Abstract
Aims: Elevated low-density lipoprotein-cholesterol (LDL-C) is regarded as one of major risks of cardiovascular diseases and atherosclerotic events. It has been previously reported that peroxisome proliferator-activated receptors (PPARs) play an important role in the regulation of lipid metabolism. In this study, we aimed to investigate the influence of PPARα/δ/γ gene polymorphisms on LDL-C level. Eight hundred twenty unrelated participants were recruited. Ten single-nucleotide polymorphisms (SNPs) were genotyped to analyze the gene–gene interactions among these polymorphisms using the generalized multifactor dimensionality reduction (GMDR) method. Results: The results of single-locus analyses indicated that the genotypes with minor allele variants at the rs1800206, rs9794, rs1805192, rs709158, and rs3856806 loci are associated with higher LDL-C levels (p<0.05) after adjusting for covariates. In contrast, individuals that were homozygous for the major allele (CC) of rs10865710 had significantly higher LDL-C than those with either one or more minor type alleles (CG+GG, mean difference: −0.21 mM; 95% confidence interval [CI]: −0.37 to −0.04 mM; p=0.013). Significant gene–gene interactions among PPAR gene polymorphisms on LDL-C were identified by a generalized multifactor dimensionality reduction (GMDR) approach in 2- to 8-locus models (p<0.05). Conclusion: Our results provide evidence that multiple PPARα/δ/γ gene polymorphisms are individually associated with increased LDL-C, and that interactions, among these alleles result in additional increased risk suggesting that PPAR genes may contribute substantially to the risk of cardiovascular diseases and atherosclerosis.
Introduction
Low-density lipoprotein (LDL) refers to a class and range of lipoprotein particles, varying in their size and contents, which contain a large amount of cholesterol and cholesteryl ester, for use by various cells. Studies have indicated that elevated LDL particles can increase the risk of cardiovascular diseases (Cromwell et al., 2007) and play a key role in the initiation and progression of atherosclerotic events (Badimon and Vilahur, 2012). In the clinical context, low-density lipoprotein-cholesterol (LDL-C), the mass of cholesterol contained in LDL, is a widely used measurement to estimate LDL particles and still the primary target for LDL-lowering therapy in almost all the guidelines recommended (Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults, 2001; Catapano et al., 2011; Anderson et al., 2013).
Peroxisome proliferator-activated receptors (PPARs) are ligand-activated receptors belonging to the nuclear hormone receptor superfamily (Grygiel-Gorniak, 2014). Since the year that Issemann et al. (1990) first found PPARs, increasing evidence has proved that PPARs play an important role in lipid metabolism and could be functioned as regulators affecting the level of LDL-C (Azhar, 2010). Moreover, it has also been known that a few drugs, such as fibrates, could be used for LDL-lowering therapy by activating PPARs (Tenenbaum and Fisman, 2012).
So far, it has been acknowledged that LDL heterogeneity could be influenced by genetic factors (Krauss, 2005). Although several studies have reported the association between PPAR gene polymorphisms and LDL-C in recent years, these researchers were mainly concerned about the single-nucleotide polymorphisms (SNPs) that only had a strong main effect on the trait (Miao et al., 2010; AlSaleh et al., 2011), which might overlook the weak marginal effect of an SNP that might possibly reflect larger effects of collections (Hoh and Ott, 2003); In addition, some of the studies were just limited to the discussion of one of the isoforms of PPARs (Mazzotti et al., 2011), yet the actual fact is that traits could be affected by the effects of several genes lying on different chromosomes (Hoh and Ott, 2003).
To overcome these deficiencies, we aimed to investigate whether PPARα/δ/γ gene polymorphisms are associated with the level of LDL-C and there is a potential gene–gene interaction among these polymorphisms in a Chinese Han population.
Materials and Methods
Study participants
All participants who had undergone the survey from April 1999 to June 2004 were selected from the Prevention of Metabolic syndrome and Multimetabolic Disorders in Jiangsu province of China Study (PMMJS) (Hu et al., 2006). Four thousand eighty-three of 4582 from start-up (89.11%) accomplished the subsequent survey in 2009. Persons with experienced stroke or exhibited cardiovascular diseases (36, 11 of whom died), type 2 diabetes (289, 31 of whom died), a body–mass index (BMI) <18.5 kg/m2 (n=27, 2 of whom died), or missing data (n=133) were excluded. Eight hundred twenty unrelated individuals with complete data were ultimately obtained from the remaining cases for investigation by random sampling. Individuals selected were similar to those who were not selected in terms of age, sex, smoking, alcohol consumption, family disease history, and metabolic variables. Blood samples harvested from veins of studied people were examined for lipid measurement, genotyping, and some other biochemical tests. Participants who had an LDL-C level ≥4.14 mM were assigned to the group of high LDL-C (Joint Committee for Developing Chinese guidelines on Prevention and Treatment of Dyslipidemia in Adults, 2007). In addition, all subjects provided written informed consent, and the procedures in the present study were approved by the Ethics Committee of Soochow University in China.
