Significance
Northern corn leaf blight (NCLB) is one of the most devastating fungal diseases of maize. The Htn1 disease resistance gene confers quantitative field resistance against most NCLB isolates. Here we show that Htn1 encodes a putative wall-associated receptor-like kinase (RLK). RLKs act as important components of the first tier of the plant innate immune system by perceiving pathogen- or host-derived elicitors on the cell surface. RLKs are often associated with resistance to nonadapted pathogens and are a component of nonhost resistance. Our work demonstrates that the Htn1-RLK plays an important role in host resistance against adapted fungal pathogens.
Keywords: wall-associated receptor-like kinase, quantitative disease resistance, pattern recognition receptor, Exserohilum turcicum, maize
Abstract
Northern corn leaf blight (NCLB) caused by the hemibiotrophic fungus Exserohilum turcicum is an important foliar disease of maize that is mainly controlled by growing resistant maize cultivars. The Htn1 locus confers quantitative and partial NCLB resistance by delaying the onset of lesion formation. Htn1 represents an important source of genetic resistance that was originally introduced from a Mexican landrace into modern maize breeding lines in the 1970s. Using a high-resolution map-based cloning approach, we delimited Htn1 to a 131.7-kb physical interval on chromosome 8 that contained three candidate genes encoding two wall-associated receptor-like kinases (ZmWAK-RLK1 and ZmWAK-RLK2) and one wall-associated receptor-like protein (ZmWAK-RLP1). TILLING (targeting induced local lesions in genomes) mutants in ZmWAK-RLK1 were more susceptible to NCLB than wild-type plants, both in greenhouse experiments and in the field. ZmWAK-RLK1 contains a nonarginine-aspartate (non-RD) kinase domain, typically found in plant innate immune receptors. Sequence comparison showed that the extracellular domain of ZmWAK-RLK1 is highly diverse between different maize genotypes. Furthermore, an alternative splice variant resulting in a truncated protein was present at higher frequency in the susceptible parents of the mapping populations compared with in the resistant parents. Hence, the quantitative Htn1 disease resistance in maize is encoded by an unusual innate immune receptor with an extracellular wall-associated kinase domain. These results further highlight the importance of this protein family in resistance to adapted pathogens.
Plants are constantly attacked by potential pathogenic microbes, specifically, viruses, bacteria, and fungi. Although lacking an adaptive immune system comparable to the one found in vertebrates, plants have evolved a plethora of strategies to fend off microbial pathogens. The first tier of defense is formed by plasma membrane-anchored pattern recognition receptors (PRRs). These receptor proteins monitor the extracellular space for the presence of microbial- or host-derived elicitors, also called pathogen-associated molecular patterns (PAMP) or danger-associated molecular patterns (DAMP), respectively. Perception of PAMPs and DAMPs triggers a signaling cascade that activates numerous defense responses, called PAMP-triggered immunity and DAMP-triggered immunity, respectively. PAMPs are often highly conserved microbial structures that are characteristic for entire pathogen classes (1, 2). Examples are the fungal cell wall component chitin or bacterial flagellin. Perception of highly conserved PAMPs therefore results in broad-spectrum resistance against whole groups of nonadapted microbes, also referred to as nonhost resistance. To avoid PAMP-triggered immunity and DAMP-triggered immunity, pathogens are equipped with specific effectors that are tailored to suppress the plant’s immune response. These virulence effectors are injected into the host cytoplasm, where they can then, in turn, be recognized by cytoplasmic receptor proteins, which results in effector-triggered immunity (ETI) (3). The intracellular recognition of effector molecules is mediated by structurally related proteins belonging to the nucleotide-binding site-leucine-rich repeat (NBS-LRR) family (4). ETI forms the second tier of the plant immune response. In contrast to nonhost resistance, the ETI response against adapted pathogens is much stronger and often results in the death of the infected cell through hypersensitive reaction. Hence, there is a strong selective pressure on the pathogen to avoid ETI by evolving new virulence effectors that escape recognition by NBS-LRR immune receptors. This coevolutionary arms race is the major reason for rapid breakdown of NBS-LRR-based disease resistance in crop plants, which often occurs within only a few years (5, 6).
Cell surface PRRs are encoded by receptor-like kinase (RLK) and receptor-like protein (RLP) genes. Both RLKs and RLPs contain an extracellular elicitor-binding domain and a transmembrane domain. In contrast to RLKs, RLPs lack a cytoplasmic serine/threonine kinase domain (7). The best-studied PRR is the leucine-rich repeat (LRR) receptor-like kinase flagellin-sensitive-2 (FLS2) of Arabidopsis, which binds an epitope of bacterial flagellin. On flagellin perception, this kinase initiates a signaling cascade that results in nonhost resistance (8). Most PRRs described until today contain such an extracellular LRR domain. Not all PRRs, however, are involved in nonhost resistance. The rice LRR-RLK Xa21, for example, confers race-specific resistance against the bacterial rice blast pathogen Xanthomonas oryzae pv. oryzae (9). The effector recognized by Xa21 is still unknown. In addition, plants have evolved the ability to perceive endogenous molecules through PRRs that are produced during pathogen infection. For example, the Arabidopsis wall-associated kinase 1 (WAK1) binds cell wall-derived oligogalacturonides that are released during pathogen infection and serve as DAMPs (10). AtDORN1, an Arabidopsis lectin-RLK, binds plant-derived extracellular ATP likely produced during pathogen infection or wounding (11).
