Abstract
Significance: A constant accumulation of oxidized proteins takes place during aging. Oxidation of proteins leads to a partial unfolding and, therefore, to aggregation. Protein aggregates impair the activity of cellular proteolytic systems (proteasomes, lysosomes), resulting in further accumulation of oxidized proteins. In addition, the accumulation of highly crosslinked protein aggregates leads to further oxidant formation, damage to macromolecules, and, finally, to apoptotic cell death. Furthermore, protein oxidation seems to play a role in the development of various age-related diseases, for example, neurodegenerative diseases. Recent Advances: The highly oxidized lipofuscin accumulates during aging. Lipofuscin formation might cause impaired lysosomal and proteasomal degradation, metal ion accumulation, increased reactive oxygen species formation, and apoptosis. Critical Issues: It is still unclear to which extent protein oxidation is involved in the progression of aging and in the development of some age-related diseases. Future Directions: An extensive knowledge of the effects of protein oxidation on the aging process and its contribution to the development of age-related diseases could enable further strategies to reduce age-related impairments. Strategies aimed at lowering aggregate formation might be a straightforward intervention to reduce age-related malfunctions of organs. Antioxid. Redox Signal. 23, 239–255.
Introduction: Oxidative Stress and Aging
Aging, also described as organismal senescence, is defined as a state with decreased ability to respond to stress, decreased homeostatic balance, and an increased risk of developing age-related diseases. There are several theories explaining the origin of the aging process. Apart from the programmed theories, assuming the aging process follows a fixed timetable determined by changes in gene expression, it can be differentiated into the damage and error theories. The latter are based on environmental influences, leading to cumulative damage, such as the “free radical theory of aging.” The “free radical theory of aging” was first described by Harman in 1956 (58). This theory postulates the ability of free radicals to cause aging by a reaction with macromolecules, resulting in an accumulation of oxidatively damaged material. Reactive oxygen species (ROS) are a major reason for oxidative damage. Due to the necessity of O2 for maintenance of human life, ROS formation and oxidation processes are an integral part of life. ROS are formed not only endogenously by a large number of processes, but also exogenously through exposure to various environmental influences. ROS comprise radicals, such as superoxide anion (O2•−), hydroxyl radical (HO•), peroxyl radical (RO2•), alkoxyl radical (RO•), and hydroperoxyl radical (HO2•) as well as non-radicals, such as hydrogen peroxide (H2O2), singlet oxygen (O21Δg), and ozone (O3). The non-radicals have the ability to generate free radicals and are, therefore, also referred to as ROS. Due to their high reactivity, some ROS may react with further molecules, generating radicals that again lead to oxidative damage. For example, reactive nitrogen species, such as peroxynitrite (ONOO−), may be generated through an initial reaction of O2•− with nitric oxide (NO) (145). The status of imbalance between oxidant generation and antioxidant defenses in favor of the oxidants is generally referred to as oxidative stress. Oxidative stress leads to an elevated oxidation of macromolecules, such as DNA, lipids, and proteins. It was postulated that age-related accumulation of damaged, oxidized, and aggregated proteins might contribute to the aging process (63, 68, 146). However, the importance of protein oxidation in progression of aging is not yet fully understood. Moreover, it is still unclear as to what extent the accumulation of oxidized proteins has an effect on aging.
Formation and Fate of Oxidized Proteins
Short overview on chemistry of protein oxidation
The reactions of ROS with proteins lead to oxidative damage. The targets for oxidant reactions are either the protein backbones, resulting in fragmentation reactions, or the amino-acid side chains, leading to the formation of a variety of different oxidation products.
Oxidative protein modifications cause changes in the protein structure and a partial unfolding of the protein. Both the unfolding and the direct oxidation of functional amino-acid side chains may lead to an impaired protein function.
Damage of the protein backbone
Radical-induced reactions often rapidly lead to backbone damage. In general, an oxidative attack on the backbone first results in the abstraction of the hydrogen atom from the α-carbon, leading to formation of a stabilized carbon-centered radical. This radical may react with other carbon-centered radicals or with O2. The latter results in the formation of a peroxyl radical (121). Peroxyl radicals may either undergo an elimination reaction, by which HO2• is released, or generate hydroperoxides through hydrogen abstraction from another molecule. Both paths finally lead to protein backbone fragmentation (35).
Damage to amino-acid side chains
Since there are 20 different amino acids, the oxidation of amino-acid side chains is a more complex mechanism than the protein backbone oxidation and leads to the formation of several oxidation products. Therefore, only a brief overview on possible reactions will be given [for an extensive review, see Refs. (36, 147)]. Similar to backbone damage, oxidation of aliphatic amino-acid side chains often leads to the formation of carbon-centered radicals. There are various fates for these radicals. For instance, they may undergo dimerization due to the reaction with other carbon-centered radicals or they may be repaired by thiols, generating thiyl radicals. However, the predominantly occurring reaction under aerobic conditions is the formation of peroxyl radicals (121). As a result, alkoxyl radicals, hydroperoxides, alcohols, and carbonyls may be generated (36).
A good example for oxidation of aromatic amino-acid residues is tyrosine. Oxidative damage of tyrosine often leads to generation of tyrosyl radicals. These radicals may dimerize, generating dityrosine, a process involved in crosslinkage and protein aggregation (2). Further important products of tyrosine oxidation are 3-chlorotyrosine, 3,4-dihydroxyphenylalanine, and 3-nitrotyrosine (3-NT) (36). The latter are generated due to the reaction with ONOO−.
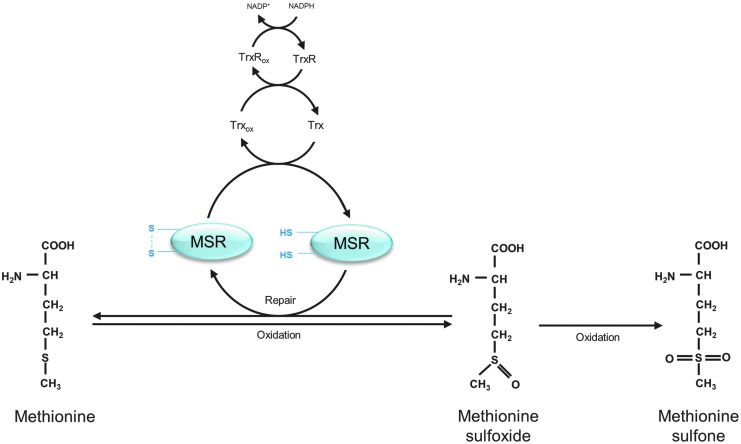
Sulfur-containing amino-acid side chains are the most sensitive targets for oxidation. The oxidation of cysteine leads to the formation of thiyl radicals. There are several fates for these thiyl radicals. Dimerization reactions give rise to disulfides, generating cystine. In addition, mixed dimers may be formed due to the reaction of thiyl radicals with other species. The reaction of thiyl radicals with O2 leads to the formation of peroxyl radicals and gives rise to oxyacids (sulfenic acid, sulfinic acid, and sulfonic acid) (11, 36). Methionine sulfoxides (MeSOs) are one of the main products, resulting from methionine oxidation (Fig. 1). They are generated in S- or R-stereoisomers. MeSOs may form sulfones after further oxidation (36) or may be reduced by methionine sulfoxide reductases (MSRs) (Fig. 1) (see Repair and Degradation of Oxidized Proteins section).
FIG. 1.
Methionine oxidation and repair of methionine sulfoxide (MeSO) via the methionine sulfoxide reductases (MSRs), modified according to Hoshi and Heinemann (71). Oxidation of methionine results in MeSO formation. MeSOs may be repaired by MSRs. Reduction of MeSOs by MSRs leads to the oxidation of the catalytic cysteines in the active sites of the MSRs and to the formation of an intramolecular disulfide. MSRs are recycled due to the thioredoxin/thioredoxin reductase system (Trx/TrxR). Further oxidation of MeSOs leads to the generation of nonrepairable methionine sulfones.
Methods to measure protein oxidation products
Two of the most commonly measured oxidative protein modifications are protein carbonyls and 3-NT, which are used to assess the cellular status of protein oxidation.
Protein carbonyls are generated due to the oxidation of proline, arginine, lysine, threonine, and other amino-acid residues and due to the oxidation of the protein backbone. Moreover, they might be the result of secondary reactions of amino acids (cysteine, histidine, and lysine) with reactive carbonyl compounds (ketones, aldehydes), emerged in lipid peroxidation or glycation/glycoxidation reactions (33). These secondary reactions may lead to the formation of advanced glycation and lipoxidation end products (AGEs/ALEs). Protein carbonyls are chemically stable and easy to detect after modification with dinitrophenylhydrazine by various methods of immunostaining; for example, immunoblot, ELISA, and fluorescence microscopy [for a review, see Ref. (154)].
The determination of 3-NT levels is widely used to investigate the role of oxidative protein modification. There are several methods available for measurement of 3-NT, including methods of immunostaining (e.g., ELISA), HPLC, and mass spectroscopy (62, 158).
Physiological formation of oxidized proteins
ROS are generated in a highly controlled way under physiological conditions. They are produced in the mitochondrial electron transport chain (mETC), which is responsible for cellular respiration, as well as in reactions of the cytochrome P450 system, involved in drug metabolism. In addition, many other enzymes are known to produce ROS, such as lipoxygenases, prostaglandin sythases, ribonucleotide reductases, xanthine oxidases, and NADPH oxidases. Besides the damaging effects of ROS, in low concentrations they may also be useful metabolites and carry out regulatory functions. Some ROS, such as H2O2, have the ability to act as second messengers due to their specificity to interact with effectors in signaling pathways. Other ROS, such as HO•, are highly reactive and interact rapidly with any target near their generation site. Thus, they do not possess the specificity to act as second messengers [for a detailed review, see Refs. (43, 101)].
Since oxidative protein modifications result in protein unfolding and the impairment of protein function, this might negatively affect cellular metabolism. However, inactivation of proteins due to protein oxidation may also have regulatory functions (100). Similar to other post-translational protein modifications, such as phosphorylation and acetylation, oxidation also seems to be involved in the regulating of a large number of metabolic processes. Especially oxidation of sulfur-containing amino acids methionine and cysteine seems to be important, as these are the only oxidative protein modifications that are reversible (see Repair and Degradation of Oxidized Proteins section). In many cases, regulatory protein oxidation is performed by enzymes, such as peroxidases, which use H2O2 as a substrate [for a review, see Refs. (43, 101)].
The function of transcription factors, protein kinases and protein phosphatases is regulated by oxidation of cysteine. Protein tyrosine phosphatases (PTPs) constitute one group of proteins that is very sensitive for oxidation. They contain catalytic cysteine residues in their active site. Oxidation of these cysteine residues leads to disulfide bond formation. Due to oxidation, the PTP is inactivated and, thus, not able to dephosphorylate the target protein kinases (127). Consequently, reversible cysteine oxidation of PTPs is implicated in tyrosine phosphorylation-dependent pathways.
Similar to cysteine oxidation, the oxidation of methionine and formation of MeSOs is often a protein inactivating post-translational protein modification. For example, the oxidation of a methionine residue of calmodulin, a protein involved in immune response, metabolism, and inflammation, leads to loss of the calcium-binding ability, responsible for activation of target enzymes. However, methionine residue oxidation may also trigger the protein activity, while reduction via MSRs (see Repair and Degradation of Oxidized Proteins section) leads to inactivation (38). To give an example, calmodulin kinase II, important for calcium homeostasis, is usually activated due to binding of calcium and calmodulin, but, in addition, shows a calcium and calmodulin-independent activation after methionine oxidation (39, 73).
Pathophysiological formation of oxidized proteins
Endogenous changes, such as inflammation, or exogenous influences, for instance UV radiation, gamma radiation, and xenobiotics, may increase ROS formation, either directly or by promoting endogenous ROS generators. As a consequence, antioxidant defenses may be overwhelmed, resulting in a cellular state referred to as oxidative stress and in enhanced macromolecule damage, including oxidative damage to proteins. One initial oxidation event may lead not only to multiple oxidative protein modifications within a protein, but also to oxidation of further proteins and other macromolecules. Thus, protein oxidation may give rise to uncontrollable chain reactions (106, 122). To protect the cells from the overall consequences of protein oxidation, there exists an effective and fast degradation and repair system to remove oxidized proteins. However, this system may be overloaded during increased protein oxidation. Therefore, oxidized proteins accumulate in the cell.
