Abstract
Non-coding variants in the human MIR137 gene locus increase schizophrenia risk at a genome-wide significance level. However, the functional consequence of these risk alleles is unknown. Here, we examined induced human neurons harboring the minor alleles of four disease-associated single nucleotide polymorphisms (SNPs) in MIR137, and observed increased MIR137 levels compared to major allele-carrying cells. We found that miR-137 gain-of-function causes downregulation of the presynaptic target genes, Complexin-1 (Cplx1), Nsf, and Synaptotagmin-1 (Syt1), leading to impaired vesicle release. In vivo, miR-137 gain-of-function results in changes in synaptic vesicle pool distribution, impaired mossy fiber-LTP induction and deficits in hippocampus-dependent learning and memory. By sequestering endogenous miR-137, we were able to ameliorate the synaptic phenotypes. Moreover, reinstatement of Syt1 expression partially restored synaptic plasticity, demonstrating the importance of Syt1 as a miR-137 target. Our data provide new insight into the mechanism by which miR-137 dysregulation can impair synaptic plasticity in the hippocampus.
Introduction
The underlying cause of neuropsychiatric disorders is suspected to be a combination of genetic predisposition and environmental factors. Genome-wide association studies have reported single nucleotide polymorphisms (SNPs) in the human microRNA-137 (miR-137) gene locus that were associated with an increased risk of schizophrenia and autism spectrum disorders 1, 2. MicroRNAs in general have the potential to modulate hundreds of messenger RNAs simultaneously by regulating mRNA translation 3. They bind to a 6- to 8-base-pair complementary ‘seed region’ typically located in the 3′ UTR of the target mRNA. This interaction either facilitates the degradation of the mRNA or delays its translation 4. Even a modest dysregulation of microRNA levels may cause cellular malfunction and could lead to disease phenotypes 5.
MicroRNA-137 is enriched in the dentate gyrus, a sub-region of the hippocampal formation 6, 7. The axonal processes of the dentate granule neurons, the mossy fibers, transmit information encoding facets of learning and memory to the proximal dendrites of pyramidal neurons in the CA3 region of the hippocampus 8. While previous studies have demonstrated that miR-137 regulates dendritic development both in vitro and in vivo in adult granule neurons 7, 9, the function of miR-137 in the presynaptic compartment remains unexplored. It is crucial to elucidate the functional consequences of psychiatric-associated gene variants as cognitive and synaptic dysfunctions are phenotypes common to schizophrenia and autism spectrum disorders 10, 11.
Here, we show that miR-137 gain-of-function downregulates in vitro and in vivo three pre-synaptic target genes causing impairment in synaptic vesicle trafficking and alterations in synaptic plasticity. Overall, we describe a novel mechanism of synaptic modulation that may be perturbed in disorders such as schizophrenia.
Results
Increased MIR137 in neurons harboring minor allele SNPs
The schizophrenia-associated SNPs (rs1198588, rs1625579, rs2660304, and rs2802535) are positioned in the non-coding region of the MIR137 gene locus (Fig. 1a). SNP rs2660304 is located only 400 base pairs upstream of the primary transcript sequence of MIR137. Variations in this region can result in defective mRNA splicing, stability, and structure 12. We observed transcriptional activity, indicated by increased luciferase expression for the genomic fragments around SNPs rs1198588 and rs2660304 (Supplementary Fig. 1)
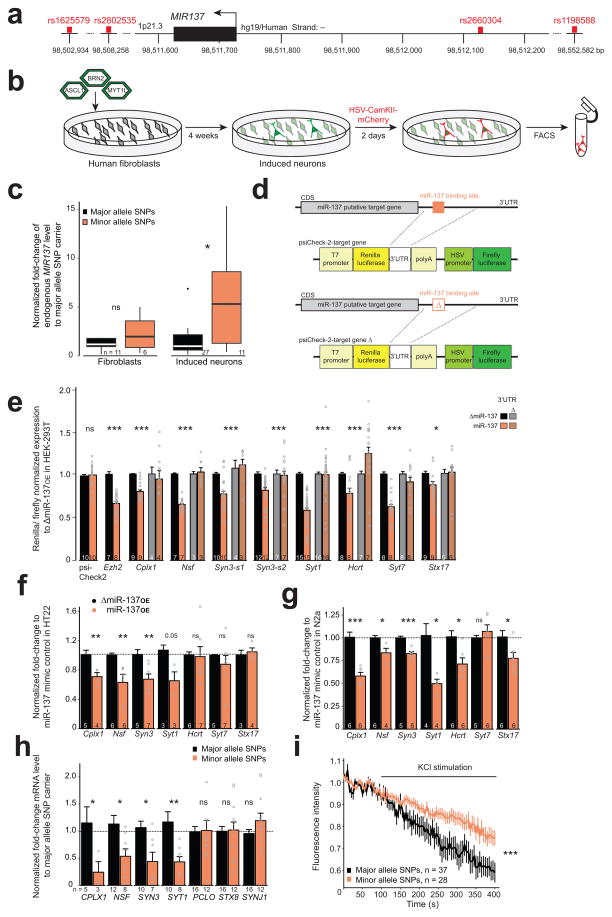
Figure 1. MIR137 gain of function affects presynaptic targets.
(a) Location of the four disease-associated SNPs (red) in the human genome. Black bar: MIR137 coding region (Chr1:98,511,626–727, UCSC genome browser, GRCh37/hg19). (b) Schematic overview for direct induction of human fibroblasts to neurons. Fluorescence-activated cell sorting (FACS). (c) Fold-change differences of MIR137 determined by qRT-PCR. WFibroblasts = 19, nsP = 0.1802, r = −0.33, WInducedNeurons = 65, *P = 0.0157, r = −0.40; box plot shows, in ascending order, the lowest maximum value, the first quartile, median, third quartile and the highest maximum value, with dots representing outliers; n, analyzed samples obtained from three reprogramming from each fibroblast line. (d–g) Confirmation of in silico predicted miR-137 presynaptic target genes. (d) Schematic overview of the luciferase assay. CDS, coding DNA sequence. (e) Luciferase activity normalized to ΔmiR-137OE. Light gray and dark orange, deleted miR-137-binding site. Syn3 has two predicted miR-137-binding sites, s1 and s2. tpsiCheck2(53.51) = 0.40, nsP = 0.6932, r = 0.06; tEzh2(40.7) = −8.94, ***P < 0.001, r = 0.81; FCplx1(3,74) = 6.73, ***P < 0.001; FNsf(3,54) = 97.32, ***P < 0.001; FSyn3-s1(3,75) = 16.67, ***P < 0.001; FSyn3-s2(3,108) = 8.47, ***P < 0.001; FSyt1(3,181) = 67.12, ***P < 0.001, FHcrt(3,85) = 14.12, ***P < 0.001, FSyt7(3,74) = 37.38, ***P < 0.001, FStx17(3,80) = 3.44, *P = 0.0207, post hoc pairwise comparison of ΔmiR-137OE-miR-137OE after Benjamini and Hochberg-adusted P value for multiple comparisons: Cplx1 (P = 0.0006), Nsf (P < 0.001), Syn3-s1 (P < 0.001), Syn3-s2 (P = 0.00016), Syt1 (P < 0.001), Hcrt (P = 0.0029), Syt7 (P < 0.001), Stx17 (P = 0.043); n, number of experiments, each in triplicate. (f,g) mRNA levels of miR-137 target genes in HT22 (f) and N2a (g) cells transfected with a commercially available miR-137 mimic normalized to a mimic-control. HT22: tCplx1(6.98) = −3.58, **P = 0.0090, r = 0.81; WNsf = 0, **P = 0.0022, r = −0.89; tSyn3(9.71) = −3.40, **P = 0.007, r = 0.74; tSyt1(3.21) = −2.99, nsP = 0.0535, r = 0.86; WHcrt = 15, nsP = 0.4452, r = −0.21; tSyt7(6.06) = −1.02, nsP = 0.3481, r = 0.38; tStx17(4.31) = 0.51, nsP = 0.6337, r = 0.24; N2a: tCplx1(9.10) = −6.18, ***P < 0.001, r = 0.90; tNsf(4.43) = −3.01, *P = 0.3453, r = 0.82; tSyn3(8.99) = −6.23, ***P < 0.001, r = 0.90; WSyt1 = 0, *P = 0.0286, r = −0.77, tHcrt(10) = −3.15, *P = 0.0103, r = −0.71; tSyt7(6.27) = 0.91, nsP = 0.3962, r = 0.34 tStx17(9.96) = −2.47, *P = 0.033, r = 0.62; n, number of experiments. (h) mRNA levels in induced neurons by qRT-PCR. tCPLX1 (5.97) = 2.56, *P = 0.043, r = 0.72; tNSF (17.89) = 2.85, *P = 0.0107, r = 0.56; tSYN3 (11.29) = 3.00, *P = 0.0118, r = 0.67; tSYT1 (13.40) = 3.45, **P = 0.004, r = 0.69; tPCLO (22.95) = 1.05, nsP = 0.3034, r = 0.22, tSTX8 (20.36) = −0.49, nsP = 0.7013, r = 0.22, tSYNJ1 (18.94) = −1.21, nsP = 0.2398, r = 0.27; n, number of samples from at least three reprogrammings of each fibroblast line. (i) FM4-64 imaging analysis, H(1) = 38.15, ***P < 0.001; n, number of analyzed cells, usually one cell per coverslip from two reprogramming from each fibroblast line. ns, not significant. Error bars, s.e.m, W, Wilcoxon-rank sum and signed rank tests, r, effect size, H, Kruskal-Wallis rank sum test.
SNPs are categorized in major and minor allele depending on the frequency of their occurrence in a given human population. To access consequences of the SNP genotype in human neurons, we obtained six human fibroblast lines, which are either homozygous for the common major allele, or for the rare minor allele for all four SNPs (Supplementary Fig. 1). We directly converted fibroblasts to neurons by transducing each line with the transcription factors ASCL1, BRN2, or MYT1L (Fig. 1b) 13. After 4 weeks, we transduced the neuron-like cells with herpes simplex virus (HSV) expressing the fluorescent protein mCherry under the CAMK2A (calcium/ calmodulin-dependent protein kinase II alpha) promoter to confirm their neuronal identity and to allow isolation of these cells from un-reprogrammed fibroblasts (Supplementary Fig. 1).
We observed a significant increase of endogenous MIR137 levels in the minor compared to the major allele SNP group (Fig. 1c). This difference was only detected in induced neurons and not in fibroblasts. As an additional control, we measured the expression levels of miR-9, miR-19b and miR-124, which are commonly described as part of the neuronal miRNA signature. We did not observe any difference in the levels between minor and the major allele SNP group (Supplementary Fig. 1). These results suggest that the minor allele SNPs likely cause MIR137 gain-of-function in induced neurons.
MiR-137 targets presynaptic proteins
MiR-137 has been shown to impact dendritic development 7, 9. However, when we performed an in silico screen for predicted miR-137 targets, we found at least 21 genes with well-known roles in presynaptic vesicle trafficking (Supplementary Fig. 2). Since a presynaptic role of microRNAs has not been described so far, we were interested whether miR-137 could influence presynaptic plasticity.
To validate the targets, we cloned the 3′UTR of each gene of interest downstream of a Renilla luciferase gene and upstream of a polyA site (Fig. 1d). In addition, we deleted the miR-137 target site in the 3′UTR to confirm miR-137-specific interaction with the predicted binding site (Δ constructs). We then transfected human embryonic kidney (HEK)-293T cells with the luciferase construct together with constructs overexpressing miR-137 or the control construct miR-CNTL as previously reported. The empty psiCheck2 vector and the 3′UTR of Ezh2 served as negative and positive controls, respectively 9. For seven of the 21 putative target genes, we observed a significant down-regulation in the presence of miR-137OE, but not ΔmiR-137OE or the 3′UTR-Δ constructs (Fig. 1e, Supplementary Fig. 2). Among these genes were well-known presynaptic proteins such as Complexin-1 (Cplx1), N-ethylmaleimide sensitive fusion protein (Nsf), and Synaptotagmin-1 (Syt1). To assess the action of miR-137 on the potential target genes in a neuronal context, we transfected HT22 and N2a neuroblastoma cell lines with miR-137 mimic, a chemically modified, commercially available double-stranded RNA, which mimic endogenous miRNA up-regulation. After 48 hours, we found that the endogenous levels of Cplx1, Nsf, Syn3, and Syt1 mRNAs were all downregulated upon expression of miR-137 mimic in both cell lines (Fig. 1f–g). Hcrt, Syt7 and Stx17 however showed inconsistent down regulation upon miR-137 mimic between the N2A and HT22 neural cell lines and were therefore excluded from further validation.
We then asked whether we could observe a similar down regulation in the mRNA levels for CPLX1, NSF, SYN3 and SYT1 in the induced human neurons showing MIR137 gain-of-function. Consistent with the increased levels of endogenous MIR137, we saw a marked decrease of endogenous mRNA levels for all four target genes in the minor allele SNP group (Fig. 1h). We also measured the mRNA levels of presynaptic genes whose expression should not be affected by MIR137, such as Piccolo (PCLO), or that were negative in our luciferase assay (synaptojanin 1 (SYNJ1), syntaxin 8 (STX8)). As expected, these mRNA levels did not differ between both groups.
Finally, we were interested whether miR-137 gain-of-function has a direct impact on vesicle trafficking in induced neurons. To test this, we monitored functional nerve terminals using the FM4–64 dye in the presence of KCl-mediated neuronal excitation (Supplementary Fig. 2). We observed that the FM4–64 dye depleted slower in induced neuron of the minor allele SNP group compared to the major allele SNPs (Fig. 1i). Altogether, these data provide evidence that miR-137 targets the mRNAs for Cplx1, Nsf, Syn3, and Syt1- presynaptic proteins implicated in vesicle trafficking and synaptic transmission.
Overexpression of miR-137 in vivo in the dentate gyrus
Next, we tested the consequences of miR-137 gain-of-function in vivo. In order to do this, we created a miR-137 overexpression construct in a lentiviral vector backbone (miR-137OE), which allows the long-term overexpression of miR-137 in adult mice. We designed the construct to contain the presumptive pri-miRNA-137, which includes 500 base pairs flanking the miR-137 coding sequence (Fig. 2a). This approach was aimed at preserving in vivo processing mechanisms, in which pri-miRNA-137 is bound by Drosha and Dicer, and then processed to the 20 nt mature miRNA-137. For the control construct, we deleted the mature miR-137 sequence (ΔmiR-137OE). In order to provide a visual confirmation of virus transduction, the miR-137OE and ΔmiR-137OE constructs co-expressed mCherry under the control of the ubiquitin promoter. We confirmed miR-137 overexpression in transduced HEK-293T cells and in primary cortical neurons (Supplementary Fig. 3).
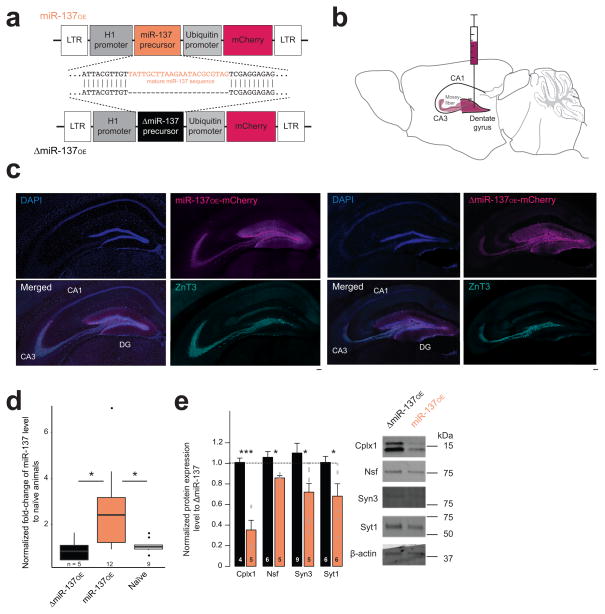
Figure 2. Recapitulation of miR-137 gain of function by overexpressing miR-137 in the mouse dentate gyrus.