Body measurements and laboratory methods
Subjects engaged in our research completed the questionnaires on diet and physical activities and had the blood drawn later. Details about the data included sociodemographic characteristics (age, sex, and occupation, etc.), personal and family history of chronic diseases, and other lifestyle habits, such as smoking and alcohol consumption. Blood samples were collected in the morning after at least 8 h of fasting, and samples were preserved at −80°C until laboratory testing. Body weight, height, and waist circumference (WC) were measured by standard procedures, and BMI was calculated by the Quetelet equation. Fasting plasma glucose was measured by an oxidase enzymatic method. Concentrations of LDL-C were determined by a homogenous direct method. Tests were all performed at the same laboratory. The methods of investigation applied at the follow-up were the same as those administered for the baseline.
Selection of SNPs and genotyping
Ten SNPs located in PPARα/δ/γ genes were selected from public databases (dbSNP, http://ncbi.nlm.nih.gov/SNP/) based on the following criteria: (i) minor allele frequency ≥0.05, (ii) reported association with multiple metabolic disorders, and (iii) loci at functional regions. Among these SNPs, rs135539, rs1800206, and rs4253778 are located in PPARα on 22q13.3; rs2016520 and rs9794 are located in PPARδ on chromosome 6p21.3; rs10865710, rs1805192, rs709158, rs3856806, and rs4684847 are located in PPARγ on chromosome 3p25. Details about the information of selected SNPs are shown in Table 1.
Table 1.
Description of the 10 Single-Nucleotide Polymorphisms in Peroxisome Proliferator-Activated Receptor Genes
| SNP ID | SNP | Chromosome | Nucleotide substitution | Chromosome position |
|---|---|---|---|---|
| PPARα | ||||
| rs135539 | 1A>C | 22 | A>C | 46163368 |
| rs1800206 | L162V | 22 | G>C | 46218377 |
| rs4253778 | 7G>C | 22 | C>G | 46234737 |
| PPARδ | ||||
| rs2016520 | −87T>C | 6 | T>C | 35411001 |
| rs9794 | 2806C>G | 6 | C>G | 35428018 |
| PPARγ | ||||
| rs709158 | Intron A>G | 3 | A>G | 12421677 |
| rs10865710 | C681G | 3 | C>G | 12311699 |
| rs1805192 | Pro12Ala | 3 | C>G | 12379739 |
| rs4684847 | Intron C>T | 3 | C>G | 12344838 |
| rs3856806 | C161T | 3 | C>T | 12434058 |
PPAR, peroxisome proliferator-activated receptor; SNPs, single-nucleotide polymorphisms.
Genomic DNA was extracted from EDTA-treated blood samples using the DNA Blood Mini Kit (Qiagen, Hilden, Germany) according to the instructions attached. Nine SNPs (rs135539, rs1800206, rs2016520, rs9794, rs10865710, rs1805192, rs709158, rs3856806, and rs4684847) were detected using fluorescent TaqMan probes, while rs4253778 was genotyped by polymerase chain reaction–restriction fragment length polymorphisms (PCR-RFLPs). A restriction enzyme was used to identify and cut specific sequences, after which PCR was performed with the following primers: forward, 5′-ACA ATC ACT CCT TAA ATA TGG TGG-3′, and reverse, 5′-AAG TAG GGA CAG ACA GGA CCA GTA-3′. A 25 μL reaction mixture was amplified by PCR, containing DNA 20 ng, 0.5 U DNA polymerase (TaKaRa, Dalian, China), 10× buffer 2.5 μL, 10 mM dNTP 0.5 μL, and 0.5 μL forward primer and reverse primer at a concentration of 100 μM. Cycling conditions were identical for all SNPs, namely at 95°C for 3 min for the initial denaturation, and 45 cycles at 95°C for 10 s, 63°C for 30 s, and extension at 72°C for 30 s. An additional cycle at 72°C for 10 min was conducted for the final extension. ABI Prism 7000 software and the allelic discrimination procedure were used for genotyping the abovementioned nine SNPs. For quality control, ∼10% of the samples were regenotyped in a blind manner, and the same results were obtained.
