Abstract
AIM: To compare kinesin family member 1B (KIF1B) expression with clinicopathologic parameters and prognosis in hepatocellular carcinoma (HCC) patients.
METHODS: KIF1B protein and mRNA expression was assessed in HCC and paracarcinomatous (PC) tissues from 68 patients with HCC using Western blot and quantitative real-time reverse transcription-PCR, respectively. Student’s t-tests were used to analyze relationships between clinicopathologic parameters and KIF1B expression, the Kaplan-Meier method was used to analyze survival outcomes, and the log-rank test was used to compare survival differences between groups.
RESULTS: Mean protein and mRNA levels of KIF1B were similar between HCC and PC tissues. However, HCC tissues with vein invasions had significantly lower KIF1B protein levels compared to those without vein invasions (2.30 ± 0.82 relative units vs 2.77 ± 0.84 relative units, P < 0.05). KIF1B protein levels in HCC tissues from patients with recurrence during the follow-up period were significantly lower than those without recurrence (2.31 ± 0.92 relative units vs 2.80 ± 0.80 relative units, P < 0.05). However, KIF1B protein and mRNA expression in HCC patients was not associated with other clinicopathologic parameters. Ratios of KIF1B mRNA expression in HCC tissues to those in PC tissues were correlated with overall survival (13.5 mo vs 20.0 mo, P < 0.05) and disease-free survival (11.5 mo vs 19.5 mo, P < 0.05).
CONCLUSION: Downregulation of KIF1B in HCC tissues is associated with poor prognosis; additional clinical studies are needed to confirm whether KIF1B can serve as a prognostic marker.
Keywords: Clinicopathologic correlation, Kinesin family member 1B, Liver cancer, Survival, Tumor progression
Core tip: Expression of kinesin family member 1B (KIF1B) protein and mRNA did not significantly differ between hepatocellular carcinoma (HCC) tissues and paracarcinomatous tissues. KIF1B protein levels in HCC tissues were inversely correlated with recurrence and tumor vein invasion. Furthermore, ratios of KIF1B mRNA in HCC tissues to paracarcinomatous tissues correlated with overall survival and disease-free survival for patients with HCC. Downregulation of KIF1B mRNA in HCC tissues was associated with poor prognosis.
INTRODUCTION
Hepatocellular carcinoma (HCC) is the fifth most common cancer and the third most common cause of cancer-related deaths worldwide[1,2]. An estimated 782500 new liver cancer cases and 745500 deaths occurred worldwide during 2012, with China alone accounting for approximately 50% of the total number of cases and deaths[3]. It imposes high social and medical burdens, especially in societies with high incidences of viral hepatitis infection[4]. Most HCC patients are diagnosed beyond the stage at which surgical options are suitable, and thus have poor prognoses. However, patients at the same stage may have different prognoses[5] because many factors affect outcomes, such as clinicopathologic parameters[6-13] and emerging biomarkers[10,14-19]. This study explores how kinesin family member 1B (KIF1B) affects the long-term survival outcomes in patients with HCC who undergo surgical treatment.
Recently, a single nucleotide polymorphism, rs17401966, located at intron 24 of KIF1B, was associated with susceptibility to hepatitis B virus (HBV)-related HCC in a genome-wide association study[20]. Several studies confirmed that KIF1B affects the progression from HBV infection to HCC[20,21]. However, expression of KIF1B protein and mRNA in tumors and paracarcinomatous (PC) tissues of HCC patients was not described in existing studies. This study retrospectively investigated the relationship between KIF1B expression and clinicopathologic parameters, and its predictive value for HCC prognosis.
MATERIALS AND METHODS
Patients and samples
Resected tumor and matched PC specimens were collected from 68 HCC patients, and were immediately frozen in liquid nitrogen and stored at -80 °C until further testing. PC tissue was defined as liver tissue collected 2-5 cm away from the tumor border. All patients were treated with surgery in the Department of Hepatobiliary Surgery, Yantaishan Hospital, between January 2012 and June 2012. The diagnosis of HCC was made based on guidelines from the Chinese Society of Hepatology, the Chinese Society of Infectious Diseases, and the Chinese Medical Association[22]. Patients who received other treatment (transarterial chemoembolization, radiofrequency ablation, or others) before surgery or had other tumor diseases were excluded from this study. The study protocol was approved by the Ethics Committee of the Yantaishan Hospital. Informed consent was obtained from each patient.
