Significance
Extracellular signal-regulated kinase (ERK) is a critical enzyme that interacts with a wide range of regulators and substrates to control cellular functions. ERK interactions have been studied using a number of biophysical and biochemical techniques, which identified two docking domains on ERK used by almost all of its substrates and regulators. However, mapping the binding interface of a new ERK interactor with little prior knowledge of the binding mechanism remains a challenge. Here we show that the introduction of a photocrosslinking amino acid into the heterodimerization interface of ERK and its substrate, Capicua, allows for precise, amino-acid–level determination of the substrate binding site using tandem mass spectrometry. This study demonstrates the utility of photocrosslinking approaches for the determination of binding sites on kinases.
Keywords: photocrosslinking, protein–protein interactions, signal transduction
Abstract
Extracellular signal-regulated kinase (ERK) coordinates cellular responses to a range of stimuli by phosphorylating its numerous substrates. One of these substrates, Capicua (Cic), is a transcriptional repressor that was first identified in Drosophila and has been implicated in a number of human diseases. Here we use a chemical biology approach to map the binding interface of ERK and Cic. The noncanonical amino acid p-azidophenylalanine (AzF) was introduced into the ERK-binding region of Drosophila Cic, and photocrosslinking and tandem mass spectrometry were used to pinpoint its binding site on ERK. We also identified the ERK-binding region of human Cic and showed that it binds to the same site on ERK despite lacking conservation with the Drosophila Cic binding region. Finally, we mapped the amino acids involved in human Cic binding to ERK using AzF-labeled ERK. These results reveal the molecular details of the ERK–Cic interaction and demonstrate that the photocrosslinking approach is complementary to existing methods for mapping kinase–substrate binding interfaces.
The transcriptional repressor Capicua (Cic) was discovered in genetic studies of pattern formation in the early Drosophila embryo and plays important roles in vertebrate development and human diseases, including spinocerebellar ataxia and cancers of neural origin (1, 2). Cic represses genes by binding directly to their regulatory regions and recruiting global corepressors. Both the DNA-binding domain of Cic and the Cic-binding sites within the regulatory DNA sequences are highly conserved across species (3–5). Another aspect of Cic regulation that appears to be conserved is its sensitivity to signaling by receptor tyrosine kinases (RTKs), which can antagonize gene repression by Cic (4–7) (Fig. 1A). Molecular mechanisms of this important regulatory effect remain poorly understood.
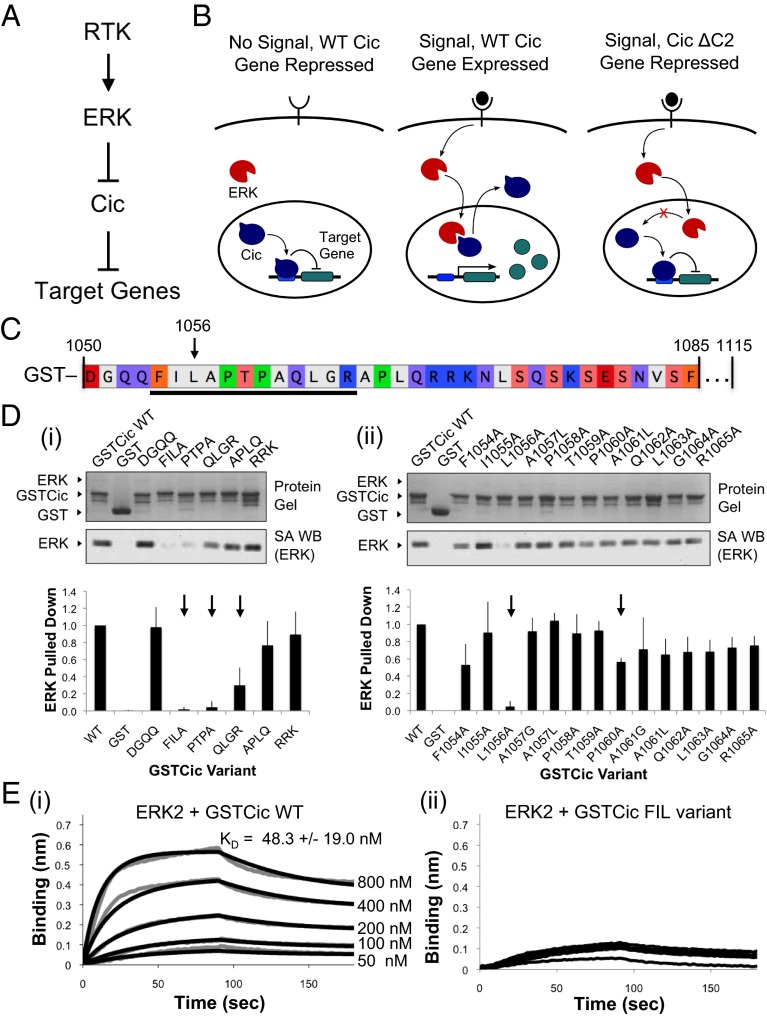
Fig. 1.
Identification of ERK-interacting residues in Drosophila Cic. (A) Antagonism of Cic repressor activity by RTK/ERK signaling. (B) In response to RTK signaling, ERK translocates to the nucleus and phosphorylates Cic, relieving repression of Cic-regulated genes. Deletion of the Cic C2 domain leads to loss of ERK binding and phosphorylation, resulting in continued Cic-mediated repression even in the presence of RTK signaling. (C) Sequence of ERK-binding region of Cic fused to GST (GSTCic). The underlined region represents groups of amino acids that, when substituted to alanine in groups of three or four, lost binding to ERK. An arrow indicates L1056, the individual position that is critical for Cic binding to ERK. (D) Drosophila Cic alanine scan. (i) Groups of three or four amino acids in GSTCic were substituted with alanine and binding to biotinylated ERK was evaluated by GST pull-down. (ii) Residues in the relevant ERK-binding regions of Cic identified in i were replaced with alanine (positions containing alanine were replaced with glycine and leucine). Substitutions that led to loss of binding to ERK are indicated with an arrow. Reported values represent an average of three experiments, and error bars correspond to 1 SD of the mean. (E) ERK–GSTCic binding by BLI. (i) Biotinylated ERK was immobilized on SA biosensors and binding to GSTCic WT at concentrations ranging from 50 to 800 nM was measured (gray curves). Data were fit globally (black curves). Reported KD is an average of fits to three datasets, and the error represents 1 SD of the mean. (ii) No interaction was detected between ERK and GSTCic with positions 1,054–1,056 replaced with alanine using concentrations up to 3,200 nM.
