Significance
Cancer research has relied on in vitro experiments with established cell lines to investigate the efficacy of chemotherapy drug panels, which limit clinical correlation. Targeting tumor-initiating cells could be a viable clinical strategy, but these cells are extremely rare, necessitating new methods for rapid and robust screening. We developed a miniaturized platform to investigate the chemosensitivity of patient-derived tumor-initiating cells using limited cell numbers. The miniature platform described herein represents, to our knowledge, the first demonstration of a cell patterning technique that combines the ability to release small molecule drugs, alone or in combination, to separate islands of cells. This device can be adopted in clinical and academic laboratories targeting cancer stem cell populations to tailor patient-specific chemotherapeutic treatment options.
Keywords: personalized medicine, chemopredictive, cancer stem cell, microarray, combination therapy
Abstract
A new paradigm in oncology establishes a spectrum of tumorigenic potential across the heterogeneous phenotypes within a tumor. The cancer stem cell hypothesis postulates that a minute fraction of cells within a tumor, termed cancer stem cells (CSCs), have a tumor-initiating capacity that propels tumor growth. An application of this discovery is to target this critical cell population using chemotherapy; however, the process of isolating these cells is arduous, and the rarity of CSCs makes it difficult to test potential drug candidates in a robust fashion, particularly for individual patients. To address the challenge of screening drug libraries on patient-derived populations of rare cells, such as CSCs, we have developed a drug-eluting microarray, a miniaturized platform onto which a minimal quantity of cells can adhere and be exposed to unique treatment conditions. Hundreds of drug-loaded polymer islands acting as drug depots colocalized with adherent cells are surrounded by a nonfouling background, creating isolated culture environments on a solid substrate. Significant results can be obtained by testing <6% of the cells required for a typical 96-well plate. Reliability was demonstrated by an average coefficient of variation of 14% between all of the microarrays and 13% between identical conditions within a single microarray. Using the drug-eluting array, colorectal CSCs isolated from two patients exhibited unique responses to drug combinations when cultured on the drug-eluting microarray, highlighting the potential as a prognostic tool to identify personalized chemotherapeutic regimens targeting CSCs.
Tumor-initiating cancer stem cells (CSCs) are being investigated as a promising therapeutic target (1). The rarity of CSCs, which constitute ∼1% of tumor cells (1, 2), limits their availability for testing, and traditional screening methods require substantial cell quantities. Industrial pharmaceutical capabilities have successfully reduced cell requirements in drug screening, but such capital-intensive facilities are typically unavailable to clinicians and pathology laboratories. The past decade has witnessed the emergence of multiple cell-based microarray platforms that address availability and cell source limitations (3–5), although these systems have inherent shortcomings. Many rely on immobilizing target molecules (6–9), limiting applicability to small molecule drug libraries, whereas others rely on robotically spotting cells (10), a technique not amenable to widespread adoption. Array platforms capable of capturing single cells have been established (11, 12), but determination of chemotherapeutic efficacy is better investigated through methods using greater cell numbers, which better capture variability in cellular responses. Furthermore, arrays of drug-loaded polymer films with an overlying cell monolayer have been developed (13), but monolayers of cells are susceptible to juxtacrine and paracrine signaling, which are particularly important for multipotent cells. In the present work, the provision of differential cell adhesion to promote seeding onto spotted drug-loaded films against a surrounding nonfouling background (i.e., a surface that resists protein adsorption and thus cell adhesion) can separate drug-eluting polymer films to create isolated culture environments. The use of programmable arraying techniques can then enable fabrication of uniquely formulated drug-eluting spots that provide prescribed drug doses and drug combinations to overlying cells for simultaneous testing on a single device.
It is becoming increasingly evident in cancer treatment that simultaneously targeting multiple critical pathways using combinations of chemotherapeutic drugs can enhance outcomes (14–17). Conventional screening of chemotherapeutics uses an established panel of cancer cell lines (18) that have been derived from bulk tumors. A recently developed clinical approach involves performing in vitro chemosensitivity testing of tumor biopsy specimens to individualize treatment (19, 20). Unfortunately, benefits have been limited, with poor correlations between bulk tumor cell sensitivity and clinical efficacy. This lack of efficacy has been attributed to patient to patient variability, owing in part to intratumor heterogeneity (21–23).
Tumors consist of multiple cell phenotypes. In the CSC model, a rare cell population of tumor-initiating cells perpetually self-renew and are responsible for tumor heterogeneity, metastasis, and disease recurrence (1, 24). Recent identification of unique cell surface markers that enrich tumor cell isolates for CSCs have led to novel techniques for isolating enriched colorectal CSC (CCSC) populations from patient tumor samples (25–28). For example, xenotransplantation of a single CCSC identified by high Wnt/Β-catenin signaling activity generates tumors that recapitulate the diverse phenotypic heterogeneity of the original tumor (29). Thus, identifying and isolating CCSCs out of the tumor bulk from an individual cancer patient and determining sensitivity to chemotherapeutic drugs in vitro is possible (30, 31). An approach such as the drug-eluting microarray, enabling use of low cell numbers, could potentiate personalized combination drug treatment screens for efficacy against patient-specific CSCs.