Viral infection, tumor size, number of tumor nodules, histopathologic classification, vein invasion, recurrence status, and patient survival time were evaluated. Vein invasion found during pathologic examination indicated tumor infiltration in the portal venous and/or hepatic veins. Recurrence was monitored with ultrasound, CT scans, and magnetic resonance imaging.
Western blot analysis
HCC and PC tissues were homogenized and treated with RIPA lysis buffer (Dingguo, Beijing, China); the extracted proteins were resolved by 4%-12% acrylamide gradient gel. After electrophoresis, samples were transferred to a polyvinylidene fluoride membrane using iBlot fast transfer electric transfer (Invitrogen of Thermo Fisher Scientific, Waltham, MA, United States). Membranes were blocked at room temperature for 1 h with 5% milk, and incubated with a primary antibody against KIF1B or GAPDH (1:1000; Abcam, Cambridge, United Kingdom) at 4 °C overnight, followed by washing with buffer three times, a secondary antibody (1:8000, Abcam) incubation at room temperature for 2 h, washing three times, and visualized with an ECL kit (Pierce of Thermo Fisher Scientific). KIF1B-specific signals were quantified from exposed X-ray films using a scanner with BandScan 4.30 densitometry software, and are expressed as integrated intensity units relative to the GAPDH signals. The results were analyzed by physicians in a blinded manner.
Quantitative real-time reverse transcription-PCR
Total RNA was extracted from HCC and PC tissues with the Trizol method. The retroviral reverse transcriptase kit (Takara, Tokyo, Japan) was used to synthesize cDNA with the reaction conditions of 37 °C for 60 min and 95 °C for 3 min. Primers were sense: 5′-TTTCCAGCACTTAATGAAAACACATAG-3′; antisense: 5′-CAAAGTTAAATTTCCCTGCTTTGAA-3′ for KIF1B, and sense: 5′-GAAGGTGAAGGTCGGAGTC-3′; antisense: 5′-GAAGATGGTGATGGGATTTC-3′ for GAPDH. Real-time PCR was performed with the 7500 real-time quantitative PCR instrument (Applied Biosystems of Thermo Fisher Scientific) with the following conditions: 95 °C for 20 s, 60 °C for 30 s, and 72 °C for 30 s for 40 cycles. Data were normalized using the GAPDH housekeeping gene and are expressed as 2∆∆Ct.
Patient follow-ups
Patient follow-up data was obtained after discharge for all 68 HCC patients by direct communication with the patients or their relatives, or by reviewing hospital records. Disease-free survival (DFS) was measured from the date of hepatectomy until tumor recurrence. Overall survival (OS) was measured from the date of hepatectomy until death or the last follow-up point. The last follow-up evaluation was censored on December 31, 2014, or up to the time of death.
Statistical analysis
Values are presented as mean ± SD or median (range). The Student’s t-test was used to evaluate differences in KIF1B protein and mRNA expression between HCC and PC tissues. Spearman correlation coefficients were used to analyze relationships between expression levels of KIF1B protein and mRNA in HCC. The Student’s t-test was used to analyze relationships between KIF1B expression level and clinicopathologic parameters. Bivariate correlations were used to analyze the association between survival time and KIF1B protein and mRNA. OS and DFS was calculated by the Kaplan-Meier method and analyzed by the log-rank test. All tests were two-tailed; P < 0.05 was considered significant. The SPSS package 13.0 (SPSS Inc., Chicago, IL, United States) was used for all analyses.
RESULTS
Patient characteristics
The mean age of the HCC patients was 58.4 ± 10.9 years; 83.8% (57/68) were male, and 80.9% (55/68) had HBV infections. Nineteen HCC patients had at least one tumor nodule larger than 5 cm. Tumors were well differentiated in 25 patients, and moderately or poorly differentiated in 32 and 11 patients, respectively. The median follow-up time was 16.5 mo (range, 1-36 mo). HCC recurred in 34 patients over a median recurrence time of 13.5 mo. During the follow-up, 31 patients died, with a mean survival time of 16.8 ± 9.4 mo.