In Drosophila, RTK activation leads to a rapid relief of gene repression by Cic, which is followed by export of Cic from the nucleus and cytoplasmic degradation (8, 9). Both of these effects rely on the extracellular signal-regulated kinase (ERK), an enzyme that is phosphorylated and activated in response to signaling by RTKs and mediates their transcriptional effects in a wide range of developmental contexts (10). In addition to being regulated by ERK signaling, Cic can control the levels of ERK phosphorylation in vivo (11). Quantitative studies in the Drosophila embryo established that RTK-dependent ERK phosphorylation is strongly attenuated in the absence of Cic, and increasing the gene dosage of Cic in the same system leads to a significant increase in ERK phosphorylation. This effect propagates through the patterning network and affects the expression of several RTK-target genes (12, 13). Cic-dependent control of ERK phosphorylation does not depend on the transcriptional activity of Cic and requires direct Cic–ERK interaction (11). The simplest model consistent with these observations is one in which Cic binding to ERK interferes with the activity of ERK phosphatases (14).
We used a chemical biology approach to test this model and characterize the interaction interface between full-length ERK and the ERK-binding regions of Drosophila and human Cic (6). We began by mutagenizing the known ERK-binding region of fly Cic to identify specific residues used to bind ERK. This knowledge enabled us to crosslink ERK and Cic by incorporating a photoactivatable noncanonical amino acid, p-azidophenylalanine (AzF), into Cic at the heterodimerization interface. Using mass spectrometry, we identified the location on ERK used for binding. Further, we discovered an ERK-binding region of human Cic and determined molecular-level details of this interaction using AzF-labeled ERK. Consistent with the proposed model, we found that both human and fly Cic bind to regions of ERK that are involved in binding to phosphatases. These studies elucidated the mechanisms of ERK-mediated regulation of Cic and demonstrate the advantages of the photocrosslinking approach to studies of enzyme–substrate interactions.
Results
Identification of ERK-Interacting Residues in Cic.
Previous studies of ERK-mediated down-regulation of Cic in Drosophila identified a small region of the protein (termed the C2 domain) that, when deleted, led to loss of nuclear exclusion of Cic and loss of binding to ERK (6) (Fig. 1B). Most ERK-interacting proteins bind to ERK at one of its two well-characterized docking domains. The first is the ERK D recruitment site (DRS) (14–17), which is used by proteins containing a D site (18–20). The D site consensus sequence is (R/K)2–3-X1–8-Φ-X-Φ, where X is any amino acid and Φ is a hydrophobic amino acid. The positively charged region of the D site makes electrostatic contacts with two negatively charged amino acids on ERK (termed the CD domain), whereas the downstream Φ-X-Φ interacts with a hydrophobic groove (15, 21). Together, these regions, located on the opposite side of ERK relative to the active site, make up the DRS. A second docking domain, termed the F recruitment site (FRS), consists of a hydrophobic pocket near the ERK active site and binds proteins containing an F site (F-X-F-P consensus sequence) (15, 22).
The ERK-binding region of Cic does not contain either consensus D site or F site sequences. So, as a first step toward characterizing the interaction between ERK and Cic, an unbiased, targeted alanine screen was performed on the Cic C2 domain. We began by expressing and purifying Cic residues 1,050–1,115 fused to GST (herein referred to as GSTCic). Previously, the deletion of residues 1,052–1,072 of Cic was found to abrogate ERK binding (6). This region is within our GSTCic construct. The interaction between GSTCic and ERK was confirmed using biolayer interferometry (BLI) and found to have a KD of ∼50 nM (Fig. 1E). A GST pull-down assay was used to screen GSTCic variants for loss of binding to ERK. Substitutions were introduced into the region corresponding to the deletion previously found to interrupt the ERK–Cic interaction.
First, groups of three or four amino acids at a time were changed to alanine. Three adjacent groups spanning Cic 1,054–1,065 had a significant effect on the amount of ERK bound to the GSTCic variant (Fig. 1 C and D). To validate the pull-down assay, BLI binding was performed between ERK and the strongest loss of binding variant (positions 1,054–1,056), and no interaction was detected (Fig. 1E). Removing clusters of positive charge from D sites in ERK-interacting proteins often results in weakened or abolished binding (14, 18, 19, 21), so it is noteworthy that mutations to the three charged residues in Cic positions 1,070–1,072 did not decrease the strength of binding to ERK. Furthermore, the F site consensus sequence (F-X-F-P) (22) is not present in relevant substituted positions.
To identify the specific residues critical for the ERK–Cic interaction, an alanine screen was performed on individual amino acids in the relevant groups (positions in this region containing alanine were changed to leucine and glycine). Of the 12 positions tested, two substitutions, L1056A and P1060A, significantly reduced binding of Cic to ERK (Fig. 1D). L1056A in particular nearly abolished binding. Thus, we identified a 12-residue region that contributes to ERK–Cic binding and an individual position, L1056, that is involved in this interaction.
Photocrosslinking Cic and ERK at the Binding Interface.
