Abstract
The goal of an HIV vaccine is to generate robust and durable protective antibody. Vital to this goal is the induction of CD4+ T follicular helper cells (TFH). However, very little is known about the TFH response to HIV vaccination and its relative contribution to magnitude and quality of vaccine-elicited antibody titers. Here, we investigated these questions in the context of a DNA/modified vaccinia virus Ankara (MVA) SIV vaccine with and without gp140 boost in alum in rhesus macaques. In addition, we determined the frequency of vaccine-induced CD4+ T cells co-expressing chemokine receptor, CXCR5 (facilitates migration to B cell follicles) in blood and whether these responses were representative of lymph node (LN) TFH responses. We show that booster MVA immunization induced a distinct and transient accumulation of proliferating CXCR5+ and CXCR5− CD4 T cells in blood at day 7-post immunization and the frequency of the former but not the latter correlated with TFH and B cell responses in germinal centers (GC) of LN. Interestingly, gp140 boost induced a skewing towards CXCR3 expression on GC TFH cells, which was strongly associated with longevity, avidity, and neutralization potential of vaccine-elicited antibody response. However, CXCR3+ cells preferentially expressed the HIV co-receptor CCR5 and vaccine-induced CXCR3+ CXCR5+ cells showed a moderate positive association with peak viremia following SIV251 infection. Taken together, our findings demonstrate that vaccine regimens that elicit CXCR3 biased TFH cell responses favor antibody persistence and avidity but may predispose to higher acute viremia in the event of breakthrough infections.
INTRODUCTION
The induction of robust and long-lived antibody responses forms the basis of protective immunity elicited by most vaccines (1). Antibody dynamics following immunization result from activation of antigen-specific B cells and their subsequent commitment into two distinct cell fates - extrafollicular plasmablasts or germinal center (GC) B cells. Plasmablasts are rapidly proliferating antibody-secreting cells (ASC) within secondary lymphoid organs that mainly contribute to peak antibody titers within the first few weeks after immunization (1, 2). Long-lived serological memory is established by GC-derived bone marrow resident plasma cells. GCs arise within B cell follicles typically 2–4 weeks after immunization and comprise of antigen-specific B cell clones of varying affinity that result from rapid B cell proliferation and receptor diversification. High affinity clones that successfully engage T cell receptor (TCR) on CD4+ T follicular helper cells (TFH) cells within the GCs receive TFH cell help in the form of cytokines such as IL-21, IL-2, and IL-4, and co-stimulatory signals such as ICOS and CD40L resulting in their survival and differentiation to plasma cells or memory B cells (3, 4).
The vital role of TFH cells in the induction of humoral immunity makes them attractive vaccine targets, and characterizing vaccine elicited TFH cell responses associated with broad and robust antibody titers will provide valuable information for vaccine design. Until recently, tracking vaccine-elicited TFH cells in humans represented a challenge due to the belief that TFH cells are exclusively localized to the GCs of secondary lymphoid organs. However, there is some evidence that TFH cells circulate transiently as CXCR5+ CD4+ T cells in blood and, based on the expression profile of activation markers, are predictive of antibody responses. For instance, HIV+ individuals that respond to H1N1 vaccine show expansion of CXCR5+ CD4+ T cells in the blood (peripheral (p) TFH), and the frequency of ICOS+ pTFH cells correlates with concurrent H1N1 titers (5). Likewise, CCR7lo PD-1+ cells within the pTFH cell pool are induced after influenza vaccination; this subset is over-represented in patients with autoimmune syndromes, and highly correlates with anti-dsDNA antibodies and disease severity (6). Together, these studies indicate that vaccine-elicited TFH cells circulate during the effector response to vaccination, these pTFH cells demonstrate an activated phenotype, and their magnitude correlates with vaccine-specific antibody titers generated within a month after vaccination. What is less understood is whether pTFH cells are predictive of long-term antibody titers and quality, and how they compare with lymph node TFH cell responses.
Recent studies have underscored the expression of chemokine receptors as a key functional attribute of TFH cells (7). Blood CXCR5+ CD4+ T cells in humans are comprised of CXCR3+ and CXCR3− subsets, which show heterogeneity in B cell helper potential. For instance, induction of CXCR3+ ICOS+ CXCR5+ pTFH cells at day 7 after influenza vaccination predicts increase in antibody titers at day 28-post immunization (8). On the other hand, in HIV infected individuals, frequency of CXCR3− PD-1+ CXCR5+ cells is associated with the development of neutralizing antibodies (9). These data indicate that phenotypic characteristics of pTFH cells may be specific to the vaccine/infectious agent and the resulting inflammatory response. In the context of HIV infection, the data suggest that CXCR3− TFH cells may be favorable for induction of antibodies. However, this paradigm has not been explored in the context of HIV vaccination.
In the present study, we examined the role of blood and lymph node CXCR5+ CD4+ T helper cells in the development of Env-specific antibody responses in the context of a DNA prime, recombinant modified vaccinia virus Ankara (MVA) boost (DNA/MVA) SIV vaccine in the presence and absence of a gp140 protein boost adjuvanted with aluminum hydroxide (alum) in rhesus macaques. We found that booster immunization with MVA induced Ki-67+ CXCR5+ CD4+ T cells, which circulated in blood (albeit at low levels) and were heterogeneous for expression of CXCR3. Blood Ki-67+ CXCR5+ CD4+ T cells at the peak effector phase were representative of GC TFH responses, and CXCR3 expression on GC TFH cells was a determinant of persistence, neutralization potential, and avidity of anti-Env antibody response. These data offer an important parameter with which to identify, track, and characterize CD4+ TFH cells in vaccine studies to gain an understanding of the types of TFH cell responses that lead to protective antibodies.
MATERIALS AND METHODS
Ethics statement
All animal protocols were approved by the Emory University Institutional Animal Care and Use Committee (IACUC) protocol YER-2002343. All experiments were conducted in strict accordance with USDA regulations and the recommendations for conducting experiments in accord with the highest scientific, humane, and ethical principles as stated in the Guide for the Care and Use of Laboratory Animals. Animals were housed in pairs in standard non-human primate cages. Animals received standard primate feed as well as fresh fruit and enrichment daily, and had free access to water. Upon infection, animals were housed singly. Immunizations, infections, blood draws, and biopsy procedures were performed under anesthesia by trained research staff. All efforts were made to schedule samples on paired animals concurrently so as to minimize potential distress.
Animals
Twenty-eight male Indian rhesus macaques were included in this study. Animals ranged in age from 2–4 years and in weight from 3 to 6 kgs at the start of the study. Animals were STLV−and SIV− at study commencement. None of the animals expressed Mamu class 1 alleles B08 and B17 and each experimental group had a total of three Mamu A*O1 animals. Animals from both experimental groups were randomized into four sampling groups for immunization and sampling of blood and tissue biopsies. Animals were monitored throughout the study period and, based on clinical parameters and complete blood counts, were reported to be healthy and immunocompetent throughout. All animals were housed at the Yerkes National Primate Research Center (YNPRC) in Atlanta, GA and treated in accordance with the YNPRC IACUC regulations.
Study Design and Immunizations
The vaccine study consisted of two experimental groups; all twenty-eight animals received two CD40L-adjuvanted DNA primes (0 and 8 weeks) followed by two MVA boosts (16 and 32 weeks; DDMM regimen). To determine whether inclusion of protein boost along with second MVA resulted in augmented antibody responses, fourteen animals were randomized to receive gp140 Env in alum along with the 2nd MVA (DDMM-Pro). The DNA immunogen expressed SIV239 Gag-Pol, Env, Tat, and Rev and was delivered at 3 mg/dose. The MVA immunogen expressed SIV239 Gag, Pol, and Env and was delivered at a 108 PFU/dose(10, 11). Protein vaccination consisted of 100µg endotoxin-free SIV239 gp140 (Immune Technology Corp, NY), which was pre-mixed with 500 µg aluminum hydroxide (Alhydrogel 2%, In vivo Gen, CA) prior to vaccination. Briefly, protein and alum were combined in a 1:1 volume ratio and were mixed by rocking at room temperature for 10 minutes. Prepared inoculum was stored on ice until the time of inoculation. All immunizations were delivered in phosphate-buffered saline intramuscularly in a single shot in the outer thigh.
SIV infection
At six months after vaccination, all vaccinated animals were challenged with SIVmac251 intrarectally on a weekly basis for a maximum of 5 weeks or until detection of plasma viremia above 250 copies/ml for two consecutive weeks. Infection with SIVmac251 (Day 8, 7.9.10 virus stock from Nancy Miller at NIH) was performed using a 1 cc slip tip syringe containing 1 ml of SIVmac251 at 200 TCID50. Syringe was inserted gently approximately 4 cm into the rectum and plunger was depressed to instill the virus and the animal was returned to the cage in a prone position.
Sample collection and processing
Peripheral blood mononuclear cells (PBMC) were isolated from whole blood collected in sodium citrate tubes and isolated by density gradient centrifugation according to standard procedures as described previously(10). PBMCs were isolated, counted, and utilized for various assays within 6 hours of blood collection. Lymph node biopsies were collected in RPMI and 10% FBS. To obtain single cell suspensions, lymph nodes were manually processed. Briefly, subsequent to removal of fat and connective tissue, biopsies were incised using scalpel and tissue segments were manually disrupted over a 100µm cell-strainer using a 10cc plunger. Cells were washed and re-suspended in complete media Biopsies and resulting suspensions were placed on ice at all times to preserve cell viability. After determination of cell counts, cells were used immediately or cryopreserved using standard techniques. Cell viability in both PBMCs and lymph node suspensions was determined to be above 90%.
ELISPOT assays
Enzyme-linked immunosorbent (ELI) spot assays were performed using fresh PBMCs using a standard ELISPOT assay(12, 13). Prior to use, PBMCs were washed upto 5 times in media to remove traces of bound antibodies that could contaminate the assay. ELISPOT plates (Millipore, MA) were coated with SIV239gp140 at 1µg/ml for detecting Ag-specific ASCs or with IgG at 10µg/ml (Rockland, PA) for detecting total IgG ASCs. After coating the plates overnight, PBMCs were incubated for upto 8 h and detected using biotinylated goat anti-monkey IgG (Rockland, PA) followed by ALP-conjugated streptavidin. (Mabtech, OH) Spots were detected using 1-step NBT substrate (Thermo scientific)
Measurement of Antibody titers
Binding antibody titers against gp140 were performed on frozen sera that was thawed and heat-inactivated as previously described(14). A NaSCN displacement ELISA as described previously was used for determining avidity(14). SIV-specific neutralization antibody was measured as a function of reduction in luciferase reporter gene expression after a single round of infection in TZM-bl cells as described previously(15).
