Abstract
O -acetylserine sulfhydrylase A (CysK) is the pyridoxal 5′-phosphate-dependent enzyme that catalyzes the final reaction of cysteine biosynthesis in bacteria. CysK was initially identified in a complex with serine acetyltransferase (CysE), which catalyzes the penultimate reaction in the synthetic pathway. This “cysteine synthase” complex is stabilized by insertion of the CysE C-terminus into the active-site of CysK. Remarkably, the CysK/CysE binding interaction is conserved in most bacterial and plant systems. For the past 40 years, CysK was thought to function exclusively in cysteine biosynthesis, but recent studies have revealed a repertoire of additional “moonlighting” activities for this enzyme. CysK and its paralogs influence transcription in both Gram-positive bacteria and the nematode C. elegans. CysK also activates an antibacterial nuclease toxin produced by uropathogenic Escherichia coli. Intriguingly, each moonlighting activity requires a binding partner that invariably mimics the C-terminus of CysE to interact with the CysK active site.
1. Introduction
Cysteine biosynthesis in bacteria occurs through two interconnected pathways. Sulfate is reduced to bisulfide (HS−) through a multistep, energy-dependent process, and L-serine is activated via O-acetylation to produce O-acetylserine (OAS). The two branches converge on the enzyme O-acetylserine sulfhydrylase (OASS) (EC 2.5.1.47), a pyridoxal 5′-phosphate (PLP)-dependent enzyme that catalyzes the replacement of the β-acetotoxy group of OAS with bisulfide (Figure 1). CysK was the first OASS isoform to be identified and was originally isolated and characterized in Salmonella typhimurium [1–11]. Those early studies showed that CysK can physically associate with serine acetyltransferase (CysE), which catalyses the preceding step in the biosynthetic pathway. CysE is a hexameric protein that catalyzes an acetyl-transfer reaction from acetyl-CoA to serine via a random-order ternary complex reaction mechanism [12]. Together, the two enzymes form a heteromeric complex known as the cysteine synthase complex (CSC) [2, 13]. Subsequent studies identified CysK and CysE homologues also in plants [14, 15] and mycobacteria [16]. CysE shows the typical left-handed β-helix domain of acetyltransferases, but its C-terminus is flexible and not resolved in available crystal structures [17, 18]. It was proposed [19], and later demonstrated [20–22], that the C-terminal tail of CysE inserts into the active site of CysK. The three-dimensional structure of the CSC has not yet been determined. However, the structure of Haemophilus influenzae CysK (HiCysK) in complex with a decapeptide corresponding to the C-terminus of HiCysE supports the notion that the CysK active site serves as the anchor point for complex formation [20]. Interestingly, CysE proteins from different organisms show a high degree of sequence variability at the C-terminus (Figure 2), yet carry an invariant C-terminal Ile residue. Ile plays a pivotal role in complex formation and its removal completely prevents CSC association [23]. From the beginning, the functional significance of the CSC was unclear because no substrate channeling was envisaged for the complex [2, 24, 25]. Additionally, only a fraction (~5–25%) of total CysK was reported to be in the complex [13, 26]. Subsequently, it has been proposed that CysK-binding provides a mechanism to protect the bacterial and plant CysE from cold-inactivation and proteolysis [27] [28].
FIGURE 1.
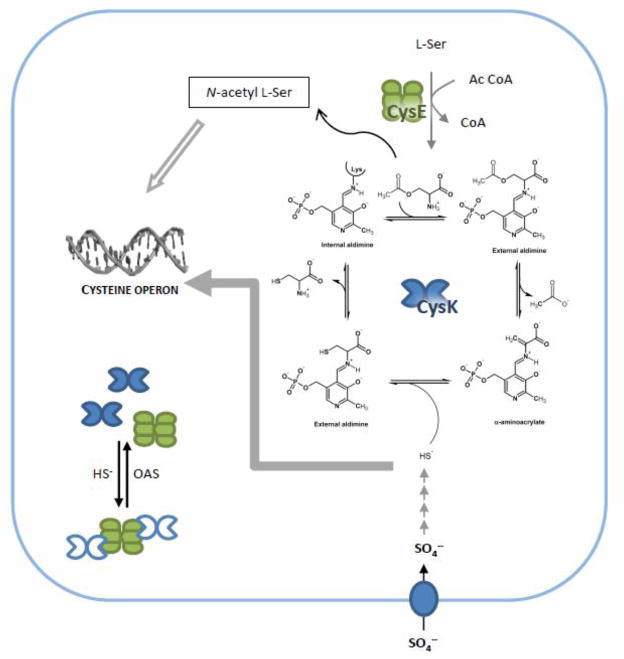
Cysteine biosynthesis in bacteria: the role of CysK, its reaction mechanism and the role of reaction intermediates in gene regulation. Closed grey arrow represents inhibition of transcription, open grey arrow represents activation of transcription. Closed symbols represent fully active enzymes, open symbols represent inhibited enzymes. OAS spontaneously converts to NAS with a pH dependent rate that is 1%/min at pH 7.6 [45].
FIGURE 2.
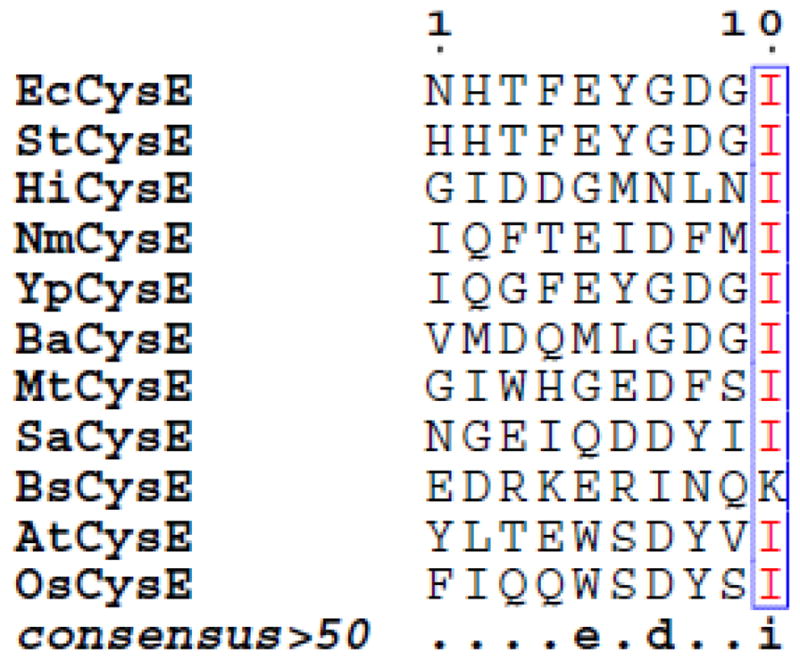
Comparison of C-terminal decapeptides of CysE from different organisms (proteobacteria: EcCysE, Escherichia coli CysE; StCysE, Salmonella typhimurium CysE; HiCysE, Haemophilus influenzae CysE; NmCysE, Neisseria menigitidis CysE; YpCysE, Yersinia pestis CysE; BaCysE, Brucella abortus CysE. Mycobacteria: MtCysE, Mycobacterium tuberculosis CysE. Gram-positive bacteria: SaCysE, Staphylococcus aureus CysE; BsCysE, Bacillus subtilis CysE. Plants: AtCysE, Arabidopsis thaliana CysE; OsCysE, Oryza sativa CysE). Similarity scores were calculated by the ESPript program [100] using the percentage of equivalent residues coloring scheme set at global score of 0.7.
A second OASS isozyme, CysM, whose biological role remains unclear [11], has been identified in several bacteria. CysM is less abundant than CysK [26] and can use alternative sulfur donors like thiosulfate [26, 29, 30]. Some reports suggest that CysM is differentially expressed during anaerobic growth conditions [26, 31], but these findings have not been supported by more recent studies (SalCom, [32]). In contrast with CysK, CysM does not interact with CysE [23, 33], suggesting that the two isozymes play distinct roles in the regulation of cysteine biosynthesis.
The interaction between CysK and CysE appears quite unique, yet very similar binding interactions have evolved between CysK and other partner proteins. These various interactions account for the different moonlighting functions of CysK (Table 1). In Bacillus subtilis, CysK modulates the affinity of an Rrf2-type transcription factor for its operator sequences, thereby regulating expression of the cysteine regulon. In E. coli CysK acts as a so-called “permissive” factor to activate an antibacterial contact-dependent growth inhibition (CDI) toxin [34]. Although alternative partners bind CysK for new functions, these interactions could also directly influence cysteine biosynthesis by inhibiting CysK activity. Here, we discuss the biological roles of known CysK complexes in the context of cysteine metabolism.
Table 1.
