Significance
In eukaryotic cells the 26S proteasome is responsible for the regulated degradation of intracellular proteins. The function of this large macromolecular machine is regulated by many cofactors, most notably the deubiquitylating enzyme ubiquitin C-terminal hydrolase 6 (Ubp6). Here, we investigate the structure of Ubp6 bound to the 26S proteasome and explore its influence on the conformational landscape of the 26S proteasome. Our structure reveals that Ubp6’s active site may contribute to a large composite active site, also formed by the mouth of the proteasomal ATPase ring and the active site of deubiquitylating enzyme regulatory particle non-ATPase 11. Moreover, Ubp6 modulates the conformational landscape of the proteasome, favoring an intermediate state, which may explain the effects of Ubp6 on proteasomal degradation.
Keywords: conformational switching, proteolysis, proteostasis, quality control
Abstract
In eukaryotic cells, the 26S proteasome is responsible for the regulated degradation of intracellular proteins. Several cofactors interact transiently with this large macromolecular machine and modulate its function. The deubiquitylating enzyme ubiquitin C-terminal hydrolase 6 [Ubp6; ubiquitin-specific protease (USP) 14 in mammals] is the most abundant proteasome-interacting protein and has multiple roles in regulating proteasome function. Here, we investigate the structural basis of the interaction between Ubp6 and the 26S proteasome in the presence and absence of the inhibitor ubiquitin aldehyde. To this end we have used single-particle electron cryomicroscopy in combination with cross-linking and mass spectrometry. Ubp6 binds to the regulatory particle non-ATPase (Rpn) 1 via its N-terminal ubiquitin-like domain, whereas its catalytic USP domain is positioned variably. Addition of ubiquitin aldehyde stabilizes the binding of the USP domain in a position where it bridges the proteasome subunits Rpn1 and the regulatory particle triple-A ATPase (Rpt) 1. The USP domain binds to Rpt1 in the immediate vicinity of the Ubp6 active site, which may effect its activation. The catalytic triad is positioned in proximity to the mouth of the ATPase module and to the deubiquitylating enzyme Rpn11, strongly implying their functional linkage. On the proteasome side, binding of Ubp6 favors conformational switching of the 26S proteasome into an intermediate-energy conformational state, in particular upon the addition of ubiquitin aldehyde. This modulation of the conformational space of the 26S proteasome by Ubp6 explains the effects of Ubp6 on the kinetics of proteasomal degradation.
Degradation of proteins that are misfolded, damaged, or no longer needed is an essential element of cellular homeostasis. In eukaryotic cells, the ubiquitin-proteasome system (UPS) is the major pathway for regulated protein degradation (1). Proteins that are processed by the UPS are marked for destruction by polyubiquitin chains, which are recognized as a degradation signal by the 26S proteasome.
The 26S proteasome consists of the core particle (CP), which degrades substrates into short peptides, and one or two 19S regulatory particles (RP), which associate with the ends of the cylinder-shaped CP to recruit substrates and prepare them for degradation (2, 3). Although the structure of the CP has been known for more than two decades (4, 5), the molecular architecture of the RP was unraveled by cryo-electron microscope (EM)–based approaches only recently (6–9). It comprises six RP triple A (AAA) ATPases (Rpt), 1–6, and 13 RP non-ATPases (Rpn), 1–3, 5–13, and 15. Similar to AAA-ATPases in prokaryotic ATP-dependent proteases, the Rpts form a hexameric ring that binds to the ends of the CP and is responsible for substrate unfolding and translocation into the CP. Unlike their prokaryotic counterparts, the Rpts are surrounded by non-ATPases. Apart from Rpn1, all Rpns form a cohesive structure, which places the RP’s catalytic subunits at their optimal positions for efficient proteasomal degradation. The deubiquitylating enzyme (DUB) Rpn11, which is responsible for the removal of polyubiquitin chains from substrates before degradation (10, 11), is positioned near the oligosaccharide-binding domain (OB) ring forming the mouth of the AAA-ATPase. The resident receptors for ubiquitin chains, Rpn10 and Rpn13, are positioned at the distal ends of the RP (12).
Recent cryo-EM analyses revealed the conformational plasticity of the RP. At least three distinct states, which we refer to as “s1–s3,” can be distinguished (13). In ATP-containing buffer, purified 26S proteasomes primarily adopt the s1 state, which is characterized by pronounced off-axis positioning of the AAA-ATPase hexamer with respect to the CP and a staircase arrangement of the Rpts with Rpt3 in the most elevated position (6–9). Under the same conditions a minority of particles (∼20%) adopts the s2 state, in which the axis of the AAA-ATPase is positioned closer to that of the CP and the Rpns concomitantly rotate largely en bloc by ∼25° (13). As a consequence of this Rpn motion, the active site of Rpn11 becomes accessible to the polyubiquitin chain of the substrate, and the ubiquitin receptor Rpn10 is positioned closer to the AAA-ATPase module. A third conformation, s3, was found in the presence of the slowly hydrolysable ATP analogue ATP-γS (14) or upon the addition of polyubiquitylated substrate to 26S proteasomes with dysfunctional Rpn11 (15). Characteristics of the s3 state are a changed staircase arrangement of the AAA-module with Rpt1 most elevated and a further translation of the Rpns compared with s2, leaving Rpn11 and Rpn10 essentially invariant with respect to the mouth of the AAA-ATPase module.
