Abstract
Obesity and diabetes arise from an intricate interplay between both genetic and environmental factors. It is well recognized that obesity plays an important role in the development of insulin resistance and diabetes. Yet, the exact mechanism of the connection between obesity and diabetes is still not completely understood. Metabolomics is an analytical approach that aims to detect and quantify small metabolites. Recently, there has been an increased interest in the application of metabolomics to the identification of disease biomarkers, with a number of well-known biomarkers identified. Metabolomics is a potent approach to unravel the intricate relationships between metabolism, obesity and progression to diabetes and, at the same time, has potential as a clinical tool for risk evaluation and monitoring of disease. Moreover, metabolomics applications have revealed alterations in the levels of metabolites related to obesity-associated diabetes. This review focuses on the part that metabolomics has played in elucidating the roles of metabolites in the regulation of systemic metabolism relevant to obesity and diabetes. It also explains the possible metabolic relation and association between the two diseases. The metabolites with altered profiles in individual disorders and those that are specifically and similarly altered in both disorders are classified, categorized and summarized.
Keywords: biomarkers, diabetes, metabolites, metabolomics, obesity
INTRODUCTION
The term “metabolites” denotes all endogenous small molecules that are involved (as substrates, intermediates or products) in common metabolic reactions in biochemical pathways essential for growth, development, reproduction and stress response mechanisms (Goodacre et al., 2004). Metabolomics is defined as a technology aimed at measuring/profiling changes in the levels of metabolites present inside a cell, tissue, or organism in response to a genetic variation or pathophysiological stimuli (Oliver et al., 1998; Scarfe et al., 1998). Another aim of metabolomics is to evaluate the relative differences between biological samples on the basis of their metabolite profiles. It can deliver an immediate snapshot of the whole physiology of an organism. Current technological developments have allowed high-throughput profiling of a large number of metabolites in biological samples, with increasing applications in disease research (Shah et al., 2012). Metabolomics comprises qualitative and quantitative analysis of intracellular and intercellular metabolites using two distinct analytical approaches: (1) non-targeted metabolite profiling, i.e. identification and characterization of a large number of metabolites and their precursors; and (2) targeted metabolite profiling, with an emphasis on quantitative variations in metabolites of interest (e.g., amino acids, carbohydrates, lipids and nucleotides) based on a prior understanding of their biological roles in metabolic pathways (Sadanala et al., 2012).
In this review, we discuss the major metabolite changes observed in obesity and diabetes. The focus is on the altered metabolite profiles in each disorder and inter-related disease conditions. We also present a short list of metabolites commonly associated with both disorders. The concept of our review is based on metabolite classes such as amino acids, lipids, carbohydrates and nucleotides, in previous studies that proposed a cross-sectional relationship between obesity and diabetes.
ANALYTICAL APPLICATIONS IN METABOLOMIC STUDIES
The diversity in the physico-chemical properties of metabolites makes metabolomics analysis very challenging. Hence, different analytical methodologies are needed and are preferably combined with each other (Goodacre et al., 2004). Among the analytical techniques that can be used for metabolomics applications, nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS) are the most commonly used. High performance liquid chromatography (HPLC), ultra performance liquid chromatography (UPLC), gas chromatography (GC) and capillary electrophoresis (CE) can detect metabolites within a few minutes. Combinations of these advanced techniques with MS include GC-MS, liquid chromatography-mass spectrometry (LC-MS) and CE-MS. Application of these techniques to metabolomics has resulted in some promising outcomes (Carr et al., 2011; Chylla et al., 2011; Junot et al., 2010; Lin et al., 2010; Zhang et al., 2012). As one of the most widely-used spectroscopic analytical techniques, NMR can identify and quantify a large number of organic compounds simultaneously in the micromolar range. GC-MS has been extensively used for metabolomics and offers effective and reproducible analysis. However, nonvolatile compounds are not suitable for GC-MS analysis. For their separation on a GC column, a derivatization reaction is needed to generate volatile compounds. This limits the applicability of GC-MS in metabolomics. LC-MS techniques have been developed that use a soft ionization method, which makes this approach more suitable for routine use. Large-scale metabolomic technologies based on LC-MS are increasingly gaining attention for their applicability in the diagnosis of human disease (Courant et al., 2012). The UPLC-MS technology is important in biomolecular research and can be used to quantify the functions of signaling and metabolic pathways in a multiplex and comprehensive manner. UPLC also permits a more rapid analysis (in comparison with HPLC) without loss of resolution. CE-MS is a potent and promising high-resolution separation technique for charged metabolites in biological fluids (Barbas et al., 2011). CE-MS, as an analytical platform, has made substantial contributions in the progress of metabolomics research. Fourier transform mass spectrometry (FTMS) instruments are popular as they provide exact quantification (Carr et al., 2011). Matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) is a rapidly evolving analytical tool that can analyze small molecules (less than 1000 Da; (van Kampen et al., 2011). Fourier transform ion cyclotron resonance mass spectrometry (FTICRMS) may become a potent new technique for superior metabolomic analysis (Cho et al., 2015). In addition to high-resolution analysis methods, such as NMR and MS, metabolomics employs chemometric statistical tools, such as partial least squares (PLS) and principal component analysis (PCA), to develop an integrated picture of both endogenous and xenobiotic metabolism.
Role of metabolomics in investigation of biomarkers in the diseases
Metabolomics studies allow identification of metabolites involved in disease mechanisms by observing metabolite level variations in diseased individuals in comparison with healthy ones (Cheng et al., 2012; Goek et al., 2012; Newgard et al., 2009; Pietiläinen et al., 2007; Rhee et al., 2011; Shaham et al., 2008; Wang et al., 2011; Zhao et al., 2010). Furthermore, the metabolic phenotypes of human populations will greatly facilitate evaluation of the metabolic response of each patient to treatment, making possible personalized clinical treatment. Sample-specific metabolomes can be studied, such as serum metabolome or liver tissue metabolome. Urine and blood (serum and plasma) are the biofluids that are most often used for metabolomics studies because both contain a multitude of detectable metabolites and can be obtained without or with minimal invasion. A number of other fluids such as saliva, bile, gut aspirate, cerebrospinal fluid, seminal fluid, amniotic fluid and synovial fluid have also been analyzed (Bala et al., 2006; Bollard et al., 2005; Gowda et al., 2006). More recently, metabolic profiling of complete tissues and their lipid and aqueous metabolites has received more attention for biomarker detection (Griffin and Kauppinen, 2007). These studies have identified potential biomarkers or revealed pathophysiology of diseases such as obesity, diabetes, cardiovascular diseases and cancers (Beger, 2013; Du et al., 2013).
Metabolomics has been used for biomarker discovery for a number of clinical conditions (Madsen et al., 2010; Vinayavekhin et al., 2010). Biomarkers are regularly used in clinical practice to measure disease severity and also to provide essential prognostic information related to survival. Biomarkers have also been used as alternatives to clinical endpoints both in clinical practice and in research settings, predominantly in novel drug development. By identifying the definite early biomarkers of disease, metabolomics provides better understanding of disease progression and metabolic pathways (Zhang et al., 2013).
Obesity
Obesity is a medical condition, in which a large amount of fat accumulates in the body to the point that it may decrease the life span and result in such health problems as diabetes, cardiovascular disease, as well as bone and joint diseases (Haslam and James, 2005; Pataky et al., 2014). Obesity is responsible for about 5% of worldwide deaths, and the total economic losses from obesity are approximately $2 trillion, or 2.8% of global GDP, equivalent to the worldwide losses from smoking or armed violence, war and terrorism. Recently, several metabolites associated with obesity have been identified by metabolomics and confirmed to be disturbed extensively in both animal models and humans (Abu Bakar et al., 2015). Obesity affects the entire body and evidently involves metabolic variations, but the definite changes in metabolism during obesity and dysfunction of specific organs or cellular organelles related to obesity are not yet well understood (Kussmann et al., 2006). As metabolomics can swiftly detect subtle alterations in the metabolic network, it is exclusively poised to improve our knowledge of obesity and diseases related to obesity.
