Abstract
The aim of this study was to further investigate the role of wild boar (Sus scrofa) as a reservoir for hepatitis E virus (HEV). Sixty-four blood and faecal samples collected from wild boar hunted in Central Italy in 2011–2012 were examined by indirect enzyme-linked immunosorbent assay and RT-PCR analysis. Positive RT-PCR samples were further examined by nucleotide sequence determination and subsequent phylogenetic analysis. Thirty-six sera (56.2%) were positive for HEV-specific antibodies, and six (9.4%) faecal samples scored RT-PCR-positive results. Four animals were positive by both enzyme-linked immunosorbent assay and RT-PCR. Phylogenetic analysis showed that the detected wild boar–derived HEV sequences clustered within genotype 3, with similarity to sequences of human origin collected in a nearby area in 2012. Our data confirm that HEV is endemic in the wild boar population in the research area and that these wild animals could play an important role in the epidemiology of HEV infection.
Keywords: Disease transmission, genetic diversity, hepatitis E virus, Italy, Sus scrofa, zoonoses
Introduction
Hepatitis E virus (HEV) is a small icosahedral nonenveloped and single-stranded positive-sense RNA virus. It has been designated as the sole member of the genus Hepevirus in the family Hepeviridae on account of the unique genomic organization [1]. HEV variants have been divided into at least four genotypes on the basis of whole or partial genome sequences of various open reading frames of the viral genome [2]. The infection course in humans is often asymptomatic or causes an acute viral hepatitis after an incubation period of 4–5 weeks; both outbreaks and individual cases have been recorded [3]. The mortality rate is generally under 0.5% but can be as high as 25% in pregnant women, especially for genotype 1 [4]. HEV infection is considered a major public health problem in low-income countries and communities living under poor hygienic conditions, where incidence of infection may be high [5] and outbreaks and sporadic cases are generally due to water contamination [6]. HEV sequences have been detected in various tissues and organs of swine, deer and wild boar and also in bivalves such as mussels, cockles and oysters [5,7,8]. Epidemiologic investigations carried out in industrialized countries demonstrated a higher incidence and prevalence in humans and animals than expected, and identified pigs and wild boar as possible source of human infection both for meat consumers or workers occupationally exposed to pigs [9–16]. Recently a case of human HEV infection has been reported in Italy, and even if the source of this infection was uncertain, the patient, who had never travelled outside Italy, butchered a previously hunted wild boar [17]. The aim of this study was to add information on the role of wild boar in HEV transmission by serologic and molecular investigations on a wild boar population in Central Italy.
Materials and Methods
Data collection
During the 2011–2012 hunting seasons, serum and faecal samples were collected from a total of 64 wild boar (35 females and 29 males) in an area of Tuscany (Central Italy) and stored at −80°C until use. Animals belonged to a free-ranging wild boar population living in an area of approximately 444 km2 within the province of Pisa. Each animal was classified by sex and divided into three age classes: young (presence of strikes; n = 2), subadult (no strikes, weight less than 20 kg; n = 32) and adult (n = 30).
Serologic analysis
Serum samples were analysed by a double antigen sandwich enzyme-linked immunosorbent assay (ELISA) detecting total antibodies to HEV (HEV Ab EIA; Axiom Diagnostic). Test procedures and interpretation of results were performed according to the manufacturer's instructions. The optical density was measured by a plate reader (Multiscan FC; Thermo Scientific) at 450 nm wavelength.