(a) Schematic overview of the lentivirus delivery constructs, expressing the miR-137 precursor under the RPPH1 promoter (miR-137OE). ΔmiR-137OE lacks mature miR-137 region. LTR, long terminal repeat. (b) Schematic overview of stereotactic virus injection into the murine dorsal dentate gyrus. (c) Tile-scan of hippocampi injected with either miR-137OE (left) or ΔmiR-137OE (right), labeled against mCherry (magenta), the mossy fiber marker zinc transporter 3 (ZnT3, green) and the nuclear dye DAPI (blue). DG, dentate gyrus. Representative images of at least three animals. Scale bars, 100 μm. (d) miR-137 expression in ΔmiR-137OE-, miR-137OE-injected, and naive animals, H(2) = 7.68, *P = 0.0215, post hoc pairwise comparison after Tukey (tΔmiR-137OE-miR-137OE = 2.53, *P = 0.0471, tmiR-137OE-naive = −2.72, *P = 0.0312, tΔmiR-137OE-naive = 0.261, nsP = 0.796), box plot shows, in ascending order, the lowest maximum value, the first quartile, median, third quartile and the highest maximum value, with dots representing outliers; n, number of dissected dorsal DG regions from at least three animals. (e) Quantified protein levels of miR-137 target genes in mossy fiber-CA3 pathway for ΔmiR-137OE and miR-137OE, tCplx1(5.43) = −6.46, ***P < 0.01, r = 0.94; tNsf(6.47) = −3.43, *P = 0.0123, r = 0.80; tSyn3(9.89) = −3.00, *P = 0.0135, r = 0.69; tSyt1(7.21) = −2.47, *P = 0.0418, r = 0.68; n, dorsal DG-CA3 region of at least three animals. Right, cropped western blot images. Full-length blots are presented in Supplementary Figure 9. ns, not significant. Error bars, s.e.m.
Structural and functional abnormalities in the hippocampus have been implicated in schizophrenia and autism phenotypes 10, 16. MiR-137 has been shown to be strongly expressed in the dentate gyrus 6, 7 and therefore we decided to assess miR-137 gain-of-function in the dorsal dentate gyrus (Fig. 2b). We successfully targeted the dorsal dentate gyrus of eight-week-old male C57BL/6 wild-type mice and found co-expression of the mCherry signal with immunoreactivity the mossy fiber marker ZnT3 17 (Fig. 2c, Supplementary Fig. 3). mCherry expression was not observed in astrocytes or microglia (Supplementary Fig. 3). Using quantitative real-time-PCR (qRT-PCR) , we found that miR-137 levels were increased approximately twofold in miR-137OE mice as compared to the endogenous levels measured in naive and ΔmiR-137OE-injected animals (Fig. 2d).
To confirm that miR-137 gain-of-function impacts presynaptic targets in vivo, we dissected mCherry-positive dentate gyrus-CA3 under an epifluorescent stereomicroscope and performed synaptosomal fractioning to enrich for synaptic proteins. All four targets showed significantly reduced protein expression in the presence of miR-137OE compared to ΔmiR-137OE (Fig. 2e) suggesting that we can recapitulate the in vitro phenotype in vivo.
Vesicle trafficking is altered in miR-137OE mice
miR-137 gain-of-function appears to affect vesicle release in human induced neurons (Fig. 1i). Since we confirmed the miR-137OE–mediated reduction of four key presynaptic proteins in vivo, we explored whether miR-137OE alters in vivo synaptic vesicle distribution. In order to do this, we performed electron microscopy at the mossy fiber synapse. We observed a strikingly vesicle-sparse region in the mossy fiber pre-synaptic terminals in the miR-137OE mice but not in control ΔmiR-137OE animals (Fig. 3a, Supplementary Fig. 4). When we quantified the number of vesicles at increasing distance from the active zone, we found a significant reduction in the vesicle number at a distance of 50–200 nm from the release site in miR-137OE synapses compared to controls (Fig. 3b). This was accompanied by increased vesicle number at 350–450 nm from the active release site in miR-137OE synapses. We did not find a difference in the overall number of vesicles across the two conditions (Supplementary Fig. 4).
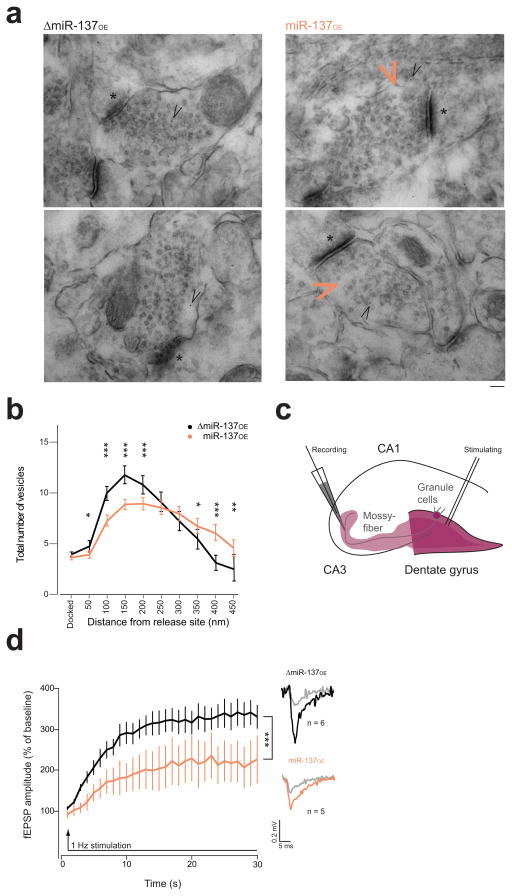
Figure 3. Morphological and functional alterations are evident at the mossy fiber synapse of miR-137OE.
(a,b) Ultrastructural analysis of the mossy fiber presynaptic terminal. (a) Representative images of virus-transduced synapse. Black arrowhead, gold-particles labeling mCherry; orange arrowhead, gap in the vesicle pool in miR-137OE synapse; *, active zone; scale bar, 100 nm. (b) Vesicle distribution for ΔmiR-137OE (black, n = 84) and miR-137OE (orange, n = 112), V = 0.26, F(11,184) = 5.81, ***P < 0.001; with post hoc analysis shown: FDocked(1,194) = 2.91, nsP = 0.0892; F50nm(1,194) = 6.83, *P = 0.0097; F100nm(1,194) = 37.7, ***P < 0.001; F150nm(1,194) = 32.5, ***P < 0.001; F200nm(1,194) = 12.4, ***P < 0.001; F250nm(1,194) = 0.87, nsP = 0.3517; F300nm(1,194) = 1.57, nsP 0.211; F350nm(1,194) = 4.00, *P = 0.0471; F400nm(1,194) = 27.3, ***P < 0.001; F450nm(1,194) = 7.46, **P = 0.0069; n, number of analyzed synapses of at least three animals. (c) Schematic overview of the electrophysiological paradigm for acute hippocampal slice recording; magenta, presumptive mCherry virus expression. (d) fEPSP amplitude for 1-Hz sustained stimulation, H(1) = 67.34, ***P < 0.001. Representative traces embedded; gray traces, response to first stimulation; n, number of analyzed hippocampal slice preparation from at least three animals. ns, not significant. Error bars, s.e.m, V, multivariate analysis of variance.
To determine the functional impact of these structural presynaptic abnormalities at the miR-137OE mossy fiber synapse, we performed frequency facilitation measurements in acute hippocampal slices. We applied sustained low frequency stimulation to dentate granule cells and recorded responses from mossy fiber synapses in CA3 (Fig. 3c). In the control ΔmiR-137OE slices, we observed a marked increase in the amplitude of synaptic responses evoked at the mossy fiber-CA3 synapse, which stabilizes around 300% and persists until the stimulation finishes (Fig. 3d). In contrast, the amplitude of the response reached a lower plateau in miR-137OE slices compared to control. This suggests that miR-137 alters presynaptic function at the mossy fiber synapse in dentate granule cells.
MiR-137 gain-of-function impacts LTP and learning behavior
To further explore the mechanism of miR-137 gain-of-function in vivo in the hippocampus, we investigated mossy fiber LTP in acute hippocampal slices prepared from miR-137OE and ΔmiR-137OE mice. In this paradigm, we placed the stimulating electrode in the granule neuron layer in the dentate gyrus, and recorded from the mossy fibers in the CA3 region (Fig. 3c). We found that high-frequency stimulation lead to less mossy fiber LTP induction in slices from miR-137OE mice, in contrast to those from ΔmiR-137OE animals (Fig. 4a). To confirm that the recorded response was mediated by mossy fibers, we applied the group II metabotropic glutamate receptor agonist DCG-IV after 60 min of recording 18. The baseline synaptic transmission was not altered (Fig. 4b). Thus, miR-137 gain-of-function in the dentate gyrus impairs synaptic plasticity at the mossy fiber synapse.
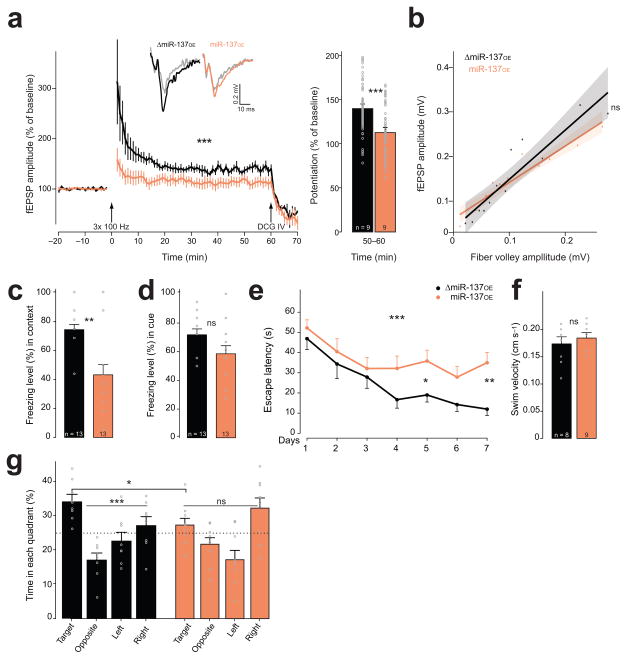
Figure 4. miR-137 overexpression in the dentate gyrus causes impairment in hippocampus-dependent learning.
(a) LTP recording for miR-137OE and control ΔmiR-137OE, F(1,1060) = 144.6, ***P < 0.001. Arrows indicate application of either high frequency stimulation or DCG-IV. Representative fEPSP traces inset; gray traces, baseline levels before stimulation. Right bar chart, LTP magnitude calculated by averaging fEPSP amplitude of the last 10 min (50–60 min) recording; W = 7283, ***P < 0.001, r = −0.42. (b) Input-output curve from ΔmiR-137OE (black, n = 12) and miR-137OE (orange, n = 14), H(1) = 0.02, nsP = 0.8774. Shaded area, 95% confidence interval; n, number of analyzed hippocampal slice preparation from at least three animals. (c,d) Fear conditioning. Percentage freezing for contextual (c; t(17.76) = 3.87, **P = 0.0011, r = 0.68) and cue response (d; t(22) = 1.90, nsP = 0.0709, r = 0.38). (e–g) Morris water maze. (e) Escape latency, F(1,117) = 14.32, ***P < 0.001; post hoc analysis shown: WDay1 = 30, nsP = 0.5535, r = −0.14; WDay2 = 28.5, nsP = 0.495, r = −0.16; WDay3 = 29, nsP = 0.5309, r = −0.15; WDay4 = 17.5, nsP = 0.0831, r = −0.41; WDay5 = 14, *P = 0.0380, r = −0.50; WDay6 = 16.5, nsP = 0.0673, r = −0.43; WDay7 = 7, **P = 0.061, r = −0.65. (f) No difference in swim velocity, t(13.51) = −0.66, nsP = 0.5224, r = 0.18. (g) Probe trial, ΔmiR-137OE mice prefer target quadrant, F(3,28) = 10.16, ***P < 0.001. This effect was not observed for miR-137OE. A significant main effect exists for the target quadrant between ΔmiR-137OE and miR-137OE, F(7, 60) = 7.24, *P = 0.0438; n, number of animals. ns, not significant. Error bars, s.e.m.
Finally, we tested the performance of miR-137OE mice in hippocampus-dependent behavior tasks. First we assessed locomotor activity and anxiety-like behaviors. We did not observe differences between miR-137OE and ΔmiR-137OE animals either in the open-field protocol or in the light-dark box or elevated-plus maze. Then, we performed the fear-conditioning paradigm, which measures memory formation following a foot shock, and memory recall in response to a conditioned stimulus consisting of either the context or a cue (tone). We found a significantly reduced context-associated freezing response in the miR-137OE mice compared to ΔmiR-137OE controls, indicating impaired hippocampus-dependent memory (Fig. 4c). In the cued fear conditioning, we did not observe this difference, which is consistent with the literature since cue-dependent learning is amygdala-dependent (Fig. 4d) 19. Both groups of mice showed equal pain sensitivity and freezing response to the initial shock (Supplementary Fig. 5).
To further assess hippocampus-dependent memory impairment in miR-137OE mice, we performed the Morris water maze test. During the training phase, the escape latency of the miR-137OE mice diminished less over time compared to the control ΔmiR-137OE mice (Fig. 4e). Both groups showed similar swim velocity (Fig. 4f). During the probe trial, miR-137OE animals showed deficits in locating the target quadrant (Fig. 4g). An increased tendency to rely on a thigmotaxis-based strategy could prevent learning; however, we did not find a difference in the percentage of trail time nor in the trail path between the two groups.
As variants in MIR137 have been associated with autism spectrum disorders in humans 1, 20, we also examined the social behavior of miR-137OE and ΔmiR-137OE mice using the three-chamber social interaction paradigm. Both groups showed a similar preference for exploring a new mouse compared to a familiar mouse, and a similar preference to spend more time with a mouse than an empty cage (Supplementary Fig. 5).
Together our data suggest that miR-137 gain of function in the dentate gyrus impairs.
Impact of miR-137 sequestration
To validate the role of miR-137 in regulating pre-synaptic vesicle trafficking, we reduced endogenous miR-137 levels using a “sponge” construct 21. The construct contained multiple miR-137 target sites to which endogenous miR-137 binds (SpongemiR-137). As control, we used an empty vector backbone expressing GFP only (SpongeControl).
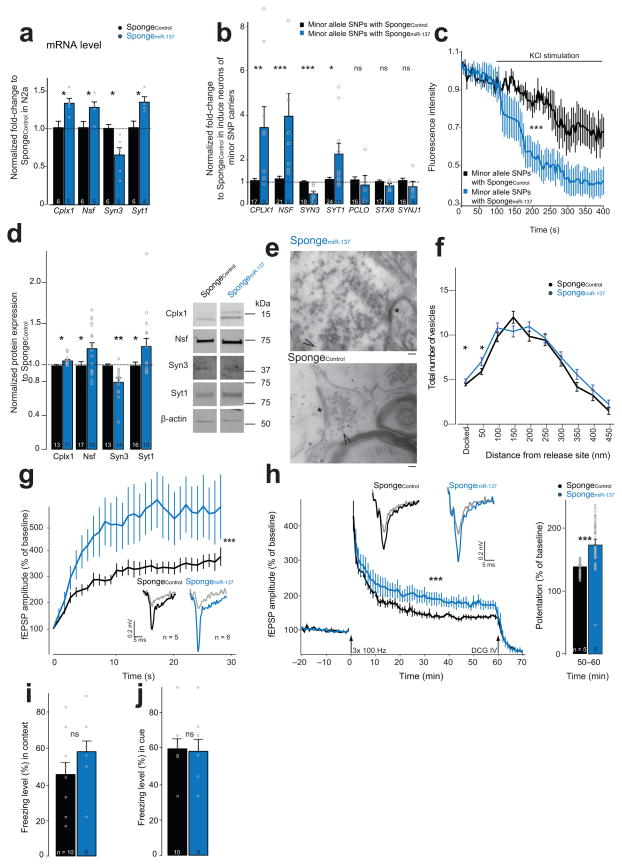
First, we transfected N2a cells with SpongemiR-137 and measured the mRNA expression of Cplx1, Nsf, Syn3, and Syt1 using qRT-PCR. We found that Cplx1, Nsf, and Syt1 mRNA levels are increased in the presence of SpongemiR-137 (Fig. 5a). Surprisingly, Syn3 mRNA levels were decreased. Subsequently, we attempted to reverse the endogenous miR-137 gain-of-function phenotype in the induced human neurons from the minor allele SNPs group by transducing those cells with the SpongemiR-137. Similar to the N2A cells, we observed increased CPLX1, NSF, and SYT1 mRNA expression in the presence of SpongemiR-137, while SYN3 is reduced (Fig. 5b). Next, we transduced induced neurons carrying the minor allele SNPs with the SpongemiR-137 to test the impact on vesicle trafficking. We observed a more rapid depletion of FM4–64 dye following KCl stimulation in the presence of the SpongemiR-137, suggesting a faster vesicle release (Fig. 5c).
Figure 5. Sequestration of endogenous miR-137 leads to phenotype reversal. SpongeControl (black), SpongemiR-137 (blue).