Diabetes
Diabetes mellitus (DM), usually called diabetes, is a collection of metabolic diseases characterized by elevated blood sugar levels over an extended period (Nyenwe et al., 2011). If untreated, diabetes can trigger many complications (Stolar, 1988). It is a chronic disease characterized by chronic hyperglycemia because of the absence or shortage of insulin, which may be due either to a gradual failure of pancreatic β-cell function and subsequently a lack of insulin production (type 1 diabetes, T1DM) or to the development of insulin resistance and consequently the loss of β-cell function (type 2 diabetes, T2DM). Using either animal models or oral glucose tolerance tests, metabolic studies have uncovered alterations in metabolites associated with pathways involved in the action of insulin, including lipolysis, ketogenesis, proteolysis and glucose metabolism (Friedrich, 2012). These results have revealed a switch from β-oxidation to glycolysis and fat storage in response to glucose intake. Moreover, metabolomics studies performed on diabetic patients and healthy subjects have revealed many significantly altered metabolic pathways and metabolic variations (Suhre et al., 2010).
METABOLOMICS IN OBESITY AND DIABETES
Metabolomics studies have offered new insights into the etiology of obesity and diabetes and individual dissimilarities. The identified metabolites and metabolite ratios might be considered as potential biomarkers useful to understand obesity-associated pathophysiological processes. A recent epidemiological study has found that plasma concentrations of metabolites, particularly branched-chain amino acids (BCAAs: isoleucine, leucine and valine), are linked to an increased risk of T2DM (Newgard et al., 2009). Other novel metabolite classes have also been associated with diabetes; these include short- and medium-chain acylcarnitines and the following lipid classes: sphingomyelins (SMs), lysophosphatidylcholines (LysoPC), phosphatidylcholines (PCs), and lysophosphatidylethanolamines (LysoPE).
Amino acid metabolism
Amino acids (AAs) are important biological compounds, which possess carboxylic and amine moieties as functional groups and serve as building blocks for proteins. They play key metabolic and physiological roles in all living organisms. It has been known that obesity is accompanied by an increase in circulating levels of several amino acids, including BCAAs (Chevalier et al., 2005; Felig et al., 1974; Krebs et al., 2007; Moore and Stein, 1954; Tremblay et al., 2007; Um et al., 2006). High fasting concentrations of BCAAs were found to be associated with obesity and low serum insulin levels.
The association between obesity and amino acid metabolism shows important physiological and clinical inferences. The concentrations of selected essential amino acids and their derivatives in blood, in specific BCAAs, tyrosine, phenylalanine and sulfur amino acids, are evidently changed with obesity (Adams, 2011). A distinctive metabolic picture related to BCAA catabolism was also found in obese subjects (Newgard et al., 2009). Reduced expression of mitochondrial branched chain amino transferase in adipose tissue is considered to be a reason for the increased levels of BCAAs in obesity (She et al., 2007). Decreased levels of glutamine and glycine are found in blood of obese individuals in comparison with lean controls (Xie et al., 2012), which was also observed in another previous study (Backman et al., 1975). Changes in the levels of a number of compounds involved in amino acid metabolism have been found in obese condition (Adams, 2011; Newgard et al., 2009). The levels of cysteine, glutamate, phenylalanine, threonine, tryptophan, tyrosine, pantothenic acid, α-hydroxyisovaleric acid, 3-methyl-2-oxobutyric acid, choline, glyceric acid, 2-ketoisocaproic acid, pipecolic acid, kynurenine, serotonin and isovaleryl carnitine are increased (Fiehn et al., 2010; Gogna et al., 2015; Moore et al., 2014; Zeng et al., 2010), whereas those of methionine, citrulline, 3-methylglutarylcarnitine and acylcarnitine are decreased (Kim et al., 2011; Mihalik et al., 2012; Oberbach et al., 2011; Wahl et al., 2012). Other metabolites such as, alanine, arginine, asparagine, glutamine, glycine, histidine, isoleucine, leucine, lysine, proline, valine, carnitines and hydroxycarnitines were increased in some studies (Fiehn et al., 2010; Kim et al., 2010; 2011; Moore et al., 2014; Oberbach et al., 2011; Wahl et al., 2012) and decreased in others (Mihalik et al., 2012).
Increased levels of amino acids can improve insulin secretion from primary islet cells and β-cell lines by diverse mechanisms (Brennan et al., 2002; Charles and Henquin, 1983; Dixon et al., 2003; Sener and Malaisse, 1980; Smith et al., 1997). The relationship between amino acids and insulin resistance has been known for decades (Felig et al., 1969; 1974; Gougeon et al., 2008), but with the arrival of comprehensive metabolomic profiling a more detailed picture emerges of how amino acids contribute to the progression of diabetes. The levels of cysteine, pantothenic acid, creatine, acetyl carnitine and butyryl carnitine are increased in diabetes patients (Bentley-Lewis et al., 2014; Fiehn et al., 2010; Guan et al., 2013; Suhre et al., 2010), whereas those of arginine, asparagine, glycine, methionine, citrulline, betaine, sarcosine, aspartic acid, benzoic acid, 4-hydroxyproline and 5-hydroxykynurenine are decreased (Bentley-Lewis et al., 2014; Diao et al., 2014; Fiehn et al., 2010; Floegel et al., 2013; Liu et al., 2013; Mihalik et al., 2012; Wang-Sattler et al., 2012). Other metabolites such as, alanine, glutamate, glutamine, histidine, isoleucine, leucine, lysine, phenylalanine, proline, serine, threonine, tryptophan, tyrosine, valine, creatinine, 2-ketocaproic acid, pyroglutamic acid, choline, ornithine, carnitines and hydroxycarnitines were increased in some studies (Bentley-Lewis et al., 2014; Diao et al., 2014; Fiehn et al., 2010; Floegel et al., 2013; Liu et al., 2013; Mihalik et al., 2012; Palmer et al., 2015; Salek et al., 2007; Wang et al., 2011; Wang-Sattler et al., 2012; Zeng et al., 2010) and decreased in others (Bentley-Lewis et al., 2014; Diao et al., 2014; Fiehn et al., 2010; Liu et al., 2013; Mihalik et al., 2012; Palmer et al., 2015; Salek et al., 2007; Zeng et al., 2010). Glutamate and BCAAs are also found increased in T1DM (Oresic et al., 2008).