The identification of this critical ERK-binding residue allowed us to use a chemical biology approach to pinpoint the location of Cic binding on ERK. The noncanonical amino acid AzF enables the covalent crosslinking of two bound polypeptides when incorporated into one of the binding partners at the heterodimerization interface (23, 24). Upon irradiation by UV light, the azide moiety of AzF is converted to a highly reactive nitrene, which can insert itself into nearby C-H or N-H bonds, either directly or through a dehydroazepine intermediate (Fig. 2A and SI Appendix, Fig. S1A) (25). We used this functionality to covalently crosslink GSTCic to ERK specifically at the binding interface by inserting AzF into sites located at the ERK binding surface of Cic identified in the alanine screen.
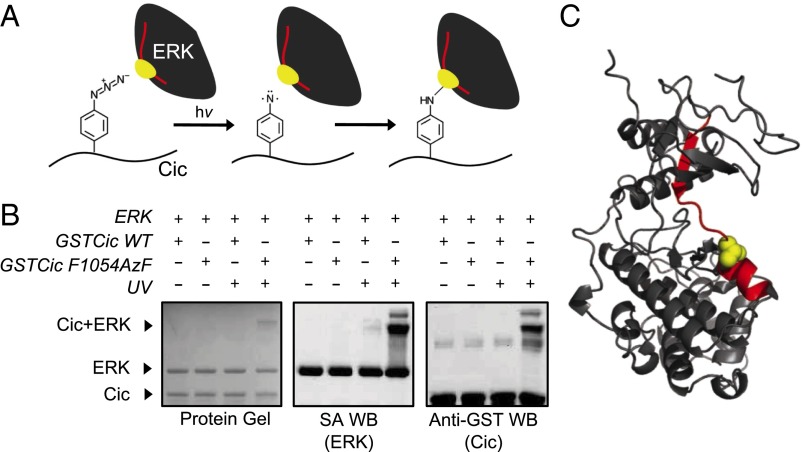
Fig. 2.
Photocrosslinking of ERK and AzF-incorporated Drosophila Cic and identification of crosslinking site by mass spectrometry. (A) Schematic representation of Cic–ERK photocrosslinking chemistry. The ERK tryptic peptide and amino acid that are crosslinked to Cic are represented in red and yellow, respectively. (B) GSTCic WT and F1054AzF were incubated with biotinylated ERK and exposed to UV light. The formation of a slow migrating, crosslinked ERK–GSTCic heterodimer was determined by protein gel, SA Western blot (detects biotinylated ERK), and anti-GST Western blot (detects GSTCic). (C) Location of crosslink on ERK. The ERK tryptic peptide found crosslinked to the Cic AzF-containing peptide is shown in red on the ERK crystal structure. The Cic-linked residue (T108) is shown in yellow in space-filling representation. Structure is drawn from Protein Data Bank (PDB) file 1ERK (27).
Three positions (F1054, L1056, and P1060) in the putative binding region of GSTCic were tested for incorporation of AzF using an orthogonal aminoacyl tRNA synthetase and tRNA derived from the Methanococcus jannaschii tyrosyl-tRNA synthetase and tRNA (23). All three AzF-substituted GSTCic variants tested expressed at levels comparable to wild-type GSTCic (SI Appendix, Fig. S1B). We next assessed the extent to which these three AzF-incorporated GSTCic variants crosslink to ERK upon exposure to UV light. The formation of a photocrosslinked heterodimer can be identified by the UV exposure-dependent appearance of a slow migrating species corresponding to the combined molecular weight of ERK and GSTCic in both anti-GST (detects GSTCic) and streptavidin (SA, detects biotinylated ERK) Western blots. For all three GSTCic variants tested, a small amount of crosslinked species appeared after 5 min of UV irradiation and increased after 30 min of exposure (SI Appendix, Fig. S1C), indicating successful photocrosslinking. The highest crosslinking efficiency was achieved with GSTCic F1054AzF. Photoreaction conditions were optimized using this variant, enabling ∼10% crosslinking efficiency with a clear band corresponding to the ERK–GSTCic crosslinked species (Fig. 2B).
Because the yield of heterologously expressed, purified mammalian ERK2 is ∼20-fold higher than the Drosophila homolog, Rolled (26), (7 mg/L and 0.3 mg/L, respectively), mammalian ERK2 was used for the GSTCic alanine screen and photocrosslinking experiments. Mammalian ERK2 and Rolled are 92.8% similar with high conservation of residues in known docking sites (SI Appendix, Fig. S2). Thus, results obtained using mammalian ERK2 are likely to be representative of the native Drosophila Cic–Rolled interaction. Nonetheless, we also incubated Rolled with GSTCic F1054AzF and exposed them to UV light in the same manner, which led to the formation of a photocrosslinked heterodimer (SI Appendix, Fig. S3A).
Mass Spectrometry-Based Identification of Cic Binding Site on ERK.
The high molecular weight, covalently crosslinked ERK–GSTCic band was excised from a Coomassie-stained protein gel of the reaction product and subjected to in-gel tryptic digestion. The resulting peptide mixture contained an AzF-incorporated GSTCic tryptic peptide covalently bound to an ERK tryptic peptide located at the Cic binding site. LC-MS/MS was performed on the mixture to identify this peptide adduct. A 4068.12-Da peptide ion was found, which corresponds to the mass of the adduct between an ERK tryptic peptide (DVYIVQDLMETDlYKLLK, 2199.16 Da) and the AzF-containing Cic tryptic peptide (GSDGQQZILAPTPAQLGR, 1896.97 Da, AzF represented by Z) including the 28.01-Da loss of N2 caused by photolysis (SI Appendix, Fig. S4). Peptide sequencing by tandem mass spectrometry confirmed the identity of the crosslinked peptide (SI Appendix, Fig. S4D and Tables S1 and S2). The same region of Rolled was crosslinked to GSTCic F1054AzF and identified by tandem mass spectrometry, confirming that results obtained using mammalian ERK2 are representative of the Drosophila ERK–Cic interaction (SI Appendix, Fig. S3 and Tables S3 and S4).