(a) Relative mRNA levels of miR-137 target genes in transfected N2a cells, tCplx1(8.80) = 3.02, *P = 0.0149, r = 0.71; tNsf(2.46) = 9.00, *P = 0.036, r = 0.64; tSyn3(7.70) = −3.24, *P = 0.0125, r = 0.76; tSyt1(7.96) = 3.00 *P = 0.0173, r = 0.73; n, number of experiments. (b) Relative mRNA levels in transduced induced minor allele SNP neurons, WCPLX1 = 27, **P = 0.0064, r = −0.53; WNSF = 13, ***P < 0.001, r = −0.70; WSYN3 = 117, ***P < 0.001, r = −0.71; WSYT1 = 66.5 *P = 0.0209, r = −0.39; tPCLO(4.82) = 0.52 nsP = 0.6282, r = 0.23; tSTX8(5.63) = 1.54 nsP = 0.1782, r = 0.54; tSYNJ1(5.56) = 1.06 nsP = 0.3338, r = 0.41; SpongeControl, n = 11; SpongemiR-137, n = 18; number of transduced minor allele SNP fibroblast lines with either construct from three reprogramming. (c) FM4-64 imaging analysis, H(1) = 152.06, ***P < 0.001; n, number of analyzed transduced induced neurons of the two minor allele fibroblast lines from three reprogramming. (d) miR-137 target genes protein level in mossy fiber-CA3 pathway, tCplx1(21.97) = −2.69, *P = 0.0140, r = 0.5; t Nsf(29.0) = −2.36, *P = 0.0251, r = 0.40; tSyn3(16.11) = 3.43, **P = 0.0034, r = 0.65; tSyt1(21.41) = −2.17, *P = 0.0415, r = 0.42; n, number of dissected dorsal DG-CA3 regions from at least three animals. Right, cropped western blot image. Full-length blots are presented in Supplementary Figure 9. (e,f) Ultrastructural analysis of mossy fiber presynaptic terminal. (e) Representative images of virus-transduced synapse labeled with gold particles (arrowhead); *, active zone; scale bars, 100 nm. (f) Vesicle distribution from the synaptic release site for SpongeControl (n = 50) and SpongemiR-137 (n = 43), V = 0.25, F(11,91) = 2.40, *P = 0.0125, post hoc analysis: FDocked(1,91) = 4.01, *P = 0.0483; F50nm(1,91) = 4.13, *P = 0.045; F100nm(1,91) = 1.68, nsP = 0.1977; F150nm(1,91) = 3.04, nsP = 0.0845; F200nm(1,91) = 2.18, nsP = 0.1429; F250nm(1,91) = 0.09, nsP = 0.7687; F300nm(1,91) = 0.29, nsP = 0.5886; F350nm(1,91) = 3.22, nsP = 0.0759; F400nm(1,91) = 0.55, nsP = 0.4619; F450nm(1,91) = 1.53, nsP = 0.2188; n, number of analyzed synapses from at least three animals. (g) fEPSP amplitude for 1 Hz sustained stimulation, F(1,305) = 77.73, ***P < 0.001. Representative traces embedded; gray trace, response to first stimulation; n, number of analyzed hippocampal slices from at least three animals. (h) LTP recording in DG-CA3 mossy fiber pathway, H(1) = 125.92, ***P < 0.001. Arrows, application of either high frequency stimulation or DCG-IV. Inset, representative fEPSP traces; gray traces, baseline before stimulation. Right bar chart, LTP magnitude for the last 10 min (50–60 min) recording, W = 341, ***P < 0.001, r = −0.67; n, number of analyzed hippocampal slices from at least three animals. (i,j) Fear conditioning. Freezing percentage for context (i; t(17.0) = −1.43, nsP = 0.1707, r = 0.33) and cue (j; t(16.21) = 0.16, nsP = 0.873, r = 0.04). n, number of animals. ns, not significant. Error bars, s.e.m.
We proceeded to test whether SpongemiR-137 can reverse previously observed miR-137OE -dependent phenotypes in vivo. We found that the protein levels of Cplx1, Nsf and Syt1 were increased upon SpongemiR-137 injection (Fig. 5d). Ultrastructural analysis showed that the vesicle number is slightly increased at the vesicle docking and at 50 nm from the synaptic release site in the SpongemiR-137 mossy fiber-synapse compared to control (Fig. 5e–f, Supplementary Fig. 6). The absolute vesicle number did not change in the presence of the SpongemiR-137 (Supplementary Fig. 6).
We then evaluated the functional impact of miR-137 sequestration on synaptic function in the mouse hippocampus by stimulating dentate gyrus granule cells and recording in the mossy fiber-CA3 region, as described in Fig. 3c. In contrast to the impaired frequency facilitation found at miR-137OE mossy fiber synapses (Fig. 3d), we observed a strong increase in the synaptic response amplitude evoked at the mossy fiber-CA3 synapse in the presence of the SpongemiR-137 construct, compared to controls (Fig. 5g). This amplitude stabilized around 500% compared to 300% for the SpongeControl. When we measured mossy fiber LTP induction, we found an enhanced induction in SpongemiR-137 compared to controls (Fig. 5h). This was not due to changes in baseline synaptic transmission (Supplementary Fig. 6).
Next, we determined the behavioral outcome of endogenous miR-137 sequestration. In the open-field arena, the SpongemiR-137 mice did not show a difference in the total distance or the time moving (Supplementary Fig. 6), but had a trend towards more time spent in the center. However, when we focused on hippocampus-dependent learning, we observed a trend toward improved associative learning in the contextual fear conditioning protocol in the SpongemiR-137 group as compared to the SpongeControl (Fig. 5i). The freezing response upon the tone in the cued condition, after context and before tone, as well as nociception did not differ between both groups (Fig. 5j, Supplementary Fig. 6). Altogether, our results suggest that the sequestration of miR-137 can significantly reverse multiple impairments caused by miR-137 gain-of-function at the mossy fiber-CA3 synapse.
Syt1 restoration ameliorates miR-137 gain-of-function
To further explore whether miR-137-associated synaptic phenotypes could be ameliorated by the expression of a single target gene, we decided to express miR-137-resistant Syt1 cDNA in the presence of miR-137OE. Syt1 is a well-known presynaptic protein located on the synaptic vesicle membrane. It has been shown to trigger calcium-dependent synaptic vesicle release and playing a role in vesicle recycling 22–26. Syt1 mRNA and protein are also prominently expressed in the dentate gyrus and hippocampal CA3 area (Supplementary Fig. 7).
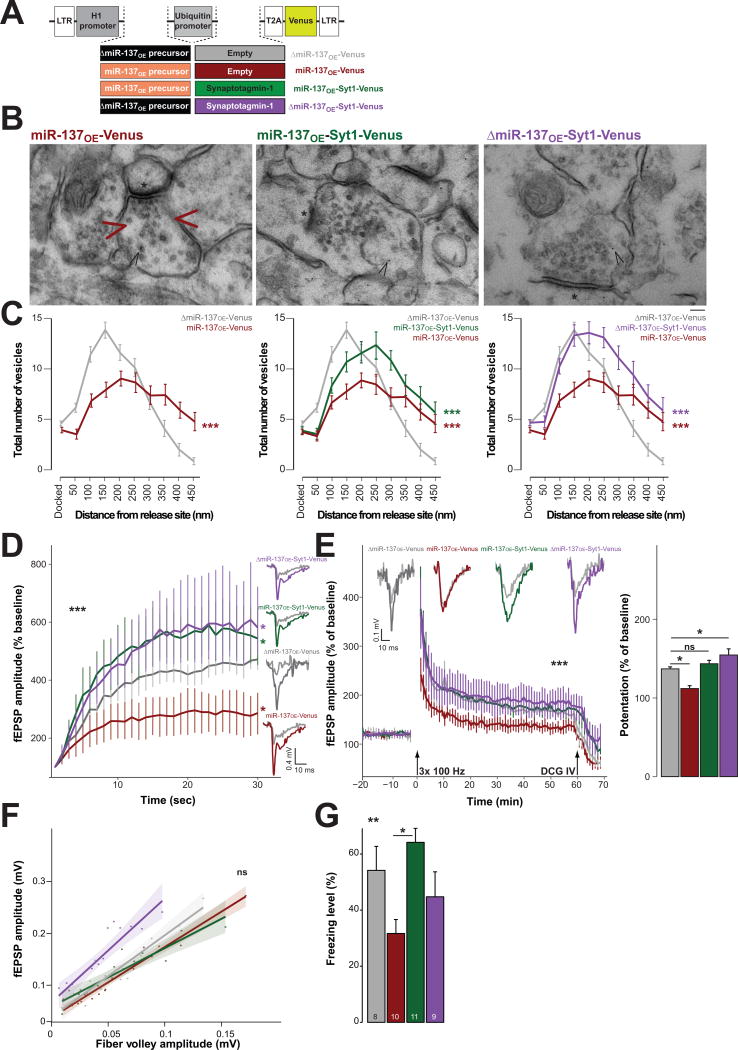
We generated a bi-cistronic lentiviral vector, which expresses miR-137OE followed by an ubiquitin promoter-driven mouse Syt1 cDNA and eYFP Venus protein (Fig. 6a). We also included a bicistronic construct overexpressing Syt1 cDNA and ΔmiR-137OE. Upon ultrastructural analysis of miR-137OE-Syt1-Venus, we observed an increase in vesicle numbers from 100–250 nm compared to miR-137OE-Venus; this, however, did not reach control levels as indicated by ΔmiR-137OE-Venus (Fig. 6b,c and Supplementary Fig. 7). In the periphery of the vesicle pool (> 350 nm), the vesicle number was not restored in the miR-137OE-Syt1-Venus-containing synapses compared to miR-137OE-Venus alone (Fig. 6c). Overall, expression of Syt1 led to an increase in total number of vesicles as compared to control (Supplementary Fig. 7e), which was also reflected in more vesicles in the periphery.
Figure 6. Impact of alteration of synaptotagmin-1 (Syt1) expression under miR-137 gain of function.
ΔmiR-137OE-Venus (gray), miR-137OE-Venus (red), miR-137-Syt1-Venus (green) and ΔmiR-137OE-Syt1-Venus (purple), (a) Schematic overview of bicistronic expression vectors. LTR, long terminal repeat. (b,c) Ultrastructural analysis of the mossy fiber terminal. (b) Representative image of Venus-immunopositive synapse (black arrowhead). Red arrowheads. gap in the vesicle pool in miR-137OE-Venus synapse; *, active zone; scale bar, 100 nm. (c) Vesicle distribution from the synaptic release site for ΔmiR-137OE-Venus (n = 46), miR-137OE-Venus (n = 47), miR-137-Syt1-Venus (n = 70) and ΔmiR-137OE-Syt1-Venus (n = 58); n, number of analyzed synapses of at least three animals, Vgray-red = 0.72, F(11,97) = 22.9, ***P < 0.001, Vgray-green = 0.66, F(11,100) = 17.7, ***P < 0.001, Vgray-purple = 0.57, F(11,102) = 12.3, ***P < 0.001. (d) fEPSP amplitude during 1 Hz sustained stimulation, H(3) = 217.81 ***P < 0.001. Multiple comparison test after Kruskal-Wallis showed *P < 0.05 for all combinations. Inset, representative traces; gray traces, baseline before stimulation; ngray = 5, nred = 7, ngreen = 4, npurple = 4; number of hippocampal slices from at least three animals. (e) Right, mossy fiber LTP recording; arrows, application of high frequency stimulation or DCG-IV. Insets, representative traces; gray, baseline before stimulation. Left, magnitude of potentiation during the last 10 min (50–60 min) recording, H(3) = 106.41, ***P < 0.001, multiple comparison two-tailed test after Kruskal-Wallis, *P < 0.05; n, number of analyzed hippocampal slices from at least 3 animals. (f) Input-output curve of fEPSP against fiber volley amplitude at the mossy fiber-CA3 synapse, H(3) = 3.85, nsP = 0.2776. Shaded area, 95% confidence interval; ngray = 15, nred = 15, npurple = 15, ngreen = 15); n, number of analyzed hippocampal slices from at least three animals. (g) Freezing level % for contextual fear conditioning, F(3,34) = 4.53, **P = 0.0089; post hoc Tukey analysis: tmiR-137-miR137-Syt1 = 3.57, **P = 0.0060; tΔmiR-137-miR137 = −2.28, nsP = 0.1236, tΔmiR-137-miR137-Syt1 = 1.03, nsP = 0.7317, tΔmiR-137-ΔmiR137-Syt1 = −0.93, nsP = 0.789, tmiR-137-ΔmiR137-Syt1 = 1.37, nsP = 0.5278, tmiR-137-Syt1-ΔmiR137-Syt1 = −2.07, nsP = 0.1825; n, number of animals. ns, not significant. Error bars, s.e.m.
We also measured frequency facilitation in the mossy fiber–CA3 pathway of miR-137OE-Syt1-Venus-transduced slices. The amplitude of the synaptic response exceeded control levels of ΔmiR-137OE-Venus and stabilized around 500%, similar to that of ΔmiR-137OE-Syt1-Venus. Facilitation in the presence of miR-137OE-Venus alone was impaired (Fig. 6d). Surprisingly, LTP induction was restored under the same conditions (Fig. 6e) and baseline synaptic transmission was not altered (Fig. 6f). When we measured the freezing response using contextual fear conditioning, we found a significant improvement in the memory performance in animals in which Syt1 was restored (miR-137OE-Syt1-Venus), as compared to those with miR-137OE-Venus or Syt1 alone (Fig. 6g). Nociception in response to shock was not altered (Supplementary Fig. 7). In conclusion, Syt1 by itself did not restore a normal vesicle distribution in the presence of miR-137OE, although the perturbation in the synaptic vesicle pool, as well as the LTP deficits, are ameliorated by its expression.
It has previously been shown that high frequency stimulation–induced LTP at the mossy fiber–CA3 synapses is independent of presynaptic mechanisms27. Since we observed LTP improvement upon Syt1 overexpression, we were interested in whether Syt1 knockdown by its own could affect LTP induction. Consistent with the miR137OE condition, we observed impairment in LTP induction at the mossy fiber synapse in the presence of Syt1 knockdown (Supplementary Fig. 8). When we measured baseline synaptic transmission, we found a decrease in synaptic strength. Frequency facilitation measurements showed a decrease in the field excitatory postsynaptic potential (fEPSP) slope in the presence of Syt1 knockdown (Supplementary Fig. 8). These data suggest that Syt1 loss-of-function results in synaptic dysfunction at the mossy fiber synapses, which likely mimics the miR-137OE phenotype..
Discussion
In this study, we investigated the functional impact of psychiatric risk SNPs in MIR137. We observed an upregulation of endogenous MIR137 in induced human neurons harboring the minor allele of four schizophrenia-associated SNPs in the putative MIR137 region. This raised the question what are the mechanistic consequences of MIR137 gain-of-function. We were intrigued to find amongst the predicted miR-137 targets multiple candidate genes with known presynaptic function. Since the role of microRNAs at the presynapse has been unexplored, we were interested in determining whether miR-137 could be a novel regulator of presynaptic function.
Here, we show that Cplx1, Nsf, and Syt1 are directly regulated by miR-137 expression: upregulation of miR-137 level caused a decrease in their protein levels (Fig. 2e) and vice versa (Fig. 5d). Remarkably, those target genes are consistently found to be downregulated in the postmortem brains of schizophrenia patients 29–33. Syn3, on the other hand, remained low even when endogenous miR-137 was sequestered, which suggests that additional endogenous factors modulate Syn3 levels. Further exploration of the role of miR-137 at the presynapse showed that miR-137 gain-of-function reduced vesicle release in induced neurons. We supported this finding by performing in vivo experiments and showing alterations in the synaptic vesicle pool, reduced frequency facilitation and LTP induction, as well as impairment in hippocampus-dependent learning.
We also provide evidence that Syt1 is an important component in inducing mossy fiber LTP27. Mossy fiber LTP has been shown to be NMDA receptor independent and is likely to be induced via presynaptic mechanism. Rab3a and RIM1α are two presynaptic proteins that have been shown to be involved in LTP induction at these synapses33,34. By Syt1 knockdown alone and in combination with miR-137OE, we have identified a new mechanism mediating mossy fiber LTP.
Previous studies have shown that loss of Cplx1, Syt1, or both, impairs Ca2+-triggered evoked neurotransmitter release independent of species and synapse type 26, 28, 36–38. Syt1 and Nsf were reported to be involved in vesicle recycling 39–41. Synaptic vesicles are depleted in synaptotagmin mutants of Caenorhabditis elegans, and hypomorphic mutation of Nsf in Drosophila melanogaster has been shown to cause paralysis within a couple of minutes after the functional vesicle pool is exhausted. Syn3 can influence recycling and recruitment of vesicles 42, 43, and Syn3 knockdown increases the number of synaptic vesicles in the reserve pool 44. Nonetheless, we should point out that those studies only knocked down one gene of interest, and not a set of genes, as we have done with a microRNA overexpression approach. Thus, microRNAs could open a new way to study closely intermingled presynaptic pathways. However, we have to stress that we only analyzed a subset of predicted miR-137 target genes at the pre-synapse. We cannot exclude that other miR-137 targets might play an additional mechanistic role at the pre-synapse. Furthermore, the relative abundance of mRNAs within a cell can impact microRNA efficiency.