Lipid metabolism
Lipids are one of the main source of energy for metabolism. Blood lipids, obtained from food intake or from adipose tissue and liver, are mainly fatty acid derivatives and cholesterol. Obesity, which is characterized by fat deposits in tissues, is usually associated with elevated levels of plasma free fatty acids (FFAs) (Golay et al., 1986). Carnitine is an important metabolite linked to obesity, due to its involvement in fatty acid metabolism. It is well recognized that provision of FFAs can stimulate fatty acid oxidation (Randle, 1998). However, fatty acids can yield energy only via β-oxidation after esterification and transfer into the mitochondrion, which requires carnitine (Cha, 2008). Thus, higher levels of plasma FFAs in obesity may require more carnitine for effective β-oxidation. Consequently, the amount of carnitine in cells is the main factor regulating β-oxidation. Choline is a crucial dietary nutrient that is essential for the maintenance of cellular structure, methyl (one-carbon) metabolism, transport and metabolism of cholesterol (Blusztajn, 1998; Zeisel, 2000). Over 95% of the total choline present in most animal tissues is used for PC synthesis by the Kennedy pathway (Gibellini and Smith, 2010). The introduction of novel analytical and information technologies for management of large volumes of data has made it practical to characterize complex mixtures of lipids in body fluids and tissues (Lagarde et al., 2003; Wenk, 2005). Obesity and associated dyslipidemias may be caused by several factors (Despres et al., 1992). In obesity, the levels of non-essential fatty acids (oleic acid, palmitic acid, palmitoleic acid, stearic acid, stearoyl carnitine, 2-hydroxybutanoic acid and 3-hydroxybutanoic acid) are increased (Fiehn et al., 2010; Gogna et al., 2015; Kim et al., 2011; Moore et al., 2014), whereas those of ethanolamine and LysoPEs are decreased (Fiehn et al., 2010; Kim et al., 2011). LysoPCs, PCs, cholesteryl esters, ceramides, SMs and total fatty acids were increased in some studies (Kim et al., 2010; 2011; Moore et al., 2014; Pietiläinen et al., 2007; Samad et al., 2006; Wahl et al., 2012) and decreased in some others (Kim et al., 2010; 2011; Moore et al., 2014; Pietiläinen et al., 2007; Samad et al., 2006; Wahl et al., 2012)
Lipid abnormalities are common in diabetic patients, in particular in those with T2DM. Both the lipid profile and body fat mass have been reported to be the main predictors of metabolic disturbances and critical medical conditions, such as dyslipidemia, hypertension, diabetes and cardiovascular diseases (Du et al., 2013). Dyslipidemias make diabetic patients susceptible to heart disease and other problems related to atherosclerosis. A number of factors are likely to be responsible for diabetic dyslipidemia: the effects of insulin on apoprotein production in the liver, regulation of lipoprotein lipase, function of cholesteryl ester transfer protein and peripheral actions of insulin on adipose tissue and muscle (Mooradian et al., 2004; 2008). In diabetes, the levels of 1-monopalmitin, 1-monostearin, 1,2-distearoyl phosphatidylserine, ceramide, diacylglycerol, diacylphosphatidylcholines, fatty-acyl CoA, linoleic acid, myristic acid, oleic acid, palmitic acid, palmitoleic acid, stearic acid, tridecane, undecane, triacylglycerol, 2-hydroxybutanoic acid, and acetoacetic acid are increased (Fiehn et al., 2010; Liu et al., 2013; Salek et al., 2007; Villarreal-Pérez et al., 2014; Xu et al., 2013; Yan et al., 2014; Zeng et al., 2011), whereas those of acyl-alkyl-phosphatidylcholines and ethanolamine are decreased (Fiehn et al., 2010; Floegel et al., 2013; Suhre et al., 2010; Xu et al., 2013). Phosphatidylcholine, phosphatidylethanolamine, sphingomyelin and 3-hydroxybutanoic acid are increased in some studies (Drogan et al., 2015; Huang et al., 2011), and decreased in others (Floegel et al., 2013; Suhre et al., 2010; Xu et al., 2013). Phosphatidylcholine was also found decreased in T1DM (Oresic et al., 2008).
Carbohydrate metabolism
The progress of obesity is often explained by excessive consumption of high-fat and high-calorie diets (Saris, 2003; Siri-Tarino et al., 2010). Yet, the role of dietary carbohydrates in the development and maintenance of obesity is now also receiving increasing attention (Boling et al., 2009; Malik et al., 2010; Siri-Tarino et al., 2010; Wylie-Rosett and Davis, 2009). Obesity is associated with a decreased glucose disposal in adipose tissue. It has been reported that obesity leads to the development of hyperglycemia (Halaas et al., 1995), hyperlipidemia (Yamamoto et al., 2002), hyperinsulinemia (Berndt et al., 2005) and insulin resistance (Heilbronn et al., 2004). The pathways that metabolize carbohydrates include glycolysis, gluconeogenesis, the pentose phosphate pathway and the TCA cycle. Carbohydrate metabolism is vital to all metabolic processes. Glucose is catabolised via glycolysis to pyruvate, which under aerobic conditions is converted into acetyl coenzyme A, the entry point into the TCA cycle. Under anaerobic conditions, pyruvate is instead converted into lactate by lactate dehydrogenase. In obesity, the levels of glucose, lactic acid, fructose, glycerol, mannose, sorbitol, xylose, gluconic acid and glucuronic acid are increased (Fiehn et al., 2010; Gogna et al., 2015; Moore et al., 2014) whereas those of 1,5-anhydroglucitol and glycerol-3-phosphate are decreased (Fiehn et al., 2010; Moore et al., 2014).
Glucose concentration in blood reflects carbohydrate metabolism both in healthy people and in diabetics. In diabetes management, reduction in carbohydrate intake is the first and the most important step (Franz, 2008). Among compounds involved in carbohydrate metabolism, the levels of glucose, fructose, glycerol, inositol, mannose, sorbitol, xylose, 2,6-anhydrogalactose, 3,6-anhydrogalactose, gluconic acid, glucuronic acid, isopropanol, fumaric acid, malic acid and cis-aconitic acid are increased (Drogan et al., 2015; Fiehn et al., 2010; Gogna et al., 2015; Salek et al., 2007; Suhre et al., 2010; Xu et al., 2013; Zeng et al., 2010), whereas those of deoxygalactose, pyruvic acid, 1,5-anhydroglucitol and glycerol-3-phosphate are decreased in diabetes (Drogan et al., 2015; Fiehn et al., 2010; Suhre et al., 2010). Other metabolites such as acetic acid, deoxyglucose, lactic acid and citric acid were increased in some studies (Diao et al., 2014; Gogna et al., 2015; Salek et al., 2007; Suhre et al., 2010; Zeng et al., 2010) and decreased in others (Diao et al., 2014; Drogan et al., 2015). One metabolite, succinic acid is decreased in T1DM, though any significant changes are not observed in T2DM (Oresic et al., 2008).
Nucleotide metabolism
The study of nucleotide metabolism is vital for understanding of energy metabolism disorders, because nucleotides take part in most metabolic reactions and also act as coenzymes. Nucleotides are secreted from several body tissues and then build up in the blood circulation and other extracellular fluids. Release from cells and extracellular degradation both contribute to the nucleotide levels in the circulation. Persistently elevated blood nucleotide levels and constant purinergic signaling may play a pathophysiological role in metabolic disorders (Di Virgilio and Solini, 2002; Sellers et al., 2009; Sparks and Chatterjee, 2012). Nucleotide levels are extremely variable in obese subjects. High nucleotide levels observed in some morbidly obese subjects seem to be associated with the initial stage of insulin resistance (Sparks et al., 2014). Uric acid and uridine are strongly increased in obesity (Fiehn et al., 2010; Kim et al., 2011).
Analysis of the relationship between circulating nucleotide concentrations and clinical measures of T2DM demonstrated that changes in nucleotide metabolism are direct metabolic consequences of the disease and do not result from secondary complications (Dudzinska, 2014). In diabetes, compounds involved in nucleotide metabolism, including AMP, GMP, GTP, IMP, adenosine, guanosine and inosine are increased in both T1DM and T2DM (Dudzinska, 2014; Huang et al., 2011). Trimethylamine, glyoxylic acid and uridine are increased only in T2DM (Fiehn et al., 2010; Guan et al., 2013; Nikiforova et al., 2014; Padberg et al., 2014; Salek et al., 2007).
Besides the metabolites of major metabolic pathways, changes in the profiles of some other metabolites have been detected in obesity and diabetes. The levels of 3,6-anhydrogalactose, 7-ketodeoxycholic acid, heptadecanoic acid, inulobiose and myoinositol are increased, whereas those of, benzylalcohol, and a few glycerophospholipids are decreased in obesity (Fiehn et al., 2010; Gogna et al., 2015; Kim et al., 2011; Moore et al., 2014; Pietiläinen et al., 2007). The levels of cholesteryl-β-d-glucoside, dimethylamine, dimethylglucosamine, fructosamine and glycocholic acid are increased (Drogan et al., 2015; Fiehn et al., 2010; Huang et al., 2011; Liu et al., 2013; Padberg et al., 2014; Salek et al., 2007; Suhre et al., 2010).
METABOLITES SHOWING ALTERED CONCENTRATIONS IN BOTH OBESITY AND DIABETES
Obesity and diabetes have many similar causes and are related. These disorders often occur together, and most patients with T2DM are obese or were previously obese (Bray, 2004; Stumvoll et al., 2005). Obesity is usually considered as to strongly increase the risk of the later development of diabetes. The question arises whether obesity is not only a risk factor but also a cause of T2DM. A strong increase in obesity initiates a parallel upward trend in the incidence of diabetes. According to Ford et al. (1997) for every kilogram of weight gain, the risk of diabetes increases by between 4.5% and 9%. The relationship between obesity and insulin resistance is probably a cause-and-effect relationship, since human and animal studies show that weight loss or gain is closely associated with an increase or decrease in insulin sensitivity, respectively (Bak et al., 1992; Freidenberg et al., 1988; Sims et al., 1973). Longitudinal studies have shown that people with a genetic predisposition to compromised insulin secretion develop diabetes when they have developed insulin resistance due to obesity. Hence, T2DM develops once insulin secretion deteriorates to a level at which it cannot further compensate for insulin resistance. Also, in a longitudinal study of normal glucose tolerance offspring of two diabetic parents, Warram et al. (1990), revealed that patients presenting insulin resistance and extreme body weight were at increased risk of developing T2DM.
