Abstract
Protein oxidation is involved in regulatory physiological events as well as in damage to tissues and is thought to play a key role in the pathophysiology of diseases and in the aging process. Protein-bound carbonyls represent a marker of global protein oxidation, as they are generated by multiple different reactive oxygen species in blood, tissues and cells. Sample preparation and stabilization are key steps in the accurate quantification of oxidation-related products and examination of physiological/pathological processes. This review therefore focuses on the sample preparation processes used in the most relevant methods to detect protein carbonyls after derivatization with 2,4-dinitrophenylhydrazine with an emphasis on measurement in plasma, cells, organ homogenates, isolated proteins and organelles. Sample preparation, derivatization conditions and protein handling are presented for the spectrophotometric and HPLC method as well as for immunoblotting and ELISA. An extensive overview covering these methods in previously published articles is given for researchers who plan to measure protein carbonyls in different samples.
Keywords: Protein carbonyls; 2,4-Dinitrophenylhydrazine; Spectrophotometry; Immunoblotting; ELISA; Sample preparation
Abbreviations: AP, alkaline phosphatase; BAL, bronchoalveolar; BCIP, 5-bromo-4-chloro-3-indolyl phosphate; BHT, butylated hydroxytoluene; DMSO, dimethyl sulfoxide; DNP, 2,4-dinitrophenyl; DNPH, 2,4-dinitrophenylhydrazine; DTT, dithiothreitol; EDTA, ethylenediaminetetraacetic acid; ELISA, enzyme-linked immunosorbent assay; HCl, hydrochloric acid; HPLC, high performance liquid chromatography; HRP, horseradish peroxidase; Hsf1, heat shock factor 1; IgG, immunoglobulin G; KLH, keyhole limpet hemocyanin; n.a., not available; NBT, nitro blue tetrazolium; PVDF, polyvinylidene difluoride; RT, room temperature; SDS, sodium dodecyl sulfate; TCA, trichloroacetic acid; TFA, trifluoracetic acid
Graphical abstract
Highlights
-
•
Derivatizing protein carbonyls with dinitrophenylhydrazine is very common.
-
•
For the measurement of protein carbonyls in plasma, cells, organ homogenates and isolated proteins are a wide variety of sample preparation and handling protocolls used.
-
•
ELISA seems best-suited for high-throughput clinical studies.
-
•
Immunoblotting is especially applicable in cell culture studies.
-
•
Existing protocols must be further improved and standardized.
Introduction
In recent years, more and more evidence has arisen that oxidative processes play a key role in the pathophysiology of many diseases and in the aging process. Besides regulatory events, a plethora of damaging effects is induced by oxidative processes, one of these damaging processes is protein oxidation [1]. Oxidative damage to proteins results in a multitude of products (for reviews see [2–6]), arising from modification of a wide range of amino acids. These include damage to sulfur-containing, aromatic, and aliphatic amino acids [7–10] (Table 1). Protein carbonyls represent an irreversible form of protein modification and have been demonstrated to be relatively stable (degradation/clearance in hours/days) in contrast to lipid peroxidation products that are removed within minutes [11,12]. In addition, protein carbonyls are formed early during oxidative stress conditions and are not a result of one specific oxidant, thus they can be called a marker of overall protein oxidation. Due to the great variety of different modifications [13], one obstacle in the detection of protein-related oxidative stress biomarkers is the requirement of complex procedures for their determination. Furthermore, the instability of some of these products as a result of repair processes (in the case of methionine sulfoxides) [14] and by peroxiredoxins and disulfide reductases [15] can contribute to difficulties in assessing and quantifying oxidation status. It is worth pointing out that some of the formed species resulting from protein oxidation (formaldehyde, acetaldehyde, acetone, etc.) do not remain protein bound but are released. Hence they are not detected by any assays that involve protein separation or precipitation. Depending on the radical treatment used, released carbonyls can however be major products [16].
Table 1.
Possible oxidation products of amino acid residues resulting in protein carbonyl formation.
| Amino acid | Oxidation products |
|---|---|
| Proline | Glutamic semialdehyde and other ring opened products |
| Arginine | Glutamic semialdehyde and other side-chain products |
| Lysine | Aminoadipic semialdehyde and other side-chain products |
| Threonine | Carbonyls formed at side chain sites |
| Methionine | Methional |
| Tryptophan | N-formylkynurenine, kynurenine |
| Histidine | 2-Oxo-histidine and ring-opened species |
| Alanine | Formaldehyde and carbonyls from methyl group |
| Valine | Acetone, formaldehyde and carbonyls on side-chain methyl groups |
| Leucine | Isobutyraldehyde, acetone, formaldehyde, and carbonyls on side-chain |
| Aspartate | Glyoxylic acid |
| Isoleucine | Formaldehyde, carbonyls on side-chain |
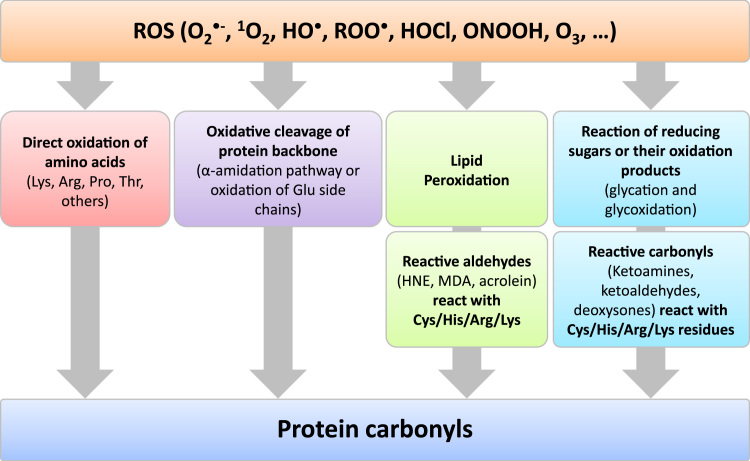
Various oxidants may attack several amino acids and are thus able to produce both protein-bound and released carbonyl groups (Table 1 and Fig. 1). The yield of these species is however oxidant dependent. Due to the structure of normal amino acids it is expected, that no carbonyl groups are part of a native protein. However, this seems to be a simplifying assumption since many proteins undergo (enzymatic) post-translational modifications where carbonyl groups might be introduced into the native, functional protein. The presence of such structures could be the reason for the high basal (not stress-induced) level of protein carbonyls found in some proteins. Furthermore, inappropriate sample handling might contribute to elevated concentrations observed in some studies.
Fig. 1.
Protein oxidation resulting in protein carbonyl formation. Reactive oxygen species (ROS) may either react directly with some amino acid residues or lead to oxidative cleavage of the protein backbone. Other possible formation routes of protein carbonyls are via the oxidation of lipids resulting in reactive aldehydes which react with cysteine (Cys), histidine (His), arginine (Arg) and lysine (Lys) residues and thus introduce carbonyl groups and furthermore via the reaction of reducing sugars or their oxidation products with the same residues.
The most commonly used marker to assess protein oxidation is via the determination of protein-bound carbonyls. Protein carbonyls can be detected by various methods, all relying on the derivatization of the carbonyl group. The reduction with radiolabeled borohydride introduces a measurable radiolabel into the protein, whereas several hydrazine derivatives, most commonly 2,4-dinitrophenylhydrazine (DNPH, Fig. 2) or biotin hydrazine, introduce detectable functional groups into the oxidized protein. So, the most often used procedure to detect protein carbonyls is after their derivatization with DNPH. During the last three decades most of these methods have referred to the basic methods described by Levine et al. [17,18] using the highly-sensitive DNP-modification of protein carbonyls followed by a detection either by spectrophotometric methods, by an HPLC-based technique or using anti-DNP antibodies in immunoblotting [19] or ELISA [20] (see Fig. 3). In addition to this, proteomic techniques have been applied to get a more detailed insight into the mechanism of protein damage, e.g. in blood [21].
Fig. 2.
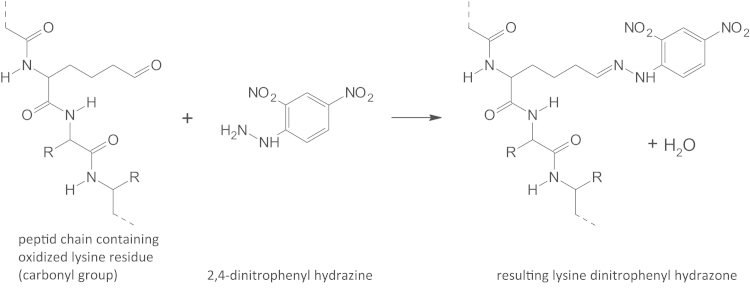
Reaction of protein carbonyl group with 2,4-dinitrophenylhydrazine. The nucleophilic addition, also called condensation reaction, resulting in a 2,4-dinitrophenyl hydrazone is shown for an oxidized lysine residue (aminoadipic semialdehyde). Note that the reaction is accompanied by the loss of one molecule of water.
Fig. 3.
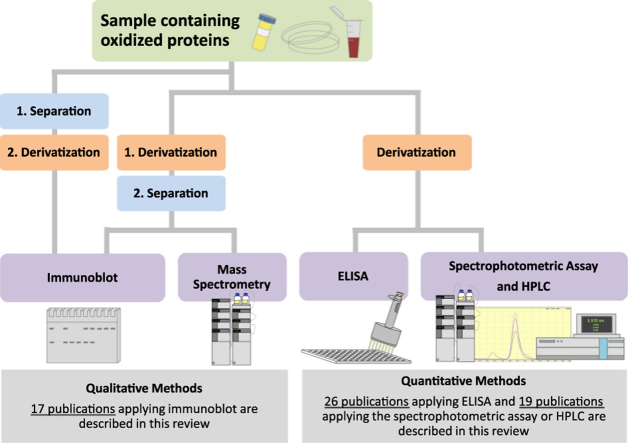
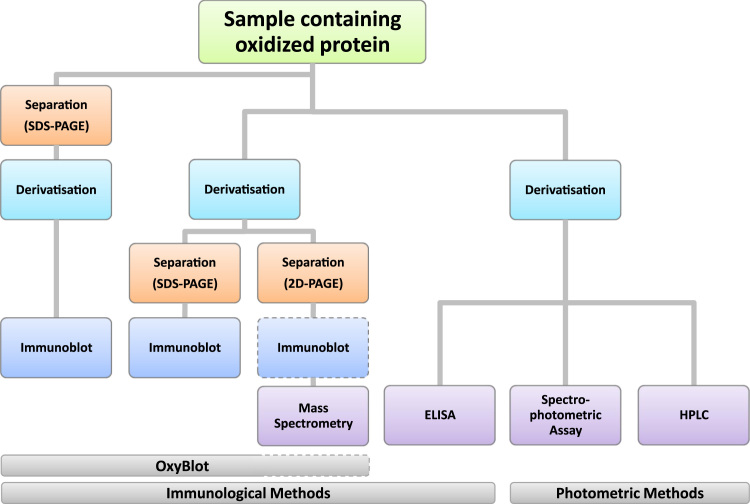
Flow chart of possibilities to analyze protein carbonyls after DNP derivatization. Several options exist to detect protein carbonyls in a sample, either quantitatively or qualitatively, depending on the research question and laboratory equipment/facilities. The left-hand side shows several ways of the so-called OxyBlot. The proteins can be separated before derivatization or the other way around. Following separation/derivatization, immunoblot is carried out. If separation is performed with 2D PAGE, the immunoblot is not mandatory and is usually followed by mass spectrometry. The right hand side depicts more quantitative methods: the spectrophotometric assay, ELISA and HPLC where standards are commonly used to assess the exact concentration of protein carbonyls in the sample.
