Abstract
The two-component regulatory system CiaRH of Streptococcus pneumoniae is involved in β-lactam resistance, maintenance of cell integrity, bacteriocin production, host colonization, virulence, and competence. The response regulator CiaR controls, among other genes, expression of five highly similar small non-coding RNAs, designated csRNAs. These csRNAs control competence development by targeting comC, encoding the precursor of the competence stimulating peptide, which is essential to initiate the regulatory cascade leading to competence. In addition, another gene product of the CiaR regulon, the serine protease HtrA, is also involved in competence control. In the absence of HtrA, five csRNAs could suppress competence, but one csRNA alone was not effective. To determine if all csRNAs are needed, reporter gene fusions to competence genes were used to monitor competence gene expression in the presence of different csRNAs. These experiments showed that two csRNAs were not enough to prevent competence, but combinations of three csRNAs, csRNA1,2,3, or csRNA1,2,4 were sufficient. In S. pneumoniae strains expressing only csRNA5, a surprising positive effect was detected on the level of early competence gene expression. Hence, the role of the csRNAs in competence regulation is more complex than anticipated. Mutations in comC (comC8) partially disrupting predicted complementarity to the csRNAs led to competence even in the presence of all csRNAs. Reconstitution of csRNA complementarity to comC8 restored competence suppression. Again, more than one csRNA was needed. In this case, even two mutated csRNAs complementary to comC8, csRNA1–8 and csRNA2–8, were suppressive. In conclusion, competence in S. pneumoniae is additively controlled by the csRNAs via post-transcriptional regulation of comC.
Keywords: Streptococcus pneumoniae, two-component regulatory system CiaRH, small non-coding csRNAs, post-transcriptional regulation, competence development
Introduction
The human pathogen Streptococcus pneumoniae shows a remarkable genomic plasticity (Croucher et al., 2012, 2014) and is a paradigm for bacteria that are able to undergo natural genetic transformation (for recent reviews see Johnston et al., 2014; Straume et al., 2014). The ability of S. pneumoniae to develop genetic competence is a highly regulated process depending on a variety of internal and external signals. Of these signals, the secreted peptide pheromone CSP (competence stimulating peptide) is absolutely essential. CSP is the processed extracellular form of the comC gene product encoded as the first gene of the comCDE operon (Håvarstein et al., 1995; Pestova et al., 1996). Pre-CSP is exported and processed by a dedicated ABC transporter ComAB (Hui and Morrison, 1991; Hui et al., 1995). CSP activates the two-component regulatory system ComDE by binding to the membrane-spanning histidine kinase ComD (Håvarstein et al., 1996). At the end of this initiating regulatory cascade, the response regulator ComE is phosphorylated (Martin et al., 2013) and helps to transcribe early competence genes, among them comAB, comCDE, and comX1/comX2 (Peterson et al., 2004). Enhanced transcription of comAB and comCDE results in an autocatalytic loop ensuring competence development throughout the population. The identical genes comX1/comX2 in turn encode the alternative sigma factor ComX, which directs transcription of late competence genes (Lee and Morrison, 1999). Proteins encoded by this class of genes are required for uptake of DNA and subsequent recombination (Claverys et al., 2009; Laurenceau et al., 2013).
The early regulatory steps constitute an extremely sensitive switch to the competence state, which is defined by the threshold to turn on the initiating autocatalytic loop. Processes interfering with CSP accumulation or CSP binding to the ComD sensor will inevitably modulate competence development. Consequently, quite a number of physiological parameters affect competence (Claverys and Håvarstein, 2002). Remarkably, stress conditions such as treatment with antibiotics may induce competence by various mechanisms (Prudhomme et al., 2006; Stevens et al., 2011; Gagne et al., 2013; Slager et al., 2014).
Besides the essential regulatory devices summarized above, competence development is modulated by other regulators such as the two-component regulatory systems CiaRH, WalRK, and the serine/threonine kinase StkP (Guenzi et al., 1994; Echenique and Trombe, 2001; Echenique et al., 2004; Saskova et al., 2007). While the regulatory link of WalRK or StkP to competence has not been established, several gene products of the CiaR regulon have been shown to interfere with CSP accumulation (Sebert et al., 2005; Cassone et al., 2012; Schnorpfeil et al., 2013). The cell surface-associated serine protease HtrA is apparently able to cleave CSP under certain conditions (Stevens et al., 2011) and five small non-coding csRNAs (cia-dependent small RNAs) regulate comC post-transcriptionally thereby reducing CSP production (Schnorpfeil et al., 2013). The csRNAs range between 87 and 151 nt in size and show a high degree of sequence similarity to each other (Halfmann et al., 2007b). Sequences potentially base-pairing to ribosome binding sites and AUG start codons are present in all csRNAs and are believed to be important for their regulatory function (Schnorpfeil et al., 2013). The csRNAs belong to an expanding class of small RNAs (sRNAs), which were designated sibling sRNAs in a recent review article (Caswell et al., 2014). Six targets for the csRNAs have been identified so far (Schnorpfeil et al., 2013). Besides comC, four genes encoding transporters and one gene coding for a DNA-binding protein are regulated. For three tested targets, among them comC, it was determined that the csRNAs act additively. Each csRNA could downregulate the respective genes, but each single csRNA was not as effective as all five csRNAs (Schnorpfeil et al., 2013). Combinations of csRNAs had not been tested in these experiments.
In this communication we determined the influence of csRNA combinations on competence gene expression. The results of these experiments showed that all five csRNAs are not needed for competence suppression. Instead, several combinations of three csRNAs were sufficient. In addition, a special role in early competence gene regulation was detected for csRNA5.
Materials and Methods
Bacterial Strains and Growth Conditions
The S. pneumoniae strains in this work are derived from S. pneumoniae R6 (Ottolenghi and Hotchkiss, 1962) and are listed in Table 1. Strains expressing single csRNAs or combinations were obtained by three approaches. First, a series of marker-less Cheshire deletions (Weng et al., 2009) were introduced as described previously (Schnorpfeil et al., 2013). Secondly, since for ccnAB, the genes encoding csRNA1 and csRNA2, only one Cheshire construct was available deleting both csRNA genes, ermAM was applied for individual ccnA or ccnB inactivation with constructions that were available from previous work (Halfmann et al., 2007b). And finally, to avoid time consuming repeated Cheshire constructions, some strains were obtained by ectopic expression of csRNA genes carried on integrative plasmids (Schnorpfeil et al., 2013). This approach results in csRNA expression levels that are indistinguishable from the wild type situation (Schnorpfeil et al., 2013). The comC8 mutant allele uncoupling competence development from csRNA control (Schnorpfeil et al., 2013) was introduced into a csRNA- and HtrA-deficient strain (RKL558, Table 1), which had been transformed with rpsl41 DNA to yield a streptomycin resistant derivative (RKL362). The subsequent Janus procedure (Sung et al., 2001) for comC8 replacement was carried out as described (Schnorpfeil et al., 2013). First, the comC::janus fragment was integrated into RKL362 to yield RKL745, which is streptomycin-sensitive and kanamycin-resistant. The final comC8 construct was introduced to RKL745 resulting in the streptomycin-resistant, kanamycin-sensitive RKL746 (Table 1). Complementing csRNA variants were then introduced into RKL746 (see below).