Results and Discussion
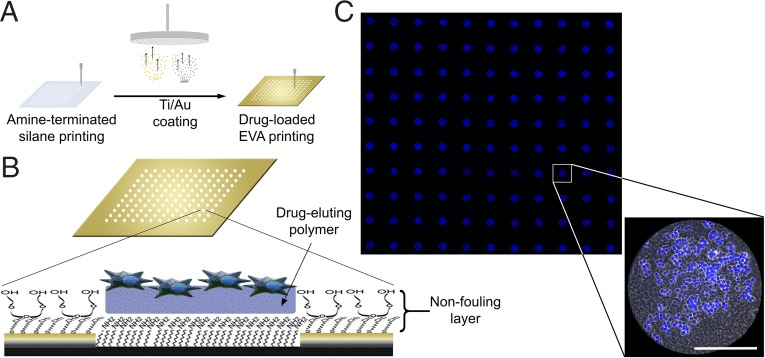
Drug-eluting microarrays were developed (Fig. 1). Fabrication (Fig. 1A) was based on a modification of our earlier cell array (32) and provided cell attachment to drug-loaded polymer islands surrounded by nonfouling surface treatment (Fig. 1B). Site-specific attachment of cells to polymer islands with minimal cell adhesion to the background was achieved (Fig. 1C). The fidelity of fabrication and cell attachment was quantified after 24 h of incubation using the following criteria (with results in parentheses): poly-d-lysine printing misalignment with polymer islands (<1.3%), proportion of islands with adherent cells (>95%), proportion of islands with <65% cell coverage (<11%), and proportion of cells on islands (>94%).
Fig. 1.
Drug-eluting cellular microarrays. (A) Fabrication. Glass coverslips are robotically printed with amine-terminated silane in isolated spots and then coated with titanium and gold. Processing exposes silane-grafted islands, whereas the gold region is passivated by the addition of a nonfouling PEG background (32). Drug-loaded EVA is then printed over the exposed silane islands, and amine groups promote polymer adhesion. Finally, poly-d-lysine is overspotted on the EVA films to promote cell adhesion before seeding. (B) Schematic of a single spot highlighting the substrate architecture, the chemistry of the nonfouling PEG coating, and the drug-eluting polymer with cells adherent (not to scale). (C) Fluorescence microscopy mosaic image of a 10 × 11 microarray seeded with HCT116 colon carcinoma cells, illustrating the fidelity of cell adhesion to isolated islands of drug-eluting polymer films. Shown is a detail of a single drug-eluting island demonstrating adherent cells (phase- contrast image overlay with nuclear staining in blue). The EVA film is fabricated using a water-oil emulsion to promote uniform film thickness during drying, and has a mottled appearance. (Scale bar: 200 μm.)
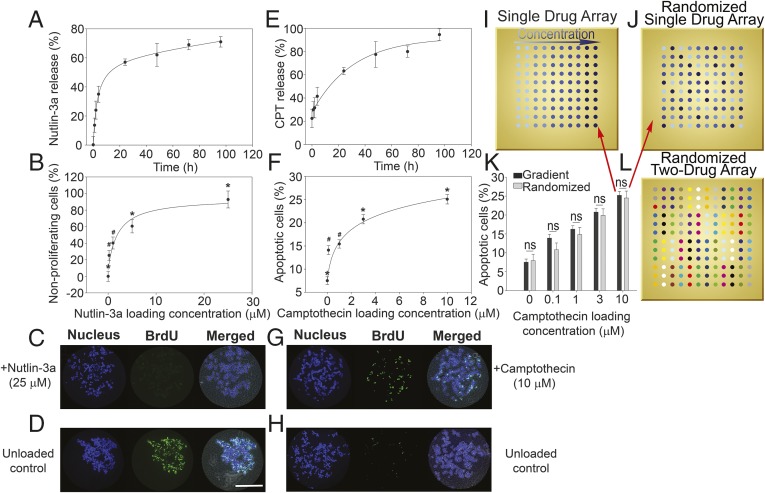
Loading efficiency (i.e., the amount of drug partitioned into the polymer as a percentage of the total amount of drug initially loaded) of small molecules from Poly(ethylene-covinyl acetate) (EVA) polymer films on microarrays was quantified (SI Appendix, Fig. S1). Release kinetics from microarrayed drug-eluting EVA films demonstrated an initial burst during the first 8–24 h, followed by a steady rate of release over 5 d (Fig. 2 A and E). Over the ranges tested, flux from the EVA films was linear in relation to initial drug loading, allowing controllable delivery of factors in a dose-dependent manner (SI Appendix, Fig. S2 A and B). Furthermore, coloading with a second drug did not alter the flux (SI Appendix, Fig. S2C), and drug release could be delayed by overspotting unloaded EVA films onto drug-loaded films, creating a diffusion barrier (SI Appendix, Fig. S2D). Cell seeding was ∼200 cells per island and was unaffected by drug loading concentration (SI Appendix, Fig. S3).
Fig. 2.