Several investigations have linked the phenotype and metabolism and identified serum or plasma metabolic markers that are involved independently in developing obesity (He et al., 2012; Williams et al., 2006) and diabetes (Bao et al., 2009; Chen et al., 2009). The occurrence of diabetes is positively associated with increased BMI across several ethnic groups (Chan et al., 2009; Maskarinec et al., 2009). Thirty nine metabolites are associated with both diabetes and high BMI or obesity (Table 1). Among compounds involved in amino acid metabolism, the levels of BCAAs, cysteine, glutamine, phenylalanine, proline, tyrosine, threonine, tryptophan, choline and pantothenic acid are increased, whereas those of asparagine, glycine, methionine and citrulline are decreased in both obesity and diabetes. Among compounds involved in lipid metabolism, the levels of 2-hydroxybutanoic acid, 3-hydroxybutanoic acid, phosphatidylcholine, stearic acid, oleic acid, palmitic acid and palmitoleic acid are increased, whereas those of ethanolamine, lysophosphatidylcholine 18:2, lysophosphatidylethanolamine and sphingomyelin are decreased. Among compounds involved in carbohydrate metabolism, the levels of fructose, glycerol, mannose, xylose, sorbitol, lactate, glucose, gluconic acid and glucuronic acid are increased, whereas those of 1,5-anhydroglucitol and glycerol-3-phosphate are decreased. Among compounds involved in nucleotide metabolism, the level of uridine is increased. All these metabolites along with the references are categorized and summarized in Table 1.
Table 1.
List of the metabolites changed in both obesity and diabetes
 , NMR;
, NMR;
 , LC/MS;
, LC/MS;
 , GC/MS
, GC/MS
STRENGH AND LIMITATIONS
This study summarizes the major metabolites, metabolite classes and pathways associated with obesity and diabetes with a focus on metabolites associated with both disorders. The study also discusses some aspects of the development of these disorders and transformation of obesity into a diabetic state. Overall, metabolomics research can provide comprehensive information about metabolite changes in the studied disease using various sample types in minimum quantities and in a very short time. The limitations of the reviewed studies were the differences in the analytical platforms used, gender, age, diet, ethnicity and the source species of the samples, which may have led to some misinterpretations in the assessment. Other unknown diseases may also affect the outcome of metabolomics studies.
CONCLUSION
Obesity and diabetes are well-known to be associated. This study provides information on all major metabolites associated with obesity and diabetes and also common metabolites associated with both disorders. The classification and changes in metabolites in each comparison have been represented in Fig. 1. These metabolites can be considered as biomarkers for metabolomics studies of obesity and diabetes and can be a focus area of future research. This study can also provide some therapeutic clues for treatment of obesity-related diabetes. Further studies are required to test whether the selected metabolites might be useful as diagnostic tools and to reveal the biological mechanisms that result in changes in the levels of certain metabolites in the pathogenesis of these metabolic diseases.
Fig. 1.
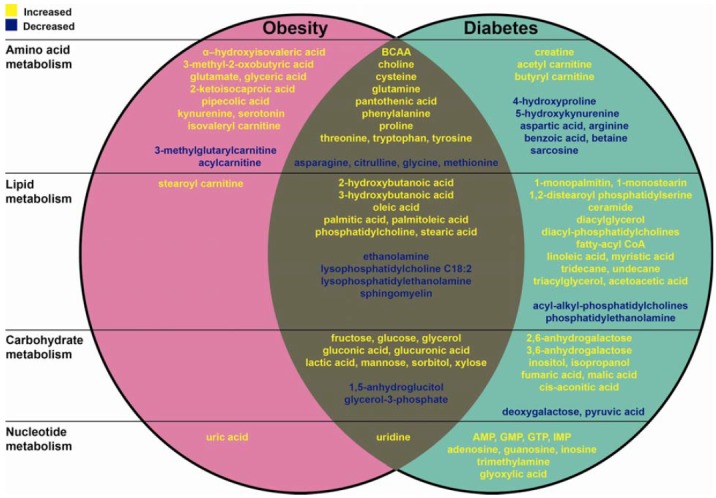
Summary of the metabolites changed in obesity and diabetes
Acknowledgments
This work was supported by the National Research Foundation (2013M3C7A1056099 and 2012M3A9C6049935) and the DGIST R&D Program of the Ministry of Science, Information and Communication Technology, and Future Planning (15-BD-0402).
REFERENCES
- Abu Bakar MH, Sarmidi MR, Cheng KK, Ali Khan A, Suan CL, Zaman Huri H, Yaakob H. Metabolomics - the complementary field in systems biology: a review on obesity and type 2 diabetes. Mol. Biosyst. 2015 doi: 10.1039/c5mb00158g. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Adams SH. Emerging perspectives on essential amino acid metabolism in obesity and the insulin-resistant state. Adv. Nutr. 2011;2:445–456. doi: 10.3945/an.111.000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backman L, Hallberg D, Kallner A. Amino acid pattern in plasma before and after jejuno-ileal shunt operation for obesity. Scand J. Gastroenterol. 1975;10:811–816. [PubMed] [Google Scholar]
- Bak JF, Moller N, Schmitz O, Saaek A, Pedersen O. In vivo insulin action and muscle glycogen synthase activity in type 2 (non-insulin-dependent) diabetes mellitus: effects of diet treatment. Diabetologia. 1992;35:777–784. doi: 10.1007/BF00429100. [DOI] [PubMed] [Google Scholar]
- Bala L, Ghoshal UC, Ghoshal U, Tripathi P, Misra A, Gowda GA, Khetrapal CL. Malabsorption syndrome with and without small intestinal bacterial overgrowth: a study on upper-gut aspirate using 1H NMR spectroscopy. Magn. Reson. Med. 2006;56:738–744. doi: 10.1002/mrm.21041. [DOI] [PubMed] [Google Scholar]
- Bao Y, Zhao T, Wang X, Qiu Y, Su M, Jia W. Metabonomic variations in the drug-treated type 2 diabetes mellitus patients and healthy volunteers. J. Proteome Res. 2009;8:1623–1630. doi: 10.1021/pr800643w. [DOI] [PubMed] [Google Scholar]
- Barbas C, Moraes EP, Villaseñor A. Capillary electrophoresis as a metabolomics tool for non-targeted fingerprinting of biological samples. J. Pharm. Biomed. Anal. 2011;55:823–831. doi: 10.1016/j.jpba.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Beger RD. A review of applications of metabolomics in cancer. Metabolites. 2013;3:552–574. doi: 10.3390/metabo3030552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley-Lewis R, Xiong G, Lee H, Yang A, Huynh J, Kim C. Metabolomic analysis reveals amino acid responses to an oral glucose tolerance test in women with prior history of gestational diabetes mellitus. J. Clin. Transl. Endocrinol. 2014;1:38–43. doi: 10.1016/j.jcte.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt J, Kloting N, Kralisch S, Kovacs P, Fasshauer M, Schon MR, Stumvoll M, Bluher M. Plasma visfatin concentrations and fat depot-specific mRNA expression in humans. Diabetes. 2005;54:2911–2916. doi: 10.2337/diabetes.54.10.2911. [DOI] [PubMed] [Google Scholar]
- Blusztajn JK. Choline, a vital amine. Science. 1998;281:794–795. doi: 10.1126/science.281.5378.794. [DOI] [PubMed] [Google Scholar]
- Boling CL, Westman EC, Yancy WS., Jr Carbohydrate-restricted diets for obesity and related diseases: an update. Curr. Atheroscler. Rep. 2009;11:462–469. doi: 10.1007/s11883-009-0069-8. [DOI] [PubMed] [Google Scholar]
- Bollard ME, Stanley EG, Lindon JC, Nicholson JK, Holmes E. NMR-based metabonomic approaches for evaluating physiological influences on biofluid composition. NMR Biomed. 2005;18:143–162. doi: 10.1002/nbm.935. [DOI] [PubMed] [Google Scholar]
- Bray GA. Medical consequences of obesity. J. Clin. Endocrinol. Metab. 2004;89:2583–2589. doi: 10.1210/jc.2004-0535. [DOI] [PubMed] [Google Scholar]