Despite the effect of miR-137 on multiple mRNAs, we attempted to rescue synaptic function by co-expressing a miR-137-insensitive Syt1 transcript. As predicted, restoration of Syt1 rescued some, but not all, of the miR-137OE phenotypes. Moreover these rescued phenotypes are distinct from Syt1 overexpression alone. For example, the vesicle distribution could not be fully restored to control levels. This suggests that reductions in Cplx1 levels, even with Syt1 restoration, may hinder vesicle recruitment to the active release site, allowing the phenotype of impaired vesicle release to persist in the presence of miR-137 gain of function. Whether a simultaneous restoration of Cplx1 and Syt1 under miR-137-mediated downregulation would rescue the phenotype remains to be determined. Additionally, other, not yet identified, miR-137 targets may be involved. For example, a recent study by Bacaj et al. 26 showed that, while Syt1 is important for fast, synchronous vesicle release, the related protein Syt7 is necessary for slow, asynchronous release. Therefore, additional factors such as Syt7, itself a potential miR-137 target (Supplementary Fig. 2), could play a role in this phenotype.
In previous studies, miR-137 has been associated with adult neurogenesis and alteration in dendritic branching 7, 9. Consistent with these findings, we observed increased adult neurogenesis in the presence of miR-137OE (Supplementary Fig. 8). It has been shown that miR-137 overexpression in adult neuronal stem cells leads to delay in maturation of these neurons 7, 9. In future studies, it will be interesting to determine how alterations in neural stem cell proliferation and maturation impact learning and memory following miR-137 overexpression. We also note that we observed normal LTP at the medial perforant path–dentate gyrus synapses suggesting that the impaired mossy fiber LTP is not caused by alteration of the input to the dentate gyrus (Supplementary Fig. 8).
Schizophrenia has long been proposed to be “a disease of the synapse” 45–47, with the underlying assumption that synaptic malfunction can severely impact brain connectivity 16. Several studies describe changes in the expression of genes functionally implicated in both pre- or post-synaptic mechanisms in schizophrenia patients 33, 48, 49. Wen et al. (2014) 49 have proposed a model in which large-scale transcriptional dysregulation in human neurons harboring alterations in disrupted in schizophrenia 1 (DISC1) could affect synaptic function and lead to major psychiatric disorders. Here, we provide a model in which a single regulatory microRNA can simultaneously control several protein targets thus regulating an entire pathway resulting in synaptic alterations. This mechanism might lead to new treatment strategies for diseases such as schizophrenia.
Methods
Animals
Animal experiments were approved from the MIT Committee on Animal Care (CAC). Mice were housed in groups of 3–5 animals in 12 h light/ 12 h dark cycle, with standard mouse chow and water ad libitum. If not otherwise indicated, at least 2 month old, male C57BL/6 mice from Jackson Laboratory were used for all experiments. Mice, which received bilateral stereotactic injection into the dorsal dentate gyrus by a lentivirus, are labeled as follows: “miR-137OE” and “ΔmiR-137OE “ for miR-137 overexpression and control, respectively; “SpongemiR-137” and “SpongeControl” for the sponge-mediated sequestration of endogenous miR-137 and control, respectively; “ΔmiR-137OE-Venus”, “miR-137OE-Venus”, “miR-137OE-Syt1-Venus” and “ΔmiR-137OE-Syt1-Venus” for miR-137 control, miR-137 overexpression, overexpression of miR-137 and synaptotagmin-1, and miR-137 control and synaptotagmin-1, respectively; “Syt1-KD” and “Syt1-KDControl” for knockdown of Syt1 only and control, respectively37. For the endogenous experiments in Figure 2d and Supplementary Figure 7a–c, naïve, age-matched C57BL/6 animals were used.
Cell lines
HEK293T/17 (CRL-11268), Neuro-2a (N2a, CCL-131) and SH-SY5Y (CRL-2266) cells were purchased from ATCC (American Type Culture Collection) and cultured as recommended. HT22 cells were a gift from S. Haggarty (MGH). Fibroblast lines were genotyped multiple times. Cell lines were frequently confirmed to be mycoplasma free.
Human fibroblasts
Human fibroblasts were obtained from the Coriell Institute, McLean Hospital, the Massachusetts General Hospital, and the American Type Culture Collection (ATCC). We selected the human fibroblasts based on four disease-associated SNPs in the MIR137 locus. There is a chance that other SNPs, further away from the gene locus, could influence miR-137 expression; however, we chose to genotype cells for SNPs that are both close to the MIR137 locus and are significant GWAS hits. Also, two of these SNPs impact transcriptional activity (Supplementary Fig. 1). Four lines were homozygous for the major allele and two lines for the minor allele at all four SNPs. Per definition, the major allele is the most common allele in a population, whereas the minor allele is less prevalent. Human fibroblasts were cultured in D15 media (DMEM with sodium pyruvate containing GlutaMAX (Invitrogen), 15% (v/v) fetal bovine serum (FBS) and penicillin/streptomycin at 37°C and 5% CO2.
Genotyping protocol for disease-associated SNPs in human fibroblasts
Genomic DNA from human fibroblasts was prepared using the DNeasy Blood & Tissue Kit (Qiagen). An approximately 1000 bp fragment was amplified using the Phusion-MasterMix according to the manufacturer’s directions (New England Biolabs; Supplementary Table 1). The PCR product was purified with NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel), then subjected to Sanger sequencing.
Direct reprogramming of human fibroblasts into induced neurons
Constructs
Human ASCL1, POU3F2 (aka BRN2), and MYT1L cDNAs were separately cloned into the lentiviral vector pHRST-IRES-GFP, which contains an IRES-EGFP cassette under the control of a CMV promoter. The lentivirus envelope plasmid pMD.2G-VSV G and the packaging plasmid pCMVdeltaR8.2 were purchased from Addgene (plasmid 12259 and 12263, respectively) 50. Lentivirus was generated for all three human plasmids as described below. The reprogramming efficiency of the different fibroblast lines should be independent of the age and gender of the patient 51.
Induction
One day before virus transduction, human fibroblasts were plated at a density of 7.5 × 105 cells per dish in 60-mm dishes in D15 medium with no antibiotics. The next day, 25 μL of each lentivirus containing human ASCL1, BRN2 and MYT1L were combined in D15 medium supplemented with 2.5 μL of 8 mg/mL Polybrene (Sigma-Aldrich). The medium was changed 8–16 h after transduction. The following day the medium was changed to N3 Induced Neuron Medium (DMEM/F12) containing GlutaMax (Invitrogen), 25 μg/mL insulin, 30 nM sodium selenite, 20 nM progesterone, 100 nM putrescine (all from Sigma-Aldrich), 50 μg/mL transferrin (Roche) and 10 ng/mL bFGF (R&D Systems). The N3 medium was changed every 3–5 d for 4 weeks. To isolate induced neurons for qRT-PCR, we used herpes simplex virus (HSV; MIT Viral Core) expressing mCherry under the control of the CAMK2A promoter (HSV-CK-mCherry). The CAMK2A promoter drives expression in glutamatergic excitatory neurons in the hippocampus and cortex52, 53. Two days after transduction with the HSV-CK-mCherry virus, cells were treated with Accutase (5 min at 25 °C) and resuspended in ice-cold FACS buffer (HBSS containing B27, 20 mM glucose, penicillin/streptomycin, 1 mM EDTA). Cultures were then centrifuged for 10 min at 300g at 4 °C and the supernatant was discarded. Cells were resuspended in cold FACS buffer and subjected to fluorescence-activated cell sorting (FACS). Cells positive for mCherry fluorescence were sorted into binding buffer provided by the PureLink miRNA isolation kit (Invitrogen) and processed as described below. The neuronal population ranged between 0.01 and 0.001% of all sorted FACS events.
Constructs
Cloning of MiR-137OE and ΔmiR-137OE
For viral expression of human miR-137, miR-137 was amplified from human genomic DNA using the miR-137_5-BamHI and miR-137_3-BstXI primers (Supplementary Table 1). The amplified DNA fragment was cloned into the lentivirus vector FH1-UGW-mCherry using the restriction enzyme sites BamHI and BstXI. The H1 promoter drives miR-137 expression.
miR-137 Construct Lentivirus Generation
Confluent (90%) HEK-293T cells were transfected with 8 μg lentivirus plasmid (see above), 4.8 μg VSV-G, and 4.8 μg of each of the packaging plasmids pRSV-Rev and pMDLg/pRRE using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s protocol. Supernatant was collected 48 h later, centrifuged for 5 min at 300g, sterile-filtered through a 0.45 μm filter, then centrifuged at 19,500 r.p.m. for 2 h at 4 °C (Optima I-90K ultracentrifuge, SW41 Ti rotor), discarded, the pellet resuspended in 20 μL cold Dulbecco’s phosphate-buffered saline (DPBS, Life Technologies) overnight at 4°C, then aliquoted and stored at −80°C. The viral titer was estimated with the Lentivirus qPCR Titer kit (ABM Inc), and typically ranged between 9 x 106–3 x 107 viral units/ mL.
FUchW-Syt1-T2A-Venus
Primers were designed to synaptotagmin-1 cDNA (NM_001252342.1) and extended by a BamHI site at the 5′ end and an AsiSI site at the 3′ end (Supplementary Table 1). Syt1 was amplified using the Phusion MasterMix PCR protocol (NEB). The vector FUchW H2KK-T2A-Venus, which contains the T2A cleavage peptide sequence, and the Syt1 PCR amplicon were digested with BamHI and AsiSI, gel-purified, ligated with Quick ligation protocol (NEB) and transformed in NEB 10-beta competent cells. Clones were picked, QuickLyse miniprep (Qiagen) performed and the plasmid send for sequencing. The positive clone was retransformed in Stable-3 cells and plasmid purified with PureLink HiPure Plasmid DNA Purification kit (Invitrogen).
miR-137OE-Syt1-Venus and ΔmiR-137OE-Syt1-Venus
The vectors miR137-FH1-UGW-mCherry and ΔmiR-137-FH1-UGW-mCherry were digested with PspXI and XcmI. The Ubiquitin-Synaptotagmin-1-T2a-Venus cassette was digested from the FUchW-Syt1-T2A-Venus using PacI and EcoRI. Both vector and insert were blunted by T4 DNA polymerase (NEB), gel-purified, ligated and transformed as described above.
Primary cortical-hippocampal neuronal culture
Primary neurons were prepared as described in ref. 54. We used embryonic day 16 embryos from timed-pregnant Swiss-Webster mice. Primary neurons cultures were used for experiments at 14 days in vitro.
Stereotactic injection
Stereotactic injections into the dentate gyrus of adult mice were performed as described 55 with the following modifications: 8 week old C57BL/6 mice were bilaterally injected with 1 μL lentivirus mixed with 0.2 μL Fast Green dye. Before loading the virus into a glass micropipette (Drummond Wiretrol 10 μL), the inside was coated with mineral oil. The stereotactic coordinates for dorsal dentate gyrus injection were anterior/posterior: −2, medial/lateral: 1.6, dorsal/ventral: −1.65. The total injection volume was 1 μL, injected at a rate of 0.125 μL/min. Overexpression of miR-137 was detectable at least four weeks after injection as quantified by RT-qPCR and by mCherry immunoreactivity (data not shown).
BrdU labeling
We stereotactically injected the lentivirus for miR-137OE and ΔmiR-137OE as described above. After one week of incubation, we injected daily 50 mg/kg of BrdU for seven days. Animals were then sacrified and perfused with 4% (w/v) PFA/PBS. The brain was dissected and post-fixed overnight. Vibratome sections were prepared and treated with 4N HCl for 2h at 25°C. The slices were washed at least three times with PBS and stained with BrdU and RFP as described in the section “Immunohistochemistry”. Images were taken, and the numbers of BrdU and RFP-positive cells were counted blinded.
Behavior experiments
Mice were housed in groups of 3–5 animals. The behavioral experiments were performed at least two months after virus injection. Mice were left to acclimate in the behavior testing rooms for 45–60 min prior to the experiments. 70% (v/v) ethanol was used to clean the experimental areas before the tests and between subjects. If the same groups of mice were used in different behavioral experiments, the tests were separated by one week. All the experiments were done during the light phase, in the second half of the day, with experimenters that were blind to the type of virus injected in the mice. The mice were consistently handled by the base of their tails and transported to the behavioral testing facility by experimenters. Data analysis was performed blinded.
Contextual and cued fear conditioning
During contextual fear conditioning, mice were placed in a conditioning chamber with Plexiglas walls and a metal grid bottom (TSE fear conditioning system, TSE Systems). They were acclimated for 3 min, then foot-shocked (2 sec, 0.8 mA constant current) and returned to their home cages. After 24 hr, mice were returned to the same chamber and freezing bouts, defined as a total lack of movement except for a heartbeat and respiration, were scored every 10 s during a 3 min period. For cued fear conditioning, animals were placed in the test chamber for 3 min followed by exposure to the auditory cue (30 s, 20 kHz, 75 dB sound pressure level) and a foot shock (2 s, 0.8 mA, constant current) and returned to the home cages. Associative learning was assessed 24 hrs later by placing the mice into modified chamber (visual, tactile, and olfactory changes), delivering the identical auditory cue for 3 min and recording freezing behavior. For measuring the freezing response before cue, we recorded the number of freezing responses for 30 s after the animal was put in the chamber.
Hot plate analgesia test
Animal was placed on a 55°C plate. The latency to respond with a nociceptive response (rear paw licking) was measured (hot plate analgesia test meter, IITC Life Sciences Inc. CA).
Morris water maze
Spatial reference memory testing55 was performed in a circular tank (diameter 1.2 m) filled with opacified water at 22 °C. The walls surrounding the tank had bright and contrasting shapes as reference cues. The tank contained a fixed platform (10 cm diameter) in a target quadrant. On day 1 of the experiment (pre-training), mice were placed into the water, guided towards a visible platform, and kept on the platform for 15 s. This procedure was repeated one more time. On the following days, the platform was submerged and the mice were placed into the maze at one of four points randomly facing the wall of the tank to avoid the development of egocentric strategies. Mice were allowed to search for the platform for 60 sec. If they did not find the platform, they were gently guided to it. Two trials a day were conducted with 2 min inter-trial intervals. Between trials, mice were warmed on a heating pad. Mouse behavior was video-recorded and analyzed using Videomot 2 software (TSE systems). The escape latency was scored for each trial. For the probe trial, 24 hr following the final training day the platform was removed and the time spent in each of the four quadrants (target, opposite, left, and right) was recorded. Thigmotaxis was automatically measured with the software.
Open field
All experimental mice were habituated in the open field arena (40 x 40 cm Plexiglas VersaMax chambers, Accusan Instruments) for 30 min 1 day before the test. To test, mice were placed into the center of the arena and allowed to explore the apparatus for 40 min. During the session, the number of beam breaks was measured automatically.
Light-Dark Exploration Test
The apparatus used for the light/dark test consisted of a cage (41 x 41 x 30 cm) divided into two sections of equal size by a partition with door. Mice are placed into the dark side and the door is opened 10 seconds afterwards. Mice are allowed to move freely between the two chambers for 10 min and the whole process was video recorded. Measurements were taken of the total number of transitions between the light and dark chambers and the total amount of time spent in the light chamber by a observer blinded to experimental condition.
Elevated plus-maze
The maze consisted of a central platform (10 x 10 cm), elevated 50 cm from the floor, with two opposing open arms (30 x 10 x 0.5 cm) and two arms enclosed by dark Plexiglas walls (30 x 10 x 30 cm). Experiments were conducted in a brightly and homogenously illuminated room, with the maze placed in the center of the room. Mice were placed in the intersection of the four arms of the elevated plus maze with the head directed toward a closed arm. The position of the animals was automatically determined and recorded for 5 min with VersaMax software (TSE systems). A test trial using a practice animal was performed before sessions with the experimental mice to make sure the recording system and test conditions were consistent across all the animals.
Social interaction paradigm
Three-chamber social interaction test 57 was performed as in 58 with minor modifications. Specifically, mice were removed from the colony and acclimated to the testing room for 1 hr. The mouse designated as “test” mouse was placed in the center chamber of the apparatus for a 10 min habituation, with the doors open on each side so that the mouse could explore all three chambers of the apparatus. After 10 min, the doors were closed to contain the mouse in the center chamber. A novel “stimulus” mouse was placed in one of the side chambers. The choice of adjacent chamber holding the “stimulus” mouse was randomly determined. The “stimulus” mouse was enclosed inside a small circular wire cage (11 cm in height, 10.5 bottom diameter, bars spaced 1 cm apart). Thus, the “stimulus” mouse could provide visual, olfactory, auditory, and some physical contact cues. An identical empty wire cage was placed in the other adjacent chamber. After that, the doors were opened and the “test” mouse was set free to examine the entire apparatus, including the “stimulus” mouse and the empty wire cage. The behavior of the “test” mouse was videotaped for this 10 min period for later scoring by an observer blind to experimental condition. The time in each compartment, as well as in close proximity to the “stimulus” mouse, was assessed from the videos. To evaluate a preference for social novelty, the “original stimulus” mouse remained in the wire cage and a second “novel stimulus” mouse was placed in the wire cage in the opposite compartment. The “test” mouse placed in the middle compartment and given a further 10 min to explore both of these “stimulus” mice. The behavior of the “test” mouse was recorded on video for this 10 min period and analyzed offline, as in the first part of the experiment.
