Abstract
The world’s crop productivity is stagnating whereas population growth, rising affluence, and mandates for biofuels put increasing demands on agriculture. Meanwhile, demand for increasing cropland competes with equally crucial global sustainability and environmental protection needs. Addressing this looming agricultural crisis will be one of our greatest scientific challenges in the coming decades, and success will require substantial improvements at many levels. We assert that increasing the efficiency and productivity of photosynthesis in crop plants will be essential if this grand challenge is to be met. Here, we explore an array of prospective redesigns of plant systems at various scales, all aimed at increasing crop yields through improved photosynthetic efficiency and performance. Prospects range from straightforward alterations, already supported by preliminary evidence of feasibility, to substantial redesigns that are currently only conceptual, but that may be enabled by new developments in synthetic biology. Although some proposed redesigns are certain to face obstacles that will require alternate routes, the efforts should lead to new discoveries and technical advances with important impacts on the global problem of crop productivity and bioenergy production.
Keywords: light capture/conversion, carbon capture/conversion, smart canopy, enabling plant biotechnology tools, sustainable crop production
Increasing demands for global food production over the next several decades portend a huge burden on the world’s shrinking farmlands. Increasing global affluence, population growth, and demands for a bioeconomy (including livestock feed, bioenergy, chemical feedstocks, and biopharmaceuticals) will all require increased agricultural productivity, perhaps by as much as 60–120% over 2005 levels (e.g., refs. 1 and 2), putting increased productivity on a collision course with environmental and sustainability goals (3). The 45 y from 1960 to 2005 saw global food production grow ∼160%, mostly (135%) by improved production on existing farmlands (4). If agricultural production is to double by 2050, it will require at least a doubling of productivity per hectare because cropland areas are shrinking, with little opportunity to sustainably reverse this trend. Alas, growth in productivity of the world’s major crops is stagnating in most of the important growing areas (5, 6).
What limits crop productivity? For most terrestrial crops it is usually the availability of water (7, 8), and, although there are some promising avenues to modestly increase water use efficiency, there are few options for dramatically reducing the amount of water required to grow a crop. The inability of photosynthesis to efficiently use high midday light intensities (9) also strongly limits productivity compared with hypothetical plants able to use high intensity light as efficiently as low intensity light. The slow catalytic rate of the carboxylation enzyme Rubisco is a major barrier to increasing the rate of carbon assimilation at the leaf level. Although current Rubisco properties may be an insurmountable barrier to increasing photosynthetic rate to make full use of direct midday sunlight at the level of an individual leaf, limitations due to Rubisco might be substantially mitigated by improved sharing of light within a crop canopy (10), by reengineering Rubisco (11), or by actively concentrating CO2 at the active site of Rubisco (ref. 12 and as discussed in Targets of Opportunity).
The remarkable gains in productivity of the Green Revolution of the late 20th century depended on improving yield potential: i.e., the yield obtained with good nutrition in the absence of pests, diseases, and drought. To a first approximation, yield potential is dependent on the amount of solar energy available during the growing season and the efficiencies with which the crop captures the photosynthetically active light, converts it to biomass, and partitions the biomass into the harvested product (13). During the Green Revolution, increases in yield potential were driven primarily by large increases in the portion of biomass partitioned into grain (i.e., harvest index), which is now near its theoretical upper limit. Improved solar energy conversion efficiency (i.e., photosynthetic efficiency) has so far played little role in improving yield potential, yet photosynthesis is the only determinant that is not close to its biological limits (14, 15). For example, the progenitors of modern crop plants evolved in, and are thus adapted to, an atmospheric [CO2] of about 240 ppm. The accelerated rate of Rubisco-catalyzed carboxylation at today’s [CO2] of >400 ppm has led to a kinetic limitation in the regeneration of the CO2 acceptor molecule ribulose-1,5-bisphosphate (RuBP), which will become increasingly limiting as [CO2] increases further (16, 17). Fortunately, there are a wide range of potential avenues to improve photosynthetic efficiency along radically different paths than those dictated by evolutionary selection on extant and emergent natural variation.
Research over many decades has produced a sophisticated continuum of understanding of photosynthesis from the initial photophysics of light absorption and excitation energy transfer to gas exchange in leaf and canopy. Recent advances in in silico modeling and increases in computational power have been critical enabling technologies for understanding photosynthetic processes, which can be used to predict the outcomes of various redesigns of photosynthesis (e.g., refs. 10, 14, and 16). The remarkable achievements in synthesizing whole genomes (18) are evidence that ambitious goals are within reach. Creative and radically new ideas for redesigning photosynthesis are therefore worth pursuing because even strategies that presently seem fanciful may inspire new thinking in unimagined directions.
Targets of Opportunity
Light Capture.
A principal limitation of efficient photosynthesis is that organisms absorb more light in full sunlight than they can use productively. The reason seems clear: high absorptivity provides effective capture at low light intensities, such as at dawn and dusk and on cloudy days, and it obviates competition from other phototrophs by absorbing the light before they do. It is generally agreed that the season-long solar energy conversion efficiencies for crop plants in both temperate and tropical zones typically do not exceed a few percent, which should be compared with the theoretical maximum yield of about 12% (15, 19). A large part of this inefficiency can be ascribed to the absorption of more photons than can be used to drive useful photochemistry. This excess energy needs to be carefully extinguished to avoid deleterious photooxidations. Plants have a variety of mechanisms for safely diverting excess absorbed energy, principally as thermal dissipation (9), but they represent a significant loss in efficiency. If plants had fewer light-harvesting pigments (e.g., chlorophylls and carotenoids) per photosystem (10) and fewer photosystems in their uppermost leaves, light might be absorbed more judiciously, and a greater proportion of absorbed photons converted to biomass. This avenue has been pursued for some years in algae and cyanobacteria, often with notable success (20, 21), but not yet convincingly in crop plants. Lowering leaf absorptivity remains a significant opportunity for improving crop yield (10) although it is worth noting that such highly efficient plants might be at a disadvantage under some conditions, compared with normally pigmented competitors, and could require careful plant husbandry to realize their increased yield.
Light Energy Conversion.