Table 1.
Streptococcus pneumoniae strains used in this study.
| Straina | Characteristics | Reference |
|---|---|---|
| RKL229 | ccnAB::lox72, ccnC::lox72, ccnD::lox72, ccnE::lox72 (ΔccnA-E) | Schnorpfeil et al. (2013) |
| RKL557 | htrA::aphlll | Schnorpfeil et al. (2013) |
| RKL558 | ccnAB::lox72, ccnC::lox72, ccnD::lox72, ccnE::lox72 (ΔccnA-E), htrA::aphlll | Schnorpfeil et al. (2013) |
| RKL362 | (ΔccnA-E), rpsL41 | This work |
| RKL688 | ccnB::ermAM, ccnC::lox72, ccnD::lox72, ccnE::lox72, htrA::aphlll | This work |
| RKL689 | ccnA::ermAM, ccnC::lox72, ccnD::lox72, ccnE::lox72, htrA::aphlll | This work |
| RKL690 | ccnAB::lox72, ccnD::lox72, ccnE::lox72, htrA::aphlll | This work |
| RKL691 | ccnAB::lox72, ccnC::lox72, ccnE::lox72, htrA::aphlll | This work |
| RKL692 | ccnAB::lox72, ccnC::lox72, ccnD::lox72, htrA::aphlll | This work |
| RKL693 | ccnAB::lox72, ccnC::lox72, htrA::aphlll | This work |
| RKL694 | ccnC::lox72, ccnD::lox72, ccnE::lox72, htrA::aphlll | This work |
| RKL695 | ccnD:: lox72, ccnE:: lox72, htrA:: aphlll | This work |
| RKL745 | ΔccnA-E, rpsL41, comC: janus | This work |
| RKL746 | ΔccnA-E, rpsL41, comC8 | This work |
| RKL913 | ccnC::lox72, ccnD::lox72, ccnE::lox72, htrA::aphlll bgaA tmp-ccnD-spr0568 | This work |
| RKL914 | ccnB::ermAM ccnC::lox72, ccnD::lox72, ccnE::lox72, htrA::aphlll, bgaA tmp-ccnC-spr0568 | This work |
| RKL915 | ccnB::ermAM ccnC::lox72, ccnD::lox72, ccnE::lox72, htrA::aphlll, bgaA tmp-ccnD-spr0568 | This work |
| RKL916 | ccnA::ermAM, ccnC::lox72, ccnD::lox72, ccnE::lox72, htrA::aphlll, bgaA tmp-ccnC-spr0568 | This work |
| RKL917 | ccnA::ermAM, ccnC::lox72, ccnD::lox72, ccnE::lox72, htrA::aphlll, bgaA tmp-ccnD-spr0568 | This work |
| RKL918 | ccnAB:: lox72, ccnD:: lox72, ccnE:: lox72, htrA:: aphlll, bgaA tmp-ccnD-spr0568 | This work |
| RKL939 | ΔccnA-E, rpsL41, comC8, htrA::aphlll | This work |
| RKL941 | ΔccnA-E, rpsL41, comC8, htrA::aphlll, bgaA tmp-ccnA-8-spr0568 | This work |
| RKL942 | ΔccnA-E, rpsL41, comC8, htrA::aphlll, bgaA tmp-ccnAB-8-spr0568 | This work |
| RKL943 | ΔccnA-E, rpsL41, comC8, htrA::aphlll, bgaA tmp-ccnB-8-spr0568 | This work |
| RKL967 | ccnBr.ermAM, ccnC::lox72, ccnD::lox72; ccnE::lox72 htrA::aphlll, bgaA tmp-ccnE-spr0568 | This work |
| RKL968 | ccnAr.ermAM, ccnC::lox72, ccnD::lox72; ccnE::lox72 htrA::aphlll, bgaA tmp-ccnE-spr0568 | This work |
| RKL969 | ccnC:: lox72, ccnD:: lox72; ccnE:: lox72 htrA::aphlll, bgaA tmp-ccnE-spr0568 | This work |
aAll strains are R6 derivatives.
For strain preservation or transformation, S. pneumoniae was grown at 37°C in C + Y medium (Lacks and Hotchkiss, 1960; Schnorpfeil et al., 2013). The competence expression tests were performed in brain–heart infusion (BHI, pH 7.4) using BHI, pH 7.0 for inoculation. BHI was purchased from Becton Dickinson, France. Growth of S. pneumoniae was monitored by measuring optical density of 600 nm (OD600).
Plasmid cloning was performed in Escherichia coli DH5α [ϕ80dlacZΔM15 Δ(lacZYA-argF) recA1 endA1 hsdR17 supE44 thi-1 gyrA96 phoA relA1]. E. coli strains were grown in LB medium.
Construction of Promoter Probe Plasmids and Transformation
To monitor competence development, two promoter fusions were constructed. One promoter originated from the genes comX1/2 and represents the early class of competence-specific promoters dependent on ComE (Peterson et al., 2004). Since comX1/2 are identical genes including identical promoter regions the resulting promoter is designated PcomX. This promoter was obtained by PCR using primers ComX1Eco (CTCGAATTCATGCACTATCCTATGAAGTAAAGTC) and comX-Prom-bam (CAGGGATCCTTTCTAGATACAGTCTAACAGATTAGAAAACAC). The resulting fragment was cloned into promoter probe plasmid pPP2 (Halfmann et al., 2007a) to yield plasmid pPPcomX. The other promoter is from the cibABC operon and constitutes a late competence promoter dependent on the alternative sigma factor ComX (Peterson et al., 2004). To clone the cibA promoter, the primers CibA-1 (GGCGCATGCAGACAAGAGTGCCCTCACTTAAC) and CibAprom (GGCGGATCCCTTGTTCACTTTTATATTCGGAAAAG) were used to yield pPP2cibA.
Plasmid pPC2 carrying another early competence promoter (PcomC) was described previously (Halfmann et al., 2007a). Upon integration of the promoter fusion plasmids, the genotype of the resulting strains was as follows: bgaA::tetM-PcomX-lacZ, bgaA::tetM-PcibA-lacZ, bgaA::tetM-PcomC-lacZ.
To introduce the plasmids into S. pneumoniae, cells were grown in C + Y medium and were transformed as described (Schnorpfeil et al., 2013). S. pneumoniae transformants with pPP derivatives were selected on plates containing D-blood agar (Alloing et al., 1996) at 37°C with 2 μg tetracycline per ml. Transformation of E. coli was carried out according to Hanahan (1983).