Quantitation of SIV RNA plasma load
The SIV copy number was determined using a quantitative real-time PCR as previously described(14). For viral load determinations in lymph node, total DNA and RNA was extracted from approximately 50,000 sorted CD4+ subsets and data were normalized to albumin or GAPDH as depicted. Sorting experiments were performed on cryopreserved lymph node samples. All PCRs were performed in duplicates with a limit of detection of 60 copies per reaction.
Flow cytometry
Staining on whole blood was done at R.T while PBMCs and LN cell (106 cells) suspensions were stained in PBS containing 2% FBS (FACS buffer) for 30 min at 4°C. Cells were stained with fluorochrome-conjugated antibodies specific for CD3 (SP34-2), CD4 (OKT4), CD8 (SK1), CD20 (2H7), CD95 (DX2), CXCR3 (1C6), CCR5 (3A9), Bcl-6 (K112-91), Ki-67 (B56), IFN-γ (B27), TNF-α (MAb11), IL-2 (MQ1-17H12) IL-21 (3A3-N2.1) from BD Pharmingen (San Jose, CA); IL-4 (7A3–3) from Miltenyi Biotec (San Diego, CA); SLAM (A12), PD-1 (EH12.2H7), T-bet (4B10), ICOS (C398.4A) from Biolegend (San Diego, CA); CXCR5 (MU5UBEE) from eBioscience, (San Diego, CA); CD127 (R34-34) from Tonbo Biosciences; and CCR7 (FAB197A) from R&D Systems. Dead cells were excluded from analysis based on staining for Live/Dead Near-IR dead cell stain from Molecular Probes, Invitrogen (Grand Island, NY). Bcl-6, T-bet, and Ki-67 stains were performed after cells were stained for surface antigens followed by permeabilization/fixation using the Foxp3 kit and protocol, followed by intracellular staining. Prior to intracellular staining for cytokines, fresh PBMCs were stimulated with peptide pools of Gag and Env for 5 h in the presence of brefeldin A (Golgi Plug; BD Biosciences, San Jose, CA), then fixed and permeabilized with CytoFix/ CytoPerm (BD Biosciences, San Jose, CA) according to the manufacturer’s instructions. Unstimulated cells from each animal served as negative control and PBMCs stimulated with phorbol myristic acetetate/ionomycin served as positive controls. Samples were acquired on a LSR Fortessa (BD Biosciences) and 500,000 total events were collected for each sample. Cell sorting was performed using the Aria II (BD Biosciences). Data was analyzed using FlowJo software v X.0.7 (Tree Star, Inc., Ashland, OR). Data was analyzed after gating out dead cells, and subsequently gating on singlet cell populations. CD3+, CD20− cell subsets were gated and all analysis with CD4+ T cells was performed on parent CD4+, CD8− T cell subsets. The minimum number of events required to score a response as positive was set at 20 events after subtraction from background.
Statistical analysis
Statistical analysis was performed using Graph Pad Prism v5.0. A two-tailed non-parametric t test was used for all comparisons unless other wise specified. Spearman correlation was used to determine associations between variables. Statistical significance was set at p<0.05.
RESULTS
DNA/MVA vaccine induces transient accumulation of PD-1 and ICOS expressing CXCR5+ and CXCR5− CD4+ T cells with B cell helper potential in peripheral blood
Twenty-eight rhesus macaques were vaccinated intramuscularly with DNA/SIV vaccine (14) at 0 and 8 weeks, and with MVA/SIV vaccine (14) at 16 and 32 weeks (DDMM group)(Figure 1A). From this group, 14 macaques also received intramuscular SIV gp140 protein boost in alum concurrently with the 2nd MVA (DDMM-Pro). Blood was sampled at baseline and peak effector and memory time points post each immunization. A single lymph node biopsy was obtained at 2 weeks post the 2nd MVA immunization.
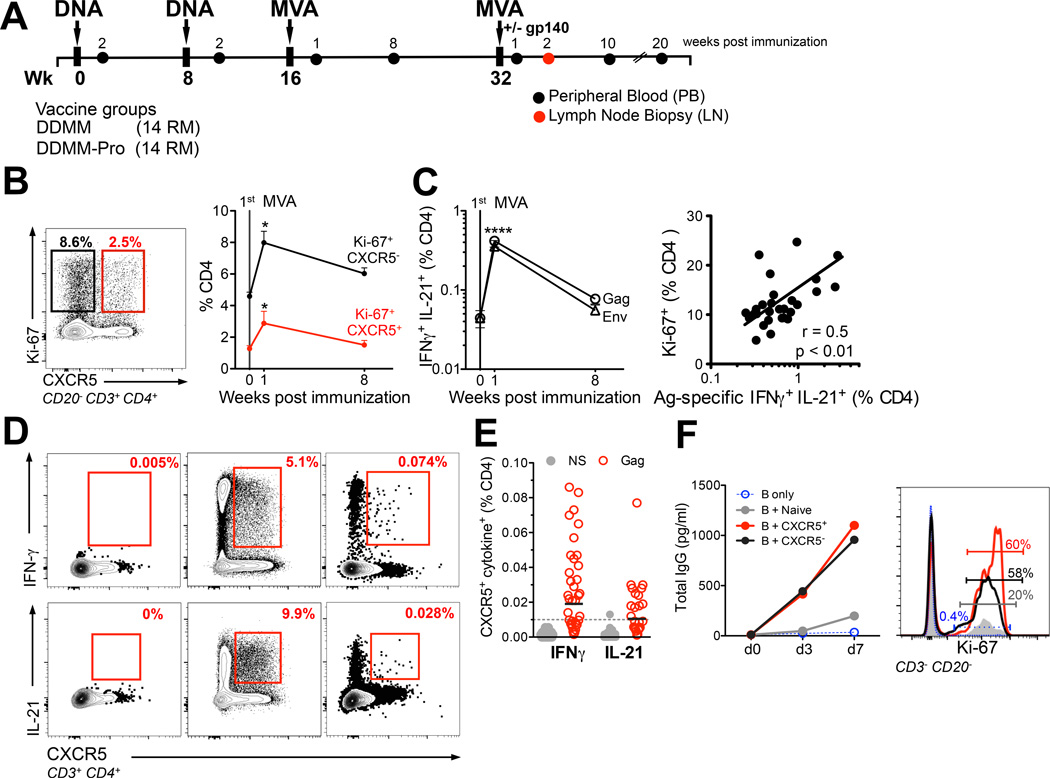
Figure 1. DNA/MVA vaccine induces transient accumulation of PD-1 and ICOS expressing CXCR5+ and CXCR5 CD4+ T cells with B cell helper potential in peripheral blood.
(A) Rhesus macaques were immunized with plasmid DNA expressing Gag, gp160 Env, Prt, RT, Tat, and Rev (3 mg at 0, 8 weeks) followed by boost with recombinant MVA expressing Gag, gp150 Env, and Prt (1×10^8 pfu at 16, 32 wks; DDMM). 14 macaques received 100µg of gp140 protein (adsorbed in 500µg Aluminum hydroxide) at week 32 concurrent with 2nd MVA immunization (DDMM-Pro). All immunizations were administered intramuscularly in the thigh. Peripheral blood (PB) and lymph node (LN) biopsies were obtained at stated time-points. (B) Examination of peripheral blood for the nuclear antigen Ki-67 together with CXCR5 showed that MVA immunization induced a proliferative burst in CXCR5+ (red line) and CXCR5− (black line) CD4+ T cells at day 7 (two-tailed paired t test showed significantly higher frequencies at week 1 compared to baseline and week 8 * p < 0.05). Data are mean ± SEM. (C) Kinetics of SIV-specific CD4 T cell response for Gag and Env in 28 RM. Data are mean ± SEM, response for Gag and Env at week 1 was significantly higher compared to baseline and week 8 using a paired two-tailed t test, **** p <0.0001). Magnitude of total Ki-67+ CD4+ T cells correlated with the magnitude of Gag + Env IFNγ+ IL-21+ CD4+ T cell response (Spearman R2 = 0.25 with a two-tailed p value of < 0.01 indicated by **). (D) Shows cytokine profile of CXCR5+ cells in blood under the following conditions: NS, PMA/Ionomycin, and Gag at 1 week post 1st MVA. Frequencies of Gag-specific CXCR5+ cells were above background for IFNγ and IL-21. (E) Scatter plot shows range of CXCR5+ IFNγ+ and CXCR5+ IL-21+ responses in NS (grey) and Gag stimulated PBMCs (red); each dot represents one animal (F) To determine B cell helper potential of blood CXCR5+ cells, these cells were co-cultured with autologous memory B cells obtained at week 1 post-1st MVA. Blood CXCR5− and CXCR5+ CD4 T cells efficiently induced IgG secretion and B cell proliferation. Data are representative of 1 of 4 independent replicates.
To determine whether DNA/MVA vaccine-elicited CD4+ T cells in blood have the potential to migrate to B cell follicles, we examined Ki-67+ (marks proliferating cells) CD4+ fraction for expression of the chemokine receptor CXCR5, a defining TFH cell marker that facilitates homing to B cell follicles/germinal centers (16)(Figure 1B). We also monitored the CXCR5− Ki-67+ CD4+ T cell fraction (presumably non-TFH fraction). About 0.1–1.7% (mean of 1%) of CD4+ T cells prior to boost were CXCR5+ Ki-67+ and this fraction increased by 2-fold at day 7 following the 1st MVA boost and returned to near baseline levels at 8 weeks post-immunization. The frequency of CXCR5− Ki-67+ CD4+ T cells was about 3–4-fold higher compared to CXCR5+ counterparts and expanded with similar kinetics. Thus, CXCR5+ CD4 T cells constituted a small proportion (approximately 15–25%) of total vaccine-elicited CD4 T cell responses in circulation. A similar phenotypic distribution was also observed within the Ki-67− memory compartment in blood (Supplemental 1A). This is consistent with data in mice demonstrating that approximately 25% of circulating OT-II effectors represent TFH cells (6), and with data in humans showing that only a small fraction (5–20%) of total tetanus-specific memory cells in blood is CXCR5+ (9).