In the following we will concentrate on the determination of protein carbonyls in plasma, cell culture, organ homogenate and isolated protein/organelle samples by the methods of Levine et al. [17,18], Shacter et al. [19], Keller et al. [22] and Buss et al. [20].
As described above, DNP-derivatized proteins can be detected by different methods; hence every laboratory should be able to detect carbonyl groups either by the simple spectrophotometric assay or by more complex procedures. The fact that no special equipment is needed for the analysis of DNP-derivatized proteins has led to the application of these methods in numerous publications. The search term “protein carbonyl” leads to more than 15,000 publications on PubMed and “protein carbonyl assay” still leads to around 6000 results (accessed in April 2015).
Search strategy
PubMed was accessed to view the publications of Levine [17,18], Shacter et al. [19], Keller et al. [22] and Buss et al. [20]. Following this, on the bottom right hand side, the link “Cited by xx PubMed Central articles” was followed to view all publications citing one of these original method publications.
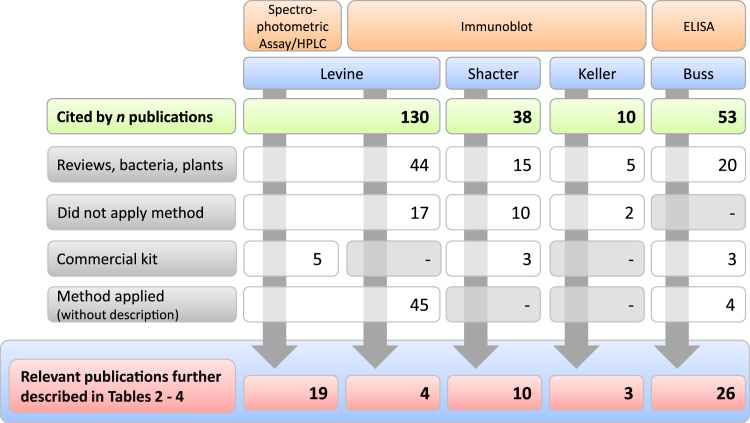
At the time PubMed was accessed in November 2013, the publication of Levine [17] was cited by 130 publications. Forty-four of these were reviews or reported on research carried out with bacteria, plants, yeast, or non-human primates. Seventeen publications referred to the method or the results in general in the introduction or in the discussion of the publication and thus did not apply the method. Five publications described using the commercial kit (OxyBlot). Another 45 publications cited the spectrophotometric, HPLC method or Western blot without any modifications or specific comments on sample preparation and analytical condition; while 19 publications described in more detail the reaction conditions and four publications the sample handling for the spectrophotometric assay and the immunoblotting, respectively (see Table 2 and Fig. 4).
Table 2.
Published sample preparation for spectrophotometric determination of protein carbonyls (n.a.: not available, ratio of sample volume to DNP volume).
| Species | Sample preparation | Derivatization conditions | Protein handling | Wave-length | Levela | Refs. |
|---|---|---|---|---|---|---|
| (Precipitation, washing, dissolving) | ||||||
| n.a. | Oxidatively modified proteins (>0.5 mg) | 10 mM DNPH (in 2 M HCl or in 6 M guanidine-HCl, pH 2.5)ratio n.a.1 hn.a. if under dark conditions | 20% TCA | Spectrum (360–390 nm) | 0.2–1.0 mol/mol protein | [17,18] |
| Ratio n.a. | Washing with ethanol–ethyl acetate (1:1) | |||||
| 1 h | 6 M guanidine | |||||
| n.a. if under dark conditions | ||||||
| Human | Heparin plasma | DNPH (concentration n.a.) | n.a. | 360 nm (280 nm) | 1.02 nmol/mg | [32] |
| −80 °C | Ratio n.a. | Washing n.a. | ||||
| Dilution n.a. | Conditions/duration n.a. | Guanidine n.a. | ||||
| Human | Serum | 10 mM DNPH (in 2.5 M HCl) | 20% TCA | n.a. | 2.4±0.21 nmol/mg | [33] |
| −80 °C | Ratio n.a. | Washing with ethanol–ethyl acetate (1:1) | 22,000 M−1/cm | |||
| 1:100 dilution | Duration n.a. | 6 M guanidine | ||||
| Decomplementation at 56 °C for 30 min | n.a. if dark | |||||
| Human | Plasma | n.a. | n.a. | n.a. | 0.48–0.7 nmol/mg | [34] |
| −80 °C | In 96-well plates | |||||
| Concentration n.a. | ||||||
| Human | Plasma of children with juvenile chronic arthritis | 20 mM DNPH | 20% TCA | 360 nm | Patients vs controls: 1.36±0.68 vs 0.807±0.16 nmol/mg | [35] |
| Undiluted plasma | Ratio (1:1) | Washing with ethanol–ethyl acetate (1:1) | ||||
| 1 h | 6 M guanidine | |||||
| Human | Mesenchymal stem cell (hMSCs) suspensions | 10 mM DNPH | 20% TCA (v/v) | 360 nm | 20–70 µmol/103 cells | [36,37] |
| 30% TCA precipitate resuspended in DNPH | 1 h (37 °C) | Washing with ethanol–ethyl acetate (1:1) | ||||
| Concentration n.a. | Guanidine in 2 mM phosphate buffer | |||||
| Human | Acute promyelocytic leukemia (APL)-derived NB-4 cells soluble protein fraction with protease inhibitors, sonicated | 10 mM DNPH | 20% TCA | 370 nm | ~3.5 nmol/mg | [38] |
| Stored at −20 °C | Ratio (1:6) | Washing with ethanol–ethyl acetate (1:1) | ||||
| Concentration n.a. | 1 h (20 °C) in the dark | 6 M guanidine | ||||
| Mouse | Neutrophil cell lysate in 50 mM potassium phosphate (pH 7.4) | High DNPH (in 2 M HCl) | 20% TCA | 370 nm | 10–30 nmol/mg | [28] |
| Concentration n.a. | Ratio n.a. | Washing with ethanol–ethyl acetate (1:1) | ||||
| 1 h in the dark | 6 M guanidine | |||||
| Mouse | Peritoneal macrophage cell lysate in 50 mM potassium phosphate (pH 7.4) | High DNPH (in 2 M HCl) | 20% TCA | 370 nm | 5–12 µmol/mg | [27] |
| Concentration n.a. | Ratio n.a. | Washing with ethanol–ethyl acetate (1:1) | ||||
| 1 h in the dark | 6 M guanidine | |||||
| Conditions n.a. | ||||||
| Mouse | Brain cortex homogenate extracts in phosphate buffer (pH 7) with DTT and EDTA | DNPH | n.a. | 370 nm in 96-well plate | 0.2–0.4 nmol/mg | [39] |
| Streptomycin substrate (10%) to remove nucleic acids | Ratio, duration and dark conditions n.a. | |||||
| Concentration n.a. | ||||||
| Mouse | Lysate of primary cortical neurons in phosphate buffer (pH 6.5) | 1× DNPH (in 2 M HCl) | 10% TCA | 375 nm | Expressed as fold-increase to controls | [40] |
| Lysis and storage n.a. | Ratio n.a. | Washing with ethanol–ethyl acetate (1:1) | ||||
| Concentration n.a. | 20 min at RT | 6 M guanidine in 20 mM sodium phosphate buffer (pH 6.5) | ||||
| Mouse | Serum and kidney homogenate (in PBS, pH 7.4, containing protease inhibitor cocktail) | 10 mM DNPH (in 2.5 M HCl) | 20% TCA | 370 nm | 3-Fold increase in protein carbonyls in mice treated with tetrachlorethene | [41] |
| Undiluted serum | Ratio (1:4) | Washing with ethanol–ethyl acetate (1:1) | ||||
| 1 h at RT in the dark | 6 M guanidine | |||||
| Rat | EDTA plasma of Sprague Dawley rats | DNPH (concentration n.a.) | 20% TCA | 380 nm | ~3 nmol/mg | [42] |
| −20 °C storage | Ratio n.a. | Washing n.a. | ||||
| Dilution n.a. | 1 h | Guanidine n.a. | ||||
| n.a. if dark | ||||||
| Cow | BSA | 10 mM DNPH | 20% TCA | 360 nm (scan 320–450 nm) | Method description | [43] |
| Concentration n.a. | Ratio (1:5) | Washing with ethanol-ethyl acetate (1:1) | ||||
| 1 h at RT | Solubilization in 0.2% (w/v) SDS/20 mM Tris–Cl, pH 6.8 | |||||
| Cow | Aortic endothelial cells | 0.2% DNPH (≙ 10 mM) (in 2 M HCl) ratio n.a.for 1 h in the dark | 20% TCA | 370 nm | µmol carbonyl/mg but expressed as percent to control | [44] |
| 800 µg protein | Ratio n.a. | Washing with ethanol–ethyl acetate (1:1) | ||||
| For 1 h in the dark | 6 M guanidine | |||||
| Cow | Oxidized BSA | 10 mM DNPH (in 2 M HCl) | 20% TCA | 376 nm | Standard curve: 2–27 nmol/mg | [22] |
| 1 mg | Ratio (1:1) | Washing with ethanol–ethyl acetate (1:1) | ||||
| 1 h at RT | 6 M guanidine | |||||
| Horse | Heart myoglobin | 10 mM DNPH (in 2.5 M HCl) | Centrifugation | 400 nm | After TEMPOL incubation the myoglobin N-terminus underwent oxidative deamination in which the Gly N-terminal amino group was replaced with a free carbonyl group | [45] |
| Concentration n.a. | Ratio (1:1) | PD-10 column | HPLC analysis | |||
| 15 min in the dark | ||||||
| Hamster | Liver supernatant | 10 mM DNPH (in 2.5 M HCl) | 20% TCA | 370 nm | 2.25–3.74 nmol/mg | [46] |
| Concentration n.a. | Ratio (1:1) | Washing with ethanol–ethyl acetate (1:1) | ||||
| 30 min at RT | Dissolved 6% SDS |
Some levels could only be drawn from figures. The concentration is therefore not the exactly measured one.