Electrophysiology
For acute hippocampal slice preparation, lentivirus-injected C57BL/6 mice (3–4 months old) were anesthetized with isoflurane and decapitated. The experimenter was blinded to which virus was injected. Transverse hippocampal slices (400 μm thick) were prepared in ice-cold dissection buffer (in mM: 211 sucrose, 3.3 KCl, 1.3 NaH2PO4, 0.5 CaCl2, 10 MgCl2, 26 NaHCO3 and 11 glucose) using a Leica VT1000S vibratome (Leica). Slices were recovered in submerged chamber with 95% O2/5% CO2-saturated artificial cerebrospinal fluid (ACSF) consisting of (mM) 124 NaCl, 3.3 KCl, 1.3 NaH2PO4, 2.5 CaCl2, 1.5 MgCl2, 26 NaHCO3, and 11 glucose for 1 hr at 28–30 °C. All recorded slices were subjected to immunohistochemistry with antibodies against mCherry to ensure that an equivalent amount of virus-transduced cells was present in each slice.
Extracellular recordings
A tungsten bipolar electrode was placed in the dentate granule cell layer to stimulate mossy fibers, and extracellular recordings were made in the stratum lucidum of CA3 using a glass microelectrode filled with ACSF (resistance of 2–3 MΩ). Frequency facilitation was elicited by low-frequency (1 Hz) stimulation for 30 sec. For mossy fiber LTP recordings, three trains of high frequency stimulation (100 Hz for 1 sec) were delivered with 20 sec intervals after observation of stable baseline. To verify mossy fiber inputs, 1 μM (2S,2′R,3′R)-2-(2′,3′ dicarboxycyclopropyl)glycine (DCG-IV; Tocris Bioscience), a group II metabotropic glutamate receptor agonist that selectively blocks mossy fiber responses, was applied at the end of each recording. The amplitude of fEPSPs was measured to quantify the strength of synaptic transmission. An AM-1800 Microelectrode amplifier (A-M systems) and a Digidata 1440A A-D converter (Axon Instruments) were used for data acquisition and data were analyzed with pClamp10 (Axon Instruments). The input-output curve was obtained by plotting the amplitude of fEPSPs against fiber volley amplitudes.
For the performant path recording, we stimulated the medial perforant path (MPP) and recorded fEPSPs within the molecular layer of the dentate gyrus as described previously 59, 60. To distinguish whether the medial or lateral perforant path was stimulated, we measured the synaptic response to paired pulses given 50 ms apart 61. LTP was induced by 4x HFS protocol (100 Hz for 0.5 sec at 20 sec intervals) 60 in the presence of picrotoxin (100 μM) to reduce GABAergic inhibition. The input-output relationship was estimated by plotting the fEPSPs slope against the stimulation intensity.
Immunohistochemistry
Following electrophysiological recording, slices were fixed in 4% (w/v) paraformaldehyde in PBS overnight at 4°C, washed in PBS several times, then transferred into 30% (w/v) sucrose in PBS and incubated overnight at 4°C for cryoprotection. To increase antibody permeability, the slices were put on a glass slide, frozen on the coverslip over dry ice, and thawed at room temperature for three cycles. Then, the slices were transferred into PBS, washed for 10 min at 25 °C, and blocked in 5% (v/v) normal donkey serum (NDS, Sigma), 0.3% (v/v) Triton X-100 (Sigma) in PBS for 1 hr at 25 °C on a shaker. The primary antibody was diluted in 2.5% (v/v) NDS in PBS and 0.3% (v/v) Triton X-100. Slices were incubated for 2–3 days at 4 °C and were afterwards washed three times for 30 min in PBS. The secondary antibody was diluted in PBS containing 0.3% (v/v) Triton X-100 and slices were incubated for 2 hr at 25 °C and then embedded in Fluoromount-G (SouthernBiotech).
Fraction of mossy fiber projections transduced by the virus
Vibratome sections (100 μm thick) from virus-injected animals were immunolabeled with antibodies against mCherry, which was expressed from the virus, as well as antibodies against the mossy fiber marker, ZnT3. CellProfiler (Version 2.1.0) software was used to quantify the degree of overlap between mCherry and ZnT3. The average amount of overlap was 70%.
Primary antibodies: rat BrdU (AbD Serotec, MCA2060GA, 1:500), rabbit GFP (Invitrogen, A11122, 1:300), rabbit Complexin-1 (Proteintech, 10246–2–AP, 1:300), mouse GFAP (G6171, 1:500), mouse Nsf-1 (MA1–12435, 1:300) and rabbit Syn3 (OSS00018W, 1:300) from Thermo Scientific, guinea pig Iba1 (234 004, 1:500), mouse Syt1 (105 011, 1:300), guinea pig ZnT3 (197 004, 1:1000) from Synaptic Systems.
Secondary antibodies. Antibodies (all obtained from Jackson Laboratory, donkey anti-guinea pig IgG (H+L) ML, #706-225-148 and 706-175-148; donkey anti-mouse IgG (H+L) ML, #715-225-151, 715-165-151, and 715-175-151; donkey anti-rabbit IgG (H+L) ML, #711-225-152, 711-165-152, and 711-175-152 were diluted 1:400. Confocal scans were carried out using a LSM 510 Imager Z.1 microscope (Zeiss) using Plan-Apochromat 63X/1.4 and EC Plan-Neofluar 40X/1.30 oil immersion objective lenses and four excitation laser lines. Tile scans were performed using a LSM 710 confocal microscope (Zeiss) with a Plan-Apochromat 20X/0.8 objective.
Electron microcopy
Tissue preparation
Mice were given an overdose of ketamine-xylazine and were transcardially perfused with cold PBS followed by ice-cold fixing solution (10% (vol/vol) formaldehyde solution (Macron) and 0.1% (vol/vol) 25% glutaraldehyde (Sigma)) for approximately 10 min. The brain was dissected and post-fixed in fixing solution overnight at 4 °C. The brain was sectioned in 100 μm slices using a vibratome. Slices were quenched with 0.2 M glycine in PBS twice for 10–15 min at 25 °C, washed several times with PBS, then permeabilized and blocked for 1 hr in 3% (w/v) bovine serum albumin (BSA, Life Technologies), 5% (v/v) NDS, 0.1% (v/v) Triton X-100 in PBS at 4°C. A rabbit anti-RFP or GFP antibody (Abcam, ab62341 or ab6556) was added at a 1:40 concentration in 3% (w/v) BSA in PBS and incubated overnight at 4 °C. Sections were washed three times for 15 min in PBS containing 0.1% (v/v) Triton X-100 and were incubated with Protein A-gold 5 nm (1:50, Utrecht University Med Ctr) for 1 hr at 25°C, washed in PBS, and then fixed in 1% (v/v) glutaraldehyde in PBS for 30 min. For Epon embedding, slices were incubated in 0.5% (w/v) osmium in ddH2O for 30 min, washed three times in ddH2O, and then stepwise dehydrated (each step for 10 min) in 70% (v/v) ethanol, 95% (v/v) ethanol, and twice in 100% (v/v) ethanol. The slices were incubated in propyleneoxide, infiltrated in 1:1 propylenoxide/TAAB Epon, embedded in fresh TAAB Epon (Marivac Canada Inc) and polymerized at 60 °C for 48 h. Each block was cut into 60 nm ultrathin sections using a Reichert Ultracut-S microtome. Sections were placed onto copper grids and stained with uranyl acetate and lead citrate. The sections were examined using a JEOL 1200EX transmission electron microscope. Images were recorded with an AMT 2k CCD camera at 20,000–30,000× magnification by an individual who was blinded to the experimental manipulation.
Image analysis was performed blind to the experimental manipulation. Images were analyzed in which clear, strong gold particles were detected within the synapse. Excitatory synapses were defined by presence of a postsynaptic density. For each section, the number of vesicles was counted using the Fiji plugin “Concentric circles” (version 1.46a). Briefly, concentric circles with a 50 nm distance to each other were created. The number of vesicles within the circle was counted using the Fiji plug-in “Cell counter”.
Compiled list of predicted miR-137 targets
MicroRNAs have been shown to bind to specific seeding sites in the 3′UTR 3. In our study, we combined the results of four different miRNA-target prediction databases, each of which uses a different algorithm, for finding the seed region and predicting potential targets. We used four miR-137 target-prediction databases, each with a different algorithm for predicting microRNA-targets 3, 62: miRBase (www.mirbase.org) 63, PicTar (http://pictar.mdc-berlin.de) 64, TargetScan (www.targetscan.org) 65, and microRNA.org (www.microRNA.org) 66. We used the standard settings. The obtained lists were downloaded and unified. To identify potential gene enrichments, we entered the list to the generic GeneOntologyTermFinder (GOTermFinder, http://go.princeton.edu/cgi-bin/GOTermFinder, Princeton University) 67 using the “Process” ontology aspect and the MGI annotation. “Synaptic transmission” was one cluster (cluster frequency: 3.2%, genome frequency: 1.9%, corrected p-value: 8.89*10–8, FDR: 0.00, False-positive: 0.00). From this list (Supplementary Fig. 2).
Dual Luciferase assay
Cloning of 3′UTR fragments
Primers were designed approximately 500 bp up- and downstream of the putative miR-137 binding site within the 3′UTR (Supplementary Table 1). If the 3′UTR was shorter, the whole 3′UTR region was cloned. Primers contained an AsiSI and a NotI site. PCR fragments were cloned with the two-step program described in the Phusion-MasterMix protocol (New England Biolabs). For the PCR template, genomic DNA was prepared from mouse embryos using the DNeasy Blood & Tissue Kit (Qiagen). The PCR product was purified with NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel) and digested with AsiSI and NotI. The vector backbone psiCheck2 (Promega) was digested with AsiSI and NotI and treated with alkaline phosphatase (CIP, New England Biolabs) for 1 hr. Vector and insert were gel-purified. Ligation was performed in the ratio 1:3 (vector: insert) using Quick Ligation protocol (New England Biolabs). 5 μL of the ligation was used to transform NEB10 beta high-efficiency cells (New England Biolabs).
Cloning of 3′UTR miR-137 deletion fragments
Site-directed mutagenesis was performed using the Agilent QuikChange XL Site-Directed Mutagenesis Kit (Agilent Technologies) based on the manufacturer’s protocol.
Dual-Luciferase assay
24 hr prior to transfection, HEK-293T cells were plated at a density of 1x105 cells/well in 24-well plates. For transfection, 0.67 μg miR-137 or ΔmiR-137 FH1-UGW-mCherry and 0.33 μg of the luciferase reporter construct were mixed with 2 μL Lipofectamine 2000 (Life Science Technology) according to the manufacturer’s instructions. 48 hr later, cells were lysed in passive lysis buffer (Promega) according to the manufacturer’s manual. Luciferase activity was measured in an EnSpire multimode plate reader (PerkinElmer) using the EnSpire Manager software. Values for each sample were normalized to the control ΔmiR-137.
Transcriptional luciferase activity assay
The putative transcriptional fragments for miR-137 were cloned into the pGL3 promoter vector (Promega). Primers contained either a MluI or a KpnI on the 5′-site and a XhoI on the 3′-site. SH-SY5Y-cells (ATCC) were transfected with different concentrations of the promoter construct using Lipofectamine. The luciferase assay was performed as described above.
RNA isolation
Small and total RNA were isolated using PureLink miRNA isolation Kit (Invitrogen). RNA quantity and quality was measured using a NanoDrop spectrometer (ThermoScientific).
Quantitative real-time PCR (qRT-PCR)
miR-137 qRT-PCR
For reverse transcription, the TaqMan MicroRNA reverse transcription kit (Applied Biosystems) with a probe set for miR-137 (Applied Biosystems, ID 001129) and for snoRNA202 probe set (ID 001232). qRT-PCR was performed using the TaqMan Universal PCR Master Mix, No AmpErase (Applied Biosystems) in a Bio-Rad CFX96 real-time PCR detection system with the following protocol: 95 °C for 10 min, 45 cycles of 95 °C for 15 s and 60 °C for 1 min.
qRT-PCR of targets
Reverse transcription was performed with the iScript cDNA synthesis kit (Bio-Rad Laboratories). For qRT-PCR, we used 10 μL SsoFast EvaGreen Supermix per reaction (Bio-Rad Laboratories), 0.8 μL primer (10 μM) and 1 μL cDNA together with a Bio-Rad CFX96 real-time PCR detection system using the following protocol: 95 °C for 30 s, 39 cycles of 95 °C for 5 s and 57 °C for 5 s. In addition, the Bio-Rad PrimePCR SYBR Green Assay was been used to measure the expression of CPLX1 (qHsaCED0003596), NSF (qHsaCID0021934), SYN3 (qHsaCID0014702), SYT1 (qHsaCID0012794), SYNJ1 (qHsaCED0045839), PCLO (qHsaCID0018129), and STX8 (qHsaCED0038901) together with the positive control assay (qHsaCtlD0001003) according to the manufacturer’s protocol (Bio-Rad Laboratories).
Analysis was performed using the ΔΔCT method with β-actin (Actb) as the calibrator gene 68. Primer sequences are β-actin (GGGAAATCGTGCGTGACATT, GGGAAATCGTGCGTGACATT), Cplx1 (GGAAGATGCTTGGGGGTGAT, GGAAGATGCTTGGGGGTGAT), Nsf (GATCGGCGGTCTGGATAAGG, GATCGGCGGTCTGGATAAGG), Syn3 (CTCGCATTCAGCATCTTGCC, CATACTGGAGGCCGATGACC), Syt1 (AAGTCCACCGGAAAACCCTC, AAGTCCACCGGAAAACCCTC)
MiR-137 mimic experiment
One day prior to transfection, 60,000 N2a or HT22 cells were seeded per well of a 12-well plate. Transfection was performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. 20 μM miRIDIAN Mimic mmu-miR-137 (Dharmacon) and 0.5 μg/μL control plasmid expressing GFP were transfected. 48 h later, RNA was isolated and qRT-PCR performed.
Western blot
Tissue preparation
Fluorescent-positive cells in the dentate gyrus-CA3 were dissected under a stereofluorescence microscope (Olympus, SZX16) and snap-frozen in liquid nitrogen.
Synaptosome preparation
500 μL ice-cold homogenization buffer (10 mM Hepes pH 7.4, 2 mM EDTA, 2 mM EGTA, 320 mM sucrose supplemented with one PhosphoSTOP tablet, Roche) was added, and the brain samples homogenized with a 26G needle/syringe on ice. For the input sample, 150 μL was set aside, supplemented with 1% (w/v) SDS (final concentration), and boiled at 95°C for 5 min. The rest of the sample was centrifuged for 10 min at 800 x g at 4 °C to remove nuclei and cell debris. The supernatant was then re-spun at 12,000 x g for 15 min at 4 °C to enrich for synaptic components. The pellet was resuspended in 60 μL 1% (w/v) SDS and boiled at 95 °C for 5 min. Protein concentrations were measured with the BCA protein assay kit (Thermo Scientific).
SDS gel
30–50 μg protein were loaded on a 4–15% mini protean TGX SDS-gradient gel (Bio-Rad Laboratories) together with Precision Plus protein pre-stained standard dual color (Bio-Rad Laboratories) ladder and run at 200V for 30 min in running buffer (25 mM Tris base, 190 mM glycine, 0.1% (w/v) SDS, pH around 8.3).
Transfer
Protein was transferred to Immobilion PVDF transfer membrane (Millipore) in transfer buffer (25 mM Tris, 192 mM glycine, 20% (v/v) methanol) for 1 hr at 100 V constant, 350 mA. The membrane was blocked for at least 1 h in 5% (wt/vol) non-fat dry milk in Tris-buffered saline (25 mM Tris, 150 mM NaCl, 2 mM KCl, pH 8.0, 15 mM NaCl, 10 mM Tris, pH 8.0). Primary rabbit Complexin-1 (Proteintech, 10246–2–AP), mouse Nsf-1 (MA1–12435) and rabbit Syn3 (OSS00018W, 1:300) from Thermo Scientific, mouse Syt1 (Synaptic Systems, 105 011, mouse β-actin (Sigma Aldrich, A5316) were added 1:2,000 in blocking solution and incubated for 2–3 days on a shaker at 4 °C. The membrane was washed 3× for 30 min in Tris-buffered saline and incubated with secondary antibody (1:10,000, Li-Cor: IRDye 680 RD donkey anti-mouse IgG (H+L), #925-68072; IRDye 680RD donkey anti-rabbit IgG (H+L), #925-68073; IRDye 800CW donkey anti-mouse IgG (H+L), #925-32212; IRDye 800 CW donkey anti-rabbit IgG (H+L), #925-32213. Detection was performed using an Odyssey machine (Li-Cor). Image Studio 2.0 software was used for image analysis. The membrane was stripped with NewBlot PVDF 5x stripping buffer (Li-Cor) according to manufacturer’s protocol. We normalized and corrected for uneven loading using the intensity of the internal control band, beta-Actin.