KIF1B protein and mRNA expression in HCC and PC tissues
The mean KIF1B protein level in HCC tissues was 2.55 ± 0.87 relative units (RU), which was higher, but not significantly so, than in PC tissues (2.38 ± 0.92 RU). The mean KIF1B mRNA level in HCC tissues (1.47 ± 0.29 RU) was also similar to that in PC tissues (1.48 ± 0.29 RU). KIF1B protein and mRNA expression did not significantly differ between HCC and PC tissues (Figure 1).
Figure 1.
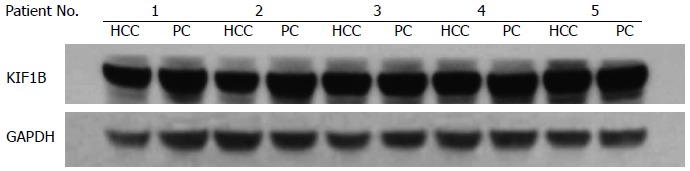
Kinesin family member 1B (KIF1B) protein expression. Representative Western blot for KIF1B expression in hepatocellular carcinoma (HCC) and paracarcinomatous (PC) tissues. GAPDH was used as the internal loading control.
Correlation between KIF1B expression and clinicopathologic features
KIF1B protein expression in HCC tissues with vein invasion was significantly lower than in those without vein invasion (2.30 ± 0.82 RU vs 2.77 ± 0.84 RU, P = 0.033). KIF1B mRNA expression in HCC tissues with vein invasion (1.41 ± 0.25 RU) was also lower than in those without vein invasion (1.50 ± 0.30 RU), but not significantly so. However, ratios of KIF1B mRNA expression in HCC/adjacent PC tissues with vein invasions were significantly lower than those without vein invasions (0.89 ± 0.29 RU vs 1.11 ± 0.33 RU, P = 0.009).
KIF1B protein levels in HCC tissues from patients who experienced recurrence during the follow-up were significantly lower than in those without recurrence (2.31 ± 0.92 RU vs 2.80 ± 0.80 RU, P = 0.022). KIF1B mRNA levels were slightly lower, but not significantly so, in the recurrence group. Ratios of HCC/PC KIF1B mRNA expression in patients with recurrence during the follow-up were significantly lower than those without recurrence (0.96 ± 0.31 vs 1.11 ± 0.29, P = 0.043).
There was no association between KIF1B protein or mRNA expression and other clinicopathologic parameters, including patient age, sex, hepatitis B virus, liver function, tumor differentiation, tumor size, and number of tumor nodules (Table 1).
Table 1.
Kinesin family member 1B expression in cancer and paracarcinomatous tissues according to clinicopathologic features in 68 patients with hepatocellular carcinoma
| Clinicopathologic feature | n |
KIF1B protein, relative units |
KIF1B mRNA, relative units |
||||
| HCC tissue | PC tissue | HCC/PC | HCC tissue | PC tissue | HCC/PC | ||
| Age (yr) | |||||||
| ≥ 50 | 54 | 2.59 ± 0.53 | 2.50 ± 0.43 | 1.19 ± 0.10 | 1.44 ± 0.24 | 1.49 ± 0.23 | 1.01 ± 0.05 |
| < 50 | 14 | 2.63 ± 0.92 | 2.26 ± 0.80 | 1.31 ± 0.63 | 1.48 ± 0.30 | 1.44 ± 0.26 | 1.07 ± 0.35 |
| Sex | |||||||
| Male | 57 | 2.62 ± 0.88 | 2.39 ± 0.90 | 1.27 ± 0.63 | 1.49 ± 0.29 | 1.48 ± 0.29 | 1.05 ± 0.31 |