Statistical analysis
Normally distributed variables were tested using the t-test and expressed as mean±standard deviation (SD). The Wilcoxon rank-sum test was used for skewed distributed traits and expressed as the median (Interquartile Range). Frequencies of categorical variables were tested using a chi-square test. All the tests were performed using SPSS v16.0. The criterion for significance was set at p<0.05 for all tests. Hardy–Weinberg equilibrium (HWE) of the SNPs was evaluated by the chi-square test. Pairwise linkage disequilibrium (LD) was calculated by the Haploview program from the HapMap website.
The association between each SNP in PPAR genes and the level of LDL-C was analyzed by linear regression models in SNPStats (Sole et al., 2006) (http://bioinfo.iconcologia.net/SNPStats) to assess the proportion of variation in the response explained by the SNPs. To investigate the possible gene–gene interactions among the SNPs, a generalized linear model was applied in the program of generalized multifactor dimensionality reduction (GMDR) (Lou et al., 2007). Cross-validation consistency, prediction accuracies, and empirical p-values were listed in the results of GMDR. The cross-validation consistency score is a measure of the degree of consistency with which the selected interaction is identified as the best model among all possibilities considered. Testing balanced accuracy is a measure of the degree to which the interaction is accurately predicted. The empirical p-values for prediction accuracy are used to measure the significance of the models with different loci. In this study, we analyzed the gene–gene interactions among the SNPs in PPAR genes by GMDR with both continuous and dichotomous phenotypes (LDL-C).
Results
The basic characteristics of study participants
The clinical and biochemical characteristics of the participants are exhibited in Table 2. Eight hundred twenty individuals (270 males and 550 females) were included in our study. The levels of BMI and WC were higher in the high LDL-C group than in the control (p<0.05). The percentage of subjects who consumed alcohol was lower in individuals with higher LDL-C (p<0.01). The distribution of sex and smoking status was not significantly different between individuals with and without high LDL-C (p>0.05). Mean (±SD) LDL-C in the high LDL-C group was 5.00±0.78, while the level was 2.71±0.75 in controls (p<0.001)
Table 2.
The Basic Characteristics of Study Participants with Different Levels of Low-Density Lipoprotein-Cholesterol
| Variables | Total (n=820) | Control (n=629) | High LDL-C (n=191) | p-Values |
|---|---|---|---|---|
| Age (years) | 50.05±9.41 | 49.89±9.34 | 50.57±9.63 | 0.384 |
| Males | 270 (32.9) | 200 (31.8) | 70 (36.6) | 0.211 |
| Smoking (n, %) | 207 (25.2) | 152 (24.2) | 55 (28.8) | 0.197 |
| Alcohol (n, %) | 205 (25.0) | 143 (17.4) | 62 (7.6) | 0.007 |
| BMI (kg/m2) | 22.96±3.12 | 22.81±3.19 | 23.43±2.85 | 0.016 |
| WC (cm) | 77.62±9.05 | 76.84±9.09 | 80.22±8.43 | <0.001 |
| Fasting plasma glucose (mM) | 5.01±0.75 | 4.99±0.77 | 5.09±0.69 | 0.107 |
| LDL-C (mM) | 3.24±1.23 | 2.71±0.75 | 5.00±0.78 | <0.001 |
BMI, body–mass index; LDL-C, low-density lipoprotein-cholesterol; WC, waist circumferences.
Genotype distribution and allele frequencies of 10 SNPs in PPARα/δ/γ genes, as well as the association between the genotypes and the level of LDL-C
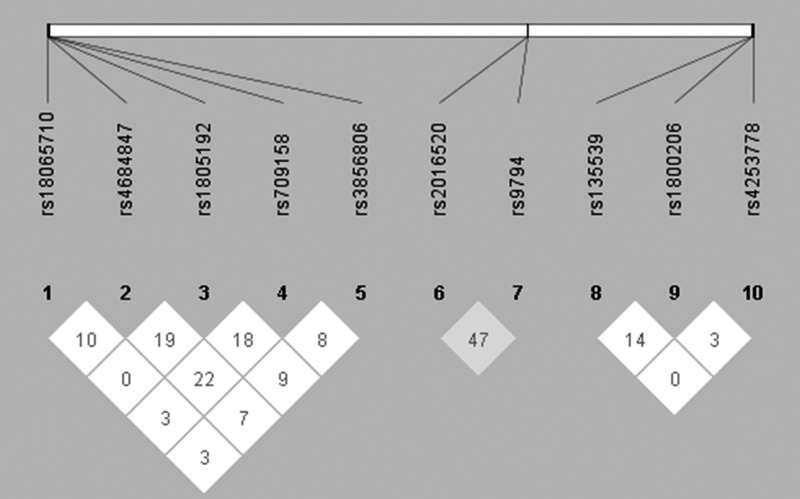
All the SNPs included in our study were in HWE (p>0.05). Linkage analysis showed that there was no significant LD between these polymorphisms (D′<0.75, Fig. 1). Genotypes and allele frequencies of the SNPs in PPAR genes are presented in Table 3. The genotype of variants in rs1800206, rs1805192, rs709158, and rs3856806 was associated with a higher LDL-C level (p<0.05), while the genotype of variants in rs10865710 was associated with lower LDL-C (mean difference was −0.21 mM; 95% confidence interval [CI] −0.37 to −0.04 mM; p=0.013). Compared with the wild-type carriers of rs9794 (CC), LDL-C was marginally significantly higher in individuals with mutants (CG+GG, mean difference was 0.16; 95% CI −0.01 to 0.33 mM; p=0.066). After adjustment for alcohol, BMI, and WC, the significant associations between these SNPs and LDL-C levels were not modified (all p<0.05). The other SNPs (rs135539, rs4253778, rs2016520, and rs4684847) did not show significant association with LDL-C (p<0.05).