The identified ERK tryptic peptide spans the N- and C-terminal lobes of the ERK structure (Fig. 2C) (27). The major peaks present on the tandem mass spectrum of the crosslinked peptide correspond to b- and y-series fragments of ERK linked to the full-length AzF-containing Cic peptide (SI Appendix, Fig. S4D and Table S1). Masses of b- and y-series fragments of Cic with the full-length ERK peptide were also detected (SI Appendix, Fig. S4D and Table S2). The fragmentation pattern suggests that the covalent link between Cic and ERK peptides occurs on ERK at T108 (Fig. 2C and SI Appendix, Fig. S4D). T108 is directly adjacent to the DRS hydrophobic groove with the side chain oriented toward the pocket formed by DRS residues L113, L119, H123, and L155 (Fig. 3A).
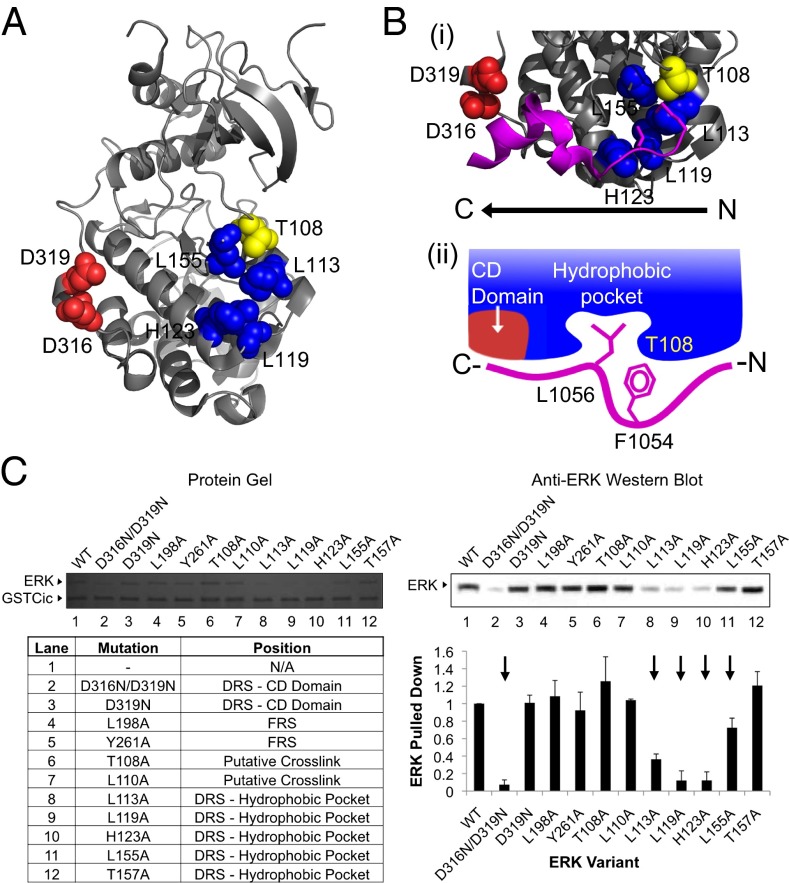
Fig. 3.
Drosophila Cic binds to the ERK DRS. (A) Location of the ERK DRS relative to Cic crosslinking site. The ERK DRS is composed of a negatively charged region (CD domain, red) and a hydrophobic pocket (blue). The location of Cic crosslinking (T108, yellow) is adjacent to the hydrophobic pocket of the DRS. ERK DRS and Cic crosslinking residues are labeled. ERK structure is drawn from PDB file 2Y9Q (32). (B) Cic binds to the DRS using a reverse D site. (i) Crystal structure of the MNK1 reverse D site peptide (magenta) bound to the ERK DRS. Shown in stick representation are a leucine side chain buried in the DRS hydrophobic pocket and, two amino acids upstream, a methionine side chain pointing toward ERK T108. ERK DRS and Cic crosslinking residues are labeled. Structure is drawn from PDB file 2Y9Q (32). (ii) Proposed ERK–Cic binding mechanism. F1054—the site of AzF incorporation located two amino acids upstream of the critical ERK-binding L1056—is aligned with T108, whereas L1056 occupies the ERK hydrophobic pocket. (C) ERK variants with substitutions in the DRS, FRS, and crosslinking site were tested for loss of binding to GSTCic by GST pull-down. Five variants with alanine substitutions in the DRS lost binding to GSTCic (indicated with an arrow). Reported values represent an average of three experiments, and error bars correspond to 1 SD of the mean.
Cic Binds to the ERK DRS with a Reverse D Site.
The structures of ERK in complex with a number of D sites have been solved (28–33). In a canonical D site–ERK interaction, an N-terminal cluster of positively charged amino acids in the D site form electrostatic interactions with the negatively charged portion of the DRS. Downstream, a C-terminal hydrophobic motif forms contacts with the DRS hydrophobic groove (15, 29, 30). Notably, in most D site–ERK interactions, a leucine residue in this C-terminal hydrophobic motif is buried in the DRS hydrophobic pocket. This suggests that the critical ERK-binding residue in Cic, L1056, may occupy this site when in complex with ERK.
The position of AzF in GSTCic F1054AzF relative to the critical L1056 is inconsistent with a classical DRS–D site interaction. AzF is located two amino acids upstream of L1056, however the ERK site at which AzF crosslinks with ERK is located downstream of the hydrophobic pocket-interacting leucine in a would-be classical D site peptide. Thus, Cic is likely extended on ERK’s surface in a reversed orientation relative to canonical D site peptides (Fig. 3B).
Other ERK-interacting proteins have been found to bind the DRS in this reversed orientation. Recently, the structures of ERK in complex with protein substrates PEA15 and RSK1 were solved and found to involve a DRS–D site interaction using a reverse-orientation D site (28, 33). The structures of peptides corresponding to the MNK1 and RSK1 reverse D sites in complex with ERK have also been solved (32). Notably, all of these interactions feature a leucine buried in the hydrophobic pocket.