Interactors of CysK and CysK paralogs
2. Regulation of cysteine biosynthesis in bacteria
Cysteine plays important roles in diverse cellular processes like bacterial swarming motility and oxidative-stress responses [37–39]. Because of the central role played by cysteine in bacterial physiology, it has been proposed that enzymes involved in its biosynthesis [40, 41], and OASS isozymes in particular [42, 43], could be exploited as targets for the development of new antibiotics. However, high levels of intracellular cysteine are potentially toxic to the cell. Therefore, the biosynthesis and degradation of this amino acid must be carefully balanced. Excess cysteine is degraded to pyruvate, ammonia and bisulfide by cysteine desulfhydrases [29, 44], which are induced by cysteine in the growth medium [29]. Cysteine biosynthesis is strictly controlled at three levels: i) regulation of gene expression, ii) feed-back inhibition of CysE activity and iii) CysE/CysK complex formation. All genes involved in cysteine biosynthesis, with the exception of cysE and cysB, are organized in the cysteine regulon, whose transcription is under the control of the transcriptional activator CysB [45]. CysB is a LysR-type transcription factor whose binding to DNA is stimulated by the inducer N-acetylserine (NAS). NAS is generated non-enzymatically from OAS through spontaneous O- to N-acetyl migration [45] (Figure 1). Thus, increased NAS concentrations reflect the accumulation of OAS, which is in turn determined by the intracellular concentration of bisulfide. The positive effect of NAS on cysteine regulon expression is counteracted by bisulfide and thiosulfate, which both act as anti-inducers [45]. The two OASS isozymes are encoded in separate transcriptional units in E. coli and S. typhimurium. The cysM gene is the last cistron of the cysPUWAM operon, which encodes proteins for the import of sulfate and thiosulfate. In contrast, cysK constitutes an independent monocistronic transcriptional unit. Interestingly, bioinformatic analysis of the E. coli cysPUWAM operon with the RegulonDB resource (http://regulondb.ccg.unam.mx) suggests that cysM is subject to transcriptional attenuation, which would allow independent regulation with respect to the other upstream genes (BC, unpublished observations).
Regulation of cysteine biosynthesis at the post-transcriptional level is focused on the activities of CysK and CysE. Cysteine acts as a competitive feed-back inhibitor to block CysE activity [12, 46]. The structural basis of this regulation has been elucidated for HiCysE, which shows a C-terminal rearrangement that positions residues Gln254 – Ile257 into the enzyme acetyl-CoA binding site [18]. This conformational change prevents unproductive acetylation of enzyme-bound cysteine. For cytosolic CysE in plants, the Ki for cysteine is 2 μM, which is well below typical intracellular cysteine concentrations (10–20 μM) [47]. The Ki increases substantially to 70 μM when CysE is bound to CysK [47], indicating that the enzyme is far less sensitive to feedback inhibition when found in the CSC. The role of CSC in cysteine metabolism is less clear and is likely to differ between bacteria and plants. The CSC is stabilized by bisulfide and destabilized by OAS [2, 48] and thus is responsive to the sulfur-status of the cell. OAS dissociates the complex by direct competition with CysE for binding to the active site of CysK [20]. Bisulfide stabilizes the complex, even in the presence of OAS, presumably by occupying an allosteric binding site that was first described for Salmonella typhimurium CysK (StCysK) [10]. CysK within the complex is inhibited because its active site is occluded by the C-terminus of CysE. However, the complex protects CysE from aggregation and proteolysis [25, 27]. Complex formation only affects the enzyme kinetics of plant CysE and allows fine-tuning of OAS production to match sulfur availability (vide infra). Importantly, cysE expression is independent of CysB and the cysteine regulon. Therefore, induction of the regulon by sulfur starvation influences CSC formation by altering the cellular ratio of CysK to CysE (Figure 1).
3. Structural features of the CysK/CysE interaction
The three-dimensional structure of the CSC is unknown. However, structural and biochemical features of the complex have been elucidated by surface plasmon resonance, steady-state fluorescence, site-directed mutagenesis, two-hybrid system, pre-steady state kinetics, molecular modeling and peptide-binding studies [19–23, 25, 27, 28, 47, 49–61]. Mino and co-workers first identified the C-terminal ten amino acid residues of CysE as critical for CysK-binding [22]. This finding was confirmed by crystallography of a complex between HiCysK and the isolated C-terminal decapeptide of HiCysE [20]. Only the last four residues of the HiCysE peptide were detected in the electron density map, and similar results were later obtained with the analogous complex from Mycobacterium tuberculosis [16]. Herein, we refer to the four CysE residues that make contact with CysK as P1-P2-P3-P4. Site-directed mutagenesis and deletion analysis show that the C-terminal Ile (residue P4) is fundamental for the CysE/CysK binding interaction [22, 23]. Deletion of the C-terminal Ile, or substitution with Ala or Glu, consistently impair complex formation [23]. However, whereas the lack of a C-terminal Ile residue precludes CSC formation, the reverse is not always true. Indeed, there are CysE proteins that carry a canonical C-terminal Ile, yet do not form a CSC (vide infra the example of E. histolytica CysE). Computational energy mapping of the HiCysE peptide interaction with HiCysK shows that the P4 Ile residue accounts for about 80% of total binding energy. The P2 and P3 positions (Asn and Leu for HiCysE, respectively) account for about 10% each, and the P1 residue negatively impacts binding [60]. A map of the interactions between the last four residues of HiCysE and HiCysK active site is shown in Figure 3A. As expected from the energy mapping, most of the interactions involve the C-terminal Ile residue. The carboxylic moiety of Ile binds to the carboxylate subsite of CysK active site, and its side-chain fits into the hydrophobic pocket normally occupied by the β-acetotoxy group of OAS.
FIGURE 3.
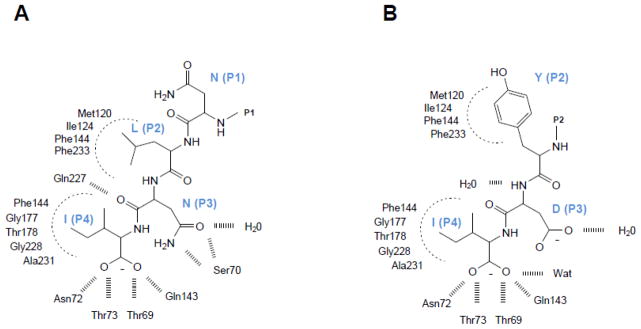
Map of interactions of HiCysE C-terminal MNLNI wild-type peptide (A) and the mutant MNYDI (B) with HiCysK active site. Modified from [60]. In the crystallographic structure only the last four and three residues are visible for MNLNI and MNYDI complexes, respectively.
Computational docking of a virtual library of four hundred peptides of general formula MNX1X2I (X = any amino acid) into the CysK active site suggested that high binding affinity is associated with an aromatic residue at position X1 (P2) and a strong H-bonding donor at position X2 (P3) [56]. Subsequent experimental measurement of peptide binding affinities for HiCysK, E. coli CysK (EcCysK) and StCysK confirms the predicted increases in binding affinity [53, 60]. However, the key energetic contribution to binding arises from an aromatic residue, specifically Tyr, at position P2, irrespective of residues at P3. Inspection of the three-dimensional structure of MNYDI peptide in complex with HiCysK shows that the aromatic P2 side-chain occupies a large hydrophobic pocket that is left partially unoccupied by the side chain of Leu in the wild-type peptide (Figures 3A and 3B). Aromatic residues at position P2 were also reported to increase the affinity of Entamoeba hystolytica CysE (EhCysE) for EhCysK [61]. In E. hystolytica, the CSC is quite unstable and probably does not form under physiological conditions. However, Ser→Asp and Pro→Trp mutations at positions P1 and P2 (respectively) significantly increase the affinity of EhCysE for EhCysK [61]. Interestingly, proteobacterial CysE enzymes never carry an aromatic residue at position P2, whereas nearly all CysK moonlighting partners have Tyr, Phe or His residues at this position. The one exception is a contact-dependent inhibition (CDI) toxin from Pseudomonas that has an Ala residue in this position (Table 1 and Figure 6). Together, these observations suggest that moonlighting partners have a higher affinity for CysK than CysE and therefore could compete with the latter enzyme for binding to the CysK active site. The only available structure of plant CysK bound to a CysE C-terminal peptide shows some important differences with respect to the bacterial complex [55]. Eight of ten Arabidopsis thaliana CysE (AtCysE) residues are resolved in the structure with the five C-terminal residues making contacts with residues in CysK active site. The five N-terminal residues bind within a cleft at the active site opening and the last two N-terminal residues are solvent exposed. In contrast to the H. influenzae complex, negative co-operativity was observed with decapeptide binding to AtCysK. While these structural studies have allowed an in-depth mapping of the interactions between CysE and the CysK active site, very little is known about interactions outside the active site that contribute to CSC stabilization. Three AtCysK residues were identified by site-directed mutagenesis as fundamental for CSC formation [51]. These residues lie within a loop that bulges at the entrance of the active site and is not conserved between bacterial CysK and CysM [62]. This loop probably interacts directly with CysE, but nothing is known about of the contribution of residues more distal to the active site. In the absence of a comprehensive interaction network, the most remarkable finding is the observation that substitution of the C-terminal Ile of CysE with Ala completely prevents CSC formation. Upon binding the active-site pocket, the C-terminal Ile triggers a conformational change that probably closes the CysK active site, thereby promoting high-affinity binding to CysE [59, 63]. This conformational transition could be obligatory for the formation of high-affinity complexes with CysE and other binding partners.