Expression of ccn Genes Encoding csRNAs Complementary to comC8
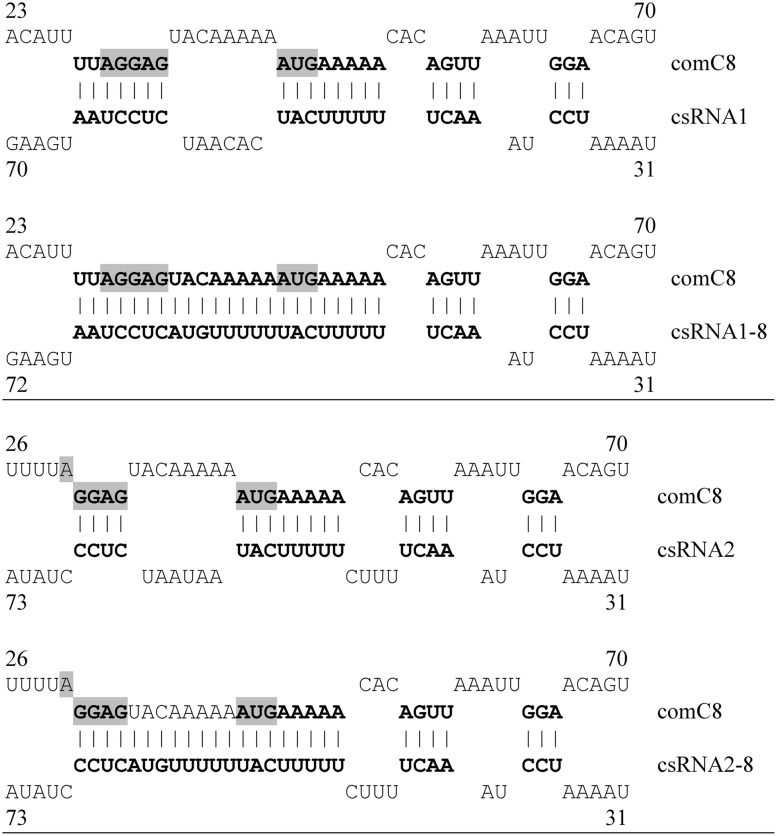
The comC8 gene variant relieved competence regulation from csRNA-mediated control (Schnorpfeil et al., 2013). Mutations were introduced into csRNA1 and csRNA2, that restored complementarity to comC8 (Figure 3). The mutated csRNA genes, csRNA1–8 and csRNA2–8, were obtained after gene synthesis cloned into pUC57 (Genewiz, South Plainfield, NJ, USA). Two plasmids contained single mutated csRNA genes, the third contained csRNA1–8 and csRNA2–8 in tandem as found for the wild type ccnAB genes in the S. pneumoniae genome (Halfmann et al., 2007b). The mutated csRNA genes were moved alone or in combination to the integrative plasmid pSW1 (Denapaite and Hakenbeck, 2011) to yield plasmids pCcnA-8, pCcnB-8, and pCcnA-8-ccnB-8. Plasmids were introduced to the csRNA- and HtrA-deficient strain carrying the comC8 allele RKL746 by selection with 100 μg/ml trimethoprim. Correct integration was determined by PCR and sequencing.
FIGURE 3.
Predicted interactions of comC mRNA with wild type csRNA1, csRNA2, and mutant csRNA1–8, csRNA2–8. Base-pairing of the csRNAs with comC8 mRNA according to IntaRNA predictions are shown (Busch et al., 2008). Base pairs are shown in bold face, the SD sequence and the start codon of comC are shaded. Complementarity of comC8 to csRNA1 and csRNA2 is abolished between the SD sequence and AUG. Mutations in csRNA1–8 and csRNA2–8 restored complementarity. The numbers refer to the bases counted from the transcriptional start sites of the csRNAs and comC8.
Determination of β-Galactosidase Activity
To follow the onset of competence gene expression, strains harboring promoter-β-galactosidase gene (lacZ) fusions in the genome (Halfmann et al., 2007b) were assayed for β-galactosidase activity during growth as described (Weng et al., 2013). To estimate the significance of the different time points of β-galactosidase induction, Student’s t-tests were performed. Differences were considered significant, when p < 0.05 was given.
Results
Competence Gene Expression in the Presence of Single csRNAs
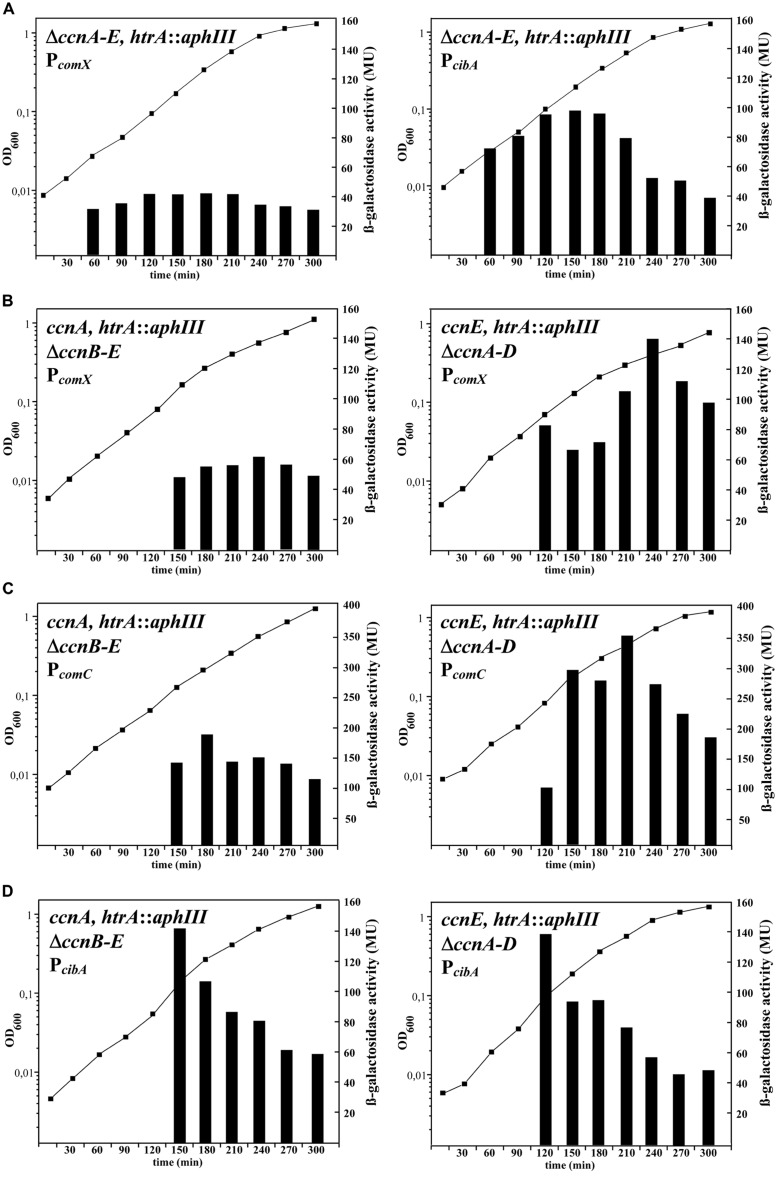
As detailed above, the two-component regulatory system CiaRH is one of the most important additional regulatory factors modulating competence development (Sebert et al., 2005; Schnorpfeil et al., 2013). Depending on the growth conditions and the expression state of CiaRH, the protease HtrA, and/or the five small csRNA are able to suppress competence. Transformation experiments performed in competence non-permissive BHI medium showed that inactivation of htrA and the csRNA genes ccnA-D is needed to allow competence to develop (Schnorpfeil et al., 2013). To monitor competence development more conveniently, promoter fusions to the E. coli β-galactosidase gene lacZ were constructed using promoters of comX and the cibABC operon as representatives for early (comX) and late (cibA) competence genes (Peterson et al., 2004). After integration of these promoter-lacZ fusions into the genome of S. pneumoniae, β-galactosidase activities were measured during growth in BHI medium. As predicted from the previous transformation experiments, lacZ expression driven by both promoters was only detectable in the absence of the csRNAs and the protease HtrA (Figure 1A). Expression of the competence genes started very early in growth at an OD600 around 0.03 (Figure 1A; Table 2). Due to the 30 min interval of measurements, the time difference between early and late gene expression of about 5–7 min (Peterson et al., 2004) is not detected (Figure 1A). In addition, competence shut-off is not clearly seen in these experiments, because of the rather stable β-galactosidase, which is still active after transcription of its gene ceases. Since CiaRH is involved in controlling early steps of competence gene expression, only their induction is relevant and, as shown in Figure 1A, clearly detected.