Molecular analysis
Faecal samples were pooled (pools of 3–5 samples) according to sampling site and animal age. Total RNA was extracted from 140 μL of faecal suspension (1–3 g of faeces in 10% w/v PBS pH 7.2) using QIAamp Viral RNA kit (Qiagen) according to the manufacturer's instructions. Template cDNAs were obtained using QuantiTect Reverse Transcription kit (Qiagen). A 347 bp fragment of HEV open reading frame 2 (ORF2) was amplified from cDNAs by nested RT-PCR (Hotstart Taq PCR master mix; Qiagen), as described by Meng et al. [18]. Samples from positive pools were analysed individually using the same nested RT-PCR amplification protocol. The limit of detection of the protocol was evaluated when preparing pools samples; HEV-positive wild boar sample were detected by nested RT-PCR from RNA extracted from 0.1 g of faeces. Nested RT-PCR products were visualized on a 2% agarose gel, and DNAs of the correct size were purified by the MiniElute Gel Extraction Kit (Qiagen). Nucleotide sequence analysis on positive PCR products was performed by BMR Genomics (Padova). A panel of HEV ORF2 GenBank-available sequences of human, swine and wild boar origin representative of all HEV genotypes and subtypes of genotype 3 according to the classifications proposed by Lu et al. [19] were aligned with six wild boar–derived HEV sequences obtained in this study using BioEdit software [20]. Evolutionary distances were estimated within the six wild boar–derived HEV sequences obtained and between a set of 14 Italian ORF2 HEV sequences of human and swine origin available in GenBank. Further phylogenetic analysis was performed by neighbour-joining and maximum-likelihood methods as available in the MEGA6 software package [21]. Phylogenetic trees were generated and genotypes and subtypes identified. The number of bootstrap replicates was 100.
Statistical analysis
Chi-square testing with the Yates correction was incorporated to evaluate the relationships between seroprevalence with age or sex. A p value of <0.05 was considered statistically significant. Statistical analysis was performed using the statistical package SPSS Advanced Statistics 13.0 (IBM Corp.).