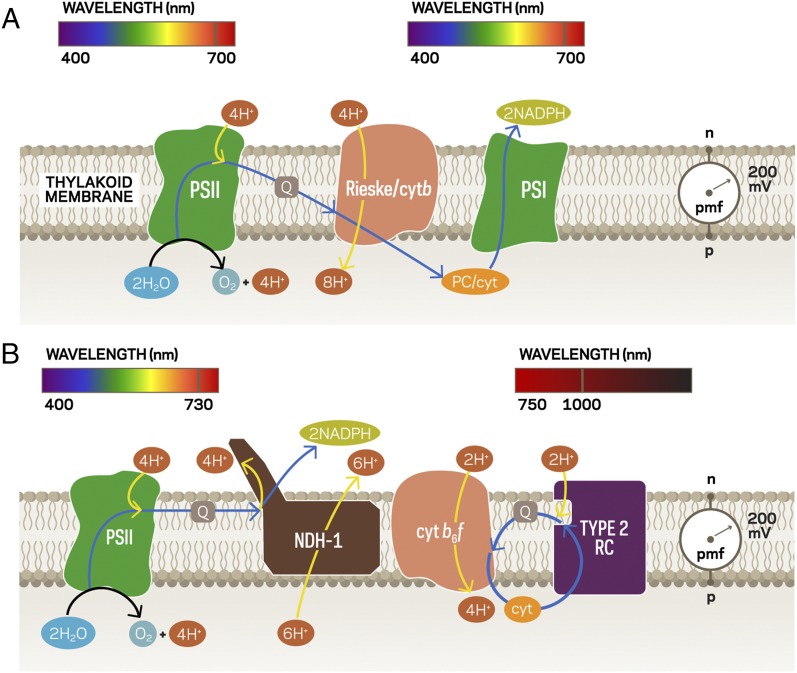
The fundamental thermodynamic principles controlling the conversion of energy from sunlight to chemical or electrical potential energy have been defined and demonstrated in the last 60 y. For reasons having more to do with biological success than thermodynamic efficiency, water-oxidizing (oxygenic) photosynthesis uses only about one half of the sun’s energetically competent photons (<700 nm) and uses two photosystems operating in series (Fig. 1A) to span the redox space from water oxidation to NADP+ reduction and to pump protons to provide proton motive force (PMF). The two photosystems compete with each other for photons rather than one of them taking advantage of the near-IR region of the solar spectrum as do many photosynthetic bacteria (19).
Fig. 1.
Schematics of tandem-photosystem, energy-coupling membranes for water-oxidizing photosynthesis and synthesis of NAD(P)H. The region of the solar spectrum driving each reaction center (RC) is indicated above it. Black arrows indicate reactions, blue arrows represent electron flow, and yellow arrows represent proton flow. (A) In current oxygenic photosynthesis, photosystem II (PSII) uses visible light (400–700 nm) to drive water oxidation and quinone reduction whereas photosystem I (PSI) oxidizes plastocyanin (PC, or in some cases cytc) and reduces NADP+ (via ferredoxin:NAD(P)+ oxidoreductase). The Rieske/cytb complex connects the two photosystems by catalyzing quinol oxidation and PC reduction in a Q-cycle mechanism, resulting in additional PMF generation (represented by the yellow arrow through the complex). PMF is consumed by ATP synthase to drive phosphorylation of ADP (not shown). Cyclic electron flow involving PSI and the cyt b6f complex is not shown. (B) The PSII RC from A. marina (using light from 400 to 730 nm) delivers electrons from water oxidation for quinone reduction whereas the BChl b-containing type 2 RC (operating out to 1,075 nm) and the cyt b6f complex collaborate to maintain PMF via a cyclic electron transfer loop. The NDH-1 complex consumes some proton-motive force (PMF) to drive electron transfer from quinol to NAD(P)+ whereas the rest is used by ATP synthase (not shown).
Could a more efficient arrangement be engineered? One option would be to replace photosystem I (PSI) (Fig. 1A) with a reaction center and its associated cyclic electron transport machinery from a purple photosynthetic bacterium that uses bacteriochlorophyll b in place of the chlorophyll a used by oxygenic organisms. Blastochloris viridis is one example, with its antenna system and reaction center acting in concert with the cytochrome (cyt) b6f complex that would use light up to 1,075 nm (19) to provide PMF. Further, if the chlorophyll a in photosystem II (PSII) were replaced by chlorophyll d (as occurs in the cyanobacterium Acaryochloris marina, which uses light up to 730 nm) and if the reduced quinone produced by this photosystem delivered electrons directly to a NADH dehydrogenase operating “backwards” as it does in purple bacteria (using PMF to drive the production of NADPH from reduced quinone), one can envision the system of Fig. 1B. A photosynthetic membrane having this arrangement of energy-transducing complexes would have more than twice the energetic efficiency of current oxygenic photosynthesis (19, 22) if the appropriate wavelengths of light were directed to the two now very distinct photosystems. Indeed, as our understanding of quantum coherence develops (22–24), it can be imagined that specific antenna could be “wired” to appropriate reaction centers. Of course other possibilities can be envisioned, but even before the powerful potential of synthetic biology is brought into play, we can initiate the research required to reengineer photosynthesis because, as illustrated in Fig. 1B, nature has provided us with energy-transducing complexes that, in thought experiments, could be combined in new tandem configurations to achieve a near-optimum match of the solar spectrum to water oxidation and carbohydrate/lipid synthesis.
Carbon Capture and Conversion.
Improving carbon uptake.
A major limiting factor in carbon fixation is CO2 transport from the intercellular airspace of leaves to the site of carboxylation within the mesophyll chloroplast, entailing movement across several compartments (25, 26). Although carbonic anhydrases are expected to play a critical role, their function in CO2 conductance and delivery to Rubisco has yet to be clearly defined. Algae and photosynthetic bacteria have various CO2 channels and bicarbonate transporters to increase the inorganic carbon flux into the cells, but these inorganic transport devices are not present in the chloroplast membranes of terrestrial plants. Introduction of CO2 channels and bicarbonate transporters into photosynthetic cells of C3 plants and even into the mesophyll cells of C4 plants is a logical strategy to improve photosynthetic performance (12, 27). Achieving a higher CO2 concentration in the chloroplast would allow the endogenous Rubisco to be replaced by a Rubisco with a lower CO2 affinity, but a higher kcat (16), thus reducing the amount of Rubisco required by the plant and improving nitrogen use efficiency. The lower intercellular CO2 concentration that would result from higher photosynthesis would likely speed its diffusive replacement from the outside air, allowing stomata to be open less, with the added benefit of improving water use efficiency.
Improving carbon conversion.