Next, we examined SIV-specific CD4+ T cell responses to study how kinetics and phenotype of antigen-specific CD4+ T cells compared to that of Ki-67+ CD4+ T cells. To enumerate vaccine-induced SIV-specific CD4 responses, PBMCs were stimulated with overlapping peptide pools (15-mer overlapping by 11aa) derived from SIVmac239 Gag and Env sequences and examined for the production of IFN-γ and IL-21 using flow cytometry. The majority of IL-21 producing CD4 T cells co-expressed IFN-γ (data not shown) and peaked at week 1 post MVA boost (Figure 1C). The frequency of SIV-specific IFN-γ+ IL-21+ CD4 T cells showed a positive association with the frequency of total Ki-67+ CD4+ T cells at day 7 following MVA boost (Figure 1C, right panel) suggesting that proliferating cells were a reasonable indicator of vaccine-induced CD4+ T cells at the peak of the effector response.
To more formally identify Ag-specific CXCR5+ CD4+ T cells in blood, we set out to examine CXCR5 expression on responding cells after stimulation. Down-regulation of CXCR5 after 6 hrs of culture at 37°C necessitated staining of cells with CXCR5 prior to stimulation. With this strategy, we observed that CXCR5+ CD4+ T cells made robust amounts of IFN-γ and IL-21 in response to PMA/Ionomycin stimulation (Figure 1D). IFN-γ and IL-21 responses in CXCR5+ cells after Gag peptide stimulation were modest but discernable. The frequency of Gag-specific CXCR5+ CD4 T cells expressing either IFN-γ or IL-21 ranged from 0.004 to 0.15% of total CD4s (Figure 1E) and correlated positively with the frequency of Ki-67+ CXCR5+ CD4 T cells (r=0.42; p<0.05)(data not shown). About 0.5 to 20% of IFN-γ+ or IL-21+ CD4 cells expressed CXCR5 (data not shown).
Notably, CXCR5− CD4+ T cells were the major producers of IFN-γ and IL-21 in response to PMA/Ionomycin and Gag. While this could be due in part, to our inability to capture all CXCR5 expressing cells these data reveal that akin to human CD4 T cells IL-21 production is not an exclusive feature of CXCR5+CD4 T cells (26). Together, the data point to the conclusion that DNA/MVA vaccine-induced antigen-specific CXCR5+ CD4+ T cells circulate, albeit at very low frequencies, in peripheral blood during the peak effector response.
To determine whether these circulating CXCR5+ CD4+ T cells possessed B cell helper potential, we co-cultured FACS sorted CXCR5+ and CXCR5− memory CD4+ T cells along with autologous memory B cells, and cell supernatants were collected at day 3 and day 7 to examine secreted IgG (Figure 1F). At day 7, cells were also harvested and examined for B cell proliferation. Both CXCR5+ and CXCR5− CD4+ T cells efficiently induced IgG secretion and B cell proliferation compared to naive CD4+ T cells indicating that circulating CXCR5+ and CXCR5− CD4+ T cells possess B cell helper potential in vitro. This helper potential is consistent with the expression of ICOS by these cells (Figure 2C). Together, these data indicated that the 1st MVA booster immunization resulted in the transient accumulation of Ki-67+ CD4 T cells in blood at the peak of the effector response. These cells were phenotypically heterogeneous for expression of CXCR5; but both CXCR5− and CXCR5+ cells demonstrated B cell help functionality in vitro.
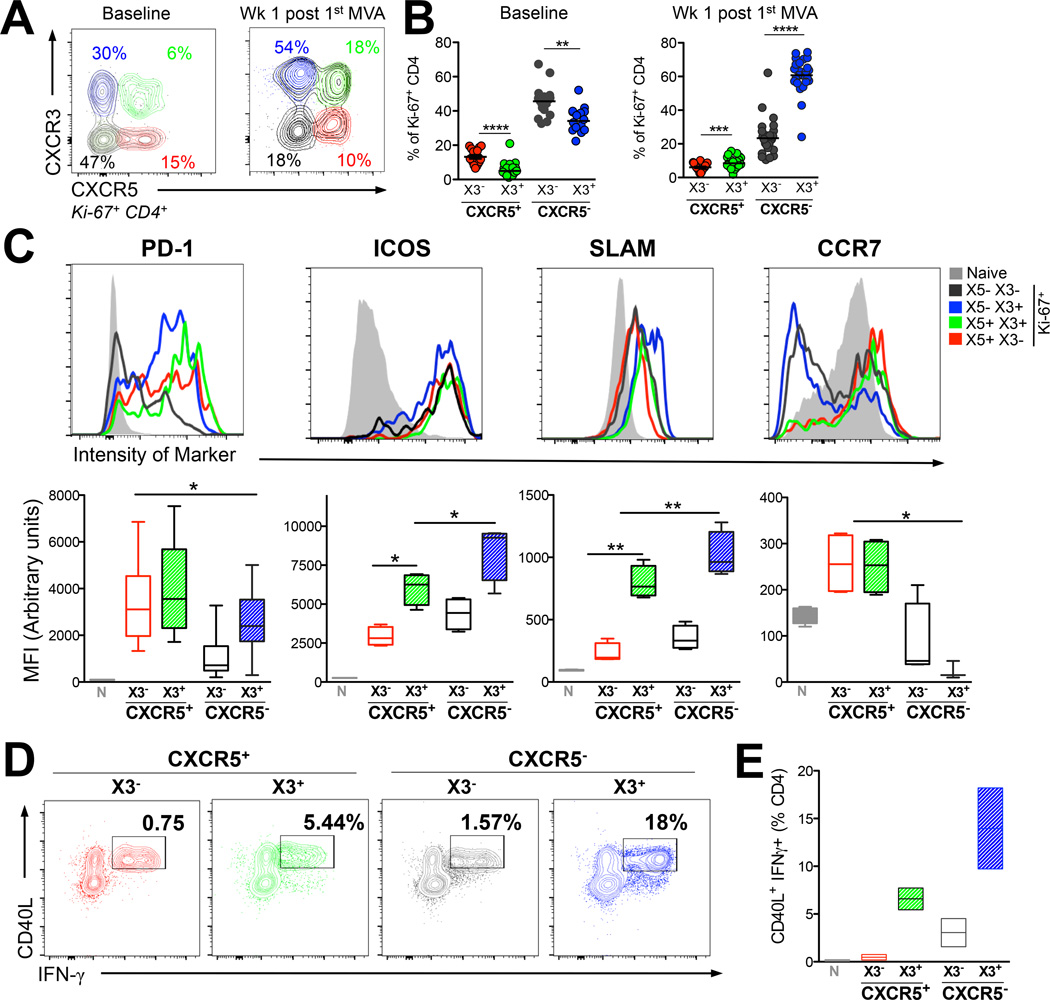
Figure 2. DNA/MVA vaccine induced CXCR5+ CD4+ T cells in peripheral blood demonstrate a TH1 propensity.
(A) Ki-67+ CD4+ T cells were distinguished into four subsets based on expression of CXCR5 (X5) and CXCR3 (X3). (B) Proportion of Ki-67+ CD4+ subsets for expression X5 and X3 at stated time points (****, p < 0.00001; ***, p < 0.001;**, p< 0.01 computed using a paired two-tailed t test). (C) Histograms show relative expression of phenotypic markers in naive (grey) X5+ X3− (red); X5+ X3+ (green), X5− X3− (tungsten), X5− X3+ (blue) subsets in blood at week 1 post 1st MVA. Box plots show median fluorescence intensity (MFI) of respective markers in Ki-67+ CD4 T cells for several markers; PD-1 (X5+ subsets showed significantly higher expression compared to X3 SP cells *, p < 0.05), X3+ cells expressed significantly higher levels of ICOS, SLAM and did not express CCR7. Data shown are from 4 animals and are representative of 3 independent replicates. (D, E) expression of CD40L and IFNγ on FACS sorted cell subsets memory cells after stimulation with PMA/Ionomycin.
DNA/MVA vaccine induced CXCR5+ and CXCR5− Ki-67+ CD4+ T cells in peripheral blood demonstrate a TH1 propensity
The cytokine profile of SIV specific CD4+ T cells indicated the induction of TH1-like CXCR5+ CD4+ T cells. Therefore, we wanted to determine the CXCR3 expression profile of Ki-67+ cells. Flow plots in Figure 2A show distribution of CXCR3 (X3) and CXCR5 on Ki-67+ CD4+ T cells at baseline and at one week after the 1st MVA. Prior to MVA boost, X3+ cells were present in both CXCR5− (X3 SP, blue population) and CXCR5+ (DP, green population) fractions and the frequency of X3+ cells were lower compared to X3− cells within the respective CXCR5+ or CXCR5− subsets. However, at one week following the MVA boost, the frequency of X3+ cells increased significantly in both CXCR5− and CXCR5+ subsets indicating generation of TH1 biased non-TFH and TFH cell subsets (Figure 2B). Non-cycling (Ki-67−), memory CD4+ T cells showed a variable composition and phenotype of these subsets likely a result of diverse antigenic history and quiescent phenotype (Supplemental Figure 1B).
To better understand the nature of cycling blood CXCR5+ and CXCR5− CD4+ T cells in the context of CXCR3, we performed a phenotypic characterization of these cells with respect to markers associated with LN resident TFH cells (Figure 2C). A majority of blood Ki-67+ CD4+ T cells within both the CXCR5− and CXCR5+ compartments were positive for programmed death (PD)-1 with CXCR5+ CD4+ T cells expressing relatively higher levels of PD-1. Interestingly, both CXCR5+ and CXCR5− CD4 T cells expressed inducible costimulator (ICOS) and ICOS expression was higher on CXCR3+ cells compared to CXCR3− cells. CXCR5− CXCR3+ cells expressed the highest levels of ICOS. This phenotype contrasts with blood CD4+ effectors in humans where CXCR5− CD4 T cells do not express ICOS (8). Expression of the activation receptor associated with IFN-γ production, signaling lymphocyte activation marker (SLAM) was lower on CXCR5+ Ki-67+ CD4+ T cells relative to CXCR5− counterparts, reflective of the SLAMlo phenotype of LN CXCR5+ cells (19, 20). Similar to ICOS expression, SLAM was expressed at higher levels on X3+ cells. Unlike, lymph node resident TFH cells which downregulate CCR7 to exit the T cell zone and gain access to the germinal centers (21), blood CXCR5+ CD4+ T cells were positive for CCR7 irrespective of X3 expression. In contrast, a majority of X3− CXCR5− CD4+ T cells had downregulated CCR7 (21).