Cumulative drug release and HCT116 cell responses to drug-loaded microarrays. (A) Nutlin-3a release profile from microarray revealed a burst release of ∼8 h, followed by a steady release rate over 5 d. Release profiles show mean ± SD of three replicates, and data are fitted using an exponential decay model. (B) Percent of nonproliferative HCT116 cells on nutlin-3a–loaded microarray increases with increasing drug loading concentration. Proliferation was quantified via BrdU incorporation, and data are normalized to an unloaded control. Significant differences were determined by ANOVA [F(4,138) = 19.068; P < 0.05], followed by Tukey’s post hoc analysis. (C) Representative fluorescence micrographs of nonproliferating cells on a 25 μΜ nutlin-3a–loaded polymer island (evidenced by low BrdU staining). (D) Representative fluorescence micrographs of an unloaded control island with highly proliferative cells (demonstrating high BrdU staining). (E) Camptothecin release profile from microarray revealing a burst release of ∼24 h, followed by a steady release rate over 5 d. Release profiles show mean ± SD of three replicates, and data are modeled using exponential decay. (F) Percent of apoptotic cells on camptothecin-loaded microarray increases with increasing drug loading concentrations. Apoptosis was quantified by annexin V staining, and significant differences were determined by ANOVA [F(4,479) = 52.778; P < 0.05], followed by Tukey’s post hoc analysis. (G) Representative fluorescence micrographs displaying high levels of cells undergoing apoptosis on a 10 μΜ camptothecin-loaded polymer island (demonstrating high annexin V staining). (H) Representative fluorescence micrographs of an unloaded control island with low levels of apoptotic cells (showing low annexin V staining). (I) Schematic of a single factor dosing array layout with increasing drug loading concentrations. (J) Schematic of a randomized single factor array with loading concentrations configured in randomized fashion. (K) Statistical comparison of cell apoptosis between randomized and nonrandomized single drug array configurations demonstrating the results are independent of array configuration (n = 3). This indicates that there is negligible cellular cross-talk and drug interaction between neighboring islands. (L) Schematic of a randomized two-factor dosing array used in combinatorial microarrays. Patterns represent the 16 different combinations of two drugs (four concentrations per drug). *P < 0.05 compared with all other conditions; #P < 0.05 compared with control. (Scale bar: 200 μM.)
The feasibility of eliciting dose-dependent responses to a model hydrophilic agent, azide, was demonstrated using the HCT116 colon carcinoma cell line (SI Appendix, Fig. S4A). Consequently, further investigation was carried out using nutlin-3a and camptothecin, representing two classes of clinically relevant drugs. Nutlin-3a, a hydrophobic drug that inhibits human double minute 2 (HDM2), is currently under clinical investigation in combination with numerous therapeutic agents (33, 34). Nutlin-3a binding to HDM2 disrupts turnover of the tumor-suppressor protein p53, increasing p53 protein levels and inducing cells to enter into either a state of cell cycle arrest or apoptosis at higher concentrations (35). HCT116 cells were cultured on nutlin-3a–loaded microarrays for 24 h, and proliferation was quantified. With increasing concentrations of nutlin-3a, the percentage of nonproliferating cells increased (Fig. 2 B–D). Correspondingly, cell numbers were diminished after 72 h of incubation (SI Appendix, Fig. S4B). Thus, the HCT116 cell line clearly evidenced dose-dependent cell cycle arrest when cultured on nutlin-3a–loaded microarrays. Camptothecin, a hydrophobic topoisomerase inhibitor that induces apoptosis, is also of interest, with various analogs used in chemotherapy (36, 37). As expected, the proportion of HCT116 cells undergoing apoptosis was greater with increasing concentrations of camptothecin after 72 h of incubation on the microarray (Fig. 2 F–H).
Seminal cell-based microarray studies have demonstrated that experimental design can control for undesirable interactions between islands through island spacing, randomized configurations, and robust statistical analysis (4). Previous work on a related type of microarray configuration has suggested that a 1.5-mm spacing between islands is sufficient to isolate cell populations from agents released from neighboring polymer spots (13). To determine whether paracrine signaling or diffusion of drugs from adjacent polymer islands was a factor in our drug-eluting microarray using this 1.5-mm (center to center) island spacing, we analyzed camptothecin-loaded arrays in various configurations (Fig. 2 I and J). No differences in apoptosis were noted between the configurations at different drug loading amounts, indicating negligible interaction with cells or drugs from neighboring islands (Fig. 2K).
Chemotherapy for colorectal cancer is often a combination of two drugs. Dual-drug microarrays were developed to investigate possible interaction effects of nutlin-3a and camptothecin on the HCT116 cell line. Ranges of six loading concentrations for the two drugs were combinatorially encapsulated and printed in randomized microarray configurations, resulting in 36 unique conditions (Fig. 2L). The results demonstrate the feasibility of the drug-eluting array platform for inducing dose-dependent nonproliferation (SI Appendix, Fig. S5). Furthermore, hyperbolic curves were generated to model dose–responses from one drug in the presence of a fixed amount of a second drug for each combination (SI Appendix, Fig. S5 B–M) using the equation E = E0 + Emax ⋅C/(C + D50), where E0 is the basal response, Emax is the maximum response obtainable, (1/D50) is the sensitivity (increasing values indicate higher sensitivity; a lower dose is required to approach Emax), and C is the concentration (38). Using this model, Emax and sensitivity (1/D50) values can be used to compare the interaction effects of combined drug administration. It was observed that for both camptothecin and nutlin-3a, increasing the loading concentration of one drug increased sensitivity of HCT116 cells to the other drug (SI Appendix, Fig. S6). In addition, reliability of the drug-eluting microarray was established, with a coefficient of variation (CV) of 12% between arrays and 12% within arrays (SI Appendix, Fig. S7). Apoptotic responses were also investigated, which revealed an antagonistic relationship between camptothecin and nutlin-3a (SI Appendix, Figs. S8 and S9), an expected finding (39). Reliability was confirmed, with a CV of 15% between arrays and 8% within arrays (SI Appendix, Fig. S10), and drug-eluting microarray results were corroborated using soluble drugs in standard 96-well plates (SI Appendix, Fig. S11). Comparing soluble doses to drug loading amounts at equivalent cell response values provides an approximation of the local effective drug concentration to which the cells were cumulatively exposed on the microarrays over 24 h (SI Appendix, Fig. S12). Comparing cellular responses between 6 × 6 and 4 × 4 microarray configurations revealed comparable trends in Emax and sensitivity values. Consequently, subsequent experiments implemented smaller (4 × 4) arrays to reduce cell requirements when using rare cells.