A major productivity limitation for many phototrophs is photorespiration, an energetically expensive process in which the undesirable oxygenase activity of Rubisco leads to a net loss of fixed carbon and metabolic energy. Nature has evolved several strategies to suppress oxygenation by sequestering Rubisco into compartments in which CO2 is concentrated. C4 plants [for example, tropical grasses such as sugarcane (Saccharum officinarum) and corn (Zea mays)] use oxygen-insensitive phosphoenolpyruvate carboxylase to initially capture CO2 as malate in mesophyll cells. The malate is subsequently decarboxylated in bundle sheath cells that contain Rubisco. Although this concentrating of CO2 requires an additional investment of ATP, there is an adequate return on this investment in the form of more biomass. A coordinated international effort to introduce this ability into rice has already produced exciting results (28). Although significant hurdles remain, success will open the door to dramatically improving the efficiency and yield of many of the world’s most important staple crops. We note that, although classical C4 plants exhibit substantial spatial separation of the carbon concentrating and carbon fixation pathways within the leaf, there are plants that do this C4 metabolism (albeit somewhat less efficiently) (29) within a single cell (30), which has structurally simpler requirements and thus may be easier to install in C3 leaves.
An alternative approach would copy cyanobacterial CO2-concentrating mechanisms, which are characterized by bicarbonate transporters and the occurrence of Rubisco-containing microcompartments called carboxysomes. Although plant chloroplasts clearly originated from a cyanobacterial progenitor through endosymbiosis, none contain these devices, making their introduction an attractive target for improving plant photosynthetic efficiency. An important advance toward this goal was the introduction of a carboxysome protein and functional cyanobacterial Rubisco into a land plant that led to the aggregation of Rubisco within chloroplasts as occurs during carboxysome biogenesis (31).
Attempts to reengineer Rubisco with increased activity and substrate specificity have failed, despite being guided by well-resolved crystal structures and detailed understanding of the reaction mechanism (11, 32, 33). With seemingly little prospect for improving the kinetic and catalytic features of native Rubiscos, it may be worthwhile to try to “evolve” a new carboxylase with different kinetic constraints from a different (perhaps smaller) protein scaffold. A smaller carboxylase would have the additional benefit of reducing the nitrogen requirement of the plant, a goal that might also be achieved by editing the existing Rubisco to remove high-nitrogen content amino acids (such as arginine) (34).
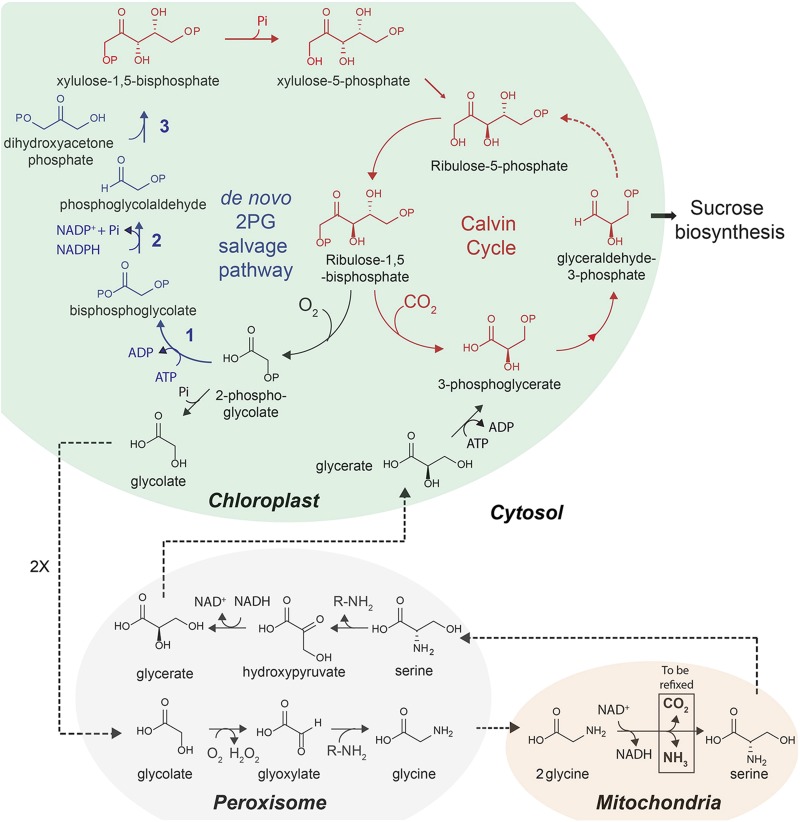
If Rubisco’s oxygenation reaction cannot be significantly reduced, an alternative approach would replace the native photorespiratory pathway with more efficient synthetic pathways. Three different pathways that metabolize glycolate have been explored so far (35, 36). Among them, the conversion of glycolate into glycerate via glyoxylate and tartronic semialdehyde can increase leaf photosynthetic CO2 uptake due to both the decreased energy demand of photorespiration and increased CO2 concentration in chloroplasts (36). Another promising experimental strategy is to introduce new anaplerotic pathways to recycle glycolate without an associated CO2 loss, such as the 3-hydroxypropionate pathway that converts glyoxylate to pyruvate in some bacteria (37, 38). It might also be possible to design entirely novel pathways, including the hypothetical one shown in Fig. 2, to reduce phosphoglycolate to phosphoglycolaldehyde and then combine that product with dihydroxyacetonephosphate to make xylulose bisphosphate, which could be dephosphorylated to form the carbon reduction cycle intermediate xylulose-5-phosphate.
Fig. 2.
Proposed de novo 2-phosphoglycolate salvage pathway. Calvin cycle in red, photorespiratory pathway in black, de novo 2-phosphoglycolate (2PG) salvage pathway in blue fonts, respectively. In the C3 cycle (not all steps shown), Rubisco fixes CO2 to ribulose-1,5-bisphosphate to generate two molecules of 3-phosphoglycerate. Approximately 25% of the time, Rubisco oxygenates ribulose-1,5-bisphosphate, generating 2PG and 3-phosphoglycerate. The photorespiratory pathway consumes ATP to regenerate 3-phosphoglycerate from two molecules of 2PG. Photorespiration is metabolically inefficient, requiring eight enzymatic steps (here the amino donor/acceptor is designated R), taking place across four different subcellular compartments, and resulting in an overall loss of carbon dioxide and ammonia that needs to be refixed at the cost of substantial additional energy (3 ATP and 2 NADPH per CO2, and 1 ATP and 1 NADPH per NH3). The proposed de novo 2PG salvage pathway would efficiently convert 2PG to xylulose-1,5-bisphosphate in three steps in a single compartment using only one ATP and one NADPH, without loss of fixed carbon. The three enzymes needed in the proposed de novo 2PG salvage pathway are as follows: kinase (step 1), reductase (step 2), and aldolase (step 3). Xylulose-1,5-bisphosphate is a stromal metabolite that is converted to the C3 cycle intermediate xylulose-5-phosphate by the cbbY protein (63). The capacity of the cbbY may not be sufficient to sustain the flux and may need to be up-regulated.