FIG. 1.
Pairwise linkage disequilibrium between 10 single-nucleotide polymorphisms (SNPs) of peroxisome proliferator-activated receptor (PPAR) genes as measured by D′ according to the Haploview.
Table 3.
Genotype and Allele Frequencies of 10 Single-Nucleotide Polymorphisms in Peroxisome Proliferator-Activated Receptor Genes as well as the Association with the Level of Low-Density Lipoprotein-Cholesterol
| SNPs | Alleles and genotypes | Frequencies, n (%) | Mean difference (95% CI)a | p-Valuea | Mean difference (95% CI)b | p-Valueb |
|---|---|---|---|---|---|---|
| rs135539 | AA | 484 (59) | 0 | 0.29 | 0 | 0.12 |
| AC+CC | 336 (41) | 0.09 (−0.08 to 0.26) | 0.13 (−0.03 to 0.30) | |||
| A | 1242 (75.7) | |||||
| C | 398 (24.3) | |||||
| rs1800206 | LL | 622 (75.9) | 0 | <0.0001 | 0 | <0.0001 |
| LV+VV | 198 (24.1) | 1.20 (1.02 to 1.38) | 1.16 (0.99 to 1.34) | |||
| L | 1435 (87.5) | |||||
| V | 205 (12.5) | |||||
| rs4253778 | GG | 615 (75) | 0 | 0.13 | 0 | 0.11 |
| GC+CC | 205 (25) | −0.15 (−0.35 to 0.04) | −0.15 (−0.34 to 0.03) | |||
| G | 1404 (85.6) | |||||
| C | 236 (14.4) | |||||
| rs2016520 | TT | 388 (47.3) | 0 | 0.91 | 0 | 0.94 |
| TC+CC | 432 (52.7) | −0.01 (−0.18 to 0.16) | 0.01 (−0.16 to 0.17) | |||
| T | 1142 (69.6) | |||||
| C | 498 (30.4) | |||||
| rs9794 | CC | 498 (60.7) | 0 | 0.066 | 0 | 0.018 |
| CG+GG | 322 (39.3) | 0.16 (−0.01 to 0.33) | 0.20 (0.03 to 0.37) | |||
| C | 1278 (77.9) | |||||
| G | 362 (22.1) | |||||
| rs10865710 | CC | 367 (44.8) | 0 | 0.03 | 0 | 0.013 |
| CG+GG | 453 (55.2) | −0.19 (−0.36 to −0.02) | −0.21 (−0.37 to −0.04) | |||
| C | 1099 (67) | |||||
| G | 541 (33) | |||||
| rs1805192 | PP | 459 (56) | 0 | <0.0001 | 0 | 0.0002 |
| PA+AA | 361 (44) | 0.39 (0.22 to 0.56) | 0.32 (0.15 to 0.48) | |||
| P | 1206 (73.5) | |||||
| A | 434 (26.5) | |||||
| rs4684847 | CC | 519 (63.3) | 0 | 0.81 | 0 | 0.78 |
| CT+TT | 301 (36.7) | 0.02 (−0.15 to 0.20) | 0.02 (−0.14 to 0.19) | |||
| C | 1295 (79) | |||||
| T | 345 (21) | |||||
| rs709158 | AA | 410 (50) | 0 | 0.0009 | 0 | 0.0025 |
| AG+GG | 410 (50) | 0.28 (0.12 to 0.45) | 0.25 (0.09 to 0.41) | |||
| A | 1154 (70.4) | |||||
| G | 486 (29.6) | |||||
| rs3856806 | CC | 418 (51) | 0 | <0.0001 | 0 | 0.0001 |
| CT+TT | 402 (49) | 0.37 (0.21 to 0.54) | 0.32 (0.16 to 0.48) | |||
| C | 1162 (70.9) | |||||
| T | 478 (29.1) |
Without adjustment.