4. The regulatory function of CysK/CysE interaction in plants
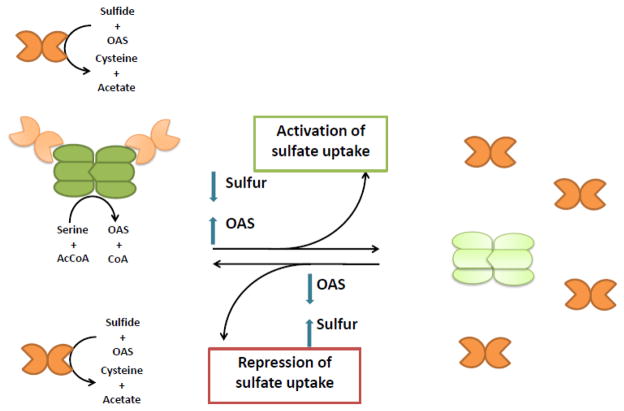
Despite insights into the structural features of the bacterial CysE/CysK interaction, the biological role of the CSC and its dynamics under different nutritional states is poorly understood. By contrast, the regulatory role of plant CSC is much better characterized. In plants, CysK is present in large excess over CysE (about 300 times more abundant in spinach chloroplasts) [15, 64, 65], and CysE is activated upon complex formation [2, 19, 25, 28]. Very recent data on Glycine max CysE indicate a 20-fold increase in catalytic efficiency upon binding to CysK. This is the result of both decreased KM for acetyl-CoA and increased kcat. Free CysE enzyme is less active and more susceptible to aggregation, inactivation and proteolysis [25]. However, because CysK is competitively inhibited within the CSC, productive cysteine biosynthesis should require a high CysK to CysE ratio. In this framework, CysE-bound CysK, though inactive, increases CysE stability and OAS production, while free CysK catalyzes cysteine biosynthesis. Biochemical experiments support this view, and the maximum rate of cysteine production is achieved in vitro when CysK is added in large excess over CysE [25]. Feed-back inhibition of CysE activity limits the overall concentration of cysteine that can be produced, but the Ki for cysteine in the plant system increases about 70-fold upon CSC formation [2, 19, 52]. Notably, this effect is not observed with the bacterial complex [2, 19, 52]. Thus, overall cysteine synthase activity is modulated by CSC assembly in plants. The stability of the CSC in turn depends upon the relative concentrations of OAS, which can dissociate the complex at concentrations around 100 μM [66], and bisulfide, which stabilizes the complex [25]. Within this framework, the CSC was proposed by Hell and colleagues to act as a switch controlling cysteine biosynthesis in plants [48] (Figure 4). When sulfur supply is adequate, OAS levels are low due to rapid conversion into cysteine by free CysK. When sulfur becomes scarce, OAS accumulates because the cell lacks the bisulfide required for cysteine synthesis. There are at least two consequences of this model. First, there is a short-term effect, in which CSC dissociation leads to partial inactivation of CysE, thereby sparing the valuable intermediate acetyl-CoA. A second, long-term effect is the OAS(NAS)-dependent transcriptional activation of genes related to sulfate transport, activation and reduction. This model was further developed with the observation that the relatively high KM of CysK for OAS (ranging from about 300 – 700 μM), allows OAS to modulate multiple stages of sulfur metabolism. OAS acts as an inducer in the form of NAS, as a substrate for CysK and as a dissociation factor for the CSC [66]. Thus, relatively low concentrations of OAS dissociate the CSC and higher levels are required to maximally stimulate CysK activity. This strict regulation of plant CysK activity is consistent with the enzyme’s key position in cysteine metabolism [67].
FIGURE 4.
CSC as a sensor for sulfur supply in plants (modified from [48]).
CysE hexamer is represented in green (dark green: fully active; light green: inactive), CysK dimer is represented in orange (dark orange: fully active; light orange: partially inhibited).
Although several aspects of CSC function and regulation are shared between the bacterial and plant enzymes, there are some key differences suggesting distinct functions in different organisms. For example, CysE within the bacterial CSC is more sensitive to cysteine-mediated feed-back inhibition than the plant enzyme. It has been postulated that this effect counterbalances the stimulatory effect of high free CysK concentrations on cysteine production [19]. However, maximal rates of cysteine production have been reported when bacterial CysK is added in large excess over CysE [25]. Moreover, increased CysE activity within the CSC would only increase cysteine production if the limiting step of cysteine biosynthesis is catalyzed by CysE because CysK has higher catalytic efficiency. This has been established for the plant enzymes [67] [25] and suggested for the bacterial enzymes [58]. However, there are no studies that report detailed comparisons between the plant and bacterial systems carried out under the same conditions. Pioneering work by Becker and Tomkins suggests a possible role for bacterial CSC in the regulation of cysteine biosynthesis [68]. CysE mutations that alter the interaction with CysK are able to induce cysteine auxotrophy in S. typhimurium [54]. More data collected on stable and homogeneous preparations of bacterial CSC are required to confirm or reject a regulatory role for CSC in bacteria similar to that observed in plants.
5. CysK as a transcriptional regulator
5.1 Interaction with CymR in Gram-positive bacteria
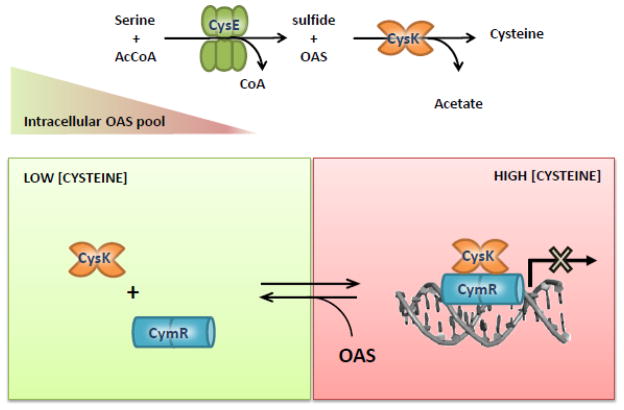
All cells, from bacteria to higher eukaryotes, adapt to their environment by sensing nutrient availability and transmitting that information into appropriate transcriptional responses. Because enzymes have evolved exquisite substrate specificity and are often regulated allosterically, they are in principle well suited to act as nutrient sensors. However, there are only a few examples of biosynthetic enzymes that directly regulate transcription. One example is the poorly characterized MocR family, in which an enzyme-like PLP-binding domain is fused to the DNA-binding domain of a transcription factor. The PLP-binding domain may retain its ancestral catalytic activity, although in all instances examined to date, the reactions do not proceed beyond initial aldimine formation [69–72]. In other instances, trigger enzymes physically interact with a transcriptional regulator depending on substrate or product availability [73]. In Gram-positive bacteria like B. subtilis and Staphylococcus aureus, the expression of key proteins in the control of cysteine metabolism is repressed by an Rrf2-type transcription factor, CymR [74–76]. CymR is dimeric in solution and binds to DNA as a dimer or a tetramer using a common helix-turn-helix motif [36]. The affinity of the CymR/DNA interaction increases up to seven-fold in the presence of CysK [75], concomitant with a decrease in the dissociation rate constant from about 10−3 s−1 to 10−4 s−1. Although the molecular details of the CymR/CysK interaction and the mechanism by which CysK alters DNA-binding affinity are poorly characterized, the role of CysK in transcription regulation has been investigated in detail. Two types of oligomeric organization have been described for the CysK/CymR complex in vitro. CysK dimers bind to CymR dimers, forming a tetrameric complex with a dissociation constant of 100 nM. This complex can further oligomerize to yield a dimer of tetramers with a dissociation constant of 4 μM [75]. Because the intracellular CymR concentration is ~0.4 μM in the presence of methionine and cystine, the latter octameric complex is unlikely to form under physiological conditions. Interestingly, CysK/CymR complex formation is inhibited by micromolar concentrations of OAS, but the complex is unaffected by millimolar NAS [75]. The concentration of OAS required for half-maximal dissociation (200 μM) is comparable to the intracellular OAS concentration in B. subtilis under cysteine-starvation conditions and about 10-fold higher than the concentration in cells grown in cysteine-rich medium. Therefore, the CysK/CymR complex can respond to changes in cysteine availability by sensing variations in OAS concentration. Low sulfur results in an accumulation of OAS to levels that are high enough to disrupt the CysK/CymR complex. Consequently, CymR dissociates from its operator sequences and genes that promote sulfur assimilation are de-repressed (Figure 5). When sulfur is in surfeit, OAS is rapidly converted into cysteine. Low OAS levels favor CysK/CymR complex formation and transcription of the target genes is downregulated. Although the physical interaction between CymR and the active site of CysK has not been directly demonstrated by structural and/or mutagenesis studies, indirect evidence suggests mimicry of the CysK/CysE interaction. First, the complex is dissociated by OAS with an apparent dissociation constant that is lower than the KM of CysK for this amino acid. Second, CymR carries a C-terminal Ile residue and an aromatic residue at P2, both of which are structural features required for stable binding to the CysK active site (Table 1). Finally, the C-terminal sequence of CymR is conserved between B. subtilis, S. aureus and Listeria innocua [36], which would be unusual for an unstructured sequence unless it plays a functional role. In contrast of Gram-negative bacteria, BsCysK does not bind CysE which, in fact, carries a Lys residue at the very C-terminus instead of the conserved Ile (Figure 2). These observations suggest that BsCysE does not compete with CymR for binding to the BsCysK active site, and this allows CymR to have a quite high dissociation constant (~100 nM) for CysK. CymR from S. aureus was suggested to bind to CysM instead of CysK, although the protein identified as CysM in S. aureus has a higher sequence identity with EcCysK (45%) than with EcCysM (35%) [77]. At present, the transcriptional regulator function of BsCysK is the only moonlighting function of CysK reported in the MoonProt database (http://www.moonlightingproteins.org).
