Significance
Although entomopathogenic fungi and their invertebrate hosts share a >300 million year co-evolutionary history, little is known concerning the biochemical and genetic basis of insect defensive tactics and the countermeasures evolved and evolving by the pathogen to thwart these defenses. Our results provide a molecular mechanism to help explain why some insects are more resistant to broad host-range entomopathogenic fungi. We identify beetle cuticular secretions and a fungal detoxifying enzyme as components of an arms race between insects and the fungal pathogen, suggesting an evolving role for the quinone reductase enzyme as a specific virulence factor for host quinone detoxification. As races have winners and losers, this paper captures a snapshot where the host is leading the race.
Keywords: entomopathogenic fungi, quinone reductase, insect cuticle, Tribolium castaneum, evolutionary arms race
Abstract
Entomopathogenic fungi and their insect hosts represent a model system for examining invertebrate-pathogen coevolutionary selection processes. Here we report the characterization of competing components of an arms race consisting of insect protective antimicrobial compounds and evolving fungal mechanisms of detoxification. The insect pathogenic fungus Beauveria bassiana has a remarkably wide host range; however, some insects are resistant to fungal infection. Among resistant insects is the tenebrionid beetle Tribolium castaneum that produces benzoquinone-containing defensive secretions. Reduced fungal germination and growth was seen in media containing T. castaneum dichloromethane extracts or synthetic benzoquinone. In response to benzoquinone exposure, the fungus expresses a 1,4-benzoquinone oxidoreductase, BbbqrA, induced >40-fold. Gene knockout mutants (ΔBbbqrA) showed increased growth inhibition, whereas B. bassiana overexpressing BbbqrA (Bb::BbbqrAO) displayed increased resistance to benzoquinone compared with wild type. Increased benzoquinone reductase activity was detected in wild-type cells exposed to benzoquinone and in the overexpression strain. Heterologous expression and purification of BbBqrA in Escherichia coli confirmed NAD(P)H-dependent benzoquinone reductase activity. The ΔBbbqrA strain showed decreased virulence toward T. castaneum, whereas overexpression of BbbqrA increased mortality versus T. castaneum. No change in virulence was seen for the ΔBbbqrA or Bb::BbbqrAO strains when tested against the greater wax moth Galleria mellonella or the beetle Sitophilus oryzae, neither of which produce significant amounts of cuticular quinones. The observation that artificial overexpression of BbbqrA results in increased virulence only toward quinone-secreting insects implies the lack of strong selection or current failure of B. bassiana to counteradapt to this particular host defense throughout evolution.
Coevolution between hosts and their parasites is considered to be a major driving force of natural selection, and significant literature exists on theoretical and ecological outcomes derived from such interactions. The Red Queen hypothesis, eponymously named after the character in Lewis Carroll’s book Through the Looking Glass, who says to Alice, “…it takes all the running you can do, to keep in the same place…” serves as a concise metaphor for the selection pressures on hosts to constantly change defense mechanisms against pathogens and the counterselection on the pathogens for continuously developing means for overcoming such evolving defenses, resulting in the appearance of both organisms “running in place” (1). Variations of the Red Queen as well as alternative theories, most unified around the centrality of biotic interactions as a driving force, have been proposed as explanations for (i) taxon extinction [Van Valen’s original Red Queen’s hypothesis (2)]; (ii) the evolution and maintenance of sex, genetic recombination, and immune systems (3–5); (iii) as a framework for understanding invasions of exotic species (6); and (iv) as mechanism(s) selecting for host–pathogen coevolution [i.e., broad versus specific host ranges and/or host tolerance (7–9)].
For fungal pathogens, the best-known examples of coevolutionary interactions are the pathogen effector and corresponding plant host target genes that have defined a gene-for-gene disease susceptibility model in which the outcome of infection is based on avirulence (Avr) genes in the pathogen and dominant resistance (R) genes in the host (10, 11). The fungal protein can trigger immunity due to R protein effector recognition, thus limiting or preventing disease. In these systems, certain Avr mutations or lack of the appropriate R gene product leads to infection. Avr systems, however, form only part of the landscape of interactions between plant fungal pathogens and their hosts. Invading microbes must overcome exterior defenses, e.g., the plant epicuticle and cell wall, and interior defenses, e.g., immune-related defenses that are both preformed and induced (11–13). Within a broader context, resistance to host antimicrobial compounds, host-induced oxidative and/or other stress, and host immune evasion can also be considered as points of interactions that can result in an evolutionary arms race. Despite the ubiquity of fungal–arthropod interactions, there are few examples in which the underlying molecular and biochemical mechanism(s) that define the competing aspects of a coevolutionary arms race have been described. Intriguingly, although effector-like sequences have been found in the genome of entomopathogenic fungi, gene-for-gene avirulence mechanisms have yet to be reported for insect pathogenic fungi (14).
Ecologically important as regulators of insect populations, and distributed throughout almost all ecosystems, entomopathogenic fungi are also of significant interest as potential biological control agents against different insect pests, in particular, as more environmentally friendly alternatives to chemical pesticides (15). Several isolates of the Ascomycetes Beauveria bassiana and Metarhizium anisopliae have been successfully used worldwide to control insects of both agricultural and medical importance (16–19). B. bassiana is known as a broad host range arthropod pathogen capable of infecting a wide range of target hosts. Infection occurs via attachment and penetration of the host cuticle, which represents the first and likely most important line of defense against microbial pathogens (20, 21). Mortality typically occurs 3–10 d postinfection, after which the fungus sporulates on the host cadaver. However, for B. bassiana, completion of its life cycle does not require parasitism of a host because the fungus is a facultative pathogen that can grow as a saprophyte as well as form intimate associations with plants (22, 23).
Despite its broad host range, an enduring mystery has been the observation that select insect species are recalcitrant to infection by B. bassiana, even though other closely related species are susceptible. Tenebrionid beetles including Tribolium castaneum (the red flour beetle) and Ulomoides dermestoides are two such resistant insect species. The former is of particular importance as a worldwide pest of stored products, resulting in significant agricultural losses per year that often disproportionately affect developing nations (24, 25). The latter, curiously enough, is consumed as an alternative medicine for the treatment of a range of illnesses including diabetes and cancer (26). The genome of T. castaneum is available, and it has become an important model system for insect development (27, 28). Both beetle species are known to produce quinone-containing cuticular secretions hypothesized to act as antimicrobial defense compounds (29, 30). These tenebrionids use prothoracic and abdominal glands to produce quinone-containing defensive secretions originally identified as displaying repellent and/or irritant against predators. Secreted quinone derivatives are also involved in cuticle tanning and sclerotization, and these compounds have been shown to possess potent antimicrobial activities (31–33). The chemical structures of the major components of these beetle secretions have been shown to consist of methyl-1,4-benzoquinone and ethyl-1,4-benzoquinone, together with the carrier alkene 1-pentadecene (34, 35).
Biodegradation of quinone derivatives by fungi is a well-known process in the lignin-degrading fungi, where quinones are the product of the oxidation of lignin-related aromatic compounds by ligninases (36). The pathway involves the induction of NAD(P)H:quinone reductases, which catalyze the conversion of cytotoxic benzoquinone to (nontoxic) hydroquinone. Several quinone reductases have been purified and characterized and their genes cloned in both white- and brown-rot fungi (basidiomycetes) as part of lignin degradation pathways (37–40). However, entomopathogenic fungi are incapable of lignin degradation, and no information is available regarding the presence and/or function of these enzymes in this group of fungi. However, entomopathogenic fungi are able to assimilate various hydrocarbon and lipid components of the insect cuticle, responding to surface cues on their hosts to initiate programs for infection (41–43).
In this work we demonstrate the antifungal properties of tenebrionid cuticular secretions and characterize a counteracting fungal NAD(P)H:1,4-benzoquinone oxidoreductase (BbBqrA) in B. bassiana. Gene expression and enzyme activity assays revealed induction of BbbqrA expression and increased BbBqrA activity in the presence of exogenous benzoquinone. Heterologous expression and characterization of the BbbqrA enzyme confirmed NADP(H)-dependent benzoquinone reductase activity. Fungi containing a targeted disruption of BbbqrA showed decreased ability to infect T. castaneum, but no changes in virulence when tested against nonquinone-producing beetles (Sitophilus oryzae, rice weevil) or the Lepidopertan host, the greater wax moth Galleria mellonella. Conversely, a B. bassiana fungal strain engineered to overexpress BbbqrA displayed increased virulence against T. castaneum with, again, no changes in virulence when tested against G. mellonella or S. oryzae. These results link BbbqrA to degradation of tenebrionid quinone-containing defensive secretions, identifying a host-specific virulence factor that is part of a coevolutionary arms race between a pathogen and its host. These data reveal a biochemical mechanism that can act as one factor to limit host range.
Results
Molecular Identification and Expression Analyses of a Benzoquinone Reductase in B. bassiana.
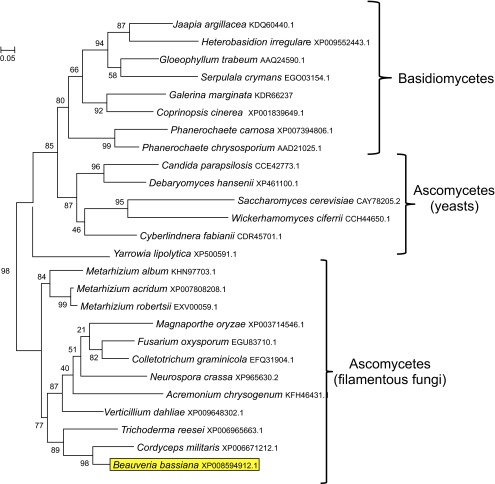
A gene fragment displaying homology to benzoquinone reductases was identified in a B. bassiana expressed sequence tagged (EST) library (44, 45). The gene fragment was used to query the B. bassiana genome (14), leading to the identification of the full-length genomic sequence of BbbqrA (gene locus ID: XM_008596690). Analyses of the 5′-upstream region of the open reading frame (ORF) revealed the presence of TATA and CCAAT boxes defining a putative promoter beginning at positions −57 and −600 relative to the AUG start site, respectively. In addition to these two elements, an inverted GC box was found at nucleotide −308 and a stress response element (STRE) was found at position −288. A xenobiotic response element (XRE), also present in the promoters of a set of B. bassiana cytochrome P450 monooxygenases implicated in insect cuticle degradation (43), was identified at positions −235, −267, −326, −539, −599, −606, −616, −1,079, and −1,181 (Fig. S1). The region surrounding the start codon contained the eukaryotic consensus (G/A)NNATGG sequence, including a purine (A) in position −3, as has been reported for highly expressed genes in Saccharomyces cerevisiae (46) and has also been found in B. bassiana P450 genes (43). Variants of the tripartite signal [5′-TAG...TA(T/A)GT…TTT], indicating potential transcription termination, were found in the 3′-flanking region, although no consensus polyadenylation formation signal (AATAAA-3′ or its common hexameric variants) was present in this region. The gene contained two introns of 17 and 67 bp; beginning at 24 and 446 bp from the start codon, respectively. The exon/intron boundaries conformed to the canonical GT/AG donor/acceptor rule. The deduced polypeptide sequence of BbBqrA contained 201 amino acids with an estimated molecular weight of 21,402 Da and an isoelectric point of 6.05. No potential glycosylation or signal peptide cleavage sites were found, and PSORT analysis suggested cytosolic localization of the protein. A phylogenetic tree was constructed using NADH:benzoquinone reductase and NADPH-dependent flavin mononucleotide (FMN) reductases found in yeasts and filamentous fungi. BLAST analysis showed a single gene encoding for a benzoquinone reductase in B. bassiana and other filamentous fungi and yeasts. A distantly related NADPH-dependent FMN reductase (gene locus ID XP_008596471) was identified in B. bassiana, which shares 30% similarity with BbBqrA. This gene appears to have true orthologs in other filamentous fungi and yeasts. Phylogenetic analyses of BqrA revealed two distinct clusters (Fig. S2), one composed mainly of yeast sequences and Basidiomycetes (e.g., Phanerochaete chrysosporium and Gloeophyllum trabeum) and a second cluster containing Ascomycetes sequences. B. bassiana BqrA subclustered together with proteins from Cordyceps militaris and Tricoderma reesei. The BqrA derived from Metarhizium species, e.g., M. album, M. robertsii, and M. acridum, fungi that are also entomopathogens, subclustered separately.