Inspection of ERK-bound reverse D site structures reveals features consistent with the results of our mutagenesis and crosslinking experiments. The MNK1 reverse D site peptide has a leucine residue buried in the ERK2 hydrophobic pocket (Fig. 3B) (32). Two residues upstream—in the position analogous to F1054 in Cic relative to the ERK-binding leucine—a methionine side chain is aligned with ERK T108—the amino acid in ERK that was shown to crosslink to GSTCic F1054AzF. This finding suggests that Cic binds to the ERK DRS hydrophobic groove in a manner similar to known reverse D site-containing peptides: A critical leucine residue occupies the pocket formed by ERK residues L113, L119, H123, and L155 with the position two amino acids upstream in-line with ERK T108 (Fig. 3B).
To validate the results of the crosslinking and MS experiments, amino acid substitutions in ERK were tested for their effects on binding to GSTCic. Substitutions were made at the crosslinking site (T108A), the charged region of the DRS (D319N, D316N/D319N), the hydrophobic groove of the DRS (L113A, L119A, H123A, L155A, and T157A), and to the FRS (L198A and Y261A) as a negative control. ERK variants were screened for loss of binding to GSTCic using the pull-down assay described above.
As expected, substitutions in the hydrophobic pocket had a major effect on the amount of ERK pulled down (Fig. 3C). In particular, the L113A, L119A, and H123A variants nearly abolished ERK binding signal. L155A also decreased binding affinity, although to a lesser extent. These amino acids make up the interior of the DRS hydrophobic groove. In the ERK-bound MNK1 D site peptide structure, L113, L119, and H123 all make contacts with the key leucine side chain (Fig. 3B). This provides further evidence that Cic binds to the DRS hydrophobic groove of ERK. The substitution to T108—the residue that crosslinks to GSTCic F1054AzF—did not have an effect on binding, indicating that, whereas it is readily accessible for crosslinking at the ERK–Cic binding interface, T108 does not participate directly in the interaction.
Surprisingly, the ERK variant that removed all of the charge in the DRS (D316N/D319N) also had a significant effect on binding (Fig. 3C). This finding was unexpected, as alanine replacements of positively charged residues in the GSTCic alanine screen did not reduce binding. Furthermore, the D319N substitution alone did not show any effect. This mutation, termed the sevenmaker mutation, was first identified in a screen for gain-of-function mutants in Drosophila that rescue ERK-dependent eye development in the absence of upstream components of the ERK signaling pathway (34). This mutation has been shown to weaken the interaction between ERK and DRS-interacting proteins, especially regulators of ERK, including a number of phosphatases (14, 21, 35–40). Thus, it appears that both Cic and ERK phosphatases recognize overlapping surfaces in ERK, although possibly involving different amino acid contacts. Taken together, these experiments show that Cic binds to the ERK DRS, mainly through an interaction of Cic L1056 with the hydrophobic pocket formed by ERK L113, L119, H123, and L155.
Human Cic Binds to the ERK DRS.
In contrast to fly Cic, where the C2 domain had been previously identified as important for ERK binding, no ERK binding site is known for human Cic. Thus, we decided to scan the C-terminal half of human Cic for ERK binding. Seventy-five amino acid fragments of human Cic spanning residues 800–1,608 were fused to GST and tested for binding to ERK by GST pull-down. The GST-fused human Cic fragment containing residues 1,325–1,399 (herein referred to as GSThCic, Fig. 4C) pulled down the largest amount of ERK and was confirmed to bind to ERK by BLI (Fig. 4A). Interestingly, this fragment of human Cic does not share significant sequence similarity to the Drosophila Cic ERK-binding region.
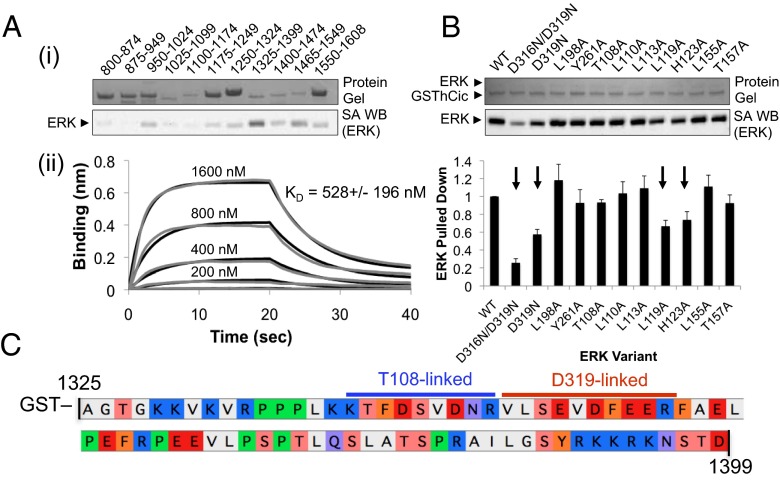
Fig. 4.
Human Cic binds to the ERK DRS. (A) Identification of the ERK-binding region of human Cic. (i) Seventy-five amino acid fragments of human Cic 800–1,608 were fused to GST and assayed for binding to ERK by GST pull-down. The GST-fused Cic fragment corresponding to positions 1,325–1,399 (GSThCic) pulled down more ERK than any other fragment. (ii) GSThCic binding to ERK was confirmed by BLI. GSThCic was immobilized on anti-GST biosensors and binding to ERK at concentrations ranging from 100 to 1,600 nM was measured (gray curves). Data were fit globally (black curves). Reported KD is an average of global fits to three datasets, and the error represents 1 SD of the mean. (B) ERK variants with substitutions in the DRS, FRS, and crosslinking site were tested for loss of binding to GSThCic by GST pull-down. Four variants with alanine substitutions in the DRS lost binding to GSThCic (indicated with arrows). Reported values represent an average of three experiments, and error bars correspond to 1 SD of the mean. (C) Sequence of the ERK-binding region of human Cic (residues 1,325–1,399) fused to GST (GSThCic). Tryptic peptides found crosslinked to ERK T108AzF and ERK D319AzF variants are indicated with blue and red lines, respectively.