More ambitious would be to import oxygen-insensitive pathways for the key carbon fixation reaction to bypass Rubisco altogether. One could be the 3-hydroxypropionate/4-hydroxybutyrate cycle of aerobic Archaea (39) and another the 4-hydroxybutyrate/dicarboxylate pathway of anaerobic Archaea [although this later pathway has at least one O2-sensitive enzyme (40), and directed evolution may be required to increase the O2 tolerance of selected enzymes]. The hypothetical C4/glyoxylate cycle (41) may be easier to implement because it requires only four enzymes, although a pathway to convert glyoxylate, the product of this cycle, into a metabolically useful intermediate would also be required. In fact, the second cycle of the recently demonstrated 3-hydroxypropionate bicycle produces a metabolically useful product by converting glycolate to pyruvate (38). Another exciting approach would be to design a completely orthogonal pathway involving intermediates that are not shared by any existing pathway in the organism. By avoiding the intermediates of native metabolism until the final step, this strategy should simplify regulatory networks and minimize metabolic interference with native pathways.
Initial work on new carbon-fixation pathways may be best performed with algae and cyanobacteria because they grow faster than higher plants, they are easier to manipulate, and their homogeneous cell cultures simplify metabolite and other analyses. Some potential carbon-fixation pathways produce acetyl-CoA, which could also be used for lipid (biofuel) production. If algae are to host new pathways, there is a choice between introducing enzyme-genes into the chloroplast genome for higher expression and better control of gene placement via homologous recombination, or into the nuclear genome where newly emerging gene-editing tools may be poised for breakthrough impact (more on gene editing in Enabling Technologies). Inserted enzymes might be engineered via directed evolution or global and targeted mutagenesis, but high-throughput enzyme assays will be essential to screen or select for desired catalysis outputs (42, 43). Systems biology, metabolic flux, and control analysis will identify bottlenecks in pathways, and expression of appropriate enzymes will be increased through an iterative process (44, 45).
Metabolic modeling will be critical for identifying bottlenecks and unanticipated diversion of metabolites, as well as for suggesting possible solutions. If new intermediates are toxic, consecutive enzymes might be tethered in a scaffold to encourage channeling and minimize release of problematic intermediates. If unusual cofactors are in high demand (e.g., bicarbonate-dependent carboxylases use biotin), then appropriate cofactor synthesis would need to be up-regulated or even introduced to avoid cofactor limitation.
Smart Canopy.
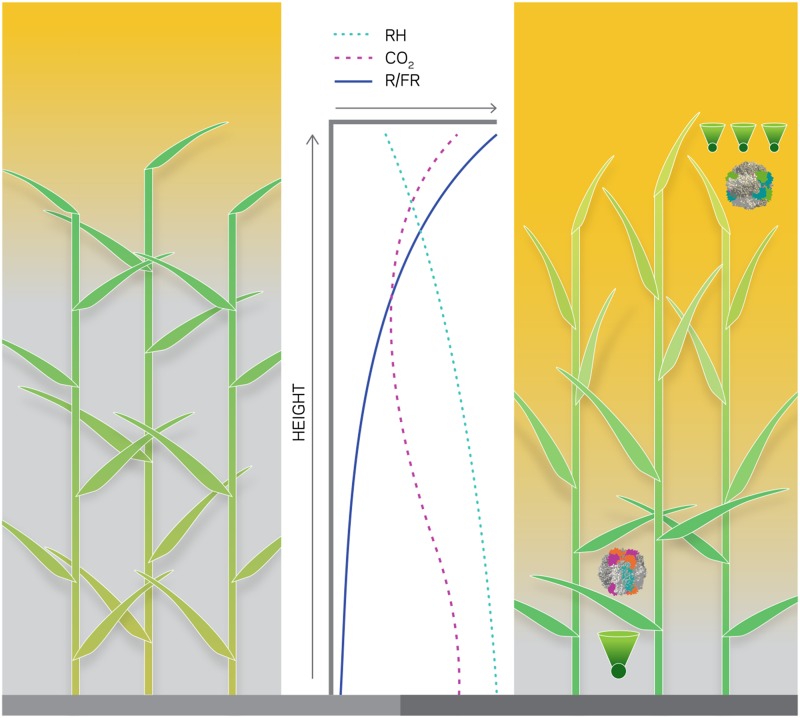
The smart canopy concept envisions an assemblage of plants that interact cooperatively (rather than competitively) at the canopy level to maximize the potential for light harvesting and biomass production per unit land area. Although selection by plant breeders for traits such as yield in a monoculture may act at the population level, natural selection acts at the level of the individual plant to maximize the genetic fitness of the individual rather than the fitness of the canopy as a whole. Therefore, engineering a smart canopy will require a detailed understanding of opportunities for cooperativity among crop plants. How could leaves in a “smart” canopy be engineered to behave optimally for productivity, as opposed to success of the individual? Light flux and spectrum, wind speed, and humidity vary with depth into a canopy but also vary minute by minute, depending on cloud movement, time of day, and wind gusts. One variable within canopies that plants are able to sense is the ratio of red to far-red radiation (R/FR) (Fig. 3). Leaves absorb strongly in the red but transmit most far-red radiation, and this absorptivity ratio is scarcely affected by variation in the amount of incoming sunlight. The phytochrome system senses this ratio (46) so that promoters downstream of phytochrome signaling could be used to make smart canopies wherein leaves are adapted to the prevailing light conditions to benefit total crop production.
Fig. 3.
The heterogeneity of microenvironments inside a canopy. Light and wind speed decline toward the base of a crop canopy, generating gradients in relative humidity (RH), CO2, and the red to far-red ratio (R/FR). In a typical canopy (Left), most light is absorbed by the upper leaves, which are dark green and often tend toward being horizontal. An optimized canopy (Right) would have lighter green upright leaves at the top of the canopy and dark green horizontal leaves at the base. A hypothesized smart canopy would also be more efficient if the Rubisco in the upper canopy (protein diagram) had a high catalytic rate that is replaced by a Rubisco with a high specificity in the lower canopy. The canopy would also be more efficient with fewer antenna pigments per photosystem (cones) in the upper canopy, replaced by large antennas serving fewer reaction centers in the lower canopy.