Fig. S1.
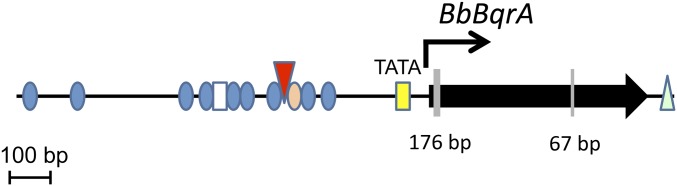
Schematic structure of BbbqrA and phylogenetic tree. Diagram of BbbqrA; the ORF is represented by an arrow. Vertical lines (gray) within the ORF and numbers identify the two introns and intron lengths, respectively. Genetic elements identified (5′- and 3′- upstream and downstream sequences, respectively) are represented as follows: yellow square, TATA box (−57 bp); empty square CCAAT box (−600), inverted red triangle, inverted GC box (−308); blue circles (XRE) (−235, −267, −326, −539, −599, −606, −616, −1,079, and −1,181); pale orange circle (STRE) (−288); light green triangle: potential transcription termination.
Fig. S2.
Maximum-likelihood phylogeny. The analysis includes amino acid sequences of quinone reductases from different yeast and filamentous fungi. The JTT amino acid substitution model was used to build the tree. Numbers at nodes indicate SH-like branch support values.
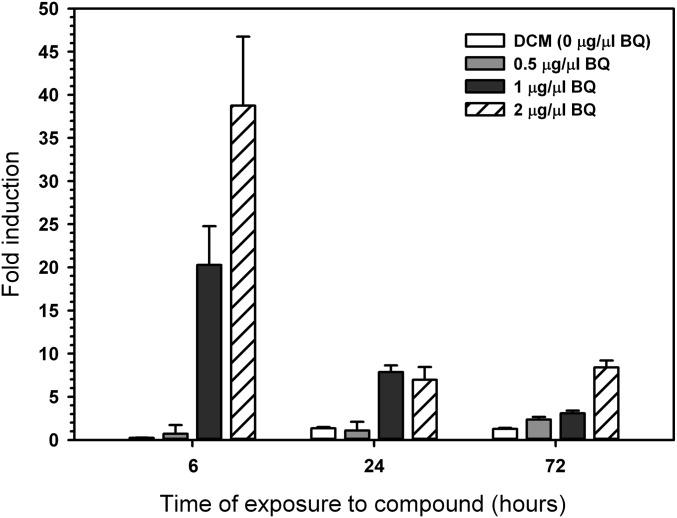
The presence of the TATA box and regulatory sequences suggested that BbbqrA was an inducible gene. To examine whether its expression was specifically induced by benzoquinone, BbbqrA transcript levels were measured by quantitative RT-PCR (qRT-PCR) in B. bassiana cultures exposed to benzoquinone using the expression of ribosomal 5.8S RNA as an internal control. Bbbqr gene expression levels showed a moderate dose-dependent increase in response to benzoquinone levels, rapidly increasing 20- and 40-fold above control levels in the presence of 9.25 and 18.5 µM benzoquinone (1 and 2 µg/µL), respectively, after 6 h of coincubation (Fig. 1). After this initial burst, BbbqrA levels decreased within 24–72 h to between three- and fivefold above controls. Little to no induction was detected when fungal cells were grown in either 4.6 μM benzoquinone (0.5 µg/µL) or in dichloromethane (solvent) compared with controls.
Fig. 1.
B. bassiana BbbqrA gene expression analysis. A time course of BbbqrA expression levels was determined by qRT-PCR after growth in the presence (0.5, 1, and 2 μg/mL) and absence of benzoquinone (BQ). Expression was normalized to fungal growth on the same media in the absence of BQ. Values are means ± SD. DCM, dichloromethane.
Generation of BbbqrA Disruption Mutant and Overexpression Strains.
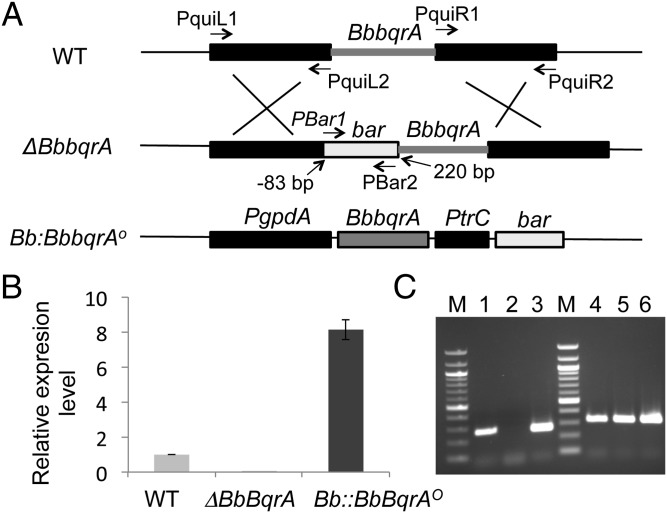
To probe the function of BbbqrA, a targeted gene deletion strain was constructed. BbbqrA was disrupted via homologous recombination, leading to the insertion of the phosphinothricin resistance marker (bar) into the ORF of the target gene as detailed in Materials and Methods. A 303-bp fragment of the gene was deleted, and the insertion event occurred between −83 bp in the 5′-UTR region and 220 bp in the ORF (Fig. 2A). A BbbqrA overexpression strain designated as Bb::BbbqrAO was constructed by cloning of the entire BbbqrA ORF from cDNA in front of the constitutive glyceraldehyde 3-phosphate dehydrogenase promoter (gpdA) and subsequent transformation of the construct into the wild-type strain. The correct integration events for construction of the targeted gene knockout and overexpression strains were confirmed by PCR (Fig. S3). To confirm BbbqrA overexpression, 10 (PCR-verified) transformants were chosen, and BbbqrA expression was examined in potato dextrose agar (PDA) media by qRT-PCR. Clones varied in BbbqrA expression, with one clone displaying a more than eightfold increase in BbbqrA transcript levels compared with the wild-type parent (Fig. 2 B and C). This strain was used in all subsequent studies reported below.
Fig. 2.
Construction and verification of B. bassiana BbbqrA deletion and overexpression strains. (A) Schematic diagram of the LBbqrA-bar-RBbqrA and pUC-bar-PgpdA:BQR constructs used to construct the ΔBbbqrA and the Bb::BbbqrAO strains, respectively. (B) Relative transcript abundance levels in the ΔBbbqrA and Bb::BbBqrAO strains normalized to the wild type as determined by qRT-PCR. The actin and/or 5.8S rRNA gene products were used as internal reference genes. (C) Agarose gel electrophoresis of RT-PCR products of BbbqrA transcripts in wild-type (lanes 1 and 4) and mutant strains (lanes 2 and 5, deletion mutant, and lanes 3 and 6, overexpression strain, respectively). M, 10-kb ladder; lanes 1–3, BbbqrA-specific primers; lanes 4–6, actin primers. Primers used in the study are listed in Table S1.
Fig. S3.
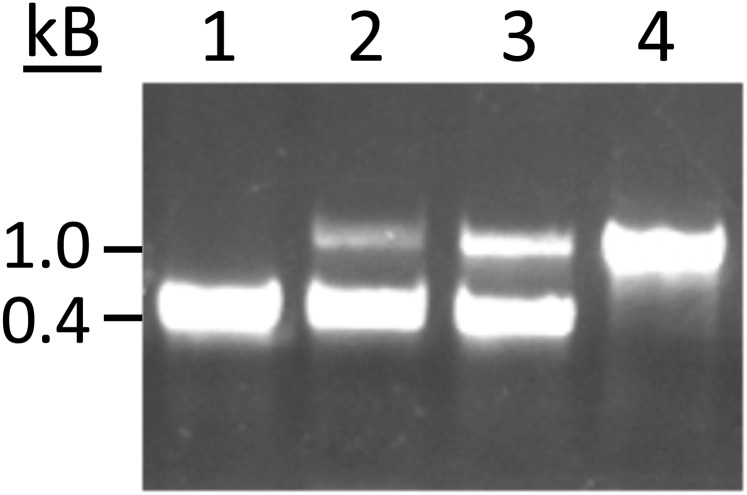
Confirmation of the integration event (ΔBbBqrA strain construction). PCR analysis of transformants using primers Pqui-U and Pqui-D. Agarose gel electrophoresis of PCR reactions: lane 1, wild type; lanes 2 and 3, ectopic insertions showing two bands; lane 4, clone with correct integration event. Increase in size is due to deletion of ∼300 bp of the BbbqrA gene and insertion of the 900-bp bar gene into the BbbqrA ORF.
Tenebrionid Secretions Have Antifungal Properties.