Fig. 4.
Final number of publications highlighted in Tables 2–4. As described in the search strategy, PubMed was accessed in November 2013 to view the publications of Levine, Shacter et al., Keller et al., and Buss et al. as well as all publications citing one of these original method publications. The extracted publications were grouped into categories (review, bacteria, plants; did not apply method; commercial kit; method applied but not described; relevant publication describing sample preparation etc.).
In November 2013, the method description for immunoblotting by Emily Shacter et al. was cited by 38 publications. Fifteen of these publications were reviews or did not relate to human or cell culture studies. Ten publications referred to results but not to the method itself. Three publications applied a commercial kit. Ten publications described the method with or without slight modifications (see Table 3 and Fig. 4).
Table 3.
Measurements of protein carbonyls by immunoblotting.
| Species | Sample preparation | Derivatization | Gel, PAGE, membrane proteins amount, transfer/blot | Antibody | Level/result | Refs. |
|---|---|---|---|---|---|---|
| Ratio (sample/DNP) | ||||||
| Conditions | ||||||
| Human | Heparin plasma | 10 mM DNPH (in 10% TCA) | SDS-PAGE (4–12% gradient) | Monoclonal anti-DNP IgE (Sigma), biotinylated rat anti-mouse IgE | Method description, different oxidized proteins | [19] |
| Nitrocellulose membrane | (Southern Biotechnologies), and biotin–avidin-peroxidase complex (Vector Laboratories) | |||||
| Blotting by standard procedures | ||||||
| Human | Serum | Post-electrophoresis derivatization | 10% SDS-PAGE | Anti-DNP (Molecular Probes) | To compare pre- and post-electrophoresis derivatization | [47] |
| 0.5 mM DNPH | Protein amount n.a. | Goat anti-rabbit HRP-conjugate (Sigma) | ||||
| Nitrocellulose and PVDF membranes | ||||||
| Human | Lysed SH-SY5Y cell supernatant | 20 mM DNPH (in 2 M HCl) sample in 12% SDS (1:1) | SDS-PAGE (10% and 15%) | Polyclonal rabbit anti-DNP (Invitrogen) | Exposure of Sphingosine Kinase 1 to TNFα caused substantial carbonylation | [48] |
| Ratio (1:1) | 30 µg soluble and 5 µg purified protein | Anti-rabbit HRP-conjugate | ||||
| For 45 min at 25 °C | PVDF membrane | |||||
| Precipitation of protein in chloroform/methanol | ||||||
| Rabbit | Purified muscle actin, total protein of SH-SY5Y cell extracts, and actin immune-precipitated from SH-SY5Y cells centrifuged soluble protein from cell extracts | 20 mM DNPH (in 2 M HCl) | SDS-PAGE and 2D Nu-PAGE (4–12%) | Polyclonal rabbit anti-DNP (Invitrogen) | Considerable actin carbonylation in SH-SY5Y cells acutely exposed to TNFα or IL-1β | [49] |
| Sample in 12% SDS (1:1) | 30–50 µg soluble and 5 µg purified protein | Secondary Ab n.a. | ||||
| (30 min, 25 °C) | PVDF membrane | |||||
| Ratio (1:1) | ||||||
| Human | Acute promyelocytic leukemia (APL)-derived NB-4 cells soluble protein fraction | 10 mM DNPH | 12% gel | Anti-DNP | Treatment of NB4 cells with increasing concentrations (0.5–6 M) of As2O3 leads to increased protein carbonyls | [38] |
| Sample in lysis buffer containing 6% SDS (pH 7.2) | Ratio (1:1, 15 µl:14 µl) | Nu-PAGE | AP-conjugated anti-rabbit IgG | |||
| For 10 min at RT | 25 µg protein | |||||
| Nitrocellulose membrane | ||||||
| Human | HeLa cell extract | 10 mM DNPH | Dot-Blot microfiltration apparatus and TransBlot transfer membrane (Bio-Rad) | Anti-DNP (Dako) | Slightly increased carbonylation in cells lacking glutaredoxin 1 | [50] |
| 1 µg/µl | Ratio (1:4) | AP-conjugated anti-rabbit (Dako) | ||||
| 30 min at RT | ||||||
| Human | Alzheimer’s disease brain homogenates | 20 mM DNPH | % n.a. | Rabbit anti-DNP (Millipore) | Different proteins carbonylated in AD on comparison to age-matched controls | [51] |
| Ratio (1:4) | SDS-PAGE | Goat anti-rabbit IgG (Sigma) | ||||
| 20 min at 25 °C | 150 µg protein | |||||
| Nitrocellulose membrane | ||||||
| Semi-dry transfer | ||||||
| Human | Bronchoalveolar lavage (BAL) fluid | OxyBlot kit (Chemicon Inc.) | 10% SDS-PAGE | Anti-DNP | Amount of carbonylated albumin per mg total albumin in BAL fluid was four times higher in older current smokers and three times higher in older former smokers than in age matched non-smokers | [52] |
| Human | Colon cells (SW620) | Post-electrophoresis derivatization: 10 µg/ml DNPH (≙0.5 mM) in 2 M HCl | 5 µg protein/slot | Anti-DNP-KLH, rabbit IgG (H&L) fraction (Molecular Probes) | Development of an immunochemical technique for the quantification of carbonyl groups in protein samples from small tissue samples and cell cultures | [24] |
| min at RT | PVDF membrane | Peroxidase-conjugated F(ab′)2 fragment donkey anti-rabbit IgG (H&L) fraction (Jackson Immunoresearch Laboratories) | ||||
| Slot-blot | ||||||
| Mouse and human | C57BL mouse tissue and human fibroblasts | DNPH/TFA from OxyBlot Kit (Oncor) | % n.a. | From the OxyBlot Kit | Improve methods | [53] |
| Ratio (1:1) | SDS-PAGE | |||||
| 15 min at RT | Protein amount n.a. | |||||
| Nitrocellulose and PVDF membrane | ||||||
| Semi-dry blotting | ||||||
| Mouse | Cortical neuron homogenate and lysate | n.a. | 12.5% SDS-PAGE | Anti-DNP kit (Chemicon Inc.) | Relative to control | [40] |
| 20 µg | 20 µg | |||||
| Nitrocellulose membrane | ||||||
| Mouse | Isolated collagen | 0.5 mM DNPH | 6% slab gel | Rabbit anti-DNP (Sigma) | Induced damage to collagen after treatment with pefloxacin | [54,55] |
| Ratio (1:1) | SDS-PAGE | AP-conjugated goat anti-rabbit IgG (Sigma) | ||||
| 1 h at RT | 50, 15 or 7 µg collagen | |||||
| PVDF membrane (Sigma) | ||||||
| Trans-Blot apparatus | ||||||
| Mouse | Mitochondria from Heat Shock factor 1 (Hsf1) knockout mice | 10 mM DNPH | % n.a. | Rabbit anti-DNP (Sigma) | Higher extent of carbonylation in mitochondrial proteins of Hsf1 knockout mice | [56] |
| Ratio n.a. | SDS-PAGE | HRP-labeled goat anti-rabbit IgG | ||||
| 60 min at RT | Protein amount n.a. | |||||
| PVDF membrane | ||||||
| Mini trans-blot electrophoretic transfer cell (Bio-Rad) | ||||||
| Rat | Serum | 20 mM DNPH (in 10% TFA) | % n.a. | Goat anti-DNP (Bethyl Laboratories Inc.) | Changes in serum levels of total protein carbonylation correspond to cardio-protective activity | [57] |
| Ratio (1:1) | SDS-PAGE | Donkey anti-goat IRDye 800CW (Li-COR) | ||||
| 10 min at RT | 5 µg protein | |||||
| PVDF membrane | ||||||
| Rat | Mitochondria from head homogenate (male, weanling Sprague-Dawley rats) | OxyBlot Kit | 10% SDS-PAGE | Anti-DNP (OxyBlot Kit) | Comparison between rats with copper deficient diets (reduced carbonyls in copper deficiency) | [58] |
| 6 µg protein | HRP-coupled anti-sheep IgG (Amersham) | |||||
| PVDF membrane | ||||||
| Semi-dry transfer | ||||||
| Rat | Ovary tissue sections | 10 mM DNPH (in 10% TFA) | 12% SDS-PAGE | Rabbit anti-DNP | Increased protein carbonyls were detected in ovaries of rats exposed to tetrachlorethylene water for 2 weeks compared to controls | [59] |
| Ratio n.a. | 25 µg protein | Biotinylated anti-rabbit (Vectastain ABC-AP kit) | ||||
| 45 min | PVDF membrane | |||||
| Tank blot | ||||||
| Rat and cow | Histones from PC12 cells (rat) and thymus, liver, spleen (bovine) | 10 mM DNPH (in 10% TFA) | % n.a. | Anti-DNP (Dako) | Higher carbonylation in untreated histone H1 in comparison to core histone | [60] |
| Ratio (1:1) | SDS-PAGE | Goat anti-rabbit conjugated with HRP (Jackson Immunoresearch Laboratories) | ||||
| 20 min at RT | 5 µg protein | |||||
| Nitrocellulose membrane | ||||||
| Semi-dry blot | ||||||
| Cow | Oxidized BSA | 0.5 mM DNPH | % n.a. | Anti-DNP antisera (Dako) | Development of an immunochemical assay | [22] |
| Ratio (1:1) | SDS-PAGE | Mouse anti-rabbit IgG conjugated with AP (Jackson Immunoresearch Laboratories) | ||||
| 1 h at RT | Protein amount n.a. | |||||
| Nitrocellulose membrane | ||||||
| E. coli | Glutamine synthetase | 20 mM DNPH and 0.5% TFA in 92.5% DMSO | Dot-blot and Western blot (14% and 8–16%) | Goat anti-DNP (Bethyl Laboratories Inc.) | Comparison between dot blot and Western blot | [61] |
| Ratio n.a. | PVDF membrane | Donkey anti-goat IRDye 800CW (Li-COR) | ||||
| 15 min at RT |
The method by Keller et al. [22] was cited by 10 publications. Five of these were reviews or used yeast/housefly models. Two publications did not apply the method. Three publications described applying the method of Keller et al. [22] and Shacter et al. [19].