FIGURE 5.
Model for regulation of cysteine biosynthesis in Gram-positive bacteria. Modified from [75].
5.2 The special case of the CYSL-1 paralog
Recent research focusing on the role of H2S as a neurotransmitter in nematodes has led to the identification of novel O-acetylserine sulfhydrylases (CYSL-1/4), whose biological roles are currently under investigation [35, 78]. Nematode CysK paralogs are cytosolic proteins whose sequences are more closely related to the plant enzymes [78]. Recombinant CYSL-1, -2 and -3 have been preliminarily characterized and all show low, though detectable, O-acetylserine sulfhydrylase activity with kcat/KM values in the range of 6–51 mM−1 s−1. These enzymes have high KM values for bisulfide, suggesting that this sulfhydrylase activity may have no biological significance [35, 78]. However, the dissociation constant of OAS for C. elegans CYSL is in the high micromolar range, suggesting that OAS might play a regulatory role similar to that in plants. In C. elegans, CYSL-1 interacts with EGL-9, a member of the proline-4-hydroxylase domain (PHD) enzymes that regulate the transcriptional activity of hypoxia inducible factor (HIF) through hydroxylation. Hydroxylated HIF is targeted for degradation and genes under its transcriptional control are thus downregulated [35]. In C. elegans, hypoxic conditions lead to increased H2S levels, which are known to activate HIF [79]. The molecular mechanism underlying this response to hypoxia is mediated by CYSL-1. EGL-1 binds CYSL-1 through a mechanism that was co-opted from the CysE/CysK interaction. In fact, the Glu-Tyr-Tyr-Ile C-terminal sequence of EGL-1 resembles the Asp-Tyr-Val-Ile C-terminal sequence of A. thaliana CysE (Table 1) and deletion or substitution of Ile with Ala disrupts binding to CYSL-1. An interesting feature of the EGL-1/CYSL-1 complex is the stabilizing effect of H2S, which is very similar to the effects observed with the bacterial and plant CSCs [2, 59] [48]. Here, H2S-dependent complex stabilization is exploited to fine-tune the inhibitory activity of EGL-1 on HIF and to coordinate the response to hypoxia with increased H2S inside the neurons. It has been speculated that CYSL-1 has an allosteric bisulfide-binding site, similar to that observed in StCysK, whose occupation by bisulfide stabilizes a partially closed conformation of the enzyme that also shows increased affinity for CysE [2, 10, 59]. Notably, both in the case of CymR and EGL-1 the interaction with CysK (or CysK paralog) is modulated by ligands, i.e. OAS and bisulfide, that fulfill a distinct function from that of enzyme substrates.
6. Toxin activation by CysK
Recent studies on bacterial contact-dependent growth inhibition (CDI) have revealed another unexpected link to CysK. CDI systems are found in many important Gram-negative pathogens, including Yersinia pestis, Neisseria meningitidis, Acinetobacter baumanii and Burkholderia pseudomallei, where they function in inter-cellular competition by delivering protein toxins into neighboring target bacteria [80, 81]. CDI is orchestrated by the CdiB/CdiA family of two-partner secretion proteins (TPS), which comprise a subset of type V secretion systems. CdiB is an outer-membrane transport protein that exports and assembles CdiA effector proteins onto the surface of CDI+ inhibitor cells (Fig. 6A). CdiA proteins are large – ranging from ~180 kDa in Moraxella catarrhalis to over 600 kDa in some Pseudomonads – and are predicted to form β-helical filaments that emanate several hundred angstroms from the cell surface [82]. CdiA binds to specific receptors on susceptible bacteria, then delivers its C-terminal toxin domain (CdiA-CT) into the target cell through a poorly understood translocation pathway. CDI+ bacteria also exchange CdiA-CT toxins with each other, but sibling cells are protected by specific CdiI immunity proteins, which bind to the CdiA-CT and neutralize toxin activity (Figure 6A) [80, 83–86]. Thus, CDI is used to suppress the growth of competing non-isogenic bacteria, while providing complete immunity to isogenic sibling cells.
FIGURE 6. Role for CysK during bacterial contact-dependent growth inhibition (CDI).
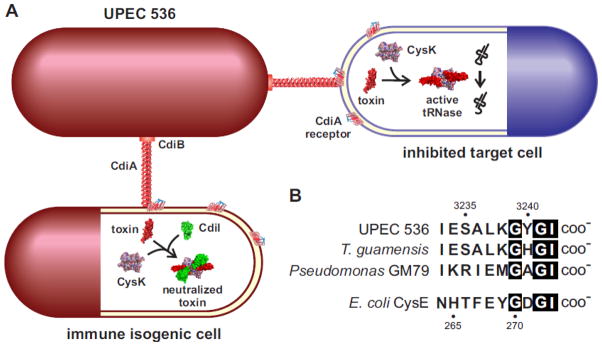
A. The CDI system from uropathogenic E. coli 536 (UPEC 536) deploys a CysK-dependent nuclease. CdiB secretes and assembles the filamentous CdiA protein on the surface of UPEC 536 cells. CdiA binds to a receptor on other E. coli cells and delivers a toxin domain derived from its C-terminus. The delivered toxin binds specifically to CysK in target cells, thereby activating the nuclease to cleave tRNA molecules. UPEC 536 also deliver toxins to sibling cells, but the toxin complex is inactivated by the CdiI immunity protein. The resulting ternary complex has no tRNase activity and isogenic sibling cells are not inhibited.
B. CdiA proteins from UPEC 536 and other bacteria carry a C-terminal peptide motif that resembles the C-terminus of CysE. The C-terminal sequences of CdiA proteins from UPEC 536 (Uniprot: Q0T963), Trabulsiella guamensis ATCC 49490 (A0A085AAB6) and Pseudomonas sp. GM79 (J3D0E2) are presented. Amino acid sequences are given in one-letter code and residue numbering corresponds to CdiAUPEC536 (top) and E. coli CysE (bottom).
CDI toxin and immunity protein sequences are highly variable between bacterial species and even between different strains of the same species [80, 81]. For example, current databases show at least 21 toxin/immunity classes encoded by the cdi loci of Escherichia coli strains. Biochemical analysis has revealed that this sequence diversity corresponds to distinct toxin activities. The CdiA-CTEC93 toxin from E. coli isolate EC93 dissipates the proton-motive force in target cells and is likely to form pores in the membrane [87]. By contrast, CdiA-CTEC869 from enterohemorrhagic E. coli EC869 is a specific tRNase that cleaves near the 3′-end of tRNAGln molecules [88]. The CDI toxin from uropathogenic E. coli strain 536 (UPEC 536) has anticodon nuclease activity in cell-free extracts [80], but surprisingly the purified CdiA-CTUPEC536 domain has no activity when incubated with tRNA substrates in vitro [34]. These latter data suggest that UPEC 536 toxin requires a cellular co-factor for activity – a conclusion supported by experiments showing that purified CdiA-CTUPEC536 is active in reactions supplemented with E. coli cell extract. Based on these observations, Diner et al. took a biochemical approach and identified CysK as the CdiA-CTUPEC536-activating factor in E. coli cell lysates [34]. EcCysK is necessary for toxin activation, because CdiA-CTUPEC536 has no detectable nuclease activity in lysates prepared from E. coli ΔcysK mutants. Moreover, purified EcCysK is sufficient to activate the toxin in completely defined reactions. E. coli ΔcysK mutants are completely resistant to growth inhibition by UPEC 536 cells in competition co-cultures. Together, these results demonstrate an integral role for EcCysK in the CDIUPEC536 growth inhibition pathway (Figure 6A). These phenomena reflect a CysK moonlighting function, because a catalytically inactive EcCysK carrying the Lys42Ala mutation, which prevents PLP-binding, can still activate the CdiA-CTUPEC536 toxin [34].
