Abstract
Two genotypes, assemblage A and B, of the pathogenic, gut protozoan parasite Giardia lamblia infect humans. Symptoms of infection range from asymptomatic to chronic diarrhea. Giardia chromosomes have long been characterized but not until the publication of the first Giardia genome sequence was chromosome mapping work, commenced nearly two decades ago, completed. Initial mapping studies identified and ordered Not I chromosome segments (summating to 1.8 Mb) of the estimated 2 Mb chromosome 3. The resulting map was confirmed with the release of the Giardia genome sequence which prompted revitalization of mapping. The result is 93% of the WB isolate genome sequence assigned to one of five major chromosomes with community access to these data made available through GiardiaDB, the database for Giardia genomes.
Giardia: from disease to chromosomes
Giardia lamblia (synonymous with G. intestinalis and G. duodenalis), the cosmopolitan, intestinal, flagellated protozoan parasite carpets the lining of the small intestine causing erosion of microvilli with severe infections resulting in malabsorption [1]. Other symptoms include diarrhea, bloating, nausea, loss of appetite, vomiting, cognitive impairment and weight loss with chronic infections but may also be asymptomatic [1]. Environmentally resistant cysts form as the trophozoite, the replicative stage, which move from the small intestine through the large intestine [2], and infection (or re-infection) results following ingestion of fecal cysts and excystation of trophozoites in response to stomach acid [2].
Giardia has proved to be a fascinating and amenable or ganism to study [3]. Originally thought to be an ancient eukaryote which evolved prior to acquisition of mitochondria [4], it is now generally agreed to have acquired and secondarily lost generalized mitochondrial function with vestigial mitochondrial organelles termed mitosomes [5]. In addition it has gained (by lateral transfer of genes from coexisting organisms [6]) the capacity to exist as a microaerophile rather than an obligate anaerobe. There is some evidence for it being sexual [7] including the presence of meiotic gene homologues [8] and recombination between assemblages [7]. However, the plasticity (variable genome structure) and aneuploidy (uneven numbers of whole chromosomes or parts thereof) of its genome [9] are inconsistent with Giardia being a conventional sexual organism. These latter characteristics complicated interpretation of early genomic studies on chromosome number and identity as well as variation among isolates. Nonetheless, the advent of pulsed field gel electrophoresis (PFGE) for separating large DNA molecules up to 5 Mb (Figure 1) [10] and rapid large scale sequencing techniques [11] have allowed a better understanding of the Giardia genome. What has been missing until recently was the integration of PFGE data and the genome sequence [12,13]. This review addresses how whole chromosome separation and publically available Giardia genome sequence data were used to construct a working map of the chromosomes of G. lamblia assemblage A isolate WB.
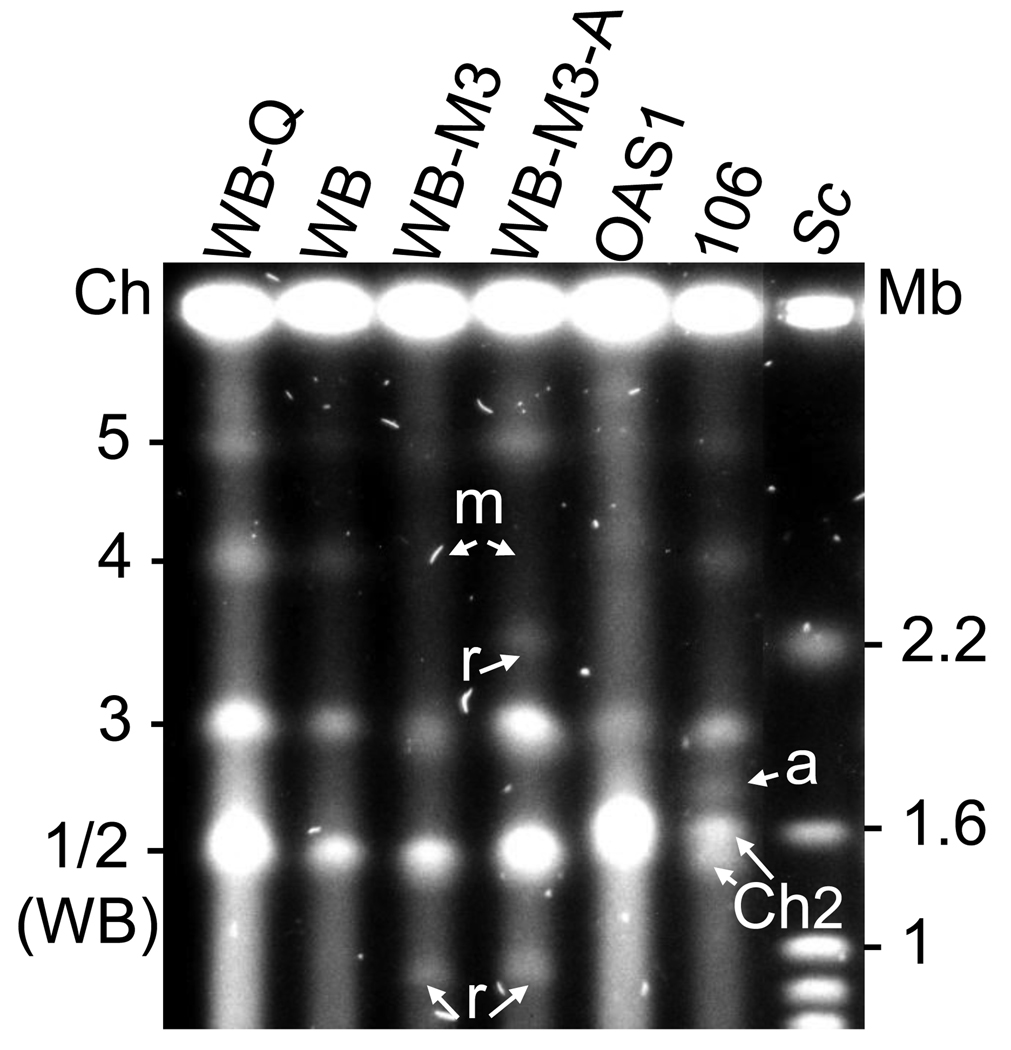
Figure 1.