Proteasome function is modulated by transiently binding cofactors, the proteasome-interacting proteins (PIPs) (16–19), which typically are found in substoichiometric amounts in purified proteasomes (20). Of these, ubiquitin C-terminal hydrolase 6 [Ubp6, human ubiquitin-specific protease 14 (Usp14)] is most abundant. It consists of an N-terminal ubiquitin-like (UBL) domain, which associates primarily with the RP via Rpn1 (21, 22), a disordered linker of ∼25 residues, and an ubiquitin-specific protease (USP) domain. The DUB activity of Ubp6 is low in isolation but increases dramatically upon binding to the 26S proteasome (16, 23–25). One main function of Ubp6 seems to be to delay the degradation of polyubiquitylated substrates by progressively deubiquitylating them (25, 26). Ubp6 thus seems to serve as a timing device. A pharmacological agent that inhibits deubiquitylation by the human Ubp6 homolog Usp14, enhances the chance that the protein is degraded (26). Consequently, Usp14 is an attractive drug target, e.g., to prevent the accumulation of protein aggregates associated with neurodegenerative diseases. Interestingly, the catalytic activity is not required for inhibiting the degradation of folded substrates (17, 25), but the structural basis is unclear. On the other hand, binding of Ub-conjugates or the USP inhibitor ubiquitin-aldehyde (UbAld) to proteasome-bound Ubp6 enhances the degradation of short unfolded peptides (27) and activates the proteasomal ATPases (28), both indicating a conformational change of the 26S proteasome. To gain structural insights into the mechanisms of the proteasomal regulation by Ubp6, we determined the position of Ubp6 in complex with UbAld bound to the 26S proteasome and its consequences for the conformation of the RP.
Results
3D Reconstructions of 26S–Ubp6, 26S–Ubp6–UbAld, and 26S–UbAld.
In previous cryo-EM studies Ubp6 was not localized because it typically is a substoichiometric component of 26S proteasome preparations. According to label-free mass spectrometry quantification, the amount of Ubp6 is ∼30% of the canonical RP subunits in our preparations (Fig. S1). Therefore, we added recombinant Ubp6 to 26S proteasome preparations (26S–Ubp6) to achieve a higher occupancy of Ubp6 and thereby facilitate the localization of Ubp6 in the holocomplex. We verified catalytic activity of the recombinant protein in the presence of purified 26S proteasomes and ensured its binding to the 26S proteasome by pulldown analysis (Fig. S2). For the cryo-EM experiments a 10-fold excess of Ubp6 was added.
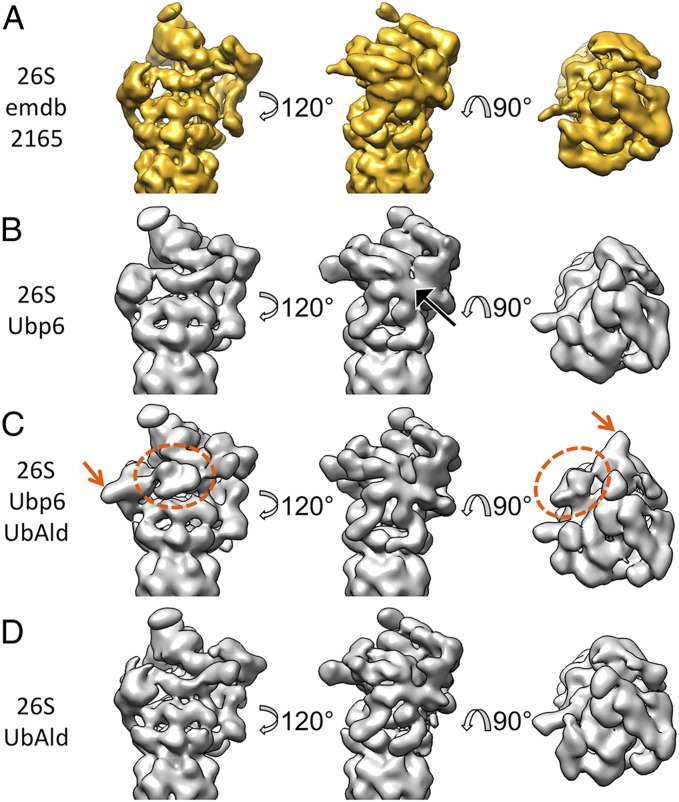
Previous analysis revealed that the conformational states of the RPs at the two CP ends are not correlated (6, 13, 15). Therefore, the following analysis was performed using the holocomplex cut into two halves (6, 13), i.e., the two RPs of the double-capped 26S proteasomes were treated as separate particles. The reconstruction from ∼160,000 26S–Ubp6 particles (each corresponding to a half of a 26S proteasome) yields a blurred RP density, indicative of a mixture of multiple conformation states (Fig. 1B). In particular, the horseshoe-shaped scaffold assembled from the six proteasome–COP9–initiation factor (PCI) subunits is smeared out. Compared with the previously published density of 26S proteasomes alone (6, 7), no globular extra density could be detected upon the addition of Ubp6 (Fig. 1 A and B). Thus, the overall blurriness of the RP is the only detectable difference.
Fig. 1.