The precise mechanism of CdiA-CTUPEC536 activation is unknown, but the toxin forms a stable complex with EcCysK and this interaction is critical for nuclease activity. Interestingly, the toxin does not bind to EcCysM and is not activated by this isozyme. This specificity is reminiscent of the relative binding affinities of the isozymes for CysE, suggesting that the CdiA-CTUPEC536 domain interacts with CysK in a similar manner. Indeed, CdiA-CTUPEC536 carries a C-terminal Gly-Tyr-Gly-Ile peptide motif that is similar to the C-terminal Gly-Asp-Gly-Ile sequence found in EcCysE (Figure 6B). This sequence motif – and the C-terminal Ile residue in particular – is required for high-affinity binding to EcCysK. Addition of peptide epitope tags to the C-terminus of CdiA-CTUPEC536 also prevents the interaction with EcCysK and ablates nuclease activity. Collectively, these observations suggest that the C-terminal tail of CdiA-CTUPEC536 mimics EcCysE to bind within the active site of EcCysK. Consistent with this model, formation of the EcCysK-CdiA-CTUPEC536 complex is blocked by OAS, but not by serine [34]. These data indicate that CdiA-CTUPEC536 competes with EcCysE for the same binding site on EcCysK. Equilibrium binding studies show that EcCysK binds to CdiA-CTUPEC536 with higher affinity than EcCysE (B.C. & C.S.H., unpublished data), suggesting that the toxin could displace EcCysE from the CSC. The CysK requirement also has implications for the immunity mechanism. In principle, CdiIUPEC536 could neutralize the toxin by disrupting the CysK-CdiA-CTUPEC536 interaction. However, CdiA-CTUPEC536 and CdiIUPEC536 both bind to EcCysK simultaneously and form a high-affinity ternary complex that lacks nuclease activity [34]. Clearly, high-resolution structural studies are needed to uncover the molecular basis of toxin activation and neutralization.
The CdiA-CTUPEC536 domain is a member of the larger Tox_52 family (Pfam PF15605) of RNase toxins [89]. This domain is commonly found at the C-terminus of CdiA proteins from uropathogenic E. coli strains, where it is encoded on pathogenicity island II [90]. Closely related toxin domains are also found at the C-termini of CdiA proteins from Shigella sp. D9, Trabulsiella guamensis ATCC 49490 and Pseudomonas sp. GM79, and these proteins all share a similar C-terminal motif (Fig. 6B). Given that the CysE-CysK interaction is highly conserved in bacteria, it seems likely that the latter CdiA-CT toxins are also activated by CysK. Indeed, CysK enzymes from other species, such as B. subtilis, bind CdiA-CTUPEC536 with high-affinity and promote nuclease activity in vitro and in vivo (Elie J. Diner & C.S.H., unpublished data). Tox_52/CdiA-CTUPEC536 domains are also found at the C-termini of proteins that are unrelated to CdiA. These include RHS core-domain containing proteins from Ruminococcus lactaris ATCC 29176 and Sandaracinus amylolyticus, as well as several predicted type VII secretion proteins from Paenibacillus species. The RHS core domain is often found in secreted toxic effector proteins [89, 91, 92], and type VII secretion systems (T7SS) are hypothesized to mediate inter-cellular competition between Gram-positive bacteria [93, 94]. Therefore, bacteria appear to deploy Tox_52 domains through different secretion systems, presumably to gain a competitive advantage over neighbors. Interestingly, many Tox_52 domains lack the C-terminal GYGI motif, suggesting that they do not interact with CysK. Preliminary analysis of the R. lactaris toxin indicates that it has CysK-independent tRNase activity in E. coli cells (C.M.B. & C.S.H., unpublished data). Thus, there are at least two sub-families of Tox_52 domains. All members of the CDI-associated sub-family probably require CysK for activity, whereas the toxins found in RHS and T7SS proteins appear to be CysK-independent. If some Tox_52 domains are intrinsically active, then why have the CDI systems co-opted CysK for toxin activation? One possibility is that the mechanism of translocation imposes structural constraints on the delivered toxin domain. CDI toxin translocation is incompletely understood, but CdiA-CT domains are thought to unfold as they cross the target-cell inner membrane. If true, then there should be a selective pressure for small toxin domains with low energy barriers to the unfolded state. Such a domain could require a binding partner to stabilize its tertiary structure once inside the target cell cytoplasm. Notably, the Tox_52 domains found in RHS and T7SS proteins contain an insertion of eight to nine residues and are therefore somewhat larger than the CDI-associated toxins. Perhaps these extra residues contribute to the overall stability of the toxin domain. Alternatively, they could participate in tRNA substrate binding and their function has been assumed by CysK for the CDI toxins. These models are actively being explored through biochemical and structural studies of CysK-dependent and -independent Tox_52 domains.
7. Conclusion
In recent years, the number of newly identified moonlighting proteins has increased dramatically and in many cases these proteins are highly-conserved metabolic enzymes [95, 96]. This is clearly the case for CysK, which is capable of interacting with a number of alternative binding partners using interactions that have been co-opted from the CSC. Although the CysK moonlighting activities are quite diverse, structural and functional themes are shared amongst all CysK interactors identified to date. First, most CysK binding partners carry a C-terminal sequence with two invariant structural features: a C-terminal Ile and an antepenultimate aromatic residue. This feature seems to have evolved to insure high-affinity binding to CysK, and in some cases it probably allows the moonlighting partner to outcompete CysE for binding to the CysK active site. Second, all moonlighting complexes are modulated by at least one of CysK substrates, OAS and bisulfide. OAS competitively displaces interactors from the active site and can thus modulate complex formation depending on the sulfur status of the cell. In some instances, bisulfide is able to stabilize complexes in the presence of OAS, presumably by binding to an allosteric site and favoring a closed conformation of the enzyme.
8. Perspective
In addition to the biological functions played by moonlighting interactors, each partner invariably inhibits the biosynthetic activity of CysK. In principle, moonlighting interactions could also significantly impact CysE activity by dissociating the CSC and thereby influencing enzyme stability, aggregation and feed-back inhibition. Of course, the abundance of CysK with respect to CysE and its other interactors will dictate whether moonlighting activities actually impinge on cysteine metabolic pathways. It is known that CysK expression varies substantially with growth conditions, and thus it is conceivable that the consequences of CysK inhibition by interactors also vary with changing environmental conditions. In the case of the CymR/CysK complex, which functions within the framework of global cysteine metabolism, it seems likely that all activities are balanced to obtain a concerted physiological output. The possible effects of CDI toxin exchange on cysteine metabolism in UPEC 536 cells has not been explored. Because the non-toxic CdiA-CT/CdiIUPEC536 complex binds with high affinity to EcCysK, it has the capacity to regulate cysteine biosynthesis. In this speculative model, the CDI toxin would have a signaling function when delivered into sibling cells that express the immunity protein. The immunity protein would first bind the toxin to form a neutralized complex lacking toxic tRNase activity. Further interaction with EcCysK would provide the opportunity to modulate cysteine metabolism. It is not clear whether enough toxin is exchanged to affect CysK activity, but delivered CdiA-CTUPEC536 antigen is sufficiently abundant to detect by immunofluorescence microscopy [97]. Intriguingly, cysteine biosynthesis is tightly linked to bacterial biofilms [98, 99], and UPEC 536 mutants that lack the CDI system are defective in biofilm formation. Swarming motility is another bacterial social behavior that is associated with cysteine metabolism. S. typhimurium cysteine auxotrophs are impaired in swarming motility in absence of added cysteine [37, 38]. Together, these observations suggest that CysK moonlighting partners have the potential to significantly impact bacterial physiology.
Highlights.
CysK catalyzes the last step of cysteine biosynthesis in bacteria and plants.
CysK interacts with CysE to form the cysteine synthase complex.
Recently, additional non-biosynthetic functions have been discovered for CysK.
CysK moonlighting activities require interactions with binding partners.
Moonlighting partners invariably mimic CysE to form complexes with CysK.