FIGURE 1.
Time course of competence gene expression in Streptococcus pneumoniae strains expressing different csRNAs. The strains were grown in brain heart infusion medium and β-galactosidase activities were determined in 30 min. intervals. β-galactosidase activities of representative experiments are shown along with the growth curve determined by measuring OD600. All strains contained a genomic integration of a competence promoter fused to the promoterless β-galactosidase gene lacZ of Escherichia coli and a disrupted protease gene htrA (htrA::aphIII). Relevant genetic characteristics are indicated. (A) Activity of early and late competence promoters in strains without csRNAs. The early competence gene promoter of comX and the late competence gene promoter of cibA were fused to lacZ and integrated into RKL 558 (ccnA:: lox72, ccnB:: lox72, ccnC::lox72, ccnD::lox72, ccnE::lox72, htrA::aphIII). The promoter fusion integrations were: bgaA::tetM-PcomX-lacZ (left), bgaA::tetM-PcibA-lacZ (right). (B) Activity of the early comX promoter in strains with csRNA1 or csRNA5. The early competence gene promoter of comX was fused to lacZ and integrated into RKL688 (ccnB::ermAM, ccnC::lox72, ccnD::lox72, ccnE::lox72, htrA::aphIII, left) or RKL692 (ccnAB::lox72, ccnC::lox72, ccnD::lox72, htrA::aphIII, right). Both strains contained the integrated PcomX-lacZ fusion: bgaA::tetM-PcomX-lacZ. (C) Activity of the early comC promoter in strains with csRNA1 or csRNA5. The early competence gene promoter of comC was fused to lacZ and integrated into the same strains as in (B). (RKL688; ccnB::ermAM, ccnC::lox72, ccnD::lox72, ccnE::lox72, htrA::aphIII, left, or RKL692; ccnAB::lox72, ccnC::lox72, ccnD::lox72, htrA::aphIII, right). Both strains contained the integrated PcomC-lacZ fusion: bgaA::tetM-PcomX-lacZ. (D) Activity of the late cibA promoter in strains with csRNA1 or csRNA5. The late competence gene promoter of cibA was fused to lacZ and integrated into the same strains as in (B,C). (RKL688; ccnB::ermAM, ccnC::lox72, ccnD::lox72, ccnE::lox72, htrA::aphIII, left, or RKL692; ccnAB::lox72, ccnC::lox72, ccnD::lox72, htrA::aphIII, right). Both strains contained the integrated PcibA-lacZ fusion: bgaA::tetM-PcomX-lacZ.
Table 2.
Expression of competence genes in S. pneumoniae strains with different csRNA genes.
| straina | csRNA genes | csRNAs | comX expression | induction of comX expression at OD600b |
|---|---|---|---|---|
| RKL557 | ccnABCDE | csRNAl,2,3,4,5 | -c | na |
| RKL558 | - | - | + | 0.03 ± 0.02 |
| RKL688 | ccnA | csRNAl | + | 0.15 ± 0.01 |
| RKL689 | ccnB | csRNA2 | + | 0.17 ± 0.002 |
| RKL690 | ccnC | csRNA3 | + | 0.21 ± 0.001 |
| RKL691 | ccnD | csRNA4 | + | 0.30 ± 0.001 |
| RKL692 | ccnE | csRNA5 | + | 0.09 ± 0.005 |
| RKL694 | ccnAB | csRNAl,2 | + | 0.36 ± 0.07 |
| RKL914 | ccnAC | csRNAl,3 | + | 0.36 ± 0.05 |
| RKL915 | ccnAD | csRNAl,4 | + | 0.31 ± 0.08 |
| RKL967 | ccnAE | csRNAl,5 | + | 0.18 ± 0.05 |
| RKL916 | ccnBC | csRNA2,3 | + | 0.30 ± 0.07 |
| RKL917 | ccnBD | csRNA2,4 | + | 0.36 ± 0.01 |
| RKL968 | ccnBE | csRNA2,5 | + | 0.19 ± 0.03 |
| RKL918 | ccnCD | csRNA3,4 | + | 0.31 ± 0.03 |
| RKL693 | ccnDE | csRNA4,5 | + | 0.25 ± 0.04 |
| RKL695 | ccnABC | csRNAl,2,3 | -c | na |
| RKL913 | ccnABD | csRNAl,2,4 | -c | na |
| RKL969 | ccnABE | csRNAl,2,5 | + | 0.38 ± 0.1 |
aAll strains are serine protease HtrA deficient (htrA::aphIII) and contain a PcomX-lacZ fusion in the genome (bgaA::tetM-PcomX-lacZ). They were grown in BHI medium.
bThe onset of comX expression was defined as the time point when more than 10 Miller units (MU) β-galactosidase were reached. Uninduced PcomX-meditaed expression was below 1 MU. Values of at least two independent cultures are shown along with SDs. Timing of β-galactosidase expression was significantly different (p < 0.05) in all strains compared to RKL558.
cExpression around or below β-galactosidase activity detection level (∼1 MU). Na, not applicable.