| Female | 11 | 2.20 ± 0.70 | 2.30 ± 0.97 | 1.16 ± 0.63 | 1.39 ± 0.28 | 1.50 ± 0.26 | 0.98 ± 0.37 |
| Child classification | |||||||
| A | 54 | 2.59 ± 0.91 | 2.50 ± 0.89 | 1.19 ± 0.63 | 1.44 ± 0.29 | 1.49 ± 0.28 | 1.01 ± 0.32 |
| B | 14 | 2.40 ± 0.64 | 1.91 ± 0.85 | 1.4 8± 0.60 | 1.58 ± 0.27 | 1.45 ± 0.28 | 1.13 ± 0.27 |
| Hepatitis B virus | |||||||
| Positive | 55 | 2.55 ± 0.90 | 2.43 ± 0.93 | 1.28 ± 0.62 | 1.45 ± 0.28 | 1.48 ± 0.28 | 1.02 ± 0.30 |
| Negative | 13 | 2.59 ± 0.70 | 2.16 ± 0.84 | 1.38 ± 0.65 | 1.58 ± 0.34 | 1.50 ± 0.28 | 1.10 ± 0.37 |
| Differentiation | |||||||
| Well | 25 | 2.53 ± 0.95 | 2.22 ± 0.86 | 1.31 ± 0.68 | 1.54 ± 0.28 | 1.46 ± 0.30 | 1.11 ± 0.33 |
| Moderately | 32 | 2.52 ± 0.76 | 2.47 ± 0.98 | 1.20 ± 0.58 | 1.43 ± 0.28 | 1.49 ± 0.27 | 0.99 ± 0.30 |
| Poorly | 11 | 2.69 ± 0.90 | 2.45 ± 0.77 | 1.27 ± 0.66 | 1.42 ± 0.32 | 1.51 ± 0.28 | 0.98 ± 0.30 |
| Tumor size (cm) | |||||||
| > 3 | 48 | 2.56 ± 0.85 | 2.42 ± 0.91 | 1.24 ± 0.64 | 1.46 ± 0.31 | 1.47 ± 0.27 | 1.03 ± 0.32 |
| ≤ 3 | 20 | 2.55 ± 0.90 | 2.27 ± 0.93 | 1.28 ± 0.63 | 1.49 ± 0.23 | 1.51 ± 0.31 | 1.05 ± 0.31 |
| Tumor nodule | |||||||
| Solitary | 45 | 2.57 ± 0.91 | 2.35 ± 0.93 | 1.27 ± 0.64 | 1.50 ± 0.28 | 1.48 ± 0.29 | 1.06 ± 0.31 |
| Multiple | 23 | 2.52 ± 0.77 | 2.44 ± 0.87 | 1.21 ± 0.62 | 1.42 ± 0.31 | 1.48 ± 0.27 | 1.00 ± 0.32 |
| Vein invasion | |||||||
| Positive | 22 | 2.30 ± 0.82 | 2.33 ± 0.97 | 1.21 ± 0.60 | 1.41 ± 0.25 | 1.50 ± 0.31 | 0.89 ± 0.29 |
| Negative | 46 | 2.77 ± 0.84a | 2.40 ± 0.89 | 1.27 ± 0.65 | 1.50 ± 0.30 | 1.47 ± 0.27 | 1.11 ± 0.33b |
| Recurrence status | |||||||
| Yes | 34 | 2.31 ± 0.92 | 2.59 ± 0.93 | 1.14 ± 0.63 | 1.42 ± 0.27 | 1.55 ± 0.31 | 0.96 ± 0.31 |
| No | 34 | 2.80 ± 0.80c | 2.17 ± 0.85 | 1.36 ± 0.61 | 1.52 ± 0.30 | 1.42 ± 0.24 | 1.11 ± 0.29c |
P < 0.05,
P < 0.01 vs positive vein invasion;
P < 0.05 vs recurrence. KIF1B: Kinesin family member 1B; PC: Paracarcinomatous; HCC: Hepatocellular carcinoma.
Bivariate correlations were used to analyze the association between survival time and KIF1B protein and mRNA. Ratios of KIF1B mRNA expression in HCC/adjacent PC tissues were correlated with OS and DFS, with respective Pearson correlations of 0.941 and 0.988 (P < 0.001 for both). Kaplan-Meier survival curves and log-rank tests showed that downregulation of KIF1B mRNA in HCC tissues was associated with poor prognosis. Median OS for the downregulated group was 13.5 mo, significantly shorter than for the upregulated group at 20 mo (P < 0.05; Figure 2A). Median DFS for down- and upregulated groups were 11.5 mo and 19.5 mo, respectively (P < 0.05; Figure 2B).
Figure 2.
