Abstract
Rotavirus is the major etiologic factor of severe diarrheal disease. Natural infection provides protection against subsequent rotavirus infection and diarrhea. This research presents a new vaccine designed based on computational models. In this study, three types of epitopes are considered—linear, conformational, and combinational—in a proposed model protein. Several studies on rotavirus vaccines have shown that VP6 and VP4 proteins are good candidates for vaccine production. In the present study, a fusion protein was designed as a new generation of rotavirus vaccines by bioinformatics analyses. This model-based study using ABCpred, BCPREDS, Bcepred, and Ellipro web servers showed that the peptide presented in this article has the necessary properties to act as a vaccine. Prediction of linear B-cell epitopes of peptides is helpful to investigate whether these peptides are able to activate humoral immunity.
Introduction
Diarrhea is the second leading cause of death around the world for children younger than 5 years of age (6). Rotavirus is the major etiologic factor of severe diarrheal disease, often associated with fever, nausea, and dehydration in infants and young children (13,34). It has been estimated that rotaviral diarrhea may be responsible for 527,000 child deaths every year, the majority of which (>85%) occur in developing countries (35). Rotavirus is a double-stranded RNA (dsRNA) virus in the Reoviridae family. The retrovirus genome consists of 11 segments of dsRNA and encodes six structural proteins (VP1–4, VP6, VP7) and six nonstructural proteins (NSP1–NSP6). The capsid contains three layers of protein (core, inner, and outer capsid) (12). For the most part, each gene segment codes for a single protein. During coinfection with more than one rotavirus strain in a patient, gene segments from the parental viruses may reassert, which may be a mechanism of viral genome diversity (38). The word rotavirus is derived from the Latin word rota, meaning wheel, which is characteristic of the morphology of this virus (16). Diarrhea caused by the virus usually occurs in children aged 6–36 months.
Retroviruses have three antigenic indexes, based on group, subgroup, and serotype. The group is determined by the VP6 protein and is divided into seven groups (A–G). Groups A, B, and C have been found in both humans and animals, and groups D, E, and F have been found only in animals (7). Rotaviral group A is a common cause of rotavirus diarrhea in humans, and it is the first-choice candidate for vaccine development. Serotype is determined by the VP4 and VP7 proteins. These two proteins are the main target for neutralizing antibodies. Rotaviruses are divided to G genotypes (glycoprotein VP7) and P (protease sensitive, VP4) based on the antigenic structure of the outer capsule; there are 24 G genotypes and 33 P genotypes. Of the genotypes described to date, 12 G and 15 P genotypes are known to be involved in human infections (9,16,32,38). Genotypes P [8] G1, p [4] G2, P [8] G3, and P [8] G4 are common rotavirus strains in human infections in many countries (21,37). Despite high diversity among the subgroups, VP6 protein is stringently conserved and very immunogenic among all group A rotaviruses. However, capsid proteins VP7 and VP4 have very high diversity among strains (17,23).
Studies indicate that natural infection provides protection against subsequent rotavirus infection and diarrhea (36). On August 31, 1998, the first rotavirus vaccine (RotaShield) was licensed by Witt et al. from the American Food and Drug Administration, and finally entered the market. The tetravalent rotavirus vaccine (RotaShield) based on attenuated virus was produced and was given orally three times a month to 2-, 4-, and 6-month-old infants. However, because of the correlation with intussusception and other intestinal diseases, as well as poor affectivity in children with poor health, RotaShield was taken off the market (43). RotaTeq and Rotarix are two live oral vaccines already available worldwide. They have been licensed in more than 85 countries and are part of the routine vaccination programs in many countries, including America, Brazil, Panama, Venezuela, Australia, and Belgium (12,22). RotaTeq is a live attenuated pentavalent vaccine isolated from five distinct human-bovine reassortant virus strains, which together express the human rotavirus G1, G2, G3, G4, and P antigens (11,52). In July 2004, a new rotavirus vaccine (51) composed of the live rotavirus strain RIX4414 was licensed in Mexico by GlaxoSmithKline. Rotarix is a monovalent live attenuated vaccine with G1P genotype (51). Yet, there is no effective vaccine program in many developing countries. Global vaccination could prevent 43% of the 527,000 rotaviral deaths in children. The World Health Organization (14,44,49) has included rotavirus vaccination in the global vaccination program (39). This study proposes a new generation vaccine against rotavirus designed using in silico analyses. In this study, three types of epitopes are considered—linear, conformational, and combinational—in a synthetic protein.