Results
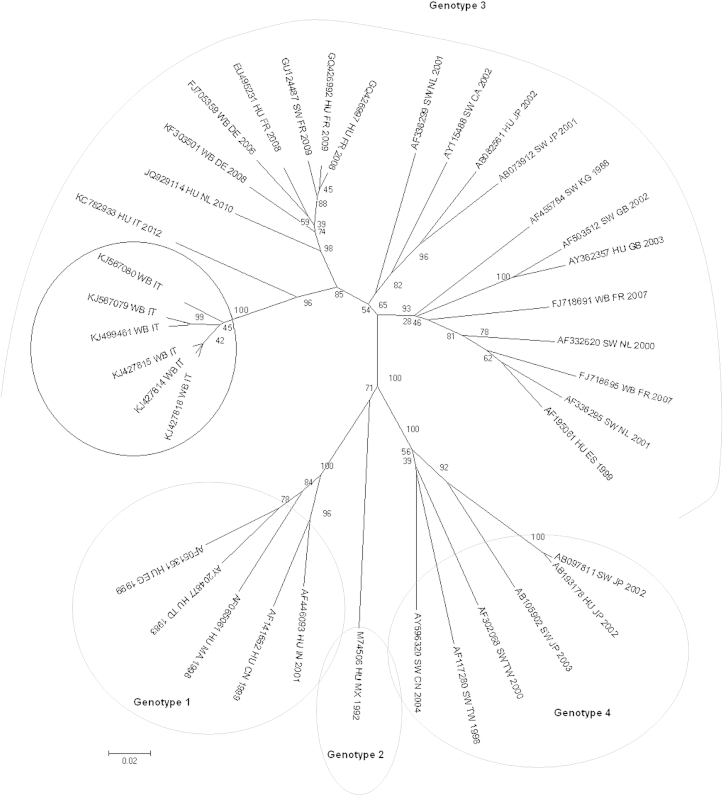
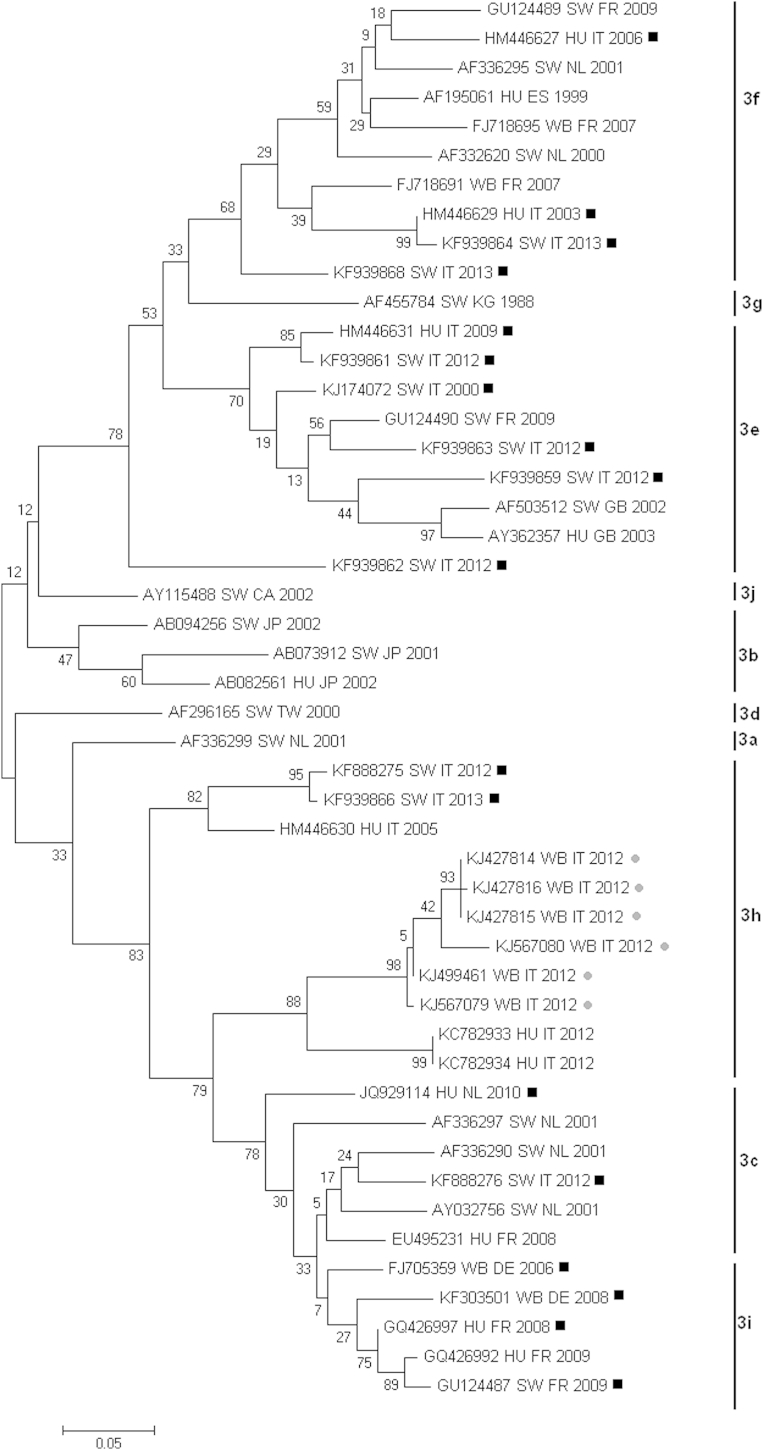
Thirty-six (56.2%) of 64 sera scored positive for anti-HEV antibodies. These were one of two young subjects, ten of 32 subadults and 25 of 30 adults. A statistically significant difference (p <0.001) was found between seropositive adult and subadult wild boar when compared. Fourteen of 29 males and 22 of 35 females were HEV seropositive, indicating that there was no statistical difference between sexes. Nested RT-PCR performed on the faecal samples followed by sequence analysis detected six positive samples, with an overall prevalence of 9.4%. The positive animals were two young animals, two subadults and two adults. Four animals (two adult females, one subadult female and one young male) were found to be positive by both ELISA and RT-PCR. Sequence analysis demonstrated a high nucleotide and amino acid sequence similarity of the amplified fragments (Table 1). Further phylogenetic analysis showed that all the Italian wild boar–derived HEV sequences (GenBank accession numbers KJ427814, KJ427815, KJ427816, KJ499461, KJ567079, KJ567080) clustered within genotype 3 (Figs. 1 and 2). BLAST analysis performed on sequence KJ567079 indicated the maximum alignment score and the lowest e-value (6e-115) with sequences of human origin (GenBank accession numbers KC782933, KC782934) collected from a case of human infection that occurred in a nearby area of Tuscany during the same year [10]. The human sequences refers to a case occurred in the province of Barberino del Mugello, a municipality bordering our sampling area presumably sharing the same wild boar population studied. The estimated evolutionary distances between a selected wild boar–derived HEV sequence and human and swine HEV sequences of Italian origin classified by host and year are shown in Table 2. Phylogenetic trees constructed by both the maximum-likelihood method and the neighbour-joining method also confirmed the nucleotide similarity among wild boar sequences reported in this study and human-derived HEV sequences from Italy in 2012 (Figs. 1 and 2).
Table 1.