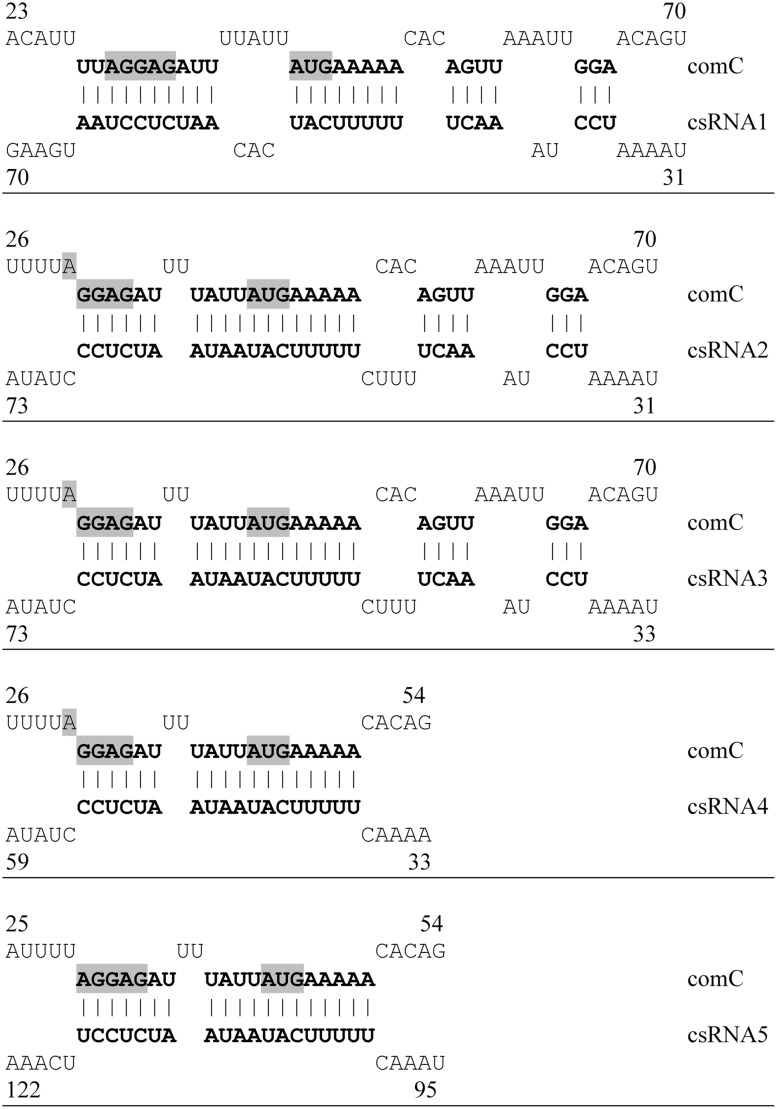
To determine which and how many csRNAs are needed to block competence, strains were constructed expressing single csRNAs and various combinations (Table 1). Each of the csRNAs is predicted to bind to comC (Figure 2) and could reduce expression from a comC′–′lacZ translational fusion (Schnorpfeil et al., 2013). Competence gene expression was measured during growth in BHI medium as shown for the PcomX-lacZ fusion and strains expressing csRNA1 or csRNA5 as examples in Figure 1B. Individual csRNAs were not able to suppress competence gene expression, which is in accordance with previous transformation tests (Schnorpfeil et al., 2013). However, the cell density, at which induction occurred, varied, and was substantially higher than in the strain without csRNAs (Figures 1A,B; Table 2). With csRNA4, expression started at OD600 of 0.3, with csRNA5 already at 0.09, and with the other csRNAs in between these cell densities (Table 2). Thus, although single csRNAs are not able to turn down competence, they are able to delay its start.
FIGURE 2.
Predicted interactions of csRNAs with comC mRNA. Base-pairing of the csRNAs with comC mRNA according to IntaRNA predictions are shown (Busch et al., 2008). Predicted base pairs are shown in bold face, the Shine Dalgarno (SD) sequence and the start codon of comC are shaded in gray. The numbers refer to the bases counted from the transcriptional start sites of the csRNAs and comC. Putative interactions of all csRNAs with comC mRNA are shown.
Competence Gene Expression in the Presence of Multiple csRNAs
Next we asked how subsets of csRNAs could affect competence in S. pneumoniae. To start with that analysis, strains were constructed expressing combinations of two csRNAs (Table 1). As shown in Table 2, none of the combinations was able to stop competence gene expression, not even the two csRNAs, csRNA3 and csRNA4, which alone could maximally delay competence induction. With most of the combinations, competence gene expression started at higher cell densities than with single csRNAs, moving competence to the mid-exponential growth phase.
Combining three csRNAs, for example csRNA1,2,3 or csRNA1,2,4 blocked competence completely. Therefore, not all csRNAs are needed and more than one combination can be effective. Combination of csRNA1,2, and 5 did not stop PcomX promoter activity, corroborating the weak negative effect of csRNA5 observed in cells with csRNA5 alone.
A Special Role for csRNA5 on the Level of Early Competence Gene Expression
During the experiments detailed above, a remarkable effect on comX promoter activity was detected when csRNA5 was present (Figure 1B). The strength of PcomX-mediated β-galactosidase expression was about twofold enhanced in the presence of csRNA5 compared to a strain with csRNA1. Strains expressing csRNA2 or csRNA3 showed the same PcomX-mediated β-galactosidase expression level as the csRNA1 containing strain (data not shown). Only expression timing was different (Table 2). In S. pneumoniae with csRNA4, PcomX promoter activity mediated β-galactosidase was slightly higher (1,4-fold). Therefore, besides acting negatively on the timing of competence, csRNA5 and to a lesser extent csRNA4 play a positive role in PcomX expression. Enhanced PcomX promoter activity was also detected when csRNA5 was expressed with other csRNAs, e.g., with csRNA2,4, or 1,2, however, not with csRNA1 alone.
To determine if the csRNA5 effect is also detected with other early competence promoters, a comC promoter fusion was subjected to the same analysis as the comX promoter. As shown in Figure 1C, PcomC activity was affected by the presence of csRNA1 or csRNA5 in the same way as PcomX. It appears therefore, that the strength of early competence promoters are positively affected by the presence of csRNA5. Therefore, the role of csRNA5 on early competence gene expression is both, negative and positive.
A different picture emerged, when late competence gene expression was tested in strains with csRNA1 and csRNA5. Measuring cibA promoter activity as an example for a late competence gene did not reveal a significant difference in the presence of csRNA1 or csRNA5 (Figure 1D). The observed positive effect of csRNA5 on the level of early competence gene expression is not passed on to late competence gene expression, most likely by elaborate feed-back control implemented in competence development (Johnston et al., 2014).
Complementation of a csRNA-Insensitive comC Variant by Mutated csRNAs
In our previous work, a mutant comC gene, comC8, was constructed, which showed greatly diminished post-transcriptional control by the csRNAs (Schnorpfeil et al., 2013). Replacing comC by comC8 resulted in strains that were able to develop competence even in the presence of all csRNAs (Schnorpfeil et al., 2013). Complementarity of comC8 to the csRNAs had been reduced by mutations between the Shine-Dalgarno sequence (Shine and Dalgarno, 1974) and the start codon AUG (Figure 3). If regulation of competence by the csRNAs is indeed a direct consequence of csRNA–comC interaction, mutations in csRNAs restoring complementarity to comC8 should re-establish csRNA-mediated competence control.