To identify the region of ERK that binds to human Cic, the same ERK variants used to study the Drosophila Cic–ERK interaction were screened for loss of binding to GSThCic. Despite the lack of conservation between ERK-binding regions, the human Cic–ERK binding affinity was reduced by many of the same DRS substitutions that affected Drosophila Cic (Fig. 4B). Both of the ERK variants with substitutions in the charged region of the DRS—the D319N and D316N/D319N variants—had a significant effect on the amount of ERK pulled down by GSThCic, as opposed to Drosophila Cic, where the effect was only observed when both aspartic acid residues were substituted. The two substitutions in the hydrophobic pocket that had the greatest effect on the binding of Drosophila Cic to ERK—L119A and H123A—also decreased binding to GSThCic, although to a lesser extent, whereas the L113A and L155A substitutions only affected the interaction with the Drosophila protein.
In experiments with Drosophila Cic, we used amino acid-level knowledge about which Cic residues are used to bind to ERK to photocrosslink these interaction partners and identify a binding site on ERK. We applied the same approach using information gleaned from ERK mutagenesis to identify where it binds to human Cic. AzF was incorporated into ERK in four positions in the regions of the ERK that play a role in binding to GSThCic (T108, H123, L155, and D319). Upon irradiation with UV light, both ERK T108AzF and ERK D319AzF formed an adduct with GSThCic, as shown by protein gel, anti-ERK Western blot, and anti-GST Western blot (SI Appendix, Fig. S5). It is notable that efficient crosslinking to human Cic was observed when AzF was inserted into the ERK site found linked to fly GSTCic F1054AzF. Analysis by LC-MS/MS identified the human Cic tryptic peptides that were photocrosslinked to ERK (SI Appendix, Figs. S6 and S7 and Tables S5–S7). Two adjacent peptides were crosslinked to the two AzF-incorporated ERK variants in a reverse D-site orientation, consistent with Drosophila Cic. The N-terminal peptide (human Cic residues 1,340–1,348) crosslinked to AzF in the T108 position of ERK and the next tryptic peptide downstream (human Cic residues 1,349–1,358) crosslinked to AzF in the D319 position (Fig. 4C). This finding was further confirmed by testing five 35-amino-acid fragments of human Cic shifted by 10 amino acids that span the 75 amino acids of human Cic present in GSThCic (1,325–1,359, 1,335–1,369, 1,345–1,379, 1,355–1,389, and 1,365–1,399) for binding to ERK. The two fragments of GSThCic that bound to ERK in a GST pull-down (1,325–1,359 and 1,335–1,369) both contain the two crosslinked human Cic peptides in their overlapping regions (SI Appendix, Fig. S8).
Discussion
A number of methods have been used to probe protein–protein interactions in the ERK pathway. Techniques that look at ERK binding or catalysis have been used alongside mutagenesis of ERK (41) or competition with molecules known to bind at certain ERK sites (19, 42). These approaches have uncovered a great deal about ERK binding interactions. However, these studies rely on prior knowledge of docking sites on ERK. Furthermore, individual point mutations within a known docking site may have different effects on different binding partners. This challenge was illustrated in this study—notably, the ERK D319N substitution had very different effects on the interaction with human and Drosophila Cic.
Crystal structures of ERK in complex with its binding partners (28–33) provide invaluable insights, many of which formed the basis of this study. However, the ability to form crystals of a complex is not guaranteed, and most of the structures of ERK complexes use linear peptides derived from the ERK-interacting molecules instead of structured proteins. Hydrogen–deuterium exchange mass spectrometry (15) has been used to map regions of ERK rendered inaccessible to solvent by a binding partner. This approach provides a picture of large regions of ERK that are involved in binding and revealed, among other things, the areas of ERK that form the FRS, but cannot distinguish between residues that are directly or peripherally involved in binding.
Chemical crosslinking and analysis of crosslinked species have been used extensively to identify binding partners and locate crosslinking sites in other systems (43–46). Chemical crosslinking relies on the natural availability of particular amino acids (usually lysine or cysteine) on the surface of interacting proteins and the use of crosslinking reagents with spacers of varying length. If none of the required side chains are present at the binding interface, crosslinking may not occur. If a crosslink is formed, the use of spacers could mean that the location of the covalent linkage is not representative of the binding interface.
Photoactivatable, crosslinking noncanonical amino acids also enable the formation of a covalent link between complexed proteins and can be analyzed in a similar manner to determine where they bind (47, 48). In this study, we began by site-specifically inserting AzF into Drosophila Cic at the ERK–Cic binding interface and used UV crosslinking and analysis by mass spectrometry to identify the site on ERK that was covalently linked to Cic. Because the photoactivatable group was inserted specifically at the binding interface using stop codon suppression, and no additional reagents or spacers were used, the crosslink we identified was formed on ERK precisely at the binding site. Furthermore, this technique did not require any assumptions as to which regions of ERK are involved in the interaction. We illustrated the robustness of this technique by using it to perform the reverse experiment, identifying where ERK binds to a newly discovered fragment of human Cic by modifying ERK positions with AzF. We propose that the photocrosslinking ERK variants we have generated here may also be used for the discovery of new ERK substrates that engage the DRS.
Materials and Methods
All recombinant proteins were expressed in Escherichia coli BL21 and purified using standard affinity chromatography techniques. Pull-down assays were carried out using glutathione Sepharose resin to bind to GST in the GST–Cic fusions. Proteins substituted with AzF were produced in E. coli BL21 using an engineered aminoacyl-tRNA synthetase. Photocrosslinking was carried out by irradiating protein mixture with a transilluminator (365 nm) at 4 °C. Details for all experiments are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Mona Singh and Alexey Veraksa for helpful discussions throughout the course of this work and for comments on the manuscript, Tharan Srikumar for assistance with mass spectrometry, Wai Ling Cheung for assistance with protein expression, and Carol MacKintosh for the clone of human Cic. This work was supported by Grant GM086537 from the National Institute of General Medical Sciences. A.J.L. is a DuPont Young Professor and a Sloan Research Fellow.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1501373112/-/DCSupplemental.