The cancer stem cell hypothesis states that rare tumor-initiating cells, constituting ∼1% of the tumor, are responsible for both the heterogeneity and the hierarchy within the tumor (22, 23). Recently, CCSCs have been linked to tumor initiation and potentiation, as well as to the genesis of metastatic deposits (1). Such primary tumor cell subpopulations are challenging to isolate in substantial numbers, and thus are a good target for determining the feasibility of the drug-eluting microarray by delineating the potency of chemotherapeutic agent combinations while using low cell numbers. In brief, CCSCs were isolated and enriched for high-level aldehyde dehydrogenase (ALDHhigh) activity, a known stem cell enrichment marker, from patients with sporadic colorectal cancer. The ALDHhigh cells have the definitive CSC property of self-renewal, as demonstrated by the gold standard assay of limiting dilution tumor xenograft analysis (26, 27, 29, 40–43). In addition, tumor heterogeneity is maintained, as defined by the ability to recapitulate all cellular aspects of the primary tumor (27, 29). The CCSC isolates can be serially propagated as tumor xenografts or as nonadherent spheres in vitro. Adherent cell growth was established using these CCSC isolates.
We investigated two patient-derived populations of CCSCs, one from a 70-y-old patient with stage IV cancer, labeled CA1, and the other from a 60-y-old patient with stage III colorectal cancer, labeled CA2. In contrast to the HCT116 cell line, which expresses wild type p53, CA1 and CA2 each have a single base pair transition substitution at amino acid 273 of the DNA-binding domain (arginine to histidine) (SI Appendix, Fig. S13). For compatibility with drug-eluting microarrays, adherent cell growth was established and serially propagated. Phenotypes were compared with those maintained as spheroid cultures with regard to expression of ALDH, CD44 (a commonly used stem cell marker) (43, 44), and mucin 2 (MUC2, which delineates differentiation along the goblet cell lineage) (SI Appendix, Fig. S14). ALDH, CD44, and MUC2 expression was maintained in adherent monolayer culture at levels equivalent to spheroid culture in CA1 cells. In contrast, there was a detectable change in expression of phenotypic markers for adherent CA2 cells compared with spheroid culture. The proportion of CA2 cells expressing ALDH declined in adherent cells, and MUC2 expression increased somewhat compared with spheroid, whereas CD44 expression was maintained.
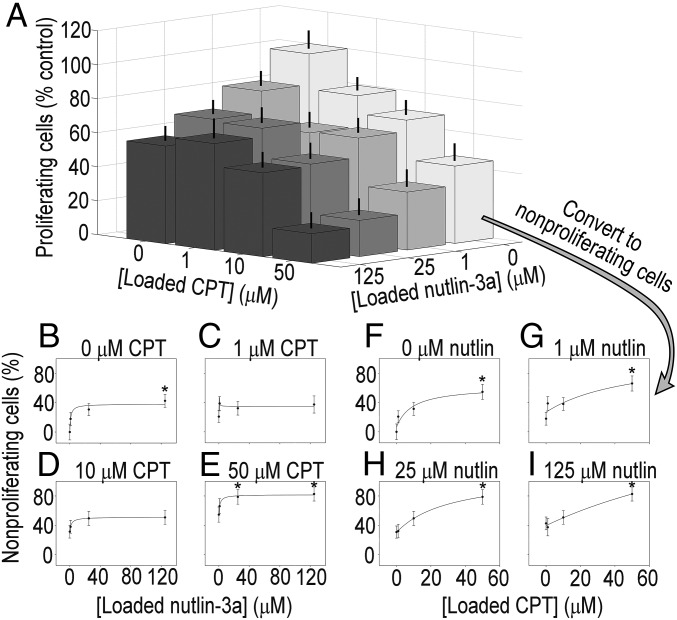
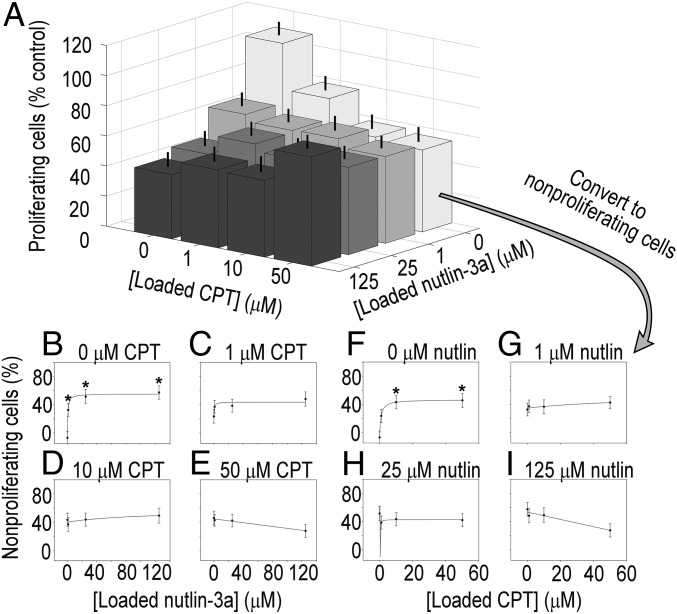
Microarrays used to screen CCSCs were seeded with ∼200 cells per island, and cell counts after 24 h across all drug combinations did not change significantly (P > 0.1). Different levels of antiproliferative efficacy were observed for the CCSCs from each patient when exposed to combinations of camptothecin and nutlin-3a on the microarrays (Figs. 3A and 4A), with both displaying adequate consistency (CV of 13% between arrays and 15% within arrays for CA1, and CV of 17% between arrays and 17% within arrays for CA2) (SI Appendix, Figs. S16 and S18). Both CA1 and CA2 cells exhibited decreasing proliferation with increasing exposure to either nutlin-3a or camptothecin alone (Figs. 3A and 4A; data expressed as percentage of nonproliferating cells with accompanying curve fits in Figs. 3 B and F and 4 B and F). However, response curves and associated fit parameters Emax and sensitivity (1/D50) indicate sizable differences between CA1 and CA2 in terms of efficacy of drug combinations (Figs. 3 B–I and 4 B–I, and SI Appendix, Figs. S15 and S17).