Therefore, mutated csRNA genes were constructed with changes complementary to comC8 (Figure 3). As a start, csRNA1 and csRNA2 were mutated, since it could not be expected that a single modified csRNA could block competence alone. The mutated csRNAs, csRNA1–8 and csRNA2–8, were cloned alone and in combination to an integrative plasmid, which was used for ectopic expression of these csRNAs in a csRNA-deficient background (Table 1). To monitor competence development the late cibA promoter fusion was applied. With single mutated csRNAs, at least a delay in competence gene expression was expected and combinations could perhaps block competence.
As shown in Table 3, csRNA1–8 as well as csRNA2–8 could clearly delay competence gene expression. While expression started at OD600 of 0.09 without any csRNA, competence gene expression occurred at OD600 of 0.38 and 0.32 with csRNA1–8 or csRNA2–8, respectively. Thus, both mutated csRNAs similarly delayed competence, when expressed alone. In combination, they could even prevent competence development (Table 3), which is in contrast to the wild type, where three csRNAs were needed. Enhanced downregulation by csRNA1–8 and csRNA2–8 may be due to the enlarged region of complementarity introduced into the mutated csRNAs (Figure 3). The results of these experiments are completely consistent with the previous identification of the csRNAs as mediators of comC control and as one link of the CiaRH system to competence regulation (Schnorpfeil et al., 2013).
Table 3.
Expression of competence genes in S. pneumoniae comC8 strains with mutated csRNA genes.
| straina | csRNA genes | csRNAs | cibA expression | Induction of cibA expression at OD600b |
|---|---|---|---|---|
| RKL939 | - | - | + | 0.09 ± 0.04 |
| RKL941 | ccnA-8 | csRNAl– | + | 0.38 ± 0.08 |
| RKL943 | ccnB-8 | csRNA2– | + | 0.32 ± 0.1 |
| RKL942 | ccnA-8, ccnB-8 | csRNAl–8,2–8 | -c | na |
aAll strains are serine protease HtrA deficient (htrA::aphIII), have the comC8 allele, which is no longer subject to csRNA control, and contain a PcibA-lacZ fusion in the genome (bgaA::tetM-PcibA-lacZ). They were grown in BHI medium.
bThe onset of cibA expression was defined as the time point when more than 10 MUs β-galactosidase were reached. Uninduced PcibA-meditaed expression was below 1 MU. Values of at least two independent cultures are shown along with standard deviations. Timing of β-galactosidase expression in strains RKL 941 and 943 was significantly different (p < 0.05) compared to RKL939. The difference between RKL941 and RKL943 was not significant.
cExpression around or below β-galactosidase activity detection level (∼1 MU). na, not applicable.
Discussion
The two-component regulatory system CiaRH acts negatively on competence by controlling CSP accumulation by two mechanisms. One relies on the serine protease HtrA, which is able to cleave secreted CSP (Stevens et al., 2011), and the second on five csRNAs to reduce CSP production. Expression of the protease gene htrA as well as the csRNA genes are absolutely dependent on CiaR (Halfmann et al., 2007b). Which of the two mechanisms is more important strongly depends on the growth conditions. In BHI medium, HtrA or the csRNAs are able to suppress transformability (Schnorpfeil et al., 2013). In cells grown in C + Y medium, however, none of these components is able to stop competence development. Upon overexpression of the csRNAs and HtrA due to a hyperactive CiaRH system (Müller et al., 2011), competence is abolished even in C + Y medium (Schnorpfeil et al., 2013). Under these conditions, only the csRNAs are able to stop competence development. The level of csRNA production is clearly one parameter defining the regulatory outcome, but is not always sufficient to explain csRNA effects. In BHI and C + Y media, the CiaRH system is similarly active (Halfmann et al., 2011), but competence is only blocked in BHI medium (Schnorpfeil et al., 2013). These differences may be explained assuming that the CSP threshold to initiate competence could vary under these conditions. How this threshold level is set and influenced by growth conditions, remains one of the unsolved mysteries of competence regulation in S. pneumoniae.
The results of the comC8 complementation experiments clearly identify comC regulation as the cause for csRNA-mediated competence suppression. They also point to the region where base pairing between csRNAs and comC occurs (Figures 2 and 3). The csRNAs bind around the comC translation initiation region, which is fully consistent with their negative effect on CSP production. Predicted csRNA-comC interaction is identical for csRNA2 and csRNA3 (Figure 2), almost identical except for one A–U pair for csRNA4 and csRNA5, and unique for csRNA1 (Figure 2). Especially for csRNA4 and csRNA5, predicted comC binding and their effect on competence regulation are not in accordance. If the delay in cell density when competence gene expression starts in strains expressing only one csRNA is taken as a measure of their effectiveness of comC downregulation (Table 2), csRNA4 and csRNA5 act at opposite ends: csRNA4 is the most (OD600 0.3) and csRNA5 the least (OD600 0.09) efficient, although predicted binding to comC is slightly better for csRNA5 (Figure 3). Likewise, csRNA4 could best reduce expression from a comC′–′lacZ translational fusion, while csRNA5 was least effective (Schnorpfeil et al., 2013). Interestingly, ranking of the effects of the csRNAs on comC′–′lacZ or the timing of competence gene expression produced the same order: csRNA4 acted best, followed by csRNA1, csRNA2, csRNA3, and at the end csRNA5 (Schnorpfeil et al., 2013), (Table 2). Thus, comC regulation by the csRNAs is indeed indicative for competence development, but the strength of regulation by individual csRNAs is apparently not correlated with predicted complementarity.
At the moment, we cannot offer a final explanation for this discrepancy. Although the region of csRNA–comC interaction is roughly defined by the comC8 mutations, base pairing may not be precisely predicted by the applied program (Busch et al., 2008). In addition, it has been demonstrated in other sRNA-mediated regulations that only a core within a larger region of complementarity is critical (Balbontin et al., 2010; Papenfort et al., 2012). Structural probing in vitro and more detailed mutagenesis in vivo will be required to define residues important for csRNA control of comC.
Another aspect of sRNA-mediated regulation is certainly the relative abundance of both partners. Since it is generally assumed that sRNAs are eliminated in the course of regulation (Gottesman and Storz, 2011; Storz et al., 2012), their expression level is of outstanding importance (Levine and Hwa, 2008). Based on previous northern blot analyses, a strong difference in expression was not detected for individual csRNAs (Halfmann et al., 2007b; Schnorpfeil et al., 2013), but subtle changes cannot be ruled out.
A further complication is also related to the consumption of sRNAs during regulation. Many sRNAs have been found to bind more than one target (Gottesman and Storz, 2011; Storz et al., 2012). All five csRNAs in S. pneumoniae regulate at least two more genes besides comC (Schnorpfeil et al., 2013) and it is most likely that there are further targets for the csRNAs. Depending on the expression level of these additional targets and the efficiency of csRNA binding to their mRNAs, single csRNAs may be titrated away from comC control.
While the reason of differential regulation of comC by single csRNAs remains elusive, it is clear that more than one csRNA is needed to prevent S. pneumoniae from reaching the competent state. Even two csRNAs were not enough, at least three csRNAs are needed. Of the three tested combinations, csRNA1,2,3 or csRNA1,2,4 were sufficient to block competence, but not the combination of csRNA1,2,5. It appears that each combination of three csRNAs among csRNA1,2,3, and 4 will be able to prevent natural transformation. Therefore, the role of each of these four csRNAs in competence regulation is clearly negative.