Acknowledgments
C. S. H. was supported by grants GM078634 and GM102318 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kredich NM, Tomkins GM. The enzymic synthesis of L-cysteine in Escherichia coli and Salmonella typhimurium. J Biol Chem. 1966;241:4955–4965. [PubMed] [Google Scholar]
- 2.Kredich NM, Becker MA, Tomkins GM. Purification and characterization of cysteine synthetase, a bifunctional protein complex, from Salmonella typhimurium. J Biol Chem. 1969;244:2428–2439. [PubMed] [Google Scholar]
- 3.Cook PF, Wedding RT. A reaction mechanism from steady state kinetic studies for O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2. J Biol Chem. 1976;251:2023–2029. [PubMed] [Google Scholar]
- 4.Cook PF, Wedding RT. Overall mechanism and rate equation for O-acetylserine sulfhydrylase. J Biol Chem. 1977;252:3459. [PubMed] [Google Scholar]
- 5.Hara S, Payne MA, Schnackerz KD, Cook PF. A rapid purification procedure and computer-assisted sulfide ion selective electrode assay for O-acetylserine sulfhydrylase from Salmonella typhimurium. Protein Expr Purif. 1990;1:70–76. doi: 10.1016/1046-5928(90)90048-4. [DOI] [PubMed] [Google Scholar]
- 6.Tai CH, Nalabolu SR, Jacobson TM, Minter DE, Cook PF. Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 using the natural and alternative reactants. Biochemistry. 1993;32:6433–6442. doi: 10.1021/bi00076a017. [DOI] [PubMed] [Google Scholar]
- 7.Tai CH, Nalabolu SR, Simmons JW, 3rd, Jacobson TM, Cook PF. Acid-base chemical mechanism of O-acetylserine sulfhydrylases-A and -B from pH studies. Biochemistry. 1995;34:12311–12322. doi: 10.1021/bi00038a027. [DOI] [PubMed] [Google Scholar]
- 8.Burkhard P, Rao GS, Hohenester E, Schnackerz KD, Cook PF, Jansonius JN. Three-dimensional structure of O-acetylserine sulfhydrylase from Salmonella typhimurium. J Mol Biol. 1998;283:121–133. doi: 10.1006/jmbi.1998.2037. [DOI] [PubMed] [Google Scholar]
- 9.Burkhard P, Tai CH, Ristroph CM, Cook PF, Jansonius JN. Ligand binding induces a large conformational change in O-acetylserine sulfhydrylase from Salmonella typhimurium. J Mol Biol. 1999;291:941–953. doi: 10.1006/jmbi.1999.3002. [DOI] [PubMed] [Google Scholar]
- 10.Burkhard P, Tai CH, Jansonius JN, Cook PF. Identification of an allosteric anion-binding site on O-acetylserine sulfhydrylase: Structure of the enzyme with chloride bound. J Mol Biol. 2000;303:279–286. doi: 10.1006/jmbi.2000.4109. [DOI] [PubMed] [Google Scholar]
- 11.Mozzarelli A, Bettati S, Campanini B, Salsi E, Raboni S, Singh R, Spyrakis F, Kumar VP, Cook PF. The multifaceted pyridoxal 5′-phosphate-dependent O-acetylserine sulfhydrylase. Biochim Biophys Acta. 2011;1814:1497–1510. doi: 10.1016/j.bbapap.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 12.Hindson VJ, Shaw WV. Random-order ternary complex reaction mechanism of serine acetyltransferase from Escherichia coli. Biochemistry. 2003;42:3113–3119. doi: 10.1021/bi0267893. [DOI] [PubMed] [Google Scholar]
- 13.Becker MA, Kredich NM, Tomkins GM. The purification and characterization of O-acetylserine sulfhydrylase-A from Salmonella typhimurium. J Biol Chem. 1969;244:2418–2427. [PubMed] [Google Scholar]
- 14.Smith IK, Thompson JF. Purification and characterization of L-serine transacetylase and O-acetyl-L-serine sulfhydrylase from kidney bean seedlings (Phaseolus vulgaris) Biochim Biophys Acta. 1971;227:288–295. doi: 10.1016/0005-2744(71)90061-1. [DOI] [PubMed] [Google Scholar]
- 15.Droux M, Martin J, Sajus P, Douce R. Purification and characterization of O-acetylserine (thiol) lyase from spinach chloroplasts. Arch Biochem Biophys. 1992;295:379–390. doi: 10.1016/0003-9861(92)90531-z. [DOI] [PubMed] [Google Scholar]
- 16.Schnell R, Oehlmann W, Singh M, Schneider G. Structural insights into catalysis and inhibition of O-acetylserine sulfhydrylase from Mycobacterium tuberculosis. Crystal structures of the enzyme alpha-aminoacrylate intermediate and an enzyme-inhibitor complex. J Biol Chem. 2007;282:23473–23481. doi: 10.1074/jbc.M703518200. [DOI] [PubMed] [Google Scholar]
- 17.Pye VE, Tingey AP, Robson RL, Moody PC. The structure and mechanism of serine acetyltransferase from Escherichia coli. J Biol Chem. 2004;279:40729–40736. doi: 10.1074/jbc.M403751200. [DOI] [PubMed] [Google Scholar]
- 18.Olsen LR, Huang B, Vetting MW, Roderick SL. Structure of serine acetyltransferase in complexes with CoA and its cysteine feedback inhibitor. Biochemistry. 2004;43:6013–6019. doi: 10.1021/bi0358521. [DOI] [PubMed] [Google Scholar]
- 19.Mino K, Yamanoue T, Sakiyama T, Eisaki N, Matsuyama A, Nakanishi K. Effects of bienzyme complex formation of cysteine synthetase from escherichia coli on some properties and kinetics. Biosci Biotechnol Biochem. 2000;64:1628–1640. doi: 10.1271/bbb.64.1628. [DOI] [PubMed] [Google Scholar]
- 20.Huang B, Vetting MW, Roderick SL. The active site of O-acetylserine sulfhydrylase is the anchor point for bienzyme complex formation with serine acetyltransferase. J Bacteriol. 2005;187:3201–3205. doi: 10.1128/JB.187.9.3201-3205.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campanini B, Speroni F, Salsi E, Cook PF, Roderick SL, Huang B, Bettati S, Mozzarelli A. Interaction of serine acetyltransferase with O-acetylserine sulfhydrylase active site: evidence from fluorescence spectroscopy. Protein Sci. 2005;14:2115–2124. doi: 10.1110/ps.051492805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mino K, Hiraoka K, Imamura K, Sakiyama T, Eisaki N, Matsuyama A, Nakanishi K. Characteristics of serine acetyltransferase from Escherichia coli deleting different lengths of amino acid residues from the C-terminus. Biosci Biotechnol Biochem. 2000;64:1874–1880. doi: 10.1271/bbb.64.1874. [DOI] [PubMed] [Google Scholar]
- 23.Zhao C, Moriga Y, Feng B, Kumada Y, Imanaka H, Imamura K, Nakanishi K. On the interaction site of serine acetyltransferase in the cysteine synthase complex from Escherichia coli. Biochem Biophys Res Commun. 2006;341:911–916. doi: 10.1016/j.bbrc.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 24.Cook PF, Wedding RT. Initial kinetic characterization of the multienzyme complex, cysteine synthetase. Arch Biochem Biophys. 1977;178:293–302. doi: 10.1016/0003-9861(77)90194-1. [DOI] [PubMed] [Google Scholar]
- 25.Droux M, Ruffet ML, Douce R, Job D. Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants - structural and kinetic properties of the free and bound enzymes. Eur J Biochem. 1998;255:235–245. doi: 10.1046/j.1432-1327.1998.2550235.x. [DOI] [PubMed] [Google Scholar]
- 26.Hulanicka MD, Hallquist SG, Kredich NM, Mojica AT. Regulation of O-acetylserine sulfhydrylase B by L-cysteine in Salmonella typhimurium. J Bacteriol. 1979;140:141–146. doi: 10.1128/jb.140.1.141-146.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mino K, Imamura K, Sakiyama T, Eisaki N, Matsuyama A, Nakanishi K. Increase in the stability of serine acetyltransferase from Escherichia coli against cold inactivation and proteolysis by forming a bienzyme complex. Biosci Biotechnol Biochem. 2001;65:865–874. doi: 10.1271/bbb.65.865. [DOI] [PubMed] [Google Scholar]
- 28.Yi H, Dey S, Kumaran S, Lee SG, Krishnan HB, Jez JM. Structure of soybean serine acetyltransferase and formation of the cysteine regulatory complex as a molecular chaperone. J Biol Chem. 2013;288:36463–36472. doi: 10.1074/jbc.M113.527143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kredich NM. Regulation of L-cysteine biosynthesis in Salmonella typhimurium. I. Effects of growth of varying sulfur sources and O-acetyl-L-serine on gene expression. J Biol Chem. 1971;246:3474–3484. [PubMed] [Google Scholar]
- 30.Nakamura T, Iwahashi H, Eguchi Y. Enzymatic proof for the identity of the S-sulfocysteine synthase and cysteine synthase B of Salmonella typhimurium. J Bacteriol. 1984;158:1122–1127. doi: 10.1128/jb.158.3.1122-1127.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Filutowicz M, Wiater A, Hulanicka D. Delayed inducibility of sulphite reductase in cysM mutants of Salmonella typhimurium under anaerobic conditions. J Gen Microbiol. 1982;128:1791–1794. doi: 10.1099/00221287-128-8-1791. [DOI] [PubMed] [Google Scholar]