Fig. 3.
Patient-derived CA1 colorectal cancer stem-like cell proliferation and dose–response curves from combinatorially loaded drug-eluting microarrays. (A) Proliferation of CA1 cells on drug-eluting cellular microarrays. Loading concentrations of nutlin-3a [F(3,233) = 5.762; P < 0.05] and camptothecin [F(3,233) = 16.884; P < 0.05] affected antiproliferative activity, as determined by ANOVA. Subadditive effects were observed from combination treatments, as evidenced by the greater decrease in proliferation from higher concentration combinations compared with the highest concentrations of either nutlin-3a or camptothecin alone. Error bars represent SEM. (B–E) Dose–response curves for fixed camptothecin concentrations of 0, 1, 10, and 50 μM over a range of nutin-3a concentrations. There was no significant change in Emax values of the nutlin-3a response with the added presence of camptothecin; however, there was a significant increase (by 75%) in the sensitivity to nutlin-3a when combined with 10 μΜ camptothecin compared with nutlin-3a alone (28.6 vs. 50.0; SI Appendix, Fig. S15), indicative of an increase in antiproliferative activity. (F–I) Dose–response curves for fixed nutlin-3a concentrations of 0, 1, 25, and 125 μM, with a range of camptothecin concentrations. Although Emax values generally increased with added nutlin-3a, the values were not significantly different. Similarly, differences in sensitivity to camptothecin owing to the addition of nutlin-3a were not observed. Proliferation data were transformed to nonproliferation data by subtracting the former from 100%. *P < 0.05 compared with 0 μM drug.
Fig. 4.
Patient-derived CA2 CCSC proliferation and dose–response curves from combinatorially loaded drug-eluting microarrays. (A) Proliferation of CA2 cells on drug-eluting cellular microarrays. Antiproliferative activity was increased from exposure to camptothecin [F(3,81) = 5.987; P < 0.05] and nutlin-3a [F(3,82) = 7.525; P < 0.05] as revealed by ANOVA, with a significant interaction effect between drugs [F(9,329) = 2.382; P < 0.05]. Combination treatments did not improve antiproliferative activity. In fact, an antagonistic effect was observed from combination treatments, in which increasing the concentrations of both drugs reversed drug-induced nonproliferation compared with high doses of individual drugs. Error bars represent SEM. (B–E) Dose–response curves for fixed camptothecin concentrations of 0, 1, 10, and 50 μM over a range of nutin-3a concentrations. There was a 90% decrease in the Emax values of the response to nutlin-3a in the presence of 10 μM camptothecin compared with 0 μM camptothecin (5.7 vs. 59.0; SI Appendix, Fig. S17), and a decrease of 114% in the Emax values with the addition of 50 μM (−8.3 vs. 59.0; SI Appendix, Fig. S17), indicating a decrease in antiproliferative activity. (E) The negative slope of the response curve for 50 μM camptothecin is indicative of an antagonistic drug interaction. (F–I) Dose–response curves for fixed nutlin-3a concentrations of 0, 1, 25, and 125 μM with a range of camptothecin concentrations. The response curves to ranges of camptothecin in the presence of fixed amounts of nutlin-3a reveal decreased Emax values, but the values were not significantly different (SI Appendix, Fig. S17). (H) Notably, the sensitivity of CA2 CCSCs to camptothecin decreased by 100-fold with the addition of just 1 μM nutlin-3a (133 for 0 µM nutlin compared with 1.6 for 1 µM nutlin; SI Appendix, Fig. S17). (I) The negative slope of the response curve for 50 μM camptothecin is indicative of an antagonistic drug interaction. Proliferation data were transformed to nonproliferation data by subtracting the former from 100%. *P < 0.05 compared with 0 μM drug.
Drug combinations were more effective for CA1 cells. Notably, sensitivity increased by 75% when 10 μΜ camptothecin was present compared with nutlin-3a alone (50.0 vs. 28.6) (SI Appendix, Fig. S15). In contrast, Emax (the maximum percentage of nonproliferating cells) for the nutlin-3a concentration response curve remained unchanged by the addition of camptothecin. Trends were observed in the Emax and sensitivity values to camptothecin in the presence of fixed amounts of nutlin-3a that suggest increasing antiproliferative effects, but these values were not significantly different. In stark contrast, for CA2 cells, combining drugs decreased efficacy. A significant interaction effect between drugs was observed [F(9,329) = 2.382; P < 0.05]. Specifically, there was a decrease in the Emax values for the concentration response curve to nutlin-3a with the addition of camptothecin compared with nutlin-3a alone (P < 0.1 for 10 μM campothecin and P < 0.05 for 50 μM campothecin) (SI Appendix, Fig. S17). Differences in sensitivity to nutlin-3a owing to the addition of camptothecin were undetectable; however, the addition of 1 μΜ nutlin-3a decreased the sensitivity of CA2 cells to camptothecin by 100-fold. At the highest concentration, Emax decreased to a negative value, signifying that addition of the second drug actually increased proliferation. These results are indicative of an antagonistic effect of combination treatments, given that the antiproliferative effect was less than that seen with either drug alone. The differential responses to drug combinations measured for these two patient-derived CCSCs support the premise that personalizing chemotherapeutic treatment will be valuable.