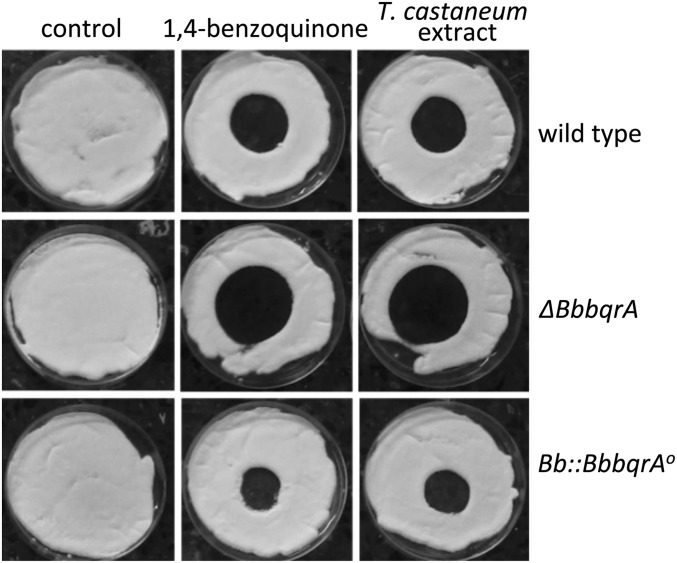
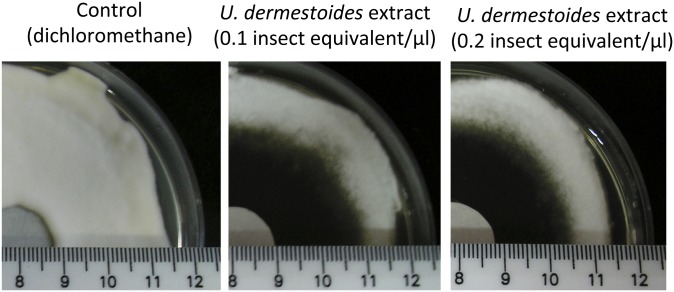
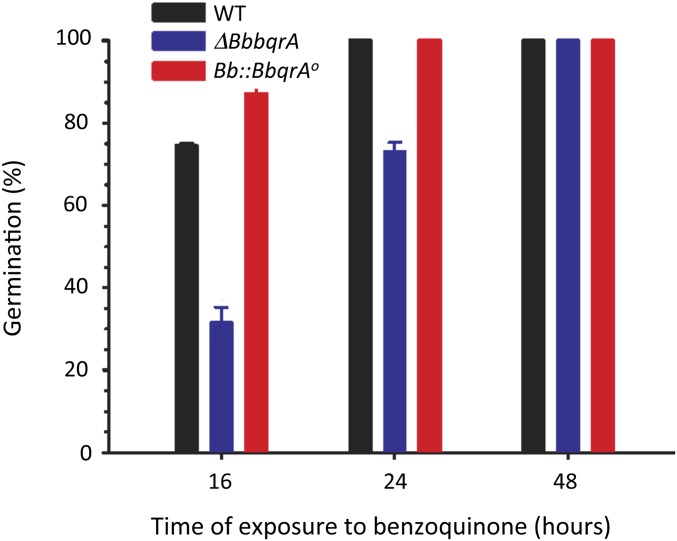
Pure benzoquinone and dichloromethane extracts of T. castaneum were used to examine the effects of these compounds on fungal growth of B. bassiana wild-type, ΔBbbqrA (deletion mutant), and Bb::BbbqrAO (overexpression) strains. Addition of benzoquinone or tenebrionid extracts resulted in concentration-dependent inhibition of wild-type B. bassiana growth compared with growth on control media (Fig. 3 and Table 1). Fungal growth media (Sabouraud dextrose agar) supplemented with insect extracts resulted in zones of growth inhibition, i.e., radii of 18.9 ± 1.2 mm for 0.3 insect equivalents/µL and 24.1 ± 1.8 mm for 0.6 insect equivalents/µL, respectively. Similar results were obtained with dichloromethane extracts derived from the tenebrionid beetle, U. dermestoides (Fig. S4). Zones of growth inhibition for wild type in the presence of benzoquinone were 7.6 ± 1.3 mm (37 µM or 4 µg/µL), 9.7 ± 1.5 mm (111 µM or 12 µg/µL), and 12.1 ± 2.3 mm (231.2 µM or 25 µg/µL), respectively (Table 1). Neither hydroquinone nor pentadecene (C15:1) inhibited fungal growth at similar concentrations. A significant decrease in growth in the presence of benzoquinone or T. castaneum extracts was seen for the ΔBbbqrA strain, and, conversely, greater resistance to these compounds was seen for the Bb::BbbqrAO overexpression strain. The ΔBbbqrA strain showed a relative inhibition (Ri) ratio [(radii of inhibition of test strain)/(radii of inhibition of wild type)] between 1.3–1.7 (P < 0.01) for the range of benzoquinone concentrations tested and Ri = 1.4 (P < 0.01) versus the tenebrionid extracts. In contrast, the Bb::BbbqrAO overexpression strain resulted in Ri = 0.4–0.7 (P < 0.01) for benzoquinone and 0.7–0.8 (P < 0.05) for the insect extracts. Fungal germination was assayed in the presence of 9.25 µM benzoquinone (1 µg/µL) (Fig. 4). The presence of benzoquinone significantly delayed germination in the wild-type, ΔBbbqrA, and Bb::BbbqrAO strains. Germination rates were calculated after 12, 16, 24, and 48 h of exposure to benzoquinone. After 12 h of exposure to benzoquinone, germination was dramatically affected for the mutant (ΔBbbqrA) and the wild-type strains, which exhibited germination rates equal to 1.7 ± 0.2% and 19.9 ± 0.5%, respectively. Twelve hours of exposure to benzoquinone, however, did not impair germination in the overexpression strain (Bb::BbbqrAO) to the same extent as in the knockout and wild-type strains (germination rate 46.4 ± 4.0%). At 16 h, the respective germination rates for wild-type, ΔBbbqrA, and Bb::BbbqrAO strains were 74.5 ± 0.5%, 31.5 ± 3.8%, and 87.0 ± 1.9%, respectively. At 24 h, the germination rate reached 100% for the wild-type and the overexpression strains, whereas for the ΔBbbqrA mutant germination was only 73.1 ± 2.2%. Finally, at 48 h, germination rates were 100% for all of the strains.
Fig. 3.
Quinone and beetle cuticular extract inhibition of B. bassiana growth. Lawns of wild type, ΔBbbqrA, and Bb::BbbqrAO were spotted with different concentrations of either pure benzoquinone or T. castaneum cuticular extracts. Zones of growth inhibition were quantified as described in Materials and Methods and Results.
Table 1.
Inhibition of B. bassiana growth by 1,4-benzoquinone
| Inhibition radium (mm ± SE) | |||
| BQ (µM) | Wild type | ΔBbbqrA | Bb::BbbqrAO |
| 0 | 0 | 0 | 0 |
| 37 (4 µg/µl) | 6.6 ± 0.8a* | 8.3 ± 0.7a | 2.7 ± 0.3b |
| 111 (12 µg/µl) | 9.7 ± 0.7a | 13.2 ± 0.2b | 4.3 ± 0.4c |
| 231 (25 µg/µl) | 12.2 ± 1.3a | 20.3 ± 0.7b | 8.2 ± 0.4c |
For each dose, different lowercase letters indicate significant differences between columns (P < 0.05, Tukey’s test).
Fig. S4.
Growth inhibition of B. bassiana by U. dermestoides cuticular extracts. Conidia derived from wild-type B. bassiana were spread onto plates inoculated with disks containing the indicated compounds.
Fig. 4.
Time course of B. bassiana germination in the presence of 1,4-benzoquinone. Conidia from wild-type, ΔBbbqrA, and Bb::BbqrAO strains were incubated in the presence of media containing 1 μg/μL (9.25 µM) 1,4-benzoquinone, and the percentage of germination was calculated at 16, 24, and 48 h postincubation. The bars represent percentages ± SD.
NAD(P)H-1,4-Benzoquinone Reductase Activity in Wild-Type, ΔBbbqrA, and Bb::BbbqrAO Strains.
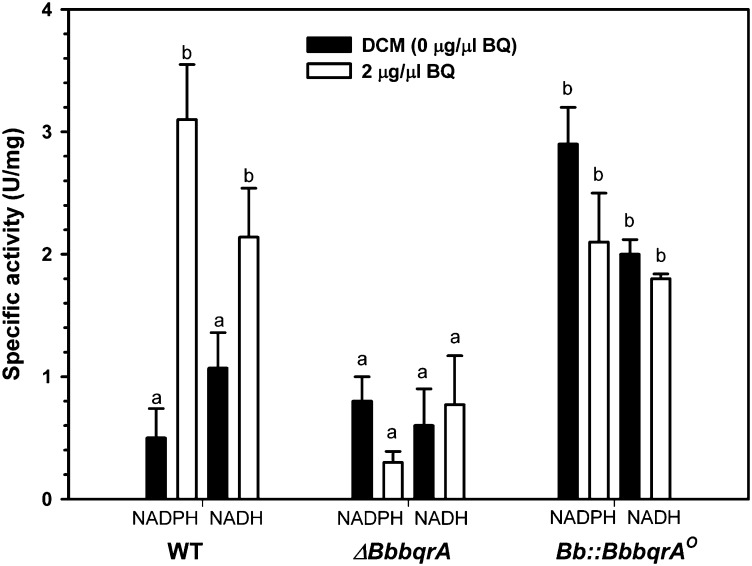
Benzoquinone reductase activity was measured in fungal extracts derived from the wild-type, ΔBbbqrA, and Bb::BbbqrAO strains grown in the presence and absence of benzoquinone (Fig. 5). Enzyme activity was measured in fungal extracts after 6 h of exposure to 18.5 μM (2 µg/µL) benzoquinone or dichloromethane as a control using either NADH or NADPH as the electron donor as described in Materials and Methods. For the wild type, BqrA activity increased six- to sevenfold after exposure to benzoquinone reaching ∼3 U/mg total protein (Fig. 5). As expected for a FMN reductase, enzyme activity was seen in the presence of either NADPH or NADH. No increase/induction of BqrA activity above background levels (0.5–1 U/mg) was seen in the ΔBbbqrA mutant strain. Constitutive BqrA activity was seen in the Bb::BbbqrAO strain (2–3 U/mg), although no further induction of activity was seen in the presence of benzoquinone.
Fig. 5.
Determination of 1,4-benzoquinone reductase activity in B. bassiana. Enzyme activity was determined by measuring either NADH or NADPH consumption in B. bassiana partially purified fractions derived from fungal cells exposed to 2 μg/μL (18.5 µM) benzoquinone for 6 h before harvesting. Bars represent means of 5–10 replicates ± SD. Different lowercase letters indicate significant differences (P < 0.05, Tukey’s test).
Heterologous Expression and Characterization of BbBqrA.