Adjusted for alcohol, BMI, and WC.
CI, confidence interval.
Gene–gene interactions among PPARα/δ/γ polymorphisms for LDL-C
The results of GMDR (from 2- to 10-locus models) are exhibited in Table 4. When quantitative traits (the level of LDL-C) were chosen as the outcome, significant gene–gene interactions could be observed in the 2- to 8-locus models (p<0.05). Among these sets of models, we pick the 3-locus model (including rs1800206, rs1805192, and rs709158) as the best model, which has the highest testing accuracy and/or maximum cross-validation consistency (cross-validation consistency=9/10, testing accuracy=0.7388, p=0.107). When the dichotomous trait (whether the individual had higher LDL-C) was selected as the outcome, similar results were obtained and the best model changed to the 2-locus model (including rs1800206 and rs1805192, cross-validation consistency=10/10, testing accuracy=0.7042, p=0.0107).
Table 4.
Multilocus Interaction Model for Low-Density Lipoprotein-Cholesterol by Generalized Multifactor Dimensionality Reduction Method
| No. of loci | Best combination | Testing accuracy | Cross-validation consistency | p-Value |
|---|---|---|---|---|
| Model Aa | ||||
| 2 | rs1800206 rs1805192 | 0.7257 | 9/10 | 10 (0.0010) |
| 3 | rs1800206 rs1805192 rs709158 | 0.7388 | 9/10 | 10 (0.0010) |
| 4 | rs1800206 rs1805192 rs709158 rs3856806 | 0.7355 | 6/10 | 10 (0.0010) |
| 5 | rs1800206 rs2016520 rs9794 rs1805192 rs4684847 | 0.6791 | 4/10 | 10 (0.0010) |
| 6 | rs1800206 rs9794 rs10865710 rs1805192 rs709158 rs3856806 | 0.6478 | 3/10 | 10 (0.0010) |
| 7 | rs1800206 rs2016520 rs9794 rs10865710 rs1805192 rs709158 rs3856806 | 0.6231 | 4/10 | 10 (0.0010) |
| 8 | rs135539 rs1800206 rs2016520 rs9794 rs10865710 rs1805192 rs709158 rs3856806 | 0.6084 | 6/10 | 9 (0.0107) |
| 9 | rs135539 rs1800206 rs2016520 rs9794 rs10865710 rs1805192 rs4684847 rs709158 rs3856806 | 0.5707 | 6/10 | 8 (0.0547) |
| 10 | rs135539 rs1800206 rs4253778 rs2016520 rs9794 rs10865710 rs1805192 rs4684847 rs709158 rs3856806 | 0.5614 | 10/10 | 6 (0.3770) |
| Model Bb | ||||
| 2 | rs1800206 rs1805192 | 0.7042 | 10/10 | 10 (0.0010) |
| 3 | rs1800206 rs1805192 rs4684847 | 0.6938 | 5/10 | 10 (0.0010) |
| 4 | rs1800206 rs1805192 rs4684847 rs3856806 | 0.6846 | 4/10 | 10 (0.0010) |
| 5 | rs1800206 rs9794 rs1805192 rs709158 rs3856806 | 0.6344 | 6/10 | 10 (0.0010) |
| 6 | rs135539 rs1800206 rs9794 rs10865710 rs1805192 rs3856806 | 0.5868 | 5/10 | 10 (0.0010) |
| 7 | rs1800206 rs2016520 rs9794 rs10865710 rs1805192 rs709158 rs3856806 | 0.6252 | 10/10 | 10 (0.0010) |
| 8 | rs135539 rs1800206 rs2016520 rs9794 rs10865710 rs1805192 rs709158 rs3856806 | 0.5763 | 6/10 | 9 (0.0107) |
| 9 | rs135539 rs1800206 rs2016520 rs9794 rs10865710 rs1805192 rs4684847 rs709158 rs3856806 | 0.5244 | 8/10 | 8 (0.0547) |
| 10 | rs135539 rs1800206 rs4253778 rs2016520 rs9794 rs10865710 rs1805192 rs4684847 rs709158 rs3856806 | 0.5683 | 10/10 | 8 (0.0547) |
Adjusted for alcohol, BMI, and WC.
LDL-C was represented as a continuous variable.
LDL-C was represented as a dichotomous variable (the cutoff point is 4.14 mM).