While the focus of the present study was on technology development, mechanisms for these patient-specific differences in combination drug responsiveness need to be explored separately. It is interesting that both CA1 and CA2 CCSCs were sensitive to nutlin-3a, which could suggest either that the p53 mutation in these cells (SI Appendix, Fig. S13) did not interfere with drug action, or that p73 (a p53 family member that can similarly induce cell cycle arrest, senescence, and apoptosis) could be involved, as has been shown previously (45). Differences in stemness markers seen on the introduction of adherent culture conditions may be responsible for the altered reactions to the agents evaluated here; however, genetic heterogeneity between the patients, confirmed by short tandem repeat analysis, also likely contributed to these unique responses. Although gene mutational analyses may provide functional predictions, these alterations do not always correspond to predictable cellular behavior. Drug-eluting microarrays, on the other hand, can provide functional readouts that complement genetic sequencing to better define the effect of mutations on CCSC function.
In summary, we have created a platform capable of performing chemosensitivity screens on patient-derived CCSCs using limited cell numbers. This drug-eluting microarray comprises a nonfouling background substrate with hundreds of isolated drug-loaded polymer islands acting as drug depots to create unique and defined culture environments. Given the current array island spacing, a large drug-eluting microarray could screen more than 4,800 unique conditions within the footprint of a standard microtiter plate, thus reducing the requisite cells and reagents needed for statistically robust investigation. Minimizing the number of cells allowed testing of a rare cell population of CCSCs, thereby providing a strategy for tailoring patient-specific cancer treatments that target this critical cell population. Platform feasibility was demonstrated by constructing and testing combinatorial chemotherapeutic drug-eluting microarrays. Application of the platform was highlighted using rare CCSCs from two different patients, and the differential responses measured support the idea that personalizing chemotherapeutic treatments could improve treatment. The drug-eluting microarray represents an enabling diagnostic tool for academic and clinical laboratories targeting CSC populations to define chemotherapeutic treatment options.
Methods
Polymer Formulation.
EVA (Sigma-Aldrich; 40% vinyl acetate by weight) was dissolved in cyclohexanol (Acros) at a 5% (wt/wt) concentration. To embed molecules into the polymer matrix, the particles were first dissolved in an appropriate solvent. A stock solution of azide, a hydrophilic molecule, was dissolved in distilled H2O, whereas nutlin-3a and camptothecin, both hydrophobic molecules, were dissolved in DMSO. Once dissolved, drug solutions were added to 5% (wt/wt) EVA in cyclohexanol at a 1:10 ratio [e.g., 50 μL of camptothecin in DMSO was added to 500 μL of 5% (wt/wt) EVA/cyclohexanol]. Water was then added to the nutlin-3a and camptothecin polymer formulations at 5% (vol/vol) final concentration (i.e., 27.5 μL in the above example), to create an emulsion that stabilized the uniformity of film thickness. Before printing, polymer mixtures were heated to 60 °C, followed by vortexing for 30 s. Mixtures were then homogenized for 60 s before being loaded onto the source plate of the robotic mini-arrayer.
Array Fabrication.
Arrays with polyethylene glycol (PEG)-based nonfouling backgrounds and amine-terminated silane adhesion islands were manufactured as reported previously (32) (Fig. 1A). Then drug-loaded EVA films were printed over the amine islands. Poly-d-lysine (0.1%) was then overspotted onto the EVA films to promote cell attachment. The arrays were placed in 35-mm Petri dishes containing PBS with 2% (vol/vol) penicillin and 2% (vol/vol) streptomycin for 30 min to rehydrate the nonfouling PEG background and as a noncaustic sterilization step.
Human Subjects.
Tissues from patients with colon cancer were retrieved under pathological supervision with Institutional Review Board approval from the University of Michigan and the University of Florida as described previously (42).
Cells and Microarray Seeding.
HCT116 (p53+/+; American Type Culture Collection) human colon cancer cells were maintained in McCoy’s 5a Medium supplemented with 10% (vol/vol) FBS (Thermo Scientific), 1% penicillin G, and 1% streptomycin (Thermo Scientific). The cells were cultured at 37 °C in a humidified incubator containing 5% CO2.
Colon cancer stem cells were generated by isolating and enriching for high-level aldehyde dehydrogenase (ALDHhigh) spheres from colon cancer tumor cells obtained from patients with sporadic colorectal cancer (26, 27, 29, 42). Isolated cells were cultured in serum-free media as described previously (26). Heterogeneity (as defined by the ability to recapitulate all cellular aspects of a tumor) in CCSCs was maintained (27, 29). The isolates maintained in culture originated directly from patients, were propagated and expanded as patient-derived xenografts in mice, and were maintained in vitro. Using these cultures, adherent cell growth was established with 0.1% gelatin (Millipore) coatings on tissue culture plates (TPP), and the cells were serially propagated. For CA1 and CA2 cells, 25,000 cells were tested per array.
Immunocytochemistry and Image Analysis.
Apoptosis was evaluated using Annexin V (BD Biosciences). Proliferation was quantified via BrdU incorporation (BD Biosciences) according to the manufacturer’s directions. All microarrays were counterstained with Hoechst 34580 (Invitrogen) to quantify total cell number and imaged with a Zeiss Axiovert 200M microscope. Analysis was performed with Axiovision (Zeiss) by quantifying the area of fluorescence per channel in each drug-eluting island, reported as relative fluorescence intensity.
Cross-Talk.
Multiple arrays were printed in randomized configurations as well as gradient configurations (Fig. 2). Data were then analyzed using the Student t test to determine whether pairs with significant cross-talk existed between the same groups (i.e., the outcome changed when the pairs were arranged differently on the array).