The accumulation of oxidized proteins results in crosslinking reactions and the increased formation of nondegradable protein aggregates. Protein aggregates are initially the result of hydrophobic interactions due to the increased surface hydrophobicity of oxidized proteins, resulting from their partial unfolding (52, 128). Furthermore, the oxidized proteins in these aggregates become chemically crosslinked by covalent reactions; for example, dityrosine formation, interaction of two carbon-centered radicals, disulfide formation, and the reaction of protein carbonyls with lysine and arginine residues (see Short Overview on Chemistry of Protein Oxidation section).
Protein aggregates are insoluble and nondegradable (79). Therefore, protein aggregates accumulate within the cell, which seems to have various consequences for the cellular metabolism and viability. It was shown that the accumulation of protein aggregates leads to inhibition of the proteasomal system and, thus, to further aggregation (66, 141) (see Repair and Degradation of Oxidized Proteins section).
Protein aggregates may accumulate in various structures, such as inclusion bodies, lysosomes, aggresomes, or plaques. “Protein aggregate” is, therefore, a very simple term that only describes the existence of crosslinked proteins. Plaques are extracellular occurring protein aggregates. Inclusion bodies describe intracellular accumulations of protein aggregates. The term aggresome was described in 1998 by the group of Kopito as a “pericentriolar, membrane-free, cytoplasmic inclusion containing misfolded, ubiquitinated proteins ensheeted in a cage of intermediate filaments, formed specifically at the microtubule organization center (MTOC)” (79). Thus, the term describes the cellular location and the composition of the aggregates, as well as the involvement of filaments. In general, aggresome formation seems to be a protective mechanism, because the smaller intermediate aggregates have a larger surface and, therefore, a higher potential for interaction with cellular macromolecules and membranes compared with mature aggregates caged in aggresomes (125). Aggresomes might contain several forms of oxidized proteins and otherwise misfolded proteins, which might be ubiquitinated but for some reason are not degraded by the 26S proteasome.
Existing protein aggregates may undergo further direct oxidation reactions or secondary reactions with products of lipid peroxidation, for example, 4-hydroxynonenal (HNE) and malondialdehyde (45), resulting in the formation of highly crosslinked fluorescent material referred to as lipofuscin (see Pathophysiological Roles of Accumulated and Aggregated Proteins section).
Repair and degradation of oxidized proteins
Repair of oxidized proteins
The majority of oxidative protein modifications are nonrepairable, making a fast and effective degradation system essential to protect the cells from accumulation of oxidized proteins and related consequences. However, a few enzymes exist, which are able to repair modifications of sulfur containing amino-acid residues (disulfides and MeSOs). These enzymes are of biological importance due to their involvement in regulatory functions of disulfide and MeSO formation (see Physiological Formation of Oxidized Proteins section).
Disulfides may be repaired either by thioredoxin (Trx) or by the protein disulfide isomerase (PDI) (24, 160). Both enzymes contain cysteine residues in their active sites, which are able to reduce the disulfides being oxidized themselves, resulting in the formation of intramolecular disulfides. Afterward, Trxs may be recycled by thioredoxin reductase (TrxR) using NADPH. PDIs may be reduced by glutathione.
A group of enzymes, called MSRs, is able to reduce the two stereoisomers of MeSO. MSRAs, which are able to reduce S-stereoisomers, reduce free MeSO and MeSO that are embedded in proteins. In contrast, MSRBs are able to reduce R-stereoisomers and almost exclusively in proteins (38). Similar to Trxs and PDIs, reduction of MeSO via MSRs is catalyzed by cysteine residues, which may form intramolecular disulfides (Fig. 1). Next to these catalytic cysteines, there are other cysteine residues in the active site of MSR, referred to as recycling cysteines, which are besides the Trx/TrxR system involved in the reduction of the catalytic cysteines (Fig. 1) (105).
Degradation by the proteasomal system
As mentioned earlier, degradation systems have a large share in the removal of protein oxidation products. The most important system responsible for degradation of oxidized proteins is the proteasomal system and, in particular, the 20S proteasome (Fig. 2) [for a review, see Refs. (34, 82)]. The 20S proteasome is a barrel-shaped multi-enzyme complex (Fig. 2), with a molecular weight of 700 kDa. It is composed of four homologous rings. Each ring is divided into seven subunits. The inner rings, which form the catalytic center of the 20S proteasome, consist of seven different β-subunits (β1–β7). Three of them possess a proteolytic activity. β1 has a peptidyl-glutamyl-peptide-hydrolizing activity, β2 has a trypsin-like activity, and β5 possesses a chymotrypsin-like activity. The outer rings consist of seven different α-subunits (α1–α7) that have a function in recognition and access of the substrates to the catalytic center of the 20S proteasome (83).
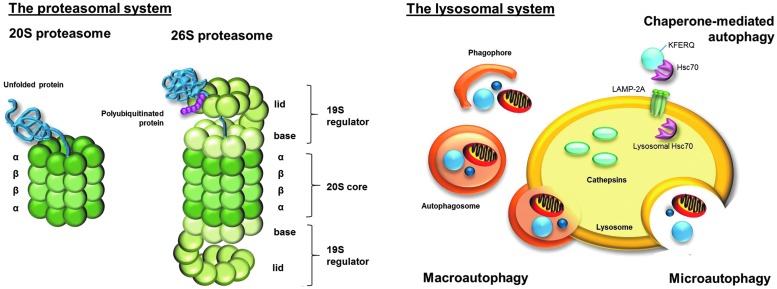
FIG. 2.
The proteasomal and the lysosomal degradation system. Although there are several proteasome forms, the attention in this review focuses on 20S proteasome and 26S proteasome. The 20S proteasome consists of four homologous rings, with each divided into seven subunits. The two inner rings are formed by the β-subunits and build the catalytic center of the proteasome; the two outer rings are formed by the α-subunits, which enable recognition and access of substrate proteins to the catalytic center. The 20S proteasome is able to recognize unfolded and, therefore, oxidized proteins due to their increased surface hydrophobicity. Binding of two 19S regulators to the 20S proteasome leads to the formation of the 26S proteasome. Each 19S regulator consists of lid and base and is composed of 19 Rpt- and Rpn-subunits. Polyubiquitination of the substrates, via the ubiqutination system, is necessary for the 26S proteasome to recognize the substrate proteins. In contrast to the 20S proteasome, the 26S proteasome degrades substrate proteins in an ATP-dependent way. ATP hydrolysis is responsible for unfolding of substrate proteins. Lysosomal degradation due to cathepsins is a less selective process than the proteasomal degradation. However, the uptake of substrate proteins via autophagy is a somewhat selective and highly controlled process. While microautophagy describes the direct intake of proteins and organelles into the lysosomal lumen, macroautophagy describes the inclusion first into a phagophore, generating an autophagsome that fuses with the lysosome. Proteins that contain a KFERQ-pentapeptide motive are selectively recognized by Hsc70 and transported to the lysosome. Via lysosomal LAMP-2A receptor and lysosomal Hsc70, proteins are translocated into the lysosomal lumen.
Another proteasomal form is the 2 MDa large 26S proteasome, which consists of the 20S core and two bound 19S regulators (Fig. 2). In contrast to the 20S proteasome, the 26S proteasome degrades substrate proteins in an ATP-dependent way and it requires a polyubiquitin sequence to recognize the substrate proteins. Due to the involvement of the ubiquitin-targeting machinery in degradation of 26S substrate proteins, this system is also referred to as the ubiquitin-proteasome system (UPS) (81, 96). The 19S regulator (also known as PA700) consists of some 19 monomers (Rpt- and Rpn-subunits). The Rpn-subunits functions in recognition of the polyubiquitinated substrate proteins. The Rpt-subunits have an ATPase activity. ATP hydrolysis causes the substrate intake through the resulting conformational change of the 26S proteasome and, in addition, the unfolding of the substrate proteins (83). The major function of 26S proteasome is the degradation of functional proteins, for example, responsible for cell cycle regulation, apoptosis, and signal transduction, which are no longer needed in the cellular metabolism and, therefore, marked by the ubiquitination machinery for degradation [for a review, see Refs. (96, 119)]. Moreover, it is postulated that newly synthesized, but misfolded proteins are degraded by the 26S proteasome (93).
For a long time, it was a controversial issue whether oxidized proteins are degraded by 20S or 26S proteasomes. However, the predominant role of the 20S proteasome was confirmed by many research groups. On the one hand, oxidized proteins seems to be degraded independently from ATP and without the necessity of a polyubiquitin-tag (34, 56, 57, 87, 140). In contrast to the 26S proteasome, the 20S proteasome is able to recognize unfolded proteins and, therefore, oxidized proteins, due to their increased surface hydrophobicity (Fig. 2) (52, 95, 128). On the other hand, the 20S proteasome seems to be more resistant to oxidative stress than the 26S proteasome (137, 138). It is very likely that during oxidative stress, the 26S proteasome dissociates into the 20S core and the 19S regulators. The increased availability of the 20S proteasome results in an elevated degradation of oxidized proteins. Hsp70 seems to be involved in this process by keeping the 19S regulator in its active form until re-association to the 26S proteasome (55) (Fig. 2).
Heat shock proteins (HSPs) are of importance for protein pool maintenance. Due to their function as molecular chaperones, they are involved in refolding of misfolded and damaged proteins (Fig. 3). Although oxidized proteins cannot be repaired by HSPs, due to covalent modifications of oxidized proteins, chaperones have other important functions during oxidative stress. Thus, they are possibly involved in the functionality of the proteasomal degradation system. For example, Conconi et al. showed that Hsp90 leads to an enhanced activity of the 20 proteasome after FeCl3-induced ROS formation in vitro (25). Whittier et al. showed that Hsp90 is selectively involved in 20S proteasome-related degradation of oxidized calmodulin, but not in degradation of native calmodulin (159). In addition, a direct participation of HSPs (Hsp70 and Hsp90) in the degradation by the 26S proteasome is postulated (Fig. 3). HSP-bound substrate proteins are polyubiquitinated by the ubiquitin E3 ligase CHIP, which is an important co-chaperone of Hsp70 and Hsp90 (117). A further co-chaperone BAG1 mediates binding of HSP-substrate complex to the 26S proteasome (4). However, the degree to which HSPs are involved in degradation of oxidized proteins by the 20S proteasome and exact mechanisms of the involvement have not yet been clarified.
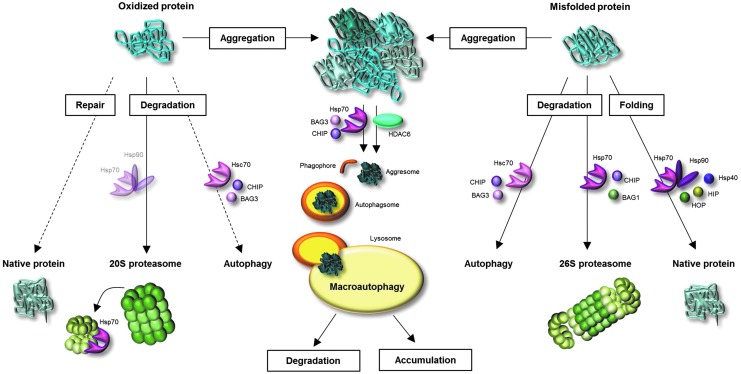
FIG. 3.
Fates of oxidized and misfolded proteins and involvement of molecular chaperones. Once a protein is oxidatively modified, several pathways might deal with this oxidized protein. A few oxidative protein modifications may be repaired. However, the large part of oxidized proteins undergoes degradation by the proteasomal system. Since oxidized proteins are partially unfolded, they are a suitable substrates for the 20S proteasome. During oxidative stress, the 26S proteasome dissociates into 20S proteasome and 19S regulators. Hsp70 binds the 19S regulator until re-association of 26S proteasome. The involvement of heat shock proteins (HSPs) in transport of oxidized proteins to the 20S proteasome is assumed, but not proved until now. Moreover, there is some evidence for degradation of oxidized proteins via autophagy. Misfolded (not-covalently modified) proteins are recognized by molecular chaperones, leading to protein refolding or degradation. Folding is accomplished due to Hsp70, Hsp90 and co-chaperones Hsp40, HIP, and HOP. Co-chaperones CHIP and BAG1 direct the Hsp70-bound protein to 26S proteasome, where it is degraded. Misfolded proteins may furthermore undergo autophagy. If oxidized and misfolded proteins are not degraded, repaired, or refolded, they undergo aggregation reactions. Protein aggregates are poor substrates for the proteasome. They generate pericentriolar located aggresomes due to involvement of either histone deacetylase 6 (HDAC6) or Hsp70 and BAG3. The aggresomes may be enclosed by the phagophore, generating the autophagosome, which merges with the lysosome. Degradable proteins are then destroyed by the lysosomal proteases, whereas crosslinked, nondegradable material undergoes further aggregation.