Materials and Methods
Based on epidemiological studies and previous findings (9,16,32,38), serotype A rotaviruses were selected as a good model for antigenic epitope prediction study. The sequence of VP4 and VP6 proteins (GenBank accession number GQ225829.1 and GQ496209.1, respectively) belonging to rotavirus common strains were retrieved from the NCBI database (www.ncbi.nlm.nih.gov/). Using homology analyses by NCBI protein–protein BLAST server, conserved motifs of these proteins were selected for construction of a model peptide:
NH3+-ANYVETARNTIDYFVDNVCMDEMVRESQRNGIAPNIFPYSKTSLWKEMQYNRDIIIRFKIDFKTLKNLNDNACTEYINNGLPPIQNTRNVVP-COOH
Physical and chemical properties of the peptide were analyzed with ProtParam (www.expasy.ch/tools/protparam.html). The secondary structure was predicted by phyre2 (www.sbg.bio.ic.ac.uk/phyre2/) (24), PHD (http: //npsa-pbil.Ibcp. Fr/cgibin/npsa_automat. Pal? age=/NPSA/npsa_phd.html) (45), PSIPRED (http://bioinf.cs.ucl. Ac.uk/psipred) (31), and Jpred (www.compbio.dundee.ac.uk/www-jpred/) (10). The tertiary structure of the model peptide was predicted by phyre2 (www.sbg.bio.ic.ac.uk/phyre2/) and m4t servers (18). PDB structure was studied topologically using Hex 6.3 software.
Prediction of B-cell epitopes
In order to identify linear epitopes, the primary sequence of the peptide was submitted to ABCpred (www.imtech.res.in/Raghava/abcpred/) (46), BCPREDS (http://ailab.cs.iastate.edu/ bcpreds/) (15), Bcepred (www.imtech.res.in/raghava/bcepred/) (48), and Ellipro (http://tools.immuneepitope.org/tools/ElliPro/iedb_input) (40).
Prediction of conformational B-cell epitopes:
Ellipro (http://tools.immuneepitope.org/tools/ElliPro/iedb_input) (40) and Discotope 1.2 (www.cbs.dtu.dk/services/DiscoTope/( (26) were used to predict conformation of diagnosable epitopes for B lymphocytes.
Prediction of T-cell epitopes
To predict T-cell-specific epitopes, primary sequence of the model antigenic peptid was submitted to CTLPred) www.imtech.res.in/raghava/ctlpred/) (5) NetCTL 1.2 (www.cbs.dtu.dk/services/NetCTL/) (27), EpiJen (www.ddg-pharmfac.net/epijen/) (14), and TAPpred (www.imtech.res.in/raghava/tappred/) (4). nHLAPred (www.imtech.res.in/raghava/nhlapred/) (1) and ProPed I (www.imtech.res.in/raghava/propred1/) (49) were used for a comparative analysis of outputs, which can identify MHC binding regions by various matrices. HLA-DR4Pred (www.imtech.res.in/raghava/hladr4pred) (3) was used for investigation and identification of T-helper epitopes in the peptide.