- 32.Kroger C, Colgan A, Srikumar S, Handler K, Sivasankaran SK, Hammarlof DL, Canals R, Grissom JE, Conway T, Hokamp K, Hinton JC. An infection-relevant transcriptomic compendium for Salmonella enterica Serovar Typhimurium. Cell Host Microbe. 2013;14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 33.Zhao C, Kumada Y, Imanaka H, Imamura K, Nakanishi K. Cloning, overexpression, purification, and characterization of O-acetylserine sulfhydrylase-B from Escherichia coli. Protein Expr Purif. 2006;47:607–613. doi: 10.1016/j.pep.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 34.Diner EJ, Beck CM, Webb JS, Low DA, Hayes CS. Identification of a target cell permissive factor required for contact-dependent growth inhibition (CDI) Genes Dev. 2012;26:515–525. doi: 10.1101/gad.182345.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma DK, Vozdek R, Bhatla N, Horvitz HR. CYSL-1 interacts with the O2-sensing hydroxylase EGL-9 to promote H2S-modulated hypoxia-induced behavioral plasticity in C. elegans. Neuron. 2012;73:925–940. doi: 10.1016/j.neuron.2011.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shepard W, Soutourina O, Courtois E, England P, Haouz A, Martin-Verstraete I. Insights into the Rrf2 repressor family - the structure of CymR, the global cysteine regulator of Bacillus subtilis. FEBS J. 2011;278:2689–2701. doi: 10.1111/j.1742-4658.2011.08195.x. [DOI] [PubMed] [Google Scholar]
- 37.Turnbull AL, Surette MG. L-Cysteine is required for induced antibiotic resistance in actively swarming Salmonella enterica serovar Typhimurium. Microbiology. 2008;154:3410–3419. doi: 10.1099/mic.0.2008/020347-0. [DOI] [PubMed] [Google Scholar]
- 38.Turnbull AL, Surette MG. Cysteine biosynthesis, oxidative stress and antibiotic resistance in Salmonella typhimurium. Res Microbiol. 2010;161:643–650. doi: 10.1016/j.resmic.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 39.Oguri T, Schneider B, Reitzer L. Cysteine catabolism and cysteine desulfhydrase (CdsH/STM0458) in Salmonella enterica serovar typhimurium. J Bacteriol. 2012;194:4366–4376. doi: 10.1128/JB.00729-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Campanini B, Pieroni M, Raboni S, Bettati S, Benoni R, Pecchini C, Costantino G, Mozzarelli A. Inhibitors of the Sulfur Assimilation Pathway in Bacterial Pathogens as Enhancers of Antibiotic Therapy. Curr Med Chem. 2015;22:187–213. doi: 10.2174/0929867321666141112122553. [DOI] [PubMed] [Google Scholar]
- 41.Paritala H, Carroll KS. New targets and inhibitors of mycobacterial sulfur metabolism. Infect Disord Drug Targets. 2013;13:85–115. doi: 10.2174/18715265113139990022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schnell R, Sriram D, Schneider G. Pyridoxal-phosphate dependent mycobacterial cysteine synthases: Structure, mechanism and potential as drug targets. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbapap.2014.11.010. in press. [DOI] [PubMed] [Google Scholar]
- 43.Spyrakis F, Singh R, Cozzini P, Campanini B, Salsi E, Felici P, Raboni S, Benedetti P, Cruciani G, Kellogg GE, Cook PF, Mozzarelli A. Isozyme-specific ligands for O-acetylserine sulfhydrylase, a novel antibiotic target. PLoS One. 2013;8:e77558. doi: 10.1371/journal.pone.0077558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kredich NM, Keenan BS, Foote LJ. The purification and subunit structure of cysteine desulfhydrase from Salmonella typhimurium. J Biol Chem. 1972;247:7157–7162. [PubMed] [Google Scholar]
- 45.Kredich NM. The molecular basis for positive regulation of cys promoters in Salmonella typhimurium and Escherichia coli. Mol Microbiol. 1992;6:2747–2753. doi: 10.1111/j.1365-2958.1992.tb01453.x. [DOI] [PubMed] [Google Scholar]
- 46.Johnson CM, Huang B, Roderick SL, Cook PF. Kinetic mechanism of the serine acetyltransferase from Haemophilus influenzae. Arch Biochem Biophys. 2004;429:115–122. doi: 10.1016/j.abb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 47.Kumaran S, Yi H, Krishnan HB, Jez JM. Assembly of the cysteine synthase complex and the regulatory role of protein-protein interactions. J Biol Chem. 2009;284:10268–10275. doi: 10.1074/jbc.M900154200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hell R, Hillebrand H. Plant concepts for mineral acquisition and allocation. Curr Opin Biotechnol. 2001;12:161–168. doi: 10.1016/s0958-1669(00)00193-2. [DOI] [PubMed] [Google Scholar]
- 49.Mino K, Yamanoue T, Sakiyama T, Eisaki N, Matsuyama A, Nakanishi K. Purification and characterization of serine acetyltransferase from Escherichia coli partially truncated at the C-terminal region. Biosci Biotechnol Biochem. 1999;63:168–179. doi: 10.1271/bbb.63.168. [DOI] [PubMed] [Google Scholar]
- 50.Bogdanova N, Hell R. Cysteine synthesis in plants: protein-protein interactions of serine acetyltransferase from Arabidopsis thaliana. Plant J. 1997;11:251–262. doi: 10.1046/j.1365-313x.1997.11020251.x. [DOI] [PubMed] [Google Scholar]
- 51.Bonner ER, Cahoon RE, Knapke SM, Jez JM. Molecular basis of cysteine biosynthesis in plants: structural and functional analysis of O-acetylserine sulfhydrylase from Arabidopsis thaliana. J Biol Chem. 2005;280:38803–38813. doi: 10.1074/jbc.M505313200. [DOI] [PubMed] [Google Scholar]
- 52.Wirtz M, Birke H, Heeg C, Muller C, Hosp F, Throm C, Konig S, Feldman-Salit A, Rippe K, Petersen G, Wade RC, Rybin V, Scheffzek K, Hell R. Structure and function of the hetero-oligomeric cysteine synthase complex in plants. J Biol Chem. 2010;285:32810–32817. doi: 10.1074/jbc.M110.157446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spyrakis F, Felici P, Bayden AS, Salsi E, Miggiano R, Kellogg GE, Cozzini P, Cook PF, Mozzarelli A, Campanini B. Fine tuning of the active site modulates specificity in the interaction of O-acetylserine sulfhydrylase isozymes with serine acetyltransferase. Biochim Biophys Acta. 2013;1834:169–181. doi: 10.1016/j.bbapap.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 54.Wirtz M, Berkowitz O, Droux M, Hell R. The cysteine synthase complex from plants. Mitochondrial serine acetyltransferase from Arabidopsis thaliana carries a bifunctional domain for catalysis and protein-protein interaction. Eur J Biochem. 2001;268:686–693. doi: 10.1046/j.1432-1327.2001.01920.x. [DOI] [PubMed] [Google Scholar]
- 55.Francois JA, Kumaran S, Jez JM. Structural basis for interaction of O-acetylserine sulfhydrylase and serine acetyltransferase in the Arabidopsis cysteine synthase complex. Plant Cell. 2006;18:3647–3655. doi: 10.1105/tpc.106.047316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu X, Yamaguchi T, Masada M. Complexes of serine acetyltransferase and isozymes of cysteine synthase in spinach leaves. Biosci Biotechnol Biochem. 1998;62:947–952. doi: 10.1271/bbb.62.947. [DOI] [PubMed] [Google Scholar]
- 57.Kumaran S, Jez JM. Thermodynamics of the interaction between O-acetylserine sulfhydrylase and the C-terminus of serine acetyltransferase. Biochemistry. 2007;46:5586–5594. doi: 10.1021/bi7001168. [DOI] [PubMed] [Google Scholar]
- 58.Wirtz M, Hell R. Functional analysis of the cysteine synthase protein complex from plants: structural, biochemical and regulatory properties. J Plant Physiol. 2006;163:273–286. doi: 10.1016/j.jplph.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 59.Salsi E, Campanini B, Bettati S, Raboni S, Roderick SL, Cook PF, Mozzarelli A. A two-step process controls the formation of the bienzyme cysteine synthase complex. J Biol Chem. 2010;285:12813–12822. doi: 10.1074/jbc.M109.075762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salsi E, Bayden AS, Spyrakis F, Amadasi A, Campanini B, Bettati S, Dodatko T, Cozzini P, Kellogg GE, Cook PF, Roderick SL, Mozzarelli A. Design of O-acetylserine sulfhydrylase inhibitors by mimicking nature. J Med Chem. 2010;53:345–356. doi: 10.1021/jm901325e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar S, Raj I, Nagpal I, Subbarao N, Gourinath S. Structural and biochemical studies of serine acetyltransferase reveal why the parasite Entamoeba histolytica cannot form a cysteine synthase complex. J Biol Chem. 2011;286:12533–12541. doi: 10.1074/jbc.M110.197376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Westrop GD, Goodall G, Mottram JC, Coombs GH. Cysteine biosynthesis in Trichomonas vaginalis involves cysteine synthase utilizing O-phosphoserine. J Biol Chem. 2006;281:25062–25075. doi: 10.1074/jbc.M600688200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang T, Leyh TS. Three-stage assembly of the cysteine synthase complex from Escherichia coli. J Biol Chem. 2012;287:4360–4367. doi: 10.1074/jbc.M111.288423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ruffet ML, Droux M, Douce R. Purification and Kinetic Properties of Serine Acetyltransferase Free of O-Acetylserine(thiol)lyase from Spinach Chloroplasts. Plant Physiol. 1994;104:597–604. doi: 10.1104/pp.104.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakamura K, Tamura G. Isolation of Serine Acetyltransferase Complexed with Cysteine Synthase from Allium tuberosum. Agric Biol Chem. 1990;54:649–656. [Google Scholar]
- 66.Berkowitz O, Wirtz M, Wolf A, Kuhlmann J, Hell R. Use of biomolecular interaction analysis to elucidate the regulatory mechanism of the cysteine synthase complex from Arabidopsis thaliana. J Biol Chem. 2002;277:30629–30634. doi: 10.1074/jbc.M111632200. [DOI] [PubMed] [Google Scholar]