Chromosomes of G. lamblia isolates and drug-resistant lines frequently have duplicated, partially duplicated or rearranged chromosomes. Giardia chromosomes were separated by pulsed field gel electrophoresis [12] with Saccharomyces cerevisiae (Sc) chromosomes as markers (their sizes are indicated in Mb). The human isolate WB has four chromosome bands with the smallest, apparently 1.5 Mb band, carrying two different chromosomes, chromosome 1 and 2 (see Figure 2). The metronidazole-resistant line (WB-M3) derived from WB [41] and the albendazole-resistant line (WB-M3-A) derived from WB-M3 [40] have clear chromosome rearrangements of chromosome 4 [40]. Chromosome 4 appears to be missing (m) in both WB-M3 and WB-M3-A but a chromosome 4-specific probe hybridizes to material retained in the well of WB-M3 and to the chromosome 3 and 5 ethidium bromide stained bands of WB-M3-A [40]. Chromosome bands of < 1 Mb, arising from the chromosome rearrangements (r), are apparent in both WB-M3 and WB-M3-A. A quinacrine-resistant line (WB-Q) also derived from WB [55] has no obvious chromosome rearrangements. The sheep isolate, OAS1 [56] appears to have similarly sized chromosomes to WB but the human isolate BRIS/83/HEU/106 has two chromosome bands of approximately 1.5 Mb. These two bands are not different chromosomes as in the case of the isolate WB, but are duplications of chromosome 2 (Ch2) as determined by hybridization with a number of WB chromosome 2-specific probes [9,12,22]. Chromosome 1 in isolate BRIS/83/HEPU/106 is not present in the 1.5 Mb chromosome band as is the case with WB [22]. An accessory chromosome (a) is indicated in isolate BRIS/83/HEPU/106. This figure is reproduced in part from Ref. [40].
About the genome of G. lamblia
G. lamblia has five typically eukaryotic chromosomes capped with telomere repeats in its two nuclei (Figure 1; Box 1) [9,14,15]. In addition to these five major chromosomes, variable minor (in copy number) or accessory chromosomes are present [15]. These accessory chromosomes appear to be duplications (or partial duplications) of major chromosomes, in some cases carrying long rRNA gene arrays [16,17]. Two G. lamblia assemblages (a term which has received strong support to describe two major groups of genetically diverse isolates not confined to any geographic location [18]), A and B, infect humans, with the best studied assemblage A isolate WB used to sequence the assemblage A, 11.7 Mb haploid genome (Box 1) [19,20]. Trophozoites, the replicative stage of the parasite, are mostly tetraploid (four complete copies of the genome split between two nuclei) (Box 1) [20,21] although there is considerable variation among different isolates and in content of minor, or accessory chromosomes [22,23]. The ploidy (the number of complete genome copies) is not uniform among isolates with some DNA sequences repeated more than others including the long tandem repeats of rRNA genes located sub-telomerically at one end (arbitrarily referred to here as the downstream end) of each chromosome [14,16,24–26]. The accessory or minor chromosomes which are extremely variable in length may be duplicated several fold, and are the location of long tandem rRNA gene arrays [9,16,17]. These rRNA gene-carrying minor chromosomes also carry conserved regions [26]. However, the focus of this review is the major WB Giardia chromosomes, the two smallest of which are of similar size (1.5 Mb) and distinguished by two different collections of probes [22,27]. The remaining three major chromosomes are clearly separated by PFGE as shown in Figure 1.
Box 1. Giardia lamblia: a flagellated, gut protozoan parasite of humans.
Human Giardia isolates
There are two major groups of genetically diverse Giardia isolates which infect humans: assemblages A and B [7,18].
Assemblage A isolates (with WB, actually the cloned line WBC6 derived from WB, being the representative sequenced isolate [20]) typically have chromosomes of 1.5, 2, 3 and ~ 3.5 Mb.
Assemblage B isolates (with isolate GS the representative sequenced isolate [21]) appear to be missing the smallest 1.5 Mb chromosome(s), the smallest assemblage B chromosome which hybridizes with a chromosome 2-specific probe, the PFOR1 gene (Figure 2), is around 1.8 Mb [9,23, 43,44].
Assemblage B isolates are notoriously difficult and slow growing in culture [43,45] but more readily infect adult mice than assemblage A isolates [46].
Genomes of assemblage B isolates appear to be more variable than the A isolates with Not I cleavage patterns among A isolates being very similar and often identical, while B isolate Not I cleavage patterns are distinctly different from each other [12,23].
Giardia genomes
The WBC6 genome sequence (assemblage A) (found at http://www.giardiadb.org [32]) comprises 306 contigs (a group of cloned or copied segments of DNA representing overlapping regions of a particular chromosome). These contigs have been compiled as 92 scaffolds or supercontigs (defined in genomic mapping as a series of contigs that are in the right order). Of the 306 contigs, 200 are larger than 1000 bp, with the largest being ctg02_01 at 870 956 bp [20]. Seventeen scaffolds are represented in the chromosome ma ps covering 93% of the genome. The largest of the unassigned scaffolds is 82 kb but 71 are less than 6 kb in length.
The GS genome (assemblage B) shares 77% identity with WBC6 [21].
Assemblages A and B are so different as to be regarded by many experts as constituting separate species (GiardiaDB discussion group found at http://www.giardiadb.org) [47].
The genomes of both WB and GS isolates are claimed to be tetraploid [15,21,48,49].
Gene probes used to distinguish assemblage A from B isolate include the rRNA gene spacer region [50], glutamate dehydrogenase, triose phosphate isomerase, ferredoxin and histone genes [7] and beta-giardin [51] with multilocus genotyping proving most informative [52]. However, a higher level of allelic sequence divergence in assemblage B genomes [52] may complicate chromosome mapping in assemblage B isolates.
Five other discrete Giardia assemblages infect animals: (i) C and D (dogs), (ii) E (livestock), (iii) F (cats) and (iv) G (rodents) [18,23,50,53].
Each major chromosome downstream end comprises a sub-telomeric region termed telomere gene unit (TGU), conserved arrangements of a cysteine-rich protein gene (or variable-surface protein (VSP) gene [28]), protein kinase (gPK) and ankyrin (ANK) genes followed by a highly conserved breakpoint into the LSrRNA gene (refer to Figure 2b) [9,14]. rRNA genes and terminal telomere repeats, (TAGGG)n, are separated by hyper-variable sequences including transposable elements [29]. Each major chromosome carries rRNA gene sequences at the downstream end only [16,17] but these may not constitute complete genes [16] and therefore will not give rise to functional ribosomes; also, the number of rRNA gene repeats on major chromosomes appears to be restricted to a maximum of four [14]. Telomeric repeat sequences are present at both ends of major chromosomes [16,17].