Maize (Zea mays L.) is the most widely grown crop in the world and represents an important source of food, feed, biofuel, and industrial products. Fungal diseases are a major threat to maize production and can result in severe crop losses. Northern corn leaf blight (NCLB) is the most devastating foliar disease of maize. It is caused by the hemibiotrophic ascomycete fungus Exserohilum turcicum (teleomorph Setosphaeria turcica) (12). The fungus spreads biotrophically during the initial infection process before switching to a necrotrophic lifestyle. Infections manifest as local lesions and necrosis, which lead to reduced photosynthetically active leaf area and yield losses. The disease occurs prevalently under conditions of high humidity and moderate temperatures and can be found in most regions where maize is grown (13–15). The Htn1 locus confers quantitative and partial resistance against NCLB by delaying lesion formation. Htn1 was originally introgressed into modern maize cultivars from the Mexican landrace Pepitilla in the 1970s (16). The Htn1 resistance reaction is different from the other known major NCLB resistance genes Ht1, Ht2, and Ht3, which confer qualitative resistance, resulting in chlorotic-necrotic lesions. In contrast, Htn1 leads to a delay of sporulation without chlorotic lesions (17, 18). The dominant and race-specific Htn1 gene is effective against the most prevalent NCLB races, but virulent isolates have been found (18). The strength of the Htn1 resistance depends on environmental conditions and maize genotype (19). Htn1 has been mapped to maize chromosome 8 ∼10 cM distal to the NCLB resistance gene Ht2 (20–23).
Here, we describe the map-based isolation of the Htn1 gene, which encodes for a pattern recognition receptor with a putative extracellular, wall-associated domain.
Results and Discussion
High-Resolution Genetic Mapping of Htn1.
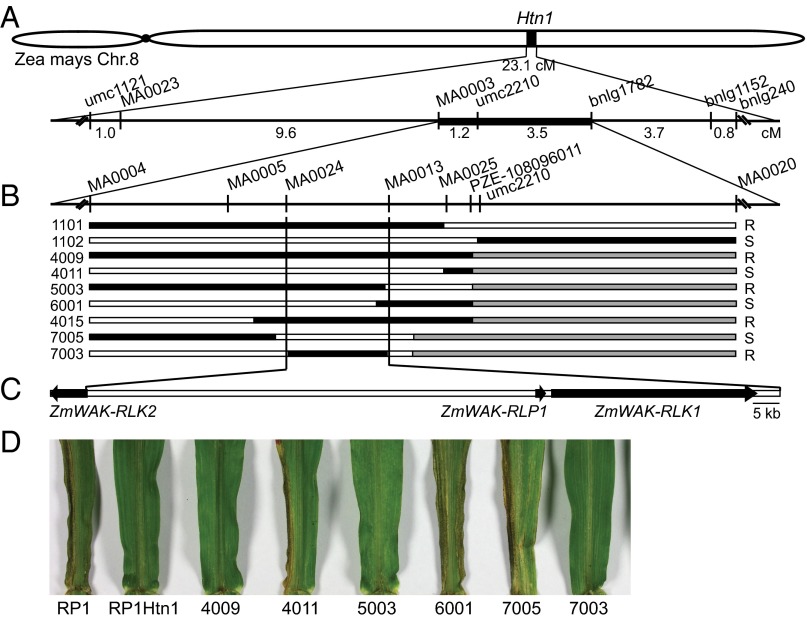
The Htn1 locus from the Mexican landrace Pepitilla was introgressed into the NCLB susceptible maize line B37. B37Htn1 (17) served as the donor to develop three pairs of near isogenic lines for map-based isolation of Htn1. The three recurrent parents RP1, RP3, and RP4 were moderately to highly susceptible to NCLB in the field (SI Appendix, Tables S1 and S2). All three Htn1 introgression lines, RP1Htn1, RP3Htn1, and RP4Htn1, were more resistant than their respective recurrent parent and only developed few and small lesions typical for the effect of Htn1. Using six microsatellite markers and two single nucleotide polymorphism (SNP) markers, Htn1 was initially mapped at low resolution to a 23.1-cM interval on the short arm of chromosome 8 in a F2 population consisting of 528 progeny of a cross between the resistant parent RP1Htn1 and the susceptible parent RP1 (Fig. 1A and SI Appendix, Fig. S1). In parallel, we established a NCLB greenhouse seedling infection assay and phenotyped an additional 262 F2 progeny of the same cross. By including additional markers, we reduced the Htn1 region to a 4.7-cM target interval flanked by SNP marker MA0003 and microsatellite marker bnlg1782. Microsatellite marker umc2210 cosegregated with Htn1 (Fig. 1A).
Fig. 1.
Map-based isolation of Htn1. (A) Initial mapping located Htn1 to a 4.7-cM region on the long arm of maize chromosome 8. (B) High-resolution mapping delimited Htn1 to a 131.7-kb physical interval flanked by SNP markers MA0024 and MA0013. Recombination breakpoints for critical recombinants are shown. Black segments represent the resistant RP1Htn1 genotype, and white segments the susceptible RP1 genotype. Gray segments indicate regions that were not tested with markers. Only critical markers around the recombination breakpoints are indicated for simplicity. Phenotypic scores (R = resistant, S = susceptible) are indicated on the right. (C) The Htn1 target interval contains three predicted candidate genes: ZmWAK-RLK2, ZmWAK-RLP1, and ZmWAK-RLK1. (D) Leaves of critical recombinants and parents of the mapping population infected with E. turcicum in a seedling greenhouse assay. Infections can be seen as black sporulating lesions on the leaves.