Discussion
In the present study, we aimed to investigate the association between PPARα/δ/γ gene polymorphisms and LDL-C in a Chinese Han population and discovered that several SNPs (including rs1800206 in PPARα, rs9794 in PPARδ, and rs1805192, rs709158, and rs3856806 in PPARγ) were associated with higher LDL-C after adjustment for alcohol, BMI, and WC (p<0.05), whereas rs10865710 in PPARγ was associated with lower LDL-C (p=0.013), which has been reported by Barbosa et al. (2012). Previous studies have also discovered that mutants at rs1800206, rs9794, rs1805192, rs709158, and rs3856806 are associated with elevated LDL-C, total cholesterol (TC), or dyslipidemia (Tanaka et al., 2007; Lu et al., 2010; Gu et al., 2014). Some of these SNPs were located in exons, while others located in introns. It is intriguing that mutants in introns could also be able to affect the trait, such as the intronic SNPs, rs709158 and rs10865710, included in our study. So far, increasing evidence has proved that introns can affect the expression level of their host genes in many ways (Chorev and Carmel, 2012). The human genome contains an average of 8.4 introns per gene (Rodriguez-Trelles et al., 2006), most of whose lengths are much longer than exons. Moreover, it is acknowledged that a great deal of cis regulatory elements were also contained in these sequences, which might render some effects on the transcriptional initiation and splicing processes, and then affect the expression levels of some genes so as to trigger some diseases or other phenotypes. As Clark et al. (1993) demonstrated, indeed, some introns are efficient in boosting expression levels and exist to guarantee high expression. Although there was no evidence that directly reported the association between rs709158 and the LDL-C level, Lu et al. have reported that the SNP was associated with TC, which suggested a possible relationship between this SNP and LDL-C or HDL-C. Besides, another intronic SNP, rs10865710, included in this study has been reported to be associated with the LDL-C level that supported our findings (Rudkowska et al., 2013). What is presented above may give a provable view that mutants even in introns of PPAR genes could also affect the LDL-C level of humans.
The results of GMDR revealed that when a continuous variable was used as the phenotype, the 3-locus model holds the highest testing accuracy and maximum cross-validation consistency among all the specific models, which indicated that there was a significant gene–gene interaction among the SNPs (rs1800206 in PPARα, rs1805192, and rs709158 in PPARγ), while the best model turned to the 2-locus model, including rs1800206 and rs1805192, when a dichotomous variable was selected. Although similar results were obtained when using different types of traits, details differed between the consequences. A few reasons could be available to explain the phenomenon: on the one hand, some information will be lost after translating quantitative traits into the binary phenotypes (Yi and Xu, 1999), on the other hand, simply transforming a quantitative trait into a binary one can usually result in loss of power if an inappropriate threshold is used (Dawn Teare and Barrett, 2005), which has been confirmed in our results that the testing accuracies within different locus models are generally lower in using the binary trait than using the quantitative.
Besides the best model, there are several models with more loci that also showed significance of genetic interactions, such as the 8-locus model (including rs135539, rs1800206, rs2016520, rs9794, rs10865710, rs1805192, rs709158, and rs3856806), which indicated that there is a potential gene–gene interaction among these polymorphisms. As existing evidence has proved that dyslipidemia is a complex disease that could be influenced by multiple genetic factors, the phenotype LDL-C level might be under the impact of multiple and possibly interacting genes as well, among which it only owns a rather small effect on the trait (Hoh and Ott, 2003); however, the actions of SNPs might not have been observed when the single-locus approach was performed because of the epistasis existing among PPAR genes, which could render the impact of a few SNPs with small effects overshadowed by the neighboring loci (Phillips, 1998).
As the so-called lipid and insulin sensors, PPARs do cast a great impact on the homeostasis of lipids and glucose in humans (Grygiel-Gorniak, 2014). Hansen et al. (2011) and Olson et al. (2012) have already found that a dual PPARα/γ agonist, aleglitazar, and GW501516, a PPARδ agonist, can significantly decrease LDL-C levels in humans, which suggested a potential LDL-lowering effect triggered by activating different isoforms of PPARs. The mechanism of increased removal of LDL particles could be possibly attributed to the formation of LDL with a higher affinity for the LDL receptor, followed by a more rapid catabolic rate when PPARα agonist, such as fibrate, was used for treatment (Staels et al., 1998), whereas a possible mechanism for PPARδ could be due to the reduction of the Niemann-Pick C1-like 1 (NPC1L1) protein by PPARδ activation in the intestine as the inhibition of NPC1L1 would lead to reduced cholesterol absorption (Riserus et al., 2008). PPARγ is highly expressed in the adipose tissue and can be functioned as a regulator linked to lipid metabolism as well. Chui et al. (2005) have found that activation of PPARγ can stimulate the uptake of oxidized LDL (oxLDL) into the adipocyte, along with the cholesterol content and fatty acid. Additionally, PPARγ activation also promotes oxLDL uptake in the monocyte and macrophage where foam cells are initiated to induce plaque buildup or atherosclerosis (Tontonoz et al., 1998; Yu et al., 2013). What is presented in this study has suggested that individuals with specific mutants of the SNPs mentioned above might be less benefited from the LDL-lowering therapy of such agonists.