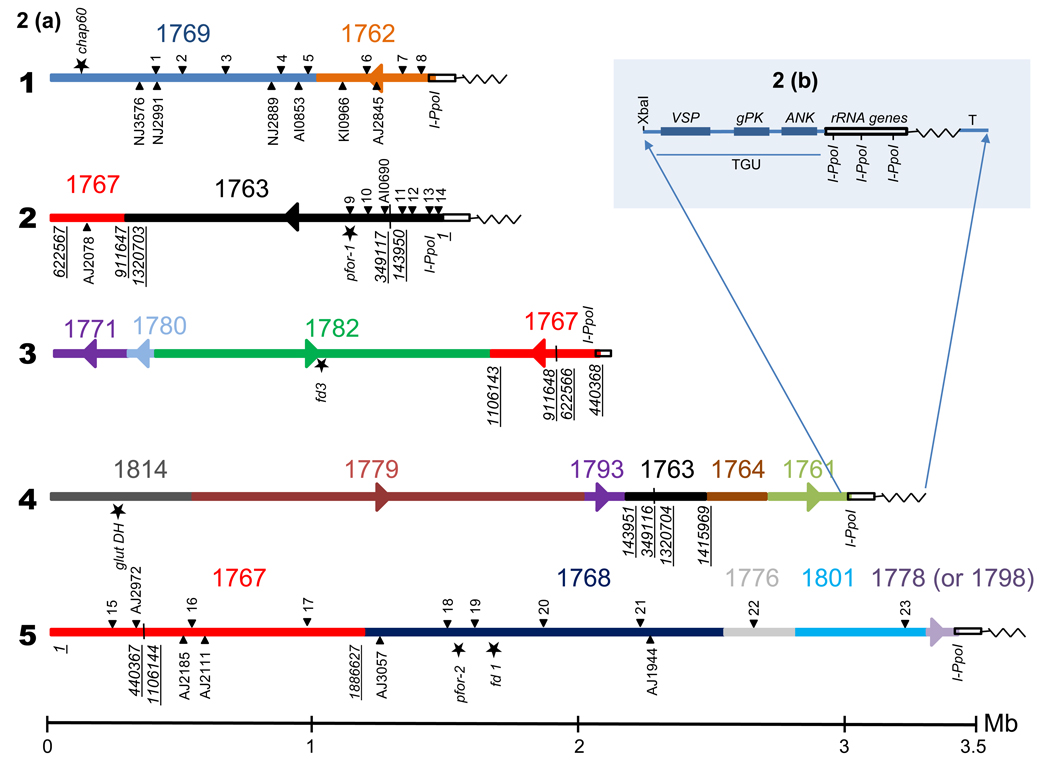
Figure 2.
Map of WBC6 chromosome sequences. (a) Refer to Box 1 for details of the genome of WBC6. Maps were compiled using chromosome-specific probes, Not I segments and I-Ppo I as a marker of the LSrRNA genes on downstream chromosome ends. Sizes of the chromosomes are indicated by the horizontal bar below the maps but sizes are only roughly drawn to scale with chromosome 5 represented as a ~ 3.5 Mb chromosome although only 3.1 Mb of sequence has been assigned to it. Maps show scaffolds (numbers and sizes defined by colours along the chromosome) associated with each chromosome. Arrow heads on chromosomes indicate scaffold orientation (where known) relative to the presentation in GiardiaDB. Chromosome-specific probes (closed arrow heads) are indicated as numbers and are detailed in Table 1. Mixed lettered and numbered sites below and above the maps are chromosome-specific probe sequences [33]. Underlined numbers in italics indicate the start and end of scaffold segments where scaffolds are believed to have been erroneously constructed. I-Ppo I sites are indicated on downstream terminal scaffolds but not in scaffold CH991779. Stars indicate chromosome marker genes. The open, bold rectangles at the end of each chromosome represent rRNA genes and the zigzagged line indicates sub-telomeric sequences and telomere repeats. The rectangles and lines are not drawn to scale. (b) Representation of the order of genes on the major chromosome downstream sub-telomeric ends. The map is not drawn to scale, e.g., gPK and ANK are juxtaposed in at least one sub-telomeric region [14]. The cysteine-rich protein gene (referred to as VSP gene [28]), protein kinase gene (gPK) and ankyrin gene (ANK) constitute a telomere gene unit (TGU) [14] although these genes may not as yet be annotated on all of the scaffolds which define the ends of the major chromosomes. The breakpoint into the LSrRNA gene is highly conserved but the sequence between the rRNA genes and telomere repeats (T) (TAGGG)n is highly variable [29]. Each LSrRNA gene has one I-Ppo I site [16].
Physical mapping of the genome
It is relatively routine to visualize ethidium bromide stained, PFGE separated Giardia chromosomes [23], but compilation of a physical chromosome map (the complete chromosome sequence) is much more difficult. These days, optical mapping is the method of choice for mapping chromosomes [30]. This technique uses fluorescence microscopy to produce high-resolution restriction maps rapidly by directly imaging restriction digestion cleavage events occurring on single deproteinized DNA molecules. Ordered maps are then constructed by noting fragment order and size. Nevertheless, two decades ago generation of a collection of chromosome-specific markers derived from cloned DNA was the basis of chromosome mapping. Such markers were used, initially, to map 13 Not I segments (totaling 1.8 Mb) of the 2 Mb chromosome [31]. When the genome sequence of WBC6 (a cloned line of the WB isolate) was released and the Giardia genome database (GiardiaDB) was made available at http://www.giardiadb.org [32], the 2 Mb chromosome map, with the help of one representative probe sequence from each Not I segment [13,31] (GenBank accession numbers L49329–L49335) was confirmed, and this inspired further mapping [12]. The 2 Mb and the other chromosomes have been variously numbered in the literature (Box 2). Compilation of the chromosome sequence maps has used other resources in conjunction with GiardiaDB and the role of three key restriction enzymes, Xba I, I-Ppo I and Not I, and a collection of chromosome-specific probes [9,12,13,22,33] in mapping G. lamblia isolate WB chromosomes are described below. The strategy used to map the chromosomes is outlined in Box 3.
Box 2. Giardia chromosome names.
WB has 5 major chromosomes [27].
Chromosome names have varied in the literature because of different separation conditions, apparatus and isolates used by various researchers [12,22].
The 1.5 Mb band of electrophoretically separated, ethidium bromide stained chromosomes (which carries chromosomes 1 and 2 in WB) (as shown in Figure 1) has been called chromosome 3 [22], since, in early field inversion gel electrophoretic chromosome separations [22] it was the third largest chromosome band. The 1.5 Mb band has also been referred to as chromosome 3/4 because of the two distinct bands of this size observed in isolate BRIS/83/HEPU/106 (Figure 1). These two bands were later shown to represent duplicated chromosomes (in fact duplications of chromosome 2) [9,12,22].
The 2 Mb band (which carries chromosome 3) (Figure 1) has previously been called chromosome 5 [31,13,54].
The 3 Mb band (carrying chromosome 4) (Figure 1) was referred to as chromosome 6 [12].
What appear to be duplicated chromosomes of around 3.5 Mb in isolate BRIS/83/HEPU/106, have been referred to as chromosomes 7/8 [9].
Box 3. How Giardia isolate WB chromosomes were mapped.
The summary below refers to Refs. [12,13,22,31]:
Intact chromosomes from G. lamblia isolate WB were separated by pulsed field gel electrophoresis in low melting temperature agarose.
The chromosome of interest was extracted from an entire row of chromosome separations.
A library of cloned inserts was constructed from purified, restriction enzyme cleaved chromosome.
Cloned inserts were selected and inserts (probes) of desired length (100–500 bp) were radio-labeled.
- Probes were hybridised against a pot of membranes carrying Southern transferred DNA. These membranes carried:
- Whole WB chromosome separations to identify chromosome-specific probes (chromosome markers).
- Chromosome separations of other isolates and drug-resistant lines.