For this flanking interval, a 1.3-Mb physical scaffold was established and sequenced, using a BAC library of the resistant parent RP4Htn1. Sequence conservation of the 1.3-Mb RP4Htn1 scaffold with the B73 reference genome sequence (24) was rather weak across the entire region, and gene content differed between the two lines (SI Appendix, Figs. S2 and S3A). The RP4Htn1 sequences were then used to saturate the target interval with SNP markers. To increase recombination events in the target region, heterozygous F2 progeny plants of the RP1 × RP1Htn1 population were backcrossed to RP1 and selfed. New recombinants were phenotyped in the field and in the greenhouse. This strategy allowed us to ultimately delimit Htn1 to a 131.7-kb physical interval flanked by SNP markers MA0024 and MA0013 (Fig. 1 B and D). The region contained three putative candidate genes in the RP4Htn1 sequences: two wall-associated receptor-like kinases (ZmWAK-RLK1 and ZmWAK-RLK2) and one wall-associated receptor-like protein that lacked a cytoplasmic kinase domain (ZmWAK-RLP1; Fig. 1C). Genes encoding WAK-RLKs and WAK-RLPs are often found as clusters in plant genomes (25). The left-flanking marker MA0024 was located in the 5′ region of the ZmWAK-RLK2 coding sequence, 296 bp downstream of the start codon. One critical recombinant, line 7003, had the susceptible allele for MA0024 but was clearly resistant in the seedling NCLB infection assay (Fig. 1 C and D), thereby excluding the sequence downstream of nucleotide 296 in ZmWAK-RLK2 as a Htn1 candidate region. We still considered ZmWAK-RLK2 a candidate gene because we could not exclude that the recombination in line 7003 occurred in the promoter region or the first 296 bp of this gene. Recombinants 5003 and 6001 excluded the region to the right of ZmWAK-RLK1 (Fig. 1 C and D), resulting in the three candidate genes mentioned earlier.
Characterization of the Three Candidate Genes Found in the Htn1 Target Interval.
All three candidate genes were expressed in line RP1Htn1 and have ORFs possibly encoding functional proteins. A conserved domain search revealed high structural similarity among the three predicted proteins. Their N-terminal, putatively extracellular domain contained a cysteine-rich galacturonan-binding domain (GUB-WAK_bind; pfam13947), followed by a wall-associated receptor kinase domain (WAK_assoc domain; pfam14380). These two cysteine-rich domains are characteristic for wall-associated kinases (26). Furthermore, the three proteins contained a predicted transmembrane domain (Fig. 2). The coding sequence of ZmWAK-RLP1 consists of a single, 878-bp exon. The predicted protein lacks a cytoplasmic kinase domain, which classifies ZmWAK-RLP1 as a receptor-like protein. Receptor-like proteins are thought to interact with other kinases for signaling (11). The 7,162-bp ZmWAK-RLK2 gene has five exons and is located in reverse orientation compared with the other two genes (Fig. 1C). The genomic sequence of ZmWAK-RLK1 is nearly 39 kb in size (Fig. 2). This massive increase in gene size is a result of the insertions of several transposons in the first intron (SI Appendix, Fig. S3B). cDNA amplification showed that this intron was correctly spliced, despite its large size. In addition to the extracellular and transmembrane domain, ZmWAK-RLK1 and ZmWAK-RLK2 contain a predicted cytoplasmic serine/threonine protein kinase domain at the C terminus. Receptor-like kinases can be further divided into RD and non-RD kinases, depending on the presence or absence of an arginine residue at the catalytic site of the kinase domain. ZmWAK-RLK2 contains an RD kinase and ZmWAK-RLK1 has a non-RD kinase domain (Fig. 2). Most receptor-like kinases involved in plant immunity identified so far belong to the non-RD kinases, whereas RD kinases are thought to play a role in other processes such as development (27). Interestingly, although the kinase domains of the two candidate proteins only showed 27% amino acid identity, their extracellular domains were highly similar (86% amino acid identity), indicating a recent gene conversion event. Despite having the same domain features, ZmWAK-RLP1 only showed weak sequence conservation to the extracellular domains of the two ZmWAK-RLKs. Compared with dicotyledonous plants, the WAK gene family has considerably expanded in cereals. Although Arabidopsis contains 26 WAK and WAK-like genes, more than 100 WAK genes have been identified in the genomes of maize and rice (28, 29). A Southern blot analysis with probes derived from the conserved extracellular domains of ZmWAK-RLK1 and ZmWAK-RLK2 revealed that there is no additional gene with a similar wall-associated kinase domain in the genomes of RP1Htn1 and RP1 (SI Appendix, Fig. S4). Despite the large number of WAK genes in the maize genome, the extracellular domains of ZmWAK-RLK1 and ZmWAK-RLK2 are therefore unique.
Fig. 2.
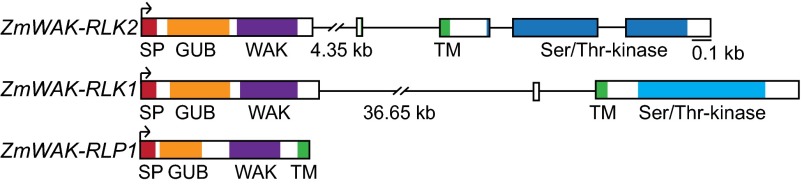
Gene structure and protein domain prediction of the three candidate genes, ZmWAK-RLK2, ZmWAK-RLK1, and ZmWAK-RLP1, from the resistant Htn1 locus. Predicted domains are in color. Exons are represented as rectangular bars, and introns as black lines between the exons. GUB (GUB_WAK), wall-associated receptor kinase galacturonan-binding; Ser/Thr-kinase, catalytic domain of the serine/threonine kinases and interleukin 1 receptor-associated kinases; SP, signal peptide; TM, transmembrane domain; WAK (WAK_assoc), wall-associated receptor kinase C-terminal. Dark blue, RD-kinase; bright blue, non-RD kinase.