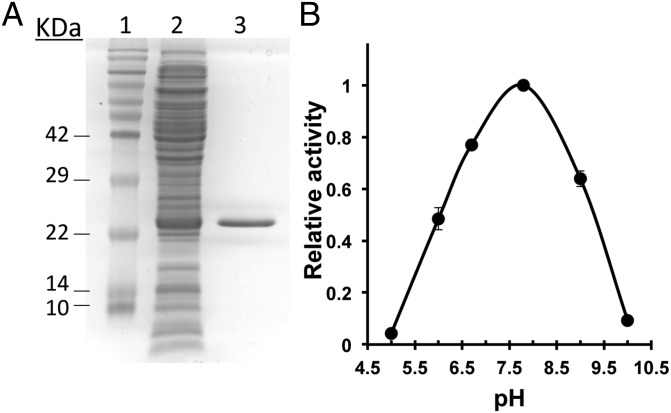
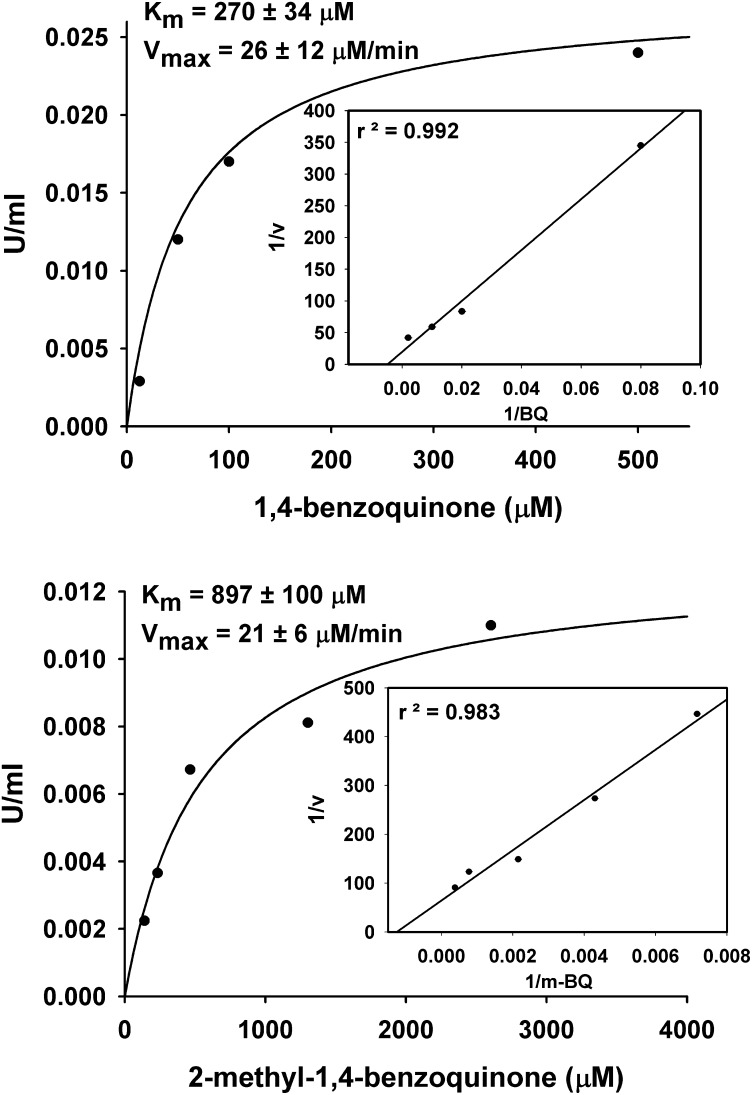
To confirm the biochemical activity of the bqrA protein product, the ORF of the gene was cloned into an Escherichia coli recombinant expression plasmid vector (pET28a) as detailed in Materials and Methods that included an N-terminal His-tag on the protein. The protein was purified by Ni2+ metal-ion affinity chromatography, and the molecular weight of the protein was estimated to be 24 kDa (Fig. 6A, theoretical 22.92 kDa). The optimum pH for enzyme activity was determined to be pH = 7.8 (Fig. 6B). The enzyme was functional using either NADH or NADPH as a cofactor, and activity was linear with respect to time and protein concentration under the conditions examined. BbBqrA was able to use both 1,4-benzoquinone and 2-methyl-1,4-benzoquinone as substrates. Kinetic analyses of the purified enzyme showed that it exhibited a Km = 270.3 μM with a Vmax = 26 µM/min (0.026 U/mL) and a KCAT = 5.99 min−1 (Fig. 7, Upper). The overall catalytic efficiency (CAT = KCAT/Km) for the enzyme with respect to 1,4-benzoquinone was 0.01 µM−1⋅min−1. Kinetic analyses using 2-methyl-1,4-benzoquinone indicated a Km = 897 μM, Vmax = 21 µM/min (0.021 U/mL), KCAT = 4.5 min−1, and CAT= 0.005 µM−1⋅min−1 (Fig. 7, Lower). The overall catalytic activity of the enzyme toward conversion of 1,4-benzoquinone was two times greater than that for 2-methyl-1,4-benzoquinone.
Fig. 6.
Heterologous expression and purification of BbBqrA. (A) Sixteen-percent SDS-polyacrylamide gel of recombinant BbBqrA: lane 1, molecular weight standards; lane 2, E. coli BbBqrA expression crude extract; lane 3, purified recombinant BbBqrA (3 µg). (B) Relative benzoquinone reductase activity at different pH values using 1,4-benzoquinone as the substrate in the presence of NADPH.
Fig. 7.
BbBqrA enzyme kinetics. (Upper) Kinetics of recombinant BbBqrA with 1,4-benzoquinone. KCAT and the catalytic efficiency (CAT = KCAT/Km) were calculated to be 5.99 min−1 and 0.01 min−1⋅µM−1, respectively. (Lower) Kinetics of recombinant BbBqrA with 2-methyl-1,4-benzoquinone. Kcat and the catalytic efficiency were calculated to be 4.50 min−1 and 0.005 min−1⋅µM−1, respectively. The ratio of catalytic efficiency of BqrA for 1,4-benzoquinone to 2-methyl-1,4–benzoquinone, CATbq/CATmbq = 2.
BbbqrA Contributes to Virulence Against Quinone-Secreting Tenebrionids.
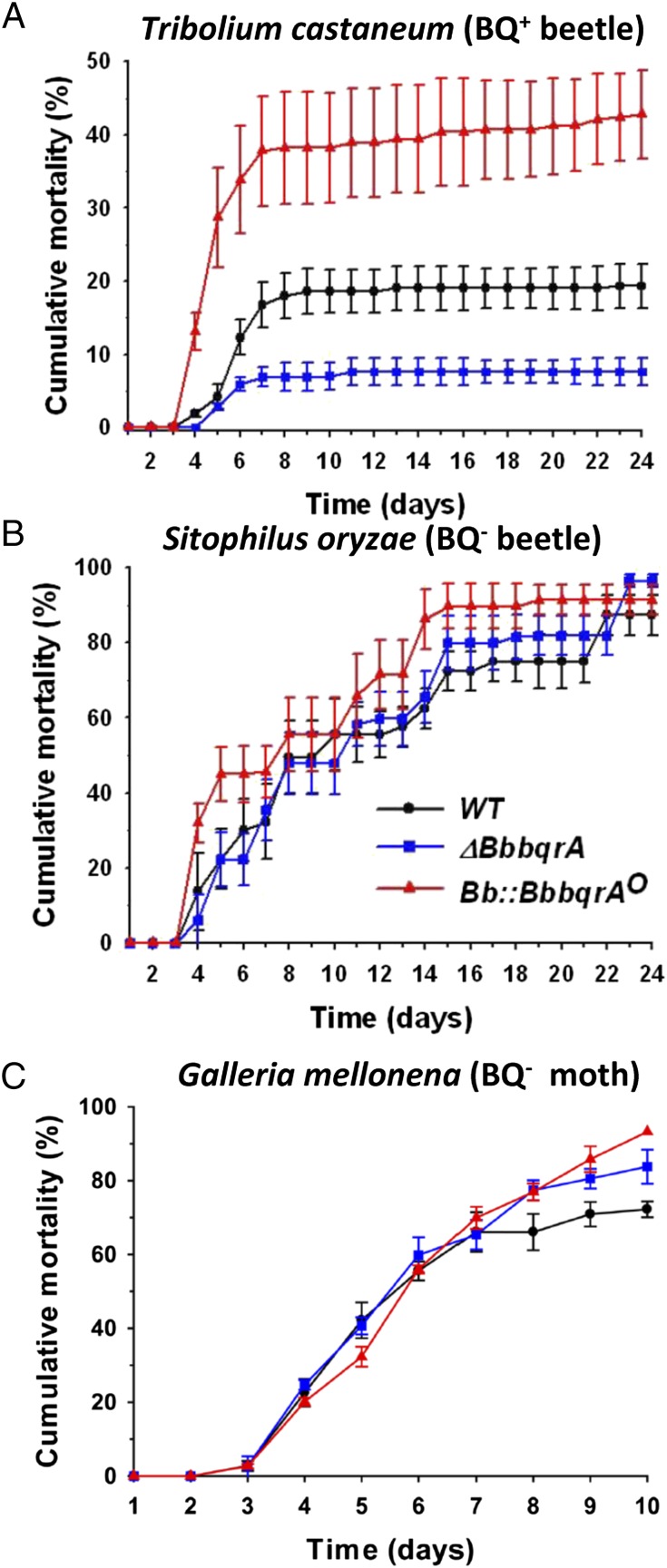
T. castaneum, a benzoquinone-secretor beetle species, was a poor host for wild-type B. bassiana, which was able to kill only 15–20% of the target insects over a 22-d time course, with infection leveling off 8 d postinoculation (Fig. 8A). Loss of BbBqrA decreased the ability of B. bassiana to successfully infect T. castaneum below that of wild type, resulting in ∼9–10% (P < 0.001) mortality against this host. In contrast, overexpression of BbBqrA (Bb::BbbqrAO) increased virulence, resulting in 35–40% mortality of the treated beetles (P < 0.001). In all instances, however, mortality reached a plateau 7–8 d postinoculation, and due to overall mortality rates <50%, accurate mean lethal time to kill 50% of target host (LT50) values could not be calculated. In contrast, insect bioassays using the nonbenzoquinone-producing rice weevil, S. oryzae (also an important stored-grain pest), revealed susceptibility of the insect to the wild-type B. bassiana parental strain, showing a cumulative mortality of 87.4 ± 5.4% after 4 wk (LT50 = 10.2 ± 2.6 d). No significant differences in virulence were seen between wild type and infections using either the ΔBbbqrA or Bb::BbbqrAO strains (F = 1.289, P = 0.2835), with LT50 = 8.9 ± 2.7 d for ΔBbbqrA and 7.6 ± 1.7 d for Bb::BbbqrAO (Fig. 8B). Similar results were obtained using the greater wax moth, G. mellonella (Lepidoptera), in which neither deletion nor overexpression of BbbqrA affected fungal virulence compared with the wild type. In this instance, infection with all strains tested reached >80% mortality within 10 d (Fig. 8C), and no significant differences were noted between the wild-type, ΔBbbqrA, and Bb::BbbqrAO strains. LT50 values were calculated to be 5.4 ± 0.3, 5.5 ± 0.1, and 6.1 ± 0.1 d for the wild-type, ΔBbbqrA, and Bb::BbbqrAO strains, respectively.
Fig. 8.
Insect bioassays using B. bassiana wild-type, ΔBbbqrA, and Bb::BbbqrAO strains. Survival curves of the quninone-producing beetle T. castaneum (A), the nonquinone-producing beetle S. oryzae (B), and the lepidopteran moth G. mellonella (C) infected with spores derived from the wild-type (black lines), ΔBbbqrA (blue), and Bb::BbbqrAO (red) strains, respectively. Data represent mean cumulative mortality percentage ± SE from four independent experiments. Control insects were immersed in water containing 0.01% Tween 80. Mortality data were corrected for control mortality using the Abbott’s formula.
Discussion
Fungal pathogens have evolved a variety of strategies for successful infection of target hosts (47, 48). Fungal spores (conidia) of the insect pathogenic fungus, B. bassiana, will attempt to attach and initiate infection essentially anywhere on host cuticle, although preferential infection sites on some hosts have been observed (21, 49, 50). The infection program is initiated by surface cues, and the fungus is capable of assimilating long-chain alkanes and other lipids that are often found as major constituents of the insect epicuticle (41). The subsequent production of cuticle-degrading enzymes and penetrating hyphae results in growth of the fungus through the integument into the hemocoel. Once in the hemocoel, B. bassiana undergoes a dimorphic transition elaborating cells that are able to evade the host immune system (51, 52). In almost all cases, if the infection progresses to the hemocoel, mortality of the host will ensue. Thus, the major “battlefield” for thwarting infection has been hypothesized to be the host cuticle (20). A significant puzzle in insect pathology is the observation that some insect species, even if closely related, show greater resistance to broad host range fungal pathogens than others. The underlying biochemical mechanism(s) behind such resistance and counterattempts by the fungus to overcome such defenses has remained obscure. For some insects, resistance has been attributed to the production of antimicrobial cuticular compounds (53–55). More recently there is growing evidence that some of these compounds are not produced by the insects themselves but by endogenous microbial flora, adding an interesting twist to coevolutionary scenarios (56, 57). In other insects, specific behaviors, including heat seeking (behavioral fever), aggregation, and, in the case of social insects, grooming, have also been shown to be strategies for impeding or minimizing microbial infections (58–60).