Involvement of the lysosomal system in the degradation of oxidized proteins
Besides the proteasomal degradation system, there exists a further degradation machinery that might be involved in proteolysis of oxidized proteins: the lysosomal system (Fig. 2). Lysosomes contain a wide range of degrading enzymes that are responsible not only for degradation of proteins (cathepsins) but also for carbohydrates and lipids. Due to the high activity of hydrolyzing enzymes in the lysosomes, the uptake of material into the lysosomes is strongly regulated. However, the lysosomal degradation of proteins is a less selective process than the proteasomal degradation.
Lysosomal uptake is accomplished via three different processes: phagocytosis, endocytosis, and autophagy. In particular, autophagy is important for degradation of endogenous proteins. There are three types of autophagy: microautophagy, macroautophagy, and chaperon-mediated autophagy (CMA) (Fig. 2) (152). Direct intake of degradable material into lysosomes is called microautophagy (115). In contrast, macroautophagy describes the inclusion of organelles and soluble proteins, first into double-membrane structures (phagophores), generating double-membrane vesicles, called autophagosomes, which afterward merge with the lysosomes (65, 76). Compared with microautophagy and macroautophagy, substrate uptake via CMA is a more substrate-specific process. Proteins, containing a KFERQ-pentapeptide motive, may be selectively recognized by the molecular chaperone Hsc70. Afterward, the Hsc70 co-chaperones CHIP and BAG3 mediate ubiquitination and transport of Hsc70-bound substrate protein to the lysosomal membrane. The substrate protein is translocated into the lysosomal lumen via LAMP-2A receptor and lysosomal Hsc70 (Fig. 2) (29, 32, 150). Even if there is evidence for an upregulation of LAMP-2A during oxidative stress, indicating a role of CMA in degradation of oxidized proteins (89), the selective degradation of oxidized proteins is not confirmed. Nevertheless, it is possible that oxidation-related unfolding might expose the KFERQ motive for selective recognition.
Macroautophagy does not seem to have a large share in degradation of oxidized proteins, due to the fact that the selectivity of this process for oxidized proteins is still under discussion. However, the role of macroautophagy in the lysosomal uptake of protein aggregates, also called aggrephagy (94), seems to be proved (Fig. 3). According to this idea, the accumulation of oxidized proteins in pericentriolar aggresomes seems to be an intermediate step in the removal of soluble oxidized proteins, followed by aggresome degradation (44, 79, 162). In 2000, Kopito describes that aggresome formation may facilitate the lysosomal degradation of aggregates (92). Thus, protein aggregates, transported to the MTOC, might be included into phagophores, generating autophagosomes, which fuse with lysosomes. The aggresome-autophagy pathway is considered a protective “back up” mechanism when the molecular chaperones and the proteasomal system are overwhelmed. The transport of protein aggregates to the aggresomes is realized either by histone deacetylase 6 (HDAC6) or by Hsp70 and co-chaperone BAG3 (94) (Fig. 3). Protein aggregates are coupled to dynein motor complex and along microtubules, they are transported to the MTOC (125).
Protein Oxidation in Human Aging
Model systems demonstrating protein oxidation in cellular senescence and aging
The investigation of the role of oxidized proteins in human aging is very difficult, on the one hand, due to the low availability of human material, on the other hand due to the difficulty to determine whether changes are a result of aging or of other processes, such as diseases, environmental exposures, or lifestyle. Since it is assumed that aging first takes place at a cellular level, human cells are often used to study processes which contribute to aging and the role of protein oxidation in aging. Differentiation is conducted between three cellular model systems: (i) in vitro aged primary cells, also referred to as postmitotic senescent cells; (ii) cells aged in vitro due to chronic oxidant treatment; and (iii) primary cells from old donors (in vivo aged) (Fig. 4).
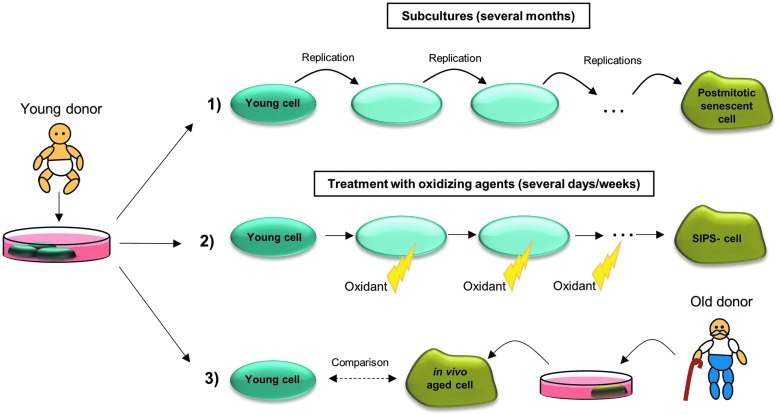
FIG. 4.
Human cellular model systems in aging research. Primary cells are often isolated from a young donor. (1) After several subcultures, the young cells reach their replicative limit and are then referred to as senescent postmitotic cells. This process might last several months. The senescent cell stops to divide, but does not immediately die and can be used as a model for aging of nondividing cells. (2) Treatment of young cells with oxidants or stressors for several days/weeks leads to stress-induced premature senescence (SIPS). These cells mirror many features of cells described under (1) and (3). (3) Primary cells isolated from a young donor can be compared with primary cells isolated from an old donor.
Cellular senescence defines the state when primary mitotic cells reach their replicative limit. To differentiate, organismal senescence describes the biological aging of the whole organism. The cellular replicative limit, also called “Hayflick limit,” was first postulated by Hayflick and Moorhead (59). They described the characteristic of primary cells, grown in culture, only to experience a finite number of cell divisions. Cells that achieve the replicative limit undergo no further division cycles and are referred to as senescent cells. These cells do not die immediately and can, therefore, be used as a model for aging of nondividing cells. Senescent cells are characterized by changes in morphology (increased cell surface and volume), short telomeres, growth arrest, apoptosis resistance, and altered gene expression (20). The occurrence of senescent cells was also observed in vivo in mitotic tissues of humans and primates, and it was shown that the number of senescent cells increases with age (20, 70, 77). Baker et al. have shown that the removal of senescent cells may delay the formation of age-related diseases (6). Thus, the usage of in vitro aged senescent cells in aging research may reflect organismal aging.
To study cellular aging without waiting for the replicative limit, which lasts several months, there is the possibility of modeling the characteristic phenotype of senescent cells due to stress-induced premature senescence (SIPS) [for a review, see Ref. (153)]. This premature aging may be induced via repeated treatments of young primary cells with nonlethal concentrations of H2O2, paraquat, or other stressors (23, 66, 67, 69, 84). The oxidant treatment is repeated every day or every 2 days, leading to chronic oxidative stress, which seems to be the main reason for premature aging (153).
One further cellular model system in aging research represents the isolation and cultivation of primary cells from old donors, although several difficulties have to be considered. The properties of these in vivo aged primary cells may be compared with those of young primary cells.
Although the usage of different cellular model systems seems to be a good tool for investigating aging at the cellular level, it is a matter of debate whether in vitro results are representative for what happens in vivo. Therefore, short living non-mammalians, such as Caenorhabditis elegans, are used as model organisms for aging studies.
In Saccharomyces cerevisiae, two types of aging can be studied: the replicative and the chronological aging. Yeast cells divide by asymmetric budding, building a daughter cell that is smaller than the mother cell. To analyze replicative aging, the daughter cells should be manually separated from the mother cells. The mother cells go only through a limited number of cell divisions (around 20–25) before they enter a short post-replicative phase and die. It is hypothesized that the replicative aging may reflect aging of proliferating cells in complex organisms. In contrast, the chronological aging may be used as a model for aging of nondividing cells. In chronological aging, also referred to as conditional senescence, the cells are limited in one nutrient and, therefore, leave the cell cycle and stop dividing. The chronological lifespan describes the time of survival of nondividing yeast cells [for a review, see Ref. (104)].
The nematode C. elegans is being used since the 1960s as a model organism in research. Since the genome is completely sequenced (27) and there is the possibility to work with techniques, such as RNAi [86% of the 20,000 genes can be knocked down (86)] and cloning, it is an extensively used model for studying molecular and developmental biology. C. elegans has a maximum lifespan of 20–25 days, which is a great advantage in aging research. Next to the short lifespan, there is a further advantage for studying aging in C. elegans: Since they are self-fertilizing hermaphrodites, there are no inbreeding effects on lifespan, which would lead to potential experimental complications (49). An adult C. elegans consists of exactly 959 somatic cells, all of which are postmitotic. Therefore, C. elegans is frequently used to study age-related cellular alterations (72), such as the accumulation of oxidized proteins. It was shown that with age there is an accumulation of fluorescent material (47, 50), comparable to lipofuscin, which is a marker for accumulation of oxidized protein during human aging (see Pathophysiological Roles of Accumulated and Aggregated Proteins section). For studying oxidation-related aging processes, oxidation-sensitive mutants, for example, mev-1, are often used. mev-1 encodes for a subunit of complex II of mETC; mutation of mev-1 leads to increased mitochondrial ROS generation and to decreased lifespan (49).
To achieve a better comparability to human aging, there are also studies in which mammals, such as rodents (102, 142, 144) and monkeys (118, 144, 163), were used to study the aging process and protein oxidation in aging.
Accumulation of oxidized proteins during aging
In humans, ROS are generated during the whole life, either endogenously or through exposure to exogenous ROS generators. As a consequence, oxidized proteins are also generated constantly during lifetime. Aging is accompanied by accumulation of damaged DNA, misfolded proteins, and oxidized proteins (124, 143, 148). The accumulation of oxidized proteins is limited in proliferating cells due to the fact that permanent cell divisions lead to a dilution of damaged molecules. In contrast, postmitotic cells have the potential to accumulate oxidatively damaged proteins. It is still unclear whether the age-related accumulation of oxidized proteins is a product of increased protein oxidation, decreased degradation of oxidized proteins, or a combination of both mechanisms.
An age-related impaired functionality of the proteasomal system has already been described. Friguet and colleagues showed an age-related increase of post-translational oxidative modifications (HNE and glycoxidaton adducts) of proteasome subunits and decreased peptidase activities in peripheral blood lymphocytes and in human lens cells (18, 21, 155). In addition, some authors described an age-related decline of the expression of proteasomal subunits (19, 88, 97, 130), which finally leads to a decreased proteasome formation. A reduced content of 19S regulators was observed in aged rat muscle cells (41). Thus, the ability of the 19S regulators to bind the 20S core and, therefore, the 26S proteasome formation is decreased. This might result in a decreased protein turnover. Reduced 20S activity, in turn, gives an explanation for increased accumulation of oxidized proteins. In addition, 20S subunits may be oxidized and activity of the 20S proteasome seems to be reduced during aging (26, 88, 114, 130).
The age-related impairment of the lysosomal system, described in “The Mitochondrial-Lysosomal Axis Theory of Aging,” defined by Brunk and Terman (16), seems to be a further explanation for age-related accumulation of oxidized proteins. With aging, there is a dysfunctional regulation of lysosomal pH, decreased lysosomal stability, as a consequence of the high sensitivity of lysosomal membrane toward oxidative damage, and an impaired protein targeting to the lysosomes (31). This may lead to a decreased lysosomal degradation of oxidized proteins. In addition, accompanied with aging, a reduced CMA activity was reported as the result of decreased levels of lysosomal membrane receptors (30, 89). However, probably only a small part of oxidized proteins is degraded by lysosomal proteases. A greater contribution to increased accumulation of oxidized proteins seems to come from the elevated ROS formation due to lysosomal impairment, which is, in particular, a result of age-related lipofuscin accumulation in lysosomes (see Pathophysiological Roles of Accumulated and Aggregated Proteins section).
Pathophysiological roles of accumulated and aggregated proteins
Due to the accumulation of oxidized proteins during aging, there is an extensive formation of protein aggregates over the whole lifespan. As mentioned earlier, these aggregates undergo secondary modifications, for example, due to lipid peroxidation products, resulting in the formation of highly crosslinked oxidized material. One important example is lipofuscin, also called “age-related fluorophore,” “age pigment,” or “ceroid.” It consists of approximately 30%–70% crosslinked proteins and 20%–50% lipids, but carbohydrates were also identified to be a component of lipofuscin (9, 37).