The role of csRNA5, however, appears to be more complex. The largest of the csRNAs acts positively on the level of early competence gene expression and only slightly suppressive on the timing of competence gene expression. This positive effect cannot be explained by regulation of comC, but is most likely due to the regulation of unidentified target(s) for csRNA5. Early competence gene expression is under feed-back control of a late competence gene product DprA (Mirouze et al., 2013; Weng et al., 2013). Inactivation of dprA results in a strong increase of early gene expression, but additionally late genes are also overexpressed. Quite in contrast, the observed effect of csRNA5 on competence gene expression is restricted to early competence genes (Figures 1B–D). Therefore, it appears unlikely that dprA is the unknown target for csRNA5. It has also been described that an intrinsic mechanism dependent on non-phosphorylated ComE could restrict especially transcription from the comCDE promoter (Martin et al., 2013). However, the effect of csRNA5 was virtually identical for both, the comC and the comX promoter (Figures 1B,C). It appears therefore, that csRNA5 regulates a novel process in early competence gene regulation.
The special role of csRNA5 in early competence gene expression adds an interesting twist to csRNA-mediated competence regulation. Due to the downregulation of CSP production, the csRNAs are involved in setting the threshold for competence induction. Once threshold is reached, csRNA5 may ensure a robust response to CSP by enhancing early gene expression.
Conclusion
The csRNAs are not a simple device to turn down competence, but interfere with competence regulation in a complex manner.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Regine Hakenbeck for her continuous interest and support. We also acknowledge the help of Sabrina Kaiser in strain constructions. The work was sponsored in part by the Priority Program “Sensory and regulatory RNAs in Prokaryotes (SPP 1258) of the Deutsche Forschungsgemeinschaft DFG (BR974/5-1), by DFG 947/8-1, and by a fellowship for AL by the Stipendienstiftung Rheinland-Pfalz.
References
- Alloing G., Granadel C., Morrison D. A., Claverys J. P. (1996). Competence pheromone, oligopeptide permease, and induction of competence in Streptococcus pneumoniae. Mol. Microbiol. 21 471–478. 10.1111/j.1365-2958.1996.tb02556.x [DOI] [PubMed] [Google Scholar]
- Balbontin R., Fiorini F., Figueroa-Bossi N., Casadesus J., Bossi L. (2010). Recognition of heptameric seed sequence underlies multi-target regulation by RybB small RNA in Salmonella enterica. Mol. Microbiol. 78 380–394. 10.1111/j.1365-2958.2010.07342.x [DOI] [PubMed] [Google Scholar]
- Busch A., Richter A. S., Backofen R. (2008). IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics 24 2849–2856. 10.1093/bioinformatics/btn544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassone M., Gagne A. L., Spruce L. A., Seeholzer S. H., Sebert M. E. (2012). The HtrA protease from Streptococcus pneumoniae digests both denatured proteins and the competence-stimulating peptide. J. Biol. Chemistry 287 38449–38459. 10.1074/jbc.M112.391482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell C. C., Oglesby-Sherrouse A. G., Murphy E. R. (2014). Sibling rivalry: related bacterial small RNAs and their redundant and non-redundant roles. Front. Cell. Infect. Microbiol. 4:151 10.3389/fcimb.2014.00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverys J. P., Håvarstein L. S. (2002). Extracellular-peptide control of competence for genetic transformation in Streptococcus pneumoniae. Front. Biosci. 7:d1798-1814. 10.2741/claverys [DOI] [PubMed] [Google Scholar]
- Claverys J. P., Martin B., Polard P. (2009). The genetic transformation machinery: composition, localization, and mechanism. FEMS Microbiol. Rev. 33 643–656. 10.1111/j.1574-6976.2009.00164.x [DOI] [PubMed] [Google Scholar]
- Croucher N. J., Coupland P. G., Stevenson A. E., Callendrello A., Bentley S. D., Hanage W. P. (2014). Diversification of bacterial genome content through distinct mechanisms over different timescales. Nat. Commun. 5 5471 10.1038/ncomms6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher N. J., Harris S. R., Barquist L., Parkhill J., Bentley S. D. (2012). A high-resolution view of genome-wide pneumococcal transformation. PLoS Pathog. 8:e1002745 10.1371/journal.ppat.1002745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denapaite D., Hakenbeck R. (2011). A new variant of the capsule 3 cluster occurs in Streptococcus pneumoniae from deceased wild chimpanzees. PLoS ONE 6:e25119 10.1371/journal.pone.0025119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echenique J., Kadioglu A., Romao S., Andrew P. W., Trombe M. C. (2004). Protein serine/threonine kinase StkP positively controls virulence and competence in Streptococcus pneumoniae. Infect. Immun. 72 2434–2437. 10.1128/IAI.72.4.2434-2437.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echenique J. R., Trombe M. C. (2001). Competence repression under oxygen limitation through the two-component MicAB signal-transducing system in Streptococcus pneumoniae and involvement of the PAS domain of MicB. J. Bacteriol. 183 4599–4608. 10.1128/JB.183.15.4599-4608.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne A. L., Stevens K. E., Cassone M., Pujari A., Abiola O. E., Chang D. J., et al. (2013). Competence in Streptococcus pneumoniae is a response to an increasing mutational burden. PLoS ONE 8:e72613 10.1371/journal.pone.0072613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S., Storz G. (2011). Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 3 a003798 10.1101/cshperspect.a003798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenzi E., Gasc A. M., Sicard M. A., Hakenbeck R. (1994). A two-component signal-transducing system is involved in competence and penicillin susceptibility in laboratory mutants of Streptococcus pneumoniae. Mol. Microbiol. 12 505–515. 10.1111/j.1365-2958.1994.tb01038.x [DOI] [PubMed] [Google Scholar]
- Halfmann A., Hakenbeck R., Brückner R. (2007a). A new integrative reporter plasmid for Streptococcus pneumoniae. FEMS Microbiol. Lett. 268 217–224. 10.1111/j.1574-6968.2006.00584.x [DOI] [PubMed] [Google Scholar]
- Halfmann A., Kovács M., Hakenbeck R., Brückner R. (2007b). Identification of the genes directly controlled by the response regulator CiaR in Streptococcus pneumoniae: five out of 15 promoters drive expression of small non-coding RNAs. Mol. Microbiol. 66 110–126. 10.1111/j.1365-2958.2007.05900.x [DOI] [PubMed] [Google Scholar]
- Halfmann A., Schnorpfeil A., Müller M., Marx P., Günzler U., Hakenbeck R., et al. (2011). Activity of the two-component regulatory system CiaRH in Streptococcus pneumoniae R6. J. Mol. Microbiol. Biotechnol. 20 96–104. 10.1159/000324893 [DOI] [PubMed] [Google Scholar]