Within this context, some tenebrionid beetles, e.g., T. castaneum, are resistant to B. bassiana infection under most conditions. Therefore, dose-mortality response curves, i.e., LT50 and mean lethal dose (LD50) values, cannot accurately be determined (30, 61, 62). A comparative study of the chemical defense system of tenebrionids, examining 147 different species, revealed that all such beetles contain abundant amounts of soluble quinones, i.e., not cross-linked to other macromolecules as can occur to quinones during cuticle sclerotization, that are produced and stored within the organism and secreted via both prothoracic and abdominal glands (63). Such insect quinone-containing secretions are typically sprayed from the various glands toward predators and intruders and have been extensively studied due to their role in deterring insects and other animals, with their participation in other chemical communication processes also reported (64–67). T. castaneum is a particularly prolific soluble quinone producer, secreting a quinone mixture that, in addition to acting as a noxious discharge to repel threats, displays antimicrobial activities against a number of bacteria and yeasts (33, 68). Our data extend the antimicrobial activities of these secretions to filamentous fungi, including insect pathogens, demonstrating inhibition of B. bassiana germination and growth. This observation suggests that production of cuticular quinones represents an adaptation that can inhibit microbial infections, even of specialized pathogens, by blocking their ability to gain a foothold on the organism and initiate infection. As rapid fungal growth and germination has been shown to correlate with virulence (69), likely due to an ensuing increased rate of fungal penetration of the insect cuticle, limiting germination on the cuticle via production of toxic compounds by the insect would be an effective strategy for resisting infection. However, quinones appeared to be fungistatic toward B. bassiana, and over time, in growth media containing quinones, the fungus is able to germinate to a similar extent as the wild type, bearing in mind that the chemical oxidation of quinones to a nontoxic derivative also occurs over time. On the insect, however, defensive/cuticular quinones are constantly being replenished. No differences were observed in terms of sporulation on the cadavers of those insects killed by any of the fungal strains examined, i.e., wild type, ΔBbbqrA, and Bb::BbBqrAO. As quinones are oxidized over time, the concentrations of quinones on a dead insect appear to drop to below fungal inhibitory levels within several days. Sporulation takes between 2 and 4 d, a time period in which quinone levels are likely to have decreased substantially on the cadaver.
In response to benzoquinone, the fungus produces a benzoquinone reductase (BbBqrA) that acts to detoxify the compound. The enzyme is widely distributed in fungi, and the B. bassiana BbbqrA gene product showed high sequence identity to quinone reductases found, but not yet characterized, in other filamentous fungi including C. militaris and T. reesei, as well as in M. roberstii, the latter another broad host range insect pathogen. BbBqrA clustered relatively far from orthologous proteins in the brown- and white-rot basidiomycetes, G. trabeum and P. chrysosporium, whose quinone reductases have been implicated in lignin biodegradation processes. Unlike these enzymes, however, B. bassiana BbBqrA does not contain a 5′-putative leader sequence for protein secretion (37, 39). The secreted enzymes target cross-linked quinones present in extracellular lignin substrates (70). In contrast, soluble quinones can readily enter cells, where they are cytotoxic in part because they rapidly undergo intracellular one-electron reduction to semiquinones, which react rapidly with O2 to produce superoxide (71). In contrast, hydroquinones are relatively nontoxic because they undergo one-electron oxidation to semiquinones only in the presence of transition metal oxidants, such as Fe3+, which are generally unavailable inside cells because they are sequestered in redox-inactive complexes.
Expression of the BbbqrA gene was induced in the presence of benzoquinone, and, as expected, an increase in benzoquinone reductase activity in fungal extracts was noted. Deletion of BbbqrA resulted in a fungal strain with increased susceptibility to benzoquinone. Although mortality of the wild-type strain against T. casteneum was poor (∼18–20%), the mutant strain was even worse, resulting in no more than 10% mortality even after 24 d. In contrast, the ΔBbbqrA mutant showed no significant alternation in virulence toward the nonquinone-secreting weevil S. oryzae or the greater wax moth G. mellonella. These data suggest that BbBqrA acts as a host-specific virulence factor for host quinone detoxification. To provide further evidence for such a hypothesis, a fungal strain overexpressing the benzoquinone reductase was produced. Overexpression of the enzyme increased fungal resistance to benzoquinone, and the virulence of the BqrA-overexpressing strain (Bb::BbBqrAO) was increased compared with the wild type against T. castaneum, reaching from 25 to 40% of the infected population. As seen for the deletion strain, no change in virulence was seen for the Bb::BbBqrAO strain when tested against S. oryzae or G. mellonella. The observation that mortality still did not reach >50% suggests that T. castaneum contains additional barriers to thwart infection by B. bassiana and/or that the reductase levels were still insufficient to completely overcome the amounts of toxic quinones present in and/or on the insect. The fact that overexpression did increase virulence suggests that the fungal response to the host defense is not yet optimized, supporting the idea that the host and pathogen are involved in an evolutionary arms race, in which, at present, T. castaneum is in the lead. Our results can possibly be viewed as capturing a snapshot of the selective pressures on a specific biochemical enzyme expressed by the pathogen. As the benzoquinone reductase is found in (nonpathogenic) fungi that do not interact with quinone-producing beetles, this indicates the potential for B. bassiana to redirect a general (quinone) detoxification enzyme to a specialized function, an adaptation that has not yet occurred.
The fact that expression of the detoxifying enzyme by the pathogen is not yet optimized can be explained by several factors. First, as a broad host range pathogen, the targets of B. bassiana are manifold, and therefore selection pressures on any given single target, which, if representing a small fraction of potential hosts, will be diluted by the availability of other more susceptible targets. As significant concentrations of antimicrobial-acting quinones are rare on most potential hosts, any selective pressure may be small within a larger environmental context. This would be especially true if, as our data show, selection for the trait does not confer a more generalized benefit, i.e., increased virulence toward other targets. Second, although we could not detect any obvious phenotypic differences in addition to BqrA activity, a potential trade-off might exist; i.e., increased BqrA activity may decrease some aspect of fungal fitness that we did not detect. This is often seen because any counteradaptive change entails some costs and most often reduces adaptation to some other factor; e.g., even a small decrease in growth rate can constrain environmental selection for higher enzyme activity. In addition, the selective pressure on T. castaneum to avoid fungal infection may be stronger because in most cases infection by the fungus results in death, whereas the fungus only loses one opportunity to grow and disperse in that particular host, a potential example of the “life-dinner principle” (72).
Significant efforts have been made to unveil the molecular mechanisms behind the strategies that entomopathogenic fungi use to overcome host defenses. These findings have led to several genetic engineering approaches to improve mycoinsecticide performance by targeting specific defenses, e.g., cuticular compounds and insect immune pathways (73–76). This paper adds benzoquinones to the list of insect cuticular defenses and presents BbBqrA as a potential adaptive response of B. bassiana to counteract the insect’s attempt to escape fungal infection. By revealing the competing components of an arms race between B. bassiana and T. castaneum, we have discovered a way to increase the virulence of the fungal biological control agent against a previously recalcitrant target and demonstrated that, although infection of insects by B. bassiana is a multifactorial process, in some instances a single host specificity virulence factor can be identified.
Materials and Methods
Cultivation of Fungi.
B. bassiana (ATCC 90517) was routinely grown on PDA, Czapek–Dox, or complete medium agar (CMA) plates. Plates were incubated at 26 °C. Except for inhibition and germination assays, fungi were grown on the surface of a cellophane sheet in CMA plates with and without addition of dichloromethane extracts of T. castaneum adults or synthetic 1,4-benzoquinone, hydroquinone, and 1-pentadecene (C15:1) (Sigma-Aldrich). Fungi were first cultured for 4 d on cellophane/CMA plates until abundant mycelia were observed, after which the sheet was transferred to CMA plates containing the test extracts/compounds. Plates were returned for incubation over the indicated time course (typically up to an additional 4 d). Dichloromethane extracts were prepared from laboratory-reared adults of T. castaneum, as previously described (26). The extracts contained mainly ethyl- and methyl-1,4-benzoquinones and 1-pentadecene, as verified by thin-layer chromatography. Solvents and other reagents were obtained from Fisher Scientific and Carlo Erba Reagents SAS.
Antimicrobial Assay.
Suspensions of conidia were prepared from PDA plates grown for 6–10 d by rubbing the sporulating surface with a bent needle. After filtering through a 60-µm mesh sieve to remove debris, sterile distilled water containing 0.01% Tween 80 was added to the suspension to reach desired cell concentrations based upon cell counts using a heamocytometer. Suspensions of 4 × 108 con/mL were homogeneously dispersed with a bent glass rod on CMA plates. After 30 min, a sterile paper dish (20-mm diameter) containing the insect cuticular extract was placed on the center of the plate. Doses assayed ranged from 0.3 to 0.6 insect equivalents/μL. Additional assays were performed on agar plates containing solutions of (reagent grade) benzoquinone in dichloromethane, hydroquinone in ethanol, or C15:1 in hexane. Concentrations of the pure chemical compounds tested included the following: (i) 37, 111, and 231.2 μM (4, 12, and 25 µg/µL) benzoquinone; (ii) 36.3, 108.9, and 227 µM (4, 12, and 25 µg/µL) hydroquinone; and (iii) 19, 57, and 118.8 µM (4, 12, and 25 µg/µL) pentadecene, spotted onto the agar plates with corresponding solvents used as controls. The zones of inhibition (diameters in millimeters) around the spotted compounds on plates containing uniformly spread fungal spores were measured after 3 d incubation at 26 °C. Each experiment used four replicate plates and was repeated at least two times with different batches of conidia.
Fungal Germination Assay.
A 40-μL drop of the conidial suspension was inoculated onto the center of CMA plates containing 1 μg/mL benzoquinone. Controls included spotting of conidial solution on CMA plates containing no additional reagents or containing the solvent dichloromethane. After inoculation, plates were incubated for 12, 16, 24, and 48 h in the dark at 26 °C. Conidial germination was assessed microscopically, and conidia were considered to be germinated when the length of the germ tube was at least half of the length of the diameter of the conidium. For each of three plates inoculated for each concentration, at least 300 conidia per plate were evaluated and the percentage of germination was calculated.
Molecular Manipulations: Construction of BbbqrA-Targeted Gene Knockout and Overexpression Strains.
A list of primers used in this study is given in Table S1. A putative benzoquinone oxidoreductase gene was identified in B. bassiana EST libraries, and the genomic DNA sequence was obtained from the B. bassiana whole genome (14). The sequences were analyzed using programs implemented at the Biology WorkBench website (workbench.sdsc.edu/). Introns were detected by comparison between both cDNA and DNA sequences. Coding sequences were translated to amino acid sequences using the ExPASy tools (ExPASy Proteomics Server, www.expasy.org). Phylogenetic analysis was performed with the Phylogeny.fr platform (77), which uses the programs MUSCLE for multiple alignment, Gblocks for automatic alignment curation, PhyML for tree building, and TreeDyn for tree drawing. The protein sequence is annotated in GenBank under accession no. XP_008594912.1.