Estimates of evolutionary divergence between wild boar–derived hepatitis E virus sequences
| Accession no. | KJ567079 | KJ499461 | KJ427814 | KJ427815 | KJ427816 | KJ567080 |
|---|---|---|---|---|---|---|
| KJ567079 | 0.000a | 0.000a | 0.000a | 0.011a | 0.011a | |
| KJ499461 | 0.004b | 0.000a | 0.000a | 0.011a | 0.011a | |
| KJ427814 | 0.029b | 0.025b | 0.000a | 0.011a | 0.011a | |
| KJ427815 | 0.029b | 0.025b | 0.000b | 0.011a | 0.011a | |
| KJ427816 | 0.032b | 0.029b | 0.004b | 0.004b | 0.022a | |
| KJ567080 | 0.036b | 0.039b | 0.036b | 0.036b | 0.039b |
Shown are the number of abase amino acid differences and bdifferences per nucleotide and per site between 6 Italian wild boar–derived hepatitis E virus sequences. Coding data were translated assuming standard genetic code table. There were a total of 279 nt and 93 aa positions in the final data set.
Fig. 1.
Evolutionary relationships of taxa. Phylogenetic tree for a set of ORF2 HEV sequence of genotypes 1, 2, 3 and 4. Evolutionary history was inferred by the neighbour-joining method. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Evolutionary distances were computed using the p-distance method and are in the units of the number of base differences per site. The analysis involved 38 nucleotide sequences; the six Italian wild boar–derived HEV sequences are circled. There were a total of 282 positions in the final data set. Evolutionary analyses were conducted in MEGA6 [21]. HEV, hepatitis E virus; ORF, open reading frame.
Fig. 2.
Molecular phylogenetic analysis by maximum-likelihood method. Phylogenetic tree on a set of ORF2 HEV genotype 3 sequences of human, wild boar and swine origin classified in subgroups. Evolutionary history was inferred by the maximum-likelihood method based on the Tamura-Nei model. The tree with the highest log likelihood (−3521.7122) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 48 nucleotide sequences. Evolutionary analyses were conducted in MEGA6 [21]. Black squares indicate assignment of putative subgroup; grey dots indicate wild boar–derived HEV sequences identified in this work. CA, Canada; CN, China; DE, Germany; EG, Egypt; ES, Spain; FR, France; GB, United Kingdom; HEV, hepatitis E virus; HU, human; IN, India; IT, Italy; JP, Japan; KG, Kyrgyzstan; MA, Morocco; MX, Mexico; NL, Netherlands; ORF, open reading frame; SW, swine; TD, Chad; TW, Taiwan; WB, wild boar.
Table 2.
Estimates of evolutionary divergence between swine and human sequences of Italian origin
| Accession no. | WB 2012 KJ567079 | HU 2012 KC782933 | HU 2005 HM446630 | SW 2012 KF888276 | SW 2012 KF888275 | SW 2013 KF939866 | SW 2013 KF939868 | SW 2012 KF939859 | SW 2012 KF939861 | SW 2012 KF939863 | HU 2006 HM446627 | HU 2003 HM446629 | HU 2009 HM446631 | SW 2000 KJ174072 | SW 2012 KF939862 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KJ567079 | |||||||||||||||
| KC782933 | 0.126 | ||||||||||||||
| HM446630 | 0.170 | 0.157 | |||||||||||||
| KF888276 | 0.179 | 0.178 | 0.145 | ||||||||||||
| KF888275 | 0.199 | 0.171 | 0.099 | 0.145 | |||||||||||
| KF939866 | 0.199 | 0.180 | 0.091 | 0.136 | 0.014 | ||||||||||
| KF939868 | 0.205 | 0.211 | 0.171 | 0.213 | 0.177 | 0.190 | |||||||||
| KF939859 | 0.219 | 0.266 | 0.217 | 0.240 | 0.210 | 0.215 | 0.167 | ||||||||
| KF939861 | 0.239 | 0.246 | 0.196 | 0.221 | 0.178 | 0.183 | 0.158 | 0.152 | |||||||
| KF939863 | 0.239 | 0.256 | 0.257 | 0.243 | 0.247 | 0.262 | 0.178 | 0.143 | 0.095 | ||||||
| HM446627 | 0.239 | 0.233 | 0.237 | 0.223 | 0.195 | 0.200 | 0.145 | 0.157 | 0.155 | 0.173 | |||||
| HM446629 | 0.252 | 0.261 | 0.237 | 0.278 | 0.209 | 0.229 | 0.127 | 0.165 | 0.154 | 0.176 | 0.127 | ||||
| HM446631 | 0.254 | 0.261 | 0.206 | 0.226 | 0.187 | 0.192 | 0.180 | 0.164 | 0.025 | 0.111 | 0.168 | 0.148 | |||
| KJ174072 | 0.254 | 0.277 | 0.220 | 0.231 | 0.211 | 0.225 | 0.154 | 0.118 | 0.075 | 0.091 | 0.155 | 0.127 | 0.063 | ||
| KF939862 | 0.264 | 0.251 | 0.220 | 0.248 | 0.227 | 0.232 | 0.182 | 0.192 | 0.189 | 0.213 | 0.182 | 0.214 | 0.202 | 0.184 | |