The limitations of the present study also need to be considered. First, as the interactions among PPAR gene polymorphisms are rather a complex issue, the exact regulatory mechanism of PPAR gene polymorphisms on LDL-C metabolism also needs to be investigated for further understanding the questions behind the observed association. Second, independent replication in other studies with a larger sample size is required to detect whether the interaction could be found in other races.
In conclusion, we discovered that a few PPARα/δ/γ gene polymorphisms were associated with the level of LDL-C in the Chinese Han population. Besides, interactions among these polymorphisms were also found to be having combined effects on the LDL-C level. Our findings may be helpful to illustrate the genetic basis of PPARs affecting the level of LDL as well as some complex diseases, including cardiovascular diseases and atherosclerosis, caused by elevated LDL particles. Since only 10 SNPs were investigated, other SNPs that may be involved with complex gene–gene interactions remain worthy of further exploration.
Acknowledgments
This study was supported by the grants from the Scientific Research Fund of the National Ministry of Health, Republic of China (WKJ 2004-2-014), and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Author Disclosure Statement
No competing financial interests exist.
References
- AlSaleh A, O'Dell SD, Frost GS, et al. (2011) Interaction of PPARG Pro12Ala with dietary fat influences plasma lipids in subjects at cardiometabolic risk. J Lipid Res 52:2298–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson TJ, Gregoire J, Hegele RA, et al. (2013) 2012 update of the Canadian Cardiovascular Society guidelines for the diagnosis and treatment of dyslipidemia for the prevention of cardiovascular disease in the adult. Can J Cardiol 29:151–167 [DOI] [PubMed] [Google Scholar]
- Azhar S. (2010) Peroxisome proliferator-activated receptors, metabolic syndrome and cardiovascular disease. Future Cardiol 6:657–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badimon L, Vilahur G. (2012) LDL-cholesterol versus HDL-cholesterol in the atherosclerotic plaque: inflammatory resolution versus thrombotic chaos. Ann N Y Acad Sci 1254:18–32 [DOI] [PubMed] [Google Scholar]
- Barbosa EJL, Glad CAM, Nilsson AG, et al. (2012) Genotypes associated with lipid metabolism contribute to differences in serum lipid profile of GH-deficient adults before and after GH replacement therapy. Eur J Endocrinol 167:353–362 [DOI] [PubMed] [Google Scholar]
- Catapano AL, Reiner Z, De Backer G, et al. (2011) ESC/EAS Guidelines for the management of dyslipidaemias The Task Force for the management of dyslipidaemias of the European Society of Cardiology (ESC) and the European Atherosclerosis Society (EAS). Atherosclerosis 217:3–46 [DOI] [PubMed] [Google Scholar]
- Chorev M, Carmel L. (2012) The function of introns. Front Genet 3:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chui PC, Guan HP, Lehrke M, et al. (2005) PPARgamma regulates adipocyte cholesterol metabolism via oxidized LDL receptor 1. J Clin Invest 115:2244–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AJ, Archibald AL, McClenaghan M, et al. (1993) Enhancing the efficiency of transgene expression. Philos Trans R Soc Lond B Biol Sci 339:225–232 [DOI] [PubMed] [Google Scholar]
- Cromwell WC, Otvos JD, Keyes MJ, et al. (2007) LDL particle number and risk of future cardiovascular disease in the Framingham Offspring Study-Implications for LDL management. J Clin Lipidol 1:583–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawn Teare M, Barrett JH. (2005) Genetic linkage studies. Lancet 366:1036–1044 [DOI] [PubMed] [Google Scholar]
- Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (2001) Executive Summary of The Third Report of The National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III). JAMA 285:2486–2497 [DOI] [PubMed] [Google Scholar]
- Grygiel-Gorniak B. (2014) Peroxisome proliferator-activated receptors and their ligands: nutritional and clinical implications—a review. Nutr J 13:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu SJ, Guo ZR, Zhou ZY, et al. (2014) PPAR alpha and PPAR gamma polymorphisms as risk factors for dyslipidemia in a Chinese Han population. Lipids Health Dis 13:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen BC, Tigno XT, Benardeau A, et al. (2011) Effects of aleglitazar, a balanced dual peroxisome proliferator-activated receptor alpha/gamma agonist on glycemic and lipid parameters in a primate model of the metabolic syndrome. Cardiovasc Diabetol 10:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoh J, Ott J. (2003) Mathematical multi-locus approaches to localizing complex human trait genes. Nat Rev Genet 4:701–709 [DOI] [PubMed] [Google Scholar]