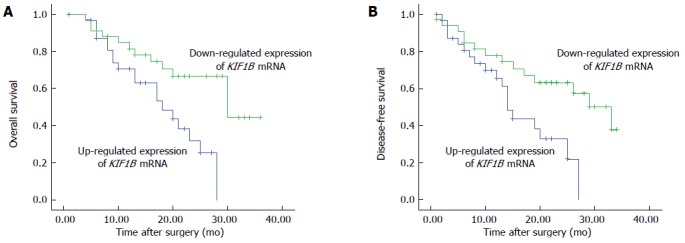
Survival of patients with hepatocellular carcinoma after surgical resection according to kinesin family member 1B (KIF1B) mRNA expression. A: Overall survival; B: Disease-free survival (P < 0.05; log-rank test).
DISCUSSION
KIF1B is located on chromosome 1p36, and belongs to the kinesin superfamily of intermediate filaments that is responsible for intracellular vesicular transport[23]. KIF1B encodes two alternatively spliced isoforms, KIF1Ba and KIF1Bb, and both isoforms form homodimers and transport mitochondria and synaptic vesicle precursors, respectively[24]. KIF1B has been shown to act as a tumor suppressor in multiple cancers, including aggressive neuroblastoma, pheochromocytoma, colon, liver, brain, breast, and other cancers, by acting on various inhibitors of cell proliferation and activators of apoptosis[25,26]. KIF1B knockdown in rat sympathetic neurons prevents apoptosis following nerve growth factor withdrawal, indicating that KIF1B plays a crucial role in neuronal apoptosis upon nerve growth factor limitation[27].
KIF1B is reportedly associated with gastric cancer invasion[28], which suggests that KIF1B has a function in cancer progression. Zhang et al[20] showed KIF1B contributed distinctly to the progression from chronic HBV infection to HCC. However, the exact function of KIF1B in HCC is unclear; conditional knockout models may be necessary to further investigate its role in hepatocarcinogenesis.
KIF1B expression in HCC tissues and its relationship with tumor progression and prognosis are not widely reported. The present study found that KIF1B protein is expressed in cancer tissues and PC tissues of patients with HCC, with no significant differences in expression levels. There is no correlation between the expression level of KIF1B protein and mRNA in HCC samples. Genetic polymorphisms and epigenetic factors may have contributed to the difference.
An important result of the current study is that KIF1B protein expression is associated with vein invasion and tumor recurrence status. These factors are highly correlated with invasion and metastasis of HCC[29-31]. The results indicate that more invasive tumors have lower KIF1B protein expression, and by extension, that KIF1B has a suppressive function in HCC. The results also show that there is no pattern for KIF1B protein expression when the subjects and their specimens are stratified by sex, age, liver function, HBV, number of tumor nodules, tumor size, and tumor differentiation. These factors might not affect KIF1B expression, or might do so only subtly.
Unlike the protein results, KIF1B mRNA expression had no correlation to any tested clinicopathologic features. Interestingly, ratios of KIF1B mRNA expression in HCC/PC pairs are negatively correlated with OS and DFS. HCC patients were divided into two groups: patients with downregulated expression (higher KIF1B mRNA expression in PC tissues than in HCC tissues) and those with upregulated expression (higher KIF1B mRNA expression in HCC tissues than in PC tissues). The downregulated KIF1B mRNA group had longer DFS than the upregulated KIF1B mRNA group. In addition, patients with downregulated KIF1B mRNA had increased risk of recurrence and significantly reduced OS.
The results of the present study show that KIF1B is a liver cancer suppressor gene. KIF1B mRNA levels may be prognostic biomarkers.
In conclusion, this is the first investigation of KIF1B expression at both the protein and mRNA levels in association with clinicopathologic features of HCC. Expression of KIF1B protein and mRNA do not differ between HCC tissues and PC tissues. KIF1B protein levels in HCC tissues from patients with recurrence during the follow-up are significantly lower than in those without recurrence. HCC tissues with vein invasions have significantly lower KIF1B protein levels than those without vein invasions. Ratios of KIF1B mRNA relative expression in HCC tissues to PC tissues correlate with OS and DFS. Based on the downregulation of KIF1B mRNA in HCC, we propose that the manipulation of KIF1B expression in HCC patients might have therapeutic implications. However, related reports, especially on KIF1B functions and mechanisms of regulation in normal and HCC tissues, are limited and warrant further study. Further large-scale clinical studies are needed to confirm whether KIF1B could serve as a liver cancer prognostic marker.