Results
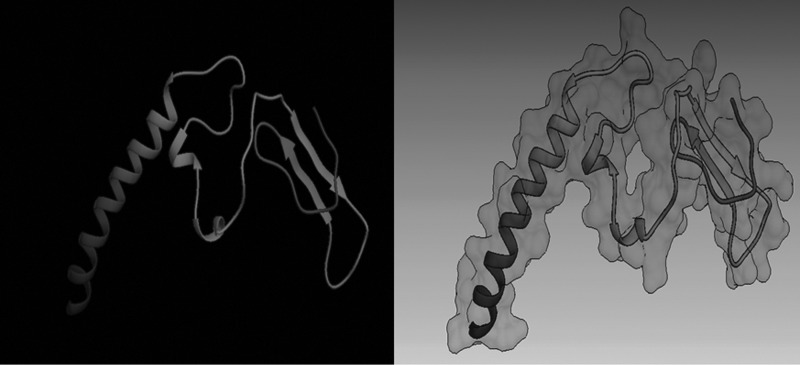
Epitopic motifs of a model were predicted. This peptide comprised 95 amino acid residues with 1,169.6 kD molecular weight. Based on the algorithm ProtParam, the peptide is stable and hydrophobic (GRAVY: −0.529) and its PI (isoelectric pH) is 5.30. As shown in Figure 1, the 2D structure of this peptide included 28.4% α-helix, 10.5% β-sheet, 4.2% turn, and 66.5% coil.
FIG. 1.
Tertiary structure of fusion peptide. The results were prepared by Pyre2 and displayed with Hex. This peptide includes 28.4% α-helix, 10.5% β-sheet, 4.2% turn, and 66.5% coil.
In addition to linear epitopes (Table 1), conformational B-cell epitopes were predicted with Ellipro, as shown in Table 2 and Figure 2. Analyses with Discotope 1.2 r showed more details of tertiary structure (Fig. 3).
Table 1.
B-Cell Linear Motif Prediction of Model Peptide Presented in This Study
| Motif | Sequence | Start position | Length | GRAVY score | Mw (Da) | pI | Software |
|---|---|---|---|---|---|---|---|
| 1* | DEMVRESQRNG | 24 | 11 | −1.964 | 1,320.4 | 4.68 | 1,2,3,4 |
| 2 | IDYFVDFVDNCVC | 11 | 13 | 0.95 | 1,551.7 | 3.42 | 1,2,3 |
| 3* | KNLNDNACT | 69 | 9 | −1.167 | 992.0 | 5.83 | 1,2,3 |
| 4* | NDNACTEY | 72 | 8 | −1.462 | 928.9 | 3.67 | 1,2 |
| 5* | PPIQNTRNVVP | 85 | 11 | −0.691 | 1,234.4 | 10.18 | 1,2,3,4 |
| 6* | FPYSKTSLWKEMQ | 40 | 13 | −0.954 | 1,644.9 | 8.50 | 1,2,3 |
| 7* | NYVETARNTID | 2 | 11 | −0.973 | 1,295.3 | 4.37 | 1,4 |
| 8 | VINNGLPPI | 79 | 9 | 0.711 | 936.1 | 5.49 | 2 |
| 9* | DIIIRRFKIDFKTLK | 56 | 15 | −0.067 | 1,906.3 | 10.28 | 1,2,3 |
| 10* | QRNGIAPNIFPYSKTS | 31 | 16 | −0.781 | 1,793.0 | 9.99 | 2 |
| 11* | TSLWKEMQYNRDIIIR | 46 | 16 | −0.713 | 2,066.4 | 8.28 | 2 |
| 12* | IDFKTLKNLNDNACTE | 63 | 16 | −0.688 | 1,839.0 | 4.56 | 1,2 |
Antigenic motifs were confirmed by different programs; hydrophilic motifs are marked with an asterisk. Servers used for prediction are shown as number described as follows: Bcepred, 1; ABCpred, 2; BCPred, 3; and Ellipro, 4. For each peptide, sequence, position, molecular weight, isoelecteric pH, and GRAVY score are indicated. The GRAVY index indicated the hydrophobicity of the peptide and was calculated as the sum of the hydropathy values of the composing amino acids divided by the number of residues in the sequence. Peptides with a negative GRAVY index are hydrophilic, whereas peptides with a positive GRAVY index are hydrophobic.