FM4–64 imaging of human direct induced neurons
We performed the experiments after consolidation of the method papers 69, 71. Four to six week old cultures of human direct induced neurons were loaded with 10 μM FM4–64 (Invitrogen) for 2 min in saline solution containing 170 mM NaCl, 3.5 mM KCl, 0.4 mM KH2PO4, 5 mM NaHCO3, 1.2 mM Na2SO4, 1.2 mM MgCl2, 1.3 mM CaCl2, 5 mM glucose, 20 mM N-tris(hydroxymethyl)-methyl-2-aminoethane-sulfonic acid, pH 7.4) supplemented with 60 mM KCl. The cells were rinsed with saline solution only and then incubated with 10 μM FM4–64 in just saline solution. The cells were washed three times with saline solution for a total of 5 min, followed by a wash for 1 min with 1 mM ADVASEP-7 (Sigma-Aldrich) in saline solution. FM imaging was performed at the LSM 710 confocal microscope (Zeiss) with a Plan-Apochromat 20X/0.8 objective with image taken every 5 s at 25°C. A one-minute baseline was recorded, followed by stimulation with 60 mM KCl in saline solution for 4 min. Cells were excited at 558 nm and the emission measured at 734 nm. Images were analyzed blinded in Fiji using the “plot z-axis profile” plug-in. We analyzed potential functional nerve terminals located along the GFP-positive cell (Supplementary Fig. 2). The FM4–64 signal was determined by F = (F1–B1)/(F0–B0). Signal was normalized to mean fluorescence intensity measured at baseline condition.
Statistical methods
All statistical analysis was performed in RStudio (version 0.98.501) and R (version 3.0.2). All charts show the mean and standard error of the mean (s.e.m). Each data set was analyzed for its ability to meet the statistical assumptions for equality of the variance, for normal distribution, and for sphericity. The effect size r is indicated, where applicable. The assumption of the parametric test was calculated using the Levene test (leveneTest, package car version 2.0–19). If the assumption was met, the following tests were used: Welch two sample t-test (t, t-test, package, stats version 3.0.2), independent t-test (Ty, yuen, package WRS version 0.24), analysis of variance for one or more fitted model objects (F, ANOVA, stats version 3.0.2), multivariate analysis of variance (V, MANOVA, package, stats version 3.0.2, Pillai-Bartlett trace). If the assumption was validated, the following tests were performed: Wilcoxon-rank sum and signed rank tests (W, wilcox.test, package stats version 3.0.2) and Kruskal-Wallis rank sum test (H, kruskalmc, package pgirmess 1.5.8). Posthoc analysis was performed using either pairwise comparisons between group levels (pairwise.t.test, package stats version 3.0.2, Benjamini & Hochberg adjusted p-value for multiple comparisons), multiple comparisons of means: Tukey contrasts (glht, package multcomp version 1.3–2, mcp and “Tukey”), or multiple comparison test following Kruskal-Wallis rank sum test. nsp; not significant, * < 0.05, ** < 0.01, *** < 0.001.
Sample size
No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those reported in previous publications. For biochemical (immunohistochemistry, western blotting) and molecular (qRT-PCR, luciferase) the minimum number of biological replicates needed for non-parametric statistical analysis is three animals per condition, per experiment. To improve our statistical power and ensure that our hypotheses are rigorously tested, we try to include 4 to 5 biological replicates in each experiment, and to replicate each experiment at least once, when possible. Behavior and in vivo experiments have a higher variability inherent to behavioral experiments. It is customary to include in > 8 animals to achieve appropriate statistical power. For behavior, these experiments are typically repeated at least once with a separate cohort of animals. The number of animals used for survival surgery is dictated by the subsequent experiments that those animals will be engaged in (behavior, tissue harvesting, both).
A supplementary methods checklist is available.
Supplementary Material
Supplementary Figure 1. Supplementary data of human induced neurons. a. Transcriptional luciferase assay for the four SNPs in human neuroblastoma cell line SH-SY5Y, F(4) = 7.87, ***p < 0.001, posthoc analysis: tpGL3-rs2660304 (4.482), ***p < 0.001, r = 0.91; trs2660304-rs2802535 (−4.57), ***p < 0.001, r = 0.92; trs2660304-rs1625579 (−4.57), ***p < 0.001, r = 0.92; tpGL3-rs2660304 (1.65), nsp = 0.4781, r = 0.64, n: three experiments. b. Fold-change differences of MIR137 determined by qRT-PCR from Fig. 1c, separated by its individual lines, WiN= 65, *p = 0.0157, r = −0.40, box plot shows, in ascending order, the lowest maximum value, the first quartile, median, third quartile and the highest maximum value, dots are outlier. c. Summary table of the human fibroblasts and their genotype alleles for the four disease-associated SNPs. First four are homozygous for common major alleles; the last two are homozygous for the minor alleles. d. Examples of successfully induced neurons before FACS. Left: bright field, middle: transduced cells, right: expression of the neuron-specific HSV-CamKII-mCherry construct. Scale bar: 100 μm. Before any iN experiments, cells were visually controlled for successful reprogramming. e. Relative microRNA levels in induced neurons from common major and minor allele SNPs determined by RT-qPCR, tmiR-9(7.87) = −0.2, nsp = 0.8475, r = 0.07, tmiR-19b(8.28) = 0.12, nsp = 0.9047, r = 0.04, tmiR-124(13.89) = −0.16, nsp = 0.8773, r = 0.04, box plots show, in ascending order, the lowest maximum value, the first quartile, median, third quartile and the highest maximum value, dots are outlier, n: number of samples from at least two reprogramming, s.e.m.
Supplementary Figure 2. Supplementary data of presynaptic target genes validation. a. Summary table of putative in silico predicted miR-137 target genes involved in “synaptic transmission”. Red: targets tested in this study. b. Luciferase assay results for additional putative miR-137 targets, light grey and dark orange: miR-137 binding site deletion construct (Δ), WCalb1 = 170, *p = 0.0180, r = −0.43; tCamK2(14.58) = −0.14, nsp = 0.8888, r = 0.04; FNrxn1(3, 80) = 13.89, ***p < 0.001; FStx8(3, 56) = 0.60, nsp = 0.6159; WStxbp5 = 187, nsp = 0.4381, r = −0.13; FSv2a(3, 20) = 0.05, nsp = 0.9831; WSyn2a = 201, nsp = 0.6326, r = −0.07; FSyn2b(3, 74) = 0.72, nsp = 0.545; FSynj1(3, 56) = 1.30, nsp = 0.2845; WSynpr = 278, nsp = 0.1515, r = −0.22; WSyt9 = 121, nsp = 0.2983, r = −0.18; WVamp1 = 119, nsp = 0.1786 r = −0.22; FVamp2(3, 81) = 3.92, *p = 0.0115; WVamp7 = 172, nsp = 0.7637, r = −0.05. c. Example images of FM4–64 recording, white: induced neuron, green: FM4–64, arrow: terminal. ns: not significant, s.e.m.
Supplementary Figure 3. miR-137OE validation and in vivo expression. a–b. Relative miR-137 expression level determined by qRT-PCR. a. in HEK-293T cells, Ty(2) = 44.89, ***p < 0.001, r = 1; b. in primary cortical neuronal culture at DIV14, Ty(3.97) = 4.48, *p = 0.0112, r = 0.91, n: number of experiments. c. Additional examples of hippocampi tile-scans injected with the lentivirus expressing the ΔmiR-137OE or miR-137OE constructs, immunostained for mCherry (magenta) and the nuclei dye DAPI (blue). CA3: Cornu Ammonis region 3. Staining was reliably throughout this study. d. Immunostaining of ΔmiR-137OE or miR-137OE-transduced dentate gyrus for DAPI (blue), mCherry (magenta), glial fibrillary acidic protein (GFAP), and ionized calcium-binding adapter molecule (Iba1). This experiment was repeated in at least three different animals. Scale bars: 100 μm. s.e.m.
Supplementary Figure 4. a. Supplementary data of ultrastructural analysis for miR-137OE and ΔmiR-137OE mice. Black arrow: mCherry gold-particle. Orange arrows: gap within the vesicle pool in miR-137OE synapses, * active zone, scale bar: 100 nm. b. Total number of vesicles for ΔmiR-137OE (black, n = 84) and miR-137OE (orange, n = 112), W = 4294, nsp = 0.2973, r = −0.07, box plot shows, in ascending order, the lowest maximum value, the first quartile, median, third quartile and the highest maximum value, dots are outlier, n: number of analyzed synapses of at least three animals. ns: not significant, s.e.m.
Supplementary Figure 5. Supplementary behavior analysis for miR-137OE and ΔmiR-137OE. a–c. Equivalent levels of locomotion and a trend for anxiety in miR-137OE as measured by a. Open-field arena paradigm: margin distance: t(12.94) = −0.61, nsp = 0.5547, r = 0.17; margin time: t(12.20) = −1.89, nsp = 0.083, r = 0.48; center distance: t(13.83) = 2.55,*p = 0.023, r = 0.57; center time: t(12.20) = 1.89, nsp = 0.083, r = 0.48; total distance: t(12.89) = 1.31, nsp = 0.2117, r = 0.34; horizontal activity: t(13.52) = 1.98, nsp = 0.0685, r = 0.47; vertical activity: t(13.15) = 1.36, nsp = 0.1952, r = 0.35; stereotype: t(13.87) = 2.13, nsp = 0.05167, r = 0.50. b. Light-dark box paradigm: Frequency of exploration: t(9.90) = 0.94, nsp = 0.3709, r = 0.29, time spend in light: t(11.88) = 0.60, nsp = 0.5585, r = 0.17. c. Elevated-plus maze paradigm. Panel from left to right: time spent in closed or open arm, no significant main effect, F(1, 26) = 2.09, nsp = 0.161, tTime ΔmiR-137 (13.66) = 42.0, ***p < 0.001, r = 1, tTime miR-137 (9.86) = 39.68, ***p < 0.001, r = 1; Frequency of entering did not differ between open-closed arm and the condition, F(1, 26) = 0.40, nsp = 0.535, tFrequency ΔmiR-137 (11.49) = 2.49, *p < 0.05, r = 0.59, tFrequency miR-137 (11.49) = 2.49, *p < 0.05, r = 0.59). Distance and velocity, tdistance(12.98) = 0.36, nsp = 0.7259, r = 0.10, tvelocity(12.97) = 0.35, nsp = 0.7298, r = 0.10. d. Nociception response, W = 20.5, nsp = 0.7471, r = −0.09. e. Freezing response after context and before cue in %, t(41.97) = −1.20, nsp = 0.2381, r = 0.18. f. Thigmotaxis, percentage of the trail time and path that were spent in the defined thigmotaxis band (0.8) relative to the diameter of the pool, tTime(24.07) = −0.83, nsp = 0.4165, r = 0.17; tPath(25.57) = −0.15, nsp = 0.8838, r = 0.03. g. Three-chamber social interaction paradigm. Amount of time that the mouse spends to explore, left panel: new versus familiar mouse, F(2, 24) = 22.41, ***p < 0.001, right panel: mouse versus object, F(2, 24) = 45.5, ***p < 0.001. No significant difference for ΔmiR-137OE and miR-137OE in both experimental set-ups, Ffamiliar mouse (2, 48) = 2.41, nsp = 0.10 and Fempty cage (2, 48) = 2.86, nsp = 0.0673. Social paradigm for the familiar paradigm: for ΔmiR-137OE: tfamiliar-middle (−3.81), **p = 0.0023, tfamiliar-new (2.86), *p = 0.02245, tmiddle-new (6.67), ***p < 0.001; for miR137OE: tfamiliar-middle (−5.89), ***p < 0.001, tfamiliar-new (3.44), **p = 0.00574, tmiddle-new (9.33), ***p < 0.001; for the empty paradigm for ΔmiR-137OE: tempty-middle (−6.06), ***p < 0.001, tempty-mouse (3.35), **p = 0.0718, tmiddle-mouse (9.41), ***p < 0.001; for miR-137OE: tempty-middle (−4.03), **p = 0.0013, tempty-mouse (3.19), *p = 0.0106, tmiddle-mouse (7.22), ***p < 0.001, n: number of animals used for each experiment. ns: not significant, s.e.m.
Supplementary Figure 6. Supplementary data for Sponge miR-137 and Sponge Control. a. Additional ultrastructural images of the mossy fiber presynaptic terminals in SpongeControl and SpongemiR-137 mice. Black arrow: gold-particle labeled GFP, * active zone, scale bar: 100 nm. b. Total vesicle number for SpongeControl (black, n = 50) and SpongemiR-137 (blue, n = 43) mice in the presynaptic compartments at the mossy fiber synapse, t(88.82) = −1.80, nsp = 0.0751, r = −0.19, box plot shows, in ascending order, the lowest maximum value, the first quartile, median, third quartile and the highest maximum value, dots are outlier, n: number of analyzed synapses of at least three animals. c. Input-output curve estimated from fEPSP against the fiber volley amplitude at the mossy fiber-CA3 synapse in hippocampal slices from SpongeControl (black, n = 4) and SpongemiR-137 (blue, n = 4), F(1, 129) = 1.46, nsp = 0.229, shaded area: 95% confidence interval, n: number of analyzed hippocampal slices from at least three animals. d. Open-field arena paradigm. Total distance: t(16.5) = −0.72, nsp = 0.4809, r = 0.18; time moving: t(16.33) = −1.01, nsp = 0.3285, r = 0.24; moves/counts: t(17.51) = −0.88, nsp = 0.389, r = 0.21; distance periphery: t(16.37) = −0.18, nsp = 0.8617, r = 0.04; time periphery: t(16.19) = 2.62, *p = 0.0182, r = 0.55; distance center: t(12.89) = −2.80, *p = 0.0151, r = 0.62; time center: t(16.21) = −2.63, *p = 0.0182, r = 0.55. e. Nociception response, t(13.47) = 0.30, nsp = 0.7664, r = 0.08. f. Freezing response after context and before cue in %, t(17.12)= 0, nsp = 1, r = 0, n: number of animals. ns: not significant, s.e.m.
Supplementary Figure 7. Supplementary data of endogenous expression of synaptic target genes and Syt1 restoration (miR-137-Syt1-Venus). a–c. Endogenous target gene expression in the dentate gyrus and the mossy fiber-CA3 pathway of naïve C57BL/6 animals. a. Absolute endogenous mRNA levels measured by RT-qPCR. b–c. Endogenous protein levels of Cplx1, Nsf, Syn3 and Syt1 by b. western blot (cropped. Full-length blots are presented in Supplementary Fig. 9), and c. immunohistochemistry. Vibratome sections, immunolabeled against the mossy fiber pathway marker zinc transporter 3 (ZnT3, green), the target gene (magenta), and stained with nuclei dye DAPI (blue). Scale bar: 10 μm. The observation was repeated in at least three different animals. d. Additional ultrastructural images of the mossy fiber presynaptic terminal of ΔmiR-137OE-Venus (grey), miR-137OE-Venus (red), miR-137OE-Syt1-Venus (green), and ΔmiR-137OE-Syt1-Venus (purple) mice, black arrow: gold particles staining of Venus, red arrows: gap within the vesicle pool in miR-137OE-Venus, * active zone, scale bar: 100 nm. e. Total vesicle number for ΔmiR-137OE-Venus (grey, n=56), miR-137OE-Venus (red, n=53), miR-137OE-Syt1-Venus (green, n=56), and ΔmiR-Syt1-Venus (purple, n=58), H(3) = 77.75, ***p < 0.001. Multiple comparison two-tailed test after Kruskal-Wallis, *p < 0.05, box plot shows, in ascending order, the lowest maximum value, the first quartile, median, third quartile and the highest maximum value, dots are outlier. f. Nociception response measured by mean speed of movement during shock in (cm s–1), H(3) = 6.10, nsp = 0.1068. ns: not significant, s.e.m.