| KF939864 | 0.264 | 0.262 | 0.243 | 0.275 | 0.206 | 0.225 | 0.128 | 0.157 | 0.150 | 0.177 | 0.119 | 0.011 | 0.145 | 0.119 | 0.206 |
| Evolutionary divergence between groups | ||
|---|---|---|
| SW | HU | |
| SW | ||
| HU | 0.181 | |
| WB | 0.226 | 0.208 |
Number of base differences per site from between sequences and between groups are shown. Analysis involved 16 nucleotide sequences. There were a total of 280 positions in the final data set. Average values within human and swine sequences is 0.203 and 0.182, respectively.
Discussion
HEV infection has a worldwide distribution, with a high prevalence in low-income countries. The infection in industrialized countries was previously recognized among travellers returning from endemic countries [22]. During the last decade, an increasing proportion of reported human HEV infections were demonstrated to be autochthonous [23], being linked to ingestion of raw or undercooked meat from pigs, wild boar and deer [24–26]. HEV infection has been well documented in European countries, and studies indicate a high similarity between wild boar, pigs and human HEV sequences, suggesting that both pigs and wild boar could represent an important source of infection for humans [12,27–31]. The case of human infection reported in Italy by Giordani et al. [17] was supposed to be of wild boar origin because the patient had never travelled outside Italy and because 2 months before the onset of acute hepatitis he had butchered a wild boar hunted in Barberino di Mugello (Tuscany). Our findings add new information supporting this hypothesis: our study area is close to Barberino di Mugello, and we may assume that these areas share the same wild boar population; autochthonous human sequences described in the work by Giordani and colleagues and the wild boar–derived HEV sequences identified in this research show sequence similarity. Furthermore, both studies were carried out in the same period of time. Our research revealed a high seroprevalence (56.2%) in the wild boar population and the presence of virus RNA in almost 10% of the tested animals. A recent study performed on a wild boar population ranging in a nearby confined area of Central Italy (Parco Migliarino San Rossore, Pisa, Tuscany) recorded a similar seroprevalence of 48.7%, while no HEV RNA was detected in liver and serum samples [32]. Our data confirm that HEV infection is endemic in the wild boar population of Central Italy and that these animals could play a role as a reservoir of the virus. Phylogenetic analysis also demonstrated that the wild boar–derived HEV sequences clustered within genotype 3 and were related to an Italian human HEV sequence, supporting the zoonotic potential of wild boar strains. Concerning the possibility of cross-infection between pig and wild boar populations, it is worth noting that a particular breed of autochthonous pig (Cinta Senese, Sus scrofa domesticus) is raised outdoors in some areas of Tuscany on extensive plots of natural pasture, where they share their habitat with wildlife. In such a context, direct contact between domestic and wild animals are frequent; moreover, possible HEV transmission between wildlife and domestic pigs may occur indirectly through water and feeding spots contaminated by infected wild boar faeces.
These findings could be even more relevant when considering the consumption of raw or undercooked meat in Tuscany of pig or wild boar sausages containing liver tissue. Moreover, the widespread practice of wild boar hunting in such territories increases the risk of infection encountered during the slaughtering of infected animals [13,15]. In conclusion, wild boar as well as domestic pigs may play a role in the epidemiology of HEV as a possible source of infection for humans through slaughtering and consumption. Our study and previous studies [12,28,30–32] suggest that HEV is commonly present in the wild boar population within defined geographic areas.