To further corroborate the TH1-like functionality of DP CD4+ cells in blood, we wanted to examine cytokine production upon stimulation. Because dynamic changes in expression of CXCR3 and CXCR5 occur upon stimulation, we sorted CXCR5+ and CXCR5− cells based on expression CXCR3 to accurately discern differences in cytokine profile among these subsets. Sorted subsets were stimulated with PMA/Ionomycin and examined the expression for CD40L and IFN-γ. Consistent with their TH1 phenotype, X3+ cells showed higher frequency of CD40L+ IFN-γ+ cells within both CXCR5− and CXCR5+ compartments (Figure 2D). In terms of MFI, CXCR5+ cells had a higher MFI of CD40L, while CXCR5−, X3+ subset showed highest expression of IFN-γ. Together, the data demonstrate that DNA/MVA induced blood CXCR5+ CD4+ T cells show phenotypic and functional heterogeneity based on CXCR3 expression, and vaccination significantly enhances the frequency of TH1 like CXCR5+ and CXCR5− cells during the effector phase in blood.
Blood CXCR5+ CD4+ T cells predict germinal center TFH cell and B cell responses in lymph node
We next determined the relationship between CXCR5+ CD4+ T cells in blood and LN, and examined how inclusion of alum-adjuvanted gp140 protein along with 2nd MVA boost impacted the magnitude of these CD4+ T cell responses. Examination of blood Ki-67+ CD4+ T cells at week 1 following the 2nd MVA boost showed that MVA+gp140 immunization resulted in about 2-fold higher frequencies of Ki-67+ CD4 T cells in circulation. The frequency of Ki-67+ CD4+ T cells ranged from 5 – 13% of total CD4s in MVA only compared to 15 – 33% of CD4s in MVA+gp140 group (Figure 3A). We also observed an increase for SIV Env-specific IFNγ+, IL-21+, and IL-4+ CD4+ T cells (Figure 3B) in the protein group and, as expected, this increase was not observed for Gag-specific CD4+ T cells (Figure 3C). Together, the data showed that concurrent gp140 immunization augmented magnitude of Env specific CD4+ T cell responses.
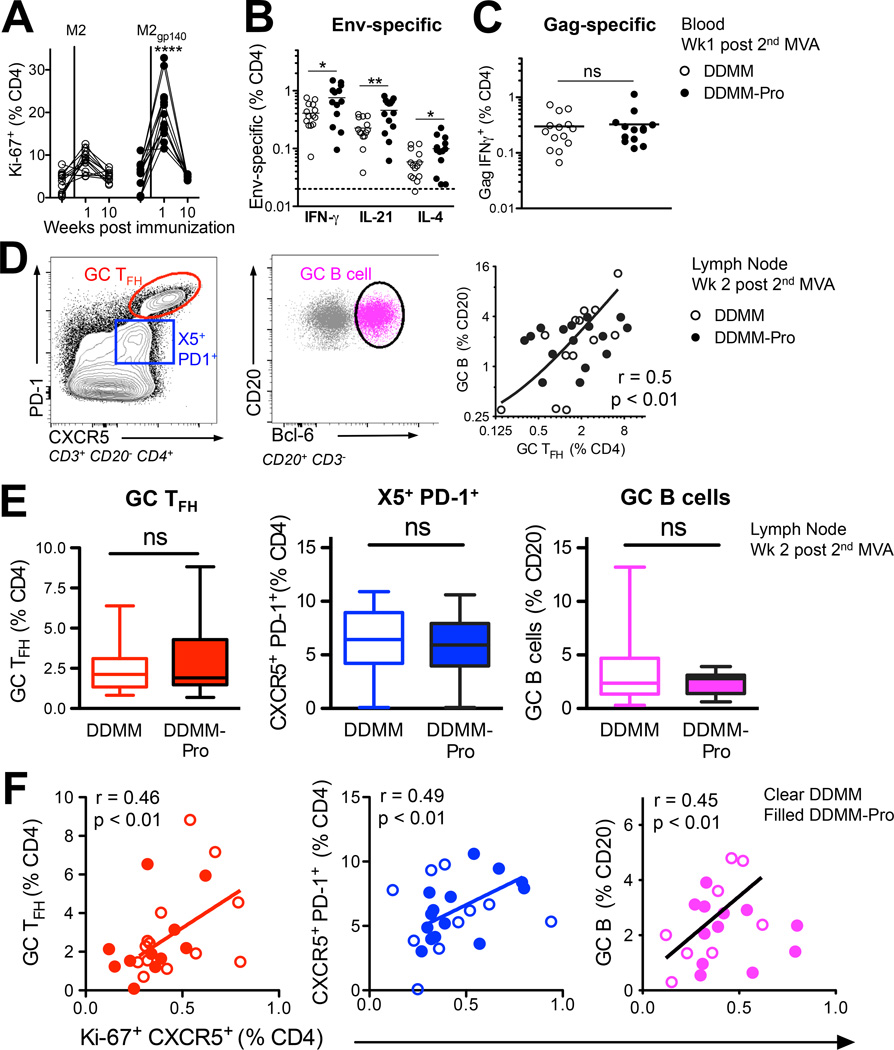
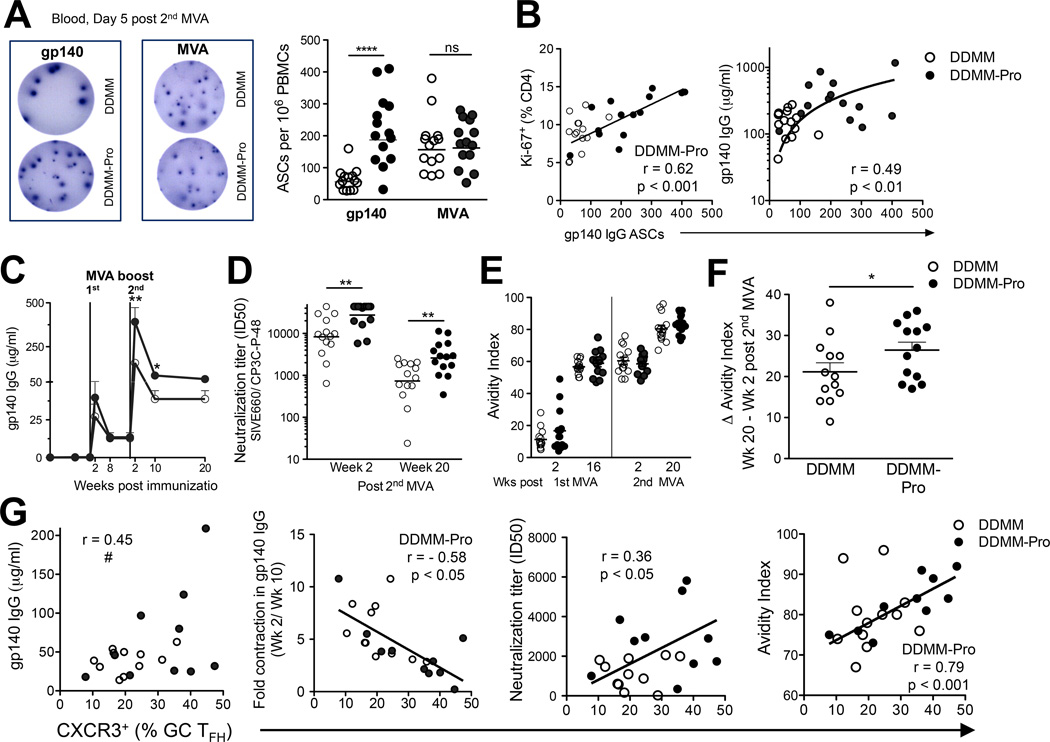
Figure 3. Blood CXCR5+ CD4+ T cells predict germinal center TFH cell and B cell responses.
(A) Higher frequency of Ki-67+ CD4+ T cells after 2nd MVA in DDMM-Pro group (****, p < 0.0001 at week 1 with a two-tailed, non-parametric t test). (B) To determine if gp140 boost resulted in higher Env-specific CD4+ T cells, responses were compared using a one-tailed t test, which showed increased Env IFN-γ+, IL-21+, and IL-4+ frequencies in DDMMgp140 group at week 1 post 2nd MVA immunization and no change in (C) Gag responses. (D) Inguinal lymph nodes were biopsied from the lymph node non-draining to immunization site at 2 weeks post-2nd MVA immunization (E) Flow plot shows presence of distinct subsets of CXCR5+ CD4+ T cells in lymph node ; PD-1++GC TFH (red) , PD-1+, (blue) , and PD-1 cells. Middle panel shows GC B cells identified by Bcl-6 and scatter lot shows correlation between GC B cells and GC TFH cells. (D) Inclusion of gp140 boost did not alter frequencies of GC TFH cells, CXCR5+PD-1+, and GC B cells. (F) To determine if responses in blood (at week 1 post 2nd MVA) were positively correlated with magnitude of GC responses, these indices were correlated using a one-tailed Spearman correlation (*, p < 0.05). Blood Ki-67+ CXCR5+ cells correlated with GC TFH cells (R2 = 0.21), CXCR5+ PD-1+ TFH cells (R2 = 0.24), and GC B cells (R2 = 0.20) in lymph node.
Having identified that DNA/MVA induced CXCR5+ cells circulate in blood during the effector response, we next examined TFH cells in the lymph nodes and compared them with blood CXCR5+ cells. We chose to biopsy the right lymph node, which was non-draining to the 2nd MVA immunization so as not to interfere with antigen complexes sequestered within the draining inguinal lymph node after immunization. It is conceivable that VLPs derived from MVA infected cells in vivo could engage memory cells in distal lymphoid organs including the right lymph node, which was the draining lymph node for the preceding DNA primes and for the 1st MVA boost.
Examination of lymph node sections by immunohistochemistry showed presence of distinct clusters of proliferating B cells within B cell follicles (data not shown). Single cell suspensions stained for flow cytometry showed presence of three distinct populations of CXCR5+ CD4+ T cells showing graded levels of PD-1 expression; GC TFH cells (CXCR5+ PD-1++), CXCR5+ PD-1+, and CXCR5+ PD-1− cell subsets (Figure 3D). The frequency of GC TFH cells correlated directly with GC B cells identified by expression of Bcl-6 as shown and with CXCR5+ PD-1+, cells but not with CXCR5− PD-1+ cells or with CXCR5+ PD-1− cells. (Supplemental Figure 2A). Examination of the frequency of GC responses revealed that the gp140 boost did not alter either the frequency of total GC TFH cells or GC B cells (Figure 3E). This is not surprising since total GC TFH cells and GC B cells comprise of polyclonal T and B cell responses directed against epitopes of different antigenic specificities including MVA and SIV Gag; a modest increase in Env specific GC responses elicited by gp140 protein boost would be difficult to discern.