GRAVY, grand average of hydropathy.
Table 2.
Conformational B-Cell Epitopes Were Predicted by the ElliPro Server
| Number | Residues | Length | Score |
|---|---|---|---|
| 1 | _:E29, _:S30, _:Q31, _:R32, _:N33, _:G34 | 6 | 0.809 |
| 2 | _:K62, _:I63, _:D64, _:F65, _:K66, _:T67, _:L68 | 7 | 0.765 |
| 3 | _:A1, _:Y3, _:V4, _:E5, _:T6, _:A7, _:R8, _:N9, _:T10, _:I11, _:D12, _:F14, _:V15, _:D16 | 14 | 0.762 |
| 4 | _:M26, _:V27, _:R28, _:S46 | 4 | 0.616 |
| 5 | _:K69, _:N70, _:L71, _:N72, _:D73, _:A75, _:P86, _:I87, _:Q88, _:N89, _:R91, _:V93, _:V94, _:P95 | 14 | 0.598 |
ElliPro associates each predicted epitope with a score, defined as a PI (Protrusion Index) value averaged over epitope residues. For each residue, a PI value is defined as percentage of the protein atoms enclosed in the ellipsoid, which approximates the protein surface, at which the residue first becomes lying outside the ellipsoid; for example, all residues that are outside the 90% ellipsoid will have PI=9 (or 0.9 in ElliPro).
FIG. 2.

Tertiary structure and position of conformational B-cell epitopes. These structures were predicted by ElliPro as shown in Table 2. (A) Motif 1 (_:E29, _:S30, _:Q31, _:R32, _:N33, _:G34) with a score of 0.809. (B) Motif 2 (_:K62, _:I63, _:D64, _:F65, _:K66, _:T67, _:L68) with a score of 0.765. (C) Motif 3 (_:A1, _:Y3, _:V4, _:E5, _:T6, _:A7, _:R8, _:N9, _:T10, _:I11, _:D12, _:F14, _:V15, _:D16) with a score of 0.762. (D) Motif 4 (_:M26, _:V27, _:R28, _:S46) with a score of 0.616. (E) Motif 5 (_:K69, _:N70, _:L71, _:N72, _:D73, _:A75, _:P86, _:I87, _:Q88, _:N89, _:R91, _:V93, _:V94, _:P95) with a score of 0.598. Scores were calculated as described in the legend to Table 2.
FIG. 3.

Epitopic residues colored by binary code. Threshold=1.900. Light, predicted epitope residues. Residues colored by DiscoTope score >1.9 (0.17 sensitivity; 0.95 specificity). Dark gray, high score; darker gray, low score. DiscoTope predicts discontinuous B-cell epitopes from protein three-dimensional structures. The method utilizes calculation of surface accessibility (estimated in terms of contact numbers) and a novel epitope propensity amino acid score. The final scores are calculated by combining the propensity scores of residues in spatial proximity and the contact numbers. A specificity of 0.95 means that 5% of the nonepitope residues were predicted as part of epitopes. A sensitivity of 0.17 means that 17% of the epitope residues were predicted as part of nonepitopes.
Motifs E29, _:S30, _:Q31, _:R32, _:N33, _:G34 and K62, _:I63, _:D64, _:F65, _:K66, _:T67, _: L68 were identified as both linear and conformational B cell epitopes by the different programs.
Different programs were used to predict T-cell epitopes in the proposed peptide (Table 3). The various programs detected identical motifs.
Table 3.