On the basis of our mapping data and the protein structure, we considered the non-RD ZmWAK-RLK1 as the best candidate gene for Htn1. A similar target interval for NCLB resistance on chromosome 8 has been identified in maize line PH99N. The PH99N NCLB interval contains a homolog of ZmWAK-RLK1 with identical predicted cDNA sequence but different intron sequences (patent international publication number WO 2011163590 A1). However, no functional candidate gene validation is described in this patent application.
Analysis of TILLING Mutants for Functional Identification of Htn1.
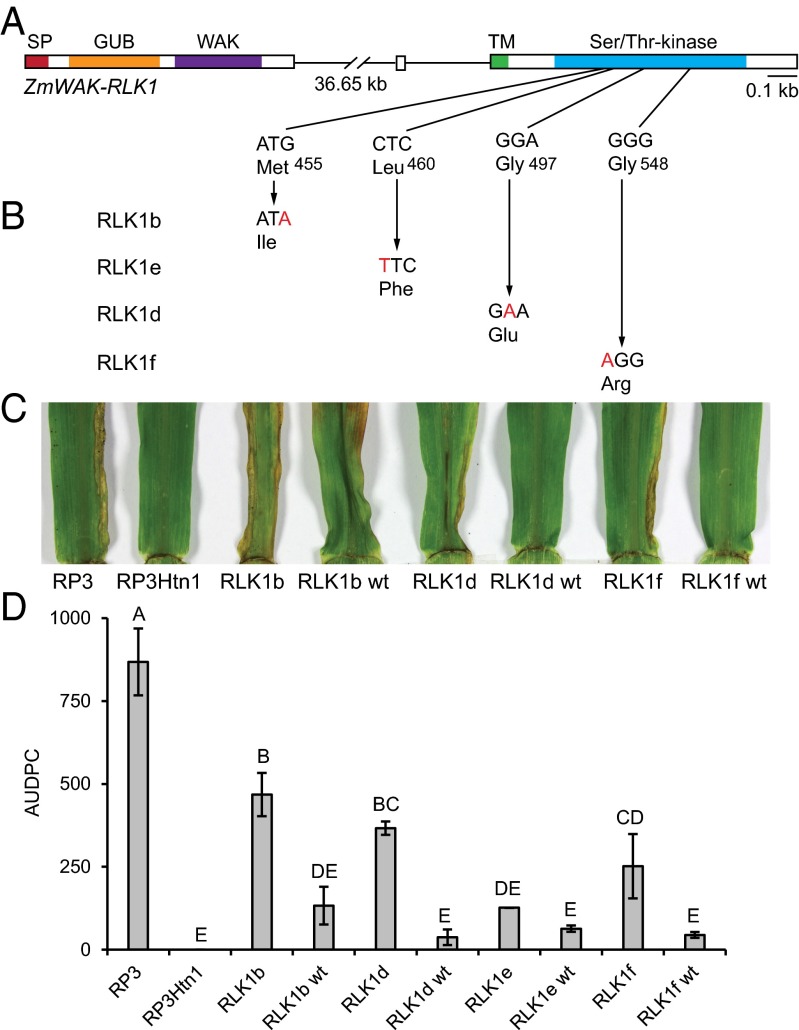
For functional validation of the three candidate genes, we developed a targeting induced local lesions in genomes (TILLING) population consisting of 10,000 plants of the resistant parent RP3Htn1. The TILLING population was screened with PCR primers targeting the transmembrane and kinase domains of ZmWAK-RLK1 and ZmWAK-RLK2 (SI Appendix, Table S3). Four mutant lines for ZmWAK-RLK1 and three mutants for ZmWAK-RLK2 were identified, all of them containing SNPs that resulted in amino acid substitutions (Fig. 3 A and B and SI Appendix, Table S4). Mutant lines, along with their respective segregating wild-type sister lines, were tested for resistance to NCLB in greenhouse infection tests. The TILLING parent RP3Htn1 was completely resistant. The three mutants in ZmWAK-RLK2, RLK2b, RLK2d, and RLK2e, showed no increase in susceptibility compared with RP3Htn1 and their corresponding wild-type segregants (SI Appendix, Fig. S5). In contrast, three of the four mutant lines in ZmWAK-RLK1 (mutants RLK1b, RLK1d, and RLK1f) were more susceptible than line RP3Htn1 and the corresponding wild-type segregants (Fig. 3 C and D). One ZmWAK-RLK1 mutant, RLK1e, showed wild-type levels of disease resistance (Fig. 3D). Identical results were obtained in field infection tests. Again, the ZmWAK-RLK1 mutant lines RLK1b, RLK1d, and RLK1f were more susceptible than wild-type (SI Appendix, Fig. S6). These results demonstrate that the NCLB disease resistance in the TILLING parent RP3Htn1 is conferred by the wall-associated kinase gene ZmWAK-RLK1. The coding sequence of ZmWAK-RLK1 was identical in the three near-isogenic lines, RP1Htn1, RP3Htn1, and RP4Htn1, used for map-based cloning and in the two independent Htn1 introgression lines B37Htn1 and W22Htn1 (16). Marker analysis showed that all Htn1 introgression lines carry the same haplotype in the target interval (SI Appendix, Table S5). The NCLB resistance gene Ht2 also maps on maize chromosome 8 to a similar interval to that of Htn1 (16, 20–23). We amplified the ZmWAK-RLK1 homolog from the Ht2 introgression line B37Ht2. The gene is different from the one found in the Htn1 near isogenic lines, showing only 79% amino acid identity in the extracellular domain. In addition, the E. turcicum isolate used in this study is avirulent on B37Htn1 and B37Ht3, but virulent on B37, B37Ht1, and B37Ht2, therefore revealing a difference of B37Htn1 and B37Ht2 in resistance reaction to E. turcicum. Therefore, we conclude that the ZmWAK-RLK1 gene identified in this study indeed represents Htn1.
Fig. 3.