Some authors cited more than one reference/method when describing the immunoblotting technique thus resulting in less relevant publications in Table 3 than observed in Fig. 4.
The method description of Hendrikje Buss et al. [20] was cited by 53 publications when accessed in November 2013. Twenty of these publications only referred to the publication in the introduction or discussion or did not describe using plasma/serum or cell culture samples. Three publications described using a commercial kit, while four did not go into further detail concerning the application of the method. Twenty-six publications described the method in detail with no modifications or only slight changes (see Table 4).
Table 4.
Determination of protein carbonyls by ELISA.
| Species | Sample preparation | Derivatization | Antibody | Plate producer | Level* | Ref | |
|---|---|---|---|---|---|---|---|
| Human | Plasma from healthy and critically ill patientsdiluted to 4 mg/ml | 10 mM DNPH (in 6 M guanidine HCl, 0.5 M potassium phosphate buffer, pH 2.5) | Biotinylated anti-DNP (Molecular Probes) | MaxiSorp (Nunc) | 0.06–0.75 nmol/mg | [20,62] | |
| Ratio sample/DNPH (1:4) | Streptavidin-biotinylated-HRP (Amersham) | ||||||
| 45 min at RT | |||||||
| Human | Plasma from sepsis patients vs controls | n.a. | Biotin-conjugated polyclonal anti-DNP IgG | n.a. | 0.32–0.45 nmol/mg | [63] | |
| Dilution n.a. | Streptavidin-conjugated HRP | ||||||
| Human | Citrate-plasma from schizophrenia patients and healthy volunteers | n.a. | n.a. | n.a. | 0.178–0.482 nmol/mg | [64] | |
| Dilution n.a | |||||||
| Human | Heparin and EDTA plasma and urine | n.a. | n.a. | n.a. | Significantly higher concentration in blood of subway workers and bus drivers than in office workers (17 and 18 vs. 15 nmol/ml) | [65] | |
| Dilution n.a. | |||||||
| Human | Plasma from 71 hepatocellular carcinoma patients and 694 controls | n.a. | n.a. | n.a. | 0.11–1.41 nmol/mg | [66] | |
| Dilution n.a. | |||||||
| Human | Plasma of chronic obstructive pulmonary disease (COPD) patients | n.a. | Polyclonal rabbit anti-DNP (Molecular Probes) | MaxiSorp (Nunc) | 17.9±2.9 nmol/mg | [67] | |
| 4 mg/ml | HRP-conjugate (Amersham) | ||||||
| Human | Plasma from participants of the New York Early Lung Cancer Action Project | n.a. | Polyclonal rabbit anti-DNP (Molecular Probes) | n.a. | ~17 nmol/ml | [68] | |
| 4 mg/ml | HRP-conjugate (Amersham) | ||||||
| Human | Plasma | 10 mM DNPH (in 6 M guanidine–HCl, 0.5 M potassium phosphate, pH 2.5) | Biotinylated anti-DNP (Molecular Probes) | ELISA (Corning Costar) | ~11–15.3 nmol/mg | [69] | |
| 4 mg/ml | Ratio and conditions n.a. | Streptavidin–biotin HRP (Amersham) | |||||
| Human | Serum from middle-aged obese subjects | Concentration and conditions n.a. | n.a. | n.a. | 0.12 arb. u. | [70] | |
| 4 mg/ml | Ratio (1:4) | ||||||
| Human | Heparin plasma | 0.05 mM DNPH (in H3PO4, pH 6.2) | Anti-DNP (Sigma) | n.a. | Method for measuring protein carbonyl in samples with low amounts of protein | [25] | |
| 5 µg/ml | 45 min at RT in the dark | Anti-rabbit HRP-linked IgG (H&L, Upstate Cell Signaling) | |||||
| NOTE: derivatization after samples have adsorbed to the plate! | |||||||
| Human | Plasma and LDL | n.a. | Anti-DNP (Dako) | n.a. | Modification of method | [71] | |
| Dilution n.a. | Goat anti-rabbit IgG peroxidase conjugate (Sigma) | ||||||
| Human | Plasma | n.a. | n.a. | n.a. | 0.1–0 5 nmol carbonyl groups fibrinogen/mg of plasma proteins | [72] | |
| Dilution n.a. | |||||||
| Human | Plasma from women with preeclampsia and controls | 45 min at RT | Rabbit anti-DNP-KLH (Invitrogen) | n.a. | Significantly higher concentration in cases than in controls | [73] | |
| Dilution n.a. | HRP-conjugated porcine anti-rabbit IgG (Dako A/S) | ||||||
| Human | Plasma | n.a. | Biotinylated anti-DNP (Molecular Probes) | n.a. |
|
[74] | |
| 4 mg/ml | Streptavidin-biotinylated HRP-conjugate (Amersham) | ||||||
| Human | K562 cell lysates (human chronic myelogenous leukemia) | n.a. | Rabbit anti-DNP IgG antiserum (Sigma) | n.a. | Relation to control | [75] | |
| 1 mg/ml in lysis buffer with 1 mM BHT | Monoclonal anti-rabbit peroxidase-conjugated IgG (Sigma) | ||||||
| Human | Parenchymal lung tissue of current smokers with chronic obstructive pulmonary disease | n.a. | Plate was pre-incubated with mouse anti-HSA before addition of samples | n.a. | 0.5–5 carbonyl residues/HSA molecule | [76] | |
| Dilution n.a. | rabbit anti-DNP | ||||||
| Anti-rabbit HRP-conjugate | |||||||
| Human | MRC-5-fibroblast cell lysates (fetal lung) | n.a. | Anti-DNP rabbit-IgG-antiserum (Sigma) | n.a. | ~1.5–3.2 nmol/mg | [77] | |
| 4 mg/ml | Monoclonal anti-rabbit-IgG-peroxidase conjugated (Sigma) | ||||||
| Mouse | Brain, liver, heart and spleen homogenates from 6-week-old male DDY mice | 10 mM DNPH (in 2 M HCl) | n.a. | n.a. | Relation to control | [78] | |
| 1 mg/ml | For 1 h at RT | ||||||
| Ratio n.a. | |||||||
| Mouse | Heart tissue homogenate of SAMP8 mice | n.a. | Rabbit anti-DNP IgG-antiserum (Sigma Aldrich) | n.a. | 664±37 nmol/g (high-polyphenol diet) and 958±70 (low-polyphenol diet) nmol/g | [79] | |
| Dilution n.a. | Monoclonal anti-rabbit IgG peroxidase conjugate (Sigma Aldrich) | ||||||
| Mouse | Mesenchymal stem cells derived from adipose tissue of C57/Black6 mice | n.a. | n.a. | n.a. | 4–32 Carbonyls/mg protein | [80] | |
| Dilution n.a. | |||||||
| Mouse | Brain homogenate from ApoD-knockout mice | n.a. | Biotinylated anti-DNP (Molecular Probes) | n.a. | Relation to control | [81] | |
| Dilution n.a. | Streptavidin-biotinylated HRP (Amersham) | ||||||
| Mouse | Isolated mitochondria from livers of Bcs1lG/G mice | Concentration and ratio n.a. | Anti-DNP (Invitrogen) | n.a. | 1.09±0.36 relative amount | [82] | |
| Dilution n.a. | 45 min at RT | Swine anti-rabbit IgG-HRP (Dako A/S) | |||||
| Mouse | Retinal pigmented epithelial cell lysates | 10 mM DNPH (in 6 M guanidine–HCl, 0.5 M potassium phosphate, pH 2.5) | Polyclonal rabbit anti-DNP (Molecular Probes) | MaxiSorp (Nunc) | 0.6–1.2 nmol | [83] | |
| 4 mg/ml | Ratio (1:4) | HRP-conjugate (Amersham) | |||||
| 45 min at RT | |||||||
| Mouse | RAW264.7 murine macrophage-like cells | Kit from Cell Biolabs | Kit from Cell Biolabs | Kit from Cell Biolabs | Results not significant | [84] | |
| Dilution n.a. | |||||||
| Mouse | HT22 cell lysates4 mg/ml in lysis buffer with 1 mM BHT | n.a. | Rabbit anti-DNP IgG antiserum (Sigma) | n.a. | 6.5–9.0 pmol/mg | [85] | |
| Monoclonal peroxidase-conjugated anti-rabbit IgG (Sigma) | |||||||
| Rat | Ileal mucosa of salmonella infected rats | n.a. | Biotinylated anti-DNP (Molecular Probes), streptavidin-biotinylated HRP (Amersham) | MaxiSorp (Nunc) | 0.1–0.2 nmol/mg | [86] | |
| 4 mg/ml | |||||||
| n.a. | Ferritin | (10 mM in 6 M guanidine HCl, 0.5 M potassium phosphate, pH 2.5) | Biotinylated anti-DNP | MaxiSorp(Nunc) | 0.55–1.0 nmol/mg | [87] | |
| Dilution n.a. | Ratio (1:3) | Anti-rabbit-IgG-peroxidase (γ-chain specific) | |||||
| 45 min at RT |
These relevant publications were further examined for sample origin and derivatization conditions etc. to allow better comparison of results as well as for finding an appropriate protocol for scientists planning future experiments.
Spectrophotometric and HPLC determination of protein carbonyls after dinitrophenyl hydrazine modification
The most-cited (original) method for derivatizing carbonyl groups with different agents to determine the carbonyl content was described by Levine in 1990 [17]. The publication discusses the reaction of carbonyls with borohydride, DNPH, fluorescein thiosemicarbazide and fluorescein amine.
In this protocol different oxidatively modified proteins (>0.5 mg protein) are treated with 10 mM DNPH for 1 h. The ratio of sample volume to DNPH volume is not stated. The protein is precipitated with 20% TCA followed by a washing step with ethanol–ethyl acetate (1:1) and dissolving in 6 M guanidine. The protocol for DNP derivatization is completed by the spectrum measurement from 360 to 390 nm.
Another publication of Levine, published 4 years later (1994), also describes the derivatization with DNP followed by HPLC measurement and/or immunoblotting [18].