COMMENTS
Background
An estimated 782500 new liver cancer cases and 745500 deaths occurred worldwide during 2012, with China alone accounting for approximately 50% of the total number of cases and deaths. Most hepatocellular carcinoma (HCC) patients are diagnosed beyond the stage at which surgical options are suitable, and thus have poor prognoses. However, patients at the same stage may have different prognoses because many factors affect outcomes, such as clinicopathologic parameters and emerging biomarkers. In this study, we explore how kinesin family member 1B (KIF1B) affects the long-term survival outcomes in patients with HCC who undergo surgical treatment.
Research frontiers
A single nucleotide polymorphism of KIF1B, rs17401966, located at intron 24, is associated with susceptibility to hepatitis B virus-related HCC in a genome-wide association stud, and several studies confirm that KIF1B affects the progression from infection to HCC. However, expression of KIF1B protein and mRNA in tumors and paracarcinomatous tissues of HCC patients was not described in existing studies. The authors therefore retrospectively investigated the relationship between KIF1B expression and clinicopathologic parameters, and its predictive value for HCC prognosis.
Innovations and breakthroughs
KIF1B expression in HCC tissues and its relationship with tumor progression and prognosis are not widely reported. The present study found that KIF1B protein is expressed in cancer and paracarcinomatous tissues of patients with HCC, with no significant differences in expression levels. The authors observed no significant correlation between the expression level of KIF1B protein and mRNA in HCC samples. Genetic polymorphisms and epigenetic factors may have contributed to the difference. An important result of the current study is that KIF1B protein expression is associated with vein invasion and tumor recurrence status. These factors are strongly correlated with invasion and metastasis of HCC. The results indicate that more invasive tumors have lower KIF1B protein expression, and by extension, that KIF1B has a suppressive function in HCC. Unlike the protein results, KIF1B mRNA expression is not correlated to any tested clinicopathologic feature. Interestingly, correlation analysis showed that ratios of KIF1B mRNA expression in HCC/paracarcinomatous pairs are negatively correlated with overall and disease-free survival. The downregulated KIF1B mRNA group had longer disease-free survival than the upregulated KIF1B mRNA group. In addition, patients with downregulated KIF1B mRNA had increased risk of recurrence and significantly reduced overall survival.
Applications
This study suggests that KIF1B is a liver cancer suppressor gene. KIF1B mRNA levels may be prognostic biomarkers for HCC.
Peer-review
This is a good retrospective study in which the authors investigated the relationship between KIF1B expression and clinicopathologic parameters, and its predictive value for HCC prognosis. The results are interesting and suggest that KIF1B mRNA levels may be prognostic biomarkers for HCC.
Footnotes
Institutional review board statement: The study protocol was approved by the Ethics Committee of the Yantaishan Hospital.
Informed consent statement: Informed consent was obtained from each patient.
Conflict-of-interest statement: No potential conflicts of interest relevant to this article were reported.