T-Cell Motifs Were Predicted by Different Programs
| Motif | Sequence | Start position | Length | GRAVY score | Mw (Da) | pI | Software |
|---|---|---|---|---|---|---|---|
| 1* | YNETARNTIDY | 4 | 11 | −1.473 | 1,359.4 | 4.37 | 1,2 |
| 2 | NTIDYFVDFV | 8 | 10 | 0.600 | 1,232.3 | 3.56 | 1,2,4 |
| 3 | YFVDFVDNVC | 12 | 10 | 0.890 | 1,220.3 | 3.56 | 2,4 |
| 4* | QRNGIAPNIFPY | 30 | 12 | −0.525 | 1,389.5 | 8.75 | 1,2,3 |
| 5* | WKEMQYNRDI | 47 | 10 | −1.820 | 1,382.5 | 6.07 | 2,4 |
| 6* | RFKIDFKTL | 59 | 9 | −0.289 | 1,167.4 | 9.99 | 2,3 |
| 7* | TLKNLNDNA | 66 | 9 | −1.022 | 1,002.0 | 5.50 | 2,4 |
| 8* | CTEYINNGLPPI | 75 | 12 | −0.067 | 1,333.5 | 4.00 | 1,2 |
| 9* | GIPPIQNTRNVVPG | 85 | 14 | −0.279 | 1,461.6 | 9.75 | 2,4 |
Antigenic motifs were confirmed by different programs; hydrophilic motifs are marked with an asterisk. Programs used for prediction are shown as number described as follows: Bcepred, 1; ABCpred, 2, BCPred, 3; and Ellipro, 4. For each peptide, sequence, position, molecular weight, isoelecteric pH, and GRAVY score was indicated.
Furthermore, MHC-I binding motifs were predicted and similar results were found in both programs. Interestingly, this motif (IDFKTLKNL) is identified by seven MHC_I in both programs, but the exact molecules vary. Five types of HLA (HLA-B*3701 HLA-B40, HLA-B60, HLA-B61, HLA-Cw*0602) were shared between both programs. RFKIDFKTL was identified by HLA (HLA-B*0702, MHC-Kb, MHC-Kd, HLA-Cw*0401) in ProPed I rand HLA-G in nHLAPred. Ten T-helper epitopes were identified by HLA-DR4Pred. The results are shown in Table 4.
Table 4.
The Results of the HLA-DR4Pred Program
| Peptide | Sequence | Start position | Score | Prediction |
|---|---|---|---|---|
| 1 | MDEMVRESQ | 23 | 0.663 | Binder |
| 2 | DFVDNVCMD | 16 | 0.555 | Binder |
| 3 | IDYFVDFVD | 11 | 0.490 | Binder |
| 4 | FVDNVCMDE | 17 | 0.485 | Binder |
| 5 | TEYINNGLP | 77 | 0.470 | Binder |
| 6 | YFVDFVDNV | 13 | 0.442 | Binder |
| 7 | SKTSLWKEM | 43 | 0.432 | Binder |
| 8 | IDFKTLKNL | 63 | 0.418 | Binder |
| 9 | LNDNACTEY | 71 | 0.410 | Binder |
| 10 | VETARNTID | 4 | 0.395 | Binder |
The HLA-DR4Pred is a support vector machine (SVM) and artificial neural network (ANN)–based HLA-DRB1*0401 (MHC class II alleles) binding peptides prediction method. The accuracy of the SVM- and ANN-based methods is ∼86% and ∼78%, respectively. The performence of the methods was tested through five-set cross-validation.
Discussion
The fusion antigenic protein introduced here may be a starting point for designing effective rotaviral vaccines. Rotavirus infection results in high child mortality rates and is a significant economic burden in the developing world. Improvements in healthcare have not had a great impact on the incidence of rotavirus infection; vaccination is the best and most effective way to control this infection (39).
The role of humoral immunity against rotavirus has been demonstrated in multiple studies (33,50,53). Prediction of linear B-cell epitopes is helpful to investigate whether these peptides are able to activate humoral immunity. Bcpred (55), Bcepred (8), ABCpred (47), Ellipro (40,42,54), and Discotope 1.2 server (30) scored the peptide in this study high for humeral immunity.