Lipofuscin accumulation is thought to be one of the most important factors limiting the lifespan of a cell (67). Lipofuscin is not degradable by the proteasomal or the lysosomal system. Therefore, with age, lipofuscin accumulates in postmitotic and slowly dividing cells. However, not only the amount of lipofuscin increases during aging, but also the rate of lipofuscin accumulation is positively correlated with the rate of aging (72, 118). Thus, liposfuscin is widely used as a biomarker of aging. Although no specific antibodies against lipofuscin are available, the content of lipofuscin is easy to detect due to its autofluorescence (85).
Liposfuscin accumulates, especially in the lysosomal lumen (84). One possible mechanism of lipofuscin formation in lysosomes was proposed by Brunk and Terman (15). ROS, generated as a consequence of age-related oxidative stress, diffuse into the lysosomes, filled with autophagocytosed material, including protein aggregates (92). Since there is a high amount of iron in lysosomes, released during intralysosomal degradation of metalloproteins, highly reactive HO• are generated via Fenton reaction. These radicals may react with protein aggregates and other macromolecules, which are engulfed in the lysosomes, resulting in crosslinkage and lipofuscin formation. However, low amounts of lipofuscin are also found in the cytosol. Cytosolic lipofuscin may be, on the one hand, a result of lysosomal membrane permeabilization (LMP), which is induced due to a variety of chemicals; for example, ROS, generated in an intralysosomal iron-catalyzed Fenton reaction (5, 129, 151). As a consequence of LMP, lysosomal content is released into the cytosol; this may finally lead to cell death [for a review, see Ref. (12)]. It is hypothesized that the accumulation of lipofuscin may be a possible reason for LMP (15); in fact, accumulation of lipofuscin leads to increased ROS formation. However, currently there is no evidence for this theory.
Although the bulk of lipofuscin is located in the lysosomes, it is not clarified whether it is exclusively generated in the lysosomes or additionally in the cytosol. Höhn et al. have shown that neither macroautophagy nor lysosomal activity is necessary for lipofuscin formation (69). Thus, lipofuscin accumulates in the cytosol after inhibition of the macroautophagy pathway.
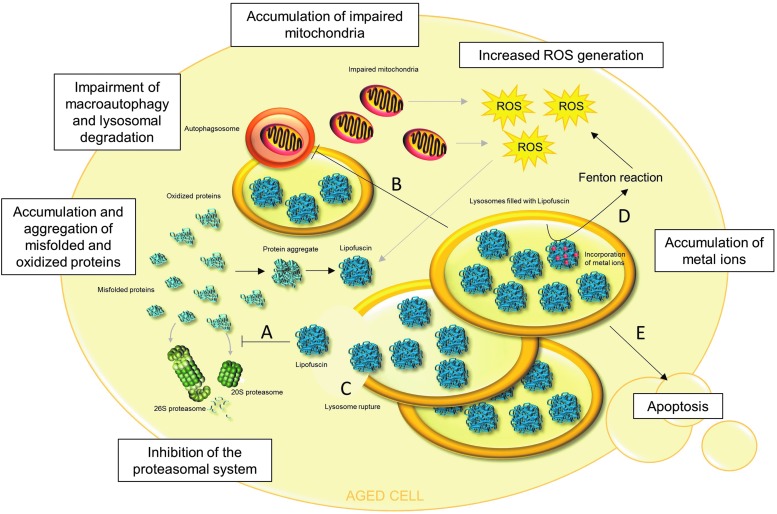
Lipofuscin is able to inhibit the proteasome. Therefore, in the presence of high amounts of lipofuscin, degradation of oxidized proteins is decreased, resulting in further intracellular accumulation of oxidized proteins and their aggregation (Fig. 5). The mechanism of proteasome inhibition was proposed by Höhn et al. (66). The 20S proteasome binds to hydrophobic patches of unfolded oxidized proteins, which are located on the lipofuscin surface. Although degradation is impossible, because of crosslinks in the lipofuscin structure, 20S proteasome remains bound to the surface, repeatedly trying to degrade the structure, and is, therefore, not available for degradation of other substrates. Moreover, impairment of autophagy and lysosomal degradation due to the lysosomal accumulation of lipofuscin is proposed by Brunk and Terman (15) (Fig. 5). As a consequence, this might cause decreased lysosomal degradation of protein aggregates and organelles, such as impaired mitochondria, finally resulting in increased ROS generation, protein oxidation, further aggregation, and more lipofuscin formation (Fig. 5).
FIG. 5.
Pathophysiological roles of lipofuscin in aged cells. In many aged cells, the presence of lipofuscin was observed. Lipofuscin is not an inert waste product of cellular metabolism, but exerts multiple effects. (A) Lipofuscin inhibits the 20S and 26S proteasome, resulting in decreased degradation and further accumulation of oxidized and misfolded proteins. This leads to a facilitation of protein aggregate formation and further lipofuscin formation. (B) It is hypothesized that accumulation of lipofuscin in lysosomes may impair autophagy and lysosomal degradation, which may result in decreased degradation of various proteins, protein aggregates, and organelles. Thus, nonfunctional, impaired mitochondria may not be degraded and accumulate, generating increased amounts of reactive oxygen species (ROS). This leads to further protein oxidation, protein aggregation, and lipofuscin formation. (C) Lysosomal membranes are very susceptible to oxidative damage, which may lead to lysosome rupture. This releases lipofuscin into the cytosol, resulting in the consequences described under (A), (D), and (E). (D) Lipofuscin has the ability to incorporate metal ions, which generate highly reactive ROS via the Fenton reaction. (E) An overall consequence of lipofuscin formation and accumulation is apoptotic cell death.
In addition to proteins, lipids, and carbohydrates, lipofuscin contains approximately 2% of metals, including Fe, Cu, Zn, Al, Mn, and Ca (80). Especially the ability of lipofuscin to incorporate iron was shown to contribute to intracellular ROS generation via Fenton reaction and, therefore, to elevated protein oxidation and lipofuscin formation (67) (Fig. 5).
Cells that are overloaded with cytosolic and lysosomal lipofuscin undergo apoptotic cell death. One possible mechanism by which protein aggregates and, in particular, lipofuscin lead to apoptosis is proposed by Powell et al. (132). Lipofuscin-related inhibition of the proteasomal system inhibits the degradation of proapoptotic proteins, including c-jun, Bax, and p27, leading to the initiation of the apoptotic cascade. In addition, proteasome inhibition blocks activation of NFκB signal transduction. NFκB, usually located in the cytosol, is inhibited by binding of IκB. In cellular stress situations, IκB is phosphorylated, ubiquitinated, and degraded by the 26S proteasome. Afterward, NFκB is translocated into the nucleus where it activates genes, in particular responsible for expression of anti-apoptotic proteins. Therefore, proteasomal inhibition leads to apoptosis (65). Castro et al. have shown that the presence of protein aggregates is sufficient to stop cell division (22), and Höhn et al. have demonstrated the induction of apoptosis due to enhanced cytosolic protein aggregate accumulation (67).
Finally, it is important to note that accumulation of protein aggregates with age is not only caused by oxidative stress. Many other processes lead to the aggregation of proteins, such as an impairment of 26S proteasome and lysosomal system. Another important cause, often involved in age-related neurodegenerative diseases (NDs), could be attributed to mutations, which may lead to abnormal fording of proteins and, therefore, to aggregation.
Protein Oxidation in Age-Related Disease
Protein oxidation in NDs
The nervous system seems to be very susceptible to oxidative damage due to the high oxygen consumption (20%–25% of the oxygen consumption of the whole body). Furthermore, the brain has a high content of polyunsaturated fatty acids, which are very susceptible to oxidation, and of iron, which is responsible for the generation of highly reactive ROS via the Fenton reaction [for a review, see Refs. (42, 48, 109)]. In addition, neurons are postmitotic cells and are, therefore, prone to accumulation of damaged molecules. Thus, various markers of oxidative stress, including the accumulation of oxidatively damaged macromolecules, are often elevated in NDs. These facts suggest a possible involvement of oxidative stress and oxidative damage, including protein oxidation, in the pathogenesis of NDs.
To investigate the role of protein oxidation in NDs, in most cases, post-mortem tissues of patients with NDs were analyzed, showing in general an increased content of protein carbonyls and 3-NT as markers for protein oxidation. Moreover, some studies have demonstrated in detail which proteins are oxidatively damaged (110). Oxidized proteins detected in NDs can be divided into different functional groups: (i) glycolysis and energy metabolism, (ii) mitochondrial proteins, (iii) cytoskeleton, (iv) chaperones, and (v) members of the UPS. The oxidative modification of proteins involved in proteostasis (group iv and v) and their impairment may result in accumulation of nonfunctional proteins. Since NDs are associated with the accumulation of aggregated proteins and protein aggregation is often a result of an imbalance between protein unfolding and the removal of these proteins, the impairment of the proteasomal system seems to play a major role in development of NDs.
Alzheimer's disease
Alzheimer's disease (AD) is an irreversible progressive brain disease that is characterized by progressive loss of memory and other cognitive functions (74). AD is the main reason for dementia in older people, and it mostly occurs after an age of 65. The formation of amyloid plaques (APs) and neurofibrillary tangles (NFTs) is characteristic for the disease. APs and NFTs are products of the accumulations of abnormal folded and aggregated beta-amyloid (Aβ) and hyperphosphorylated tau proteins (75).
Tau, the term derived from “tubulin associated unit,” is a protein that is highly expressed in the human brain and which functions in binding and stabilization of microtubules (91, 107). Phosphorylation ensures the biological activity of tau. Tau turnover is catalyzed by the 20S proteasome (54). However, in AD, tau undergoes hyperphosphorylation, which reduces the proteasomal susceptibility of the protein (131). Hyperphosphorylation results in conformational changes of tau. It twists into pairs of helical filaments, which again twist into tangles. Thus, NFTs consist of aggregated and hyperphosphorylated tau proteins, displaying one of the most important markers for AD. Since microtubules are not able to function correctly, aggregation of tau finally leads to an impaired neuronal transport (17, 91).
APs, also called senile plaques, display a further important histological marker of AD. APs consist mainly of aggregated Aβ, and emerge due to an alternative enzymatic cleavage of the amyloid precursor protein (APP). Usually, α- and γ-secretase cleave the APP into the short fragments sAPPα and P3. In AD, the cleavage is carried out by the β- and γ-secretase, leading to the generation of the sAPPβ and Aβ. The latter accumulates in the extracellular space between the neurons. It is known that the formation of APs may be promoted by mutations of three different genes: APP, PS1, and PS2. The transmembrane proteins presenilin-1 (PS1) and presenilin-2 (PS2) are a part of the γ-secretase complex and are involved in the activity of the secretase.
There is evidence supporting the role of oxidative stress in AD, including increased levels of markers for DNA oxidation, lipid peroxidation, and protein oxidation as well as altered activities and expression of antioxidant enzymes (164). Especially the fact that oxidative stress is increased already in the stage of mild cognitive impairment, an early stage of dementia, may indicate a crucial role of oxidative stress during initiation and development of AD (133). It was shown that APs contribute to the formation of oxidative stress in AD brains. For example, Aβ is able to interact with Cu2+ and Fe3+ via its metal-binding domain and it catalyzes the reduction of Cu2+ and Fe3+ and the generation of H2O2 (78, 126), which may be converted to HO• in a Fenton reaction. As mentioned earlier, in AD brains, there are increased levels of protein carbonyls and 3-NT (3, 61, 143), which serve as reliable markers for protein oxidation. Especially in AD hippocampus and inferior parietal lobule, regions that are rich in APs, protein carbonyl levels were significantly higher than in the same regions of control brains (61).