We next evaluated possible associations between blood CXCR5 + cells and PD-1 expressing GC TFH cells and GC B cells within the LN and found a positive correlation between Ki-67+ CXCR5+ cells in blood and all three LN subsets (Figure 3F). We next asked whether the Ki-67+ CXCR5− CD4 T cell subset was also positively associated with GC responses. The data showed that CXCR5− cells did not correlate with any of the measured GC responses (Supplemental Figure 2B). Therefore, the magnitude of responding blood CXCR5+ CD4+ T cells but not CXCR5− CD4+ T at peak post-vaccination was predictive of subsequent germinal center responses. Studies showing that gene expression profile of blood PD-1+ CXCR5+ cells but not of CXCR5− cells closely matches that of GC TFH cells are consistent with this possibility (9).
Alum-adjuvanted gp140 protein boost enhances the frequency of CXCR3+ CD4+ TFH cells in lymph node
Next, we wanted to ascertain whether inclusion of gp140 + Alum resulted in discernable effects on phenotype of CXCR5+ cells in blood and LN. Consistent with the 1st MVA boost, the majority of Ki-67+ cells post the 2nd MVA boost comprised of X3 SP CD4+ T cells, and the inclusion of protein boost further enhanced the frequency of these cells (data not shown). The protein boost did not significantly increase the frequency of CXCR5+ Ki-67+ cells beyond that induced by 2nd MVA alone. Interestingly, however, gp140 protein boost resulted in a significant induction of X3 expression on GC TFH cells and CXCR5+ PD-1+ cells in the LN (Figure 4A). While the relative contribution of gp140 antigen versus alum or a combination thereof to this phenotype cannot be discerned, these results demonstrate that alum did not skew against a TH1 CD4+ helper recall response established by the preceding DNA/MVA immunizations. Interestingly, we observed a direct positive association between the magnitude of GC B cell responses and proportion CXCR3+ GC TFH cells within lymph nodes in DDMM-Pro group (right panel). As anticipated, CXCR3− GC TFH cells showed an inverse correlation (Supplemental Figure 2A)
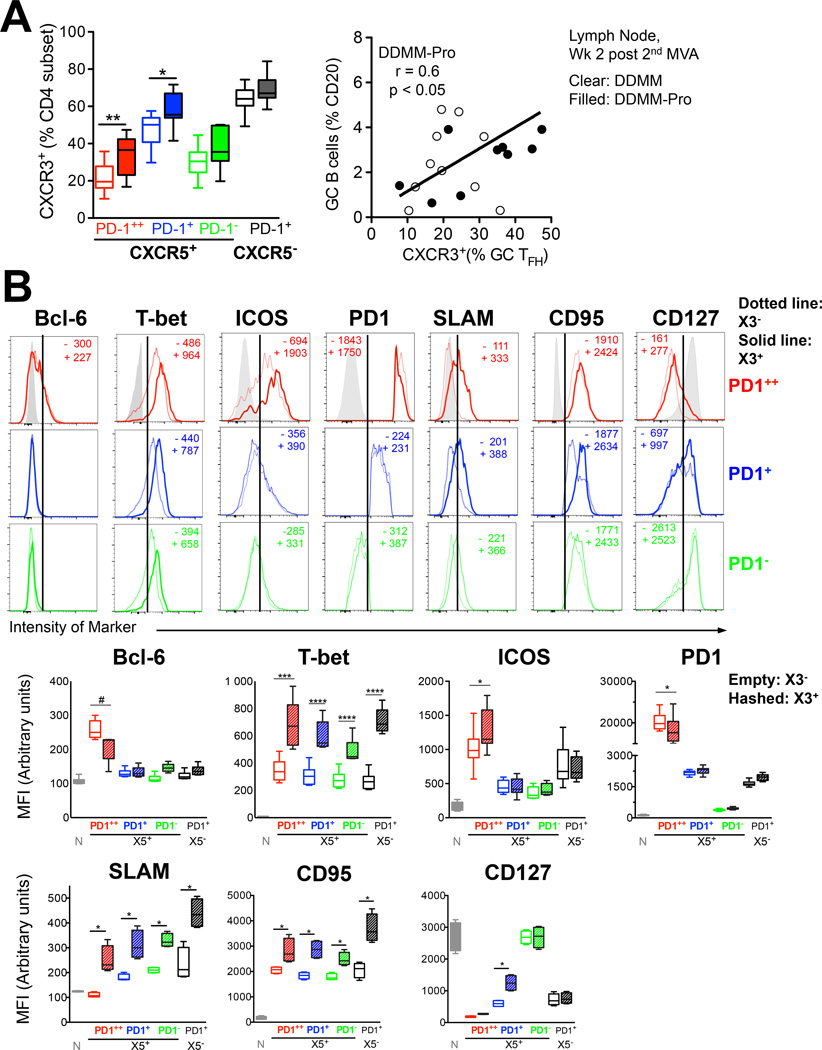
Figure 4. Alum-adjuvanted gp140 protein boost enhances the frequency of CXCR3+ CD4+ TFH cells in lymph node.
Inclusion of protein boost in alum skewed the phenotype of responding CD4+ T cells towards CXCR3 in (A) lymph node GC TFH and CXCR5+ PD1+ cells examined at 2 weeks post-2nd MVA boost. Clear bars DDMM group, filled bars DDMM-Pro group. Comparisons were made using a two-tailed non-parametric t test, **, p < 0.001; *, p < 0.01. Scatter plot shows association of % CXCR3+ GC TFH cells with magnitude of GC B cell response (B) Histograms show expression of markers on GC TFH cells (red), CXCR5+ PD1+ cells (blue), and CXCR5+ PD1 cells (green) in lymph node. Solid lines denote expression in CXCR3+ subsets, dotted line CXCR3 subset and whisker plots show Median Fluorescence intensity for X3 (empty plots) and X3+ (hashed plots) TFH subsets for the transcription factors Bcl-6, T-bet; activation markers ICOS, PD-1, SLAM, CD95 and memory marker CD127 (#, p = 0.08; ****, p< 0.0001; ***, p < 0.001, * p < 0.05 computed using a paired two-tailed t test)
The increase in X3 expression on CXCR5+ cells following MVA boost led us to examine phenotypic differences among X3− and X3+ subsets in the LN to understand possible functional differences among these two subsets. We started by examining expression of the transcription factor Bcl-6, a unique and characteristic marker of GC cells (Figure 4B). PD-1++ GC TFH cells exclusively expressed Bcl-6 albeit at lower levels compared to GC B cells (data not shown). Within GC TFH cells, the CXCR3+ subset expressed lower levels of Bcl-6 on a per cell basis in most of the animals. As expected, decreased levels of Bcl-6 were associated with higher relative expression of T-bet in CXCR3+ GC TFH cells. This dichotomy in T-bet expression was true for all CXCR5+ and CXCR5− PD-1+ subsets. These data suggest that X3 expression identifies TH1-like TFH cells in GCs and extrafollicular regions.
Because Bcl-6 positively regulates expression of PD-1 (9), we next examined PD-1 expression and found that X3+ cells also expressed lower amounts of PD-1 on a per cell basis. This difference was only observed within the GC TFH subset. We asked if the same was true for expression of another activation/co-stimulation marker, ICOS. Unlike PD-1, ICOS was expressed at higher levels by CXCR3+ GC TFH cells reflecting expression profile of X3+ and X3− Ki-67+ CXCR5+ cells in blood. Higher magnitude expression of ICOS on CXCR3+ GC TFH cells is supportive of enhanced B cell helper potential. PD-1+ and PD-1− CXCR5+ cells expressed lower levels of ICOS compared to GC TFH cells and demonstrated no difference in expression in the context of X3. Expression of an IFNγ-related marker SLAM was consistently higher on X3+ subsets; X3− GC TFH cells did not express SLAM showing expression levels comparable to naive cells. X3+ subsets showed higher levels of CD95. Frequency of Ki-67 among X3+ and X3− CXCR5+ PD-1+ subsets was comparable, indicating that the phenotypic differences were not due to cell cycle status of these subsets.
Levels of another memory marker, CD127 were low on GC TFH cells but relatively higher in the X3 compartment and the same was true for the CXCR5+ PD-1+ compartment raising the possibility that X3+ GC TFH cells represent memory precursors that differentiate into CXCR5+ PD-1− cells after antigen clearance.
To determine whether these phenotypic differences resulted in differential ability to provide B cell help, we sorted GC TFH and CXCR5+ PD-1+ cells from the lymph node based on X3 expression (Supplemental Figure 2C). Sorted subsets were co-cultured with autologous memory B cells in the presence of SEB. Determination of IgG in supernatant after 7 days revealed that both X3+ and X3− GC TFH subsets were able to provide B cell help. The relative ability was variable; two animals RGf14 and RVz12 showed 1.5 to 8-fold higher IgG in X3+ GC TFH B cell co-cultures compared to X3- counterparts while in two animals RPq13 and RWf114, the X3− GC TFH subset showed higher B cell helper potential. CXCR5+ PD-1+ cell subsets also showed similar variability among X3+ and X3− compartments. In all the data indicate that LN CXCR3+ GC TFH cells are phenotypically distinct from CXCR3− GC TFH cells, but both subsets demonstrate potential to help B cells in vitro.
CXCR3+ GC TFH cells correlate with longevity and avidity of gp140 antibody titers
Among the first B cell effectors are plasmablasts/antibody secreting cells (ASCs), derived either from activation of naive B cells during a primary response or from memory B cells during antigen re-exposure (28, 29). After the 2nd MVA boost, gp140-specific ASC responses were observed in 100% of immunized macaques (Figure 5A). These responses ranged from 30–160 (mean 63) ASCs per million PBMCs in the DDMM group. Concurrent immunization of gp140 with the 2nd MVA increased the magnitude of ASC response by 3-fold which ranged from 30–400 (mean 200) ASCs per million PBMCs. MVA-specific ASC responses were comparable between the two groups and were not inhibited as a result of gp140 immunization. To explore the role of CD4+ T cells in fostering plasmablast responses, we examined peripheral Ki-67+ CD4+ T cell responses at day 7, as a surrogate for T cell responses in the lymph node on or prior to day 5. After the 2nd MVA boost, the magnitude of total Ki-67+ CD4+ T cells (Figure 5B) in the DDMM-Pro group correlated with higher gp140 ASC responses. Correspondingly, Env IFNγ+ CD4+ T cells were also positively correlated with ASC responses (Supplemental Figure 3A). The gp140 ASC response at day 5 was a key determinant of peak gp140 titers at 2 weeks following the 2nd MVA boost .
Figure 5. CXCR3+ GC TFH cells correlate with longevity and avidity of gp140 antibody titers.