Supplementary Figure 8. Supplementary data of Syt1-KD and Syt1-KDControl in the DG-CA3 synapse, and miR-137OE and ΔmiR-137OE at the entorhinal cortex-DG synapse. a–c. Electrophysiological properties of Syt1 knockdown in acute hippocampal slices, black: Syt1-KDcontrol, brown: Syt1-shRNA. n: number of hippocampal slice preparation from at least 3 animal. a. LTP recording for Syt1-shRNA (n = 5) and Syt-1 control (n = 5), H(1) = 129,44 ***p < 0.001. Arrows: application of either high frequency stimulation or DCG-IV, embedded representative fEPSP traces, grey traces: baseline level before stimulation. Right bar chart: LTP magnitude calculated by averaging fEPSP amplitude during the last 10 min (50–60 min) recording, W = 2681, ***p < 0.001, r = −0.39. b. Input-output curve from fEPSP against the fiber volley amplitude at the mossy fiber-CA3 synapse from Syt1-shRNA (brown, n=5) and Syt-1 control (black, n=5), H(1) = 6.63, *p = 0.0100, shaded area: 95% confidence interval. c. fEPSP amplitude for 1 Hz sustained stimulation for Syt1-shRNA (n = 5) and Syt-1 control (black, n = 5), H(1) = 47.32, ***p < 0.001. Embedded: representative traces, grey traces: response to the 1st stimulation. d. BrdU incorporation in miR-137OE (orange) compared to ΔmiR-137OE (black) in the dentate gyrus, t(4.77) = −3.18, *p = 0.0264. Right panel: immunostaining for BrdU (green) and virus expression of mCherry (red), n: number of animals. b. LTP recording at the molecular layer of the dentate gyrus with stimulation of medial perfornant path in miR-137OE and control ΔmiR-137OE, H(1) = 3.19, nsp = 0.0741, arrows: application of stimulation paradigm, right bar chart: LTP magnitude during the last 10 min (50–60 min) recording, W = 1528, nsp = 0.9285, r = −0.01. c. Input-output curve estimated from fEPSP against the fiber volley amplitude measured at a range of stimulus intensities at the molecular layer of the dentate gyrus in hippocampal slices from ΔmiR-137OE (black, n=12) and miR-137OE (orange, n = 14), F(1,66) = 0.91, nsp = 0.4543, shaded area: 95% confidence interval. n: number of analyzed hippocampal slices from at least three animals, ns: not significant, s.e.m.
Supplementary Figure 9. Full length western blot images. Samples were run on a 4–15% TGX SDS-gradient gel (Bio-Rad Laboratories). Molecular weight (M, in kDa): BLUEstain 2 Protein ladder, 5–245 kDA (Goldbio). Images were taken with the infrared imaging system from Odyseey (Li-cor). This allows staining for two proteins at the same time on the same plot. Light-blue bands indicate oversaturation. a. for Fig. 2e, Left and right panel show the same plot, but with adjustment of brightness and contrast; b. for Fig. 2d, the second and the fourth row are adjustment in brightness and contrast from the plots in the first and the third row, respectively; c. and d. for Supplementary Fig. 7. For Syt1, we always observed in varying intensity a band at ~75 kDa. This band is likely to be unstripped Nsf. We tried to optimize the stripping procedure, however we could never remove the strong signal band for Nsf completely. For consistency, we decided to stain always Syt1 after Nsf.
Acknowledgments
We thank B. Cohen (McLean Hospital) and J. Madison (Broad Institute/Stanley Center) for providing the human fibroblasts; N. Nadif Kasri (Radbound University Medical Center Nijmegen) and A. Aschrafi (Radbound University Medical Center Nijmegen) for sharing the SpongeControl and SpongemiR-137 plasmids; T. Südhof (Stanford School of Medicine) for providing us the Syt1-KD and Syt1-KDControl plasmid28; K. Jones for the synaptosomal preparation protocol; W. Xu, J. Penney and M. Benevento for comments on the manuscript; and the members of the Tsai laboratory for their overall comments on the project.
S.S. was supported by Human Frontier Science Program (HFSP) long-term postdoctoral fellowship and a Swiss National Science Foundation fellowship for prospective researchers. E.J.K. was supported by a Simons Foundation Postdoctoral Fellowship. A.R. was supported by NARSAD Young Investigator Award. This work was supported by a Seed Grant from the Simons Center for the Social Brain and US National Institutes of Health grant RO1 MH 091115 to L.-H.T.
Footnotes
Author contributions
The study was designed by S.S., E.J.K, and L.-H.T., and directed and coordinated by L.-H.T. S.S. and L.-H.T. designed the experiments and wrote the manuscript. E.J.K. initiated the study and contributed to the preliminary data. J.S. and S.C. performed electrophysiological experiments. A.R. and W.Y.W. performed behavior experiments. W.Y.W. and E.J.K. cloned the miR-137OE and ΔmiR-137OE constructs. Z.F and A.J.M provided technical support. M.E. performed electron microscopy. A.E.M contributed to the induced neuron experiments and helped write the manuscript.
References
- 1.Smoller JW, et al. Identification of risk loci with shared effects on five major psychiatric disorders: a genome–wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ripke S, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nature Genetics. 2013;45:1150–U1282. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kosik KS. The neuronal microRNA system. Nature Reviews Neuroscience. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- 5.Esquela-Kerscher A, Slack FJ. Oncomirs - MicroRNAs with a role in cancer. Nature Reviews Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 6.Herzer S, Silahtaroglu A, Meister B. Locked nucleic acid-based in situ hybridisation reveals miR-7a as a hypothalamus-enriched microRNA with a distinct expression pattern. J Neuroendocrinol. 2012;24:1492–1504. doi: 10.1111/j.1365-2826.2012.02358.x. [DOI] [PubMed] [Google Scholar]
- 7.Smrt RD, et al. MicroRNA miR-137 Regulates Neuronal Maturation by Targeting Ubiquitin Ligase Mind Bomb-1. Stem Cells. 2010;28:1060–1070. doi: 10.1002/stem.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kesner RP, Lee I, Gilbert P. A behavioral assessment of hippocampal function based on a subregional analysis. Reviews in the Neurosciences. 2004;15:333–351. doi: 10.1515/revneuro.2004.15.5.333. [DOI] [PubMed] [Google Scholar]
- 9.Szulwach KE, et al. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. The Journal of Cell Biology. 2010;189:127–141. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heckers S, et al. Impaired recruitment of the hippocampus during conscious recollection in schizophrenia. Nature Neuroscience. 1998;1:318–323. doi: 10.1038/1137. [DOI] [PubMed] [Google Scholar]
- 11.Won H, Mah W, Kim E. Autism spectrum disorder causes, mechanisms, and treatments: focus on neuronal synapses. Frontiers in molecular neuroscience. 2013;6:19–19. doi: 10.3389/fnmol.2013.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunt R, Sauna ZE, Ambudkar SV, Gottesman MM, Kimchi-Sarfaty C. Silent (synonymous) SNPs: should we care about them? Single Nucleotide Polymorphisms: Methods and Protocols. (2nd) 2009;578:23–39. doi: 10.1007/978-1-60327-411-1_2. [DOI] [PubMed] [Google Scholar]
- 13.Vierbuchen T, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Busskamp V, et al. Rapid neurogenesis through transcriptional activation in human stem cells. Molecular Systems Biology. 2014;10 doi: 10.15252/msb.20145508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwon E, Wang W, Tsai LH. Validation of schizophrenia-associated genes CSMD1, C10orf26, CACNA1C and TCF4 as miR-137 targets. Molecular Psychiatry. 2013;18:11–12. doi: 10.1038/mp.2011.170. [DOI] [PubMed] [Google Scholar]
- 16.Weinberger DR. Cell biology of the hippocampal formation in schizophrenia. Biological Psychiatry. 1999;45:395–402. doi: 10.1016/s0006-3223(98)00331-x. [DOI] [PubMed] [Google Scholar]
- 17.Wenzel HJ, Cole TB, Born DE, Schwartzkroin PA, Palmiter RD. Ultrastructral localization of zinc transporter-3 (ZnT-3) to synaptic vesicle membranes within mossy fiber boutons in the hippocampus of mouse and monkey. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:12676–12681. doi: 10.1073/pnas.94.23.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamiya H, Shinozaki H, Yamamoto C. Activation ef metabotropic glutamate receptor type 2/3 suppresses transmission at rat hippocampal mossy fibre synapses. J Physiol-London. 1996;493:447–455. doi: 10.1113/jphysiol.1996.sp021395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phillips RG, LeDoux JE. Differential Contribution of Amygdala and Hippocampus to Cued and Contextual Fear Conditioning. Behavioral Neuroscience. 1992;106:274–285. doi: 10.1037//0735-7044.106.2.274. [DOI] [PubMed] [Google Scholar]
- 20.Devanna P, Vernes SC. A direct molecular link between the autism candidate gene RORa and the schizophrenia candidate MIR137. Sci Rep. 2014;4:3994. doi: 10.1038/srep03994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nature Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chapman ER. How does synaptotagmin trigger neurotransmitter release? Annual Review of Biochemistry. 2008;77:615–641. doi: 10.1146/annurev.biochem.77.062005.101135. [DOI] [PubMed] [Google Scholar]
- 23.Tang J, et al. A complexin/synaptotagmin 1 switch controls fast synaptic vesicle exocytosis. Cell. 2006;126:1175–1187. doi: 10.1016/j.cell.2006.08.030. [DOI] [PubMed] [Google Scholar]
- 24.Xu J, Pang ZPP, Shin OH, Sudhof TC. Synaptotagmin-1 functions as a Ca2+ sensor for spontaneous release. Nature Neuroscience. 2009;12:759–U111. doi: 10.1038/nn.2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Südhof TC. Calcium control of neurotransmitter release. Cold Spring Harb Perspect Biol. 2012;4:a011353. doi: 10.1101/cshperspect.a011353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bacaj T, et al. Synaptotagmin-1 and Synaptotagmin-7 trigger synchronous and asynchronous phases of neurotransmitter release. Neuron. 2013;80(4):947–959. doi: 10.1016/j.neuron.2013.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicoll RA, Schmitz D. Synaptic plasticity at hippocampal mossy fibre synapses. Nature Reviews Neuroscience. 2005;6:863–876. doi: 10.1038/nrn1786. [DOI] [PubMed] [Google Scholar]
- 28.Xu W, et al. Distinct neuronal coding schemes in memory revealed by selective erasure of fast synchronous synaptic transmission. Neuron. 2012;73:990–1001. doi: 10.1016/j.neuron.2011.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Q, et al. Association and expression study of synapsin III and schizophrenia. Neurosci Lett. 2009;465:248–251. doi: 10.1016/j.neulet.2009.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sawada K, et al. Hippocampal complexin proteins and cognitive dysfunction in schizophrenia. Archives of General Psychiatry. 2005;62:263–272. doi: 10.1001/archpsyc.62.3.263. [DOI] [PubMed] [Google Scholar]
- 31.Vawter MP, et al. Reduction of synapsin in the hippocampus of patients with bipolar disorder and schizophrenia. Molecular Psychiatry. 2002;7:571–578. doi: 10.1038/sj.mp.4001158. [DOI] [PubMed] [Google Scholar]
- 32.Eastwood SL, Cotter D, Harrison PJ. Cerebellar synaptic protein expression in schizophrenia. Neuroscience. 2001;105:219–229. doi: 10.1016/s0306-4522(01)00141-5. [DOI] [PubMed] [Google Scholar]
- 33.Mirnics K, Middleton FA, Marquez A, Lewis DA, Levitt P. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron. 2000;28:53–67. doi: 10.1016/s0896-6273(00)00085-4. [DOI] [PubMed] [Google Scholar]
- 34.Castillo PE, et al. Rab3A is essential for mossy fibre long-term potentiation in the hippocampus. Nature. 1997;388:590–593. doi: 10.1038/41574. [DOI] [PubMed] [Google Scholar]
- 35.Lonart G, Janz R, Johnson KM, Sudhof TC. Mechanism of action of rab3A in mossy fiber LTP. Neuron. 1998;21:1141–1150. doi: 10.1016/s0896-6273(00)80631-5. [DOI] [PubMed] [Google Scholar]
- 36.Cao P, Yang XF, Südhof TC. Complexin activates exocytosis of distinct secretory vesicles controlled by different synaptotagmins. J Neurosci. 2013;33:1714–27. doi: 10.1523/JNEUROSCI.4087-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin JA, Hu ZT, Fenz KM, Fernandez J, Dittman JS. Complexin Has Opposite Effects on Two Modes of Synaptic Vesicle Fusion. Current Biology. 2011;21:97–105. doi: 10.1016/j.cub.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jorquera RA, Huntwork-Rodriguez S, Akbergenova Y, Cho RW, Littleton JT. Complexin Controls Spontaneous and Evoked Neurotransmitter Release by Regulating the Timing and Properties of Synaptotagmin Activity. Journal of Neuroscience. 2012;32:18234–18245. doi: 10.1523/JNEUROSCI.3212-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geppert M, et al. Synaptotagmin-1 - A major Ca2+ sensor for transmitter release at a central synapse. Cell. 1994;79:717–727. doi: 10.1016/0092-8674(94)90556-8. [DOI] [PubMed] [Google Scholar]
- 40.Jorgensen EM, et al. Defective recycling of synaptic vesicles in Synaptotagmin mutants of Caenorhabditis elegans. Nature. 1995;378:196–199. doi: 10.1038/378196a0. [DOI] [PubMed] [Google Scholar]
- 41.Littleton JT, et al. SNARE-complex disassembly by NSF follows synaptic-vesicle fusion. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:12233–12238. doi: 10.1073/pnas.221450198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cesca F, Baldelli P, Valtorta F, Benfenati F. The synapsins: Key actors of synapse function and plasticity. Progress in Neurobiology. 2010;91:313–348. doi: 10.1016/j.pneurobio.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 43.Pieribone VA, et al. Expression of synapsin III in nerve terminals and neurogenic regions of the adult brain. Journal of Comparative Neurology. 2002;454:105–114. doi: 10.1002/cne.10417. [DOI] [PubMed] [Google Scholar]
- 44.Feng J, et al. Regulation of neurotransmitter release by synapsin III. Journal of Neuroscience. 2002;22:4372–4380. doi: 10.1523/JNEUROSCI.22-11-04372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kao HT, Porton B. Synaptic vesicle associated proteins and schizophrenia. In: Javitt D, Kantrowitz J, editors. Schizophrenia. 3rd. Springer; 2009. pp. 267–284. [Google Scholar]
- 46.Gordon Frankle W, Lerma J, Laruelle M. The synaptic hypothesis of schizophrenia. Neuron. 2003;39(2):205–216. doi: 10.1016/s0896-6273(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 47.Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Molecular Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. [DOI] [PubMed] [Google Scholar]
- 48.Föcking M, et al. Proteomic and genomic evidence implicates the postsynaptic density in schizophrenia. 1–9. Mol Psychiatry. 2015;20(4):424–432. doi: 10.1038/mp.2014.63. [DOI] [PubMed] [Google Scholar]
- 49.Wen Z, et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature. 2014;515:414–418. doi: 10.1038/nature13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshimizu T, et al. Functional implications of a psychiatric risk variant within CACNA1C in induced human neurons. Mol Psychiatry. 2015;20(2):162–169. doi: 10.1038/mp.2014.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pfisterer U, et al. Efficient induction of functional neurons from adult human fibroblasts. Cell Cycle. 2011;10:3311–3316. doi: 10.4161/cc.10.19.17584. [DOI] [PubMed] [Google Scholar]
- 52.Dittgen T, et al. Lentivirus-based genetic manipulations of cortical neurons and their optical and electrophysiological monitoring in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:18206–18211. doi: 10.1073/pnas.0407976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Hooijdonk LWA, et al. Lentivirus-mediated transgene delivery to the hippocampus reveals sub-field specific differences in expression. BMC Neurosci. 2009;10:2. doi: 10.1186/1471-2202-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vicario-Abejón C. Long-term culture of hippocampal neurons. Current Protocols in Neuroscience. 2004:3.2.1–3.2.13. doi: 10.1002/0471142301.ns0302s26. [DOI] [PubMed] [Google Scholar]
- 55.Cetin A, Komai S, Eliava M, Seeburg PH, Osten P. Stereotaxic gene delivery in the rodent brain. Nature Protocols. 2006;1:3166–3173. doi: 10.1038/nprot.2006.450. [DOI] [PubMed] [Google Scholar]
- 56.Morris R. Developments of a water-maze procedure for studying spatial-learning in the rat. Journal of Neuroscience Methods. 1984;11:47–60. doi: 10.1016/0165-0270(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 57.Moy SS, et al. Sociability and preference for social novelty in five inbred strains: an approach to assess autistic-like behavior in mice. Genes Brain and Behavior. 2004;3:287–302. doi: 10.1111/j.1601-1848.2004.00076.x. [DOI] [PubMed] [Google Scholar]
- 58.Peca J, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–U534. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reid CA, Clements JD. Postsynaptic expression of long-term potentiation in the rat dentate gyrus demonstrated by variance-mean analysis. Journal of Physiology-London. 1999;518:121–130. doi: 10.1111/j.1469-7793.1999.0121r.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Colino A, Malenka RC. Mechanisms underlying induction of long-term potentiation in rat medial and lateral perforate paths in vitro. Journal of Neurophysiology. 1993;69:1150–1159. doi: 10.1152/jn.1993.69.4.1150. [DOI] [PubMed] [Google Scholar]
- 61.McNaughton BL. Evidence for 2 physiologically distinct perforant pathways to the fascia dentata. Brain Research. 1980;199:1–19. doi: 10.1016/0006-8993(80)90226-7. [DOI] [PubMed] [Google Scholar]
- 62.Pasquinelli AE. NON-CODING RNA MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nature Reviews Genetics. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 63.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Research. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krek A, et al. Combinatorial microRNA target predictions. Nature Genetics. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 65.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 66.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Research. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boyle EI, et al. GO::TermFinder - open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(−Delta Delta C) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 69.Gaffield MA, Betz WJ. Imaging synaptic vesicle exocytosis and endocytosis with FM dyes. Nature Protocols. 2006;1:2916–2921. doi: 10.1038/nprot.2006.476. [DOI] [PubMed] [Google Scholar]
- 70.Cousin MA. Use of FM1–43 and other derivatives to investigate neuronal function. In: Crawley Jacqueline N, et al., editors. Current protocols in neuroscience / editorial board. Unit 2.6–Unit 2.6. Chapter 2. 2008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Supplementary data of human induced neurons. a. Transcriptional luciferase assay for the four SNPs in human neuroblastoma cell line SH-SY5Y, F(4) = 7.87, ***p < 0.001, posthoc analysis: tpGL3-rs2660304 (4.482), ***p < 0.001, r = 0.91; trs2660304-rs2802535 (−4.57), ***p < 0.001, r = 0.92; trs2660304-rs1625579 (−4.57), ***p < 0.001, r = 0.92; tpGL3-rs2660304 (1.65), nsp = 0.4781, r = 0.64, n: three experiments. b. Fold-change differences of MIR137 determined by qRT-PCR from Fig. 1c, separated by its individual lines, WiN= 65, *p = 0.0157, r = −0.40, box plot shows, in ascending order, the lowest maximum value, the first quartile, median, third quartile and the highest maximum value, dots are outlier. c. Summary table of the human fibroblasts and their genotype alleles for the four disease-associated SNPs. First four are homozygous for common major alleles; the last two are homozygous for the minor alleles. d. Examples of successfully induced neurons before FACS. Left: bright field, middle: transduced cells, right: expression of the neuron-specific HSV-CamKII-mCherry construct. Scale bar: 100 μm. Before any iN experiments, cells were visually controlled for successful reprogramming. e. Relative microRNA levels in induced neurons from common major and minor allele SNPs determined by RT-qPCR, tmiR-9(7.87) = −0.2, nsp = 0.8475, r = 0.07, tmiR-19b(8.28) = 0.12, nsp = 0.9047, r = 0.04, tmiR-124(13.89) = −0.16, nsp = 0.8773, r = 0.04, box plots show, in ascending order, the lowest maximum value, the first quartile, median, third quartile and the highest maximum value, dots are outlier, n: number of samples from at least two reprogramming, s.e.m.