Conflict of Interest
None declared.
Acknowledgements
This study was supported in part by a grant from the province of Pisa, Italy. We thank Mauro Pistello, University of Pisa, for revision and helpful suggestions.
References
- 1.Emerson S.U., Purcell R.H. Hepatitis E virus. Rev Med Virol. 2003;13:145–154. doi: 10.1002/rmv.384. [DOI] [PubMed] [Google Scholar]
- 2.Panda S.K., Thakral D., Rehman S. Hepatitis E virus. Rev Med Virol. 2007;17:151–180. doi: 10.1002/rmv.522. [DOI] [PubMed] [Google Scholar]
- 3.Reyes G.R., Yarbough P.O., Tam A.W., Purdy M.A., Huang C.C., Kim J.S. Isolation of a cDNA from the virus responsible for enterically transmitted non-A, non-B hepatitis. Science. 1990;247:1335–1339. doi: 10.1126/science.2107574. [DOI] [PubMed] [Google Scholar]
- 4.Kumar A., Beniwal M., Kar P., Sharma J.B., Murthy N.S. Hepatitis E in pregnancy. Int J Gynaecol Obstet. 2004;85:240–244. doi: 10.1016/j.ijgo.2003.11.018. [DOI] [PubMed] [Google Scholar]
- 5.Kumar S., Subhadra S., Singh B., Panda B.K. Hepatitis E virus: the current scenario. Int J Infect Dis. 2013;17:228–233. doi: 10.1016/j.ijid.2012.11.026. [DOI] [PubMed] [Google Scholar]
- 6.Purcell R.H., Emerson S.U. Hepatitis E: an emerging awareness of an old disease. J Hepatol. 2008;48:494–503. doi: 10.1016/j.jhep.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 7.Crossan C., Baker P.J., Craft J., Takeuchi Y., Dalton H.R., Scobie L. Hepatitis E virus genotype 3 in shellfish, United Kingdom. Emerg Infect Dis. 2012;18:2085–2087. doi: 10.3201/eid1812.120924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donia D., Dell'Amico M.C., Petrinca A.R., Martinucci I., Mazzei M., Tolari F. Presence of hepatitis E RNA in mussels used as bio-monitors of viral marine pollution. J Virol Methods. 2012;186:198–202. doi: 10.1016/j.jviromet.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 9.Li T.C., Chijiwa K., Sera N., Ishibashi T., Etoh Y., Shinohara Y. Hepatitis E virus transmission from wild boar meat. Emerg Infect Dis. 2005;11:1958–1960. doi: 10.3201/eid1112.051041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teo C.G. Hepatitis E indigenous to economically developed countries: to what extent a zoonosis? Curr Opin Infect Dis. 2006;19:460–466. doi: 10.1097/01.qco.0000244052.61629.49. [DOI] [PubMed] [Google Scholar]
- 11.Adlhoch C., Wolf A., Meisel H., Kaiser M., Ellerbrok H., Pauli G. High HEV presence in four different wild boar populations in East and West Germany. Vet Microbiol. 2009;139:270–278. doi: 10.1016/j.vetmic.2009.06.032. [DOI] [PubMed] [Google Scholar]
- 12.Kaba M., Davoust B., Marié J.L., Colson P. Detection of hepatitis E virus in wild boar (Sus scrofa) livers. Vet J. 2010;186:259–261. doi: 10.1016/j.tvjl.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 13.Krumbholz A., Mohn U., Lange J., Motz M., Wenzel J.J., Jilg W. Prevalence of hepatitis E virus–specific antibodies in humans with occupational exposure to pigs. Med Microbiol Immunol. 2011;201:239–244. doi: 10.1007/s00430-011-0210-5. [DOI] [PubMed] [Google Scholar]
- 14.Widén F., Sundqvist L., Matyi-Toth A., Metreveli G., Belák S., Hallgren G. Molecular epidemiology of hepatitis E virus in humans, pigs and wild boars in Sweden. Epidemiol Infect. 2011;139:361–371. doi: 10.1017/S0950268810001342. [DOI] [PubMed] [Google Scholar]