- Not I cleaved whole chromosomes to identify sizes of Not I segments.
- Partially Not I cleaved chromosomes to aid in nearest neighbor identification.
- Giardia DNA cleaved into specific, small segments to identify unique DNA sequence probes.
All unique copy, chromosome-specific markers were sequenced and the sequence identified in GiardiaDB. Not I sites were matched from the estimated size of a specific Not I segment with sites in GiardiaDB. The number of markers is critical for fine mapping. Fewer markers per chromosome may limit the map resolution and leave sequence data unassigned as is the case here with 7% of the G. lamblia sequence not assigned to a specific chromosome.
The target contig and scaffold of interest were identified.
Scaffolds were aligned and oriented using a variety of techniques, e.g. I-Ppo I sites to define downstream ends and one internal I-Ppo I site in chromosome 4, rRNA genes sites at downstream ends, Not I cleavage site data and partial Not I chromosome cleavage data, rearranged chromosome data from drug-resistant lines, etc.
The logic (in the simplest outcome) behind partial cleavage hybridisation data relies on a specific marker identifying the smallest DNA band in PFGE separations as the segment of interest with the second smallest band being the segment of interest together with one of its adjoining neighbours. The latter can then be matched with a similar band in a hybridization with the marker for the nearest neighbour segment.
Where obvious errors in scaffold construction were apparent, contigs were kept intact.
XbaI, rRNA arrays and minor chromosomes
Sequence analysis of the VSP, gPK and ANK genes (constituting the TGU blocks) and rRNA genes revealed no sites for the restriction enzyme Xba I, which recognizes the 6 bp sequence TCTAGA [14,24]. Thus, Xba I-cleaved G. lamblia chromosomes maintain sub-telomeric regions including VSP, gPK, ANK, rRNA genes and telomeric repeat sequence intact [9,14]. The most downstream Xba I site in all chromosomes is located upstream of the TGUs [9,14]. Thus Xba I cleavage of intact Giardia chromosomes results in bands of sub-telomeric sequences including rRNA genes [14]. Long arrays of rRNA genes are apparent following rRNA gene hybridization of Xba I cleaved chromosomes with shorter rRNA gene sequences associated with the major chromosomes [14]. Telomere repeats do not appear to be associated with the long arrays of rRNA genes found on accessory chromosomes [14]. It has been proposed that the lack of telomere repeats and their replacement by transposable elements (or rRNA genes, for example) leads to length instability (see Ref. [14]) possibly explaining the instability of minor accessory chromosomes.
I-Ppo I defined the downstream end of Giardia chromosomes
I-PpoI is a rare-cutting intron-encoded endonuclease isolated from Physarum polycephalum. It has a 13 to15 bp recognition sequence which appears to be conserved in the LSrRNA genes of higher eukaryotes [34]. In G. lamblia it also cleaves the LSrRNA gene once, and as previously reported [9,12,16], is a useful tool to remove rRNA gene tandem arrays located at the downstream end of each chromosome. The removal of the arrays allows chromosomes to more readily enter agarose gels leading to improved chromosome separation [12]. However, I-Ppo I also cleaves the 3 Mb chromosome 4, and this was a useful mapping tool [12]. The I-Ppo I site within chromosome 4 (on scaffold CH991779) (refer to Box 1 for scaffold and contig definitions) (Figure 2(a)) is embedded in approximately 500 bp of rRNA gene sequence with the upstream sequence carrying an ankyrin gene, found in TGUs [14]. However, similarity with sub-telomeric and rRNA gene sequences deteriorates upstream of the ankyrin stop codon and downstream from the 500 bp of rRNA gene sequence [12]. The internal I-Ppo I site in chromosome 4 (on scaffold CH991779) was the only internal chromosome I-Ppo I site observed in gel separations of I-Ppo I cleaved chromosomes. All other I-Ppo I sites are sub-telomeric [12]. Thus when the association of chromosome 4 with the internally located scaffold CH991763 carrying another I-Ppo I site was identified it was deduced that scaffold CH991763 had been erroneously constructed [12]. It was concluded that the majority of scaffold CH991763, including the I-Ppo I site, was associated with chromosome 2 (Figure 2).
GiardiaDB has 7 scaffolds which carry an I-PpoI site, one for each downstream end of each of the major chromosomes, one for the internal scaffold CH991779 of chromosome 4 and one other. The latter possibly belongs to chromosome 3 since this single linkage group [13,31] appears to have two different downstream sub-telomeric ends [16].
Not I untied the knots
Not I cleaves an 8 bp GC rich sequence which is represented twice in the LSrRNA G. lamblia gene [16,24]. It removes rRNA gene arrays from chromosomes and cleaves the remaining chromosome into a range of sizes, the largest around 500 kb [12,23]. This range of Not I segment sizes gave rise to a relatively small number of segments per chromosome which facilitated mapping [12,13,22, 23,31], but Not I segments alone were of no specific mapping value without appropriate markers. The latter came from cloned Giardia sequences hybridized against Giardia DNA cleaved into small fragments to identify unique sequences and then against chromosome separations to distinguish chromosome-specific markers [12,13,31,33]. Initially, partially cleaved chromosomes were used to identify neighbouring Not I segments (Box 3) [23,31], a technique which proved remarkably successful when GiardiaDB was released and the maps and the orientation of some scaffolds based on the Not I sites in GiardiaDB were confirmed [12,13]. The exception is the large ~3.5 Mb chromosome, to which only 3.1 Mb of sequence has been assigned, as there are not enough markers to either orient or map the scaffolds for this chromosome. The reason for this was the difficulty in obtaining enough clean material of the large chromosome to make a comprehensive chromosome-specific library.
The chromosome maps
G. lamblia isolate WB has four clear, ethidium bromide stained chromosome bands of 1.5, 2, 3 and ~3.5 Mb in pulsed field gel electrophoretic separations (PFGE) when compared with chromosome size markers (Figure 1) [27]. These chromosomes have been variously referred to depending on a number of factors as explained in Box 2. Other isolates belonging to assemblage A may have variations in size and position of the designated WB chromosomes as shown in Figure 1.
WB chromosome 1 is identified by a collection of probes previously referred to as Group 2 probes [12,22], four additional probes (Table 1) and a collection of probes published by Yu et al. [33]. These chromosome 1-specific probe sequences are found on scaffolds CH991769 (958263 bp) and CH991762 (439942 bp) (Figure 2) the sum of which is 1 398 205 Mb. The difference between this length and the estimated 1.5 Mb by PFGE is likely to be accounted for by telomeric and sub-telomeric sequences [14,29]. Scaffold CH991762 is downstream of CH991769 with the I-Ppo I site defining the downstream end of the chromosome, but the orientation of CH991762 is uncertain.
Table 1.