Supplementary Figure 2. Supplementary data of presynaptic target genes validation. a. Summary table of putative in silico predicted miR-137 target genes involved in “synaptic transmission”. Red: targets tested in this study. b. Luciferase assay results for additional putative miR-137 targets, light grey and dark orange: miR-137 binding site deletion construct (Δ), WCalb1 = 170, *p = 0.0180, r = −0.43; tCamK2(14.58) = −0.14, nsp = 0.8888, r = 0.04; FNrxn1(3, 80) = 13.89, ***p < 0.001; FStx8(3, 56) = 0.60, nsp = 0.6159; WStxbp5 = 187, nsp = 0.4381, r = −0.13; FSv2a(3, 20) = 0.05, nsp = 0.9831; WSyn2a = 201, nsp = 0.6326, r = −0.07; FSyn2b(3, 74) = 0.72, nsp = 0.545; FSynj1(3, 56) = 1.30, nsp = 0.2845; WSynpr = 278, nsp = 0.1515, r = −0.22; WSyt9 = 121, nsp = 0.2983, r = −0.18; WVamp1 = 119, nsp = 0.1786 r = −0.22; FVamp2(3, 81) = 3.92, *p = 0.0115; WVamp7 = 172, nsp = 0.7637, r = −0.05. c. Example images of FM4–64 recording, white: induced neuron, green: FM4–64, arrow: terminal. ns: not significant, s.e.m.
Supplementary Figure 3. miR-137OE validation and in vivo expression. a–b. Relative miR-137 expression level determined by qRT-PCR. a. in HEK-293T cells, Ty(2) = 44.89, ***p < 0.001, r = 1; b. in primary cortical neuronal culture at DIV14, Ty(3.97) = 4.48, *p = 0.0112, r = 0.91, n: number of experiments. c. Additional examples of hippocampi tile-scans injected with the lentivirus expressing the ΔmiR-137OE or miR-137OE constructs, immunostained for mCherry (magenta) and the nuclei dye DAPI (blue). CA3: Cornu Ammonis region 3. Staining was reliably throughout this study. d. Immunostaining of ΔmiR-137OE or miR-137OE-transduced dentate gyrus for DAPI (blue), mCherry (magenta), glial fibrillary acidic protein (GFAP), and ionized calcium-binding adapter molecule (Iba1). This experiment was repeated in at least three different animals. Scale bars: 100 μm. s.e.m.
Supplementary Figure 4. a. Supplementary data of ultrastructural analysis for miR-137OE and ΔmiR-137OE mice. Black arrow: mCherry gold-particle. Orange arrows: gap within the vesicle pool in miR-137OE synapses, * active zone, scale bar: 100 nm. b. Total number of vesicles for ΔmiR-137OE (black, n = 84) and miR-137OE (orange, n = 112), W = 4294, nsp = 0.2973, r = −0.07, box plot shows, in ascending order, the lowest maximum value, the first quartile, median, third quartile and the highest maximum value, dots are outlier, n: number of analyzed synapses of at least three animals. ns: not significant, s.e.m.
Supplementary Figure 5. Supplementary behavior analysis for miR-137OE and ΔmiR-137OE. a–c. Equivalent levels of locomotion and a trend for anxiety in miR-137OE as measured by a. Open-field arena paradigm: margin distance: t(12.94) = −0.61, nsp = 0.5547, r = 0.17; margin time: t(12.20) = −1.89, nsp = 0.083, r = 0.48; center distance: t(13.83) = 2.55,*p = 0.023, r = 0.57; center time: t(12.20) = 1.89, nsp = 0.083, r = 0.48; total distance: t(12.89) = 1.31, nsp = 0.2117, r = 0.34; horizontal activity: t(13.52) = 1.98, nsp = 0.0685, r = 0.47; vertical activity: t(13.15) = 1.36, nsp = 0.1952, r = 0.35; stereotype: t(13.87) = 2.13, nsp = 0.05167, r = 0.50. b. Light-dark box paradigm: Frequency of exploration: t(9.90) = 0.94, nsp = 0.3709, r = 0.29, time spend in light: t(11.88) = 0.60, nsp = 0.5585, r = 0.17. c. Elevated-plus maze paradigm. Panel from left to right: time spent in closed or open arm, no significant main effect, F(1, 26) = 2.09, nsp = 0.161, tTime ΔmiR-137 (13.66) = 42.0, ***p < 0.001, r = 1, tTime miR-137 (9.86) = 39.68, ***p < 0.001, r = 1; Frequency of entering did not differ between open-closed arm and the condition, F(1, 26) = 0.40, nsp = 0.535, tFrequency ΔmiR-137 (11.49) = 2.49, *p < 0.05, r = 0.59, tFrequency miR-137 (11.49) = 2.49, *p < 0.05, r = 0.59). Distance and velocity, tdistance(12.98) = 0.36, nsp = 0.7259, r = 0.10, tvelocity(12.97) = 0.35, nsp = 0.7298, r = 0.10. d. Nociception response, W = 20.5, nsp = 0.7471, r = −0.09. e. Freezing response after context and before cue in %, t(41.97) = −1.20, nsp = 0.2381, r = 0.18. f. Thigmotaxis, percentage of the trail time and path that were spent in the defined thigmotaxis band (0.8) relative to the diameter of the pool, tTime(24.07) = −0.83, nsp = 0.4165, r = 0.17; tPath(25.57) = −0.15, nsp = 0.8838, r = 0.03. g. Three-chamber social interaction paradigm. Amount of time that the mouse spends to explore, left panel: new versus familiar mouse, F(2, 24) = 22.41, ***p < 0.001, right panel: mouse versus object, F(2, 24) = 45.5, ***p < 0.001. No significant difference for ΔmiR-137OE and miR-137OE in both experimental set-ups, Ffamiliar mouse (2, 48) = 2.41, nsp = 0.10 and Fempty cage (2, 48) = 2.86, nsp = 0.0673. Social paradigm for the familiar paradigm: for ΔmiR-137OE: tfamiliar-middle (−3.81), **p = 0.0023, tfamiliar-new (2.86), *p = 0.02245, tmiddle-new (6.67), ***p < 0.001; for miR137OE: tfamiliar-middle (−5.89), ***p < 0.001, tfamiliar-new (3.44), **p = 0.00574, tmiddle-new (9.33), ***p < 0.001; for the empty paradigm for ΔmiR-137OE: tempty-middle (−6.06), ***p < 0.001, tempty-mouse (3.35), **p = 0.0718, tmiddle-mouse (9.41), ***p < 0.001; for miR-137OE: tempty-middle (−4.03), **p = 0.0013, tempty-mouse (3.19), *p = 0.0106, tmiddle-mouse (7.22), ***p < 0.001, n: number of animals used for each experiment. ns: not significant, s.e.m.
Supplementary Figure 6. Supplementary data for Sponge miR-137 and Sponge Control. a. Additional ultrastructural images of the mossy fiber presynaptic terminals in SpongeControl and SpongemiR-137 mice. Black arrow: gold-particle labeled GFP, * active zone, scale bar: 100 nm. b. Total vesicle number for SpongeControl (black, n = 50) and SpongemiR-137 (blue, n = 43) mice in the presynaptic compartments at the mossy fiber synapse, t(88.82) = −1.80, nsp = 0.0751, r = −0.19, box plot shows, in ascending order, the lowest maximum value, the first quartile, median, third quartile and the highest maximum value, dots are outlier, n: number of analyzed synapses of at least three animals. c. Input-output curve estimated from fEPSP against the fiber volley amplitude at the mossy fiber-CA3 synapse in hippocampal slices from SpongeControl (black, n = 4) and SpongemiR-137 (blue, n = 4), F(1, 129) = 1.46, nsp = 0.229, shaded area: 95% confidence interval, n: number of analyzed hippocampal slices from at least three animals. d. Open-field arena paradigm. Total distance: t(16.5) = −0.72, nsp = 0.4809, r = 0.18; time moving: t(16.33) = −1.01, nsp = 0.3285, r = 0.24; moves/counts: t(17.51) = −0.88, nsp = 0.389, r = 0.21; distance periphery: t(16.37) = −0.18, nsp = 0.8617, r = 0.04; time periphery: t(16.19) = 2.62, *p = 0.0182, r = 0.55; distance center: t(12.89) = −2.80, *p = 0.0151, r = 0.62; time center: t(16.21) = −2.63, *p = 0.0182, r = 0.55. e. Nociception response, t(13.47) = 0.30, nsp = 0.7664, r = 0.08. f. Freezing response after context and before cue in %, t(17.12)= 0, nsp = 1, r = 0, n: number of animals. ns: not significant, s.e.m.
Supplementary Figure 7. Supplementary data of endogenous expression of synaptic target genes and Syt1 restoration (miR-137-Syt1-Venus). a–c. Endogenous target gene expression in the dentate gyrus and the mossy fiber-CA3 pathway of naïve C57BL/6 animals. a. Absolute endogenous mRNA levels measured by RT-qPCR. b–c. Endogenous protein levels of Cplx1, Nsf, Syn3 and Syt1 by b. western blot (cropped. Full-length blots are presented in Supplementary Fig. 9), and c. immunohistochemistry. Vibratome sections, immunolabeled against the mossy fiber pathway marker zinc transporter 3 (ZnT3, green), the target gene (magenta), and stained with nuclei dye DAPI (blue). Scale bar: 10 μm. The observation was repeated in at least three different animals. d. Additional ultrastructural images of the mossy fiber presynaptic terminal of ΔmiR-137OE-Venus (grey), miR-137OE-Venus (red), miR-137OE-Syt1-Venus (green), and ΔmiR-137OE-Syt1-Venus (purple) mice, black arrow: gold particles staining of Venus, red arrows: gap within the vesicle pool in miR-137OE-Venus, * active zone, scale bar: 100 nm. e. Total vesicle number for ΔmiR-137OE-Venus (grey, n=56), miR-137OE-Venus (red, n=53), miR-137OE-Syt1-Venus (green, n=56), and ΔmiR-Syt1-Venus (purple, n=58), H(3) = 77.75, ***p < 0.001. Multiple comparison two-tailed test after Kruskal-Wallis, *p < 0.05, box plot shows, in ascending order, the lowest maximum value, the first quartile, median, third quartile and the highest maximum value, dots are outlier. f. Nociception response measured by mean speed of movement during shock in (cm s–1), H(3) = 6.10, nsp = 0.1068. ns: not significant, s.e.m.
Supplementary Figure 8. Supplementary data of Syt1-KD and Syt1-KDControl in the DG-CA3 synapse, and miR-137OE and ΔmiR-137OE at the entorhinal cortex-DG synapse. a–c. Electrophysiological properties of Syt1 knockdown in acute hippocampal slices, black: Syt1-KDcontrol, brown: Syt1-shRNA. n: number of hippocampal slice preparation from at least 3 animal. a. LTP recording for Syt1-shRNA (n = 5) and Syt-1 control (n = 5), H(1) = 129,44 ***p < 0.001. Arrows: application of either high frequency stimulation or DCG-IV, embedded representative fEPSP traces, grey traces: baseline level before stimulation. Right bar chart: LTP magnitude calculated by averaging fEPSP amplitude during the last 10 min (50–60 min) recording, W = 2681, ***p < 0.001, r = −0.39. b. Input-output curve from fEPSP against the fiber volley amplitude at the mossy fiber-CA3 synapse from Syt1-shRNA (brown, n=5) and Syt-1 control (black, n=5), H(1) = 6.63, *p = 0.0100, shaded area: 95% confidence interval. c. fEPSP amplitude for 1 Hz sustained stimulation for Syt1-shRNA (n = 5) and Syt-1 control (black, n = 5), H(1) = 47.32, ***p < 0.001. Embedded: representative traces, grey traces: response to the 1st stimulation. d. BrdU incorporation in miR-137OE (orange) compared to ΔmiR-137OE (black) in the dentate gyrus, t(4.77) = −3.18, *p = 0.0264. Right panel: immunostaining for BrdU (green) and virus expression of mCherry (red), n: number of animals. b. LTP recording at the molecular layer of the dentate gyrus with stimulation of medial perfornant path in miR-137OE and control ΔmiR-137OE, H(1) = 3.19, nsp = 0.0741, arrows: application of stimulation paradigm, right bar chart: LTP magnitude during the last 10 min (50–60 min) recording, W = 1528, nsp = 0.9285, r = −0.01. c. Input-output curve estimated from fEPSP against the fiber volley amplitude measured at a range of stimulus intensities at the molecular layer of the dentate gyrus in hippocampal slices from ΔmiR-137OE (black, n=12) and miR-137OE (orange, n = 14), F(1,66) = 0.91, nsp = 0.4543, shaded area: 95% confidence interval. n: number of analyzed hippocampal slices from at least three animals, ns: not significant, s.e.m.
Supplementary Figure 9. Full length western blot images. Samples were run on a 4–15% TGX SDS-gradient gel (Bio-Rad Laboratories). Molecular weight (M, in kDa): BLUEstain 2 Protein ladder, 5–245 kDA (Goldbio). Images were taken with the infrared imaging system from Odyseey (Li-cor). This allows staining for two proteins at the same time on the same plot. Light-blue bands indicate oversaturation. a. for Fig. 2e, Left and right panel show the same plot, but with adjustment of brightness and contrast; b. for Fig. 2d, the second and the fourth row are adjustment in brightness and contrast from the plots in the first and the third row, respectively; c. and d. for Supplementary Fig. 7. For Syt1, we always observed in varying intensity a band at ~75 kDa. This band is likely to be unstripped Nsf. We tried to optimize the stripping procedure, however we could never remove the strong signal band for Nsf completely. For consistency, we decided to stain always Syt1 after Nsf.