ZmWAK-RLK1 mutant plants are more susceptible to NCLB than wild-type segregants. (A) Gene structure of ZmWAK-RLK1. (B) Nucleotide and resulting amino acid changes in the four ZmWAK-RLK1 mutants. (C) Representative images of the second leaves of maize plants 21 d postinfection with E. turcicum in the greenhouse. Necrotic, sporulating NCLB areas are spreading from the leaf base toward the leaf apex along leaf borders in the susceptible control line RP3 and in three of the four ZmWAK-RLK1 mutant lines (RLK1b, RLK1d, and RLK1f). The TILLING parent RP3Htn1 and the wild-type segregants (RLK1b wt, RLK1d wt, and RLK1f wt) are free of disease. (D) Area under the disease progress curve values of the four ZmWAK-RLK1 mutants and wild-type. Values represent means of two to three biological replicates, and error bars represent SEs. Different letters on top of bars denote significant differences in area under the disease progress curve values (Student’s t test, P < 0.05).
Comparison of ZmWAK-RLK1 Sequence and Gene Expression Between RP1Htn1 and RP1.
Both the resistant line RP1Htn1 and the susceptible recurrent parent RP1 contain an expressed ZmWAK-RLK1 allele. The predicted amino acid sequence of the extracellular WAK domain was highly divergent between the two proteins (SI Appendix, Figs. S3C and S7). The ZmWAK-RLK1 protein version of the susceptible parent RP1 showed 46 amino acid changes, 19 amino acid deletions, and 2 amino acid insertions in the extracellular domain compared with in the resistant protein version of RP1Htn1, resulting in an amino acid identity of only 79%. The kinase domain of ZmWAK-RLK1, in contrast, was 98.5% identical between the two near-isogenic lines. In the B73 reference genome, multiple sequences showed homology to the ZmWAK-RLK1 gene. However, none of the sequences comprised a complete gene with the same structure as ZmWAK-RLK1 (SI Appendix, Fig. S3B). The closest homolog of ZmWAK-RLK1 in maize variety B73 is gene GRMZM2G164612. Homology was restricted to the kinase domain, and the extracellular domain was only weakly conserved. The ZmWAK-RLK1 ortholog in rice (LOC_Os01g49614) showed 83% amino acid identity in the kinase domain but only 49% amino acid identity in the wall-associated kinase domain. These sequence comparisons indicate that the extracellular domain of ZmWAK-RLK1 is highly diverse among different maize genotypes and is not conserved in closely related species such as rice.
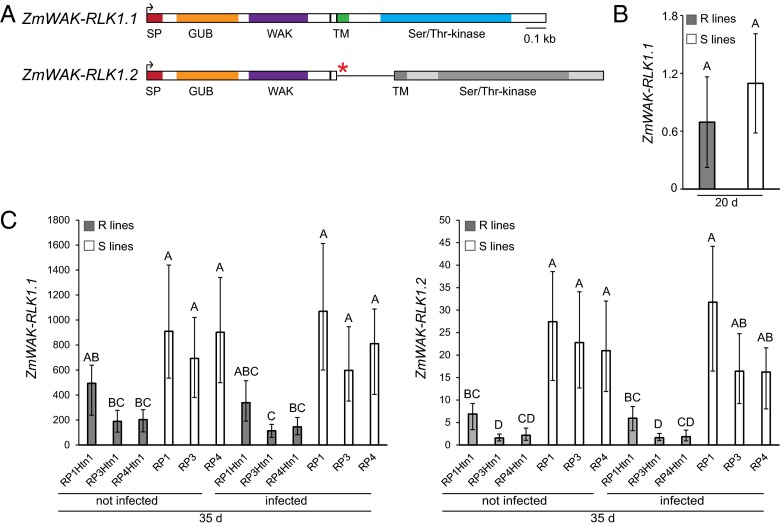
Full-length cDNA amplification from the susceptible line RP1 revealed an alternative splice variant of ZmWAK-RLK1 that retained the second intron (ZmWAK-RLK1.2; Fig. 4A). This results in a premature stop codon, which would lead to a truncated protein version that only contained the extracellular domain, not the transmembrane and kinase domains. Expression analysis of the correctly spliced ZmWAK-RLK1.1 variant showed no difference in transcript level between susceptible RP1 and resistant RP1Htn1 parents in 20-d-old seedlings without infection (Fig. 4B). Surprisingly, ZmWAK-RLK1.1 expression was 1.8–5.6 times higher in the three susceptible lines, RP1, RP3, and RP4, in both infected and uninfected 35-d-old seedlings compared with in the three Htn1 introgression lines, RP1Htn1, RP3Htn1, and RP4Htn1 (Fig. 4C). Infection did not induce ZmWAK-RLK1.1 expression in either of the parents (Fig. 4C). Only trace amounts of the alternatively spliced variant ZmWAK-RLK1.2 were detectable in the resistant parents, RP1Htn1, RP3Htn1, and RP4Htn1 (Fig. 4C). In comparison, the susceptible parents, RP1, RP3, and RP4, showed a 4.0–14.4 times higher expression of ZmWAK-RLK1.2 in both infected and uninfected 35-d-old seedlings (Fig. 4C). The correctly spliced ZmWAK-RLK1.1 transcript variant was 57–120 times and 30–50 times higher expressed compared with the incorrectly spliced transcript ZmWAK-RLK1.2 in the resistant and susceptible lines, respectively.
Fig. 4.