Parkinson's disease
Parkinson's disease (PD) is a movement disorder, associated with a progressive loss of dopaminergic neurons in the substantia nigra pars compacta and with a decrease of neurons in the locus coeruleus. It is the second most prevalent ND after AD. PD is associated with the occurrence of intraneuronal cytoplasmic inclusion bodies, referred to as Lewy bodies. Components of these Lewy bodies are synphilin-1, α-synuclein, parkin, members of the UPS, and others. Although α-synuclein is the major constituent of Lewy bodies, its function is not known. It is highly expressed in the brain and is associated with membrane and vesicular structures of presynaptic terminals (157). Thus, it may be involved in regulation of dopamine release (1). α-synuclein may form oligomeric and amyloid fibrils, which represents the major component of the Lewy bodies. Synphilin-1 is an α-synuclein binding protein. Parkin is an ubiquitin E3 ligase that seems to be involved in turnover of α-synuclein. In most cases, PD occurs sporadic; however, there are some cases of PD that are familial linked. It has been shown that missense mutations in α-synuclein genes, as well as gene duplications and triplications increase the aggregation of α-synuclein and may cause familial PD. PD is characterized by a decreased activity of mitochondrial complex I, which leads to an increased mitochondrial ROS generation and elevated levels of oxidatively damaged lipids, DNA, and proteins. Thus, there is evidence for the role of oxidative stress in pathogenesis of PD.
It is believed that decreased activity of mitochondrial complex I may contribute to aggregation of α-synuclein (10, 108). Furthermore, nitration of tyrosine residues of α-synuclein may play a role in α-synuclein aggregation and its deposition in Lewy bodies (51, 64). Other oxidative modifications are also known to contribute to α-synuclein aggregation and pathogenesis of PD. For example, oxidation of dopamine leads to the generation of oxidized species, such as dopaminochrome, which may interact with α-synuclein. These dopamine adducts on α-synuclein are able to stabilize the toxic α-synuclein protofibrils and inhibit the further aggregation into less toxic mature fibrils (112, 123).
Huntington's disease
Huntington's disease (HD) is an autosomal dominant inherited ND, characterized by cognitive impairment, neuropsychiatric symptoms, and premature death. Further symptoms, which are often associated, are motor skill impairments, such as uncontrollable twitching and spasms. HD is associated with aggregation of the protein Huntingtin. This aggregation is a result of a mutation-driven prolongation of a CAG-motive in the Huntington gene. These abnormal Huntingtin proteins tend to aggregate, leading to the formation of inclusion bodies. Similar to other NDs, markers of oxidative stress are also increased in HD (13). For example, elevated levels of 3-NT in the cortex and striatum have been shown (14).
Protein oxidation in other age-related diseases
Atherosclerosis
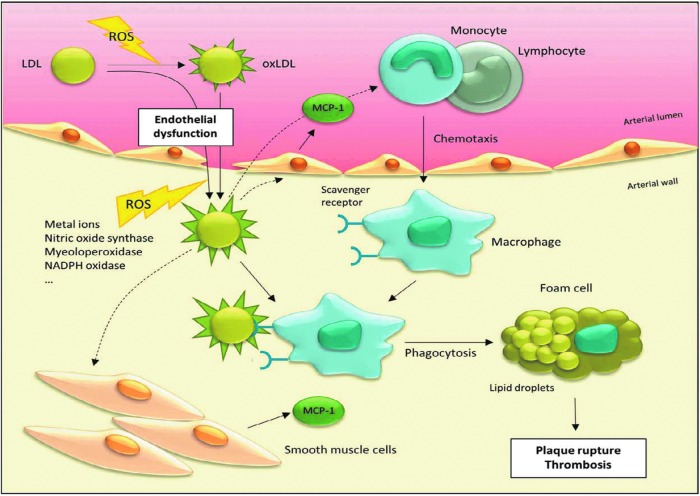
Atherosclerosis is a chronic inflammatory disease that is characterized by the accumulation of modified low-density lipoproteins (LDL) in the arterial wall, which leads to proliferation of different cell types. It is one of the major causes for mortality in the Western world (149). The risk to become diseased increases with age. Other risk factors are, for example, increased plasma cholesterol levels, smoking, hypertension, and diabetes. The pathogenesis of atherosclerosis is initiated by an endothelial dysfunction of arteries, resulting in an extravasation of (modified) LDLs, responsible for cholesterol and triglycerides transport, into the subendothelial space (Fig. 6). These LDLs become further modified, for example, by oxidation. Expression of adhesion molecule VCAM-1 is increased, and monocytes and lymphocytes are recruited. Monocytes differentiate into macrophages. Phagocytosis of modified LDLs by macrophages results in intracellular lipid accumulation and foam cell formation, what appears as a “fatty streak.” Furthermore, smooth muscle cells, which are usually localized in other layers of the artery wall (media and adventitia), proliferate and migrate into the intima, forming a fibrous capsule over the “fatty streak.” This process can progress unnoticed for years until the atherosclerotic plaque becomes ruptured, exposing the core of the plaque in the arterial lumen, leading to fatal or nonfatal thrombosis (40, 149).
FIG. 6.
Role of oxidized low-density lipoprotein (oxLDL) in development of atherosclerosis. Due to an endothelial dysfunction, (modified) low-density lipoprotein (LDL) immigrates from arterial lumen into the arterial wall. In the arterial wall, ROS are generated due to a number of enzymes, including nitric oxide synthase, myeloperoxidase, and NADPH oxidase, and via metal ion-catalyzed Fenton reaction, what results in LDL oxidation (oxLDL). Furthermore, it is still a matter of debate whether circulating oxLDL may also immigrate into the arterial wall. OxLDL stimulates endothelial cells and smooth muscle cells to secrete monocyte chemotactic protein-1 (MCP-1). MCP-1 stimulates arterial monocytes and lymphocytes to immigrate into the arterial wall, where monocytes are converted to macrophages. These macrophages have specific scavenger receptors for oxLDL. Phagocytosis of oxLDL contributes to foam cell formation, which results in plaque formation and thrombosis.
The oxidation of LDL is thought to play a central role in development of artherosclerosis (Fig. 6), although it is widely unknown where, how, and to what extent oxidized LDL (oxLDL) is generated during atherogenesis. It was shown that oxLDL is involved in recruitment of monocytes and lymphocytes (113, 134); for example, via stimulating endothelial cells and smooth muscle cells to secrete monocyte chemotactic protein-1 (MCP-1) (Fig. 6) (120, 136). Furthermore, oxLDL is involved in foam cell formation (103, 116). One major source for ROS that is responsible for subendothelial oxidation processes is the NADPH oxidase complex of leukocytes, which converts O2 into O2•−. Studies with mice deficient in NADPH oxidase complex activity show either no changes or a decrease in lesion development (7, 90). Other ROS generators in the arterial wall are enzymes, such as xanthine oxidase, nitric oxide synthase, myeoloperoxidase and liopxygenase, and redox-active metal ions (156) (Fig. 6). LDL, oxidized by HO•, exhibits increased levels of ortho-tyrosine and meta-tyrosine, products of phenylalanine residue oxidation. Increased levels of these oxidative modifications of LDL were not found in early atherosclerosis, but in advanced atherosclerosis (99). NO is generated by endothelial cells and nitric oxide synthase, and it mediates a variety of potentially anti-atherogenic effects. However, the highly reactive ONOO−, a product of a reaction of NO and O2•−, generates 3-NT modifications on LDL (98). Myeloperoxidase generates HOCl, which also may react with tyrosine residues, generating 3-chlorotyrosine. LDLs of atherosclerotic plaques contain elevated levels of 3-NT and 3-chorotyrosine (46). Furthermore, these oxidative protein modifications seem to contribute to foam cell formation (53, 60).
These results show a probable participation of protein oxidation in the conversion of native LDL into oxLDL and thus in foam cell formation and development of atherosclerosis. However, it is important to mention that oxLDL is not only a result of protein oxidation; there are also ongoing lipid peroxidation processes, leading to modification of LDL apoproteins (149).
Age-related macular degeneration
Age-related macular degeneration (AMD) is characterized by the progessive loss of vision, which results from the degeneration of the macula. The macula, consisting of rods and cones, is a part of the retina, that functions in high resolution and color vision (161). Similar to other age-related diseases, markers of oxidative stress, including oxidative damage to DNA, lipids, and proteins, are also increased in AMD. It is postulated that the cumulative oxidative damage may contribute to pathogenesis of AMD (8). The retina seems to be very susceptible to oxidative stress. Besides the exposure to visible light, there is a high consumption of oxygen and a high content of polyunsaturated fatty acids, comparable to neuronal tissues. Oxidative protein modifications were found in retina (139) and drusen (28). Drusen are accumulations of extracellular material in the macula. The increased levels of drusen in the macula are correlated with AMD. Analyses of drusen from AMD patients show, in particular, an increased occurrence of protein crosslinks, which are a result of reactions with products of lipid peroxidation and glycation/glycoxidation (28). Furthermore, there is evidence indicating the connection between the development of AMD and the occurrence of lipofuscin. Lipofuscin accumulates in the lysosomes of the retinal pigment epithelium (RPE). RPE cells are responsible for phagocytosis and lysosomal degradation of the discs. In this degradation process, the fluorophore A2E is generated, which accumulates in the lysosomes and represents a preliminary stage of lipofuscin formation and a component of lipofuscin in the macula (161). It was demonstrated that humans with Stargardt disease, which accumulate A2E in the RPE, show degeneration of the macula and loss of vision, similar to AMD (111, 135).
Conclusion
The investigation of the role of potentially contributing factors to the aging process is a very difficult task, as aging is a complex process involving a whole array of contributing factors. However, a multitude of data indicate that protein oxidation, accumulation of oxidized proteins, and protein aggregation as well as the impairment of the proteasomal system play a major role in the aging process and in the development of some, if not all, age-related diseases. In particular, the accumulation of lipofuscin during aging is an indication of the involvement of protein oxidation in the aging process. More importantly, the accumulation of lipofuscin has serious consequences for cell functionality and metabolism and is an important factor determining the lifespan of a cell. Accumulation and aggregation of oxidized proteins and impairment of the proteasomal system are two mechanisms that are interdependent. It remains to be clarified whether an early intervention on the process of oxidation-related protein aggregation would potentially slow down the aging process or the progression of diseases related to aging.