- 15.Carpentier A., Chaussade H., Rigaud E., Rodriguez J., Berthault C., Boué F. High hepatitis E virus seroprevalence in forestry workers and in wild boars in France. J Clin Microbiol. 2012;50:2888–2893. doi: 10.1128/JCM.00989-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berto A., Grierson S., Hakze-van der Honing R., Martelli F., Johne R., Reetz J. Hepatitis E virus in pork liver sausage, France. Emerg Infect Dis. 2013;19(2):264–266. doi: 10.3201/eid1902.121255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giordani M.T., Fabris P., Brunetti E., Goblirsch S., Romanò L. Hepatitis E and lymphocytic leukemia in Man, Italy. Emerg Infect Dis. 2013;12:2054–2056. doi: 10.3201/eid1912.130521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng X.J., Purcell R.H., Halbur P.G., Lehman J.R., Webb D.M., Tsareva T.S. A novel virus in swine is closely related to the human hepatitis E virus. Proc Natl Acad Sci U S A. 1997;94:9860–9865. doi: 10.1073/pnas.94.18.9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu L., Li C., Hagedorn C.H. Phylogenetic analysis of global hepatitis E virus sequences: genetic diversity, subtypes and zoonosis. Rev Med Virol. 2006;16:5–36. doi: 10.1002/rmv.482. [DOI] [PubMed] [Google Scholar]
- 20.Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 21.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis, version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bader T.F., Krawczynski K., Polish L.B., Favorov M.O. Hepatitis E in a US traveler to Mexico. N Engl J Med. 1991;325:1659. doi: 10.1056/NEJM199112053252321. [DOI] [PubMed] [Google Scholar]
- 23.Teshale E.H., Hu D.J., Holmberg S.D. The two faces of hepatitis E virus. Clin Infect Dis. 2010;51:328–334. doi: 10.1086/653943. [DOI] [PubMed] [Google Scholar]
- 24.Matsuda H., Okada K., Takahashi K., Mishiro S. Severe hepatitis E virus infection after ingestion of uncooked liver from a wild boar. J Infect Dis. 2003;188:944. doi: 10.1086/378074. [DOI] [PubMed] [Google Scholar]
- 25.Dalton H.R., Bendall R., Ijaz S., Banks M., Hepatitis E. an emerging infection in developed countries. Lancet Infect Dis. 2008;8:698–709. doi: 10.1016/S1473-3099(08)70255-X. [DOI] [PubMed] [Google Scholar]
- 26.Romanò L., Paladini S., Tagliacarne C., Canuti M., Bianchi S., Zanetti A.R. Hepatitis E in Italy: a long-term prospective study. J Hepatol. 2011;54:34–40. doi: 10.1016/j.jhep.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 27.Colson P., Borentain P., Queyriaux B., Kaba M., Moal V., Gallian P. Pig liver sausage as a source of hepatitis E virus transmission to humans. J Infect Dis. 2010;202:825–834. doi: 10.1086/655898. [DOI] [PubMed] [Google Scholar]
- 28.de Deus N., Peralta B., Pina S., Allepuz A., Mateu E., Vidal D. Epidemiological study of hepatitis E virus infection in European wild boars (Sus scrofa) in Spain. Vet Microbiol. 2008;129:163–170. doi: 10.1016/j.vetmic.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Di Bartolo I., Martelli F., Inglese N., Pourshaban M., Caprioli A., Ostanello F. Widespread diffusion of genotype 3 hepatitis E virus among farming swine in Northern Italy. Vet Microbiol. 2008;132:47–55. doi: 10.1016/j.vetmic.2008.04.028. [DOI] [PubMed] [Google Scholar]
- 30.Kaci S., Nockler K., Johne R. Detection of hepatitis E virus in archived German wild boar serum samples. Vet Microbiol. 2008;128:380–385. doi: 10.1016/j.vetmic.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 31.Martelli F., Caprioli A., Zengarini M., Marata A., Fiegna C., Di Bartolo I. Detection of hepatitis E virus (HEV) in a demographic managed wild boar (Sus scrofa scrofa) population in Italy. Vet Microbiol. 2008;126:74–81. doi: 10.1016/j.vetmic.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 32.Martinelli N., Pavoni E., Filogari D., Ferrari N., Chiari M., Canelli E. Hepatitis E virus in wild boar in the central northern part of Italy. Transbound Emerg Dis. 2015;62:217–222. doi: 10.1111/tbed.12118. [DOI] [PubMed] [Google Scholar]