NetCTL 1.2 (20), EpiJen (41), nHLAPred (28), CTLPred (44), TAPpred (29), and HLA-DR4Pred (25) confirmed the possibility of the peptide in this study stimulating cellular immunity as well.
One of the biggest complications in producing powerful vaccines against retroviruses is the variability of their genome. In developing countries, human rotavirus strains include different combinations of G/P types. This is due to rearrangement of different rotavirus strains, including animal-specific types, and this results in diversity in strains from area to area (58). This diversity results in variable efficacy of present vaccines in different countries (59). In this study, conserved regions of the rotavirus genome were considered as a vaccine target, as rapid genomic changes may have less impact on its effectiveness.
Another advantage of the model peptide presented in this research as a vaccine is the absence of motifs that may promote an autoimmune response. The current live vaccines have side effects such as bowel obstruction, restlessness, fever, and nausea in infants (60,61). Production, transportation, and storage of current vaccines are very expensive (62). In addition, this vaccine was designed for production in a prokaryotic expression system that is simpler and cheaper than a eukaryotic system. Practical studies are needed to validate the model peptide, but this type of vaccine could improve health in developing and underdeveloped countries.
Acknowledgments
This study was supported by research funding of Shahrekord University.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Adams HP, and Koziol J. Prediction of binding to MHC class I molecules. Immunol Methods 1995;185:181–190 [DOI] [PubMed] [Google Scholar]
- 2.Aiyegbo MS, Sapparapu G, Spiller B, et al. Human rotavirus VP6-specific antibodies mediate intracellular neutralization by binding to a quaternary structure in the transcriptional pore. PLoS One 2013;8:e61101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhasin M, and Raghava G. SVM based method for predicting HLA-DRB1*0401 binding peptides in an antigen sequence. Bioinformatics 2004;20:421–423 [DOI] [PubMed] [Google Scholar]
- 4.Bhasin M, and Raghava G. Analysis and prediction of affinity of TAP binding peptides using cascade SVM. Protein Sci 2004;13:596–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhasin M, and Raghava G. Prediction of CTL epitopes using QM, SVM and ANN techniques. Vaccine 2004; 22:3195–3204 [DOI] [PubMed] [Google Scholar]
- 6.Black RE, Cousens S, Johnson HL, et al. Global, regional, and national causes of child mortality in 2008: a systematic analysis. Lancet 2010;375:1969–1987 [DOI] [PubMed] [Google Scholar]
- 7.Bridger JC, and Oldham G. Novel rotaviruses in animals and man. Clin Microbiol 1987;128:6–23 [DOI] [PubMed] [Google Scholar]
- 8.Chen P, Rayner S, and Hu K. Advances of bioinformatics tools applied in virus epitopes prediction. Virol Sin 2011;26:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark HF, Hoshino Y, Bell LM, et al. Rotavirus isolate WI61 representing a presumptive new human serotype. Clin Microbiol 1987;25:1757–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole C, Barber J, and Barton G. The Jpred 3 secondary structure prediction server. Nucleic Acids Res 2008;36:W197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dennehy PH, Vesikari T, Matson DO, et al. Efficacy of the pentavalent rotavirus vaccine, RotaTeq® (RV5), between doses of a 3-dose series and with less than 3 doses (incomplete regimen). Human Vaccines 2011;7:563–568 [DOI] [PubMed] [Google Scholar]
- 12.Donato CM, Cannana D, Bogdanovic-Sakrana N, et al. Characterisation of a G9P[8] rotavirus strain identified during a gastroenteritis outbreak in Alice Springs, Australia post Rotarix™ vaccine introduction. Vaccine 2012;30:152–158 [DOI] [PubMed] [Google Scholar]