Expression analysis of the two ZmWAK-RLK1 splice variants. (A) cDNA structure of the two splice variants ZmWAK-RLK1.1 and ZmWAK-RLK1.2. Exons are represented as rectangular bars, and the retained intron two in ZmWAK-RLK1.2 as a black line. Predicted protein domains are indicated in color. The red asterisk indicates the premature stop codon. (B and C) Relative ZmWAK-RLK1 expression levels. (B) ZmWAK-RLK1.1 in resistant lines (R lines; mix of line RP1Htn1 and three resistant recombinants) and susceptible lines (S lines; mix of RP1 and two susceptible recombinants) of 20-d-old seedlings and (C) ZmWAK-RLK1.1 (Left) and ZmWAK-RLK1.2 (Right) in resistant and susceptible mapping parents of 35-d-old seedlings, either noninfected or after E. turcicum infection. Mean (B) and untransformed mean (C) values are presented, and the 95% confidence intervals (back-transformed) are plotted. On top of the bars, different letters denote a significant difference in expression level (Tukey’s honestly significant difference test, α = 0.050).
We therefore consider it unlikely that these low levels of alternative ZmWAK-RLK1.2 transcript have an influence on the Htn1 resistance phenotype. It is more plausible that the amino acid differences found in ZmWAK-RLK1 between resistant and susceptible maize lines explain the difference in resistance phenotype.
Conclusion
In this study, we identified a wall-associated receptor-like kinase as the Htn1 NCLB disease resistance gene. Wall-associated kinases have been found to play a role in disease resistance (30, 31), phosphorus-starvation tolerance (32), and developmental processes such as root growth (33) and gametophyte development (34). A recent study reported that the maize quantitative head smut resistance gene qHSR1 also encodes a wall-associated kinase. Htn1 and qHSR1 share structural homology, but their extracellular domains have no sequence identity detectable by BLAST (28). Hence, these two genes represent very diverse members of the same protein superfamily that are involved in quantitative resistance against different fungal pathogens. Wall-associated non-RD kinases might therefore represent a novel class of quantitative immune receptors in monocots. Wall-associated kinases are the only known proteins that can physically link the cell wall to the plasma membrane, which enables them to perceive changes to the cell wall structure (10, 35). WAKs can therefore serve as DAMP receptors that recognize changes to the cell wall during pathogen entry. It is therefore possible that Htn1 perceives cell wall fragments generated during fungal penetration. In contrast, the high cultivar-specific diversity of the extracellular ZmWAK-RLK1 domain could also indicate that this protein is involved in the perception of a microbial elicitor. These results provide the basis to study the molecular function of a novel pattern recognition receptor involved in plant immunity against an adapted fungal pathogen.
Materials and Methods
Plant Material.
Line B37Htn1 (17), carrying the Htn1 locus from the Mexican landrace Pepitilla, served as donor for the Htn1 introgression into three recurrent parents from the German maize breeding program of KWS SAAT SE, RP1, RP3, and RP4. Introgressions were enriched for the recurrent genome by backcrossing three times to the recurrent parent and by checking for the presence of the Htn1 locus with flanking markers. For fine mapping, an F2-population was generated from the cross RP1 × RP1Htn1. The following generations were obtained by selfing and were genotyped and phenotyped at each selfing step. The genotyping was done in a first step with simple sequence repeat markers, but during the process of selfing and recombinant selection, new SNP markers were established as Kompetitive Allele Specific PCR-Assays markers for the region. The phenotyping was done at different field sites with two replications. Recombinant plants for the Htn1 region were selected and correlated with the phenotypic observations. The selection incorporated plants that had different segments of the target region, as well as heterozygous plants, to get new recombinants in the region. Each year, two generations of selfings lead to new recombinants. These new recombinants were phenotyped in the field and in the greenhouse. The results where then correlated with the genotypic data.
Maize lines B37, B37HtN (B37Htn1), B37Ht1, B37Ht2, B37Ht3, W22, and W22Htn1 were obtained from the Maize Genetics Cooperation Stock Center (maizecoop.cropsci.uiuc.edu/) and from KWS SAAT SE.
NCLB Field Infection Tests.
At each location, 20 kernels per row were sown for each genotype. The artificial inoculation was done with crushed infected leaf material, which was collected the year before at the same location. This method allowed for a comparable disease pressure from year to year at different locations. As a control, the near isogenic parents of the population were also planted at all locations. Infection rates were scored as described in SI Appendix, Table S1.
Maize Infection with E. turcicum in the Greenhouse.
To generate an E. turcicum spore suspension for infection, a single spore was isolated from dried leaf material infected with NCLB. The dried leaf material was collected from infected maize plants in Passau in 2005. This isolated spore was propagated on potato dextrose agar (Difco, 213400) plates. The isolate was further propagated for infection tests by inoculating fresh plates with a small piece of mycelium with an inoculation needle. Plates were incubated at room temperature in the dark, upside down, for approximately 3 wk. Spores were harvested by pouring 0.1% Tween into the plates and removing spores by scraping, followed by pouring the spore suspension through a fine mesh. The spore solution was adjusted to a final concentration of 4.5 × 103 spores/mL by counting spores in a Neubauer chamber.
Maize plants were grown each in Jiffy pots (ø 8cm) filled with standard soil (Einheitserde Classic ED 73 + BIMS). Fifteen plants of the same maize line were placed in one tray. Plants were grown in the greenhouse at cycles of 16 h at 20 °C with light and 8 h at 18 °C in the dark and around 60% relative humidity. After the second leaf had fully emerged, the newly emerging leaves were cut and removed until the end of the experiment. Maize plants were infected at 19–21 d after sowing, and a second time 1 or 2 d after the first inoculation. At both infection points, a total of 100 μL freshly made spore suspension (4.5 × 104 spores/mL) was distributed into the leaf sheath of the second leaf and into the stem whorl. Alternatively, first infection tests with recombinants were made by placing a small piece of dried leaf material infected with NCLB (field Passau 2005) on the base of the second leaf. Plastic hoods were placed on top of each tray after the first infection to keep humidity very high. Disease symptoms were evaluated between 11 and 25 d after infection. For each evaluated day and plant line, disease severity was calculated as the percentage of infected plants. From this, we calculated the area under the disease progress curve by the trapezoidal method (36).