References
- 1.Jiménez G, Shvartsman SY, Paroush Z. The Capicua repressor—a general sensor of RTK signaling in development and disease. J Cell Sci. 2012;125(Pt 6):1383–1391. doi: 10.1242/jcs.092965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiménez G, Guichet A, Ephrussi A, Casanova J. Relief of gene repression by torso RTK signaling: Role of capicua in Drosophila terminal and dorsoventral patterning. Genes Dev. 2000;14(2):224–231. [PMC free article] [PubMed] [Google Scholar]
- 3.Lam YC, et al. ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell. 2006;127(7):1335–1347. doi: 10.1016/j.cell.2006.11.038. [DOI] [PubMed] [Google Scholar]
- 4.Ajuria L, et al. Capicua DNA-binding sites are general response elements for RTK signaling in Drosophila. Development. 2011;138(5):915–924. doi: 10.1242/dev.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dissanayake K, et al. ERK/p90(RSK)/14-3-3 signalling has an impact on expression of PEA3 Ets transcription factors via the transcriptional repressor capicúa. Biochem J. 2011;433(3):515–525. doi: 10.1042/BJ20101562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Astigarraga S, et al. A MAPK docking site is critical for downregulation of Capicua by Torso and EGFR RTK signaling. EMBO J. 2007;26(3):668–677. doi: 10.1038/sj.emboj.7601532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fryer JD, et al. Exercise and genetic rescue of SCA1 via the transcriptional repressor Capicua. Science. 2011;334(6056):690–693. doi: 10.1126/science.1212673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grimm O, et al. Torso RTK controls Capicua degradation by changing its subcellular localization. Development. 2012;139(21):3962–3968. doi: 10.1242/dev.084327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim B, et al. Kinetics of gene derepression by ERK signaling. Proc Natl Acad Sci USA. 2013;110(25):10330–10335. doi: 10.1073/pnas.1303635110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Futran AS, Link AJ, Seger R, Shvartsman SY. ERK as a model for systems biology of enzyme kinetics in cells. Curr Biol. 2013;23(21):R972–R979. doi: 10.1016/j.cub.2013.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim Y, et al. Substrate-dependent control of MAPK phosphorylation in vivo. Mol Syst Biol. 2011;7:467. doi: 10.1038/msb.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim Y, et al. Gene regulation by MAPK substrate competition. Dev Cell. 2011;20(6):880–887. doi: 10.1016/j.devcel.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim Y, et al. Context-dependent transcriptional interpretation of mitogen activated protein kinase signaling in the Drosophila embryo. Chaos. 2013;23(2):025105. doi: 10.1063/1.4808157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanoue T, Adachi M, Moriguchi T, Nishida E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat Cell Biol. 2000;2(2):110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- 15.Lee T, et al. Docking motif interactions in MAP kinases revealed by hydrogen exchange mass spectrometry. Mol Cell. 2004;14(1):43–55. doi: 10.1016/s1097-2765(04)00161-3. [DOI] [PubMed] [Google Scholar]
- 16.Tanoue T, Maeda R, Adachi M, Nishida E. Identification of a docking groove on ERK and p38 MAP kinases that regulates the specificity of docking interactions. EMBO J. 2001;20(3):466–479. doi: 10.1093/emboj/20.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubinfeld H, Hanoch T, Seger R. Identification of a cytoplasmic-retention sequence in ERK2. J Biol Chem. 1999;274(43):30349–30352. doi: 10.1074/jbc.274.43.30349. [DOI] [PubMed] [Google Scholar]
- 18.Bardwell AJ, Flatauer LJ, Matsukuma K, Thorner J, Bardwell L. A conserved docking site in MEKs mediates high-affinity binding to MAP kinases and cooperates with a scaffold protein to enhance signal transmission. J Biol Chem. 2001;276(13):10374–10386. doi: 10.1074/jbc.M010271200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bardwell AJ, Abdollahi M, Bardwell L. Docking sites on mitogen-activated protein kinase (MAPK) kinases, MAPK phosphatases and the Elk-1 transcription factor compete for MAPK binding and are crucial for enzymic activity. Biochem J. 2003;370(Pt 3):1077–1085. doi: 10.1042/BJ20021806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang S-H, Yates PR, Whitmarsh AJ, Davis RJ, Sharrocks AD. The Elk-1 ETS-domain transcription factor contains a mitogen-activated protein kinase targeting motif. Mol Cell Biol. 1998;18(2):710–720. doi: 10.1128/mcb.18.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Be, Stippec S, Robinson FL, Cobb MH. Hydrophobic as well as charged residues in both MEK1 and ERK2 are important for their proper docking. J Biol Chem. 2001;276(28):26509–26515. doi: 10.1074/jbc.M102769200. [DOI] [PubMed] [Google Scholar]
- 22.Jacobs D, Glossip D, Xing H, Muslin AJ, Kornfeld K. Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev. 1999;13(2):163–175. [PMC free article] [PubMed] [Google Scholar]
- 23.Chin JW, et al. Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli. J Am Chem Soc. 2002;124(31):9026–9027. doi: 10.1021/ja027007w. [DOI] [PubMed] [Google Scholar]
- 24.Escher E, Schwyzer R. p-Nitrophenylalanine, p-azidophenylalanine, m-azidophenylalanine, and o-nitro-p-azido-phenylalanine as photoaffinity labels. FEBS Lett. 1974;46(1):347–350. doi: 10.1016/0014-5793(74)80403-5. [DOI] [PubMed] [Google Scholar]
- 25.Leyva E, Platz MS, Persy G, Wirz J. Photochemistry of phenyl azide: The role of singlet and triplet phenylnitrene as transient intermediates. J Am Chem Soc. 1986;108(13):3783–3790. [Google Scholar]