26S proteasome reconstructions for different buffers. (A) Reconstructions from 26S proteasomes alone, filtered to 15-Å resolution (6). (B–D) Reconstructions from 26S–Ubp6 (B), 26S–Ubp6–UbAld (C), and 26S–UbAld (D) datasets obtained in the present study, all filtered to ∼15-Å resolution. The black arrow in B indicates the blurred PCI horseshoe. The dotted orange ellipse and the orange arrow in C mark the extra density seen for the 26S–Ubp6–UbAld reconstruction.
In a second step, a surplus of UbAld was added to study the structural consequences of ubiquitin binding to Ubp6 (26S–Ubp6–UbAld). The simultaneous binding of Ubp6 and UbAld to the 26S proteasome was confirmed by a pulldown assay (Fig. S2). Additionally, we verified that the catalytic activity of Ubp6 was fully inhibited at the used concentration of 12 μM UbAld, indicating a nearly stoichiometric binding of UbAld to Ubp6 (Fig. S3A). Although the number of particles in the 26S–Ubp6–UbAld dataset (∼170,000) is comparable to that in the 26S–Ubp6 dataset, the reconstruction is better defined (Fig. 1C), indicating less structural heterogeneity. The overall structure differs notably from the reconstruction from 26S proteasomes alone, which predominantly adopt the s1 conformation (6), and appears more similar to the s2 conformation (13). Most importantly, two extra densities in the direct vicinity of Rpn1 are clearly distinguishable. A smaller density is attached to Rpn1, and a larger density is in contact with Rpn1 and with the ATPase OB-ring.
To verify that these effects are the result of the interaction of UbAld with Ubp6, we repeated the experiment without added Ubp6 (26S–UbAld). The reconstruction from ∼500,000 particles also yielded a well-defined structure (Fig. 1D). In contrast to the 26S–Ubp6–UbAld map, it is highly similar to the reconstruction from 26S alone (Fig. 1A) (6). Notably, the extra density of the 26S–Ubp6–UbAld reconstruction is not found in this reconstruction.
Taken together, these data strongly suggest that the extra density seen in 26S–Ubp6–UbAld can be attributed to Ubp6–UbAld. To support this interpretation further, we performed cross-linking coupled to mass spectrometry (XL-MS) of 26S–Ubp6–UbAld (Table S1). Overall roughly 100 high-confidence crosslinks between different subunits of the 26S proteasome were identified; among them six involved Ubp6. Of those six, three links were found between the USP domain and the unstructured N-terminal region of Rpt2, and two links to the C-terminal part of Rpt1. Thus, essentially all detected cross-links involving Ubp6 are in the immediate vicinity of the extra density, corroborating the findings from the EM reconstructions.
Analysis of Conformational Ensembles and a Correlation of States with Extra Density.
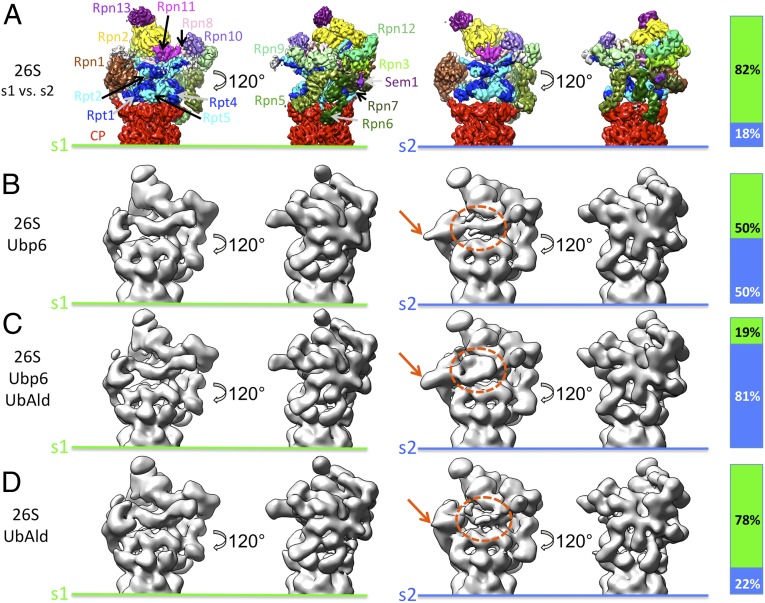
To investigate the three datasets in more detail, subsets of equal size were subjected to 3D classification, and the resultant class reconstructions were assigned to the known conformations s1, s2, and s3 (Fig. S4). Different distributions of classes in the s1 and s2 states were observed in the datasets, but no classes were assigned to s3 (Fig. 2). Although the s1/s2 ratio of 26S–UbAld particles is comparable to that in 26S datasets (S1/S2 = 4:1) (13), the other two datasets with supplemented Ubp6 exhibit different state occupancies. For the 26S–Ubp6 dataset approximately equal amounts of particles are assigned to s1 and s2 (s1/s2 = 1:1). In contrast, the majority of proteasomes are in an s2-like conformation in the 26S–Ubp6–UbAld dataset (s1/s2 = 1:5). Overall, the different state occupancies of the three datasets are consistent with the respective reconstructions before classification (Fig. 1). Thus, the binding of Ubp6 and additional UbAld to the 26S proteasome strongly increases the occupancy of the s2 state.
Fig. 2.