The general drawbacks of the spectrophotometric assay are that the method is rather work-intensive, time-consuming and high throughput measurement is not possible. Additionally, the requirement of protein and volume is relatively high, accompanied by the fact that the loss of acid-soluble proteins during washing steps (about 10–15%) must be considered and the results adjusted to the actual protein concentration. Furthermore additional carbonyl groups may also be introduced due to acidic conditions, DNP may be trapped in the protein pellet and the resolubilization may be incomplete, thus falsifying the results [23].
Another general problem is that nucleic acids may interfere as they also contain carbonyl groups, and in addition other biological compounds such as hemoglobin, myoglobin and retinoids absorb at 370 nm which result in high background readings.
Since there are no established commercially available protein standards of reduced and oxidized BSA to include as controls it is difficult to compare results obtained with the spectrophotometric assay (and other methods) in different laboratories.
Some of these drawbacks can be overcome by HPLC analysis which provides the advantages that DNP absorbance can be monitored at 366 nm in parallel to protein absorbance at 280 nm. Here DNP derivatization should be carried out in sodium dodecyl sulfate (SDS) since guanidine–HCl is not suitable for most columns.
Detection of protein carbonyls by immunoblotting
In 1994 Shacter and Levine et al. published the immunoblotting method to detect carbonyls of plasma proteins [19]. Here they describe the preparation of standards with iron and ascorbate as oxidizing agent. Heparin plasma was then treated with 10 mM DNPH (in 10% TCA) for 15–30 min at room temperature. The separation was performed with an SDS-PAGE (4–12% gradient) followed by blotting onto a nitrocellulose membrane by standard procedures. A monoclonal anti-DNP IgE, a secondary biotinylated rat anti-mouse IgE and biotin–avidin-peroxidase complex were used to detect DNP-carbonyl epitopes.
The publication is a general method description which uses different oxidized proteins. Fibrinogen was identified as a major oxidized protein in plasma. The amount of carbonyls present in untreated plasma samples was 0.6 nmol/mg. Native, as well as oxidized glutamine synthetase were run to assess the sensitivity of the assay which was found to be 30 ng of protein.
Keller et al. developed an independent immunoblotting protocol relying on the DNP-reaction [22]. Here, the authors oxidize BSA with H2O2/vanadyl and radiolysis and derivatize the samples with an equal volume of DNP for 1 h. SDS-PAGE is followed by blotting on nitrocellulose and detection with BCIP and NBT. The results show that the oxidation results in a linear increase in carbonyl concentration in the albumin molecule and the method is clearly more sensitive than the spectrophotometric assay.
Optimizing the immunoblotting method can be quite time-consuming. One must decide whether to perform pre- or post-electrophoresis derivatization and consider the different types of membranes. In terms of membrane choice, nitrocellulose membranes should only be used when derivatization is carried out before SDS-PAGE and immunoblotting because the membrane is not suitable for incubation in strong acids. The derivatization time is also crucial and varies significantly between laboratories (see Table 3). Concerning post-electrophoresis derivatization; Robinson et al. note that the membrane should be treated with DNP for exactly 5 min at RT [24], which is also our recommendation.
Both approaches, the pre- and post-electrophoresis derivatization have limitations. They both involve numerous washing steps making the approaches very time-consuming. The post-electrophoresis derivatization requires several pre-treatment steps (methanol, HCl) of the membrane. The pre-electrophoresis derivatization is not recommended for samples with a low protein concentration which will result in problems with loading the pockets and in separating/visualizing low-molecular weight proteins.
Because carbonyl groups are present in every molecule there are problems with the reproducibility and the analysis of the bands, furthermore the membranes often show a high background. The acidic derivatization affects the pI and hence complicates the identification of proteins in 2D PAGE.
An accurate determination of the carbonyl concentration is not possible with immunoblotting. Therefore the results should always be expressed in relation to appropriate controls/samples and theoretically, a control which has not been treated with DNP must be included in every assay.
Using ELISA to determine protein carbonyls
Buss et al. described the use of an anti-DNP antibody in an ELISA for the first time in 1997. Standards of HOCl-oxidized and sodium borohydride-reduced BSA are prepared for a standard curve. It is important to note that the blocking step is carried out with reduced BSA (original publication) and only with PBS in the original publication and in the erratum, respectively. The original publication contained errors concerning the preparation of reduced BSA [20], i.e. the ten-fold amount of NaBH4 and the four-fold amount of BSA are described in the original publication.
Plasma from healthy and critically ill patients was diluted to 4 mg/ml and treated with 10 mM DNPH for 45 min at RT. The ratio of sample volume to DNPH volume was 1:4. Samples were coated onto MaxiSorp plate and detected with a biotinylated anti-DNP and streptavidin-biotinylated-HRP. The fully reduced BSA showed a carbonyl concentration of 0.6 nmol/mg, this concentration has also been stated by Shacter et al. [19]. There was a linear correlation (r=0.70) between the absorbance of the spectrophotometric assay (375 nm) and the ELISA for plasma samples (n=26). After subtracting the absorbance of the blank (reduced BSA), plasma protein carbonyl concentration of healthy controls was in the range of 0.06 nmol/mg whereas that of patients was around 0.75 nmol/mg.
A modification of the protocol described by Buss et al. is the method by Alamdari [25]. Here the samples are adsorbed onto the plate prior to derivatization. This is especially interesting for samples with low protein concentrations (5 µg/ml).
Of the immunological methods, ELISA methods allow the simultaneous quantification of a great number of samples and require only small sample volumes. They can be used for the measurement of carbonyl concentrations not only in plasma, but also in tissues and cell culture samples.
Limitations of this approach include that the determination of the protein concentration is mandatory before carrying out the assay and the assay takes 2 days because the samples are usually adsorbed to the plate overnight. In addition the in-between washing steps promote the risk for loss of sample. Many companies offer ELISA plates with different binding capacities for different requirements, e.g. for molecules with hydrophobic, hydrophilic or mixed domains.
The available different monoclonal as well as polyclonal antibodies also represent a problem since they all potentially react with different epitopes. However, when the carbonyl groups of a protein are chemically reduced, e.g. by borohydride, there is less binding of DNP and hence also less antibody binding resulting in low signal intensity [26] so antibody bias is of limited relevance.
As every laboratory uses its own standards accompanied by the lack of available uniform and accepted protein standards resulting in insufficient comparability must be mentioned.
Discussion and conclusions
The described assays require no specific equipment and can be adapted to actual laboratory facilities and any methods which are applicable in the respective laboratory can be used.
As mentioned above, each method has its limitations.
Besides these, there are some other facts that must be considered. As shown in Tables 2–4, the applied protocols differ considerably in important details such as the protein concentration, ratio of sample volume to DNP volume, DNP concentration and incubation duration, temperature and conditions.
The biggest difference between these various assays lies in the derivatization step. The DNPH concentration for the spectrophotometric assay is often used according to Levine, i.e. 10 mM, equal to 0.2%, but 20 mM has also been used. Two groups state excessively high DNP derivatization concentrations [27,28]. For ELISA 10 mM seems to be the most practical concentration, while for immunoblotting 10 and 20 mM are most frequently used. For the case that the derivatization is carried out after SDS-PAGE or adsorption of samples onto ELISA plate the DNP concentration is significantly lower: 0.5 and 0.05 mM DNPH, respectively. Besides the concentration, the ratio of sample volume to DNP volume is not consistent as it varies between equal volumes, 1:3, 1:4, 1:5 and 1:6. However, often this information is not stated at all. Additionally the protein concentration or dilution of the sample is often not stated, nevertheless this should be considered as the protein concentration and DNP concentration should be adjusted/correspond.
Furthermore the influence of the incubation temperature should be considered. While some authors do not state any temperature, most authors keep their samples at RT (or close to RT: 20 °C, 25 °C) but 37 °C is also frequently used, perhaps because it is easier to keep this temperature constant. The next critical point is the duration of incubation: Levine suggests incubating the sample for 1 h. However, other authors apply incubation times of as short as 5 min. For ELISA, most authors incubate their samples between 45 and 60 min. This seem to be relatively comparable, however samples for the spectrophotometric assay are incubated between 15 and 60 min, and those for immunoblotting even differ between 5 and 60 min. We suggest incubating the samples for ELISA and spectrophotometric assay for 45–60 min and mixing the samples every 15 min (or applying constant agitation) to help minimize differences in protocols.
The results, even if theoretically measured by the same protocol, are difficult to compare, as the levels, shown in Tables 2–4, are often not given in the same concentration.
The immunoblotting method does not of course give quantitative data (i.e. absolute concentrations). The results of the spectrophotometric method, which could easily be given in as mol/mg (depending on the concentration of a native or treated sample) are frequently displayed in different units, e.g. mol/mol protein, nmol/mg, µmol/mg or µmol/10³ cells. Some authors only present their results expressed as fold-increase or percentage to controls. The units for ELISA results are given as nmol/mg, pmol/mg, nmol, nmol/ml, nmol/g, nmol carbonyl groups fibrinogen/mg of plasma proteins or carbonyl residues/HSA molecule. Some authors state their results as arbitrary units, relation to control, and relative amount. This makes it especially difficult to compare results and is also odd since the original protocol uses the unit pmol/mg and this should be easily achieved by diluting all samples to the same protein concentration and preparing standards of known carbonyl concentrations.
The protein carbonyl concentration of a given sample reflects a snapshot of generated and removed carbonyls. Protein carbonyls are not only a result of amino acid oxidation but can also arise from secondary reactions by nucleophilic addition of aldehydes or reducing sugars and their oxidation products. Scientists must evaluate whether they are interested in the oxidation of a specific cellular or circulating protein or want to compare the carbonyl content in blood of many samples, e.g. in clinical studies.
Theoretically, due to steric reasons the DNP molecule should not be able to attack all carbonyl moieties in a similar way. However, the derivatization of carbonyl compounds with DNP has been used since the late 1950s [29]. We are not aware of any publication studying the differences in terms of yield and rate of the reaction between DNP and different carbonyl moieties.
The so-called OxyBlot is semi-quantitative but has shown the best sensitivity and specificity [30], and is especially applicable in cell culture studies. On the other hand ELISA seems to be best-suited for clinical studies and the inclusion of external standards allows standardization, better comparison, as well as high-throughput. Nevertheless this decision depends on the laboratory equipment and research question.