ELISPOT assays were used to quantify gp140 and MVA ASCs. ELISPOT image for (A) gp140 and MVA at day 5 post 2nd MVA boost in DDMM (top) and DDMM-Pro (bottom) groups. Spots shown are for an input of 100,000 PBMCs per well. Graph in (A) shows concurrent immunization of gp140 with 2nd MVA increases gp140 IgG ASC responses (***, p < 0.001 with a non-parametric two-tailed t test). MVA IgG ASCs comparable in DDMM (clear circles) and DDMM-Pro groups (filled circles). (B) Correlation between gp140 ASCs and magnitude of Ki-67+ CD4 T cells (Spearman R2 = 0.38). Magnitude of gp140 ASC response correlates with scale of gp140 antibody titers at week 2 post 2nd MVA (Spearman R2 = 0.24). (C) kinetics of gp140 antibody responses after 1st MVA boost showed significant increase with gp140 boost at week 2 and week 10 post 2nd MVA (**, p< 0.001; *, p< 0.01 using a non-parametric two-tailed t test) (D) neutralization titers against tier 1 SIV E660 also increased with gp140 boost (E) kinetics of antibody avidity (F) Computation of increase in avidity (Wk 20- Wk 2) after 2nd MVA immunization revealed greater increase with gp140 boost (*, p < 0.05 with a non-parametric one-tailed t test). (G) shows a trend for association between CXCR3+ GC TFH and gp140 titers at memory measured at week 20 (#, p = 0.05); shows inverse correlation between contraction of gp140 titers and % of CXCR3+ GC TFH cells in DDMMgp140 group (Spearman R2 = 0.33; *, p < 0.05). CXCR3 expression on GC TFH cells directly correlated with neutralization titers (Spearman R2 = 0.12; *, p < 0.05) and avidity index (Spearman R2 = 0.62; **, p < 0.01).
In order to delineate CD4 determinants of peak and memory anti-gp140 antibody responses, we first examined the kinetics of antibody by ELISA (Figure 5C). Anti-gp140 antibody titers were below the limit of detection in 25 of 28 animals after the 2nd DNA immunization. The first MVA immunization induced titers ranging from 5 to 200 µg/ml at week 2 which contracted by week 8 and were potently recalled after the 2nd MVA resulting in higher magnitude gp140 titers at memory.
We next determined the neutralization potential of anti-Env antibody against tier 1 (easy to neutralize) and tier 2 (harder to neutralize) SIVE660 pseudovirus isolates. While no detectable responses against tier 2 isolates was observed, co-immunization with alum in gp140 resulted in stronger neutralization titers against tier 1 isolates at week 2 and week 20 following the 2nd MVA boost (Figure 5D). As a measure of antibody maturation, we monitored relative antibody avidity following 1st and 2nd MVA immunizations. Our analyses using an ELISA with a 1.5 M sodium thiocyanate wash showed that gp140 avidity increased over time and reached a maximal response at week 20 after the 2nd MVA boost (Figure 5E). To ascertain whether inclusion of gp140 protein boost during the 2nd MVA increased avidity of anti-Env antibody, we examined relative increase in avidity at week 20 versus week 2 and observed greater increase in avidity with inclusion of gp140 protein boost (Figure 5F).
Next, we wanted to investigate how the GC TFH response in the LN at week 2 post the 2nd MVA boost influenced the magnitude and longevity of anti-Env antibody titers. The frequency of CXCR3+ GC TFH cells showed a trend for correlation with the binding titers at week 20 (p = 0.05) or at week 10 (data not shown), and directly correlated with neutralizing activity and avidity of anti-Env antibody. We also observed an inverse correlation between the frequency of CXCR3+ GC TFH and fold-contraction in peak antibody titers (Figure 5G). These results demonstrated that CXCR3+ GC TFH cells contribute significantly for the generation of long-lived, antibody responses.
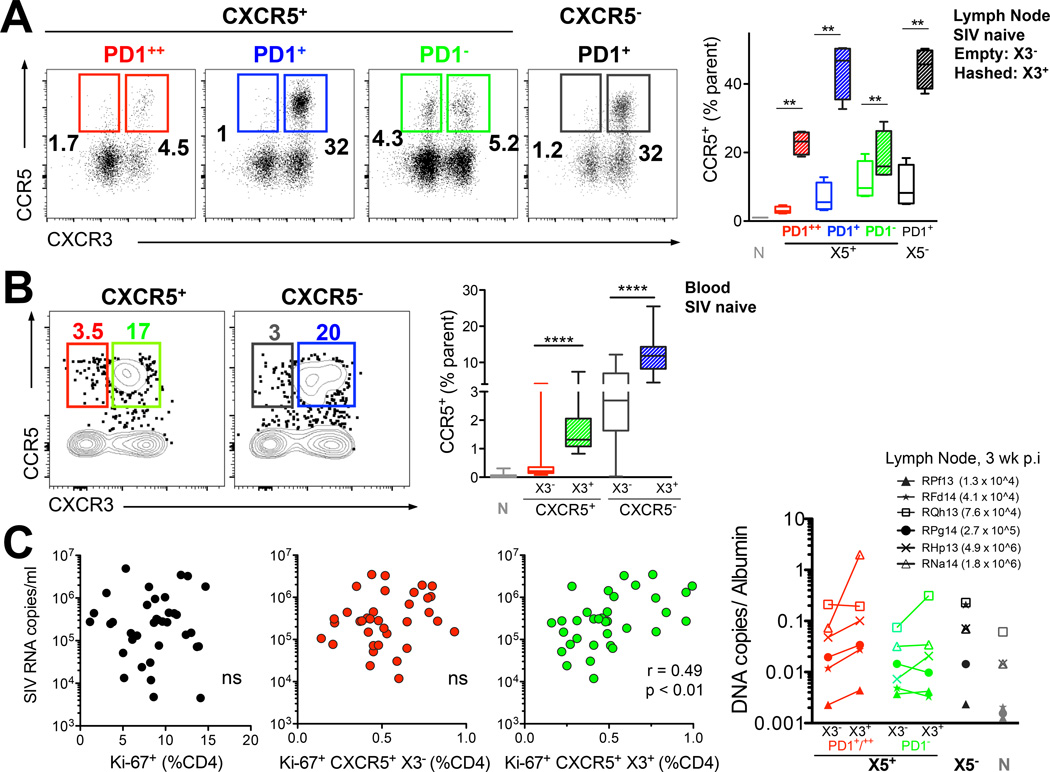
Vaccine elicited induction of CXCR5+ CXCR3+ CD4+ T cells associates with peak viral load after infection with SIV251
Studies in humans have shown that blood X3+ CCR6+ CD4+ T cells express high levels of the HIV co-receptor CCR5 (30) raising the possibility that vaccine mediated generation of CXCR3+ CXCR5+ cells could favor viral replication upon infection. To address this possibility, we first examined CCR5 expression in lymph node and blood X3+ CD4+ T cells in SIV naive animals (Figure 6A,B) and found that a significantly higher fraction of X3+ CD4+ T cells express CCR5 compared to X3− cells. This was true both in LN (Figure 6A) and blood compartments (Figure 6B), and within CXCR5+ and CXCR5− subsets. Following 5 weekly intrarectal challenges with SIVmac251, all vaccinated animals became infected but showed a 2-log-fold decrease (p< 0.01) in peak viremia compared to unvaccinated controls (data not shown). The induction of CXCR3+ CXCR5+ CD4 TFH cells following vaccination and the higher expression of CCR5 on CXCR3+ CD4 T cells prompted us to test associations between CD4+ T cell response induced at peak after 2nd MVA boost and peak viremia at 3 weeks post SIV challenge. We found a moderate but significant direct correlation with Ki-67+ CXCR5+ X3+ CD4+ T cells and peak viremia (Figure 6C, last panel). Interestingly this association was not observed with either total Ki-67+ CD4+ T cells or CXCR5+ X3− CD4+ T cells.
Figure 6. Vaccine elicited induction of CXCR3+ CXCR5+ CD4+ T cells associates with peak viral load after infection with SIV251.
Expression of CCR5 on CXCR5+ and CXCR5 CD4 T cells in SIV naive (A) lymph node and (B) blood. (C) vaccine-induced CXCR3+ CXCR5+ CD4 T cells at week 1 post 2nd MVA associate with peak viremia at 3 weeks post SIV infection (Spearman R2 = 0.2; *, p < 0.01 with a non-parametric two-tailed t test ). (D) CXCR3+ CXCR5+ PD-1+/++ TFH cells in lymph node at 3 weeks post SIV infection harbor higher levels of pro-viral DNA compared to X3 subsets. Values in parentheses are serum viral RNA copies at 3 weeks post-infection for each animal.
To determine whether permissiveness of X3+ TFH cell subsets to SIV infection facilitated seeding of reservoirs, we sorted lymph node CXCR5+ cells (PD-1++ and PD-1+) based on X3 expression at 3 weeks post-infection from several animals with a log-fold difference in viral RNA copies in plasma; the data showed that in 5 out of the 6 animals sampled X3+ PD-1+/++TFH cells harbored significantly higher levels of pro-viral DNA compared to X3− counterparts (p < 0.05; Figure 6D).
We do not, at present, fully understand the relationship between the SIV-specific CD8 T cell responses post infection and the infection status of CXCR3+ TFH cells. However, we observed that the frequency of Ki-67+ CD8 T cells associated with peak viral load and positively correlated with frequency of GC TFH cells and proportion of CXCR3+ GC TFH cells, consistent with antigen or inflammation driven immune activation of T cells (Supplementary Figure 4). More detailed studies are needed to investigate the dynamics of antigen-specific CD8 T cell responses in peripheral blood and lymphoid compartments in controlling frequency and infection status of CXCR3− and CXCR3+ TFH cell subsets.
In summary, our data raise the possibility that vaccination regimens which skew towards generation of X3+ CXCR5+ cells while favoring durable, higher avidity antibody could facilitate viral replication and seeding of reservoirs by generation of target cells that are efficiently recalled upon antigen-re-exposure.
DISCUSSION
Post-RV144, HIV vaccine strategies to augment anti-Env antibody responses have generated immense interest and concurrent immunization of Env subunit proteins with recombinant viral vectors expressing HIV immunogens is emerging as a promising HIV vaccine modality (31) . While a great deal of research has focused on antibody responses following Env subunit immunization, relatively little is known about the CD4+ helper response, and, in particular, the B cell helper CD4+ TFH cell response resulting from these immunization regimens. A better understanding of CD4 helper correlates of antibody responses will, undoubtedly, yield useful insights for rational vaccine design(32).