A greenhouse infection test with lines B37, B37Htn1, B37Ht1, B37Ht2, and B37Ht3 classified our single spore isolate as race 12 (37).
Map-Based Cloning of Htn1.
Detailed descriptions of materials and methods for BAC library screening, marker development for genetic mapping and development, and screening of a TILLING mutant population are provided in SI Appendix, Materials and Methods and SI Appendix, Tables S6 and S7.
Candidate Gene Characterization.
The proteins were analyzed with the National Center for Biotechnology Information conserved domain search tool (www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). In all three proteins, a signal peptide was predicted with the SignalP 4.0 Server (www.cbs.dtu.dk/services/SignalP/) at the aminoterminal end and a transmembrane domain with DAS (www.sbc.su.se/∼miklos/DAS), HMMTOP (www.enzim.hu/hmmtop), and TMpred (www.ch.embnet.org/software/TMPRED_form.html).
Quantitative Real-Time PCR Analysis for Detection of ZmWAK-RLK1 Expression.
Expression of ZmWAK-RLK1 was quantified in a reverse transcription, quantitative real-time PCR (RT-qPCR) assay, using a CFX96 Real-Time System C1000TM Thermal cycler (Bio-Rad). Leaf material was harvested from 20-d-old plants just before infection and 15 d later with E. turcicum-infected plants, as well as noninfected plants. RNA extraction was performed as described by Hurni et al. (38), and first-strand cDNA was synthesized from 0.5 μg RNA, using 1/2 reaction of the iScript Advanced cDNA Kit (172-5038, Bio-Rad). RT-qPCR primers and master mixes used for the targets ZmWAK-RLK1.1 and ZmWAK-RLK1.2 and the reference gene FPGS are described in SI Appendix, Table S8. RT-qPCR was performed with 4 µL of 10-fold-diluted cDNA for 20-d-old plants and with 4 µL of 20-fold-diluted cDNA for 35-d-old plants in a total reaction volume of 10 µL. Technical duplicates were made for 20-d-old plants, and technical triplicates for five biological replicates of 35-d-old plants. Thermocycling conditions were 95 °C for 20 s, followed by 40 cycles of 95 °C for 3 s, then 60 °C for 20 s (5 s for FPGS). Specificities of amplicons, RT-minus control check, and efficiency calculation were performed as described in Hurni et al. (38). To allow comparison of the expression levels between the two splice variants ZmWAK-RLK1.1 and ZmWAK-RLK1.2, the RT-qPCR data were calibrated on the basis of plasmid DNA containing the ZmWAK-RLK1.1 or ZmWAK-RLK1.2 construct, respectively. Relative quantities were calculated and normalized to the reference genes FPGS revealing the calibrated normalized relative quantities (CNRQ) values, using the program qbase+ V 3.0 (Biogazelle).
Data analysis was performed using the statistical package JMP version 11.0 (SAS Institute). In Fig. 4B, Tukey’s honestly significant difference test was done on log10 transformed expression values. The untransformed means are given in columns, and back-transformed 95% confidence intervals are plotted.
Supplementary Material
Acknowledgments
This work was supported by an Advanced Investigator Grant from the European Research Council (ERC-2009-AdG 249996, durable resistance) and by Swiss National Science Foundation Grant 310030B_144081/1.
Footnotes
Conflict of interest statement: A patent application has been filed relating to this work.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the NCBI GenBank database (accession nos. KR014666, KR014667, and KR080530).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1502522112/-/DCSupplemental.
References
- 1.Dodds PN, Rathjen JP. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat Rev Genet. 2010;11(8):539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 2.Boller T, Felix G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60(1):379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 3.Dangl JL, Horvath DM, Staskawicz BJ. Pivoting the plant immune system from dissection to deployment. Science. 2013;341(6147):746–751. doi: 10.1126/science.1236011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McHale L, Tan X, Koehl P, Michelmore RW. Plant NBS-LRR proteins: Adaptable guards. Genome Biol. 2006;7(4):212. doi: 10.1186/gb-2006-7-4-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pretorius ZA, Singh RP, Wagoire WW, Payne TS. Detection of virulence to wheat stem rust resistance gene Sr31 in Puccinia graminis. f. sp. tritici in Uganda. Plant Dis. 2000;84(2):203.2. doi: 10.1094/PDIS.2000.84.2.203B. [DOI] [PubMed] [Google Scholar]
- 6.Wilson RA, Talbot NJ. Under pressure: Investigating the biology of plant infection by Magnaporthe oryzae. Nat Rev Microbiol. 2009;7(3):185–195. doi: 10.1038/nrmicro2032. [DOI] [PubMed] [Google Scholar]
- 7.Macho AP, Zipfel C. Targeting of plant pattern recognition receptor-triggered immunity by bacterial type-III secretion system effectors. Curr Opin Microbiol. 2015;23(0):14–22. doi: 10.1016/j.mib.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 8.Chinchilla D, et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448(7152):497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- 9.Song WY, et al. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270(5243):1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- 10.Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G. A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci USA. 2010;107(20):9452–9457. doi: 10.1073/pnas.1000675107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Böhm H, Albert I, Fan L, Reinhard A, Nürnberger T. Immune receptor complexes at the plant cell surface. Curr Opin Plant Biol. 2014;20(0):47–54. doi: 10.1016/j.pbi.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Chang HS, Fan KC. Comparative studies on some biology and pathology of corn and broom corn isolates of Exserohilum turcicum (Pass) Leonard & Suggs. Bot Bull Acad Sinica. 1986;27:209–218. [Google Scholar]
- 13.Perkins JM, Pedersen WL. Disease development and yield losses associated with northern leaf blight on corn. Plant Dis. 1987;71(10):940–943. [Google Scholar]
- 14.Raymundo AD, Hooker AL. Measuring the relationship between northern corn leaf blight and yield losses. Plant Dis. 1981;65:325–327. [Google Scholar]
- 15. Pratt RC, Gordon SG (2005) Breeding for resistance to maize foliar pathogens. Plant Breeding Reviews, ed Janick J (John Wiley & Sons, Oxford), pp 119–173.