MS-based methods are inevitable when the aim is to identify specific carbonylated residues of single proteins. However, not every laboratory is able to apply MS and for some research questions it is also not practicable, e.g. in clinical/epidemiological studies where high-throughput of complex protein mixtures such as plasma is necessary. In these cases ELISA is still the method of choice. For further information on MS-based techniques to identify and quantify oxidative protein modifications on proteins and peptides see Rogowska-Wrzesinska et al. [23].
Conclusion
When immunoblotting is applied, the results cannot be expressed in a certain concentration since the method is semiquantitative. In this case appropriate controls must be included (such as untreated cells, etc.). Every researcher should act according to good laboratory practice and include appropriate controls.
When using the spectrophotometric assay or ELISA, most authors present their results in pmol/mg. However, as you can see in Tables 2 and 4, some authors only express their results in relation to controls or do not take the protein concentration into consideration. In our opinion, it is inevitable to measure the protein concentration when assessing protein-bound carbonyls since the protein concentration is the measure which best corresponds to the number of modifiable residues. Since carbonyl groups on proteins can arise from a multitude of chemical, thermal and radiation processes it is impossible to link carbonyl groups to a specific mechanism. It has been demonstrated that the carbonyl groups increased linearly in samples treated with iron/ascorbate [19]. Hence, when measured in complex mixtures such as plasma, protein carbonyls serve as a biomarker of global protein oxidation.
In a multi-center ring study by Augustyniak et al. [31], six European laboratories were invited to measure protein carbonyls according to their own protocol in homogenized liver samples (UV-radiated and not radiated). Four laboratories used ELISA, while three used immunoblotting techniques. Unexpectedly, the concentrations measured by ELISA were quite similar and results from immunoblotting were also homogenous, indicating that ELISA techniques (self-prepared standards, quantification of standards by spectrophotometry; commercial kits) represent the best available method to quantify protein carbonyl concentration, whereas immunoblotting allows the comparable detection of the molecular weight of oxidized proteins.
The ring study relies on a review by Rogowska-Wrzesinska et al. [23] which covers advantages and pitfalls of the most commonly used methods and examines commercially available oxidized proteins, oxidants used and lysis buffers. This review, together with the previous one will help researchers to make a decision which method to use and may lead to a consensus on common protocols, hence allowing better comparability of results.
Therefore, we are convinced that also in future the detection of protein carbonyls will be a valuable tool in the detection of oxidative damage, although further improvements in sample stabilization, method robustness and standard design have to be performed.
Acknowledgment
TG and MJD were part of the EU COST CM1001 action, which in part supported this article.
References
- 1.Breusing N., Grune T. Biomarkers of protein oxidation from a chemical, biological and medical point of view. Exp. Gerontol. 2010;45(10):733–737. doi: 10.1016/j.exger.2010.04.004. 20403419 [DOI] [PubMed] [Google Scholar]
- 2.Naskalski J.W., Bartosz G. Oxidative modifications of protein structures. Adv. Clin. Chem. 2000;35:161–253. doi: 10.1016/s0065-2423(01)35017-5. 11040960 [DOI] [PubMed] [Google Scholar]
- 3.Stadtman E.R. Protein oxidation and aging. Respirology. 2006;40(12):1250–1258. doi: 10.1080/10715760600918142. [DOI] [PubMed] [Google Scholar]
- 4.Hoehn A., Jung T., Grune T. Pathophysiological importance of aggregated damaged proteins. Biol. Med. 2014;71:70–89. doi: 10.1016/j.freeradbiomed.2014.02.028. [DOI] [PubMed] [Google Scholar]
- 5.Davies M.J. The oxidative environment and protein damage. Biochim. Biophys. Acta. 2005;1703(2):93–109. doi: 10.1016/j.bbapap.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 6.Hawkins C.L., Davies M.J. Generation and propagation of radical reactions on proteins. Biochim. Biophys. Acta. 2001;1504(2–3):196–219. doi: 10.1016/s0005-2728(00)00252-8. 11245785 [DOI] [PubMed] [Google Scholar]
- 7.Morin B., Bubb W.A., Davies M.J., Dean R.T., Fu S. 3-Hydroxylysine, a potential marker for studying radical-induced protein oxidation. Chem. Res. Toxicol. 1998;11(11):1265–1273. doi: 10.1021/tx980118h. 9815186 [DOI] [PubMed] [Google Scholar]
- 8.Fu S.L., Dean R.T. Structural characterization of the products of hydroxyl-radical damage to leucine and their detection on proteins. Biochem. J. 1997;324(1):41–48. doi: 10.1042/bj3240041. 9164839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu S., Hick L.A., Sheil M.M., Dean R.T. Structural identification of valine hydroperoxides and hydroxides on radical-damaged amino acid, peptide, and protein molecules. Free Radic. Biol. Med. 1995;19(3):281–292. doi: 10.1016/0891-5849(95)00021-o. 7557542 [DOI] [PubMed] [Google Scholar]
- 10.Pietzsch J. Measurement of 5-hydroxy-2-aminovaleric acid as a specific marker of iron-mediated oxidation of proline and arginine side-chain residues of low-density lipoprotein apolipoprotein B-100. Biochem. Biophys. Res. Commun. 2000;270(3):852–857. doi: 10.1006/bbrc.2000.2533. 10772915 [DOI] [PubMed] [Google Scholar]
- 11.Grune T., Reinheckel T., Davies K.J. Degradation of oxidized proteins in K562 human hematopoietic cells by proteasome. J. Biol. Chem. 1996;271(26):15504–15509. doi: 10.1074/jbc.271.26.15504. 8663134 [DOI] [PubMed] [Google Scholar]
- 12.Grune T., Reinheckel T., Joshi M., Davies K.J. Proteolysis in cultured liver epithelial cells during oxidative stress. role of the multicatalytic proteinase complex, proteasome. J. Biol. Chem. 1995;270(5):2344–2351. doi: 10.1074/jbc.270.5.2344. 7836468 [DOI] [PubMed] [Google Scholar]
- 13.Davies M.J., Fu S., Wang H., Dean R.T. Stable markers of oxidant damage to proteins and their application in the study of human disease. Biol. Med. 1999;27(11–12):1151–1163. doi: 10.1016/s0891-5849(99)00206-3. [DOI] [PubMed] [Google Scholar]
- 14.Kim G., Weiss S.J., Levine R.L. Methionine oxidation and reduction in proteins. Biochim. Biophys. Acta. 2014;1840(2):901–905. doi: 10.1016/j.bbagen.2013.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanschmann E.M., Godoy J.R., Berndt C., Hudemann C., Lillig C.H. Thioredoxins, glutaredoxins, and peroxiredoxins – molecular mechanisms and health significance: from cofactors to antioxidants to redox signaling. Antioxid. Redox Signal. 2013;19(13):1539–1605. doi: 10.1089/ars.2012.4599. 23397885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Headlam H.A., Davies M.J. Markers of protein oxidation: different oxidants give rise to variable yields of bound and released carbonyl products. Free Radic. Biol. Med. 2004;36(9):1175–1184. doi: 10.1016/j.freeradbiomed.2004.02.017. 15082071 [DOI] [PubMed] [Google Scholar]
- 17.Levine R.L., Garland D., Oliver C.N., Amici A., Climent I., Lenz A.G., Ahn B.W., Shaltiel S., Stadtman E.R. Determination of carbonyl content in oxidatively modified proteins. Methods Enzymol. 1990;186:464–478. doi: 10.1016/0076-6879(90)86141-h. 1978225 [DOI] [PubMed] [Google Scholar]
- 18.Levine R.L., Williams J.A., Stadtman E.R., Shacter E. Carbonyl assays for determination of oxidatively modified proteins. Methods Enzymol. 1994;233:346–357. doi: 10.1016/s0076-6879(94)33040-9. 8015469 [DOI] [PubMed] [Google Scholar]
- 19.Shacter E., Williams J.A., Lim M., Levine R.L. Differential susceptibility of plasma proteins to oxidative modification: examination by western blot immunoassay. Free Radic. Biol. Med. 1994;17(5):429–437. doi: 10.1016/0891-5849(94)90169-4. 7835749 [DOI] [PubMed] [Google Scholar]
- 20.Buss H., Chan T.P., Sluis K.B., Domigan N.M., Winterbourn C.C. Protein carbonyl measurement by a sensitive ELISA method. Free Radic. Biol. Med. 1997;23(3):361–366. doi: 10.1016/s0891-5849(97)00104-4. 9214571 [DOI] [PubMed] [Google Scholar]
- 21.Tammen H., Schulte I., Hess R., Menzel C., Kellmann M., Mohring T., Schulz-Knappe P. Peptidomic analysis of human blood specimens: comparison between plasma specimens and serum by differential peptide display. Proteomics. 2005;5(13):3414–3422. doi: 10.1002/pmic.200401219. 16038021 [DOI] [PubMed] [Google Scholar]
- 22.Keller R.J., Halmes N.C., Hinson J.A., Pumford N.R. Immunochemical detection of oxidized proteins. Chem. Res. Toxicol. 1993;6(4):430–433. doi: 10.1021/tx00034a007. 8374038 [DOI] [PubMed] [Google Scholar]
- 23.Rogowska-Wrzesinska A., Wojdyla K., Nedić O., Baron C.P., Griffiths H.R. Analysis of protein carbonylation – pitfalls and promise in commonly used methods. Respirology. 2014;48(10):1145–1162. doi: 10.3109/10715762.2014.944868. 25072785 [DOI] [PubMed] [Google Scholar]
- 24.Robinson C.E., Keshavarzian A., Pasco D.S., Frommel T.O., Winship D.H., Holmes E.W. Determination of protein carbonyl groups by immunoblotting. Anal. Biochem. 1999;266(1):48–57. doi: 10.1006/abio.1998.2932. 9887212 [DOI] [PubMed] [Google Scholar]
- 25.Alamdari D.H., Kostidou E., Paletas K., Sarigianni M., Konstas A.G., Karapiperidou A., Koliakos G. High sensitivity enzyme-linked immunosorbent assay (ELISA) method for measuring protein carbonyl in samples with low amounts of protein. Free Radic. Biol. Med. 2005;39(10):1362–1367. doi: 10.1016/j.freeradbiomed.2005.06.023. 16257645 [DOI] [PubMed] [Google Scholar]
- 26.Nakamura A., Goto S. Analysis of protein carbonyls with 2,4-dinitrophenyl hydrazine and its antibodies by immunoblot in two-dimensional gel electrophoresis. J. Biochem. 1996;119(4):768–774. doi: 10.1093/oxfordjournals.jbchem.a021306. 8743580 [DOI] [PubMed] [Google Scholar]