Classification of datasets into s1 and s2 states. The bars indicate the relative frequencies of s1 and s2 states. (A) The s1 and s2 reconstructions obtained from 26S proteasomes alone (13). The different subunits of the regulatory particles are indicated for the s1 state. (B–D) As in A, for the 26S–Ubp6 (B), 26S–Ubp6–UbAld (C), and 26S–UbAld (D) datasets. The dotted orange ellipses and the orange arrows mark the extra density seen for the 26S–Ubp6–UbAld reconstruction shown in Fig. 1C.
A striking property of the reconstructions from the s1- and s2-like particles is that the extra density (marked by orange ellipses and arrows in Fig. 2), which is observed in the overall 26S–Ubp6–UbAld reconstruction, is present exclusively in the s2-like conformations. This feature is discernable most clearly in the reconstruction from the s2-like 26S–Ubp6–UbAld particles. In addition, the s2-like class reconstructions from the 26S–Ubp6 and 26S–UbAld datasets show indications of this extra density in the respective areas, but none of the s1-like groups exhibit any sign of it.
To follow up on the relatively weak extra density in the s2-like states of the 26S–Ubp6 and 26S–UbAld datasets, the respective particles were subjected to a second round of classification focused on the region between Rpn1 and Rpn10 (Fig. S5). Classification analysis of 26S proteasomes did not detect density in this area (13). In contrast, for the 26S–Ubp6 dataset, 6% of all particles show an extra density, which is located at the same position as the one identified in the 26S–Ubp6–UbAld reconstruction but which appears to be slightly smaller (Fig. S5). Thus, although the population is relatively small, Ubp6 alone induces a conformation similar to that seen with the 26S–Ubp6–UbAld dataset.
The analysis of the 26S–UbAld dataset also reveals an extra density for ∼4% of the particles. In contrast to 26S–Ubp6, this extra density has approximately the same size as that seen in 26S–Ubp6–UbAld. The most likely explanation for this population is the binding of UbAld to the endogenous Ubp6 associated with the 26S proteasome in substoichiometric amounts. Because of the small percentage of particles exhibiting the extra density, it is likely that some of these particles are erroneously grouped during the multistep classification procedure. Therefore, the given percentages of particles with this feature most likely constitute a lower boundary value. The excellent agreement of the extra density-containing reconstructions from the 26S–Ubp6–UbAld and 26S–UbAld data (Fig. S5) also indicates that the N-terminal GST tag of the recombinant Ubp6 is not resolved in the 26S–Ubp6–UbAld reconstruction because it is flexibly linked to the UBL domain by an eight-residue linker. Taken together, all three datasets contain a varying fraction of s2-like particles with an extra density.
Structure of the 26S–Ubp6–UbAld Complex at 9.5-Å Resolution.
For a more detailed analysis of proteasome-bound Ubp6–UbAld, the 26S–Ubp6–UbAld data were sorted out to achieve higher homogeneity, and the corresponding reconstruction was refined (Fig. S6A). The global resolution of the refined map is 9.5 Å (Fig. S6B). Subsequently, the local resolution was computed, and the refined map was filtered to its local resolution (Fig. 3A and Fig. S6C). Although many parts of the 26S proteasome are resolved at subnanometer level, the area harboring the extra density is less well-defined (Fig. S6C). The locally reduced resolution most likely is caused by the structural heterogeneity of Ubp6, as would be consistent with the differences in the shape of the extra density found in the individual classes (Fig. S4).
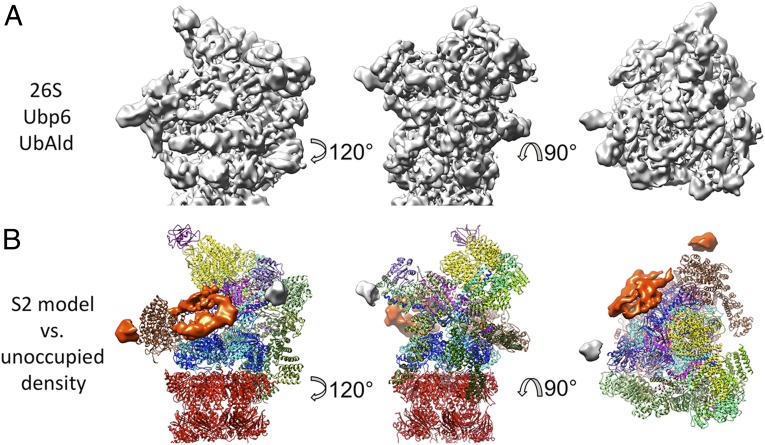
Fig. 3.
Density of proteasome-bound Ubp6–UbAld. (A) Refined density of the 26S–Ubp6–UbAld complex from 26S–Ubp6–UbAld dataset. (B) Comparison of the atomic model of the s2 state and the density. The difference between the two is rendered as an isosurface. The orange density is specific for the 26S–Ubp6–UbAld data, and the gray area corresponds to the disordered N termini of the coiled-coils of Rpt4/5, which also are present in the EM density from 26S proteasomes alone but are not included in the model.