What changes are desirable in a smart canopy as leaves become increasingly shaded? Metabolic and biophysical modeling suggests significant benefits from three distinct changes (Fig. 3). First, transitioning from vertical leaves in high light in the upper canopy to horizontal leaves in low light deeper in the canopy permits a more even distribution of sunlight and minimizes saturation of the upper leaves and light starvation of the lower leaves (47). Recent insights into the molecular basis of hinging in grass leaf blades at the ligule and auricles provide a means to produce controlled variation in leaf angles (48). Secondly, the amount of Rubisco is a major limitation to light-saturated photosynthesis, despite being as much as 50% of leaf soluble protein (16). Canopy photosynthesis would be increased by deploying a Rubisco with a high catalytic rate in the upper leaves, even at the expense of specificity for CO2 over O2, and replacing Rubisco with a high specificity form in the lower canopy where light is limiting to minimize the diversion of the limiting light energy into photorespiratory metabolism (16). Thirdly, upper canopy leaves are typically light-saturated, and reaction centers may be limiting. In a smart canopy, the leaves receiving the most light would form small antenna systems feeding many reaction centers. Reaction centers are expensive to maintain and not limiting in low light. Therefore, reaction center numbers should be decreased in low light but antenna sizes increased and pigment composition changed to maximize interception of light, which would be enriched in the green (46).
The ideal architectural and metabolic features of such a smart canopy need to be explored for different crops using systems biology (49). Although optimal properties such as height, leaf area index, leaf angle, albedo, and orientation will differ among crops or even within a single crop in different environments, a number of common features can be proposed for individuals in a smart canopy. First, although semidwarf phenotypes were a major contributing factor in the success of the Green Revolution, new molecular approaches to enhance lodging resistance should allow increased canopy height as a promising option to increase crop biomass and thus grain yields on a per area basis if the harvest index can be held constant. Furthermore, repositioning floral organs and panicles inside the canopy would avoid shading photosynthetically active leaves; indeed, this strategy is used by many elite hybrid rice lines. Decreasing leaf chlorophyll content in sun-exposed leaves is another promising approach to increase total canopy photosynthetic rate (10). This increase would be achieved both through increasing quantum yield of PSII via decreasing photoprotective energy loss and improving light energy distribution inside the canopy. This strategy could be combined with the optimization of light harvesting in the canopy by using different pigments to extend the absorption properties to the infrared spectral range and by exploiting the capacity of the proteins to tune the absorption properties of their bound pigments (Fig. 1B). Chl d, which is red-shifted compared with Chl a, could be produced in the lower leaves of the canopy, which mainly receives IR light, thus increasing the light-harvesting capacity (50). A similar effect might be obtained by mutating the protein sequence of the natural antenna complexes to influence protein–protein and protein–pigment interactions to shift the absorption of some of the Chls to the red by up to 20 nm. This second option could eliminate possible problems due to assembly and regulation with nonnative pigments because changing the absorption spectrum of native pigments requires mutations only of existing components of the photosynthetic apparatus (51).
Given the difference in the light and CO2 environments inside a canopy, it would conceptually be beneficial to use different photosynthetic machineries for leaves at the top of the canopy versus lower layers. Considering that C4 assimilation or other carbon-concentrating mechanisms require extra ATP for assimilating CO2, it is preferable to operate such carbon-concentrating mechanisms only in the top canopy layers whereas the light-limited lower region would use only the C3 pathway. Such partitioning of C3 and C4 metabolism requires the engineering of a switchable system where a leaf in a nascent canopy initially operates a light-driven CO2-concentrating mechanism and later conducts C3 photosynthesis after it is shaded during canopy development.
Enabling Technologies
Due to limited natural genetic variation in the enzymes and processes of plant photosynthesis, most of the limitations in photosynthetic efficiency will not likely be decisively overcome by conventional breeding approaches. Instead, key improvements will need to be pursued through genetic engineering and synthetic biology guided by well-validated mechanistic models. The implementation of concepts for radical redesign of the photosynthetic apparatus and/or its regulation (such as those discussed above in Targets of Opportunity) will especially call for the introduction of dozens of transgenes—possibly on synthetic chromosomes—and require genetic engineering at an unprecedented scale, as well as public discussion of the costs and benefits of such organisms (52). Genetic engineering of this large scale poses a number of significant technical challenges (Table 1).
Table 1.
Tools and technologies relevant to engineering photosynthesis, their potential applications, and current limitations
| Technology/tools | Applications | Limitations |
| Bacterial transformation | Engineering photosynthesis in cyanobacteria | No serious technical limitations in the most important cyanobacterial model species. |
| Nuclear transformation | Engineering of nucleus-encoded components of the photosynthetic apparatus; expression of novel genes and pathways. Development of synthetic chromosomes. | Lack effective strategies to avoid transgene silencing; lack effective strategies for high transgene expression levels (often significantly lower than with plastid transformation); lack sufficient characterized promoters, terminators, and chloroplast transport signals; lack mature technologies to stably transform very large DNA segments (>10 genes) into plants; lack efficient tools for site-directed engineering; lack sufficient information on centromere and telomere sequences. |
| Plastid transformation | Engineering of plastid-encoded components of the photosynthetic apparatus; expression of novel genes and pathways of carbon metabolism. | Small number of transformable species, which do not include cereals or other major crops; lack of robust and flexible regulatory strategies. |
| Mitochondrial transformation | Engineering of mitochondrially encoded components of the respiratory chain to minimize respiratory losses; expression of novel pathways of carbon metabolism. | Not yet possible in any seed plant; currently possible only in the algal species Chlamydomonas reinhardtii. |
| Multigene engineering | Engineering of protein complexes in the electron transfer chain; engineering of carbon fixation pathways. | No serious limitations; can be performed via synthetic operons in cyanobacteria and plastids and/or by combinatorial transformation in the nucleus, but mature technologies to introduce very large fragments (>10 genes) into plants are needed. |
| Protein design | Redesign of the electron transfer chain; Rubisco engineering; redesign of carbon-fixing enzymes. | Computer-based structural modeling not yet sufficiently developed; limited success with rational design and rational optimization of proteins; limited understanding of protein dynamics. |
| Synthetic genomics | Radical redesign of the photosynthetic apparatus via synthetic plastid genomes and/or artificial (mini)chromosomes in the nucleus. | Incomplete parts list of the photosynthetic apparatus, its assembly factors and regulators; incomplete knowledge about dynamic (quantitative) changes in response to environmental cues; limitations in synthesis and efficiency of assembly of very large DNA molecules (>1 Mbp) and in stability of minichromosomes. |
| Design of logic circuits; development of sensors for light intensity, light quality, temperature, and CO2 concentration | Smart canopy concept. | Incomplete parts list: insufficient understanding of the structure and organization of the genetic and metabolic networks underlying photosynthesis and its regulation; limited knowledge about pool sizes of metabolites in different cellular compartments and subcompartments; incomplete knowledge of transporters. |
| Phenotyping in the field | Evaluation of design concepts under field conditions and further optimization through mutagenesis. | Insufficient sensitivity of current imaging methods; lack of sensitive spectrometric methods for large-scale and continuous measurement of the dynamics of photosynthetic parameters, dynamics of radiation quality and quantity within the canopy, and growth of stands of many genotypes/transformants in the field. |
Thirty years of transgenic research have assembled an impressively large toolbox for genetic engineering of cyanobacteria, eukaryotic algae, and land plants. Recently developed plant biotechnology and synthetic biology tools have included synthetic promoters, artificial chromosomes, and large DNA fragment assembly (of entire bacterial genomes and eukaryotic chromosomes) (18, 53). Improved transformation methods are poised for the redesign of at least some aspects of photosynthesis for rapid progress. However, there are still critical bottlenecks and unsolved problems (Table 1). In photosynthetic eukaryotes, many of the key components of the electron transport chain (e.g., the reaction center subunits of the photosystems), as well as the catalytic large subunit of Rubisco, are encoded in the plastid genome. Thus, many strategies toward redesigning photosynthesis would be substantially enabled by technologies allowing the plastid genome to be engineered in a greater number of species. In addition, plastid genome transformation offers a number of further benefits, including (i) the capacity for high-level transgene expression, (ii) the convenient stacking of multiple transgenes in synthetic operons, (iii) the high precision of plastid engineering due to the presence of an efficient homologous recombination system, and (iv) the greatly increased transgene containment conferred by the maternal mode of plastid inheritance in most crops, which largely excludes plastid (trans)genes from pollen transmission.