- Brennan L, Shine A, Hewage C, Malthouse JP, Brindle KM, McClenaghan N, Flatt PR, Newsholme P. A nuclear magnetic resonance-based demonstration of substantial oxidative L-alanine metabolism and L-alanine-enhanced glucose metabolism in a clonal pancreatic beta-cell line: metabolism of L-alanine is important to the regulation of insulin secretion. Diabetes. 2002;51:1714–1721. doi: 10.2337/diabetes.51.6.1714. [DOI] [PubMed] [Google Scholar]
- Carr PW, Stoll DR, Wang X. Perspectives on recent advances in the speed of high-performance liquid chromatography. Anal. Chem. 2011;83:1890–1900. doi: 10.1021/ac102570t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha YS. Effects of L-carnitine on obesity, diabetes, and as an ergogenic aid. Asia Pac. J. Clin. Nutr. 2008;17:306–308. [PubMed] [Google Scholar]
- Chan JC, Malik V, Jia W, Kadowaki T, Yajnik CS, Yoon KH, Hu FB. Diabetes in Asia: epidemiology, risk factors, and pathophysiology. JAMA. 2009;301:2129–2140. doi: 10.1001/jama.2009.726. [DOI] [PubMed] [Google Scholar]
- Charles S, Henquin JC. Distinct effects of various amino acids on 45Ca2+ fluxes in rat pancreatic islets. Biochem. J. 1983;214:899–907. doi: 10.1042/bj2140899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhao X, Fritsche J, Yin P, Schmitt-Kopplin P, Wang W, Lu X, Häring HU, Schleicher ED, Lehmann R, et al. Practical approach for the identification and isomer elucidation of biomarkers detected in a metabonomic study for the discovery of individuals at risk for diabetes by integrating the chromatographic and mass spectrometric information. Anal. Chem. 2009;80:1280–1289. doi: 10.1021/ac702089h. [DOI] [PubMed] [Google Scholar]
- Cheng S, Rhee EP, Larson MG, Lewis GD, McCabe EL, Shen D, Palma MJ, Roberts LD, Dejam A, Souza AL, et al. Metabolite profiling identifies pathways associated with metabolic risk in humans. Circulation. 2012;125:2222–2231. doi: 10.1161/CIRCULATIONAHA.111.067827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier S, Marliss EB, Morais JA, Lamarche M, Gougeon R. Whole-body protein anabolic response is resistant to the action of insulin in obese women. Am. J. Clin. Nutr. 2005;82:355–365. doi: 10.1093/ajcn.82.2.355. [DOI] [PubMed] [Google Scholar]
- Cho Y, Ahmed A, Islam A, Kim S. Developments in FT-ICR MS instrumentation, ionization techniques, and data interpretation methods for petroleomics. Mass Spectrom. Rev. 2015;34:248–263. doi: 10.1002/mas.21438. [DOI] [PubMed] [Google Scholar]
- Chylla RA, Hu K, Ellinger JJ, Markley JL. Deconvolution of two-dimensional NMR spectra by fast maximum likelihood reconstruction: application to quantitative metabolomics. Anal. Chem. 2011;83:4871–4880. doi: 10.1021/ac200536b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courant F, Royer AL, Chereau S, Morvan ML, Monteau F, Antignac JP, Le Bizec B. Implementation of a semi-automated strategy for the annotation of metabolomic fingerprints generated by liquid chromatography-high resolution mass spectrometry from biological samples. Analyst. 2012;137:4958–4967. doi: 10.1039/c2an35865d. [DOI] [PubMed] [Google Scholar]
- Despres JP, Moorjani S, Lupien PJ, Tremblay A, Nadeau A, Bouchard C. Genetic aspects of susceptibility to obesity and related dyslipidemias. Mol. Cell Biochem. 1992;113:151–169. doi: 10.1007/BF00231535. [DOI] [PubMed] [Google Scholar]
- Di Virgilio F, Solini A. P2 receptors: new potential players in atherosclerosis. Br J. Pharmacol. 2002;135:831–842. doi: 10.1038/sj.bjp.0704524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao C, Zhao L, Guan M, Zheng Y, Chen M, Yang Y, Lin L, Chen W, Gao H. Systemic and characteristic metabolites in the serum of streptozotocin-induced diabetic rats at different stages as revealed by a (1)H-NMR based metabonomic approach. Mol. Biosyst. 2014;10:686–693. doi: 10.1039/c3mb70609e. [DOI] [PubMed] [Google Scholar]
- Dixon G, Nolan J, McClenaghan N, Flatt PR, Newsholme P. A comparative study of amino acid consumption by rat islet cells and the clonal beta-cell line BRIN-BD11 - the functional significance of L-alanine. J. Endocrinol. 2003;179:447–454. doi: 10.1677/joe.0.1790447. [DOI] [PubMed] [Google Scholar]
- Drogan D, Dunn WB, Lin W, Buijsse B, Schulze MB, Langenberg C, Brown M, Floegel A, Dietrich S, Rolandsson O, et al. Untargeted metabolic profiling identifies altered serum metabolites of type 2 diabetes mellitus in a prospective, nested case control study. Clin. Chem. 2015;61:487–497. doi: 10.1373/clinchem.2014.228965. [DOI] [PubMed] [Google Scholar]
- Du F, Virtue A, Wang H, Yang XF. Metabolomic analyses for atherosclerosis, diabetes, and obesity. Biomark Res. 2013;1:17. doi: 10.1186/2050-7771-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudzinska W. Purine nucleotides and their metabolites in patients with type 1 and 2 diabetes mellitus. J. Biomed. Sci. Eng. 2014;7:38–44. [Google Scholar]
- Felig P, Marliss E, Cahill GF., Jr Plasma amino acid levels and insulin secretion in obesity. N Engl. J. Med. 1969;281:811–816. doi: 10.1056/NEJM196910092811503. [DOI] [PubMed] [Google Scholar]
- Felig P, Wahren J, Hendler R, Brundin T. Splanchnic glucose and amino acid metabolism in obesity. J. Clin. Invest. 1974;53:582–590. doi: 10.1172/JCI107593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O, Garvey WT, Newman JW, Lok KH, Hoppel CL, Adams SH. Plasma metabolomic profiles reflective of glucose homeostasis in non-diabetic and type 2 diabetic obese African-American women. PLoS One. 2010;5:e15234. doi: 10.1371/journal.pone.0015234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floegel A, Stefan N, Yu Z, Mühlenbruch K, Drogan D, Joost HG, Fritsche A, Häring HU, Hrabě de Angelis M, Peters A, et al. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes. 2013;62:639–648. doi: 10.2337/db12-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford ES, Williamson DF, Liu S. Weight change and diabetes incidence: findings from a national cohort of US adults. Am. J. Epidemiol. 1997;146:214–222. doi: 10.1093/oxfordjournals.aje.a009256. [DOI] [PubMed] [Google Scholar]
- Franz MJ. Food, Nutrition and Diet therapy. Canada: Saunders; 2008. Medical nutrition therapy for diabetes mellitus and hypoglycemia of nondiabetic origin. [Google Scholar]
- Freidenberg GR, Reichart D, Olefsky JM, Henry RR. Reversibility of defective adipocyte insulin receptor kinase activity in non-insulin-dependent diabetes mellitus. Effect of weight loss. J. Clin. Invest. 1988;82:1398–1406. doi: 10.1172/JCI113744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich N. Metabolomics in diabetes research. Metabolomics in human type 2 diabetes research. J. Endocrinol. 2012;215:29–42. doi: 10.1530/JOE-12-0120. [DOI] [PubMed] [Google Scholar]
- Gibellini F, Smith TK. The Kennedy pathway--De novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life. 2010;62:414–428. doi: 10.1002/iub.337. [DOI] [PubMed] [Google Scholar]