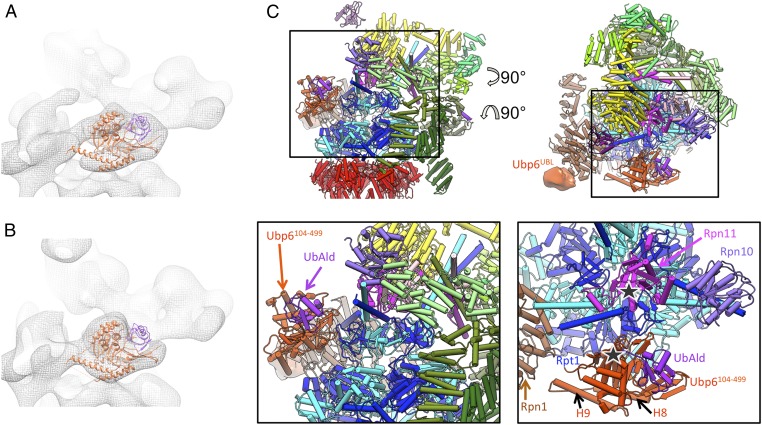
Fitting the s2 pseudoatomic model (PDB ID code 4CR3) into the map allowed us to isolate computationally the segment not accounted for by the model (Fig. 3B). In addition to the extra density, the difference between the map and model includes a small density at the tips of the coiled-coils of Rpt4/5. However, this density does not correlate with Ubp6, because it also is present in the s2 26S cryo-EM maps (Fig. 2A), although it is not explained by the atomic model. The extra density attributed to Ubp6 consists of two segments. The small one is attached to Rpn1, and its volume approximately matches that of the UBL domain of Ubp6. It contacts Rpn1 in proximity to the helices H2–H5 of its toroid domain. These helices are part of the domain that has been found to be essential for the Rpn1–UBL interaction (21). The size, shape, and volume of the larger segment are similar to the catalytic domain of Ubp6 (PDB ID code 1VJV) and Usp14 bound to UbAld (PDB ID code 2AYO) (24).
Fitting of the Catalytic Domain of Ubp6.
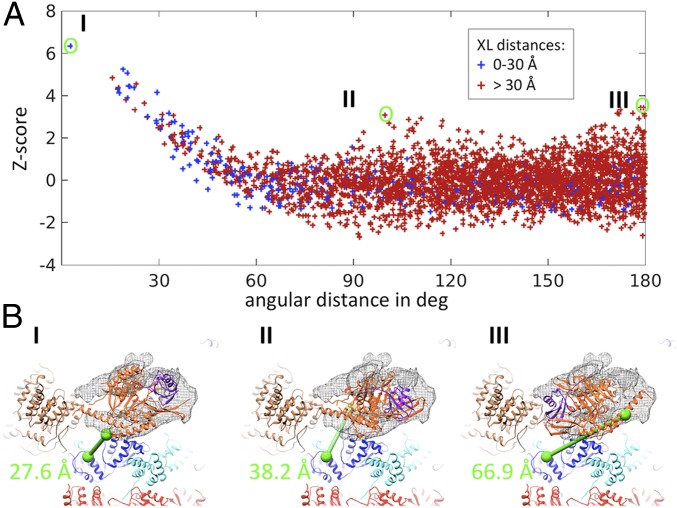
Although the UBL domain is too small for accurate fitting at the achieved resolution, we attempted to position the USP domain into the extra density based on the available crystal structures. We created a model of Ubp6104-499–UbAld by superposing the Ubp6104-499 crystal structure onto the Usp14–UbAld structure. Specifically, a six-dimensional correlation search (exhaustive translation and orientation) of the Ubp6104-499–UbAld template against the area of the extra density was performed. To assess the specificity of the orientations, the respective correlation values were transformed to Z-scores, revealing that the best-fitting orientation scores significantly better than all other solutions (Fig. 4A and Fig. S7). The fitting score decreased slightly when the search was performed for Ubp6104-499 alone, but the angular positioning remained the same (Fig. S8). In the best-fitting result (Fig. 4 B, I and Fig. 5C) UbAld points to the OB-ring of the ATPase. Interestingly, the extra density in the classified subset of the 26S–Ubp6 dataset lacks density in precisely the area assigned to UbAld (Fig. 5B), but the entire extra density is present in the subset from the 26S–UbAld dataset (Fig. S5). Taken together, the findings indicate that the extra density represents UbAld.
Fig. 4.
Fit of the Ubp6 USP domain in the refined 26S–Ubp6–UbAld density. (A) Assessment of orientation specificity of fitting. The Z-scores of the correlation are plotted against the angular distance to the best-fitting result, which is more than six standard deviations above the mean value. The coordinates are colored according to the distance of the cross-linked residues of the USP domain and Rpt1 (Table S1). (B) Visualization of the cross-link for the three best-scoring solutions in A (orange: Ubp6; purple: UbAld; blue: Rpt1; brown: Rpt1; cyan: Rpt5; red: CP).
Fig. 5.
Model of proteasome-bound Ubp6–UbAld. (A and B) Best-fitting model from Fig. 4 positioned in the extra density-containing class from the 26S–Ubp6–UbAld and 26S–Ubp6 datasets, respectively. (C, Upper) Atomic model of the 26S–Ubp6–UbAld complex seen in two different views. (Lower) The boxed region is enlarged for better visualization of Ubp6104-499 and UbAld. In the enlarged top view (Lower Right) Rpn13, Rpn2, Rpn8, and the PCI horseshoe have been removed for clarity.
The positioning of Ubp6 is further supported by the XL-MS data (Table S1). Although most of the detected crosslinks involve residues within unstructured domains, one crosslink occurs between lysines of Ubp6 and Rpt1 that are covered by the available atomic models (PDB ID codes 1VJV and 4CR3, respectively). The distance of the respective Cα-atoms is ∼28 Å for the best fit, which is below the cutoff of 30 Å previously applied for intersubunit crosslinks of the 26S proteasome (29); the distances are much larger for the majority of alternative solutions (Fig. 4). Thus, XL-MS analysis of 26S–Ubp6–UbAld provides orthogonal evidence for the suggested placement of the catalytic domain of Ubp6 bound to the 26S proteasome.