- 16.Gevers HO. A new major gene for resistance to Helminthosporium turcicum leaf blight of maize. Plant Dis Rep. 1975;59:296–299. [Google Scholar]
- 17.Raymundo AD, Hooker AL, Perkins JM. Effect of gene HtN on the development of northern corn leaf blight epidemics. Plant Dis. 1981;65(4):327–330. [Google Scholar]
- 18.Welz HG, Geiger HH. Genes for resistance to northern corn leaf blight in diverse maize populations. Plant Breed. 2000;119(1):1–14. [Google Scholar]
- 19.Thakur RP, Leonard KJ, Leath S. Effects of temperature and light on virulence of Exserohilum turcicum on corn. Phytopathology. 1989;79(6):631–635. [Google Scholar]
- 20.Chung CL, Jamann T, Longfellow J, Nelson R. Characterization and fine-mapping of a resistance locus for northern leaf blight in maize bin 8.06. Theor Appl Genet. 2010;121(2):205–227. doi: 10.1007/s00122-010-1303-z. [DOI] [PubMed] [Google Scholar]
- 21.Poland JA, Bradbury PJ, Buckler ES, Nelson RJ. Genome-wide nested association mapping of quantitative resistance to northern leaf blight in maize. Proc Natl Acad Sci USA. 2011;108(17):6893–6898. doi: 10.1073/pnas.1010894108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simcox KD, Bennetzen JL. The use of molecular markers to study Setosphaeria turcica resistance in maize. Phytopathology. 1993;83(12):1326–1330. [Google Scholar]
- 23.Simcox KD, Bennetzen JL. Mapping the HtN resistance gene to the long arm of chromosome 8. Maize Genet. Coop. Newsl. 1993;67:118–119. [Google Scholar]
- 24.Schnable PS, et al. The B73 maize genome: Complexity, diversity, and dynamics. Science. 2009;326(5956):1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 25.de Oliveira LF, et al. The Wall-associated Kinase gene family in rice genomes. Plant Sci. 2014;229:181–192. doi: 10.1016/j.plantsci.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 26.Verica JA, He Z-H. The cell wall-associated kinase (WAK) and WAK-like kinase gene family. Plant Physiol. 2002;129(2):455–459. doi: 10.1104/pp.011028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dardick C, Schwessinger B, Ronald P. Non-arginine-aspartate (non-RD) kinases are associated with innate immune receptors that recognize conserved microbial signatures. Curr Opin Plant Biol. 2012;15(4):358–366. doi: 10.1016/j.pbi.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 28.Zuo W, et al. A maize wall-associated kinase confers quantitative resistance to head smut. Nat Genet. 2015;47(2):151–157. doi: 10.1038/ng.3170. [DOI] [PubMed] [Google Scholar]
- 29.Zhang S, et al. Evolutionary expansion, gene structure, and expression of the rice wall-associated kinase gene family. Plant Physiol. 2005;139(3):1107–1124. doi: 10.1104/pp.105.069005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diener AC, Ausubel FM. RESISTANCE TO FUSARIUM OXYSPORUM 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics. 2005;171(1):305–321. doi: 10.1534/genetics.105.042218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Zhou SY, Zhao WS, Su SC, Peng YL. A novel wall-associated receptor-like protein kinase gene, OsWAK1, plays important roles in rice blast disease resistance. Plant Mol Biol. 2009;69(3):337–346. doi: 10.1007/s11103-008-9430-5. [DOI] [PubMed] [Google Scholar]
- 32.Hufnagel B, et al. Duplicate and conquer: Multiple homologs of PHOSPHORUS-STARVATION TOLERANCE1 enhance phosphorus acquisition and sorghum performance on low-phosphorus soils. Plant Physiol. 2014;166(2):659–677. doi: 10.1104/pp.114.243949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaur R, Singh K, Singh J. A root-specific wall-associated kinase gene, HvWAK1, regulates root growth and is highly divergent in barley and other cereals. Funct Integr Genomics. 2013;13(2):167–177. doi: 10.1007/s10142-013-0310-y. [DOI] [PubMed] [Google Scholar]
- 34.Wang N, et al. The rice wall-associated receptor-like kinase gene OsDEES1 plays a role in female gametophyte development. Plant Physiol. 2012;160(2):696–707. doi: 10.1104/pp.112.203943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kohorn BD, Kohorn SL. The cell wall-associated kinases, WAKs, as pectin receptors. Front Plant Sci. 2012;3:88. doi: 10.3389/fpls.2012.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madden LV, Hughes G, van den Bosch F. The study of plant disease epidemics. APS Press; St. Paul, MN: 2007. p. 432. [Google Scholar]
- 37.Leonard KJ, Levy Y, Smith DR. Proposed nomenclature for pathogenic races of Exserohilum turcicum on corn. Plant Dis. 1989;73(9):776–777. [Google Scholar]
- 38.Hurni S, et al. Rye Pm8 and wheat Pm3 are orthologous genes and show evolutionary conservation of resistance function against powdery mildew. Plant J. 2013;76(6):957–969. doi: 10.1111/tpj.12345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.