Unfortunately, efficient plastid transformation technology is presently available in very few species. Despite significant efforts, the development of plastid transformation protocols for cereals, the world’s most important staple crops, has remained unsuccessful (54, 55). Major technical challenges include the development of suitable selectable markers and efficient tissue culture and selection systems. In addition, the sometimes very high expression levels of plastid transgenes would almost certainly need to be managed to achieve optimal outcomes. Expanded research efforts will be needed to further optimize plastid transformation technology by, for example, increasing the number of transgenes that can be effectively expressed and by ensuring proper folding and posttranslational modification of proteins expressed in the plastid. Overcoming these challenges will require major investments in long-term research programs, which are presently not being made, at least not in the public sector.
In the meantime, it will be appropriate to test radical redesign concepts in established model systems, such as cyanobacteria and the green alga Chlamydomonas reinhardtii. It will also be essential to develop an intermediate land plant model as a chassis to implement strategies for improving photosynthesis and to optimize the reengineered systems for transfer to staple crops once the transgenic technologies are sufficiently developed. A possible model could be a diploid tobacco species (e.g., Nicotiana sylvestris), whose chloroplast and nuclear genomes are readily transformable. An intermediate land plant model would provide the ability to test the efficacy of different photosynthetic manipulations in a closed canopy paralleling that of major crops.
Important new technologies (18) promise to greatly improve efficiency and feasibility of nuclear transformation (Table 1). Novel technologies, such as transcription activator-like effector nucleases (TALENs) (56, 57) and the RNA-guided Cas9 system (58, 59), are enabling the precise engineering of genomes, including gene replacement and editing of genomic sequences within their authentic regulatory contexts. However, to take full advantage of these emerging tools, our knowledge about the factors and regulatory processes involved in the assembly and functioning of the photosynthetic apparatus must keep pace. For example, the catalog of protein factors that participate in the assembly, stability, and degradation of thylakoid protein complexes is incomplete, and many key players remain to be discovered. Likewise, there are significant gaps in our knowledge of the proteins and small molecules that regulate electron transfer processes and the biochemical pathways of carbon metabolism. Developments in mass spectrometry to improve scale and sensitivity for quantitative and targeted proteomics are needed to improve our knowledge of posttranslational modifications and protein degradation (60). Although advanced methods in quantitative biology, systems biology, and new modeling approaches (61, 62) will certainly improve our understanding of the architecture of the genetic and biochemical networks underlying photosynthesis and will also aid the design of novel regulatory circuits, the predictive power of these methods will remain limited by the still rather large number of unknown variables in the networks (Table 1). The mitigation of these limitations will require significant investments in basic research on primary metabolism, gene expression, and their regulation at all levels.
Outlook
Stimulated by the exciting new opportunities of synthetic biology, we have considered an array of redesigns to improve photosynthetic efficiency and performance, with a primary focus on improving the productivity of food and bioenergy plants. Our intent was neither to produce an exhaustive list of all current ideas nor to focus exclusively on a new generation of strategies. Rather, we sought to broadly consider several potential routes to improving photosynthesis. We confined our focus to the processes in chloroplasts although we are aware that important downstream processes, such as the export of photosynthate from cells and leaves, can exert strong metabolic and genetic feedback on this organelle. We have indicated where there is already proof-of-concept evidence for improved photosynthetic efficiency, as well as notions for complete redesigns that are only conceptual, but which are consistent with what we currently understand. An important conclusion is that the implementation of many key ideas is limited by our ability to introduce, position, and regulate inserted genes. Finally, we emphasize that a central challenge to improving photosynthetic efficiency is knowing how alterations made to the photosynthetic process at the level of the chloroplast will scale to a whole canopy, whose complex seasonal development ultimately determines biomass production and yield. Thus, an overarching continuum of models that span from cellular metabolism to the agricultural field will be essential for successfully redesigning photosynthesis to sustainably meet global food and bioenergy demand.