Table S1.
Primers used in this paper
| PquiL1: ATCGGGCAGCTTCACCAGAC |
| PquiL2: CAATATCATCTTCTGTCGACGGTAAACTGGCTGTGGCTACG |
| PquiR1: GCCCGTCACCGAGATCTGACAAGCAGCTGGCCGATGCCG |
| PquiR2: TCTCCATCTCTGCTTCTAATC |
| PBar1: GTCGACAGAAGATGATATTG |
| PBar2: TCAGATCTCGGTGACGGGC |
| Pqui-U: CTGCTCAGCTTCGCTCAAAG |
| Pqui-D: GATCTGGTAAAGGTCGGCGGT |
| Pquiov-1: AATCAATAACAGCGGCCGCGATGGTCAAGATTGCGATTGTC |
| Pquiov-2: TCTAGATCAAGCGCCCTTGATGGCGT |
| PgpdA1: CTCTAGAGAATTCGTTGGGT |
| PgpdA2: CGCGGCCGCTGTTATTGATT |
| quiNcoI-fwd: ATATACCATGGATGGTCAAGATTGCGATTGT |
| quiHindIII-rev: ACAACGCCATCAAGGGCGCTAAGCTTATATA |
| 5.8S-fwd: AAGAACGCAGCGAAACGCGATAAG |
| 5.8S-rev: TCGTTAAGTTCAGCGGGGTAGTCCT |
| qui-qPCR1-fwd: ACGGCTTCCTGTTTGGCATTCC |
| qui-qPCR1-rev: GTAATGGATGCCGTGGTGCGTG |
| Actin-fwd: TTGGTGCGAAACTTCAGCGTCTAGTC |
| Actin-rev: TCCAGCAAATGTGGATCTCCCAAGCAG |
| qui-qPCR2-fwd: CTACTCCATGTACGGCCACA |
| qui-qPCR2-rev: GCCAGTCTTGTCCCAGAAAG |
For construction of a targeted gene knockout strain, the upstream (LB) and downstream (RB) regions of BbbqrA were obtained by PCR using primer pairs PquiL1/L2 and PquiR1/R2 (Table S1). Primers PBar1 and PBar2 were used to amplify the bar gene expression cassette, conferring resistance to phosphinothricin. PquiL2 and PquiR1 were designed to contain a 20-bp bar gene cassette overlapping sequences. The products derived from individual PCR, i.e., LB, RB, and bar, were used to perform primer-less assembly by PCR cycling conditions as follows: 98 °C (2 min), followed by 22 cycles of 98 °C (15 s), 55 °C (30 s), and 72 °C (1.5 min), after which 1 μL of the reaction mixture was used as the template to PCR-amplify the desired full-length homologous recombinant construct (LBbqrA-bar-RBbqrA) with primers of L1 and R2. During the whole process of construction, Phusion DNA polymerase (New England Biolabs) was used. The PCR product was introduced into B. bassiana competent cells following our method (78). Screening for the correct homologous recombinant mutant was performed using primers Pqui-U and Pqui-D as described (79).
To construct a B. bassiana BbbqrA overexpressing strain, the full-length BbbqrA gene was obtained via PCR amplification using primers Pquiov-1 and Pquiov-2. The resultant PCR product was fused to a PCR fragment containing the PgpgdA promoter by primer-less assembly, taking advantage of a 20-bp PgpdA sequence overlap designed in primer Pquiov-1. The obtained construct, PgpdA::BbbqrA, was cloned into pUC-bar and transformed into B. bassiana competent cells as above. To confirm BbbqrA overexpression, qPCR analyses were performed with 10 transformants and the wild type. Fungi were grown in PDA plates with cellophane sheets for 5 d. RNA and cDNA preps were obtained with RNAeasy Plant Mini Kit (Qiagen) and a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). qPCR was performed with 10 ng of reverse-transcribed RNA/sample under the following conditions (30 s at 95 °C for denaturation, 30 s at 59 °C for annealing, and 30 s at 72 °C for polymerase elongation + melting step). The BbbqrA-specific primers, qui-qPCR2-fwd and qui-qPCR2-rev, and primers encoding for the actin gene, actin-fwd and actin-rev, were used. The experiment was performed in triplicate, and appropriate controls were included.
Gene Expression Analysis.
B. bassiana was grown in complete medium and exposed to different concentrations of benzoquinone (0, 0.5, 1, and 2 μg/μL or 0, 4.6, 9.25, and 18.5 µM). Fungal cells were examined over a time course (6, 24, 72 h) of exposure, and total RNA of each culture was extracted using an RNAeasy Plant Mini kit (Qiagen), including an on-column DNA digestion step. RNA was quantified by the Qubit fluorometer using the Quant-iT RNA assay kit (Invitrogen), and the integrity of the RNA was assessed on a 1% (wt/vol) agarose gel. Two-step reverse transcriptase real-time PCR was performed using the iScript cDNA Synthesis kit and iQ SYBR Green Supermix (Bio-Rad). Amplification was performed in a Mx3000P QPCR System (Stratagene) using 20 ng of reverse-transcribed total RNA for each sample. The following amplification program was used: denaturation at 95 °C for 10 min, followed by 40 cycles with three-segment amplification (30 s at 95 °C for denaturation, 30 s at 61 °C for annealing, and 30 s at 72 °C for polymerase elongation). To confirm that only single products were amplified, a temperature melting step was then performed. Relative expression was calculated using 5.8S rRNA as the reference gene amplified with 5.8S-fwd and 5.8S-rev. The gene-specific primers for BbbqrA were qui-qPCR1-fwd and qui-qPCR-rev (Table S1). Negative controls included samples generated without reverse transcriptase as templates. Reactions containing primer pairs without template were also included as blank controls. The assay was performed in duplicate with each of the three independent biological replicates.
Subcellular Fractionation and Benzoquinone Reductase Enzyme Activity Measurement.
B. bassiana was grown on cellophane sheets placed on CMA for 4 d after which the sheets were transferred to fresh CMA plates supplemented with or without 2 μg/μL benzoquinone and incubated for an additional 6-h induction period. Fungal mycelia were harvested from the cellophane sheets and disrupted using a Mini-Bead Beater homogenizer (BioSpec) with glass beads (0.5-mm diameter). Four cycles of 20 s each were performed, with 20-s cooling periods between bursts. Homogenization buffer was 50 mM sodium citrate (pH 6), 300 mM sucrose, 5 mM EDTA, 5 mM DTT, and 1 mM PMSF. The homogenate was centrifuged for 20 min at 10,000 × g to remove mitochondria and cell debris, and the supernatant was ultracentrifuged for 60 min at 100,000 × g. The soluble fraction (S100,000 × g) was used for determination of enzyme activity.
1,4-Benzoquionone reductase activity was measured spectrophotometrically by following the decrease in absorbance at 340 nm due to NADPH or NADH consumption. The amount of enzyme that catalyzed the oxidation of 1 μmol of NAD(P)H/min at 25 °C was defined as 1 U of activity. Standard reaction mixtures (1 mL) consisted of 50 mM sodium citrate buffer (pH 6), 100 μM 1–4 benzoquinone, and 0.1–0.2 mL of the S100,000 × g supernatant fraction. Reactions were initiated by the addition of 200 μM NAD(P)H (Sigma-Aldrich). The rate of nonenzymatic oxidation of NAD(P)H by quinones was measured in a blank containing no enzyme, and its value was subtracted from the sample containing the enzyme (80). Protein concentration was measured by the bicinchoninic acid method (Pierce), using BSA as standard.
Heterologous Expression and Characterization of BbBqrA.
The BbbqrA ORF was amplified from PgpdA::BbbqrA pCB1536 vector. The PCR primers quiNcoI-fwd and qui-HindIII-rev (Table S1) were designed from the BQR cDNA sequence with the restriction sites NcoI and HindIII appended at the 5′ end to facilitate directional cloning into PET28a (AddGene). PCR products were digested with NcoI and HindIII and ligated into the multiple cloning site of the vector, also digested with the same restriction enzymes. The integrity of the construct sequence, which included a six-amino acid histidine tag at the C terminus, was confirmed by sequencing (UF ICBR Molecular Core). The expression plasmid was then transformed in Rosetta (DE3) pLysS E. coli competent cells. For protein expression and purification, E. coli transformants cells were grown in LB media for 3.5 h using a 1:50 overnight inoculum before being induced with 0.5 mM IPTG and allowed to continue to grow overnight at 18 °C. Cells were harvested by centrifugation (8,000 × g for 20 min), resuspended in buffer (20 mM Hepes, 100 mM NaCl, 10 mM imidazole, pH 7.5), and lysed using a French press. The lysate was centrifuged at 15,000 × g for 30 min to remove cell debris. The expressed protein in the resulting supernatant was purified using Co2+-agarose beads (GoldBio). The column was equilibrated and washed with 20 mM Hepes, 100 mM NaCl, 10 mM imidazole (pH 7.5), and BbBqrA was eluted with a stepwise gradient in buffer containing 50, 100, 200, and 300 mM imidazole. Fractions were analyzed by SDS/PAGE and those containing BbBqrA were pooled, dialyzed in 20 mM Hepes and 100 mM NaCl (pH 7.5) and concentrated using a 6- to 8-kDa dialysis membrane (Pierce) via embedding in polyethylene glycol 20000 (Sigma Aldrich). Protein concentration was measured by the bicinchoninic acid method (Pierce) using BSA as standard.1,4-Benzoquionone and 2-methyl-1,4-benzoquinone reductase activity of the recombinant BbBqrA was measured spectrophotometrically by following the decrease in absorbance at 340 nm due to NADPH consumption. Two protein solutions of 9 and 4.6 µM BbBqrA containing 200 µM NADPH were titrated with 12, 50, 100, and 500 µM 1,4-benzoquinone and 140, 232, 465, 1,300, and 2,600 µM 2-methyl-1,4-benzoquinone, respectively. Data were fit to a Michael–Menten equation to calculate the respective Vmax and Km values. The catalytic efficiency for both substrates was calculated as follows: CAT = Kcat/Km, where Kcat = Vmax/[BbbqrA]total. Optimal pH conditions for BbBqrA activity was determined by assaying 1,4-benzoquinone reductase activity as described above using benzoquinone (100 µM) as the substrate and different buffers including 50 mM citrate phosphate buffer (pH 5.0), 50 mM citrate phosphate buffer (pH 6.1), 50 mM citrate buffer (pH 6.6), 50 mM phosphate buffer (pH 7.7), 50 mM Tris⋅ClH buffer (pH 8.9), and 50 mM glycine–NaOH buffer (pH 10.0).
Insect Bioassays.