In the best-fitting Ubp6104-499 model, the N terminus (residue 104) is in direct proximity to Rpn1, in line with the proposed positioning of the UBL domain also at Rpn1. The contact with Rpn1 is established further by an extension formed by helices H8 and H9 that protrude from the palm domain of Ubp6 (Fig. 5C). This segment is specific for Ubp6 and is not present in related USP enzymes, such as herpesvirus-associated ubiquitin-specific protease (24). The N terminus of H9 is most proximal to the very N-terminal helix of the toroid domain of Rpn1. H8 and H9 are connected by an unstructured linker, which also may be involved in the interaction.
The largest interaction area of Ubp6 with the 26S proteasome is located in a patch constituted by Ubp6 residues K316–V333 and E473–S488. Interestingly, the K316–V333 loop and an adjacent loop block the binding groove for the C terminus of ubiquitin in the crystal structure of free Usp14 (24). The interacting patch contacts the OB domain of Rpt1. Structured residues of Rpt1 in the interaction area include the residue ranges G158–E169 and Y181–R190. Intriguingly, Rpt1 has an extended unstructured region (residues G110–D140) in immediate proximity to this area. Because this segment is specific for Rpt1, it is conceivable that it has evolved to interact with Ubp6. The catalytic triad faces the Rpt1/2 coiled-coil (Fig. 5C), and the UbAld moiety is positioned in immediate proximity of the pore of the OB-ring of the ATPase module, between the coiled-coils of Rpt1/2 and Rpt4/5.
Discussion
Structure of the 26S–Ubp6 Complex.
In the overall reconstruction of the s2-like 26S proteasomes from the 26S–Ubp6 data, the UBL domain can be distinguished, but the USP domain is not resolved (Fig. 2). Thus, the USP domain adopts variable positions, likely caused by the unstructured linker region of ∼25 amino acids connecting the UBL and USP domains. Consistent with these findings, Ubp6 was found to be highly mobile in previous cryo-EM analyses (7).
The most abundant conformation is stabilized upon the addition of UbAld (Fig. S5), allowing us to resolve the 26S–Ubp6–UbAld structure sufficiently well to dock the USP domain accurately. It contacts Rpn1 as well as the ATPase OB-ring via Rpt1, placing it in the direct vicinity of the mouth of the ATPase as well as the ubiquitin receptor Rpn10 and the DUB Rpn11. Interestingly, the contact site of Rpt1 is accessible only in the s2 and s3 states of the 26S proteasome, not in the s1 state, because of the rotation and translation of Rpn1 with respect to the ATPase. This finding is consistent with the absence of extra density in s1-class reconstructions. Moreover, it is likely that the occupancy of a state in which the USP domain contacts Rpt1 also can be increased in the presence of ATP-γS or substrate, which induces switching to s3 (14, 15). It has been proposed that the s1 state is primarily responsible for substrate recruitment, whereas both the s2 and s3 states are engaged in dealing with substrate (13). Thus, binding of the catalytic USP domain to the OB-ring appears to correlate with substrate processing by the 26S proteasome. A notable feature of the s2 and s3 states compared with the s1 state is the repositioning of Rpn10 in immediate vicinity of the USP domain, implying that Rpn10 has a cooperative role in the substrate transfer to Ubp6.
Saccharomyces cerevisiae 26S proteasomes are equipped with two DUBs to cleave ubiquitin moieties from polyubiquitylated substrates: Ubp6 and the stoichiometric subunit Rpn11. However, these DUBs appear to have distinct but complementary roles during substrate degradation. Rpn11 enhances degradation by cleaving the polyubiquitin chain en bloc in an ATP-dependent manner (10, 11, 30, 31). In contrast, Ubp6 binds transiently to the 26S proteasome and removes short chains; this removal may extend the lifetime of the substrate (16, 26, 32). The structure of Ubp6 bound to the 26S proteasome determined here reveals that the active sites of these DUBs, Ubp6 and Rpn11, are both positioned close to the mouth of the OB-ring (∼50 Å and ∼30 Å, respectively) and to each other (∼40 Å). Whereas the catalytic site of Rpn11 is positioned right above the OB mouth, Ubp6 is placed off-axis. The close positioning of the two active sites may allow their simultaneous processing of substrates, possibly thereby increasing the efficiency of proteasomal degradation for specific types of ubiquitylation patterns (33). However, the precise structural basis and the functional implications of such a mechanism remain to be explored.
Activation of Ubp6.
The DUB activity of Ubp6 is increased dramatically upon binding to the 26S proteasome (16, 26). The crystal structures of unbound Usp14 and Ubp6 revealed that two surface loops (BL1 and BL2) are located above the binding site for the C-terminal tail of ubiquitin, presumably blocking the access to the catalytic site (24). Binding of UbAld to the USP domain of free Usp14 induces a conformational change of these surface loops (24). Intriguingly, the inhibitor ubiquitin vinyl-sulfone binds covalently to Usp14 only in the presence of the RP, indicating a proteasome-induced structural change. Our EM analysis reveals that not only the UBL domain but also the catalytic USP domain interacts with the 26S proteasome. The major interaction area of Ubp6 with the 26S proteasome is located in a patch containing BL1, with BL2 in close vicinity. Accordingly, the physical interaction of these loops with Rpt1 may activate Ubp6.