- Goek ON, Doring A, Gieger C, Heier M, Koenig W, Prehn C, Romisch-Margl W, Wang-Sattler R, Illig T, Suhre K, et al. Serum metabolite concentrations and decreased GFR in the general population. Am. J. Kidney Dis. 2012;60:197–206. doi: 10.1053/j.ajkd.2012.01.014. [DOI] [PubMed] [Google Scholar]
- Gogna N, Krishna M, Oommen AM, Dorai K. Investigating correlations in the altered metabolic profiles of obese and diabetic subjects in a South Indian Asian population using an NMR-based metabolomic approach. Mol. Biosyst. 2015;11:595–606. doi: 10.1039/c4mb00507d. [DOI] [PubMed] [Google Scholar]
- Golay A, Swislocki AL, Chen YD, Jaspan JB, Reaven GM. Effect of obesity on ambient plasma glucose, free fatty acid, insulin, growth hormone, and glucagon concentrations. J. Clin. Endocrinol. Metab. 1986;63:481–484. doi: 10.1210/jcem-63-2-481. [DOI] [PubMed] [Google Scholar]
- Goodacre R, Vaidyanathan S, Dunn WB, Harrigan GG, Kell DB. Metabolomics by numbers: acquiring and understanding global metabolite data. Trends Biotechnol. 2004;22:245–252. doi: 10.1016/j.tibtech.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Gougeon R, Morais JA, Chevalier S, Pereira S, Lamarche M, Marliss EB. Determinants of whole-body protein metabolism in subjects with and without type 2 diabetes. Diabetes Care. 2008;31:128–133. doi: 10.2337/dc07-1268. [DOI] [PubMed] [Google Scholar]
- Gowda GA, Ijare OB, Somashekar BS, Sharma A, Kapoor VK, Khetrapal CL. Single-step analysis of individual conjugated bile acids in human bile using 1H NMR spectroscopy. Lipids. 2006;41:591–603. doi: 10.1007/s11745-006-5008-7. [DOI] [PubMed] [Google Scholar]
- Griffin JL, Kauppinen RA. Tumour metabolomics in animal models of human cancer. J. Proteome Res. 2007;6:498–505. doi: 10.1021/pr060464h. [DOI] [PubMed] [Google Scholar]
- Guan M, Xie L, Diao C, Wang N, Hu W, Zheng Y, Jin L, Yan Z, Gao H. Systemic perturbations of key metabolites in diabetic rats during the evolution of diabetes studied by urine metabonomics. PLoS One. 2013;8:e60409. doi: 10.1371/journal.pone.0060409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaas JL, Gajiwala KS, Maffei M, Cohen SL, Chait BT, Rabinowitz D, Lallone RL, Burley SK, Friedman JM. Weight-reducing effects of the plasma protein encoded by the obese gene. Science. 1995;269:543–546. doi: 10.1126/science.7624777. [DOI] [PubMed] [Google Scholar]
- Haslam DW, James WP. Effect of obesity on the incidence of type 2 diabetes mellitus varies with age among Indian women. Lancet. 2005;366:1197–1209. doi: 10.1016/S0140-6736(05)67483-1. [DOI] [PubMed] [Google Scholar]
- He Q, Ren P, Kong X, Wu Y, Wu G, Li P, Hao F, Tang H, Blachier F, Yin Y. Comparison of serum metabolite compositions between obese and lean growing pigs using an NMR-based metabonomic approach. J. Nutr. Biochem. 2012;23:133–139. doi: 10.1016/j.jnutbio.2010.11.007. [DOI] [PubMed] [Google Scholar]
- Heilbronn LK, Rood J, Janderova L, Albu JB, Kelley DE, Ravussin E, Smith SR. Relationship between serum resistin concentrations and insulin resistance in nonobese, obese, and obese diabetic subjects. J. Clin. Endocrinol. Metab. 2004;89:1844–1848. doi: 10.1210/jc.2003-031410. [DOI] [PubMed] [Google Scholar]
- Huang Q, Yin P, Wang J, Chen J, Kong H, Lu X, Xu G. Method for liver tissue metabolic profiling study and its application in type 2 diabetic rats based on ultra performance liquid chromatography-mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2011;879:961–967. doi: 10.1016/j.jchromb.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Junot C, Madalinski G, Tabet JC, Ezan E. Fourier transform mass spectrometry for metabolome analysis. Analyst. 2010;135:2203–2219. doi: 10.1039/c0an00021c. [DOI] [PubMed] [Google Scholar]
- Kim JY, Park JY, Kim OY, Ham BM, Kim HJ, Kwon DY, Jang Y, Lee JH. Metabolic profiling of plasma in overweight/obese and lean men using ultra performance liquid chromatography and Q-TOF mass spectrometry (UPLC-Q-TOF MS) J. Proteome Res. 2010;9:4368–4375. doi: 10.1021/pr100101p. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Kim JH, Noh S, Hur HJ, Sung MJ, Hwang JT, Park JH, Yang HJ, Kim MS, Kwon DY, et al. Metabolomic analysis of livers and serum from high-fat diet induced obese mice. J. Proteome Res. 2011;10:722–731. doi: 10.1021/pr100892r. [DOI] [PubMed] [Google Scholar]
- Krebs M, Brunmair B, Brehm A, Artwohl M, Szendroedi J, Nowotny P, Roth E, Furnsinn C, Promintzer M, Anderwald C, et al. The Mammalian target of rapamycin pathway regulates nutrient-sensitive glucose uptake in man. Diabetes. 2007;56:1600–1607. doi: 10.2337/db06-1016. [DOI] [PubMed] [Google Scholar]
- Kussmann M, Raymond F, Affolter M. OMICS-driven biomarker discovery in nutrition and health. J. Biotechnol. 2006;124:758–787. doi: 10.1016/j.jbiotec.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Lagarde M, Géloën A, Record M, Vance D, Spener F. Lipidomics is emerging. Biochim. Biophys. Acta. 2003;1634:61. doi: 10.1016/j.bbalip.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Lin L, Yu Q, Yan X, Hang W, Zheng J, Xing J, Huang B. Direct infusion mass spectrometry or liquid chromatography mass spectrometry for human metabonomics? A serum metabonomic study of kidney cancer. Analyst. 2010;135:2970–2978. doi: 10.1039/c0an00265h. [DOI] [PubMed] [Google Scholar]
- Liu L, Wang M, Yang X, Bi M, Na L, Niu Y, Li Y, Sun C. Fasting serum lipid and dehydroepiandrosterone sulfate as important metabolites for detecting isolated post-challenge diabetes: serum metabolomics via ultra-high-performance LC-MS. Clin. Chem. 2013;59:1338–1348. doi: 10.1373/clinchem.2012.200527. [DOI] [PubMed] [Google Scholar]
- Madsen R, Lundstedt T, Trygg J. Chemometrics in metabolomics--a review in human disease diagnosis. Anal. Chim. Acta. 2010;659:23–33. doi: 10.1016/j.aca.2009.11.042. [DOI] [PubMed] [Google Scholar]
- Malik VS, Popkin BM, Bray GA, Despres JP, Hu FB. Sugar-sweetened beverages, obesity, type 2 diabetes mellitus, and cardiovascular disease risk. Circulation. 2010;121:1356–1364. doi: 10.1161/CIRCULATIONAHA.109.876185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskarinec G, Grandinetti A, Matsuura G, Sharma S, Mau M, Henderson BE, Kolonel LN. Diabetes prevalence and body mass index differ by ethnicity: the Multiethnic Cohort. Ethn. Dis. 2009;19:49–55. [PMC free article] [PubMed] [Google Scholar]
- Mihalik SJ, Michaliszyn SF, de las Heras J, Bacha F, Lee S, Chace DH, DeJesus VR, Vockley J, Arslanian SA. Metabolomic profiling of fatty acid and amino acid metabolism in youth with obesity and type 2 diabetes: evidence for enhanced mitochondrial oxidation. Diabetes Care. 2012;35:605–611. doi: 10.2337/DC11-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooradian AD, Haas MJ, Wong NC. Transcriptional control of apolipoprotein A–I gene expression in diabetes. Diabetes. 2004;53:513–520. doi: 10.2337/diabetes.53.3.513. [DOI] [PubMed] [Google Scholar]