Statistical Analyses.
Statistical analyses were performed using either a one-way ANOVA or a two-way ANOVA, using Systat version 12 (Systat Software), with the individual experimental run identifier (biological replicate) and the loaded drug condition identifier (technical replicate) as independent variables. Values were nested by experimental condition and by individual microarrays during the statistical analysis. Error bars in the figures represent the combined error of the pooled datasets, combining both the technical replicates and complete biological replicates. Post hoc pairwise comparisons were made using Tukey’s honestly significant difference test, with P < 0.05 considered significant unless stated otherwise. Curve-fitting of drug-release and dose–response curves were performed using SigmaPlot version 10 (Systat Software).
Modeling for concentration-response was performed for each concentration interval using the equation E = E0 + (Emax × C)/(C + D50), and Emax and D50 values were obtained. Drug sensitivity values were obtained by taking the inverse of the D50 value and multiplying it by 100. Those values marked with “#” indicate an R2 value of the curve fit of <0.65. N/A values are present where negative parameters were obtained.
Supplementary Material
Acknowledgments
We thank the University of Florida, the flow cytometry core at the University of Florida, Dr. Tom Rowe, and Dr. Jeremy Fields. Research reported in this publication was supported by the following grants: National Science Foundation DGE-0802270 (graduate research fellowship to M.R.C.); National Institutes of Health R01 DK091658, R01 DK098589, and R21 AI094360 (to B.G.K.); and National Institutes of Health R01 CA142808 and R01 CA157663 (to E.H.H.). Initial support was provided by the University of Florida Research Opportunity Seed Fund (M.R.C., B.G.K., and E.H.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.G.A. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1505374112/-/DCSupplemental.
References
- 1.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 2.Huang EH, Heidt DG, Li CW, Simeone DM. Cancer stem cells: A new paradigm for understanding tumor progression and therapeutic resistance. Surgery. 2007;141(4):415–419. doi: 10.1016/j.surg.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 3.Soen Y, Chen DS, Kraft DL, Davis MM, Brown PO. Detection and characterization of cellular immune responses using peptide-MHC microarrays. PLoS Biol. 2003;1(3):E65. doi: 10.1371/journal.pbio.0000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soen Y, Mori A, Palmer TD, Brown PO. Exploring the regulation of human neural precursor cell differentiation using arrays of signaling microenvironments. Mol Syst Biol. 2006;2:37. doi: 10.1038/msb4100076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flaim CJ, Chien S, Bhatia SN. An extracellular matrix microarray for probing cellular differentiation. Nat Methods. 2005;2(2):119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 6.Ziauddin J, Sabatini DM. Microarrays of cells expressing defined cDNAs. Nature. 2001;411(6833):107–110. doi: 10.1038/35075114. [DOI] [PubMed] [Google Scholar]
- 7.Stone JD, Demkowicz WE, Jr, Stern LJ. HLA-restricted epitope identification and detection of functional T cell responses by using MHC-peptide and costimulatory microarrays. Proc Natl Acad Sci USA. 2005;102(10):3744–3749. doi: 10.1073/pnas.0407019102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bailey SN, Ali SM, Carpenter AE, Higgins CO, Sabatini DM. Microarrays of lentiviruses for gene function screens in immortalized and primary cells. Nat Methods. 2006;3(2):117–122. doi: 10.1038/nmeth848. [DOI] [PubMed] [Google Scholar]
- 9.Tuleuova N, et al. Using growth factor arrays and micropatterned co-cultures to induce hepatic differentiation of embryonic stem cells. Biomaterials. 2010;31(35):9221–9231. doi: 10.1016/j.biomaterials.2010.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandes TG, et al. On-chip, cell-based microarray immunofluorescence assay for high-throughput analysis of target proteins. Anal Chem. 2008;80(17):6633–6639. doi: 10.1021/ac800848j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rettig JR, Folch A. Large-scale single-cell trapping and imaging using microwell arrays. Anal Chem. 2005;77(17):5628–5634. doi: 10.1021/ac0505977. [DOI] [PubMed] [Google Scholar]
- 12.Love JC, Ronan JL, Grotenbreg GM, van der Veen AG, Ploegh HL. A microengraving method for rapid selection of single cells producing antigen-specific antibodies. Nat Biotechnol. 2006;24(6):703–707. doi: 10.1038/nbt1210. [DOI] [PubMed] [Google Scholar]
- 13.Bailey SN, Sabatini DM, Stockwell BR. Microarrays of small molecules embedded in biodegradable polymers for use in mammalian cell-based screens. Proc Natl Acad Sci USA. 2004;101(46):16144–16149. doi: 10.1073/pnas.0404425101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee MJ, et al. Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell. 2012;149(4):780–794. doi: 10.1016/j.cell.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lake RA, Robinson BW. Immunotherapy and chemotherapy—a practical partnership. Nat Rev Cancer. 2005;5(5):397–405. doi: 10.1038/nrc1613. [DOI] [PubMed] [Google Scholar]
- 16.DeVita VT, Jr, Young RC, Canellos GP. Combination versus single agent chemotherapy: A review of the basis for selection of drug treatment of cancer. Cancer. 1975;35(1):98–110. doi: 10.1002/1097-0142(197501)35:1<98::aid-cncr2820350115>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 17.Lage H. An overview of cancer multidrug resistance: A still unsolved problem. Cell Mol Life Sci. 2008;65(20):3145–3167. doi: 10.1007/s00018-008-8111-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shoemaker RH, et al. Development of human tumor cell line panels for use in disease-oriented drug screening. Prog Clin Biol Res. 1988;276:265–286. [PubMed] [Google Scholar]