Location of Chromosome (Chrm)-specific probes
| Clone number |
Clone name |
Chromosome | Scaffold | Probe Start |
Probe end | Contig | GeneBank Accession number or Ref. |
|---|---|---|---|---|---|---|---|
| 1 | C6/14 | 1 | CH991769 | 375576 | 375965 | ctg02_23 | GF110436 |
| 2 | 855 | 1 | CH991769 | 457147 | 457254 | ctg02_23 | GF110437 |
| 3 | C6/320 | 1 | CH991769 | 614631 | 614821 | ctg02_22 | GF110438 |
| 4 | C3/29a | 1 | CH991769 | 711549 | 711908 | ctg02_11 | GF110429 |
| 5 | C3/20a | 1 | CH991769 | 953576 | 953774 | ctg02_11 | GF110430 |
| 6 | C6/50 | 1 | CH991762 | 212080 | 212349 | ctg02_04 | GF110439 |
| 7 | C3/25a | 1 | Ch991762 | 124371 | 124753 | ctg02_04 | GF110431 |
| 8 | C3–31a | 1 | Ch991762 | 48920 | 49091 | ctg02_04 | GF110432 |
| 9 | G6/1c | 2 | CH991763 | 504969 | 505158 | ctg02_06 | [12] |
| 10 | C3–19b | 2 | CH991763 | 408583 | 408877 | ctg02_06 | [12] |
| 11 | C3–28b | 2 | CH991763 | 146023 | 146718 | ctg02_28 | [12] |
| 12 | G15/4b | 2 | CH991763 | 122563 | 122796 | ctg02_28 | [12] |
| 13 | C6/318 | 2 | CH991763 | 80493 | 80695 | ctg02_28 | GF110440 |
| 14 | C3/11b | 2 | CH991763 | 49718 | 50074 | ctg02_28 | [12] |
| 15 | 811 | 5 | Ch991767 | 296551 | 296605 | ctg02_55 | GF110435 |
| 16 | 307 | 5 | Ch991767 | 1253148 | 1253279 | ctg02_14 | GF110434 |
| 17 | 833 | 5 | Ch991767 | 1684166 | 1684286 | ctg02_27 | GF110433 |
| 18 | C6/317 | 5 | CH991768 | 270364 | 270592 | ctg02_05 | GF110441 |
| 19 | 808 | 5 | CH991768 | 373767 | 373828 | ctg02_05 | GF110442 |
| 20 | 308 | 5 | CH991768 | 682967 | 683209 | ctg02_10 | GF110443 |
| 21 | 16 | 5 | CH991768 | 944658 | 944870 | ctg02_03 | GF110444 |
| 22 | 831 | 5 | CH991776 | 183364 | 183544 | ctg02_20 | GF110445 |
| 23 | 837 | 5 | CH991801 | 129299 | 129371 | ctg02_52 | GF110446 |
Chromosome 1-specific probes [22].
chromosome 2-specific probes [22]. These two groups of probes (a and b) were originally derived from the 1.5 Mb chromosome band of WB separated chromosomes [22].
G6/1 was described in [57] and when sequenced was found to encode the enzyme pyruvate:ferredoxin oxidoreductase.
The unique 1.5 Mb chromosome 2 is constituted by segments from two scaffolds, CH991767 and CH991763 (Figure 2) [12]. The nearly 300 kb segment of CH991767 (comprised largely of ctg02_13) is defined by the chromosome 2-specific probe sequence AJ2078 [33]. The remainder of chromosome 2 comprises 2 segments of CH991763 [12] with the I-Ppo I site located at the downstream end of the chromosome. The downstream CH991763 segment is defined by four chromosome 2-specific probes (Table 1) [12,22], and the second segment of CH991763 is defined by chromosome 2-specific probes [12] located on contig ctg02_06 (Table 1). It is assumed that the segment from the beginning of CH991763 is juxtaposed to the second segment from CH991763, but this may not be the case. It is possible that this second segment of CH991763 is in another orientation or indeed that it defines the upstream end of chromosome 2. The sequence length of chromosome 2, the sum of 289 080 bp from scaffold CH991767 and the two segments of CH9991763 amounting to 1 151 710 bp, is 1 440 790 Mb.
The 2 Mb chromosome 3 (Figure 2) [13,31] was first mapped as 13 Not I segments which were ordered according to analyses of Not I partially cleaved chromosomes [31]. In addition to the presentation of the Not I map, representative probes from each Not I segment were sequenced. When GiardiaDB was released, the order of the Not I segments was confirmed [13], but the total length of the scaffolds CH991771, CH991780, CH991782 and CH991767 identified by these probe sequences was > 3 Mb [13]. Because of the density of the 46 probes sequences spread along the chromosome [13], it was possible to predict that CH991767 was incorrectly compiled, with one segment of the almost 2 Mb scaffold belonging to chromosome 2 and two other segments of the scaffold belonging to the ~3.5 Mb chromosome [13]. In all cases of suspected erroneous scaffold construction, contigs were kept intact [12,13]. It was clear from the Not I and the sequence maps of the 2 Mb chromosome 3 that this chromosome is a single linkage group and that no other 2 Mb chromosome is present in WB [13,31]. However, as seen in a two-dimensional separation of Not I-cleaved chromosomes, there are two different rRNA gene carrying, sub-telomeric segments associated with chromosome 3 (arrowed spots aligning with the 2 Mb chromosome in Ref. [16]). One of these is predicted to be the I-Ppo I carrying segment of the beginning of CH991767, and the other is likely to be either CH991778 or CH991798, the only two of seven I-Ppo I carrying scaffolds not clearly associated with a specific chromosome. If there are two chromosomes of 2 Mb, then the majority of the chromosomes are the same with different downstream ends that were not detected by available probe sequences. This possibility raises the issue of similarity of the two nuclei: is it possible that there are two versions of some chromosomes and that they are located in different nuclei? The final size of chromosome 3 was determined to be 1 963 438 Mb [13].
The 3 Mb chromosome 4 was also mapped by identifying Not I segments which were later confirmed to be in that order (except two of the Not I segments) (Figure 2) [12]. Chromosome 4-specific probes identified 15 Not I segments accounting for 2.2 Mb of the chromosome, ensuring that the probe sequences were well spread along the chromosome. The probe sequences were located on scaffolds CH991814, CH991779, CH991793, CH991763, CH991764 and CH991761 unambiguously in that order with CH991761 carrying the I-Ppo I site [12]; however, it was not possible to predict the orientation of scaffolds CH991814, CH991763 and CH991764 [12]. Several anomalies associated with scaffold construction were evident including the incorrect compilation of scaffold CH991763. Removal of segments from CH991763 carrying intact contigs and sequences associated with other chromosomes resulted in a segment which was consistent with the Not I segment sizes that were mapped and also with the apparent size of chromosome 4 [12]. The final length of chromosome 4 was predicted to be 2 890 648 Mb [12]
Chromosome 5 has been difficult to map, and indeed the map shown in Figure 2 presents the scaffolds predicted by only 14 probe sequences (Table 1) to be associated with the ~3.5 Mb chromosome. These probes are made up of five reported to be chromosome 5-specific by Yu et al. [33] and nine others (Table 1). The map is presented in numerical order of the scaffolds and scaffold segments, but neither the orientation nor the order of these scaffolds has been confirmed. Nevertheless, the probe sequences have identified four scaffolds associated with chromosome 5: (i) CH991767 (two segments of this scaffold of 440 367 and 780 483 bp as shown in Figure 2), (ii) scaffold CH991768 (1 312 034 Mb), (iii) CH991776 (426 100 bp), (iv) and CH991801 (154 297 bp). The downstream end of chromosome 5 is likely to be associated with an I-Ppo I carrying scaffold two of which (CH991778 of 41 350 bp or CH991798 of 64 051 bp) have so far not been assigned to another chromosome. The sum of all sequences assigned to chromosome 5, assuming CH991778 is the terminal scaffold, is 3 113 281 Mb.