- Mooradian AD, Haas MJ, Wehmeier KR, Wong NC. Obesity-related changes in high-density lipoprotein metabolism. Obesity (Silver Spring) 2008;16:1152–1160. doi: 10.1038/oby.2008.202. [DOI] [PubMed] [Google Scholar]
- Moore S, Stein WH. Procedures for the chromatographic determination of amino acids on four per cent cross-linked sulfonated polystyrene resins. J. Biol. Chem. 1954;211:893–906. [PubMed] [Google Scholar]
- Moore SC, Matthews CE, Sampson JN, Stolzenberg-Solomon RZ, Zheng W, Cai Q, Tan YT, Chow WH, Ji BT, Liu DK, et al. Human metabolic correlates of body mass index. Metabolomics. 2014;10:259–269. doi: 10.1007/s11306-013-0574-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforova VJ, Giesbertz P, Wiemer J, Bethan B, Looser R, Liebenberg V, Ruiz Noppinger P, Daniel H, Rein D. Glyoxylate, a new marker metabolite of type 2 diabetes. J. Diabetes Res. 2014;2014:685204. doi: 10.1155/2014/685204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyenwe EA, Jerkins TW, Umpierrez GE, Kitabchi AE. Management of type 2 diabetes: evolving strategies for the treatment of patients with type 2 diabetes. Metabolism. 2011;60:1–23. doi: 10.1016/j.metabol.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberbach A, Bluher M, Wirth H, Till H, Kovacs P, Kullnick Y, Schlichting N, Tomm JM, Rolle-Kampczyk U, Murugaiyan J, et al. Combined proteomic and metabolomic profiling of serum reveals association of the complement system with obesity and identifies novel markers of body fat mass changes. J. Proteome Res. 2011;10:4769–4788. doi: 10.1021/pr2005555. [DOI] [PubMed] [Google Scholar]
- Oliver SG, Winson MK, Kell DB, Baganz F. Systematic functional analysis of the yeast genome. Trends Biotechnol. 1998;16:373–378. doi: 10.1016/s0167-7799(98)01214-1. [DOI] [PubMed] [Google Scholar]
- Oresic M, Simell S, Sysi-Aho M, Nanto-Salonen K, Seppanen-Laakso T, Parikka V, Katajamaa M, Hekkala A, Mattila I, Keskinen P, et al. Dysregulation of lipid and amino acid metabolism precedes islet autoimmunity in children who later progress to type 1 diabetes. J. Exp. Med. 2008;205:2975–2984. doi: 10.1084/jem.20081800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padberg I, Peter E, Gonzalez-Maldonado S, Witt H, Mueller M, Weis T, Bethan B, Liebenberg V, Wiemer J, Katus HA, et al. A new metabolomic signature in type-2 diabetes mellitus and its pathophysiology. PLoS One. 2014;9:e85082. doi: 10.1371/journal.pone.0085082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer ND, Stevens RD, Antinozzi PA, Anderson A, Bergman RN, Wagenknecht LE, Newgard CB, Bowden DW. Metabolomic profile associated with insulin resistance and conversion to diabetes in the insulin resistance atherosclerosis study. J. Clin. Endocrinol. Metab. 2015;100:E463–468. doi: 10.1210/jc.2014-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pataky Z, Armand S, Muller-Pinget S, Golay A, Allet L. Effects of obesity on functional capacity. Obesity (Silver Spring) 2014;22:56–62. doi: 10.1002/oby.20514. [DOI] [PubMed] [Google Scholar]
- Pietiläinen KH, Sysi-Aho M, Rissanen A, Seppänen-Laakso T, Yki-Järvinen H, Kaprio J, Oresic M. Acquired obesity is associated with changes in the serum lipidomic profile independent of genetic effects--a monozygotic twin study. PLoS One. 2007;2:e218. doi: 10.1371/journal.pone.0000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randle PJ. Regulatory interactions between lipids and carbohydrates: the glucose fatty acid cycle after 35 years. Diabetes Metab. Rev. 1998;14:263–283. doi: 10.1002/(sici)1099-0895(199812)14:4<263::aid-dmr233>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Rhee EP, Cheng S, Larson MG, Walford GA, Lewis GD, McCabe E, Yang E, Farrell L, Fox CS, O’Donnell CJ, et al. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J. Clin. Invest. 2011;121:1402–1411. doi: 10.1172/JCI44442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadanala KC, Jeongae L, Bong Chul C, Man Ho C. Targeted metabolite profiling: sample preparation techniques for GC-MS-based steroid analysis. Mass Spectrometry Lett. 2012;3:4–9. [Google Scholar]
- Salek RM, Maguire ML, Bentley E, Rubtsov DV, Hough T, Cheeseman M, Nunez D, Sweatman BC, Haselden JN, Cox RD, et al. A metabolomic comparison of urinary changes in type 2 diabetes in mouse, rat, and human. Physiol. Genomics. 2007;29:99–108. doi: 10.1152/physiolgenomics.00194.2006. [DOI] [PubMed] [Google Scholar]
- Samad F, Hester KD, Yang G, Hannun YA, Bielawski J. Altered adipose and plasma sphingolipid metabolism in obesity: a potential mechanism for cardiovascular and metabolic risk. Diabetes. 2006;55:2579–2587. doi: 10.2337/db06-0330. [DOI] [PubMed] [Google Scholar]
- Saris WH. Sugars, energy metabolism, and body weight control. Am. J. Clin. Nutr. 2003;78:850S–857S. doi: 10.1093/ajcn/78.4.850S. [DOI] [PubMed] [Google Scholar]
- Scarfe GB, Wright B, Clayton E, Taylor S, Wilson I, Lindon JC, Nicholson JK. 19F-NMR and directly coupled HPLC-NMR-MS investigations into the metabolism of 2-bromo-4-trifluoromethylaniline in rat: a urinary excretion balance study without the use of radiolabelling. Xenobiotica. 1998;28:373–388. doi: 10.1080/004982598239489. [DOI] [PubMed] [Google Scholar]
- Sellers MB, Tricoci P, Harrington RA. A new generation of antiplatelet agents. Curr. Opin. Cardiol. 2009;24:307–312. doi: 10.1097/HCO.0b013e32832e2b44. [DOI] [PubMed] [Google Scholar]
- Sener A, Malaisse WJ. L-leucine and a nonmetabolized analogue activate pancreatic islet glutamate dehydrogenase. Nature. 1980;288:187–189. doi: 10.1038/288187a0. [DOI] [PubMed] [Google Scholar]
- Shah SH, Kraus WE, Newgard CB. Metabolomic profiling for the identification of novel biomarkers and mechanisms related to common cardiovascular diseases: form and function. Circulation. 2012;126:1110–1120. doi: 10.1161/CIRCULATIONAHA.111.060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham O, Wei R, Wang TJ, Ricciardi C, Lewis GD, Vasan RS, Carr SA, Thadhani R, Gerszten RE, Mootha VK. Metabolic profiling of the human response to a glucose challenge reveals distinct axes of insulin sensitivity. Mol. Syst. Biol. 2008;4:214. doi: 10.1038/msb.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She P, Reid TM, Bronson SK. Disruption of BCATm in mice leads to increased energy expenditure associated with the activation of a futile protein turnover cycle. Cell Metab. 2007;6:181–194. doi: 10.1016/j.cmet.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims EA, Danforth E, Jr, Horton ES, Bray GA, Glennon JA, Salans LB. Endocrine and metabolic effects of experimental obesity in man. Recent Prog. Horm. Res. 1973;29:457–496. doi: 10.1016/b978-0-12-571129-6.50016-6. [DOI] [PubMed] [Google Scholar]