- 27.Mahapatra S.K., Chakraborty S.P., Das S., Roy S. Methanol extract of Ocimum gratissimum protects murine peritoneal macrophages from nicotine toxicity by decreasing free radical generation, lipid and protein damage and enhances antioxidant protection. Oxid. Med. Cell Longev. 2009;2(4):222–230. doi: 10.4161/oxim.2.4.9000. 20716908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chakraborty S.P., Pramanik P., Roy S. Staphylococcus aureus infection induced oxidative imbalance in neutrophils: possible protective role of nanoconjugated vancomycin. ISRN Pharmacol. 2012;2012:435214. doi: 10.5402/2012/435214. 22530141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pesez M. 4-Dinitrophenylhydrazine, a suitable reagent for the colorimetric determination of carbonyl compounds. J. Pharm. Pharmacol. 1959;11:475–476. doi: 10.1111/j.2042-7158.1959.tb12584.x. 14432131 [DOI] [PubMed] [Google Scholar]
- 30.Dalle-Donne I., Rossi R., Giustarini D., Milzani A., Colombo R. Protein carbonyl groups as biomarkers of oxidative stress. Clin. Chim. Acta. 2003;329(1–2):23–38. doi: 10.1016/s0009-8981(03)00003-2. 12589963 [DOI] [PubMed] [Google Scholar]
- 31.Augustyniak E., Adam A., Wojdyla K., Rogowska-Wrzesinska A., Willetts R., Korkmaz A., Atalay M., Weber D., Grune T., Borsa C., Gradinaru D., Chand Bollineni R., Fedorova M., Griffiths H.R. Validation of protein carbonyl measurement: a multi-centre study. Redox Biol. 2015;4:149–157. doi: 10.1016/j.redox.2014.12.014. 25560243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmad A., Shameem M., Husain Q. Relation of oxidant–antioxidant imbalance with disease progression in patients with asthma. Ann. Thorac. Med. 2012;7(4):226–232. doi: 10.4103/1817-1737.102182. 23189100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Al-Shobaili H.A., Al Robaee A.A., Alzolibani A., Khan M.I., Rasheed Z. Hydroxyl radical modification of immunoglobulin G generated cross-reactive antibodies: its potential role in systemic lupus erythematosus. Clin. Med. Insights Arthritis Musculoskelet. Disord. 2011;4:11–19. doi: 10.4137/CMAMD.S6793. 21487454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chevion S., Moran D.S., Heled Y., Shani Y., Regev G., Abbou B., Berenshtein E., Stadtman E.R., Epstein Y. Plasma antioxidant status and cell injury after severe physical exercise. Proc. Natl. Acad. Sci. U. S. A. 2003;100(9):5119–5123. doi: 10.1073/pnas.0831097100. 12702774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Renke J., Popadiuk S., Korzon M., Bugajczyk B., Wozniak M. Protein carbonyl groups’ content as a useful clinical marker of antioxidant barrier impairment in plasma of children with juvenile chronic arthritis. Free Radic. Biol. Med. 2000;29(2):101–104. doi: 10.1016/s0891-5849(00)00288-4. 10980398 [DOI] [PubMed] [Google Scholar]
- 36.Estrada J.C., Torres Y., Benguría A., Dopazo A., Roche E., Carrera-Quintanar L., Pérez R.A., Enríquez J.A., Torres R., Ramírez J.C., Samper E., Bernad A. Human mesenchymal stem cell-replicative senescence and oxidative stress are closely linked to aneuploidy. J. Cell Death. 2013;4:e691. doi: 10.1038/cddis.2013.211. 23807220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Estrada J.C., Albo C., Benguría A., Dopazo A., López-Romero P., Carrera-Quintanar L., Roche E., Clemente E.P., Enríquez J.A., Bernad A., Samper E. Culture of human mesenchymal stem cells at low oxygen tension improves growth and genetic stability by activating glycolysis. J. Cell Death. 2012;19(5):743–755. doi: 10.1038/cdd.2011.172. 22139129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khan M.A., Oubrahim H., Stadtman E.R. Inhibition of apoptosis in acute promyelocytic leukemia cells leads to increases in levels of oxidized protein and LMP2 immunoproteasome. Proc. Natl. Acad. Sci. U. S. A. 2004;101(32):11560–11565. doi: 10.1073/pnas.0404101101. 15284441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Falone S., D’Alessandro A., Mirabilio A., Petruccelli G., Cacchio M., Di I.C., Di L.S., Amicarelli F. Long term running biphasically improves methylglyoxal-related metabolism, redox homeostasis and neurotrophic support within adult mouse brain cortex. PLOS One. 2012;7(2):e31401. doi: 10.1371/journal.pone.0031401. 22347470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shan X., Chi L., Ke Y., Luo C., Qian S., Gozal D., Liu R. Manganese superoxide dismutase protects mouse cortical neurons from chronic intermittent hypoxia-mediated oxidative damage. Neurobiol. Dis. 2007;28(2):206–215. doi: 10.1016/j.nbd.2007.07.013. 17719231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang G., Wang J., Ma H., Khan M.F. Increased nitration and carbonylation of proteins in MRL+/+ mice exposed to trichloroethene: potential role of protein oxidation in autoimmunity. Toxicol. Appl. Pharmacol. 2009;237(2):188–195. doi: 10.1016/j.taap.2009.03.010. 19332086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abdul Hamid Z., Budin S.B., Wen Jie N., Hamid A., Husain K., Mohamed J. Nephroprotective effects of Zingiber Zerumbet Smith ethyl acetate extract against paracetamol-induced nephrotoxicity and oxidative stress in rats. J. Zhejiang Univ. Sci. B. 2012;13(3):176–185. doi: 10.1631/jzus.B1100133. 22374609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan L.J., Sohal R.S. Analysis of oxidative modification of proteins. Curr. Protoc. Protein Sci. 2001;Chapter 14: Unit 14.4 doi: 10.1002/0471140864.ps1404s20. [DOI] [PubMed] [Google Scholar]
- 44.Paixão J., Dinis T.C., Almeida L.M. Protective role of malvidin-3-glucoside on peroxynitrite-induced damage in endothelial cells by counteracting reactive species formation and apoptotic mitochondrial pathway. Oxid. Med. Cell Longev. 2012;2012:428538. doi: 10.1155/2012/428538. 22792413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lardinois O.M., Maltby D.A., Medzihradszky K.F., de Montellano P.R., Tomer K.B., Mason R.P., Deterding L.J. Spin scavenging analysis of myoglobin protein-centered radicals using stable nitroxide radicals: characterization of oxoammonium cation-induced modifications. Chem. Res. Toxicol. 2009;22(6):1034–1049. doi: 10.1021/tx9000094. 19449826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silva M., Bonomo L. de F., Oliveira R. de P., Geraldo de Lima W., Silva M.E., Pedrosa M.L. Effects of the interaction of diabetes and iron supplementation on hepatic and pancreatic tissues, oxidative stress markers, and liver peroxisome proliferator-activated receptor-alpha expression. J. Clin. Biochem. Nutr. 2011;49(2):102–108. doi: 10.3164/jcbn.10-135. 21980225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conrad C.C., Talent J.M., Malakowsky C.A., Gracy R.W. Post-electrophoretic identification of oxidized proteins. Biol. Proced. Online. 2000;2:39–45. doi: 10.1251/bpo17. 12734585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barth B.M., Gustafson S.J., Kuhn T.B. Neutral sphingomyelinase activation precedes NADPH oxidase-dependent damage in neurons exposed to the proinflammatory cytokine tumor necrosis factor-alpha. J. Neurosci. Res. 2012;90(1):229–242. doi: 10.1002/jnr.22748. 21932365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barth B.M., Stewart-Smeets S., Kuhn T.B. Proinflammatory cytokines provoke oxidative damage to actin in neuronal cells mediated by Rac1 and NADPH oxidase. Neuroscientist. 2009;41(2):274–285. doi: 10.1016/j.mcn.2009.03.007. 19344766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lillig C.H., Lönn M.E., Enoksson M., Fernandes A.P., Holmgren A. Short interfering RNA-mediated silencing of glutaredoxin 2 increases the sensitivity of HeLa cells toward doxorubicin and phenylarsine oxide. Proc. Natl. Acad. Sci. U. S. A. 2004;101(36):13227–13232. doi: 10.1073/pnas.0401896101. 15328416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sultana R., Perluigi M., Newman S.F., Pierce W.M., Cini C., Coccia R., Butterfield D.A. Redox proteomic analysis of carbonylated brain proteins in mild cognitive impairment and early Alzheimer’s disease. Antioxid. Redox Signal. 2010;12(3):327–336. doi: 10.1089/ars.2009.2810. 19686046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagai K., Betsuyaku T., Kondo T., Nasuhara Y., Nishimura M. Long term smoking with age builds up excessive oxidative stress in bronchoalveolar lavage fluid. Thorax. 2006;61(6):496–502. doi: 10.1136/thx.2005.049148. 16537669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Talent J.M., Kong Y., Gracy R.W. A double stain for total and oxidized proteins from two-dimensional fingerprints. Anal. Biochem. 1998;263(1):31–38. doi: 10.1006/abio.1998.2752. 9750139 [DOI] [PubMed] [Google Scholar]