- 13.Donato CM, Ch'ng LS, Boniface KF, et al. Identification of strains of RotaTeq rotavirus vaccine in infants with gastroenteritis following routine vaccination. J Infect Dis 2012;375:377–383 [DOI] [PubMed] [Google Scholar]
- 14.Doytchinova IA, Guan P, and Flower D. EpiJen: a server for multistep T cell epitope prediction. BMC Bioinformatics 2006;7:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El-Manzalawy Y, Dobbs D, and Honavar V. Predicting linear B-cell epitopes using string kernels. J Mol Recognit 2008;21:243–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Estes KM, and Cohen J. Rotavirus gene structure and function. Microbiol Rev 1989;53:410–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Estes MK, Knipe DM, Howley PM, et al. Rotaviruses and their replication. Virology 2007;1917–1974 [Google Scholar]
- 18.Fernandez-Fuentes N, Madrid-Aliste C, Rai B, et al. M4T: a comparative protein structure modeling server. Nucleic Acids Res 2007;35:W363–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garaicoechea L, Olichon A, Marcoppido G, et al. Llama-derived single-chain antibody fragments directed to rotavirus VP6 protein possess broad neutralizing activity in vitro and confer protection against diarrhea in mice. Virology 2008;28:9753–9764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garrison KE, Champiat S, York V, et al. Transcriptional errors in human immunodeficiency virus type 1 generate targets for T-cell responses. Clin Vaccine Immunol 2009;16:1369–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gentsch JR, Woods PA, Ramachandran M, et al. Review of G and P typing results from a global collection of rotavirus strains: implications for vaccine development. Infect Dis 1996;174:S30–S36 [DOI] [PubMed] [Google Scholar]
- 22.Grimwood K, and Lambert SB. Rotavirus vaccines: opportunities and challenges. Hum Vaccines 2009;5:57–69 [DOI] [PubMed] [Google Scholar]
- 23.Kapikian AZ, Hoshino Y, and Chanok RM. Rotaviruses. Fields Virol 2001;2:1787–1833 [Google Scholar]
- 24.Kelley LA, and Sternberg M. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 2009;4:363–371 [DOI] [PubMed] [Google Scholar]
- 25.Knutson KL, and Disis M. Tumor antigen-specific T helper cells in cancer immunity and immunotherapy. Cancer Immunol Immunother 2005;54:721–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kringelum JV, Lundegaard C, Lund O, et al. Reliable B cell epitope predictions: impacts of method development and improved benchmarking. PLoS Comput Biol 2012;8:e1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larsen MV, Lundegaard C, Lamberth K, et al. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinformatics 2007;8:424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lata S, Bhasin M, and Raghava G. Application of machine learning techniques in predicting MHC binders. Methods Mol Biol 2007;409:201–215 [DOI] [PubMed] [Google Scholar]
- 29.Li Z, Zhang M, Hu H, et al. [On predicting the T cell and B cell epitopes of platelet membrane glycoprotein II b/III a antibody from human and mice]. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi 2011;27:1146–1151 [PubMed] [Google Scholar]
- 30.Liang S, Zheng D, Zhang C, et al. Prediction of antigenic epitopes on protein surfaces by consensus scoring. BMC Bioinformatics 2009;10:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGuffin LJ, Bryson K, and Jones D. The PSIPRED protein structure prediction server. Bioinformatics 2000;16:404–405 [DOI] [PubMed] [Google Scholar]
- 32.Nakagomi T, Ohshima A, Akatani K, et al. Isolation and molecular characterization of a serotype 9 human rotavirus strain. Microbiol Immunol 1990;34:77–82 [DOI] [PubMed] [Google Scholar]
- 33.Offit PA, Hoffenberg E, Santos N, et al. Rotavirus-specific humoral and cellular immune response after primary, symptomatic infection. J Infect Dis 1993;167:1436–1440 [DOI] [PubMed] [Google Scholar]