- 19.Ugurel S, et al. Dermatologic Cooperative Oncology Group In vitro drug sensitivity predicts response and survival after individualized sensitivity-directed chemotherapy in metastatic melanoma: A multicenter phase II trial of the Dermatologic Cooperative Oncology Group. Clin Cancer Res. 2006;12(18):5454–5463. doi: 10.1158/1078-0432.CCR-05-2763. [DOI] [PubMed] [Google Scholar]
- 20.Grigsby PW, Zighelboim I, Powell MA, Mutch DG, Schwarz JK. In vitro chemoresponse to cisplatin and outcomes in cervical cancer. Gynecol Oncol. 2013;130(1):188–191. doi: 10.1016/j.ygyno.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maitland ML, DiRienzo A, Ratain MJ. Interpreting disparate responses to cancer therapy: The role of human population genetics. J Clin Oncol. 2006;24(14):2151–2157. doi: 10.1200/JCO.2005.05.2282. [DOI] [PubMed] [Google Scholar]
- 22.Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: Impact, heterogeneity, and uncertainty. Cancer Cell. 2012;21(3):283–296. doi: 10.1016/j.ccr.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shackleton M, Quintana E, Fearon ER, Morrison SJ. Heterogeneity in cancer: Cancer stem cells versus clonal evolution. Cell. 2009;138(5):822–829. doi: 10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 24.O’Brien CA, Kreso A, Jamieson CH. Cancer stem cells and self-renewal. Clin Cancer Res. 2010;16(12):3113–3120. doi: 10.1158/1078-0432.CCR-09-2824. [DOI] [PubMed] [Google Scholar]
- 25.Ricci-Vitiani L, et al. Identification and expansion of human colon cancer-initiating cells. Nature. 2007;445(7123):111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 26.Shenoy A, Butterworth E, Huang EH. ALDH as a marker for enriching tumorigenic human colonic stem cells. Methods Mol Biol. 2012;916:373–385. doi: 10.1007/978-1-61779-980-8_27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang EH, et al. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009;69(8):3382–3389. doi: 10.1158/0008-5472.CAN-08-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boman BM, Huang E. Human colon cancer stem cells: A new paradigm in gastrointestinal oncology. J Clin Oncol. 2008;26(17):2828–2838. doi: 10.1200/JCO.2008.17.6941. [DOI] [PubMed] [Google Scholar]
- 29.Shenoy AK, et al. Transition from colitis to cancer: high Wnt activity sustains the tumor-initiating potential of colon cancer stem cell precursors. Cancer Res. 2012;72(19):5091–5100. doi: 10.1158/0008-5472.CAN-12-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang EH, Wicha MS. Colon cancer stem cells: Implications for prevention and therapy. Trends Mol Med. 2008;14(11):503–509. doi: 10.1016/j.molmed.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao C, et al. Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukaemia. Nature. 2009;458(7239):776–779. doi: 10.1038/nature07737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Acharya AP, Clare-Salzler MJ, Keselowsky BG. A high-throughput microparticle microarray platform for dendritic cell-targeting vaccines. Biomaterials. 2009;30(25):4168–4177. doi: 10.1016/j.biomaterials.2009.04.032. [DOI] [PubMed] [Google Scholar]
- 33.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303(5659):844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 34.Khoo KH, Verma CS, Lane DP. Drugging the p53 pathway: Understanding the route to clinical efficacy. Nat Rev Drug Discov. 2014;13(3):217–236. doi: 10.1038/nrd4236. [DOI] [PubMed] [Google Scholar]
- 35.Vassilev LT. p53 Activation by small molecules: Application in oncology. J Med Chem. 2005;48(14):4491–4499. doi: 10.1021/jm058174k. [DOI] [PubMed] [Google Scholar]
- 36.Goldwasser F, Bae I, Valenti M, Torres K, Pommier Y. Topoisomerase I-related parameters and camptothecin activity in the colon carcinoma cell lines from the National Cancer Institute anticancer screen. Cancer Res. 1995;55(10):2116–2121. [PubMed] [Google Scholar]
- 37.Motwani M, et al. Augmentation of apoptosis and tumor regression by flavopiridol in the presence of CPT-11 in Hct116 colon cancer monolayers and xenografts. Clin Cancer Res. 2001;7(12):4209–4219. [PubMed] [Google Scholar]
- 38. (2000) Drug Synergism and Dose-Effect Data Analysis, ed Stern B (Chapman & Hall/CRC, Boca Raton, FL)
- 39.Kranz D, Dobbelstein M. Nongenotoxic p53 activation protects cells against S-phase–specific chemotherapy. Cancer Res. 2006;66(21):10274–10280. doi: 10.1158/0008-5472.CAN-06-1527. [DOI] [PubMed] [Google Scholar]
- 40.Ginestier C, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ginestier C, et al. CXCR1 blockade selectively targets human breast cancer stem cells in vitro and in xenografts. J Clin Invest. 2010;120(2):485–497. doi: 10.1172/JCI39397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carpentino JE, et al. Aldehyde dehydrogenase-expressing colon stem cells contribute to tumorigenesis in the transition from colitis to cancer. Cancer Res. 2009;69(20):8208–8215. doi: 10.1158/0008-5472.CAN-09-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dalerba P, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007;104(24):10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lau LM, Nugent JK, Zhao X, Irwin MS. HDM2 antagonist Nutlin-3 disrupts p73-HDM2 binding and enhances p73 function. Oncogene. 2008;27(7):997–1003. doi: 10.1038/sj.onc.1210707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.