Abbreviations Used
- 3-NT
3-nitrotyrosine
- AD
Alzheimer's disease
- AGE
advanced glycation end product
- ALE
advanced lipoxidation end product
- AMD
age-related macular degeneration
- AP
amyloid plaque
- APP
amyloid precursor protein
- Aβ
beta-amyloid
- CMA
chaperon-mediated autophagy
- H2O2
hydrogen peroxide
- HD
Huntington's disease
- HDAC6
histone deacetylase 6
- HNE
4-hydroxynonenal
- HSP
heat shock protein
- IκB
inhibitor of NFκB
- LDL
low-density lipoprotein
- LMP
lysosomal membrane permeabilization
- MCP-1
monocyte chemotactic protein-1
- MeSO
methionine sulfoxide
- mETC
mitochondrial electron transport chain
- MSR
methionine sulfoxide reductase
- MTOC
microtubule organization center
- ND
neurodegenerative disease
- NFκB
nuclear factor κB
- NFT
neurofibrillary tangle
- NO
nitric oxide
- oxLDL
oxidized low-density lipoprotein
- PD
Parkinson's disease
- PDI
protein disulfide isomerase
- PS1
presenilin-1
- PS2
presenilin-2
- PTP
protein tyrosine phosphatase
- ROS
reactive oxygen species
- RPE
retinal pigment epithelium
- SIPS
stress-induced premature senescence
- Trx
thioredoxin
- TrxR
thioredoxin reductase
- UPS
ubiquitin-proteasome system
References
- 1.Abeliovich A, Schmitz Y, Farinas I, Choi-Lundberg D, Ho WH, Castillo PE, Shinsky N, Verdugo JM, Armanini M, Ryan A, Hynes M, Phillips H, Sulzer D, and Rosenthal A. Mice lacking alpha-synuclein display functional deficits in the nigrostriatal dopamine system. Neuron 25: 239–252, 2000 [DOI] [PubMed] [Google Scholar]
- 2.Aeschbach R, Amado R, and Neukom H. Formation of dityrosine cross-links in proteins by oxidation of tyrosine residues. Biochim Biophys Acta 439: 292–301, 1976 [DOI] [PubMed] [Google Scholar]
- 3.Aksenov MY, Aksenova MV, Butterfield DA, Geddes JW, and Markesbery WR. Protein oxidation in the brain in Alzheimer's disease. Neuroscience 103: 373–383, 2001 [DOI] [PubMed] [Google Scholar]
- 4.Alberti S, Demand J, Esser C, Emmerich N, Schild H, and Hohfeld J. Ubiquitylation of BAG-1 suggests a novel regulatory mechanism during the sorting of chaperone substrates to the proteasome. J Biol Chem 277: 45920–45927, 2002 [DOI] [PubMed] [Google Scholar]
- 5.Antunes F, Cadenas E, and Brunk UT. Apoptosis induced by exposure to a low steady-state concentration of H2O2 is a consequence of lysosomal rupture. Biochem J 356: 549–555, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, Kirkland JL, and van Deursen JM. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 479: 232–236, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barry-Lane PA, Patterson C, van der Merwe M, Hu Z, Holland SM, Yeh ET, and Runge MS. p47phox is required for atherosclerotic lesion progression in ApoE(-/-) mice. J Clin Invest 108: 1513–1522, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beatty S, Koh H, Phil M, Henson D, and Boulton M. The role of oxidative stress in the pathogenesis of age-related macular degeneration. Surv Ophthalmol 45: 115–134, 2000 [DOI] [PubMed] [Google Scholar]
- 9.Benavides SH, Monserrat AJ, Farina S, and Porta EA. Sequential histochemical studies of neuronal lipofuscin in human cerebral cortex from the first to the ninth decade of life. Arch Gerontol Geriatr 34: 219–231, 2002 [DOI] [PubMed] [Google Scholar]
- 10.Betarbet R, Sherer TB, MacKenzie G, Garcia-Osuna M, Panov AV, and Greenamyre JT. Chronic systemic pesticide exposure reproduces features of Parkinson's disease. Nat Neurosci 3: 1301–1306, 2000 [DOI] [PubMed] [Google Scholar]
- 11.Bindoli A, Fukuto JM, and Forman HJ. Thiol chemistry in peroxidase catalysis and redox signaling. Antioxid Redox Signal 10: 1549–1564, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boya P. and Kroemer G. Lysosomal membrane permeabilization in cell death. Oncogene 27: 6434–6451, 2008 [DOI] [PubMed] [Google Scholar]
- 13.Browne SE. and Beal MF. Oxidative damage in Huntington's disease pathogenesis. Antioxid Redox Signal 8: 2061–2073, 2006 [DOI] [PubMed] [Google Scholar]
- 14.Browne SE, Bowling AC, MacGarvey U, Baik MJ, Berger SC, Muqit MM, Bird ED, and Beal MF. Oxidative damage and metabolic dysfunction in Huntington's disease: selective vulnerability of the basal ganglia. Ann Neurol 41: 646–653, 1997 [DOI] [PubMed] [Google Scholar]
- 15.Brunk UT. and Terman A. Lipofuscin: mechanisms of age-related accumulation and influence on cell function. Free Radic Biol Med 33: 611–619, 2002 [DOI] [PubMed] [Google Scholar]
- 16.Brunk UT. and Terman A. The mitochondrial-lysosomal axis theory of aging: accumulation of damaged mitochondria as a result of imperfect autophagocytosis. Eur J Biochem 269: 1996–2002, 2002 [DOI] [PubMed] [Google Scholar]
- 17.Buee L, Bussiere T, Buee-Scherrer V, Delacourte A, and Hof PR. Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res Brain Res Rev 33: 95–130, 2000 [DOI] [PubMed] [Google Scholar]
- 18.Bulteau AL, Petropoulos I, and Friguet B. Age-related alterations of proteasome structure and function in aging epidermis. Exp Gerontol 35: 767–777, 2000 [DOI] [PubMed] [Google Scholar]
- 19.Bulteau AL, Szweda LI, and Friguet B. Age-dependent declines in proteasome activity in the heart. Arch Biochem Biophys 397: 298–304, 2002 [DOI] [PubMed] [Google Scholar]
- 20.Campisi J. and d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol 8: 729–740, 2007 [DOI] [PubMed] [Google Scholar]
- 21.Carrard G, Dieu M, Raes M, Toussaint O, and Friguet B. Impact of ageing on proteasome structure and function in human lymphocytes. Int J Biochem Cell Biol 35: 728–739, 2003 [DOI] [PubMed] [Google Scholar]
- 22.Castro JP, Ott C, Jung T, Grune T, and Almeida H. Carbonylation of the cytoskeletal protein actin leads to aggregate formation. Free Radic Biol Med 53: 916–925, 2012 [DOI] [PubMed] [Google Scholar]
- 23.Chen Q. and Ames BN. Senescence-like growth arrest induced by hydrogen peroxide in human diploid fibroblast F65 cells. Proc Natl Acad Sci U S A 91: 4130–4134, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collet JF. and Messens J. Structure, function, and mechanism of thioredoxin proteins. Antioxid Redox Signal 13: 1205–1216, 2010 [DOI] [PubMed] [Google Scholar]
- 25.Conconi M, Petropoulos I, Emod I, Turlin E, Biville F, and Friguet B. Protection from oxidative inactivation of the 20S proteasome by heat-shock protein 90. Biochem J 333 (Pt 2): 407–415, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conconi M, Szweda LI, Levine RL, Stadtman ER, and Friguet B. Age-related decline of rat liver multicatalytic proteinase activity and protection from oxidative inactivation by heat-shock protein 90. Arch Biochem Biophys 331: 232–240, 1996 [DOI] [PubMed] [Google Scholar]
- 27.Consortium CeS. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282: 2012–2018, 1998 [DOI] [PubMed] [Google Scholar]
- 28.Crabb JW, Miyagi M, Gu X, Shadrach K, West KA, Sakaguchi H, Kamei M, Hasan A, Yan L, Rayborn ME, Salomon RG, and Hollyfield JG. Drusen proteome analysis: an approach to the etiology of age-related macular degeneration. Proc Natl Acad Sci U S A 99: 14682–14687, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuervo AM. and Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science 273: 501–503, 1996 [DOI] [PubMed] [Google Scholar]
- 30.Cuervo AM. and Dice JF. Age-related decline in chaperone-mediated autophagy. J Biol Chem 275: 31505–31513, 2000 [DOI] [PubMed] [Google Scholar]
- 31.Cuervo AM. and Dice JF. When lysosomes get old. Exp Gerontol 35: 119–131, 2000 [DOI] [PubMed] [Google Scholar]
- 32.Cuervo AM. and Wong E. Chaperone-mediated autophagy: roles in disease and aging. Cell Res 24: 92–104, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dalle-Donne I, Rossi R, Giustarini D, Milzani A, and Colombo R. Protein carbonyl groups as biomarkers of oxidative stress. Clin Chim Acta 329: 23–38, 2003 [DOI] [PubMed] [Google Scholar]
- 34.Davies KJ. Degradation of oxidized proteins by the 20S proteasome. Biochimie 83: 301–310, 2001 [DOI] [PubMed] [Google Scholar]
- 35.Davies MJ. Protein and peptide alkoxyl radicals can give rise to C-terminal decarboxylation and backbone cleavage. Arch Biochem Biophys 336: 163–172, 1996 [DOI] [PubMed] [Google Scholar]
- 36.Davies MJ. The oxidative environment and protein damage. Biochim Biophys Acta 1703: 93–109, 2005 [DOI] [PubMed] [Google Scholar]
- 37.Double KL, Dedov VN, Fedorow H, Kettle E, Halliday GM, Garner B, and Brunk UT. The comparative biology of neuromelanin and lipofuscin in the human brain. Cell Mol Life Sci 65: 1669–1682, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drazic A. and Winter J. The physiological role of reversible methionine oxidation. Biochim Biophys Acta 1844: 1367–1382, 2014 [DOI] [PubMed] [Google Scholar]
- 39.Erickson JR, Joiner ML, Guan X, Kutschke W, Yang J, Oddis CV, Bartlett RK, Lowe JS, O'Donnell SE, Aykin-Burns N, Zimmerman MC, Zimmerman K, Ham AJ, Weiss RM, Spitz DR, Shea MA, Colbran RJ, Mohler PJ, and Anderson ME. A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation. Cell 133: 462–474, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Falk E. Pathogenesis of atherosclerosis. J Am Coll Cardiol 47: C7–C12, 2006 [DOI] [PubMed] [Google Scholar]
- 41.Ferrington DA, Husom AD, and Thompson LV. Altered proteasome structure, function, and oxidation in aged muscle. FASEB J 19: 644–646, 2005 [DOI] [PubMed] [Google Scholar]
- 42.Floyd RA. and Carney JM. Free radical damage to protein and DNA: mechanisms involved and relevant observations on brain undergoing oxidative stress. Ann Neurol 32 Suppl: S22–S27, 1992 [DOI] [PubMed] [Google Scholar]
- 43.Forman HJ, Maiorino M, and Ursini F. Signaling functions of reactive oxygen species. Biochemistry 49: 835–842, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fortun J, Dunn WA, Jr., Joy S, Li J, and Notterpek L. Emerging role for autophagy in the removal of aggresomes in Schwann cells. J Neurosci 23: 10672–10680, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Friguet B. and Szweda LI. Inhibition of the multicatalytic proteinase (proteasome) by 4-hydroxy-2-nonenal cross-linked protein. FEBS Lett 405: 21–25, 1997 [DOI] [PubMed] [Google Scholar]
- 46.Fu S, Davies MJ, Stocker R, and Dean RT. Evidence for roles of radicals in protein oxidation in advanced human atherosclerotic plaque. Biochem J 333 (Pt 3): 519–525, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, and Kenyon C. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics 161: 1101–1112, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gemma C, Vila J, Bachstetter A, and Bickford PC. Oxidative stress and the aging brain: from theory to prevention. In: Brain Aging: Models, Methods, and Mechanisms, edited by Riddle DR. Boca Raton, FL: CRC Press, 2007, pp. 353–374 [PubMed] [Google Scholar]
- 49.Gems D. Ageing and oxidants in the nematode Caenorhabditis elegans. SEB Exp Biol Ser 62: 31–56, 2009 [PubMed] [Google Scholar]
- 50.Gerstbrein B, Stamatas G, Kollias N, and Driscoll M. In vivo spectrofluorimetry reveals endogenous biomarkers that report healthspan and dietary restriction in Caenorhabditis elegans. Aging Cell 4: 127–137, 2005 [DOI] [PubMed] [Google Scholar]
- 51.Giasson BI, Duda JE, Murray IV, Chen Q, Souza JM, Hurtig HI, Ischiropoulos H, Trojanowski JQ, and Lee VM. Oxidative damage linked to neurodegeneration by selective alpha-synuclein nitration in synucleinopathy lesions. Science 290: 985–989, 2000 [DOI] [PubMed] [Google Scholar]
- 52.Giulivi C, Pacifici RE, and Davies KJ. Exposure of hydrophobic moieties promotes the selective degradation of hydrogen peroxide-modified hemoglobin by the multicatalytic proteinase complex, proteasome. Arch Biochem Biophys 311: 329–341, 1994 [DOI] [PubMed] [Google Scholar]
- 53.Graham A, Hogg N, Kalyanaraman B, O'Leary V, Darley-Usmar V, and Moncada S. Peroxynitrite modification of low-density lipoprotein leads to recognition by the macrophage scavenger receptor. FEBS Lett 330: 181–185, 1993 [DOI] [PubMed] [Google Scholar]