Data sharing statement: No additional data are available.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: March 17, 2015
First decision: March 26, 2015
Article in press: May 7, 2015
P- Reviewer: El-Emshaty HM, Pinero F, Xu Y S- Editor: Qi Y L- Editor: AmEditor E- Editor: Zhang DN
References
- 1.World Health Organization. Mortality Database. Updated 2014. Available from: http://www.who.int/healthinfo/mortality_data/en/ [Google Scholar]
- 2.Blum HE. Hepatocellular carcinoma: therapy and prevention. World J Gastroenterol. 2005;11:7391–7400. doi: 10.3748/wjg.v11.i47.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 4.Shariff MI, Cox IJ, Gomaa AI, Khan SA, Gedroyc W, Taylor-Robinson SD. Hepatocellular carcinoma: current trends in worldwide epidemiology, risk factors, diagnosis and therapeutics. Expert Rev Gastroenterol Hepatol. 2009;3:353–367. doi: 10.1586/egh.09.35. [DOI] [PubMed] [Google Scholar]
- 5.Miao R, Luo H, Zhou H, Li G, Bu D, Yang X, Zhao X, Zhang H, Liu S, Zhong Y, et al. Identification of prognostic biomarkers in hepatitis B virus-related hepatocellular carcinoma and stratification by integrative multi-omics analysis. J Hepatol. 2014;61:840–849. doi: 10.1016/j.jhep.2014.05.025. [DOI] [PubMed] [Google Scholar]
- 6.Liu L, Miao R, Yang H, Lu X, Zhao Y, Mao Y, Zhong S, Huang J, Sang X, Zhao H. Prognostic factors after liver resection for hepatocellular carcinoma: a single-center experience from China. Am J Surg. 2012;203:741–750. doi: 10.1016/j.amjsurg.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 7.Wan S, Civan J, Rossi S, Yang H. Profiling HBV integrations in hepatocellular carcinoma. Hepatobiliary Surg Nutr. 2013;2:124–126. doi: 10.3978/j.issn.2304-3881.2012.10.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhan P, Ji YN. Prognostic significance of TP53 expression for patients with hepatocellular carcinoma: a meta-analysis. Hepatobiliary Surg Nutr. 2014;3:11–17. doi: 10.3978/j.issn.2304-3881.2014.01.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhan P, Ji YN, Yu LK. TP53 mutation is associated with a poor outcome for patients with hepatocellular carcinoma: evidence from a meta-analysis. Hepatobiliary Surg Nutr. 2013;2:260–265. doi: 10.3978/j.issn.2304-3881.2013.07.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ha SY, Song DH, Hwang SH, Cho SY, Park CK. Expression of prothymosin alpha predicts early recurrence and poor prognosis of hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2015;14:171–177. doi: 10.1016/s1499-3872(14)60326-x. [DOI] [PubMed] [Google Scholar]
- 11.Li JJ, Pan K, Gu MF, Chen MS, Zhao JJ, Wang H, Liang XT, Sun JC, Xia JC. Prognostic value of soluble MICA levels in the serum of patients with advanced hepatocellular carcinoma. Chin J Cancer. 2013;32:141–148. doi: 10.5732/cjc.012.10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu C, Zeng XH, Wang L, Tao SQ, Wu QX, Zhu P, Deng GH, Wang YM. sFRP-4, a potential novel serum marker for chronic hepatitis B-related hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2015;14:164–170. doi: 10.1016/s1499-3872(15)60352-6. [DOI] [PubMed] [Google Scholar]
- 13.Yang L, Rong W, Xiao T, Zhang Y, Xu B, Liu Y, Wang L, Wu F, Qi J, Zhao X, et al. Secretory/releasing proteome-based identification of plasma biomarkers in HBV-associated hepatocellular carcinoma. Sci China Life Sci. 2013;56:638–646. doi: 10.1007/s11427-013-4497-x. [DOI] [PubMed] [Google Scholar]
- 14.Geng M, Cao YC, Chen YJ, Jiang H, Bi LQ, Liu XH. Loss of Wnt5a and Ror2 protein in hepatocellular carcinoma associated with poor prognosis. World J Gastroenterol. 2012;18:1328–1338. doi: 10.3748/wjg.v18.i12.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun Y, Yang H, Mao Y, Xu H, Zhang J, Li G, Lu X, Sang X, Zhao H, Zhong S, et al. Increased Golgi protein 73 expression in hepatocellular carcinoma tissue correlates with tumor aggression but not survival. J Gastroenterol Hepatol. 2011;26:1207–1212. doi: 10.1111/j.1440-1746.2011.06733.x. [DOI] [PubMed] [Google Scholar]
- 16.Aida T, Bodell WJ. [Mechanism of cellular resistance to ACNU in rat brain tumor cell line] Neurol Med Chir (Tokyo) 1986;26:931–936. doi: 10.2176/nmc.26.931. [DOI] [PubMed] [Google Scholar]