Acknowledgments
We thank Haley Ahlers, who works on the Realizing Improved Photosynthetic Efficiency (RIPE) project funded by the Bill and Melinda Gates Foundation at the University of Illinois, for assistance with graphics. This paper was conceived at the workshop “Redesigning Photosynthesis–Identifying Opportunities and Novel Ideas” held at the Banbury Center, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, May 13–16, 2013. We thank the Cold Spring Harbor Laboratory Banbury Center for hosting and the Cold Spring Harbor Laboratory Corporate Sponsor Program for funding the workshop.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Tilman D, Balzer C, Hill J, Befort BL. Global food demand and the sustainable intensification of agriculture. Proc Natl Acad Sci USA. 2011;108(50):20260–20264. doi: 10.1073/pnas.1116437108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexandratos N, Bruinsma J. World Agriculture Towards 2030/2050: The 2012 Revision No. 12-03. Food and Agricultural Organization; Rome: 2012. [Google Scholar]
- 3.Sayer J, et al. Ten principles for a landscape approach to reconciling agriculture, conservation, and other competing land uses. Proc Natl Acad Sci USA. 2013;110(21):8349–8356. doi: 10.1073/pnas.1210595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burney JA, Davis SJ, Lobell DB. Greenhouse gas mitigation by agricultural intensification. Proc Natl Acad Sci USA. 2010;107(26):12052–12057. doi: 10.1073/pnas.0914216107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long SP, Ort DR. More than taking the heat: Crops and global change. Curr Opin Plant Biol. 2010;13(3):241–248. doi: 10.1016/j.pbi.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Ray DK, Ramankutty N, Mueller ND, West PC, Foley JA. Recent patterns of crop yield growth and stagnation. Nat Commun. 2012;3:1293. doi: 10.1038/ncomms2296. [DOI] [PubMed] [Google Scholar]
- 7.Connor DJ, Loomis RS, Cassman KG. Crop Ecology: Productivity and Management in Agricultural Systems. Cambridge Univ Press; Cambridge, UK: 2011. [Google Scholar]
- 8.Lobell DB, et al. Greater sensitivity to drought accompanies maize yield increase in the U.S. Midwest. Science. 2014;344(6183):516–519. doi: 10.1126/science.1251423. [DOI] [PubMed] [Google Scholar]
- 9.Murchie EH, Niyogi KK. Manipulation of photoprotection to improve plant photosynthesis. Plant Physiol. 2011;155(1):86–92. doi: 10.1104/pp.110.168831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ort DR, Zhu XG, Melis A. Optimizing antenna size to maximize photosynthetic efficiency. Plant Physiol. 2011;155(1):79–85. doi: 10.1104/pp.110.165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parry MAJ, Madgwick PJ, Carvahlo JFC, Andralojc PJ. Prospects for increasing photosynthesis by overcoming the limitations of Rubisco. J Agric Sci. 2007;145:31–43. [Google Scholar]
- 12.Price GD, et al. The cyanobacterial CCM as a source of genes for improving photosynthetic CO2 fixation in crop species. J Exp Bot. 2013;64(3):753–768. doi: 10.1093/jxb/ers257. [DOI] [PubMed] [Google Scholar]
- 13.Monteith JL. Climate and the efficiency of crop production in Britain. Philos T Roy Soc B. 1977;281:277–294. [Google Scholar]
- 14.Long SP, Zhu X-G, Naidu SL, Ort DR. Can improvement in photosynthesis increase crop yields? Plant Cell Environ. 2006;29(3):315–330. doi: 10.1111/j.1365-3040.2005.01493.x. [DOI] [PubMed] [Google Scholar]
- 15.Zhu X-G, Long SP, Ort DR. What is the maximum efficiency with which photosynthesis can convert solar energy into biomass? Curr Opin Biotechnol. 2008;19(2):153–159. doi: 10.1016/j.copbio.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 16.Zhu X-G, Portis AR, Long SP. Would transformation of C3 crop plants with foreign Rubisco increase productivity? A computational analysis extrapolating from kinetic properties to canopy photosynthesis. Plant Cell Environ. 2004;27:155–165. [Google Scholar]
- 17.Lefebvre S, et al. Increased sedoheptulose-1,7-bisphosphatase activity in transgenic tobacco plants stimulates photosynthesis and growth from an early stage in development. Plant Physiol. 2005;138(1):451–460. doi: 10.1104/pp.104.055046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibson DG, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329(5987):52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 19.Blankenship RE, et al. Comparing photosynthetic and photovoltaic efficiencies and recognizing the potential for improvement. Science. 2011;332(6031):805–809. doi: 10.1126/science.1200165. [DOI] [PubMed] [Google Scholar]
- 20.Nakajima Y, Itayama T. Analysis of photosynthetic productivity of microalgal mass cultures. J Appl Phycol. 2003;15:497–505. [Google Scholar]
- 21.Kirst H, Formighieri C, Melis A. Maximizing photosynthetic efficiency and culture productivity in cyanobacteria upon minimizing the phycobilisome light-harvesting antenna size. Biochim Biophys Acta. 2014;1837(10):1653–1664. doi: 10.1016/j.bbabio.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 22.Hanna MC, Nozik AJ. Solar conversion efficiency of photovoltaic and photoelectrolysis cells with carrier multiplication absorbers. J Appl Phys. 2006;100:074510. [Google Scholar]
- 23.Scholes GD, Fleming GR, Olaya-Castro A, van Grondelle R. Lessons from nature about solar light harvesting. Nat Chem. 2011;3(10):763–774. doi: 10.1038/nchem.1145. [DOI] [PubMed] [Google Scholar]
- 24.Fassioli F, Dinshaw R, Arpin PC, Scholes GD. 2014. Photosynthetic light harvesting: Excitons and coherence. J R Soc Interface 11:2013.0901. [DOI] [PMC free article] [PubMed]
- 25.Evans JR, Kaldenhoff R, Genty B, Terashima I. Resistances along the CO2 diffusion pathway inside leaves. J Exp Bot. 2009;60(8):2235–2248. doi: 10.1093/jxb/erp117. [DOI] [PubMed] [Google Scholar]
- 26.von Caemmerer S, Evans JR. Enhancing C3 photosynthesis. Plant Physiol. 2010;154(2):589–592. doi: 10.1104/pp.110.160952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaldenhoff R. Mechanisms underlying CO2 diffusion in leaves. Curr Opin Plant Biol. 2012;15(3):276–281. doi: 10.1016/j.pbi.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 28.von Caemmerer S, Quick WP, Furbank RT. The development of C4rice: Current progress and future challenges. Science. 2012;336(6089):1671–1672. doi: 10.1126/science.1220177. [DOI] [PubMed] [Google Scholar]
- 29.von Caemmerer S. C4 photosynthesis in a single C3 cell is theoretically inefficient but may ameliorate internal CO2 diffusion limitations of C3 leaves. Plant Cell Environ. 2003;26:1191–1197. [Google Scholar]
- 30.Sharpe RM, Offermann S. One decade after the discovery of single-cell C4 species in terrestrial plants: What did we learn about the minimal requirements of C4 photosynthesis? Photosynth Res. 2014;119(1-2):169–180. doi: 10.1007/s11120-013-9810-9. [DOI] [PubMed] [Google Scholar]