Effects of Ubp6 on 26S Proteasome Conformation and Catalytic Activity.
Our classification analysis indicates that the 26S proteasome preferably adopts the s2 conformational state in the presence of Ubp6, especially upon the addition of UbAld. The influence of Ubp6 on the 26S proteasome conformation may explain the multifunctional nature of Ubp6. Independent of its own catalytic activity, Ubp6 also modulates the activity of the 26S proteasome. For example, it increases the hydrolysis rate of small peptides upon binding of ubiquitin conjugates or UbAld (27, 28). Although initially interpreted as enhanced gate opening into the 20S CP (27, 28), the most notable effect we observe upon binding of Ubp6–UbAld is a conformational change from the s1 to the s2 state. A characteristic feature of the s2 state, as compared with the s1 state, is a better alignment of the channel axes of the CP and ATPase, suggesting that unfolded peptides can access the gated pore and catalytic cavity of the CP more easily. In a similar manner, the increased degradation of unfolded peptides by the 26S proteasome in the presence of ATP-γS (34) could be explained by a conformational change from the s1 state to the s3 state (14). In addition, binding of Ubp6 was reported to increase ATP hydrolysis (28). Our findings of the conformational change upon binding of Ubp6 to the OB domain of the ATPase imply alternations of the energy landscape of the 26S proteasome, which might lead to increased ATPase activity.
On the other hand, Ubp6 delays the degradation of folded proteins in a catalytic and noncatalytic manner (16, 25, 35). Catalytically, the placement of Ubp6 in the vicinity of Rpn10 may facilitate substrate transfer for deubiquitylation, resulting in the release of the deubiquitylated substrate from the proteasome. Noncatalytically, Ubp6 was suggested to impair ubiquitin chain removal by Rpn11 (25). Whether Ubp6 directly inhibits Rpn11 activity (e.g., by steric hindrance of substrate access to Rpn11) or whether alteration of the nucleotide cycle prevents efficient unfolding requires further investigation.
Ubp6/Usp14 is the most abundant PIP and probably the most important one in regulating and integrating diverse proteasomal processes. Taken together our structural studies on Ubp6 constitute a first step toward a mechanistic understanding of regulation of proteasomal degradation by PIPs.
Materials and Methods
Sample Preparation and Characterization.
Isolated 26S proteasomes were obtained by affinity purification via Rpn11-3FLAG from Saccharomyces cell lysate, followed by further isolation using a sucrose gradient. Ubp6 was obtained by recombinant expression in E. coli and purification by gel filtration. The samples were characterized by hydrolysis assays and label-free quantification of protein abundances using mass spectrometry. The 26S–Ubp6–UbAld sample was additionally subjected to XL-MS (36).
Cryo-EM and Image Analysis.
Transmission electron microscope images were acquired on an FEI Titan Krios equipped with a Falcon2 4k4k camera as described in ref. 6. EM data were analyzed using XMIPP (37) and the TOM toolbox (38). Fitting of densities simulated from atomic models was performed in PyTom (39) and analyzed using the TOM toolbox (38). All images of the resulting 3D reconstructions and atomic models were rendered in UCSF Chimera (40).
A detailed description of all protocols including XL-MS analysis is found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Oana Mihalache and Stefan Pfeffer for assistance with cryo-EM; Marc Wehmer for help with purifications; Claire Basquin for advice on florescence assays; Andreas Peth for helpful discussions; Radostin Danev and Maryam Khoshouei for help with data acquisition; and Hyoung Tae Kim and Ruben Fernandez-Busnadiego for reading the manuscript. The label-free protein quantification was performed by the core facility of the Max-Planck Institute of Biochemistry. This work was supported by the German Science Foundation Excellence Cluster Center for Integrated Protein Science Munich and Collaborative Research Center (SFB)-1035/Project A01 (W.B.) and SFB969 (F.S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: Crystallography, atomic coordinates, and structure factors reported in this paper have been deposited in the Protein Data Bank (ID code 5A5B) and the Electron Microscopy Data Bank (accession code 3034).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1510449112/-/DCSupplemental.