- 34.Parashar UD, Burton A, Lanata C, et al. Global mortality associated with rotavirus disease among children in 2004. Infect Dis 2009;200:9–15 [DOI] [PubMed] [Google Scholar]
- 35.Parashar UD, Gibson CJ, Bresee JS, et al. Rotavirus and severe childhood diarrhea. Emerg Infect Dis 2006;12:304–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parashar UM, Hummelman EG, Bresee GS, et al. Global illness and deaths caused by rotavirus disease in children. Emerg Infect Dis 2003;9:565–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel M, Steele D, Gentsch JR, et al. Real-world impact of rotavirus vaccination. Infect Dis J 2011;30:S1–5 [DOI] [PubMed] [Google Scholar]
- 38.Penelope H. Rotavirus vaccines: an overview. Clin Microbiol Rev 2008;21:198–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phua KB, Emmanuel SC, Goh P, et al. A rotavirus vaccine for infants: the Asian experience. Ann Acad Med Singapore 2006;35:38–44 [PubMed] [Google Scholar]
- 40.Ponomarenko J, Bui H, Li W, et al. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics 2008;9:514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rai J, Lok K, Mok C, et al. Immunoinformatic evaluation of multiple epitope ensembles as vaccine candidates: E. coli 536. Bioinformation 2012;8:272–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reimer U. Prediction of linear B-cell epitopes. Methods Mol Biol 2009;524:335–344 [DOI] [PubMed] [Google Scholar]
- 43.Rennels MB, Glass RI, Dennehy PH, et al. Safety and efficacy of high-dose rhesus-human reassortant rotavirus vaccine. Pediatrics 1996;97:7–13 [PubMed] [Google Scholar]
- 44.Rossi G, Cornaro C, Battilani M, et al. Production of IFN-gamma in feline whole blood after incubation with potential T-cell epitopes of the nucleocapsid protein of feline coronavirus. Vet Microbiol 2011;150;248 (248–256) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rost B, and Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol 1993;232:458–459 [DOI] [PubMed] [Google Scholar]
- 46.Saha S, and Raghava G. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins 2006;65:40–48 [DOI] [PubMed] [Google Scholar]
- 47.Saha S, and Raghava G. Prediction methods for B-cell epitopes. Methods Mol Biol 2007;409:387–394 [DOI] [PubMed] [Google Scholar]
- 48.Saha S, Raghava GPS. 2004. BcePred: Prediction of continuous B-cell epitopes in antigenic sequences using physico-chemical properties. In ICARIS 2004, LNCS, vol. 3239 Nicosia G., Cutello V., Bentley P.J. and Timis J. (eds). Springer, UK, 197–204 [Google Scholar]
- 49.Singh H, and Raghava G. ProPred1: prediction of promiscuous MHC Class-I binding sites. Bioinformatics 2003;19:1009–1014 [DOI] [PubMed] [Google Scholar]
- 50.Vasil'ev B, Moskvin A, Marchenko L, et al. [Indicators of humoral immunity in rotavirus infection among various age groups of St. Petersburg residents from CFT data]. Vopr Virusol 1995;40:129–130 [PubMed] [Google Scholar]
- 51.Ward RL, and Bernstein DI. Rotarix: a rotavirus vaccine for the world. Vaccine 2009;48:222–228 [DOI] [PubMed] [Google Scholar]
- 52.Widdowson MA, Steele D, Vojdani J, et al. Global rotavirus surveillance: determining the need and measuring the impact of rotavirus vaccines. Infect Dis 2009;200:S1–8 [DOI] [PubMed] [Google Scholar]
- 53.Williams MB, Rose J, Rott L, et al. The memory B cell subset responsible for the secretory IgA response and protective humoral immunity to rotavirus expresses the intestinal homing receptor, alpha4beta7. Immunol 1998;161:4227–4235 [PubMed] [Google Scholar]
- 54.Zhang W, Xiong Y, Zhao M, et al. Prediction of conformational B-cell epitopes from 3D structures by random forests with a distance-based feature. BMC Bioinformatics 2011;12:341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang W, Liu J, and Zhao M. Predicting linear B-cell epitopes by using sequence-derived structural and physicochemical features. Int J Data Min Bioinform 2012;6:557–569 [DOI] [PubMed] [Google Scholar]