- 54.Simonin M.A., Gegout-Pottie P., Minn A., Gillet P., Netter P., Terlain B. Proteoglycan and collagen biochemical variations during fluoroquinolone-induced chondrotoxicity in mice. Antimicrob. Agents Chemother. 1999;43(12):2915–2921. doi: 10.1128/aac.43.12.2915. 10582882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simonin M.A., Gegout-Pottie P., Minn A., Gillet P., Netter P., Terlain B. Pefloxacin-induced Achilles tendon toxicity in rodents: biochemical changes in proteoglycan synthesis and oxidative damage to collagen. Antimicrob. Agents Chemother. 2000;44(4):867–872. doi: 10.1128/aac.44.4.867-872.2000. 10722483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yan L.J., Christians E.S., Liu L., Xiao X., Sohal R.S., Benjamin I.J. Mouse heat shock transcription factor 1 deficiency alters cardiac redox homeostasis and increases mitochondrial oxidative damage. EMBO J. 2002;21(19):5164–5172. doi: 10.1093/emboj/cdf528. 12356732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dickey J.S., Gonzalez Y., Aryal B., Mog S., Nakamura A.J., Redon C.E., Baxa U., Rosen E., Cheng G., Zielonka J., Parekh P., Mason K.P., Joseph J., Kalyanaraman B., Bonner W., Herman E., Shacter E., Rao V.A. Mito-tempol and dexrazoxane exhibit cardioprotective and chemotherapeutic effects through specific protein oxidation and autophagy in a syngeneic breast tumor preclinical model. PLOS One. 2013;8(8):e70575. doi: 10.1371/journal.pone.0070575. 23940596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson W.T., Newman S.M., Jr. Copper deficiency: a potential model for determining the role of mitochondria in cardiac aging. J. Am. Aging Assoc. 2003;26(1–2):19–28. doi: 10.1007/s11357-003-0003-x. 23604915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu K.L., Berger T. Trichloroethylene metabolism in the rat ovary reduces oocyte fertilizability. Chem. Biol. Interact. 2007;170(1):20–30. doi: 10.1016/j.cbi.2007.06.038. 17673192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wondrak G.T., Cervantes-Laurean D., Jacobson E.L., Jacobson M.K. Histone carbonylation in vivo and in vitro. Biochem. J. 2000;351(3):769–777. 11042133 [PMC free article] [PubMed] [Google Scholar]
- 61.Wehr N.B., Levine R.L. Quantitation of protein carbonylation by dot blot. Anal. Biochem. 2012;423(2):241–245. doi: 10.1016/j.ab.2012.01.031. 22326366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buss H., Chan T.P., Sluis K.B., Domigan N.M., Winterbourn C.C. Protein carbonyl measurement by a sensitive ELISA method (vol 23, pg 361, 1997) Biol. Med. 1998;24:1352. doi: 10.1016/s0891-5849(97)00104-4. [DOI] [PubMed] [Google Scholar]
- 63.Andresen M., Regueira T., Bruhn A., Perez D., Strobel P., Dougnac A., Marshall G., Leighton F. Lipoperoxidation and protein oxidative damage exhibit different kinetics during septic shock. Mediators Inflamm. 2008;2008:168652. doi: 10.1155/2008/168652. 18566692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dietrich-Muszalska A., Malinowska J., Olas B., Głowacki R., Bald E., Wachowicz B., Rabe-Jabłońska J. The oxidative stress may be induced by the elevated homocysteine in schizophrenic patients. Neurochem. Res. 2012;37(5):1057–1062. doi: 10.1007/s11064-012-0707-3. 22270909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grass D.S., Ross J.M., Family F., Barbour J., James Simpson H., Coulibaly D., Hernandez J., Chen Y., Slavkovich V., Li Y., Graziano J., Santella R.M., Brandt-Rauf P., Chillrud S.N. Airborne particulate metals in the New York City subway: a pilot study to assess the potential for health impacts. Environ. Res. 2010;110(1):1–11. doi: 10.1016/j.envres.2009.10.006. 19926083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu Z.M., Li L.Q., Peng M.H., Liu T.W., Qin Z., Guo Y., Xiao K.Y., Ye X.P., Mo X.S., Qin X., Li S., Yan L.N., Shen H.M., Wang L., Wang Q., Wang K.B., Liang R.X., Wei Z.L., Ong C.N., Santella R.M., Peng T. Hepatitis B virus infection contributes to oxidative stress in a population exposed to aflatoxin b1 and high-risk for hepatocellular carcinoma. Cancer Lett. 2008;263(2):212–222. doi: 10.1016/j.canlet.2008.01.006. 18280645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mesia-Vela S., Yeh C.C., Austin J.H., Dounel M., Powell C.A., Reeves A., Santella R.M., Stevenson L., Yankelevitz D., Barr R.G. Plasma carbonyls do not correlate with lung function or computed tomography measures of lung density in older smokers. Biomarkers. 2008;13(4):422–434. doi: 10.1080/13547500802002859. 18484356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yeh C.C., Barr R.G., Powell C.A., Mesia-Vela S., Wang Y., Hamade N.K., Austin J.H., Santella R.M. No effect of cigarette smoking dose on oxidized plasma proteins. Environ. Res. 2008;106(2):219–225. doi: 10.1016/j.envres.2007.09.008. 17996865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zipprich J., Terry M.B., Liao Y., Agrawal M., Gurvich I., Senie R., Santella R.M. Plasma protein carbonyls and breast cancer risk in sisters discordant for breast cancer from the New York site of the Breast Cancer Family Registry. Cancer Res. 2009;69(7):2966–2972. doi: 10.1158/0008-5472.CAN-08-3418. 19339271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Venojärvi M., Aunola S., Puhke R., Marniemi J., Hämäläinen H., Halonen J.P., Lindström J., Rastas M., Hällsten K., Nuutila P., Hänninen O., Atalay M. Exercise training with dietary counselling increases mitochondrial chaperone expression in middle-aged subjects with impaired glucose tolerance. Disorders. 2008;8:3. doi: 10.1186/1472-6823-8-3. 18371210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marangon K., Devaraj S., Jialal I. Measurement of protein carbonyls in plasma of smokers and in oxidized LDL by an ELISA. Clin. Chem. 1999;45(4):577–578. 10102923 [PubMed] [Google Scholar]
- 72.Olas B., Nowak P., Kolodziejczyk J., Ponczek M., Wachowicz B. Protective effects of resveratrol against oxidative/nitrative modifications of plasma proteins and lipids exposed to peroxynitrite. J. Nutr. Biochem. 2006;17(2):96–102. doi: 10.1016/j.jnutbio.2005.05.010. 16111878 [DOI] [PubMed] [Google Scholar]
- 73.Olsson M.G., Centlow M., Rutardóttir S., Stenfors I., Larsson J., Hosseini-Maaf B., Olsson M.L., Hansson S.R., Akerström B. Increased levels of cell-free hemoglobin, oxidation markers, and the antioxidative heme scavenger alpha(1)-microglobulin in preeclampsia. Free Radic. Biol. Med. 2010;48(2):284–291. doi: 10.1016/j.freeradbiomed.2009.10.052. 19879940 [DOI] [PubMed] [Google Scholar]
- 74.Rossner P., Jr., Terry M.B., Gammon M.D., Agrawal M., Zhang F.F., Ferris J.S., Teitelbaum S.L., Eng S.M., Gaudet M.M., Neugut A.I., Santella R.M. Plasma protein carbonyl levels and breast cancer risk. J. Cell. Mol. Med. 2007;11(5):1138–1148. doi: 10.1111/j.1582-4934.2007.00097.x. 17979889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grune T., Catalgol B., Licht A., Ermak G., Pickering A.M., Ngo J.K., Davies K.J. HSP70 mediates dissociation and reassociation of the 26S proteasome during adaptation to oxidative stress. Free Radic. Biol. Med. 2011;51(7):1355–1364. doi: 10.1016/j.freeradbiomed.2011.06.015. 21767633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hackett T.L., Scarci M., Zheng L., Tan W., Treasure T., Warner J.A. Oxidative modification of albumin in the parenchymal lung tissue of current smokers with chronic obstructive pulmonary disease. Respir. Res. 2010;11:180. doi: 10.1186/1465-9921-11-180. 21176186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sitte N., Merker K., Grune T. Proteasome-dependent degradation of oxidized proteins in MRC-5 fibroblasts. FEBS Lett. 1998;440(3):399–402. doi: 10.1016/s0014-5793(98)01495-1. 9872410 [DOI] [PubMed] [Google Scholar]
- 78.Wang L., Muxin G., Nishida H., Shirakawa C., Sato S., Konishi T. Psychological stress-induced oxidative stress as a model of sub-healthy condition and the effect of TCM. Evid. Based Complement. Alternat. Med. 2007;4(2):195–202. doi: 10.1093/ecam/nel080. 17549236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bayram B., Ozcelik B., Grimm S., Roeder T., Schrader C., Ernst I.M., Wagner A.E., Grune T., Frank J., Rimbach G. A diet rich in olive oil phenolics reduces oxidative stress in the heart of SAMP8 mice by induction of Nrf2-dependent gene expression. Rejuvenation Res. 2012;15(1):71–81. doi: 10.1089/rej.2011.1245. 22236145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Efimenko A., Starostina E., Kalinina N., Stolzing A. Angiogenic properties of aged adipose derived mesenchymal stem cells after hypoxic conditioning. J. Transl. Med. 2011;9:10. doi: 10.1186/1479-5876-9-10. 21244679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ganfornina M.D., Do Carmo S., Lora J.M., Torres-Schumann S., Vogel M., Allhorn M., González C., Bastiani M.J., Rassart E., Sanchez D. Apolipoprotein D is involved in the mechanisms regulating protection from oxidative stress. Aging Cell. 2008;7(4):506–515. doi: 10.1111/j.1474-9726.2008.00395.x. 18419796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kotarsky H., Keller M., Davoudi M., Levéen P., Karikoski R., Enot D.P., Fellman V. Metabolite profiles reveal energy failure and impaired beta-oxidation in liver of mice with complex III deficiency due to a BCS1L mutation. PLOS One. 2012;7(7):e41156. doi: 10.1371/journal.pone.0041156. 22829922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lu L., Oveson B.C., Jo Y.J., Lauer T.W., Usui S., Komeima K., Xie B., Campochiaro P.A. Increased expression of glutathione peroxidase 4 strongly protects retina from oxidative damage. Antioxid. Redox Signal. 2009;11(4):715–724. doi: 10.1089/ars.2008.2171. 18823256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Paromov V., Qui M., Yang H., Smith M., Stone W.L. The influence of N-acetyl-L-cysteine on oxidative stress and nitric oxide synthesis in stimulated macrophages treated with a mustard gas analogue. BMC Cell Biol. 2008;9:33. doi: 10.1186/1471-2121-9-33. 18570648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Poppek D., Keck S., Ermak G., Jung T., Stolzing A., Ullrich O., Davies K.J., Grune T. Phosphorylation inhibits turnover of the Tau protein by the proteasome: influence of RCAN1 and oxidative stress. Biochem. J. 2006;400(3):511–520. doi: 10.1042/BJ20060463. 16939415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van Ampting M.T., Schonewille A.J., Vink C., Brummer R.J., van der Meer R., Bovee-Oudenhoven I.M. Intestinal barrier function in response to abundant or depleted mucosal glutathione in Salmonella-infected rats. BMC Physiol. 2009;9:6. doi: 10.1186/1472-6793-9-6. 19374741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Voss P., Horakova L., Jakstadt M., Kiekebusch D., Grune T. Ferritin oxidation and proteasomal degradation: protection by antioxidants. Respirology. 2006;40(7):673–683. doi: 10.1080/10715760500419357. 16983994 [DOI] [PubMed] [Google Scholar]