- Siri-Tarino PW, Sun Q, Hu FB, Krauss RM. Saturated fat, carbohydrate, and cardiovascular disease. Am. J. Clin. Nutr. 2010;91:502–509. doi: 10.3945/ajcn.2008.26285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PA, Sakura H, Coles B, Gummerson N, Proks P, Ashcroft FM. Electrogenic arginine transport mediates stimulus-secretion coupling in mouse pancreatic beta-cells. J. Physiol. 1997;499(Pt 3):625–635. doi: 10.1113/jphysiol.1997.sp021955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks DL, Chatterjee C. Purinergic signaling, dyslipidemia and inflammatory disease. Cell Physiol. Biochem. 2012;30:1333–1339. doi: 10.1159/000343322. [DOI] [PubMed] [Google Scholar]
- Sparks DL, Doelle H, Chatterjee C. Circulating nucleotide levels in health and disease. Receptors Clin. Invest. 2014;1:e344. [Google Scholar]
- Stolar MW. Atherosclerosis in diabetes: the role of hyperinsulinemia. Metabolism. 1988;37:1–9. doi: 10.1016/0026-0495(88)90180-1. [DOI] [PubMed] [Google Scholar]
- Stumvoll M, Goldstein BJ, van Haeften TW. Type 2 diabetes: principles of pathogenesis and therapy. Lancet. 2005;365:1333–1346. doi: 10.1016/S0140-6736(05)61032-X. [DOI] [PubMed] [Google Scholar]
- Suhre K, Meisinger C, Doring A, Altmaier E, Belcredi P, Gieger C, Chang D, Milburn MV, Gall WE, Weinberger KM, et al. Metabolic footprint of diabetes: a multiplatform metabolomics study in an epidemiological setting. PLoS One. 2010;5:e13953. doi: 10.1371/journal.pone.0013953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay F, Lavigne C, Jacques H, Marette A. Role of dietary proteins and amino acids in the pathogenesis of insulin resistance. Annu. Rev. Nutr. 2007;27:293–310. doi: 10.1146/annurev.nutr.25.050304.092545. [DOI] [PubMed] [Google Scholar]
- Um SH, D'Alessio D, Thomas G. Nutrient overload, insulin resistance, and ribosomal protein S6 kinase 1, S6K1. Cell Metab. 2006;3:393–402. doi: 10.1016/j.cmet.2006.05.003. [DOI] [PubMed] [Google Scholar]
- van Kampen JJ, Burgers PC, de Groot R, Gruters RA, Luider TM. Biomedical application of MALDI mass spectrometry for small-molecule analysis. Mass Spectrom Rev. 2011;30:101–120. doi: 10.1002/mas.20268. [DOI] [PubMed] [Google Scholar]
- Villarreal-Pérez JZ, Villarreal-Martínez JZ, Lavalle-González FJ, Torres-Sepúlveda MdR, Ruiz-Herrera C, Cerda-Flores RM, Castillo-García ER, Rodríguez-Sánchez IP, Villarreal LEMd. Plasma and urine metabolic profiles are reflective of altered beta-oxidation in non-diabetic obese subjects and patients with type 2 diabetes mellitus. Diabetol. Metab. Syndr. 2014;6:129–136. doi: 10.1186/1758-5996-6-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinayavekhin N, Homan EA, Saghatelian A. Exploring disease through metabolomics. ACS Chem. Biol. 2010;5:91–103. doi: 10.1021/cb900271r. [DOI] [PubMed] [Google Scholar]
- Wahl S, Yu Z, Kleber M, Singmann P, Holzapfel C, He Y, Mittelstrass K, Polonikov A, Prehn C, Römisch-Margl W, et al. Childhood obesity is associated with changes in the serum metabolite profile. Obes. Facts. 2012;5:660–670. doi: 10.1159/000343204. [DOI] [PubMed] [Google Scholar]
- Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, et al. Metabolite profiles and the risk of developing diabetes. Nat. Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang-Sattler R, Yu Z, Herder C, Messias AC, Floegel A, He Y, Heim K, Campillos M, Holzapfel C, Thorand B, et al. Novel biomarkers for pre-diabetes identified by metabolomics. Mol. Syst. Biol. 2012;8:615–625. doi: 10.1038/msb.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warram JH, Martin BC, Krolewski AS, Soeldner JS, Kahn CR. Slow glucose removal rate and hyperinsulinemia precede the development of type II diabetes in the offspring of diabetic parent. Ann. Intern. Med. 1990;113:909–915. doi: 10.7326/0003-4819-113-12-909. [DOI] [PubMed] [Google Scholar]
- Wenk MR. The emerging field of lipidomics. Nat. Rev. Drug Discov. 2005;4:594–610. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- Williams R, Lenz EM, Wilson AJ, Granger J, Wilson ID, Major H, Stumpf C, Plumb R. A multi-analytical platform approach to the metabonomic analysis of plasma from normal and Zucker (fa/fa) obese rats. Mol. Biosyst. 2006;2:174–183. doi: 10.1039/b516356k. [DOI] [PubMed] [Google Scholar]
- Wylie-Rosett J, Davis NJ. Low-carbohydrate diets: an update on current research. Curr. Diab. Rep. 2009;9:396–404. doi: 10.1007/s11892-009-0061-2. [DOI] [PubMed] [Google Scholar]
- Xie B, Waters MJ, Schirra HJ. Investigating potential mechanisms of obesity by metabolomics. J. Biomed. Biotechnol. 2012;2012:805683. doi: 10.1155/2012/805683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Tavintharan S, Sum CF, Woon K, Lim SC, Ong CN. Metabolic signature shift in type 2 diabetes mellitus revealed by mass spectrometry-based metabolomics. J. Clin. Endocrinol. Metab. 2013;98:E1060–1065. doi: 10.1210/jc.2012-4132. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Hirose H, Saito I, Tomita M, Taniyama M, Matsubara K, Okazaki Y, Ishii T, Nishikai K, Saruta T. Correlation of the adipocyte-derived protein adiponectin with insulin resistance index and serum high-density lipoproteincholesterol, independent of body mass index, in the Japanese population. Clin. Sci. 2002;103:137–142. doi: 10.1042/cs1030137. [DOI] [PubMed] [Google Scholar]
- Yan Y, Wang Q, Li W, Zhao Z, Yuan X, Huang Y, Duan Y. Discovery of potential biomarkers in exhaled breath for diagnosis of type 2 diabetes mellitus based on GC-MS with metabolomics. RSC Adv. 2014;4:25430–25439. [Google Scholar]
- Yi LZ, He J, Liang YZ, Yuan DL, Chau FT. Plasma fatty acid metabolic profiling and biomarkers of type 2 diabetes mellitus based on GC/MS and PLS-LDA. FEBS Lett. 2006;580:6837–6845. doi: 10.1016/j.febslet.2006.11.043. [DOI] [PubMed] [Google Scholar]
- Zeisel SH. Choline: an essential nutrient for humans. Nutrition. 2000;16:669–671. doi: 10.1016/s0899-9007(00)00349-x. [DOI] [PubMed] [Google Scholar]
- Zeng M, Liang Y, Li H, Wang M, Wang B, Chen X, Zhou N, Cao D, Wu J. Plasma metabolic fingerprinting of childhood obesity by GC/MS in conjunction with multivariate statistical analysis. J Pharm Biomed Anal. 2010;52:265–272. doi: 10.1016/j.jpba.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Zeng M, Liang Y, Li H, Wang B, Chen X. A metabolic profiling strategy for biomarker screening by GC-MS combined with multivariate resolution method and Monte Carlo PLS-DA. Analytical Methods. 2011;3:438–445. doi: 10.1039/c0ay00518e. [DOI] [PubMed] [Google Scholar]
- Zhang A, Sun H, Wang P, Han Y, Wang X. Modern analytical techniques in metabolomics analysis. Analyst. 2012;137:293–300. doi: 10.1039/c1an15605e. [DOI] [PubMed] [Google Scholar]
- Zhang AH, Sun H, Qiu S, Wang XJ. Recent highlights of metabolomics in chinese medicine syndrome research. Evid Based Complement Alternat Med. 2013;2013:402159. doi: 10.1155/2013/402159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Fritsche J, Wang J, Chen J, Rittig K, Schmitt-Kopplin P, Fritsche A, Haring HU, Schleicher ED, Xu G, et al. Metabonomic fingerprints of fasting plasma and spot urine reveal human pre-diabetic metabolic traits. Metabolomics. 2010;6:362–374. doi: 10.1007/s11306-010-0203-1. [DOI] [PMC free article] [PubMed] [Google Scholar]