- 26.Biggs WH, 3rd, et al. The Drosophila rolled locus encodes a MAP kinase required in the sevenless signal transduction pathway. EMBO J. 1994;13(7):1628–1635. doi: 10.1002/j.1460-2075.1994.tb06426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang F, Strand A, Robbins D, Cobb MH, Goldsmith EJ. Atomic structure of the MAP kinase ERK2 at 2.3 A resolution. Nature. 1994;367(6465):704–711. doi: 10.1038/367704a0. [DOI] [PubMed] [Google Scholar]
- 28.Mace PD, et al. Structure of ERK2 bound to PEA-15 reveals a mechanism for rapid release of activated MAPK. Nat Commun. 2013;4:1681. doi: 10.1038/ncomms2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou T, Sun L, Humphreys J, Goldsmith EJ. Docking interactions induce exposure of activation loop in the MAP kinase ERK2. Structure. 2006;14(6):1011–1019. doi: 10.1016/j.str.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 30.Liu S, Sun J-P, Zhou B, Zhang Z-Y. Structural basis of docking interactions between ERK2 and MAP kinase phosphatase 3. Proc Natl Acad Sci USA. 2006;103(14):5326–5331. doi: 10.1073/pnas.0510506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gógl G, Törő I, Reményi A. Protein-peptide complex crystallization: A case study on the ERK2 mitogen-activated protein kinase. Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 3):486–489. doi: 10.1107/S0907444912051062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garai Á, et al. Specificity of linear motifs that bind to a common mitogen-activated protein kinase docking groove. Sci Signal. 2012;5(245):ra74–ra74. doi: 10.1126/scisignal.2003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alexa A, et al. Structural assembly of the signaling competent ERK2-RSK1 heterodimeric protein kinase complex. Proc Natl Acad Sci USA. 2015;112(9):2711–2716. doi: 10.1073/pnas.1417571112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brunner D, et al. A gain-of-function mutation in Drosophila MAP kinase activates multiple receptor tyrosine kinase signaling pathways. Cell. 1994;76(5):875–888. doi: 10.1016/0092-8674(94)90362-x. [DOI] [PubMed] [Google Scholar]
- 35.Chu Y, Solski PA, Khosravi-Far R, Der CJ, Kelly K. The mitogen-activated protein kinase phosphatases PAC1, MKP-1, and MKP-2 have unique substrate specificities and reduced activity in vivo toward the ERK2 sevenmaker mutation. J Biol Chem. 1996;271(11):6497–6501. doi: 10.1074/jbc.271.11.6497. [DOI] [PubMed] [Google Scholar]
- 36.Zhou B, et al. Multiple regions of MAP kinase phosphatase 3 are involved in its recognition and activation by ERK2. J Biol Chem. 2001;276(9):6506–6515. doi: 10.1074/jbc.M009753200. [DOI] [PubMed] [Google Scholar]
- 37.Zhao Y, Zhang Z-Y. The mechanism of dephosphorylation of extracellular signal-regulated kinase 2 by mitogen-activated protein kinase phosphatase 3. J Biol Chem. 2001;276(34):32382–32391. doi: 10.1074/jbc.M103369200. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, Zhou B, Zheng C-F, Zhang Z-Y. A bipartite mechanism for ERK2 recognition by its cognate regulators and substrates. J Biol Chem. 2003;278(32):29901–29912. doi: 10.1074/jbc.M303909200. [DOI] [PubMed] [Google Scholar]
- 39.Oellers N, Hafen E. Biochemical characterization of rolledSem, an activated form of Drosophila mitogen-activated protein kinase. J Biol Chem. 1996;271(40):24939–24944. doi: 10.1074/jbc.271.40.24939. [DOI] [PubMed] [Google Scholar]
- 40.Camps M, et al. Catalytic activation of the phosphatase MKP-3 by ERK2 mitogen-activated protein kinase. Science. 1998;280(5367):1262–1265. doi: 10.1126/science.280.5367.1262. [DOI] [PubMed] [Google Scholar]
- 41.Burkhard KA, Chen F, Shapiro P. Quantitative analysis of ERK2 interactions with substrate proteins: Roles for kinase docking domains and activity in determining binding affinity. J Biol Chem. 2011;286(4):2477–2485. doi: 10.1074/jbc.M110.177899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee S, et al. Examining docking interactions on ERK2 with modular peptide substrates. Biochemistry. 2011;50(44):9500–9510. doi: 10.1021/bi201103b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rappsilber J. The beginning of a beautiful friendship: Cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J Struct Biol. 2011;173(3):530–540. doi: 10.1016/j.jsb.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leitner A, Walzthoeni T, Aebersold R. Lysine-specific chemical cross-linking of protein complexes and identification of cross-linking sites using LC-MS/MS and the xQuest/xProphet software pipeline. Nat Protoc. 2014;9(1):120–137. doi: 10.1038/nprot.2013.168. [DOI] [PubMed] [Google Scholar]
- 45.Leitner A, et al. Chemical cross-linking/mass spectrometry targeting acidic residues in proteins and protein complexes. Proc Natl Acad Sci USA. 2014;111(26):9455–9460. doi: 10.1073/pnas.1320298111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang B, et al. Identification of cross-linked peptides from complex samples. Nat Methods. 2012;9(9):904–906. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
- 47.Freinkman E, Chng S-S, Kahne D. The complex that inserts lipopolysaccharide into the bacterial outer membrane forms a two-protein plug-and-barrel. Proc Natl Acad Sci USA. 2011;108(6):2486–2491. doi: 10.1073/pnas.1015617108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grunbeck A, Huber T, Sakmar TP. Mapping a ligand binding site using genetically encoded photoactivatable crosslinkers. Methods Enzymol. 2013;520:307–322. doi: 10.1016/B978-0-12-391861-1.00014-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.