- 31.Lin MT, Occhialini A, Andralojc PJ, Parry MAJ, Hanson MR. A faster Rubisco with potential to increase photosynthesis in crops. Nature. 2014;513(7519):547–550. doi: 10.1038/nature13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tcherkez GGB, Farquhar GD, Andrews TJ. Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc Natl Acad Sci USA. 2006;103(19):7246–7251. doi: 10.1073/pnas.0600605103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Savir Y, Noor E, Milo R, Tlusty T. Cross-species analysis traces adaptation of Rubisco toward optimality in a low-dimensional landscape. Proc Natl Acad Sci USA. 2010;107(8):3475–3480. doi: 10.1073/pnas.0911663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmollinger S, et al. Nitrogen-sparing mechanisms in Chlamydomonas affect the transcriptome, the proteome, and photosynthetic metabolism. Plant Cell. 2014;26(4):1410–1435. doi: 10.1105/tpc.113.122523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peterhansel C, Blume C, Offermann S. Photorespiratory bypasses: How can they work? J Exp Bot. 2013;64(3):709–715. doi: 10.1093/jxb/ers247. [DOI] [PubMed] [Google Scholar]
- 36.Xin CP, Tholen D, Devloo V, Zhu XG. The benefits of photorespiratory bypasses: How can they work? Plant Physiol. 2015;167(2):574–585. doi: 10.1104/pp.114.248013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zarzycki J, Brecht V, Müller M, Fuchs G. Identifying the missing steps of the autotrophic 3-hydroxypropionate CO2 fixation cycle in Chloroflexus aurantiacus. Proc Natl Acad Sci USA. 2009;106(50):21317–21322. doi: 10.1073/pnas.0908356106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shih PM, Zarzycki J, Niyogi KK, Kerfeld CA. Introduction of a synthetic CO2-fixing photorespiratory bypass into a cyanobacterium. J Biol Chem. 2014;289(14):9493–9500. doi: 10.1074/jbc.C113.543132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berg IA, Kockelkorn D, Buckel W, Fuchs G. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science. 2007;318(5857):1782–1786. doi: 10.1126/science.1149976. [DOI] [PubMed] [Google Scholar]
- 40.Huber H, et al. A dicarboxylate/4-hydroxybutyrate autotrophic carbon assimilation cycle in the hyperthermophilic Archaeum Ignicoccus hospitalis. Proc Natl Acad Sci USA. 2008;105(22):7851–7856. doi: 10.1073/pnas.0801043105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bar-Even A, Noor E, Lewis NE, Milo R. Design and analysis of synthetic carbon fixation pathways. Proc Natl Acad Sci USA. 2010;107(19):8889–8894. doi: 10.1073/pnas.0907176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goldsmith M, Tawfik DS. Directed enzyme evolution: Beyond the low-hanging fruit. Curr Opin Struct Biol. 2012;22(4):406–412. doi: 10.1016/j.sbi.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 43.Bornscheuer UT, et al. Engineering the third wave of biocatalysis. Nature. 2012;485(7397):185–194. doi: 10.1038/nature11117. [DOI] [PubMed] [Google Scholar]
- 44.Mukhopadhyay A, Redding AM, Rutherford BJ, Keasling JD. Importance of systems biology in engineering microbes for biofuel production. Curr Opin Biotechnol. 2008;19(3):228–234. doi: 10.1016/j.copbio.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 45.Asadollahi MA, et al. Enhancing sesquiterpene production in Saccharomyces cerevisiae through in silico driven metabolic engineering. Metab Eng. 2009;11(6):328–334. doi: 10.1016/j.ymben.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 46.Gilbert IR, Jarvis PG, Smith H. Proximity signal and shade avoidance differences between early and late successional trees. Nature. 2001;411(6839):792–795. doi: 10.1038/35081062. [DOI] [PubMed] [Google Scholar]
- 47.Drewry DT, Kumar P, Long SP. Simultaneous improvement in productivity, water use, and albedo through crop structural modification. Glob Change Biol. 2014;20(6):1955–1967. doi: 10.1111/gcb.12567. [DOI] [PubMed] [Google Scholar]
- 48.Johnston R, et al. Transcriptomic analyses indicate that maize ligule development recapitulates gene expression patterns that occur during lateral organ initiation. Plant Cell. 2014;26(12):4718–4732. doi: 10.1105/tpc.114.132688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu X-G, Song Q, Ort DR. Elements of a dynamic systems model of canopy photosynthesis. Curr Opin Plant Biol. 2012;15(3):237–244. doi: 10.1016/j.pbi.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 50.Chen M, Blankenship RE. Expanding the solar spectrum used by photosynthesis. Trends Plant Sci. 2011;16(8):427–431. doi: 10.1016/j.tplants.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 51.Croce R, van Amerongen H. Natural strategies for photosynthetic light harvesting. Nat Chem Biol. 2014;10(7):492–501. doi: 10.1038/nchembio.1555. [DOI] [PubMed] [Google Scholar]
- 52.McClung CR. Plant science: Making hunger yield. Science. 2014;344(6185):699–700. doi: 10.1126/science.1254135. [DOI] [PubMed] [Google Scholar]
- 53.Dymond JS, et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature. 2011;477(7365):471–476. doi: 10.1038/nature10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maliga P. Plastid transformation in higher plants. Annu Rev Plant Biol. 2004;55:289–313. doi: 10.1146/annurev.arplant.55.031903.141633. [DOI] [PubMed] [Google Scholar]
- 55.Bock R. Plastid biotechnology: Prospects for herbicide and insect resistance, metabolic engineering and molecular farming. Curr Opin Biotechnol. 2007;18(2):100–106. doi: 10.1016/j.copbio.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 56.Bedell VM, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491(7422):114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li T, Liu B, Spalding MH, Weeks DP, Yang B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol. 2012;30(5):390–392. doi: 10.1038/nbt.2199. [DOI] [PubMed] [Google Scholar]
- 58.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feng Z, et al. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci USA. 2014;111(12):4632–4637. doi: 10.1073/pnas.1400822111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bensimon A, Heck AJ, Aebersold R. Mass spectrometry-based proteomics and network biology. Annu Rev Biochem. 2012;81:379–405. doi: 10.1146/annurev-biochem-072909-100424. [DOI] [PubMed] [Google Scholar]
- 61.Chang RL, et al. Metabolic network reconstruction of Chlamydomonas offers insight into light-driven algal metabolism. Mol Syst Biol. 2011;7:518. doi: 10.1038/msb.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nogales J, Gudmundsson S, Knight EM, Palsson BO, Thiele I. Detailing the optimality of photosynthesis in cyanobacteria through systems biology analysis. Proc Natl Acad Sci USA. 2012;109(7):2678–2683. doi: 10.1073/pnas.1117907109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bracher A, et al. Degradation of potent Rubisco inhibitor by selective sugar phosphatase. Nat Plants. 2015;1:14002. doi: 10.1038/nplants.2014.2. [DOI] [PubMed] [Google Scholar]