- 54.Grune T, Botzen D, Engels M, Voss P, Kaiser B, Jung T, Grimm S, Ermak G, and Davies KJ. Tau protein degradation is catalyzed by the ATP/ubiquitin-independent 20S proteasome under normal cell conditions. Arch Biochem Biophys 500: 181–188, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grune T, Catalgol B, Licht A, Ermak G, Pickering AM, Ngo JK, and Davies KJ. HSP70 mediates dissociation and reassociation of the 26S proteasome during adaptation to oxidative stress. Free Radic Biol Med 51: 1355–1364, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grune T, Reinheckel T, and Davies KJ. Degradation of oxidized proteins in mammalian cells. FASEB J 11: 526–534, 1997 [PubMed] [Google Scholar]
- 57.Grune T, Reinheckel T, Joshi M, and Davies KJ. Proteolysis in cultured liver epithelial cells during oxidative stress. Role of the multicatalytic proteinase complex, proteasome. J Biol Chem 270: 2344–2351, 1995 [DOI] [PubMed] [Google Scholar]
- 58.Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol 11: 298–300, 1956 [DOI] [PubMed] [Google Scholar]
- 59.Hayflick L. and Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res 25: 585–621, 1961 [DOI] [PubMed] [Google Scholar]
- 60.Hazell LJ. and Stocker R. Oxidation of low-density lipoprotein with hypochlorite causes transformation of the lipoprotein into a high-uptake form for macrophages. Biochem J 290 (Pt 1): 165–172, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hensley K, Hall N, Subramaniam R, Cole P, Harris M, Aksenov M, Aksenova M, Gabbita SP, Wu JF, Carney JM, et al. Brain regional correspondence between Alzheimer's disease histopathology and biomarkers of protein oxidation. J Neurochem 65: 2146–2156, 1995 [DOI] [PubMed] [Google Scholar]
- 62.Herce-Pagliai C, Kotecha S, and Shuker DE. Analytical methods for 3-nitrotyrosine as a marker of exposure to reactive nitrogen species: a review. Nitric Oxide 2: 324–336, 1998 [DOI] [PubMed] [Google Scholar]
- 63.Hipkiss AR. Accumulation of altered proteins and ageing: causes and effects. Exp Gerontol 41: 464–473, 2006 [DOI] [PubMed] [Google Scholar]
- 64.Hodara R, Norris EH, Giasson BI, Mishizen-Eberz AJ, Lynch DR, Lee VM, and Ischiropoulos H. Functional consequences of alpha-synuclein tyrosine nitration: diminished binding to lipid vesicles and increased fibril formation. J Biol Chem 279: 47746–47753, 2004 [DOI] [PubMed] [Google Scholar]
- 65.Höhn A. and Grune T. Lipofuscin: formation, effects and role of macroautophagy. Redox Biol 1: 140–144, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Höhn A, Jung T, Grimm S, Catalgol B, Weber D, and Grune T. Lipofuscin inhibits the proteasome by binding to surface motifs. Free Radic Biol Med 50: 585–591, 2011 [DOI] [PubMed] [Google Scholar]
- 67.Höhn A, Jung T, Grimm S, and Grune T. Lipofuscin-bound iron is a major intracellular source of oxidants: role in senescent cells. Free Radic Biol Med 48: 1100–1108, 2010 [DOI] [PubMed] [Google Scholar]
- 68.Höhn A, Konig J, and Grune T. Protein oxidation in aging and the removal of oxidized proteins. J Proteomics 92: 132–159, 2013 [DOI] [PubMed] [Google Scholar]
- 69.Höhn A, Sittig A, Jung T, Grimm S, and Grune T. Lipofuscin is formed independently of macroautophagy and lysosomal activity in stress-induced prematurely senescent human fibroblasts. Free Radic Biol Med 53: 1760–1769, 2012 [DOI] [PubMed] [Google Scholar]
- 70.Hornsby PJ. Cellular senescence and tissue aging in vivo. J Gerontol A Biol Sci Med Sci 57: B251–B256, 2002 [DOI] [PubMed] [Google Scholar]
- 71.Hoshi T. and Heinemann S. Regulation of cell function by methionine oxidation and reduction. J Physiol 531: 1–11, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hosokawa H, Ishii N, Ishida H, Ichimori K, Nakazawa H, and Suzuki K. Rapid accumulation of fluorescent material with aging in an oxygen-sensitive mutant mev-1 of Caenorhabditis elegans. Mech Ageing Dev 74: 161–170, 1994 [DOI] [PubMed] [Google Scholar]
- 73.Howe CJ, Lahair MM, McCubrey JA, and Franklin RA. Redox regulation of the calcium/calmodulin-dependent protein kinases. J Biol Chem 279: 44573–44581, 2004 [DOI] [PubMed] [Google Scholar]
- 74.Huang Y. and Mucke L. Alzheimer mechanisms and therapeutic strategies. Cell 148: 1204–1222, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Irvine GB, El-Agnaf OM, Shankar GM, and Walsh DM. Protein aggregation in the brain: the molecular basis for Alzheimer's and Parkinson's diseases. Mol Med 14: 451–464, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jahreiss L, Menzies FM, and Rubinsztein DC. The itinerary of autophagosomes: from peripheral formation to kiss-and-run fusion with lysosomes. Traffic 9: 574–587, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jeyapalan JC, Ferreira M, Sedivy JM, and Herbig U. Accumulation of senescent cells in mitotic tissue of aging primates. Mech Ageing Dev 128: 36–44, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiang D, Li X, Williams R, Patel S, Men L, Wang Y, and Zhou F. Ternary complexes of iron, amyloid-beta, and nitrilotriacetic acid: binding affinities, redox properties, and relevance to iron-induced oxidative stress in Alzheimer's disease. Biochemistry 48: 7939–7947, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Johnston JA, Ward CL, and Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol 143: 1883–1898, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jolly RD, Douglas BV, Davey PM, and Roiri JE. Lipofuscin in bovine muscle and brain: a model for studying age pigment. Gerontology 41 Suppl 2: 283–295, 1995 [DOI] [PubMed] [Google Scholar]
- 81.Jung T, Catalgol B, and Grune T. The proteasomal system. Mol Aspects Med 30: 191–296, 2009 [DOI] [PubMed] [Google Scholar]
- 82.Jung T. and Grune T. The proteasome and its role in the degradation of oxidized proteins. IUBMB Life 60: 743–752, 2008 [DOI] [PubMed] [Google Scholar]
- 83.Jung T. and Grune T. The proteasome and the degradation of oxidized proteins: Part I-structure of proteasomes. Redox Biol 1: 178–182, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jung T, Hohn A, Catalgol B, and Grune T. Age-related differences in oxidative protein-damage in young and senescent fibroblasts. Arch Biochem Biophys 483: 127–135, 2009 [DOI] [PubMed] [Google Scholar]
- 85.Jung T, Hohn A, and Grune T. Lipofuscin: detection and quantification by microscopic techniques. Methods Mol Biol 594: 173–193, 2010 [DOI] [PubMed] [Google Scholar]
- 86.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, and Ahringer J. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237, 2003 [DOI] [PubMed] [Google Scholar]
- 87.Kastle M. and Grune T. Proteins bearing oxidation-induced carbonyl groups are not preferentially ubiquitinated. Biochimie 93: 1076–1079, 2011 [DOI] [PubMed] [Google Scholar]
- 88.Keller JN, Huang FF, and Markesbery WR. Decreased levels of proteasome activity and proteasome expression in aging spinal cord. Neuroscience 98: 149–156, 2000 [DOI] [PubMed] [Google Scholar]
- 89.Kiffin R, Christian C, Knecht E, and Cuervo AM. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell 15: 4829–4840, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kirk EA, Dinauer MC, Rosen H, Chait A, Heinecke JW, and LeBoeuf RC. Impaired superoxide production due to a deficiency in phagocyte NADPH oxidase fails to inhibit atherosclerosis in mice. Arterioscler Thromb Vasc Biol 20: 1529–1535, 2000 [DOI] [PubMed] [Google Scholar]
- 91.Kolarova M, Garcia-Sierra F, Bartos A, Ricny J, and Ripova D. Structure and pathology of tau protein in Alzheimer disease. Int J Alzheimers Dis 2012: 731526, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol 10: 524–530, 2000 [DOI] [PubMed] [Google Scholar]
- 93.Kriegenburg F, Ellgaard L, and Hartmann-Petersen R. Molecular chaperones in targeting misfolded proteins for ubiquitin-dependent degradation. FEBS J 279: 532–542, 2012 [DOI] [PubMed] [Google Scholar]
- 94.Lamark T. and Johansen T. Aggrephagy: selective disposal of protein aggregates by macroautophagy. Int J Cell Biol 2012: 736905, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lasch P, Petras T, Ullrich O, Backmann J, Naumann D, and Grune T. Hydrogen peroxide-induced structural alterations of RNAse A. J Biol Chem 276: 9492–9502, 2001 [DOI] [PubMed] [Google Scholar]
- 96.Lecker SH, Goldberg AL, and Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol 17: 1807–1819, 2006 [DOI] [PubMed] [Google Scholar]
- 97.Lee CK, Klopp RG, Weindruch R, and Prolla TA. Gene expression profile of aging and its retardation by caloric restriction. Science 285: 1390–1393, 1999 [DOI] [PubMed] [Google Scholar]
- 98.Leeuwenburgh C, Hardy MM, Hazen SL, Wagner P, Oh-ishi S, Steinbrecher UP, and Heinecke JW. Reactive nitrogen intermediates promote low density lipoprotein oxidation in human atherosclerotic intima. J Biol Chem 272: 1433–1436, 1997 [DOI] [PubMed] [Google Scholar]
- 99.Leeuwenburgh C, Rasmussen JE, Hsu FF, Mueller DM, Pennathur S, and Heinecke JW. Mass spectrometric quantification of markers for protein oxidation by tyrosyl radical, copper, and hydroxyl radical in low density lipoprotein isolated from human atherosclerotic plaques. J Biol Chem 272: 3520–3526, 1997 [DOI] [PubMed] [Google Scholar]
- 100.Levine RL, Moskovitz J, and Stadtman ER. Oxidation of methionine in proteins: roles in antioxidant defense and cellular regulation. IUBMB Life 50: 301–307, 2000 [DOI] [PubMed] [Google Scholar]
- 101.Liochev SI. Reactive oxygen species and the free radical theory of aging. Free Radic Biol Med 60: 1–4, 2013 [DOI] [PubMed] [Google Scholar]
- 102.Liu R, Liu IY, Bi X, Thompson RF, Doctrow SR, Malfroy B, and Baudry M. Reversal of age-related learning deficits and brain oxidative stress in mice with superoxide dismutase/catalase mimetics. Proc Natl Acad Sci U S A 100: 8526–8531, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu W, Yin Y, Zhou Z, He M, and Dai Y. OxLDL-induced IL-1 beta secretion promoting foam cells formation was mainly via CD36 mediated ROS production leading to NLRP3 inflammasome activation. Inflamm Res 63: 33–43, 2014 [DOI] [PubMed] [Google Scholar]
- 104.Longo VD, Shadel GS, Kaeberlein M, and Kennedy B. Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab 16: 18–31, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lowther WT, Brot N, Weissbach H, Honek JF, and Matthews BW. Thiol-disulfide exchange is involved in the catalytic mechanism of peptide methionine sulfoxide reductase. Proc Natl Acad Sci U S A 97: 6463–6468, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Luxford C, Dean RT, and Davies MJ. Induction of DNA damage by oxidised amino acids and proteins. Biogerontology 3: 95–102, 2002 [DOI] [PubMed] [Google Scholar]
- 107.Mandelkow EM. and Mandelkow E. Tau protein and Alzheimer's disease. Neurobiol Aging 15 Suppl 2: S85–S86, 1994 [DOI] [PubMed] [Google Scholar]
- 108.Manning-Bog AB, McCormack AL, Li J, Uversky VN, Fink AL, and Di Monte DA. The herbicide paraquat causes up-regulation and aggregation of alpha-synuclein in mice: paraquat and alpha-synuclein. J Biol Chem 277: 1641–1644, 2002 [DOI] [PubMed] [Google Scholar]
- 109.Mariani E, Polidori MC, Cherubini A, and Mecocci P. Oxidative stress in brain aging, neurodegenerative and vascular diseases: an overview. J Chromatogr B Analyt Technol Biomed Life Sci 827: 65–75, 2005 [DOI] [PubMed] [Google Scholar]
- 110.Martinez A, Portero-Otin M, Pamplona R, and Ferrer I. Protein targets of oxidative damage in human neurodegenerative diseases with abnormal protein aggregates. Brain Pathol 20: 281–297, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mata NL, Tzekov RT, Liu X, Weng J, Birch DG, and Travis GH. Delayed dark-adaptation and lipofuscin accumulation in abcr+/− mice: implications for involvement of ABCR in age-related macular degeneration. Invest Ophthalmol Vis Sci 42: 1685–1690, 2001 [PubMed] [Google Scholar]