References
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Voges D, Zwickl P, Baumeister W. The 26S proteasome: A molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 3.Förster F, Unverdorben P, Sledź P, Baumeister W. Unveiling the long-held secrets of the 26S proteasome. Structure. 2013;21(9):1551–1562. doi: 10.1016/j.str.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Löwe J, et al. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science. 1995;268(5210):533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 5.Groll M, et al. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386(6624):463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 6.Beck F, et al. Near-atomic resolution structural model of the yeast 26S proteasome. Proc Natl Acad Sci USA. 2012;109(37):14870–14875. doi: 10.1073/pnas.1213333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lander GC, et al. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482(7384):186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nickell S, et al. Insights into the molecular architecture of the 26S proteasome. Proc Natl Acad Sci USA. 2009;106(29):11943–11947. doi: 10.1073/pnas.0905081106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bohn S, et al. Structure of the 26S proteasome from Schizosaccharomyces pombe at subnanometer resolution. Proc Natl Acad Sci USA. 2010;107(49):20992–20997. doi: 10.1073/pnas.1015530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verma R, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298(5593):611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 11.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419(6905):403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 12.Sakata E, et al. Localization of the proteasomal ubiquitin receptors Rpn10 and Rpn13 by electron cryomicroscopy. Proc Natl Acad Sci USA. 2012;109(5):1479–1484. doi: 10.1073/pnas.1119394109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Unverdorben P, et al. Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome. Proc Natl Acad Sci USA. 2014;111(15):5544–5549. doi: 10.1073/pnas.1403409111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Śledź P, et al. Structure of the 26S proteasome with ATP-γS bound provides insights into the mechanism of nucleotide-dependent substrate translocation. Proc Natl Acad Sci USA. 2013;110(18):7264–7269. doi: 10.1073/pnas.1305782110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matyskiela ME, Lander GC, Martin A. Conformational switching of the 26S proteasome enables substrate degradation. Nat Struct Mol Biol. 2013;20(7):781–788. doi: 10.1038/nsmb.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leggett DS, et al. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10(3):495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- 17.Verma R, et al. Proteasomal proteomics: Identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell. 2000;11(10):3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Huang L. Identifying dynamic interactors of protein complexes by quantitative mass spectrometry. Mol Cell Proteomics. 2008;7(1):46–57. doi: 10.1074/mcp.M700261-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Besche HC, Haas W, Gygi SP, Goldberg AL. Isolation of mammalian 26S proteasomes and p97/VCP complexes using the ubiquitin-like domain from HHR23B reveals novel proteasome-associated proteins. Biochemistry. 2009;48(11):2538–2549. doi: 10.1021/bi802198q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakata E, et al. The catalytic activity of Ubp6 enhances maturation of the proteasomal regulatory particle. Mol Cell. 2011;42(5):637–649. doi: 10.1016/j.molcel.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 21.Elsasser S, et al. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002;4(9):725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 22.Rosenzweig R, Bronner V, Zhang D, Fushman D, Glickman MH. Rpn1 and Rpn2 coordinate ubiquitin processing factors at proteasome. J Biol Chem. 2012;287(18):14659–14671. doi: 10.1074/jbc.M111.316323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borodovsky A, et al. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 2001;20(18):5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu M, et al. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 2005;24(21):3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanna J, et al. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127(1):99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 26.Lee BH, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467(7312):179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peth A, Besche HC, Goldberg AL. Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Mol Cell. 2009;36(5):794–804. doi: 10.1016/j.molcel.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peth A, Kukushkin N, Bossé M, Goldberg AL. Ubiquitinated proteins activate the proteasomal ATPases by binding to Usp14 or Uch37 homologs. J Biol Chem. 2013;288(11):7781–7790. doi: 10.1074/jbc.M112.441907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walzthoeni T, et al. False discovery rate estimation for cross-linked peptides identified by mass spectrometry. Nat Methods. 2012;9(9):901–903. doi: 10.1038/nmeth.2103. [DOI] [PubMed] [Google Scholar]
- 30.Pathare GR, et al. Crystal structure of the proteasomal deubiquitylation module Rpn8-Rpn11. Proc Natl Acad Sci USA. 2014;111(8):2984–2989. doi: 10.1073/pnas.1400546111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Worden EJ, Padovani C, Martin A. Structure of the Rpn11-Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation. Nat Struct Mol Biol. 2014;21(3):220–227. doi: 10.1038/nsmb.2771. [DOI] [PubMed] [Google Scholar]
- 32.Crosas B, et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127(7):1401–1413. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- 33.Lu Y, Lee BH, King RW, Finley D, Kirschner MW. Substrate degradation by the proteasome: A single-molecule kinetic analysis. Science. 2015;348(6231):1250834. doi: 10.1126/science.1250834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith DM, Fraga H, Reis C, Kafri G, Goldberg AL. ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell. 2011;144(4):526–538. doi: 10.1016/j.cell.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee MJ, Lee BH, Hanna J, King RW, Finley D. 2011. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Mol Cell Proteomics 10(5):R110 003871–003875. [DOI] [PMC free article] [PubMed]
- 36.Leitner A, Walzthoeni T, Aebersold R. Lysine-specific chemical cross-linking of protein complexes and identification of cross-linking sites using LC-MS/MS and the xQuest/xProphet software pipeline. Nat Protoc. 2014;9(1):120–137. doi: 10.1038/nprot.2013.168. [DOI] [PubMed] [Google Scholar]
- 37. doi: 10.1038/nprot.2008.62. Scheres SH, Nunez-Ramirez R, Sorzano CO, Carazo JM, Marabini R (2008) Image processing for electron microscopy single-particle analysis using XMIPP. Nat Protoc 3(6):977–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. doi: 10.1016/j.jsb.2004.10.006. Nickell S, et al. (2005) TOM software toolbox: Acquisition and analysis for electron tomography. J Struct Biol 149(3):227–234. [DOI] [PubMed] [Google Scholar]
- 39. doi: 10.1016/j.jsb.2011.12.003. Hrabe T, et al. (2012) PyTom: A python-based toolbox for localization of macromolecules in cryo-electron tomograms and subtomogram analysis. J Struct Biol 178(2):177–188. [DOI] [PubMed] [Google Scholar]
- 40. doi: 10.1016/j.jsb.2006.06.010. Goddard TD, Huang CC, Ferrin TE (2007) Visualizing density maps with UCSF Chimera. J Struct Biol 157:281–287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.