The sum of all sequences from the 11.7 Mb in GiardiaDB assigned to a specific chromosome is 10 911 763 Mb with the remainder of the genome unassigned to chromosomes being ~800 kb, i.e. 93% of the genome sequence has been assigned to one of the five major chromosomes (Box 3). Genes which are markers for each chromosome (or duplicated chromosomes as shown in isolate BRIS/83/HEPU/106 (106) in Figure 1 and also in Refs. [9,22] are: (i) chaperonin 60 [GL50803_103891] (refer to http://www.giardiadb.org for gene sequences) for chromosome 1 [35]; (ii) the pyruvate ferredoxin oxidoreductase gene pfor-1 [GL50803_114609] for chromosome 2 [9,22]; (iii) the ferredoxin 3 gene, Fd3 [GL50803_10329] for chromosome 3; (iv) NADP-specific glutamate dehydrogenase gene, glutDH [GL50803_21942 for chromosome 4 [36]; and (v) pfor-2 and Fd2 [GL50803_17063 and GL50803_9662, respectively] both on scaffold CH991768 for chromosome 5 (Figure 2).
Community access to Giardia chromosome mapping data
The data presented here, in previous publications [12,13] and together with recently available optical mapping data (Hilary Morrison and Rodney Adam, personal communication) will be integrated into GiardiaDB which is one of the genomics database resources available under the Eukaryotic Pathogen Database Resources (EuPathDB) for eukaryotic pathogens umbrella [37]. EuPathDB includes ToxoDB (http://www.toxodb.org), the genomic resource, for Toxoplasma, and PlasmoDB (http://www.plasmodb.org), for Plasmodium species. The new format for chromosome presentation in GiardiaDB will be the replacement of scaffolds or supercontigs in the GBrowse [38] view with whole chromosomes. Such a view format is available in ToxoDB and can be accessed using the Anciliary GBrowse tool. Similarly to ToxoDB and PlasmoDB, GiardiaDB has genomic sequence from several isolates or species; in addition to the reference genome of assemblage A, WBC6 isolate, assemblage B isolate GS and an assemblage E isolate are represented (Box 1). In ToxoDB, for example, users can drill down from the chromosome view, to the contig and gene view, to synteny displayed among other isolates and species compared with the reference genome. Another possible addition to GiardiaDB are chromosome-specific marker sites also in the GBrowse view with associated mouse-over popup boxes providing all relevant information. EuPath has recently undergone an upgrade to enhance data accessibility and connections between diverse data types [37] and is dedicated to integrating new data.
What next?
The map shown in Figure 2 is a working map requiring confirmation of some regions and likely to vary from isolate to isolate. Fine sequencing of some regions is required, e.g., across scaffold boundaries to ensure the correct orientation of the juxtaposed scaffolds and to ensure the scaffold boundaries fully represent that region of the intact chromosome. PCR across these scaffold boundaries is a likely starting point for sequence confirmation. The assemblage A chromosome map provides the basis from which to map the chromosomes of the assemblage B isolate GS (Box 1), the genome of which has recently been published [21]. Preliminary mapping of WB chromosome 3 homologues in the assemblage B isolate BRIS/91/HEPU/1279 and an isolate from a calf BRIS/93/HEPU/1709 showed, where partial Not I cleavage data was available, that the order of Not I segments of the chromosome homologues are maintained [23]. Further mapping using WB as a guide will facilitate the compilation of a working sequence map of assemblage B chromosomes.
Compilation of the WB chromosome maps has highlighted the genetic variation among isolates of assemblage A and while the chromosome designation for WB shown in Figure 1 has long been accepted, it does not apply to isolate 106, for example (Figure 1). The latter has duplications of chromosome 2 of approximately 1.5 Mb but chromosome 1-specific sequences hybridise to two bands significantly larger than 1.5 Mb [22]. In addition, 106 also appears to have duplications of chromosome 5 [9]. An assemblage A isolate derived from a cockatoo has duplications of chromosome 2 [9]. Why these and other isolates have duplicated chromosomes can now be addressed in context of the WB chromosome maps, e.g. fine mapping of the duplicated 106 chromosomes will inevitably reveal some differences [9,23]. In situ hybridization can then be employed to show the nuclear localization of each of the duplicated chromosome sequences since evidence is accumulating that the nuclei may not be identical [39]. Furthermore, identifying the differences between these duplicated chromosomes will lead to an understanding of the mechanism of chromosome duplication in Giardia.
Genetic variation, a hallmark of Giardia genomics can now be studied in the context of whole chromosome rearrangements which may also be useful epidemiological tools. Under the stress of drug pressure, Giardia readily rearranges its chromosomes as shown for a metronidazole-resistant and an albendazole-resistant line of WB (Figure 1) [12,31,40]. Chromosome 4 is not evident in the metronidazole-resistant line WB-M3 [40,41]. However, chromosome bands identified by chromosome 4-specific probes were evident in both the metronidazole- and albendazole-resistant lines derived from WB [40]. These chromosome rearrangements and others have been useful in mapping studies to verify contiguous regions of chromosomes [12,31]. Much more intriguing is how Giardia so readily rearranges its chromosomes. By comparing maps of the parental and drug-resistant lines, we can now identify breakpoints of the rearrangements to gain an understanding of this mechanism of survival and defense of the parasite. Integration of foreign DNA into the Giardia genome as shown for a rep-like sequence [42] can be studied with the maps as reference. A plasmid was found integrated into the genome of a Giardia isolate obtained from an extremely sick child [3]. The region of integration can now be studied more globally: is the region of integration a hotspot for foreign DNA and as such a risk for virulence production? As pointed out above, the I-Ppo I site found internally in chromosome 4 of assemblage A but not assemblage B isolates is an example of a likely evolutionary marker. Flanking regions of this site can be compared with homologous regions in assemblages not carrying the I-Ppo I site to identify any obvious breakpoints.