- 112.Mazzulli JR, Mishizen AJ, Giasson BI, Lynch DR, Thomas SA, Nakashima A, Nagatsu T, Ota A, and Ischiropoulos H. Cytosolic catechols inhibit alpha-synuclein aggregation and facilitate the formation of intracellular soluble oligomeric intermediates. J Neurosci 26: 10068–10078, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.McMurray HF, Parthasarathy S, and Steinberg D. Oxidatively modified low density lipoprotein is a chemoattractant for human T lymphocytes. J Clin Invest 92: 1004–1008, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Merker K, Sitte N, and Grune T. Hydrogen peroxide-mediated protein oxidation in young and old human MRC-5 fibroblasts. Arch Biochem Biophys 375: 50–54, 2000 [DOI] [PubMed] [Google Scholar]
- 115.Mijaljica D, Prescott M, and Devenish RJ. Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy 7: 673–682, 2011 [DOI] [PubMed] [Google Scholar]
- 116.Mori M, Itabe H, Higashi Y, Fujimoto Y, Shiomi M, Yoshizumi M, Ouchi Y, and Takano T. Foam cell formation containing lipid droplets enriched with free cholesterol by hyperlipidemic serum. J Lipid Res 42: 1771–1781, 2001 [PubMed] [Google Scholar]
- 117.Murata S, Minami Y, Minami M, Chiba T, and Tanaka K. CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep 2: 1133–1138, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nakano M. and Gotoh S. Accumulation of cardiac lipofuscin depends on metabolic rate of mammals. J Gerontol 47: B126–B129, 1992 [DOI] [PubMed] [Google Scholar]
- 119.Naujokat C. and Hoffmann S. Role and function of the 26S proteasome in proliferation and apoptosis. Lab Invest 82: 965–980, 2002 [DOI] [PubMed] [Google Scholar]
- 120.Navab M, Imes SS, Hama SY, Hough GP, Ross LA, Bork RW, Valente AJ, Berliner JA, Drinkwater DC, Laks H, et al. Monocyte transmigration induced by modification of low density lipoprotein in cocultures of human aortic wall cells is due to induction of monocyte chemotactic protein 1 synthesis and is abolished by high density lipoprotein. J Clin Invest 88: 2039–2046, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Neta P, Huie RE, and Ross AB. Rate constants for reactions of peroxyl radicals in fluid solutions. J Phys Chem Ref Data 19: 413, 1990 [Google Scholar]
- 122.Neuzil J, Gebicki JM, and Stocker R. Radical-induced chain oxidation of proteins and its inhibition by chain-breaking antioxidants. Biochem J 293 (Pt 3): 601–606, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Norris EH, Giasson BI, Hodara R, Xu S, Trojanowski JQ, Ischiropoulos H, and Lee VM. Reversible inhibition of alpha-synuclein fibrillization by dopaminochrome-mediated conformational alterations. J Biol Chem 280: 21212–21219, 2005 [DOI] [PubMed] [Google Scholar]
- 124.Oliver CN, Ahn BW, Moerman EJ, Goldstein S, and Stadtman ER. Age-related changes in oxidized proteins. J Biol Chem 262: 5488–5491, 1987 [PubMed] [Google Scholar]
- 125.Olzmann JA, Li L, and Chin LS. Aggresome formation and neurodegenerative diseases: therapeutic implications. Curr Med Chem 15: 47–60, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Opazo C, Huang X, Cherny RA, Moir RD, Roher AE, White AR, Cappai R, Masters CL, Tanzi RE, Inestrosa NC, and Bush AI. Metalloenzyme-like activity of Alzheimer's disease beta-amyloid. Cu-dependent catalytic conversion of dopamine, cholesterol, and biological reducing agents to neurotoxic H(2)O(2). J Biol Chem 277: 40302–40308, 2002 [DOI] [PubMed] [Google Scholar]
- 127.Ostman A, Frijhoff J, Sandin A, and Bohmer FD. Regulation of protein tyrosine phosphatases by reversible oxidation. J Biochem 150: 345–356, 2011 [DOI] [PubMed] [Google Scholar]
- 128.Pacifici RE, Kono Y, and Davies KJ. Hydrophobicity as the signal for selective degradation of hydroxyl radical-modified hemoglobin by the multicatalytic proteinase complex, proteasome. J Biol Chem 268: 15405–15411, 1993 [PubMed] [Google Scholar]
- 129.Persson HL, Yu Z, Tirosh O, Eaton JW, and Brunk UT. Prevention of oxidant-induced cell death by lysosomotropic iron chelators. Free Radic Biol Med 34: 1295–1305, 2003 [DOI] [PubMed] [Google Scholar]
- 130.Petropoulos I, Conconi M, Wang X, Hoenel B, Bregegere F, Milner Y, and Friguet B. Increase of oxidatively modified protein is associated with a decrease of proteasome activity and content in aging epidermal cells. J Gerontol A Biol Sci Med Sci 55: B220–B227, 2000 [DOI] [PubMed] [Google Scholar]
- 131.Poppek D, Keck S, Ermak G, Jung T, Stolzing A, Ullrich O, Davies KJ, and Grune T. Phosphorylation inhibits turnover of the tau protein by the proteasome: influence of RCAN1 and oxidative stress. Biochem J 400: 511–520, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Powell SR, Wang P, Divald A, Teichberg S, Haridas V, McCloskey TW, Davies KJ, and Katzeff H. Aggregates of oxidized proteins (lipofuscin) induce apoptosis through proteasome inhibition and dysregulation of proapoptotic proteins. Free Radic Biol Med 38: 1093–1101, 2005 [DOI] [PubMed] [Google Scholar]
- 133.Pratico D, Clark CM, Liun F, Rokach J, Lee VY, and Trojanowski JQ. Increase of brain oxidative stress in mild cognitive impairment: a possible predictor of Alzheimer disease. Arch Neurol 59: 972–976, 2002 [DOI] [PubMed] [Google Scholar]
- 134.Quinn MT, Parthasarathy S, Fong LG, and Steinberg D. Oxidatively modified low density lipoproteins: a potential role in recruitment and retention of monocyte/macrophages during atherogenesis. Proc Natl Acad Sci U S A 84: 2995–2998, 1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Radu RA, Mata NL, Nusinowitz S, Liu X, Sieving PA, and Travis GH. Treatment with isotretinoin inhibits lipofuscin accumulation in a mouse model of recessive Stargardt's macular degeneration. Proc Natl Acad Sci U S A 100: 4742–4747, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rajavashisth TB, Andalibi A, Territo MC, Berliner JA, Navab M, Fogelman AM, and Lusis AJ. Induction of endothelial cell expression of granulocyte and macrophage colony-stimulating factors by modified low-density lipoproteins. Nature 344: 254–257, 1990 [DOI] [PubMed] [Google Scholar]
- 137.Reinheckel T, Sitte N, Ullrich O, Kuckelkorn U, Davies KJ, and Grune T. Comparative resistance of the 20S and 26S proteasome to oxidative stress. Biochem J 335 (Pt 3): 637–642, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Reinheckel T, Ullrich O, Sitte N, and Grune T. Differential impairment of 20S and 26S proteasome activities in human hematopoietic K562 cells during oxidative stress. Arch Biochem Biophys 377: 65–68, 2000 [DOI] [PubMed] [Google Scholar]
- 139.Shen JK, Dong A, Hackett SF, Bell WR, Green WR, and Campochiaro PA. Oxidative damage in age-related macular degeneration. Histol Histopathol 22: 1301–1308, 2007 [DOI] [PubMed] [Google Scholar]
- 140.Shringarpure R, Grune T, Mehlhase J, and Davies KJ. Ubiquitin conjugation is not required for the degradation of oxidized proteins by proteasome. J Biol Chem 278: 311–318, 2003 [DOI] [PubMed] [Google Scholar]
- 141.Sitte N, Huber M, Grune T, Ladhoff A, Doecke WD, Von ZT, and Davies KJ. Proteasome inhibition by lipofuscin/ceroid during postmitotic aging of fibroblasts. FASEB J 14: 1490–1498, 2000 [DOI] [PubMed] [Google Scholar]
- 142.Siu PM, Pistilli EE, and Alway SE. Age-dependent increase in oxidative stress in gastrocnemius muscle with unloading. J Appl Physiol (1985) 105: 1695–1705, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Smith CD, Carney JM, Starke-Reed PE, Oliver CN, Stadtman ER, Floyd RA, and Markesbery WR. Excess brain protein oxidation and enzyme dysfunction in normal aging and in Alzheimer disease. Proc Natl Acad Sci U S A 88: 10540–10543, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Someya S, Tanokura M, Weindruch R, Prolla TA, and Yamasoba T. Effects of caloric restriction on age-related hearing loss in rodents and rhesus monkeys. Curr Aging Sci 3: 20–25, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Squadrito GL. and Pryor WA. Oxidative chemistry of nitric oxide: the roles of superoxide, peroxynitrite, and carbon dioxide. Free Radic Biol Med 25: 392–403, 1998 [DOI] [PubMed] [Google Scholar]
- 146.Stadtman ER. and Berlett BS. Reactive oxygen-mediated protein oxidation in aging and disease. Chem Res Toxicol 10: 485–494, 1997 [DOI] [PubMed] [Google Scholar]
- 147.Stadtman ER. and Levine RL. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids 25: 207–218, 2003 [DOI] [PubMed] [Google Scholar]
- 148.Starke-Reed PE. and Oliver CN. Protein oxidation and proteolysis during aging and oxidative stress. Arch Biochem Biophys 275: 559–567, 1989 [DOI] [PubMed] [Google Scholar]
- 149.Stocker R. and Keaney JF., Jr. Role of oxidative modifications in atherosclerosis. Physiol Rev 84: 1381–1478, 2004 [DOI] [PubMed] [Google Scholar]
- 150.Terlecky SR, Chiang HL, Olson TS, and Dice JF. Protein and peptide binding and stimulation of in vitro lysosomal proteolysis by the 73-kDa heat shock cognate protein. J Biol Chem 267: 9202–9209, 1992 [PubMed] [Google Scholar]
- 151.Terman A, Kurz T, Gustafsson B, and Brunk UT. Lysosomal labilization. IUBMB Life 58: 531–539, 2006 [DOI] [PubMed] [Google Scholar]
- 152.Todde V, Veenhuis M, and van der Klei IJ. Autophagy: principles and significance in health and disease. Biochim Biophys Acta 1792: 3–13, 2009 [DOI] [PubMed] [Google Scholar]
- 153.Toussaint O, Medrano EE, and von Zglinicki T. Cellular and molecular mechanisms of stress-induced premature senescence (SIPS) of human diploid fibroblasts and melanocytes. Exp Gerontol 35: 927–945, 2000 [DOI] [PubMed] [Google Scholar]
- 154.Vistoli G, De Maddis D, Cipak A, Zarkovic N, Carini M, and Aldini G. Advanced glycoxidation and lipoxidation end products (AGEs and ALEs): an overview of their mechanisms of formation. Free Radic Res 47 Suppl 1: 3–27, 2013 [DOI] [PubMed] [Google Scholar]
- 155.Viteri G, Carrard G, Birlouez-Aragon I, Silva E, and Friguet B. Age-dependent protein modifications and declining proteasome activity in the human lens. Arch Biochem Biophys 427: 197–203, 2004 [DOI] [PubMed] [Google Scholar]
- 156.Vogiatzi G, Tousoulis D, and Stefanadis C. The role of oxidative stress in atherosclerosis. Hellenic J Cardiol 50: 402–409, 2009 [PubMed] [Google Scholar]
- 157.Waxman EA. and Giasson BI. Molecular mechanisms of alpha-synuclein neurodegeneration. Biochim Biophys Acta 1792: 616–624, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Weber D, Kneschke N, Grimm S, Bergheim I, Breusing N, and Grune T. Rapid and sensitive determination of protein-nitrotyrosine by ELISA: Application to human plasma. Free Radic Res 46: 276–285, 2012 [DOI] [PubMed] [Google Scholar]
- 159.Whittier JE, Xiong Y, Rechsteiner MC, and Squier TC. Hsp90 enhances degradation of oxidized calmodulin by the 20 S proteasome. J Biol Chem 279: 46135–46142, 2004 [DOI] [PubMed] [Google Scholar]
- 160.Wilkinson B. and Gilbert HF. Protein disulfide isomerase. Biochim Biophys Acta 1699: 35–44, 2004 [DOI] [PubMed] [Google Scholar]
- 161.Wolf G. Lipofuscin and macular degeneration. Nutr Rev 61: 342–346, 2003 [DOI] [PubMed] [Google Scholar]
- 162.Zaarur N, Meriin AB, Bejarano E, Xu X, Gabai VL, Cuervo AM, and Sherman MY. Proteasome failure promotes positioning of lysosomes around the aggresome via local block of microtubule-dependent transport. Mol Cell Biol 34: 1336–1348, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zainal TA, Oberley TD, Allison DB, Szweda LI, and Weindruch R. Caloric restriction of rhesus monkeys lowers oxidative damage in skeletal muscle. FASEB J 14: 1825–1836, 2000 [DOI] [PubMed] [Google Scholar]
- 164.Zhao Y. and Zhao B. Oxidative stress and the pathogenesis of Alzheimer's disease. Oxid Med Cell Longev 2013: 316523, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]