- Hanahan D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166 557–580. 10.1016/S0022-2836(83)80284-8 [DOI] [PubMed] [Google Scholar]
- Håvarstein L. S., Coomaraswamy G., Morrison D. A. (1995). An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U.S.A. 92 11140–11144. 10.1073/pnas.92.24.11140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Håvarstein L. S., Gaustad P., Nes I. F., Morrison D. A. (1996). Identification of the streptococcal competence-pheromone receptor. Mol. Microbiol. 21 863–869. 10.1046/j.1365-2958.1996.521416.x [DOI] [PubMed] [Google Scholar]
- Hui F. M., Morrison D. A. (1991). Genetic transformation in Streptococcus pneumoniae: nucleotide sequence analysis shows comA, a gene required for competence induction, to be a member of the bacterial ATP-dependent transport protein family. J. Bacteriol. 173 372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui F. M., Zhou L., Morrison D. A. (1995). Competence for genetic transformation in Streptococcus pneumoniae: organization of a regulatory locus with homology to two lactococcin A secretion genes. Gene 153 25–31. 10.1016/0378-1119(94)00841-F [DOI] [PubMed] [Google Scholar]
- Johnston C., Campo N., Berge M. J., Polard P., Claverys J. P. (2014). Streptococcus pneumoniae, le transformiste. Trends Microbiol. 22 113–119. 10.1016/j.tim.2014.01.002 [DOI] [PubMed] [Google Scholar]
- Lacks S., Hotchkiss R. D. (1960). A study of the genetic material determining an enzyme in Pneumococcus. Biochim. Biophys. Acta 39 508–518. 10.1016/0006-3002(60)90205-5 [DOI] [PubMed] [Google Scholar]
- Laurenceau R., Pehau-Arnaudet G., Baconnais S., Gault J., Malosse C., Dujeancourt A., et al. (2013). A type IV pilus mediates DNA binding during natural transformation in Streptococcus pneumoniae. PLoS Pathog. 9:e1003473 10.1371/journal.ppat.1003473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. S., Morrison D. A. (1999). Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181 5004–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine E., Hwa T. (2008). Small RNAs establish gene expression thresholds. Curr. Opin. Microbiol. 11 574–579. 10.1016/j.mib.2008.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin B., Soulet A. L., Mirouze N., Prudhomme M., Mortier-Barriere I., Granadel C., et al. (2013). ComE/ComE˜P interplay dictates activation or extinction status of pneumococcal X-state (competence). Mol. Microbiol. 87 394–411. 10.1111/mmi.12104 [DOI] [PubMed] [Google Scholar]
- Mirouze N., Berge M. A., Soulet A. L., Mortier-Barriere I., Quentin Y., Fichant G., et al. (2013). Direct involvement of DprA, the transformation-dedicated RecA loader, in the shut-off of pneumococcal competence. Proc. Natl. Acad. Sci. U.S.A. 110 1035–1044. 10.1073/pnas.1219868110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller V. S., Jungblut P. R., Meyer T. F., Hunke S. (2011). Membrane-SPINE: an improved method to identify protein-protein interaction partners of membrane proteins in vivo. Proteomics 11 2124–2128. 10.1002/pmic.201000558 [DOI] [PubMed] [Google Scholar]
- Ottolenghi E., Hotchkiss R. D. (1962). Release of genetic transforming agent from pneumococcal cultures during growth and disintegration. J. Exp. Med. 116 491–519. 10.1084/jem.116.4.491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K., Podkaminski D., Hinton J. C., Vogel J. (2012). The ancestral SgrS RNA discriminates horizontally acquired Salmonella mRNAs through a single G-U wobble pair. Proc. Natl. Acad. Sci. U.S.A. 109 757–764. 10.1073/pnas.1119414109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova E. V., Håvarstein L. S., Morrison D. A. (1996). Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol. Microbiol. 21 853–862. 10.1046/j.1365-2958.1996.501417.x [DOI] [PubMed] [Google Scholar]
- Peterson S. N., Sung C. K., Cline R., Desai B. V., Snesrud E. C., Luo P., et al. (2004). Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 51 1051–1070. 10.1046/j.1365-2958.2003.03907.x [DOI] [PubMed] [Google Scholar]
- Prudhomme M., Attaiech L., Sanchez G., Martin B., Claverys J. P. (2006). Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313 89–92. 10.1126/science.1127912 [DOI] [PubMed] [Google Scholar]
- Saskova L., Novakova L., Basler M., Branny P. (2007). Eukaryotic-type serine/threonine protein kinase StkP is a global regulator of gene expression in Streptococcus pneumoniae. J. Bacteriol. 189 4168–4179. 10.1128/JB.01616-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorpfeil A., Kranz M., Kovacs M., Kirsch C., Gartmann J., Brunner I., et al. (2013). Target evaluation of the non-coding csRNAs reveals a link of the two-component regulatory system CiaRH to competence control in Streptococcus pneumoniae R6. Mol. Microbiol. 89 334–349. 10.1111/mmi.12277 [DOI] [PubMed] [Google Scholar]
- Sebert M. E., Patel K. P., Plotnick M., Weiser J. N. (2005). Pneumococcal HtrA protease mediates inhibition of competence by the CiaRH two-component signaling system. J. Bacteriol. 187 3969–3979. 10.1128/JB.187.12.3969-3979.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. (1974). The 3’-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. U.S.A. 71 1342–1346. 10.1073/pnas.71.4.1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slager J., Kjos M., Attaiech L., Veening J. W. (2014). Antibiotic-induced replication stress triggers bacterial competence by increasing gene dosage near the origin. Cell 157 395–406. 10.1016/j.cell.2014.01.068 [DOI] [PubMed] [Google Scholar]
- Stevens K. E., Chang D., Zwack E. E., Sebert M. E. (2011). Competence in Streptococcus pneumoniae is regulated by the rate of ribosomal decoding errors. mBio 2 e00071–e00011. 10.1128/mBio.00071-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G., Vogel J., Wassarman K. M. (2012). Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell. 43 880–891. 10.1016/j.molcel.2011.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straume D., Stamsas G. A., Håvarstein L. S. (2014). Natural transformation and genome evolution in Streptococcus pneumoniae. Infect. Genet. Evol. 33 371–380. 10.1016/j.meegid.2014.10.020 [DOI] [PubMed] [Google Scholar]
- Sung C. K., Li H., Claverys J. P., Morrison D. A. (2001). An rpsL cassette, janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 67 5190–5196. 10.1128/AEM.67.11.5190-5196.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng L., Biswas I., Morrison D. A. (2009). A self-deleting Cre-loxP-ermAM cassette, chesire, for morker-less gene deletion in Streptococus pneumoniae. J. Microbiol. Methods 9 353–357. 10.1016/j.mimet.2009.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng L., Piotrowski A., Morrison D. A. (2013). Exit from competence for genetic transformation in Streptococcus pneumoniae is regulated at multiple levels. PLoS ONE 8:e64197 10.1371/journal.pone.0064197 [DOI] [PMC free article] [PubMed] [Google Scholar]