Using the best available experimental model for HIV vaccines- the rhesus macaque, we conducted an in-depth characterization of the CD4+ T cell response and examined its effects on Env B cell responses in the context of a DNA/MVA+ gp140 vaccine regimen. Our data allow for three main conclusions; first, DNA/MVA vaccine induces CXCR5+ CD4+ T cells with B cell helper potential, which circulate at the peak of immune response. Second, vaccine-induced CXCR5+ CD4+ T cells are skewed towards expression of CXCR3, which is augmented by co-immunization with alum-adjuvanted gp140 protein. Third, CXCR3 expression on CXCR5+ lymph node resident CD4+ T cells is strongly associated with antibody persistence and with antibody avidity. The data offer an important parameter with which to predict Env memory responses and suggest that strategies to target the CXCR5+ CXCR3+ subset could provide a means to enhance Env titers. However, an association between vaccine-raised CXCR5+ CXCR3+ cells and peak viral load, albeit modest, suggests that these cells could represent targets for viral replication and predispose to higher acute viremia upon infection. Together, these results provide a strong rationale to investigate and delineate protein-adjuvant driven CXCR5+ CXCR3+ CD4+ T cell responses in HIV vaccine studies to better understand CD4 helper correlates of vaccine efficacy and viral control.
The chemokine expression profile of CD4+ T cells typifies differentiation status and is regulated by the antigenic stimulus and the resulting inflammatory response(7). MVA is a potent inducer of IFNγ production in DCs (33) and significant systemic induction of IFNγ-induced protein 10 (IP-10), one of the ligands of CXCR3, is observed after primary and booster MVA immunization in rhesus macaques (34). Because CXCR3 is induced by IFNs, the generation of CXCR3 skewed CD4 response after MVA immunization is anticipated. Notably, this phenotype was augmented by addition of alum-adjuvanted gp140 and was accompanied by an increase in Env-specific IFNγ+, IL-4+ responses. Induction of a TH1 bias with alum, which is considered largely as a TH2 adjuvant, is surprising but is not without precedence. Studies in mice show that OVA in alum immunization results in CXCR3 expression on ASCs and GC B cells which is mediated by IFNγ+ Ag-specific CD8+ T cells (35, 36). Our data show that TH1 and TH1-skewed CXCR5+ recall responses primed by preceding DNA/MVA immunizations were reacquired after the 2nd MVA boost and were not inhibited by alum adjuvant. This phenotype, likely driven by vaccine-mediated induction of T-bet in CD4 T cells, is consistent with the proposed model of TFH and TH1 CD4+ helper differentiation during recall responses (37).
The mechanisms by which CXCR3+ GC TFH cells promote antibody longevity and avidity remain to be understood. Our in vitro assays indicate that both X3− and X3+ subsets were effective at helping B cells. The dynamics of X3+ cells in vivo in fostering antibody responses could be due to differential localization and/or due to enhanced expression of B cell helper cytokines and co-stimulatory signals in the context of an IFN-γ-rich environment. With regards to HIV, there is evidence that CXCR3− CXCR5+ cells possess superior B cell help potential. Recently, Locci et al reported higher % of blood PD-1+ CXCR3− CD4+ cells (within total CXCR5+ cells) at 4 and 40 months after infection in HIV+ individuals that develop broadly neutralizing antibodies(9). Furthermore, CXCR3+ but not CXCR3− cells were effective at providing B cell help in vitro. It is possible that phenotypic attributes of TFH cells that foster antibody responses during vaccination and during infection are very different especially during chronic antigen stimulation where CD4+ T cells are viral targets. This is particularly relevant as CCR5 expression is largely confined to CXCR3+ CD4+ subsets. In our study, we found that the magnitude of vaccine-induced X3+ CXCR5+ CD4+ T cells was significantly associated with peak viral load after infection. This would suggest that rapid recall of vaccine induced X3+ CXCR5+ CD4+ T cells upon antigen-re-exposure results in generation of target cells for viral replication(38). These data indicate that in the context of a TH1 skewed immunization regimen, CXCR3+ TFH cells favor antibody responses but could skew towards higher peak viremia.
Our findings underscore a pressing need to better understand the effect of adjuvanted Env subunit protein immunizations on CD4+ T cell helper responses in humans. Protein immunizations augment CD4+ responses and typically do not raise/boost CD8 responses (39). The generation of CD4 viral target cells that are efficiently recalled upon antigen-re-exposure in absence of rapid CD8 recall responses could favor initial viral replication. There is evidence supporting this paradigm- a previous macaque study by Buge et al demonstrated that inclusion of alum-adjuvanted gp120 boost to a DDM vaccination regimen resulted higher peak viremia following a SHIV challenge compared to DDM alone. Several studies have shown that a combination regimen of Env subunit proteins and viral vectored vaccines does not improve control of viremia compared to subunit protein immunization alone(39, 40). In light of the study by Buge et al and the present findings, it would be important to include a viral vector alone vaccine arm to determine whether of addition of adjuvanted protein boost results in higher peak viremia. This is critical as extended boosting regimens with subunit proteins are being tested in RV144 vaccinees as a means to overcome waning humoral immune responses over time. While multiple booster immunizations offer the advantage of augmenting magnitude and quality of antibody responses, their potential to engender target CD4+ T cells without boosting CD8 responses could tip the balance in favor of viral replication. In this context, characterizing and understanding CD4 responses in terms of CXCR5 and CXCR3 expression within genital and rectal mucosa, the major portals of HIV entry, could provide significant insights into their roles in acquisition of infection.
In conclusion, our data present an integrated view of CD4 helper determinants of Env antibody responses in a multicomponent HIV vaccine in rhesus macaques. The study adds to our understanding of vaccine-induced immune responses; further research is needed to determine the application of our findings to other leading poxvirus vectors and to HIV vaccine outcomes in humans.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Yerkes Division of Research Resources Stephanie Ehnert, Christopher Souder, Robert Sheffield and all the animal care staff for immunizations, infections sample collections, and animal care. We thank Drs. Elizabeth Strobert, Sherrie Jean and veterinary staff for veterinary counsel. We are grateful to the Emory Flow Cytometry core, Barbara Cervasi and Kiran Gill for cell sorting. We thank Nancy Miller for SIV251challenge stock. We thank Patricia Earl and Jeffrey Americo for providing the MVA virus. We thank the Emory CFAR Virology Core for viral load assays and the NIH AIDS Research and Reference Reagent Program for the provision of peptides. We thank the CFAR virology core, Benton Lawson and Melon T Nega for assays on viral load. We are grateful to Shane Crotty and Colin Havenar-Daughton for critical input regarding CXCR5 staining in rhesus samples. We thank Chris Igbebu, Rama S. Akondy, and Vijayakumar Velu for suggestions on antibodies for flow cytometry. The authors are thankful to Michael J Sabula for technical help.
This study was supported in part by the NIH grants PO1 AI088575 to RRA, P51 OD011132 to Yerkes National Primate Research Center, P30 AI50409 to Emory Center for AIDS Research, HHSN27201100016C to DM. Partial support was also provided by the Division of Intramural Research, NIAID, NIH.
REFERENCES
- 1.Slifka MK, Matloubian M, Ahmed R. Bone marrow is a major site of long-term antibody production after acute viral infection. Journal of virology. 1995;69:1895–1902. doi: 10.1128/jvi.69.3.1895-1902.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan TD, Gatto D, Wood K, Camidge T, Basten A, Brink R. Antigen affinity controls rapid T-dependent antibody production by driving the expansion rather than the differentiation or extrafollicular migration of early plasmablasts. J Immunol. 2009;183:3139–3149. doi: 10.4049/jimmunol.0901690. [DOI] [PubMed] [Google Scholar]
- 3.Crotty S. Follicular helper CD4 T cells (TFH) Annual review of immunology. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 4.Cyster JG. Shining a light on germinal center B cells. Cell. 2010;143:503–505. doi: 10.1016/j.cell.2010.10.036. [DOI] [PubMed] [Google Scholar]
- 5.Pallikkuth S, Parmigiani A, Silva SY, George VK, Fischl M, Pahwa R, Pahwa S. Impaired peripheral blood T-follicular helper cell function in HIV-infected nonresponders to the 2009 H1N1/09 vaccine. Blood. 2012;120:985–993. doi: 10.1182/blood-2011-12-396648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He J, Tsai LM, Leong YA, Hu X, Ma CS, Chevalier N, Sun X, Vandenberg K, Rockman S, Ding Y, Zhu L, Wei W, Wang C, Karnowski A, Belz GT, Ghali JR, Cook MC, Riminton DS, Veillette A, Schwartzberg PL, Mackay F, Brink R, Tangye SG, Vinuesa CG, Mackay CR, Li Z, Yu D. Circulating precursor CCR7(lo)PD-1(hi) CXCR5(+) CD4(+) T cells indicate Tfh cell activity and promote antibody responses upon antigen reexposure. Immunity. 2013;39:770–781. doi: 10.1016/j.immuni.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Morita R, Schmitt N, Bentebibel SE, Ranganathan R, Bourdery L, Zurawski G, Foucat E, Dullaers M, Oh S, Sabzghabaei N, Lavecchio EM, Punaro M, Pascual V, Banchereau J, Ueno H. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34:108–121. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bentebibel SE, Lopez S, Obermoser G, Schmitt N, Mueller C, Harrod C, Flano E, Mejias A, Albrecht RA, Blankenship D, Xu H, Pascual V, Banchereau J, Garcia-Sastre A, Palucka AK, Ramilo O, Ueno H. Induction of ICOS+CXCR3+CXCR5+ TH cells correlates with antibody responses to influenza vaccination. Science translational medicine. 2013;5 doi: 10.1126/scitranslmed.3005191. 176ra132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Locci M, Havenar-Daughton C, Landais E, Wu J, Kroenke MA, Arlehamn CL, Su LF, Cubas R, Davis MM, Sette A, Haddad EK, Poignard P, Crotty S. Human circulating PD-(+)1CXCR3(−)CXCR5(+) memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. 2013;39:758–769. doi: 10.1016/j.immuni.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amara RR, Villinger F, Altman JD, Lydy SL, O’Neil SP, Staprans SI, Montefiori DC, Xu Y, Herndon JG, Wyatt LS, Candido MA, Kozyr NL, Earl PL, Smith JM, Ma HL, Grimm BD, Hulsey ML, McClure HM, McNicholl JM, Moss B, Robinson HL. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Vaccine. 2002;20:1949–1955. doi: 10.1016/s0264-410x(02)00076-2. [DOI] [PubMed] [Google Scholar]
