AP, Rohr J, Méndez C, Salas JA. Proc Natl Acad Sci. 2005;102:461–466. doi: 10.1073/pnas.0407809102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glenn WS, Nims E, O’Connor SE. J Am Chem Soc. 2011;133:19346–19349. doi: 10.1021/ja2089348. [DOI] [PubMed] [Google Scholar]
- 18.Poor CB, Andorfer MC, Lewis JC. Chem Bio Chem. 2014;15:1286–1289. doi: 10.1002/cbic.201300780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.a) Fasan R, Meharenna YT, Snow CD, Poulos TL, Arnold FH. J Mol Biol. 2008;383:1069–1080. doi: 10.1016/j.jmb.2008.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Savile CK, Janey JM, Mundorff EC, Moore JC, Tam S, Jarvis WR, Colbeck JC, Krebber A, Fleitz FJ, Brands J, Devine PN, Huisman GW, Hughes GJ. Science. 2010;329:305–309. doi: 10.1126/science.1188934. [DOI] [PubMed] [Google Scholar]
- 20.Heckman KL, Pease LR. Nat Protoc. 2007;2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- 21.Lang A, Polnick S, Nicke T, William P, Patallo EP, Naismith JH, van Pée KH. Angew Chem Int Ed. 2011;50:2951–2953. doi: 10.1002/anie.201007896. [DOI] [PubMed] [Google Scholar]
- 22.Idowu TO, Iwalewa EO, Aderogba MA, Akinpelu BA, Ogundaini AO. J Biol Sci. 2006;6:1029–1034. [Google Scholar]
- 23.Pähkla R, Harro J, Rägo L. Pharmacol Res. 1996;34:73–78. doi: 10.1006/phrs.1996.0066. [DOI] [PubMed] [Google Scholar]
- 24.Song H, Liu Y, Liu Y, Wang L, Wang Q. J Agri Food Chem. 2014;62:1010–1018. doi: 10.1021/jf404840x. [DOI] [PubMed] [Google Scholar]
- 25.Adla SK, Golz G, Jones PG, Lindel T. Synthesis. 2010;13:2161–2170. [Google Scholar]
- 26.a) Shimotohno A, Oue S, Yano T, Kuramitsu S, Kagamiyama H. J Biochem. 2001;129:943–948. doi: 10.1093/oxfordjournals.jbchem.a002941. [DOI] [PubMed] [Google Scholar]; b) Spiller B, Gershenson A, Arnold FH, Stevens RC. Proc Natl Acad Sci. 1999;96:12305–12310. doi: 10.1073/pnas.96.22.12305. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Romero PA, Arnold FH. Nat Rev Mol Cell Biol. 2009;10:866–876. doi: 10.1038/nrm2805. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Fasan R, Chen MM, Crook NC, Arnold FH. Angew Chem Int Ed. 2007;46:8414–8418. doi: 10.1002/anie.200702616. [DOI] [PubMed] [Google Scholar]
- 27.Tam SW, Worcel M, Wyllie M. Pharmacol Ther. 2001;91:215–243. doi: 10.1016/s0163-7258(01)00156-5. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi Y, Nakano Y, Kizaki M, Hoshikuma K, Yokoo Y, Kamiya T. Planta Med. 2001;67:628–633. doi: 10.1055/s-2001-17353. [DOI] [PubMed] [Google Scholar]
- 29.Aellig WH. Br J Clin Pharmac. 1982;13:187S–192S. doi: 10.1111/j.1365-2125.1982.tb01909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Innis RB, Correa FMA, Snyder SH. Life Sci. 1979;24:2255–2264. doi: 10.1016/0024-3205(79)90102-4. [DOI] [PubMed] [Google Scholar]
- 31.Stafylas PC, Sarafidis PA. Vasc Health Risk Manag. 2008;4:23–30. doi: 10.2147/vhrm.2008.04.01.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bitto E, Bingman CA, Singh S, Phillips GN., Jr Proteins. 2008;70:289–293. doi: 10.1002/prot.21627. [DOI] [PubMed] [Google Scholar]
- 33.a) Reetz MT, Bocoa M, Carballeira JD, Zha D, Vogel A. Angew Chem Int Ed. 2005;44:4192–4196. doi: 10.1002/anie.200500767. [DOI] [PubMed] [Google Scholar]; b) Lewis JC, Mantovani SM, Fu Y, Snow CD, Komor RS, Wong CH, Arnold FH. Chem Bio Chem. 2010;11:2502–2505. doi: 10.1002/cbic.201000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