Summary
93% of the G. lamblia assemblage A genome has been assigned to one of 5 major chromosomes ranging in size from 1.5 to 3.5 Mb. A working chromosome map of this genome has been compiled and highlights both techniques of large scale sequencing and physical mapping which prove to be complementary in understanding the genome biology of this early diverging eukaryote. With this map as reference and within GBrowse in GiardiaDB: (i) areas requiring fine sequencing, i.e. across scaffold boundaries can be addressed initially using PCR across scaffold boundaries; (ii) other genome sequences can be mapped in context using the tools available to EuPathDB; (iii) the search for functional aspects of gene arrangement such as breakpoints in chromosome rearrangements involved in drug resistance can be addressed; (iv) clues to understanding the function of the two nuclei can be sought; and (v) genetic variation, a hallmark of Giardia genomics, has a new focus.
Acknowledgements
This work was supported in part by the National Health and Medical Research Council of Australia, the Australian Research Council and by U01 Cooperative Research Agreement AI75527 from the National Institutes of Health, U.S.A. Our thanks to Hilary Morrison, Rodney Adam, GiardiaDB and EuPathDB members for helpful and enthusiastic cooperation. Thanks also to past Upcroft laboratory members involved in this work especially Mahin Abedinia and Nanhua Chen.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Buret AG. Pathophysiology of enteric infections with Giardia duodenalis. Parasite. 2008;15:261–265. doi: 10.1051/parasite/2008153261. [DOI] [PubMed] [Google Scholar]
- 2.Lauwaet T, et al. Encystation of Giardia lamblia: a model for other parasites. Curr. Opin. Microbiol. 2007;10:554–559. doi: 10.1016/j.mib.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Upcroft J, Upcroft P. My favorite cell: Giardia. Bioessays. 1998;20:256–263. doi: 10.1002/(SICI)1521-1878(199803)20:3<256::AID-BIES9>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 4.Gupta RS, Golding GB. The origin of the eukaryotic cell. Trends Biochem. Sci. 1996;21:166–171. [PubMed] [Google Scholar]
- 5.Regoes A, et al. Protein import, replication, and inheritance of a vestigial mitochondrion. J. Biol. Chem. 2005;280:30557–30563. doi: 10.1074/jbc.M500787200. [DOI] [PubMed] [Google Scholar]
- 6.Nixon JE, et al. Evidence for lateral transfer of genes encoding ferredoxins, nitroreductases, NADH oxidase, and alcohol dehydrogenase 3 from anaerobic prokaryotes to Giardia lamblia and Entamoeba histolytica. Eukaryot. Cell. 2002;1:181–190. doi: 10.1128/EC.1.2.181-190.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lasek-Nesselquist E, et al. Genetic exchange within and between assemblages of Giardia duodenalis. J. Eukaryot. Microbiol. 2009;56:504–518. doi: 10.1111/j.1550-7408.2009.00443.x. [DOI] [PubMed] [Google Scholar]
- 8.Ramesh MA, et al. A phylogenomic inventory of meiotic genes; evidence for sex in Giardia and an early eukaryotic origin of meiosis. Curr. Biol. 2005;15:185–191. doi: 10.1016/j.cub.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Upcroft P, Upcroft JA. Organization and structure of the Giardia genome. Protist. 1999;150:17–23. doi: 10.1016/S1434-4610(99)70005-2. [DOI] [PubMed] [Google Scholar]
- 10.Cantor CR, et al. Pulsed-field gel electrophoresis of very large DNA molecules. Annu. Rev. Biophys. Biophys. Chem. 1988;17:287–304. doi: 10.1146/annurev.bb.17.060188.001443. [DOI] [PubMed] [Google Scholar]
- 11.Hall N. Advanced sequencing technologies and their wider impact in microbiology. J. Exp. Biol. 2007;210(Pt 9):1518–1525. doi: 10.1242/jeb.001370. [DOI] [PubMed] [Google Scholar]
- 12.Upcroft JA, et al. Sequence map of the 3-Mb Giardia duodenalis assemblage A chromosome. Chromosome Res. 2009;17:1001–1014. doi: 10.1007/s10577-009-9084-4. [DOI] [PubMed] [Google Scholar]
- 13.Krauer KG, et al. Sequence map of the 2 Mb Giardia lamblia assemblage A chromosome. J. Parasitol. 2010;96:660–662. doi: 10.1645/GE-2328.1. [DOI] [PubMed] [Google Scholar]
- 14.Upcroft P, et al. Telomeric organization of a variable and inducible toxin gene family in the ancient eukaryote Giardia duodenalis. Genome Res. 1997;7:37–46. doi: 10.1101/gr.7.1.37. [DOI] [PubMed] [Google Scholar]
- 15.Le Blancq SM, Adam RD. Structural basis of karyotype heterogeneity in Giardia lamblia. Mol. Biochem. Parasitol. 1998;97:199–208. doi: 10.1016/s0166-6851(98)00150-9. [DOI] [PubMed] [Google Scholar]
- 16.Upcroft JA, et al. Rearranged subtelomeric rRNA genes in Giardia duodenalis. Eukaryot. Cell. 2005;4:484–486. doi: 10.1128/EC.4.2.484-486.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Blancq SM. Chromosome rearrangements in Giardia lamblia. Parasitol. Today. 1994;10:177–179. doi: 10.1016/0169-4758(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 18.Thompson RC, et al. Nomenclature and genetic groupings of Giardia infecting mammals. Parasitol. Today. 2000;16:210–213. doi: 10.1016/s0169-4758(99)01624-5. [DOI] [PubMed] [Google Scholar]
- 19.Fan JB, et al. Giardia lamblia: haploid genome size determined by pulsed field gel electrophoresis is less than 12 Mb. Nucleic Acids Res. 1991;19:1905–1908. doi: 10.1093/nar/19.8.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morrison HG, et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;317:1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- 21.Franzén O, et al. Draft genome sequencing of Giardia intestinalis assemblage B isolate GS: is human giardiasis caused by two different species? PLoS Pathog. 2009;5:e1000560. doi: 10.1371/journal.ppat.1000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Upcroft JA, et al. Chromosomal duplication in Giardia duodenalis. Int. J. Parasitol. 1993;23:609–616. doi: 10.1016/0020-7519(93)90167-w. Erratum in: Int. J. Parasitol. 23, 1091. [DOI] [PubMed] [Google Scholar]
- 23.Upcroft JA, et al. Mapping variation i n chromosome homologues of different Giardia strains. Mol. Biochem. Parasitol. 1996;76:135–143. doi: 10.1016/0166-6851(95)02554-5. [DOI] [PubMed] [Google Scholar]