- 67.Wirtz M, Droux M, Hell R. O-acetylserine (thiol) lyase: an enigmatic enzyme of plant cysteine biosynthesis revisited in Arabidopsis thaliana. J Exp Bot. 2004;55:1785–1798. doi: 10.1093/jxb/erh201. [DOI] [PubMed] [Google Scholar]
- 68.Becker MA, Tomkins GM. Pleiotrophy in a cysteine-requiring mutant of Samonella typhimurium resulting from altered protein-protein interaction. J Biol Chem. 1969;244:6023–6030. [PubMed] [Google Scholar]
- 69.Belitsky BR. Bacillus subtilis GabR, a protein with DNA-binding and aminotransferase domains, is a PLP-dependent transcriptional regulator. J Mol Biol. 2004;340:655–664. doi: 10.1016/j.jmb.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 70.Belitsky BR, Sonenshein AL. GabR, a member of a novel protein family, regulates the utilization of gamma-aminobutyrate in Bacillus subtilis. Mol Microbiol. 2002;45:569–583. doi: 10.1046/j.1365-2958.2002.03036.x. [DOI] [PubMed] [Google Scholar]
- 71.Edayathumangalam R, Wu R, Garcia R, Wang Y, Wang W, Kreinbring CA, Bach A, Liao J, Stone TA, Terwilliger TC, Hoang QQ, Belitsky BR, Petsko GA, Ringe D, Liu D. Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT. Proc Natl Acad Sci USA. 2013;110:17820–17825. doi: 10.1073/pnas.1315887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bramucci E, Milano T, Pascarella S. Genomic distribution and heterogeneity of MocR-like transcriptional factors containing a domain belonging to the superfamily of the pyridoxal-5′-phosphate dependent enzymes of fold type I. Biochem Biophys Res Commun. 2011;415:88–93. doi: 10.1016/j.bbrc.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 73.Commichau FM, Stulke J. Trigger enzymes: bifunctional proteins active in metabolism and in controlling gene expression. Mol Microbiol. 2008;67:692–702. doi: 10.1111/j.1365-2958.2007.06071.x. [DOI] [PubMed] [Google Scholar]
- 74.Albanesi D, Mansilla MC, Schujman GE, de Mendoza D. Bacillus subtilis cysteine synthetase is a global regulator of the expression of genes involved in sulfur assimilation. J Bacteriol. 2005;187:7631–7638. doi: 10.1128/JB.187.22.7631-7638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tanous C, Soutourina O, Raynal B, Hullo MF, Mervelet P, Gilles AM, Noirot P, Danchin A, England P, Martin-Verstraete I. The CymR regulator in complex with the enzyme CysK controls cysteine metabolism in Bacillus subtilis. J Biol Chem. 2008;283:35551–35560. doi: 10.1074/jbc.M805951200. [DOI] [PubMed] [Google Scholar]
- 76.Hullo MF, Martin-Verstraete I, Soutourina O. Complex phenotypes of a mutant inactivated for CymR, the global regulator of cysteine metabolism in Bacillus subtilis. FEMS Microbiol Lett. 2010;309:201–207. doi: 10.1111/j.1574-6968.2010.02043.x. [DOI] [PubMed] [Google Scholar]
- 77.Lithgow JK, Hayhurst EJ, Cohen G, Aharonowitz Y, Foster SJ. Role of a cysteine synthase in Staphylococcus aureus. J Bacteriol. 2004;186:1579–1590. doi: 10.1128/JB.186.6.1579-1590.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vozdek R, Hnizda A, Krijt J, Sera L, Kozich V. Biochemical properties of nematode O-acetylserine(thiol)lyase paralogs imply their distinct roles in hydrogen sulfide homeostasis. Biochim Biophys Acta. 2013;1834:2691–2701. doi: 10.1016/j.bbapap.2013.09.020. [DOI] [PubMed] [Google Scholar]
- 79.Budde MW, Roth MB. Hydrogen sulfide increases hypoxia-inducible factor-1 activity independently of von Hippel-Lindau tumor suppressor-1 in C. elegans. Mol Biol Cell. 2010;21:212–217. doi: 10.1091/mbc.E09-03-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Aoki SK, Diner EJ, de Roodenbeke CT, Burgess BR, Poole SJ, Braaten BA, Jones AM, Webb JS, Hayes CS, Cotter PA, Low DA. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature. 2010;468:439–442. doi: 10.1038/nature09490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ruhe ZC, Low DA, Hayes CS. Bacterial contact-dependent growth inhibition. Trends Microbiol. 2013;21:230–237. doi: 10.1016/j.tim.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kajava AV, Cheng N, Cleaver R, Kessel M, Simon MN, Willery E, Jacob-Dubuisson F, Locht C, Steven AC. Beta-helix model for the filamentous haemagglutinin adhesin of Bordetella pertussis and related bacterial secretory proteins. Mol Microbiol. 2001;42:279–292. doi: 10.1046/j.1365-2958.2001.02598.x. [DOI] [PubMed] [Google Scholar]
- 83.Aoki SK, Pamma R, Hernday AD, Bickham JE, Braaten BA, Low DA. Contact-dependent inhibition of growth in Escherichia coli. Science. 2005;309:1245–1248. doi: 10.1126/science.1115109. [DOI] [PubMed] [Google Scholar]
- 84.Beck CM, Morse RP, Cunningham DA, Iniguez A, Low DA, Goulding CW, Hayes CS. CdiA from Enterobacter cloacae delivers a toxic ribosomal RNase into target bacteria. Structure. 2014;22:707–718. doi: 10.1016/j.str.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Morse RP, Nikolakakis KC, Willett JL, Gerrick E, Low DA, Hayes CS, Goulding CW. Structural basis of toxicity and immunity in contact-dependent growth inhibition (CDI) systems. Proc Natl Acad Sci USA. 2012;109:21480–21485. doi: 10.1073/pnas.1216238110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nikolakakis K, Amber S, Wilbur JS, Diner EJ, Aoki SK, Poole SJ, Tuanyok A, Keim PS, Peacock S, Hayes CS, Low DA. The toxin/immunity network of Burkholderia pseudomallei contact-dependent growth inhibition (CDI) systems. Mol Microbiol. 2012;84:516–529. doi: 10.1111/j.1365-2958.2012.08039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Aoki SK, Webb JS, Braaten BA, Low DA. Contact-dependent growth inhibition causes reversible metabolic downregulation in Escherichia coli. J Bacteriol. 2009;191:1777–1786. doi: 10.1128/JB.01437-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ruhe ZC, Nguyen JY, Beck CM, Low DA, Hayes CS. The proton-motive force is required for translocation of CDI toxins across the inner membrane of target bacteria. Mol Microbiol. 2014 doi: 10.1111/mmi.12779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang D, de Souza RF, Anantharaman V, Iyer LM, Aravind L. Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct. 2012;7:18. doi: 10.1186/1745-6150-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dobrindt U, Blum-Oehler G, Nagy G, Schneider G, Johann A, Gottschalk G, Hacker J. Genetic structure and distribution of four pathogenicity islands (PAI I(536) to PAI IV(536)) of uropathogenic Escherichia coli strain 536. Infect Immun. 2002;70:6365–6372. doi: 10.1128/IAI.70.11.6365-6372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Poole SJ, Diner EJ, Aoki SK, Braaten BA, t’Kint de Roodenbeke C, Low DA, Hayes CS. Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems. PLoS Genet. 2011;7:e1002217. doi: 10.1371/journal.pgen.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Koskiniemi S, Lamoureux JG, Nikolakakis KC, t’Kint de Roodenbeke C, Kaplan MD, Low DA, Hayes CS. Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci USA. 2013;110:7032–7037. doi: 10.1073/pnas.1300627110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang D, Iyer LM, Aravind L. A novel immunity system for bacterial nucleic acid degrading toxins and its recruitment in various eukaryotic and DNA viral systems. Nucleic Acids Res. 2011;39:4532–4552. doi: 10.1093/nar/gkr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Holberger LE, Garza-Sanchez F, Lamoureux J, Low DA, Hayes CS. A novel family of toxin/antitoxin proteins in Bacillus species. FEBS Lett. 2012;586:132–136. doi: 10.1016/j.febslet.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Henderson B, Martin A. Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect Immun. 2011;79:3476–3491. doi: 10.1128/IAI.00179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mani M, Chen C, Amblee V, Liu H, Mathur T, Zwicke G, Zabad S, Patel B, Thakkar J, Jeffery CJ. MoonProt: a database for proteins that are known to moonlight. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Webb JS, Nikolakakis KC, Willett JL, Aoki SK, Hayes CS, Low DA. Delivery of CdiA nuclease toxins into target cells during contact-dependent growth inhibition. PLoS One. 2013;8:e57609. doi: 10.1371/journal.pone.0057609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee J, Bansal T, Jayaraman A, Bentley WE, Wood TK. Enterohemorrhagic Escherichia coli biofilms are inhibited by 7-hydroxyindole and stimulated by isatin. Appl Environ Microbiol. 2007;73:4100–4109. doi: 10.1128/AEM.00360-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ren D, Zuo R, Gonzalez Barrios AF, Bedzyk LA, Eldridge GR, Pasmore ME, Wood TK. Differential gene expression for investigation of Escherichia coli biofilm inhibition by plant extract ursolic acid. Appl Environ Microbiol. 2005;71:4022–4034. doi: 10.1128/AEM.71.7.4022-4034.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]