- Hu XS, Guo ZR, Zhou H, et al. (2006) [Study on the prevalence of metabolic syndrome among 35–74 year-olds in Jiangsu province]. Zhonghua Liu Xing Bing Xue Za Zhi 27:751–756 [PubMed] [Google Scholar]
- Issemann I, Green S. (1990) Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature 347:645–650 [DOI] [PubMed] [Google Scholar]
- Joint Committee for Developing Chinese guidelines on Prevention and Treatment of Dyslipidemia in Adults (2007) [Chinese guidelines on prevention and treatment of dyslipidemia in adults]. Zhonghua Xin Xue Guan Bing Za Zhi 35:390–419 [PubMed] [Google Scholar]
- Krauss RM. (2005) Dietary and genetic probes of atherogenic dyslipidemia. Arterioscler Thromb Vasc Biol 25:2265–2272 [DOI] [PubMed] [Google Scholar]
- Lou XY, Chen GB, Yan L, et al. (2007) A generalized combinatorial approach for detecting gene-by-gene and gene-by-environment interactions with application to nicotine dependence. Am J Hum Genet 80:1125–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YC, Feskens EJM, Boer JMA, et al. (2010) Exploring genetic determinants of plasma total cholesterol levels and their predictive value in a longitudinal study. Atherosclerosis 213:200–205 [DOI] [PubMed] [Google Scholar]
- Mazzotti DR, Singulane CC, Ota VK, et al. (2011) PPAR alpha polymorphisms as risk factors for dyslipidemia in a Brazilian population. Mol Genet Metab 102:189–193 [DOI] [PubMed] [Google Scholar]
- Miao L, Yin RX, Wu DF, et al. (2010) Peroxisome proliferator-activated receptor delta+294T>C polymorphism and serum lipid levels in the Guangxi Bai Ku Yao and Han populations. Lipids in Health and Disease 9:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EJ, Pearce GL, Jones NP, et al. (2012) Lipid effects of peroxisome proliferator-activated receptor-delta agonist GW501516 in subjects with low high-density lipoprotein cholesterol: characteristics of metabolic syndrome. Arterioscler Thromb Vasc Biol 32:2289–2294 [DOI] [PubMed] [Google Scholar]
- Phillips PC. (1998) The language of gene interaction. Genetics 149:1167–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riserus U, Sprecher D, Johnson T, et al. (2008) Activation of peroxisome proliferator-activated receptor (PPAR)delta promotes reversal of multiple metabolic abnormalities, reduces oxidative stress, and increases fatty acid oxidation in moderately obese men. Diabetes 57:332–339 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Trelles F, Tarrio R, Ayala FJ. (2006) Origins and evolution of spliceosomal introns. Annu Rev Genet 40:47–76 [DOI] [PubMed] [Google Scholar]
- Rudkowska I, Dewailly E, Hegele RA, et al. (2013) Gene-diet interactions on plasma lipid levels in the Inuit population. Br J Nutr 109:953–961 [DOI] [PubMed] [Google Scholar]
- Sole X, Guino E, Valls J, et al. (2006) SNPStats: a web tool for the analysis of association studies. Bioinformatics 22:1928–1929 [DOI] [PubMed] [Google Scholar]
- Staels B, Dallongeville J, Auwerx J, et al. (1998) Mechanism of action of fibrates on lipid and lipoprotein metabolism. Circulation 98:2088–2093 [DOI] [PubMed] [Google Scholar]
- Tanaka T, Ordovas JM, Delgado-Lista J, et al. (2007) Peroxisome proliferator-activated receptor alpha polymorphisms and postprandial lipemia in healthy men. J Lipid Res 48:1402–1408 [DOI] [PubMed] [Google Scholar]
- Tenenbaum A, Fisman EZ. (2012) Balanced pan-PPAR activator bezafibrate in combination with statin: comprehensive lipids control and diabetes prevention? Cardiovasc Diabetol 11:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tontonoz P, Nagy L, Alvarez JG, et al. (1998) PPARgamma promotes monocyte/macrophage differentiation and uptake of oxidized LDL. Cell 93:241–252 [DOI] [PubMed] [Google Scholar]
- Yi N, Xu S. (1999) Mapping quantitative trait loci for complex binary traits in outbred populations. Heredity (Edinb) 82(Pt 6):668–676 [DOI] [PubMed] [Google Scholar]
- Yu XH, Fu YC, Zhang DW, et al. (2013) Foam cells in atherosclerosis. Clin Chim Acta 424:245–252 [DOI] [PubMed] [Google Scholar]