- 24.Healey A, et al. Complete nucleotide sequence of the ribosomal RNA tandem repeat unit from Giardia intestinalis. Nucleic Acids Res. 1990;18:4006. doi: 10.1093/nar/18.13.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adam RD, et al. Telomeric location of Giardia rDNA genes. Mol. Cell. Biol. 1991;11:3326–3330. doi: 10.1128/mcb.11.6.3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adam RD. Chromosome-size variation in Giardia lamblia: the role of rDNA repeats. Nucleic Acids Res. 1992;20:3057–3061. doi: 10.1093/nar/20.12.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adam RD, et al. The Giardia lamblia trophozoite contains sets of closely related chromosomes. Nucleic Acids Res. 1988;16:4555–4567. doi: 10.1093/nar/16.10.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adam RD. Biology of Giardia lamblia. Clin. Microbiol. Rev. 2001;4:447–475. doi: 10.1128/CMR.14.3.447-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prabhu A, et al. Characterisation of the subtelomeric regions of Giardia lamblia genome isolate WBC6. Int. J. Parasitol. 2007;37:503–513. doi: 10.1016/j.ijpara.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 30.Wang YK, et al. Optical mapping of site-directed cleavages on single DNA molecules by the RecA-assisted restriction endonuclease technique. Proc. Natl. Acad. Sci. USA. 1995;92:165–169. doi: 10.1073/pnas.92.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen N, et al. Physical map of a 2 Mb chromosome of the intestinal protozoan parasite Giardia duodenalis. Chromosome Res. 1994;2:307–313. doi: 10.1007/BF01552724. [DOI] [PubMed] [Google Scholar]
- 32.Aurrecoechea C, et al. GiardiaDB and TrichDB: integrated genomic resources for the eukaryotic protist pathogens Giardia lamblia and Trichomonas vaginalis. Nucleic Acids Res. 2009;37:D526–D530. doi: 10.1093/nar/gkn631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu LZ, et al. The two nuclei of Giardia each have complete copies of the genome and are partitioned equationally at cytokinesis. Eukaryot. Cell. 2002;1:191–199. doi: 10.1128/EC.1.2.191-199.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muscarella DE, et al. Characterization of I-Ppo I, an intron-encoded endonuclease that mediates homing of a group I intron in the ribosomal DNA of Physarum polycephalum. Mol. Cell Biol. 1990;10:3386–3396. doi: 10.1128/mcb.10.7.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roger AJ, et al. A mitochondrial-like chaperonin 60 gene in Giardia lamblia: evidence that diplomonads once harbored an endosymbiont related to the progenitor of mitochondria. Proc. Natl. Acad. Sci. USA. 1998;95:229–234. doi: 10.1073/pnas.95.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monis PT, et al. Molecular genetic analysis of Giardia intestinalis isolates at the glutamate dehydrogenase locus. Parasitology. 1996;112:1–12. doi: 10.1017/s0031182000065021. [DOI] [PubMed] [Google Scholar]
- 37.Aurrecoechea C, et al. EuPathDB: a portal to eukaryotic pathogen databases. Nucleic Acids Res. 2010;38(Database issue):D415–D419. doi: 10.1093/nar/gkp941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stein LD, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4:e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Upcroft J, et al. Albendazole resistance in Giardia is correlated with cytoskeletal changes but not with a mutation at amino acid 200 in beta-tubulin. Microb. Drug Resist. 1996;2:303–308. doi: 10.1089/mdr.1996.2.303. [DOI] [PubMed] [Google Scholar]
- 41.Townson SM, et al. Induction of metronidazole and furazolidone resistance in Giardia. Trans. R. Soc. Trop. Med. Hyg. 1992;86:521–522. doi: 10.1016/0035-9203(92)90095-t. [DOI] [PubMed] [Google Scholar]
- 42.Gibbs MJ, et al. Two families of rep-like genes that probably originated by interspecies recombination are represented in viral, plasmid, bacterial, and parasitic protozoan genomes. Mol. Biol. Evol. 2006;23:1097–1100. doi: 10.1093/molbev/msj122. [DOI] [PubMed] [Google Scholar]
- 43.Upcroft JA, et al. Biological and genetic analysis of a longitudinal collection of Giardia samples derived from humans. Acta Trop. 1995;60:35–46. doi: 10.1016/0001-706x(95)00100-s. [DOI] [PubMed] [Google Scholar]
- 44.Upcroft JA, Upcroft P. Two distinct varieties of Giardia in a mixed infection from a single human patient. J. Eukaryot. Microbiol. 1994;41:189–194. doi: 10.1111/j.1550-7408.1994.tb01495.x. [DOI] [PubMed] [Google Scholar]
- 45.Karanis P, Ey PL. Characterization of axenic isolates of Giardia intestinalis established from humans and animals in Germany. Parasitol. Res. 1998;84:442–449. doi: 10.1007/s004360050427. [DOI] [PubMed] [Google Scholar]
- 46.Byrd LG, et al. Giardia lamblia infections in adult mice. Infect. Immun. 1994;62:3583–3585. doi: 10.1128/iai.62.8.3583-3585.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Monis PT, et al. Variation in Giardia: towards a taxonomic revision of the genus. Trends Parasitol. 2009;25:93–100. doi: 10.1016/j.pt.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 48.Bernander R, et al. Genome ploidy in different stages of the Giardia lamblia life cycle. Cell Microbiol. 2001;3:55–62. doi: 10.1046/j.1462-5822.2001.00094.x. [DOI] [PubMed] [Google Scholar]
- 49.Hou G, et al. Structure of a frequently rearranged rRNA-encoding chromosome in Giardia lamblia. Nucleic Acids Res. 1995;23:3310–3317. doi: 10.1093/nar/23.16.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Upcroft JA, et al. A new rDNA repeat unit in human Giardia. J. Eukaryot. Microbiol. 1994;41:639–642. doi: 10.1111/j.1550-7408.1994.tb01526.x. [DOI] [PubMed] [Google Scholar]
- 51.Cacciò SM, et al. Multilocus genotyping of Giardia duodenalis reveals striking differences between assemblages A and B. Int. J. Parasitol. 2008;38:1523–1531. doi: 10.1016/j.ijpara.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 52.Lebbad M, et al. From mouse to moose: multilocus genotyping of Giardia isolates from various animal species. Vet. Parasitol. 2010;168:231–239. doi: 10.1016/j.vetpar.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 53.Wielinga CM, Thompson RC. Comparative evaluation of Giardia duodenalis sequence data. Parasitology. 2007;134:1795–1821. doi: 10.1017/S0031182007003071. [DOI] [PubMed] [Google Scholar]
- 54.Upcroft P, Upcroft JA. Comparison of properties of agarose for electrophoresis of DNA. J. Chromatogr. 1993;618:79–93. doi: 10.1016/0378-4347(93)80028-3. [DOI] [PubMed] [Google Scholar]
- 55.Upcroft JA, et al. Quinacrine-resistant Giardia duodenalis. Parasitology. 1996;112:309–313. doi: 10.1017/s0031182000065823. [DOI] [PubMed] [Google Scholar]
- 56.Capon AG, et al. Similarities of Giardia antigens derived from human and animal sources. Int. J. Parasitol. 1989;19:91–98. doi: 10.1016/0020-7519(89)90026-x. [DOI] [PubMed] [Google Scholar]
- 57.Upcroft JA, et al. A gene associated with cell division and drug resistance in Giardia duodenalis. Parasitology. 1992;104:397–405. doi: 10.1017/s0031182000063642. [DOI] [PubMed] [Google Scholar]