Fungal insect assays were performed using G. mellonella larvae (Pet Solutions) and adults of T. castaneum and S. orzyae from colonies regularly maintained at El Instituto de Investigaciones Bioquímicas de La Plata (25). For G. mellonella, larvae were treated topically via dipping of larvae for 5–10 s in solutions of 1 × 108 conidia/mL harvested in sterile distilled H2O containing 0.01% Tween 80. Excess liquid on the insect bodies was removed by placement on a dry paper towel. Controls were treated with sterile distilled H2O/0.01% Tween 80. Experimental and control larvae were placed in large (150-mm diameter) petri dishes and incubated at 26 °C. For each experimental condition, 40–50 larvae were used, and all experiments were repeated three times. For T. castaneum and S. oryzae, groups of 10 adults were separately immersed for 6 s in a fungal suspension (1 × 109 conidia/mL) and then placed on a paper towel to remove excess water and transferred to different 200-mL glass vials covered with a muslin cloth (treated insects). Control insects were treated similarly with water containing 0.01% Tween 80. Insect diet (wheat flour for T. castaneum, durum wheat for S. oryzae) was added to the vials 1 d posttreatment. Five replicates of each treatment (fungus-treated and controls) with 10 insects per replicate were performed, and the entire experiment was repeated three more times. Insect mortality was checked daily, and dead insects were removed after each count. Mortality data were corrected for control mortality using the Abbott’s equation (81). Cumulative mortality percentage and LT50 values were calculated by Probit analysis, and differences were determined by analysis of variance and by using Tukey’s test to separate treatment mean (P < 0.05). Instat 3.05 (GraphPad Software Inc.) was used for all statistical analyses.
Acknowledgments
We thank Cecilia Fusé for insect rearing and assistance in bioassays. M.P.J. and N.P. are members of the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) Researcher’s Career. This work was supported in part by grants from Agencia Nacional de Promoción Científica y Tecnológica (PICT 2012 1964) and CONICET (PIP 0237) (to N.P.), and US National Science Foundation Grant IOS-1121392 and University of Florida, Institute of Food and Agricultural Sciences funds (to N.O.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 8519.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1504552112/-/DCSupplemental.
References
- 1.Brockhurst MA, et al. 2014. Running with the Red Queen: The role of biotic conflicts in evolution. Proc Biol Sci 281(1797):pii: 20141382.
- 2.Van Valen L. A new evolutionary law. Evol Theory. 1973;1:1–30. [Google Scholar]
- 3.Danilova N. The evolution of immune mechanisms. J Exp Zoolog B Mol Dev Evol. 2006;306(6):496–520. doi: 10.1002/jez.b.21102. [DOI] [PubMed] [Google Scholar]
- 4.Hartfield M, Keightley PD. Current hypotheses for the evolution of sex and recombination. Integr Zool. 2012;7(2):192–209. doi: 10.1111/j.1749-4877.2012.00284.x. [DOI] [PubMed] [Google Scholar]
- 5.Tobler M, Schupp I. Expanding the horizon: The Red Queen and potential alternatives. Can J Zool. 2008;86(8):765–773. [Google Scholar]
- 6.Platt V, Jeschke JM. Are exotic species red queens? Ethol Ecol Evol. 2014;26(2-3):101–111. [Google Scholar]
- 7.Clay K, Kover PX. The Red Queen Hypothesis and plant/pathogen interactions. Annu Rev Phytopathol. 1996;34:29–50. doi: 10.1146/annurev.phyto.34.1.29. [DOI] [PubMed] [Google Scholar]
- 8.Best A, White A, Boots M. The coevolutionary implications of host tolerance. Evolution. 2014;68(5):1426–1435. doi: 10.1111/evo.12368. [DOI] [PubMed] [Google Scholar]
- 9.Tellier A, Moreno-Gámez S, Stephan W. Speed of adaptation and genomic footprints of host-parasite coevolution under arms race and trench warfare dynamics. Evolution. 2014;68(8):2211–2224. doi: 10.1111/evo.12427. [DOI] [PubMed] [Google Scholar]
- 10.Ravensdale M, Nemri A, Thrall PH, Ellis JG, Dodds PN. Co-evolutionary interactions between host resistance and pathogen effector genes in flax rust disease. Mol Plant Pathol. 2011;12(1):93–102. doi: 10.1111/j.1364-3703.2010.00657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson JP, et al. Plants versus pathogens: An evolutionary arms race. Funct Plant Biol. 2010;37(6):499–512. doi: 10.1071/FP09304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maor R, Shirasu K. The arms race continues: Battle strategies between plants and fungal pathogens. Curr Opin Microbiol. 2005;8(4):399–404. doi: 10.1016/j.mib.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 13.Ferreira RB, et al. Fungal pathogens: The battle for plant infection. Crit Rev Plant Sci. 2006;25(6):505–524. [Google Scholar]
- 14.Xiao G, et al. Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Sci Rep. 2012;2:483. doi: 10.1038/srep00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glare T, et al. Have biopesticides come of age? Trends Biotechnol. 2012;30(5):250–258. doi: 10.1016/j.tibtech.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Shah PA, Pell JK. Entomopathogenic fungi as biological control agents. Appl Microbiol Biotechnol. 2003;61(5-6):413–423. doi: 10.1007/s00253-003-1240-8. [DOI] [PubMed] [Google Scholar]
- 17.Pedrini N, et al. Control of pyrethroid-resistant Chagas disease vectors with entomopathogenic fungi. PLoS Negl Trop Dis. 2009;3(5):e434. doi: 10.1371/journal.pntd.0000434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang W, et al. Development of transgenic fungi that kill human malaria parasites in mosquitoes. Science. 2011;331(6020):1074–1077. doi: 10.1126/science.1199115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirkland BH, Westwood GS, Keyhani NO. Pathogenicity of entomopathogenic fungi Beauveria bassiana and Metarhizium anisopliae to Ixodidae tick species Dermacentor variabilis, Rhipicephalus sanguineus, and Ixodes scapularis. J Med Entomol. 2004;41(4):705–711. doi: 10.1603/0022-2585-41.4.705. [DOI] [PubMed] [Google Scholar]
- 20.Ortiz-Urquiza A, Keyhani NO. Action on the surface: Entomopathogenic fungi versus the insect cuticle. Insects. 2013;4:357–374. doi: 10.3390/insects4030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holder DJ, Keyhani NO. Adhesion of the entomopathogenic fungus Beauveria (Cordyceps) bassiana to substrata. Appl Environ Microbiol. 2005;71(9):5260–5266. doi: 10.1128/AEM.71.9.5260-5266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Behie SW, Bidochka MJ. Ubiquity of insect-derived nitrogen transfer to plants by endophytic insect-pathogenic fungi: An additional branch of the soil nitrogen cycle. Appl Environ Microbiol. 2014;80(5):1553–1560. doi: 10.1128/AEM.03338-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Behie SW, Zelisko PM, Bidochka MJ. Endophytic insect-parasitic fungi translocate nitrogen directly from insects to plants. Science. 2012;336(6088):1576–1577. doi: 10.1126/science.1222289. [DOI] [PubMed] [Google Scholar]
- 24.Phillips TW, Throne JE. Biorational approaches to managing stored-product insects. Annu Rev Entomol. 2010;55:375–397. doi: 10.1146/annurev.ento.54.110807.090451. [DOI] [PubMed] [Google Scholar]
- 25. Rajendran S, and Chayakumari (2003) Insect infestation and control in stored grain sorghum and millets. J Food Sci Technol Mysore 40(5):451–457.
- 26.Crespo R, et al. Cytotoxic and genotoxic effects of defence secretion of Ulomoides dermestoides on A549 cells. J Ethnopharmacol. 2011;136(1):204–209. doi: 10.1016/j.jep.2011.04.056. [DOI] [PubMed] [Google Scholar]
- 27.Richards S, et al. Tribolium Genome Sequencing Consortium The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452(7190):949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- 28.Posnien N, Schinko JB, Kittelmann S, Bucher G. Genetics, development and composition of the insect head: A beetle’s view. Arthropod Struct Dev. 2010;39(6):399–410. doi: 10.1016/j.asd.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 29.Lord JC. Efficacy of Beauveria bassiana for control of Tribolium castaneum with reduced oxygen and increased carbon dioxide. J Appl Entomol. 2009;133(2):101–107. [Google Scholar]
- 30.Pedrini N, Villaverde ML, Fuse CB, Dal Bello GM, Juárez MP. Beauveria bassiana infection alters colony development and defensive secretions of the beetles Tribolium castaneum and Ulomoides dermestoides (Coleoptera: Tenebrionidae) J Econ Entomol. 2010;103(4):1094–1099. doi: 10.1603/ec10072. [DOI] [PubMed] [Google Scholar]
- 31.Suderman RJ, Dittmer NT, Kanost MR, Kramer KJ. Model reactions for insect cuticle sclerotization: Cross-linking of recombinant cuticular proteins upon their laccase-catalyzed oxidative conjugation with catechols. Insect Biochem Mol Biol. 2006;36(4):353–365. doi: 10.1016/j.ibmb.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 32.Eisner T. Beetle’s spray discourages predators. Nat Hist. 1966;75:42–47. [Google Scholar]
- 33.Yezerski A, Ciccone C, Rozitski J, Volingavage B. The effects of a naturally produced benzoquinone on microbes common to flour. J Chem Ecol. 2007;33(6):1217–1225. doi: 10.1007/s10886-007-9293-2. [DOI] [PubMed] [Google Scholar]
- 34.Villaverde ML, Girotti JR, Mijailovsky SJ, Pedrini N, Juarez MP. 2009. Volatile secretions and epicuticular hydrocarbons of the beetle Ulomoides dermestoides. Comp Biochem Physiol B Biochem Mol Biol 154(4):381–386. [DOI] [PubMed]
- 35.Villaverde ML, Juarez MP, Mijailovsky S. Detection of Tribolium castaneum (Herbst) volatile defensive secretions by solid phase micro extraction-capillary gas chromatography (SPME-CGC) J Stored Prod Res. 2007;43(4):540–545. [Google Scholar]
- 36.Kirk TK, Farrell RL. Enzymatic “combustion”: The microbial degradation of lignin. Annu Rev Microbiol. 1987;41:465–505. doi: 10.1146/annurev.mi.41.100187.002341. [DOI] [PubMed] [Google Scholar]