- 17.Lo RC, Ng IO. Hepatocellular tumors: immunohistochemical analyses for classification and prognostication. Chin J Cancer Res. 2011;23:245–253. doi: 10.1007/s11670-011-0245-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng L, Liang P, Li J, Huang XB, Wang WW, Wang L, Feng H. Expression of BC047440 protein in hepatocellular carcinoma and its relationship to prognosis. Chin J Cancer. 2010;29:931–936. doi: 10.5732/cjc.010.10272. [DOI] [PubMed] [Google Scholar]
- 19.Wang YL, Zhu ZJ, Teng DH, Yao Z, Gao W, Shen ZY. Glypican-3 expression and its relationship with recurrence of HCC after liver transplantation. World J Gastroenterol. 2012;18:2408–2414. doi: 10.3748/wjg.v18.i19.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, Zhai Y, Hu Z, Wu C, Qian J, Jia W, Ma F, Huang W, Yu L, Yue W, et al. Genome-wide association study identifies 1p36.22 as a new susceptibility locus for hepatocellular carcinoma in chronic hepatitis B virus carriers. Nat Genet. 2010;42:755–758. doi: 10.1038/ng.638. [DOI] [PubMed] [Google Scholar]
- 21.Wang ZC, Gao Q, Shi JY, Yang LX, Zhou J, Wang XY, Shi YH, Ke AW, Shi GM, Ding ZB, et al. Genetic polymorphism of the kinesin-like protein KIF1B gene and the risk of hepatocellular carcinoma. PLoS One. 2013;8:e62571. doi: 10.1371/journal.pone.0062571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chinese Society of Hepatology, Chinese Medical Association; Chinese Society of Infectious Diseases, Chinese Medical Association. [The guidelines of prevention and treatment for chronic hepatitis B] Zhonghua Ganzangbing Zazhi. 2005;13:881–891. [PubMed] [Google Scholar]
- 23.Nangaku M, Sato-Yoshitake R, Okada Y, Noda Y, Takemura R, Yamazaki H, Hirokawa N. KIF1B, a novel microtubule plus end-directed monomeric motor protein for transport of mitochondria. Cell. 1994;79:1209–1220. doi: 10.1016/0092-8674(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 24.MacAskill AF, Kittler JT. Control of mitochondrial transport and localization in neurons. Trends Cell Biol. 2010;20:102–112. doi: 10.1016/j.tcb.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 25.Munirajan AK, Ando K, Mukai A, Takahashi M, Suenaga Y, Ohira M, Koda T, Hirota T, Ozaki T, Nakagawara A. KIF1Bbeta functions as a haploinsufficient tumor suppressor gene mapped to chromosome 1p36.2 by inducing apoptotic cell death. J Biol Chem. 2008;283:24426–24434. doi: 10.1074/jbc.M802316200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strunze S, Engelke MF, Wang IH, Puntener D, Boucke K, Schleich S, Way M, Schoenenberger P, Burckhardt CJ, Greber UF. Kinesin-1-mediated capsid disassembly and disruption of the nuclear pore complex promote virus infection. Cell Host Microbe. 2011;10:210–223. doi: 10.1016/j.chom.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 27.Schlisio S, Kenchappa RS, Vredeveld LC, George RE, Stewart R, Greulich H, Shahriari K, Nguyen NV, Pigny P, Dahia PL, et al. The kinesin KIF1Bbeta acts downstream from EglN3 to induce apoptosis and is a potential 1p36 tumor suppressor. Genes Dev. 2008;22:884–893. doi: 10.1101/gad.1648608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong Z, Xu X, Du L, Yang Y, Cheng H, Zhang X, Li Z, Wang L, Li J, Liu H, et al. Leptin-mediated regulation of MT1-MMP localization is KIF1B dependent and enhances gastric cancer cell invasion. Carcinogenesis. 2013;34:974–983. doi: 10.1093/carcin/bgt028. [DOI] [PubMed] [Google Scholar]
- 29.Tung-Ping Poon R, Fan ST, Wong J. Risk factors, prevention, and management of postoperative recurrence after resection of hepatocellular carcinoma. Ann Surg. 2000;232:10–24. doi: 10.1097/00000658-200007000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang J. Current progress in epigenetic research for hepatocarcinomagenesis. Sci China C Life Sci. 2009;52:31–42. doi: 10.1007/s11427-009-0014-7. [DOI] [PubMed] [Google Scholar]
- 31.Shimada K, Sano T, Sakamoto Y, Kosuge T. A long-term follow-up and management study of hepatocellular carcinoma patients surviving for 10 years or longer after curative hepatectomy. Cancer. 2005;104:1939–1947. doi: 10.1002/cncr.21461. [DOI] [PubMed] [Google Scholar]