- 11.Van Rompay KK, Greenier JL, Cole KS, Earl P, Moss B, Steckbeck JD, Pahar B, Rourke T, Montelaro RC, Canfield DR, Tarara RP, Miller C, McChesney MB, Marthas ML. Immunization of newborn rhesus macaques with simian immunodeficiency virus (SIV) vaccines prolongs survival after oral challenge with virulent SIVmac251. Journal of virology. 2003;77:179–190. doi: 10.1128/JVI.77.1.179-190.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crotty S, Aubert RD, Glidewell J, Ahmed R. Tracking human antigen-specific memory B cells: a sensitive and generalized ELISPOT system. Journal of immunological methods. 2004;286:111–122. doi: 10.1016/j.jim.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 13.Silveira EL, Kasturi SP, Kovalenkov Y, Rasheed AU, Yeiser P, Jinnah ZS, Legere TH, Pulendran B, Villinger F, Wrammert J. Vaccine-induced plasmablast responses in rhesus macaques: Phenotypic characterization and a source for generating antigen-specific monoclonal antibodies. Journal of immunological methods. 2014 doi: 10.1016/j.jim.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwa S, Lai L, Gangadhara S, Siddiqui M, Pillai VB, Labranche C, Yu T, Moss B, Montefiori DC, Robinson HL, Kozlowski PA, Amara RR. CD40L–adjuvanted DNA/modified vaccinia virus Ankara simian immunodeficiency virus SIV239 vaccine enhances SIV-specific humoral and cellular immunity and improves protection against a heterologous SIVE660 mucosal challenge. Journal of virology. 2014;88:9579–9589. doi: 10.1128/JVI.00975-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montefiori DC. Coligan John E et al. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Chapter 12. Current protocols in immunology. 2005;(Unit 12):11. doi: 10.1002/0471142735.im1211s64. [DOI] [PubMed] [Google Scholar]
- 16.Schaerli P, Willimann K, Lang AB, Lipp M, Loetscher P, Moser B. CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. The Journal of experimental medicine. 2000;192:1553–1562. doi: 10.1084/jem.192.11.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iyer SS, Latner DR, Zilliox MJ, McCausland M, Akondy RS, Penaloza-Macmaster P, Hale JS, Ye L, Mohammed AU, Yamaguchi T, Sakaguchi S, Amara RR, Ahmed R. Identification of novel markers for mouse CD4(+) T follicular helper cells. European journal of immunology. 2013;43:3219–3232. doi: 10.1002/eji.201343469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kroenke MA, Eto D, Locci M, Cho M, Davidson T, Haddad EK, Crotty S. Bcl6 and Maf cooperate to instruct human follicular helper CD4 T cell differentiation. J Immunol. 2012;188:3734–3744. doi: 10.4049/jimmunol.1103246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petrovas C, Yamamoto T, Gerner MY, Boswell KL, Wloka K, Smith EC, Ambrozak DR, Sandler NG, Timmer KJ, Sun X, Pan L, Poholek A, Rao SS, Brenchley JM, Alam SM, Tomaras GD, Roederer M, Douek DC, Seder RA, Germain RN, Haddad EK, Koup RA. CD4 T follicular helper cell dynamics during SIV infection. The Journal of clinical investigation. 2012;122:3281–3294. doi: 10.1172/JCI63039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yusuf I, Kageyama R, Monticelli L, Johnston RJ, Ditoro D, Hansen K, Barnett B, Crotty S. Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150) J Immunol. 2010;185:190–202. doi: 10.4049/jimmunol.0903505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haynes NM, Allen CD, Lesley R, Ansel KM, Killeen N, Cyster JG. Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. J Immunol. 2007;179:5099–5108. doi: 10.4049/jimmunol.179.8.5099. [DOI] [PubMed] [Google Scholar]
- 22.Ballesteros-Tato A, Leon B, Graf BA, Moquin A, Adams PS, Lund FE, Randall TD. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity. 2012;36:847–856. doi: 10.1016/j.immuni.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monteiro P, Gosselin A, Wacleche VS, El-Far M, Said EA, Kared H, Grandvaux N, Boulassel MR, Routy JP, Ancuta P. Memory CCR6+CD4+ T cells are preferential targets for productive HIV type 1 infection regardless of their expression of integrin beta7. J Immunol. 2011;186:4618–4630. doi: 10.4049/jimmunol.1004151. [DOI] [PubMed] [Google Scholar]
- 24.Smith JM, Amara RR, McClure HM, Patel M, Sharma S, Yi H, Chennareddi L, Herndon JG, Butera ST, Heneine W, Ellenberger DL, Parekh B, Earl PL, Wyatt LS, Moss B, Robinson HL. Multiprotein HIV type 1 clade B DNA/MVA vaccine: construction, safety, and immunogenicity in Macaques. AIDS research and human retroviruses. 2004;20:654–665. doi: 10.1089/0889222041217419. [DOI] [PubMed] [Google Scholar]
- 25.Robinson HL, Sharma S, Zhao J, Kannanganat S, Lai L, Chennareddi L, Yu T, Montefiori DC, Amara RR, Wyatt LS, Moss B. Immunogenicity in macaques of the clinical product for a clade B DNA/MVA HIV vaccine: elicitation of IFN-gamma, IL-2, and TNF-alpha coproducing CD4 and CD8 T cells. AIDS Res Hum Retroviruses. 2007;23:1555–1562. doi: 10.1089/aid.2007.0165. [DOI] [PubMed] [Google Scholar]
- 26.Chtanova T, Tangye SG, Newton R, Frank N, Hodge MR, Rolph MS, Mackay CR. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. J Immunol. 2004;173:68–78. doi: 10.4049/jimmunol.173.1.68. [DOI] [PubMed] [Google Scholar]
- 27.Rivino L, Messi M, Jarrossay D, Lanzavecchia A, Sallusto F, Geginat J. Chemokine receptor expression identifies Pre-T helper (Th)1, Pre-Th2, and nonpolarized cells among human CD4+ central memory T cells. The Journal of experimental medicine. 2004;200:725–735. doi: 10.1084/jem.20040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng NY, Mays I, Garman L, Helms C, James J, Air GM, Capra JD, Ahmed R, Wilson PC. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wrammert J, Onlamoon N, Akondy RS, Perng GC, Polsrila K, Chandele A, Kwissa M, Pulendran B, Wilson PC, Wittawatmongkol O, Yoksan S, Angkasekwinai N, Pattanapanyasat K, Chokephaibulkit K, Ahmed R. Rapid and massive virus-specific plasmablast responses during acute dengue virus infection in humans. Journal of virology. 2012;86:2911–2918. doi: 10.1128/JVI.06075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gosselin A, Monteiro P, Chomont N, Diaz-Griffero F, Said EA, Fonseca S, Wacleche V, El-Far M, Boulassel MR, Routy JP, Sekaly RP, Ancuta P. Peripheral blood CCR4+CCR6+ and CXCR3+CCR6+CD4+ T cells are highly permissive to HIV-1 infection. J Immunol. 2010;184:1604–1616. doi: 10.4049/jimmunol.0903058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haynes BF, McElrath MJ. Progress in HIV-1 vaccine development. Current opinion in HIV and AIDS. 2013;8:326–332. doi: 10.1097/COH.0b013e328361d178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Streeck H, D’Souza MP, Littman DR, Crotty S. Harnessing CD4(+) T cell responses in HIV vaccine development. Nature medicine. 2013;19:143–149. doi: 10.1038/nm.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dai P, Wang W, Cao H, Avogadri F, Dai L, Drexler I, Joyce JA, Li XD, Chen Z, Merghoub T, Shuman S, Deng L. Modified Vaccinia Virus Ankara Triggers Type I IFN Production in Murine Conventional Dendritic Cells via a cGAS/STING-Mediated Cytosolic DNA-Sensing Pathway. PLoS pathogens. 2014;10:e1003989. doi: 10.1371/journal.ppat.1003989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teigler JE, Phogat S, Franchini G, Hirsch VM, Michael NL, Barouch DH. The canarypox virus vector ALVAC induces distinct cytokine responses compared to the vaccinia virus-based vectors MVA and NYVAC in rhesus monkeys. Journal of virology. 2014;88:1809–1814. doi: 10.1128/JVI.02386-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohr E, Cunningham AF, Toellner KM, Bobat S, Coughlan RE, Bird RA, MacLennan IC, Serre K. IFN-{gamma} produced by CD8 T cells induces T-bet-dependent and -independent class switching in B cells in responses to alum-precipitated protein vaccine. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:17292–17297. doi: 10.1073/pnas.1004879107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Serre K, Cunningham AF, Coughlan RE, Lino AC, Rot A, Hub E, Moser K, Manz R, Ferraro A, Bird R, Toellner KM, Demengeot J, MacLennan IC, Mohr E. CD8 T cells induce T-bet-dependent migration toward CXCR3 ligands by differentiated B cells produced during responses to alum-protein vaccines. Blood. 2012;120:4552–4559. doi: 10.1182/blood-2012-03-417733. [DOI] [PubMed] [Google Scholar]
- 37.Hale JS, Youngblood B, Latner DR, Mohammed AU, Ye L, Akondy RS, Wu T, Iyer SS, Ahmed R. Distinct memory CD4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection. Immunity. 2013;38:805–817. doi: 10.1016/j.immuni.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paiardini M, Cervasi B, Reyes-Aviles E, Micci L, Ortiz AM, Chahroudi A, Vinton C, Gordon SN, Bosinger SE, Francella N, Hallberg PL, Cramer E, Schlub T, Chan ML, Riddick NE, Collman RG, Apetrei C, Pandrea I, Else J, Munch J, Kirchhoff F, Davenport MP, Brenchley JM, Silvestri G. Low levels of SIV infection in sooty mangabey central memory CD(4)(+) T cells are associated with limited CCR5 expression. Nature medicine. 2011;17:830–836. doi: 10.1038/nm.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pegu P, Vaccari M, Gordon S, Keele BF, Doster M, Guan Y, Ferrari G, Pal R, Ferrari MG, Whitney S, Hudacik L, Billings E, Rao M, Montefiori D, Tomaras G, Alam SM, Fenizia C, Lifson JD, Stablein D, Tartaglia J, Michael N, Kim J, Venzon D, Franchini G. Antibodies with high avidity to the gp120 envelope protein in protection from simian immunodeficiency virus SIV(mac251) acquisition in an immunization regimen that mimics the RV-144 Thai trial. Journal of virology. 2013;87:1708–1719. doi: 10.1128/JVI.02544-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Earl PL, Wyatt LS, Montefiori DC, Bilska M, Woodward R, Markham PD, Malley JD, Vogel TU, Allen TM, Watkins DI, Miller N, Moss B. Comparison of vaccine strategies using recombinant env-gag-pol MVA with or without an oligomeric Env protein boost in the SHIV rhesus macaque model. Virology. 2002;294:270–281. doi: 10.1006/viro.2001.1345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.