- 37.Akileswaran L, Brock BJ, Cereghino JL, Gold MH. 1,4-Benzoquinone reductase from Phanerochaete chrysosporium: cDNA cloning and regulation of expression. Appl Environ Microbiol. 1999;65(2):415–421. doi: 10.1128/aem.65.2.415-421.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brock BJ, Rieble S, Gold MH. Purification and characterization of a 1,4-benzoquinone reductase from the Basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol. 1995;61(8):3076–3081. doi: 10.1128/aem.61.8.3076-3081.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jensen KA, Jr, Ryan ZC, Vanden Wymelenberg A, Cullen D, Hammel KE. An NADH:quinone oxidoreductase active during biodegradation by the brown-rot basidiomycete Gloeophyllum trabeum. Appl Environ Microbiol. 2002;68(6):2699–2703. doi: 10.1128/AEM.68.6.2699-2703.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cohen R, Suzuki MR, Hammel KE. Differential stress-induced regulation of two quinone reductases in the brown rot basidiomycete Gloeophyllum trabeum. Appl Environ Microbiol. 2004;70(1):324–331. doi: 10.1128/AEM.70.1.324-331.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pedrini N, Ortiz-Urquiza A, Huarte-Bonnet C, Zhang S, Keyhani NO. Targeting of insect epicuticular lipids by the entomopathogenic fungus Beauveria bassiana: Hydrocarbon oxidation within the context of a host-pathogen interaction. Front Microbiol. 2013;4:24. doi: 10.3389/fmicb.2013.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang S, et al. CYP52X1, representing new cytochrome P450 subfamily, displays fatty acid hydroxylase activity and contributes to virulence and growth on insect cuticular substrates in entomopathogenic fungus Beauveria bassiana. J Biol Chem. 2012;287(16):13477–13486. doi: 10.1074/jbc.M111.338947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pedrini N, Zhang S, Juárez MP, Keyhani NO. Molecular characterization and expression analysis of a suite of cytochrome P450 enzymes implicated in insect hydrocarbon degradation in the entomopathogenic fungus Beauveria bassiana. Microbiology. 2010;156(Pt 8):2549–2557. doi: 10.1099/mic.0.039735-0. [DOI] [PubMed] [Google Scholar]
- 44.Cho EM, Liu L, Farmerie W, Keyhani NO. EST analysis of cDNA libraries from the entomopathogenic fungus Beauveria (Cordyceps) bassiana. I. Evidence for stage-specific gene expression in aerial conidia, in vitro blastospores and submerged conidia. Microbiology. 2006;152(Pt 9):2843–2854. doi: 10.1099/mic.0.28844-0. [DOI] [PubMed] [Google Scholar]
- 45.Cho EM, Boucias D, Keyhani NO. EST analysis of cDNA libraries from the entomopathogenic fungus Beauveria (Cordyceps) bassiana. II. Fungal cells sporulating on chitin and producing oosporein. Microbiology. 2006;152(Pt 9):2855–2864. doi: 10.1099/mic.0.28845-0. [DOI] [PubMed] [Google Scholar]
- 46.Hamilton R, Watanabe CK, de Boer HA. Compilation and comparison of the sequence context around the AUG startcodons in Saccharomyces cerevisiae mRNAs. Nucleic Acids Res. 1987;15(8):3581–3593. doi: 10.1093/nar/15.8.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horbach R, Navarro-Quesada AR, Knogge W, Deising HB. When and how to kill a plant cell: Infection strategies of plant pathogenic fungi. J Plant Physiol. 2011;168(1):51–62. doi: 10.1016/j.jplph.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 48.Perez-Nadales E, et al. Fungal model systems and the elucidation of pathogenicity determinants. Fungal Genet Biol. 2014;70:42–67. doi: 10.1016/j.fgb.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holder DJ, Kirkland BH, Lewis MW, Keyhani NO. Surface characteristics of the entomopathogenic fungus Beauveria (Cordyceps) bassiana. Microbiology. 2007;153(Pt 10):3448–3457. doi: 10.1099/mic.0.2007/008524-0. [DOI] [PubMed] [Google Scholar]
- 50.Zhang S, Xia YX, Kim B, Keyhani NO. Two hydrophobins are involved in fungal spore coat rodlet layer assembly and each play distinct roles in surface interactions, development and pathogenesis in the entomopathogenic fungus, Beauveria bassiana. Mol Microbiol. 2011;80(3):811–826. doi: 10.1111/j.1365-2958.2011.07613.x. [DOI] [PubMed] [Google Scholar]
- 51.Wanchoo A, Lewis MW, Keyhani NO. Lectin mapping reveals stage-specific display of surface carbohydrates in in vitro and haemolymph-derived cells of the entomopathogenic fungus Beauveria bassiana. Microbiology. 2009;155(Pt 9):3121–3133. doi: 10.1099/mic.0.029157-0. [DOI] [PubMed] [Google Scholar]
- 52.Lewis MW, Robalino IV, Keyhani NO. Uptake of the fluorescent probe FM4-64 by hyphae and haemolymph-derived in vivo hyphal bodies of the entomopathogenic fungus Beauveria bassiana. Microbiology. 2009;155(Pt 9):3110–3120. doi: 10.1099/mic.0.029165-0. [DOI] [PubMed] [Google Scholar]
- 53.James RR, Buckner JS, Freeman TP. Cuticular lipids and silverleaf whitefly stage affect conidial germination of Beauveria bassiana and Paecilomyces fumosoroseus. J Invertebr Pathol. 2003;84(2):67–74. doi: 10.1016/j.jip.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 54.Lecuona R, Clement JL, Riba G, Joulie C, Juarez P. Spore germination and hyphal growth of Beauveria sp on insect lipids. J Econ Entomol. 1997;90(1):119–123. [Google Scholar]
- 55.Sosa-Gomez DR, Boucias DG, Nation JL. Attachment of Metarhizium anisopliae to the southern green stink bug Nezara viridula cuticle and fungistatic effect of cuticular lipids and aldehydes. J Invertebr Pathol. 1997;69(1):31–39. doi: 10.1006/jipa.1996.4619. [DOI] [PubMed] [Google Scholar]
- 56.Toledo AV, Alippi AM, de Remes Lenicov AM. Growth inhibition of Beauveria bassiana by bacteria isolated from the cuticular surface of the corn leafhopper, Dalbulus maidis and the planthopper, Delphacodes kuscheli, two important vectors of maize pathogens. J Insect Sci. 2011;11:29. doi: 10.1673/031.011.0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boucias DG, et al. Detection and characterization of bacterial symbionts in the Heteropteran, Blissus insularis. FEMS Microbiol Ecol. 2012;82(3):629–641. doi: 10.1111/j.1574-6941.2012.01433.x. [DOI] [PubMed] [Google Scholar]
- 58.Roy HE, Steinkraus DC, Eilenberg J, Hajek AE, Pell JK. Bizarre interactions and endgames: Entomopathogenic fungi and their arthropod hosts. Annu Rev Entomol. 2006;51:331–357. doi: 10.1146/annurev.ento.51.110104.150941. [DOI] [PubMed] [Google Scholar]
- 59.Wang YD, Yang PC, Cui F, Kang L. Altered immunity in crowded locust reduced fungal (Metarhizium anisopliae) pathogenesis. PLoS Pathog. 2013;9(1):e1003102. doi: 10.1371/journal.ppat.1003102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yanagawa A, Shimizu S. Resistance of the termite, Coptotermes formosanus Shiraki to Metarhizium anisopliae due to grooming. BioControl. 2007;52(1):75–85. [Google Scholar]
- 61.Akbar W, Lord JC, Nechols JR, Howard RW. Diatomaceous earth increases the efficacy of Beauveria bassiana against Tribolium castaneum larvae and increases conidia attachment. J Econ Entomol. 2004;97(2):273–280. doi: 10.1603/0022-0493-97.2.273. [DOI] [PubMed] [Google Scholar]
- 62.Lord JC. Enhanced efficacy of Beauveria bassiana for red flour beetle with reduced moisture. J Econ Entomol. 2007;100(4):1071–1074. doi: 10.1603/0022-0493(2007)100[1071:eeobbf]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 63.Tschinkel WR. Comparative study of chemical defensive system of Tenebrionid beetles: Chemistry of secretions. J Insect Physiol. 1975;21(4):753–783. [Google Scholar]
- 64.Eisner T, Meinwald J. Defensive secretions of arthropods. Science. 1966;153(3742):1341–1350. doi: 10.1126/science.153.3742.1341. [DOI] [PubMed] [Google Scholar]
- 65.Tschinkel WR. Comparative study of chemical defensive system of Tenebrionid beetles. 2. Defensive behavior and ancillary features (Coleoptera-Tenebrionidae) Ann Entomol Soc Am. 1975;68(3):439–453. [Google Scholar]
- 66.Ruther J, Reinecke A, Tolasch T, Hilker M. Make love not war: A common arthropod defence compound as sex pheromone in the forest cockchafer Melolontha hippocastani. Oecologia. 2001;128(1):44–47. doi: 10.1007/s004420100634. [DOI] [PubMed] [Google Scholar]
- 67.Dettner K. Dabbing and shooting of benzo- and naphthoquinone secretions: Defensive strategies of bark-inhabiting aleocharine (col.: Staphylinidae) and tenebrionid (col.: Tenebrionidae) beetle larvae. J Chem Ecol. 1993;19(7):1337–1354. doi: 10.1007/BF00984880. [DOI] [PubMed] [Google Scholar]
- 68.Prendeville HR, Stevens L. Microbe inhibition by Tribolium flour beetles varies with beetle species, strain, sex, and microbe group. J Chem Ecol. 2002;28(6):1183–1190. doi: 10.1023/a:1016281600915. [DOI] [PubMed] [Google Scholar]
- 69.Altre JA, Vandenberg JD, Cantone FA. Pathogenicity of Paecilomyces fumosoroseus isolates to diamondback moth, Plutella xylostella: Correlation with spore size, germination speed, and attachment to cuticle. J Invertebr Pathol. 1999;73(3):332–338. doi: 10.1006/jipa.1999.4844. [DOI] [PubMed] [Google Scholar]
- 70.Schoemaker HE, et al. 1989. Oxidation and reduction in lignin biodegradation. Plant Cell Wall Polymers: Biogenesis and Biodegradation, eds Lewis NG, Paice MG (ACS Publishers, Washington, DC), pp 454–471.
- 71.Halliwell B, Gutteridge JMC. Free Radicals in Biology and Medicine. 3rd Ed Oxford University Press; Oxford: 1999. [Google Scholar]
- 72.Dawkins R, Krebs JR. Arms races between and within species. Proc R Soc Lond B Biol Sci. 1979;205(1161):489–511. doi: 10.1098/rspb.1979.0081. [DOI] [PubMed] [Google Scholar]
- 73.Yang L, et al. Expression of a toll signaling regulator serpin in a mycoinsecticide for increased virulence. Appl Environ Microbiol. 2014;80(15):4531–4539. doi: 10.1128/AEM.01197-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kamareddine L, Fan Y, Osta MA, Keyhani NO. Expression of trypsin modulating oostatic factor (TMOF) in an entomopathogenic fungus increases its virulence towards Anopheles gambiae and reduces fecundity in the target mosquito. Parasit Vectors. 2013;6:22. doi: 10.1186/1756-3305-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fan Y, Pereira RM, Kilic E, Casella G, Keyhani NO. Pyrokinin β-neuropeptide affects necrophoretic behavior in fire ants (S. invicta), and expression of β-NP in a mycoinsecticide increases its virulence. PLoS ONE. 2012;7(1):e26924. doi: 10.1371/journal.pone.0026924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fan Y, Borovsky D, Hawkings C, Ortiz-Urquiza A, Keyhani NO. Exploiting host molecules to augment mycoinsecticide virulence. Nat Biotechnol. 2012;30(1):35–37. doi: 10.1038/nbt.2080. [DOI] [PubMed] [Google Scholar]
- 77.Dereeper A, et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36(Web Server issue):W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fan Y, Zhang S, Kruer N, Keyhani NO. High-throughput insertion mutagenesis and functional screening in the entomopathogenic fungus Beauveria bassiana. J Invertebr Pathol. 2011;106(2):274–279. doi: 10.1016/j.jip.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 79.Zhang S, Fan Y, Xia YX, Keyhani NO. Sulfonylurea resistance as a new selectable marker for the entomopathogenic fungus Beauveria bassiana. Appl Microbiol Biotechnol. 2010;87(3):1151–1156. doi: 10.1007/s00253-010-2636-x. [DOI] [PubMed] [Google Scholar]
- 80.Constam D, Muheim A, Zimmermann W, Fiechter A. Purification and partial characterization of an intracellular NADH-quinone oxidoreductase from Phanerochaete chrysosporium. J Gen Microbiol. 1991;137:2209–2214. [Google Scholar]
- 81.Abbott WS. A method of computing the effectiveness of an insecticide. J Econ Entomol. 1925;18(2):265–267. [Google Scholar]