Significance
The tumor suppressor protein p53 plays a critical role in protecting humans from cancer. In response to cellular stresses, such as DNA damage, p53 levels increase and induce a variety of protective biological responses. In unstressed cells, p53 is maintained at a low level by MDM2, a protein that interacts with p53 and promotes its degradation. Following DNA damage, MDM2 is degraded rapidly, resulting in increased p53 levels. Because of the essential role of MDM2 in modulating p53 function, it is critical to understand how MDM2 levels are regulated. Here we show that, following DNA damage, the F-box protein FBXO31, a potential tumor suppressor, is responsible for promoting MDM2 degradation and therefore is essential for the increase in p53 levels.
Keywords: p53, MDM2, FBXO31, DNA damage, tumor suppressor
Abstract
The tumor suppressor p53 plays a critical role in maintaining genomic stability. In response to genotoxic stress, p53 levels increase and induce cell-cycle arrest, senescence, or apoptosis, thereby preventing replication of damaged DNA. In unstressed cells, p53 is maintained at a low level. The major negative regulator of p53 is MDM2, an E3 ubiquitin ligase that directly interacts with p53 and promotes its polyubiquitination, leading to the subsequent destruction of p53 by the 26S proteasome. Following DNA damage, MDM2 is degraded rapidly, resulting in increased p53 stability. Because of the important role of MDM2 in modulating p53 function, it is critical to understand how MDM2 levels are regulated. Here we show that the F-box protein FBXO31, a candidate tumor suppressor encoded in 16q24.3 for which there is loss of heterozygosity in various solid tumors, is responsible for promoting MDM2 degradation. Following genotoxic stress, FBXO31 is phosphorylated by the DNA damage serine/threonine kinase ATM, resulting in increased levels of FBXO31. FBXO31 then interacts with and directs the degradation of MDM2, which is dependent on phosphorylation of MDM2 by ATM. FBXO31-mediated loss of MDM2 leads to elevated levels of p53, resulting in growth arrest. In cells depleted of FBXO31, MDM2 is not degraded and p53 levels do not increase following genotoxic stress. Thus, FBXO31 is essential for the classic robust increase in p53 levels following DNA damage.
One of the most fundamental and extensively studied anticancer mechanisms is the large increase in the levels of the tumor suppressor p53 that occurs following DNA damage (reviewed in ref. 1). The increased p53 then mediates growth arrest and/or apoptosis. The importance of this anticancer mechanism is highlighted by the mutation or functional inactivation of the p53 gene in more than 50% of human cancers (2, 3).
In unstressed cells, p53 is maintained at a low level. The major negative regulator of p53 is MDM2, an E3 ubiquitin ligase that interacts directly with p53 and promotes its polyubiquitination, leading to the subsequent destruction of p53 by the 26S proteasome (reviewed in ref. 4). Following DNA damage, MDM2 is degraded rapidly, resulting in increased p53 stability. Originally it was proposed that MDM2 degradation was caused by auto-ubiquitination; however, subsequent experiments showed that the E3 ubiquitin ligase activity of MDM2 is not required for its degradation (5).
We originally identified the F-box protein FBXO31 in an RNAi screen as one of 17 factors required for oncogenic BRAF to induce senescence in primary human cells (6). F-box proteins are best known for their role as the substrate-recognition components of the SKP1/CUL1/F-box protein (SCF) class of E3 ubiquitin ligases (7). The F-box motif is responsible for the ability of F-box proteins to interact with the SCF complex and to promote ubiquitination of their targets (8).
One of the other genes we isolated in our original RNAi screen was p53 (6), raising the possibility that FBXO31 and p53 function in a common pathway(s). Consistent with this idea, both FBXO31 and p53 can induce growth arrest (9, 10), and we have found that after DNA damage there is a posttranslational increase of FBXO31 levels, as there is for p53 (9). These considerations prompted us to ask whether there was a functional relationship between FBXO31 and p53.
Results
FBXO31 Is Required for Decreased MDM2 and Increased p53 Levels Following DNA Damage.
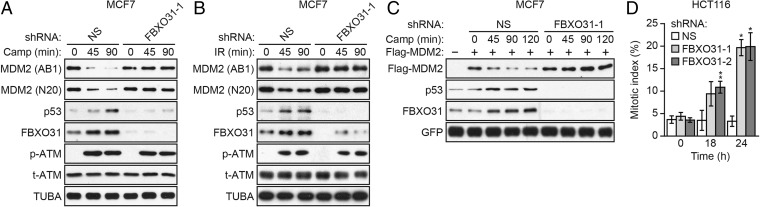
We asked whether the ability of FBXO31 to induce growth arrest results, at least in part, from the regulation of p53 levels. Toward this end, p53-positive MCF7 cells expressing either a control nonsilencing (NS) shRNA or an FBXO31 shRNA were treated with the DNA-damaging agent camptothecin or γ-irradiation, and the levels of p53 and MDM2 were analyzed by immunoblotting. Previous studies have shown that MDM2 levels decrease rapidly following genotoxic stress (4), and therefore in the first set of experiments we monitored the levels of p53 and other proteins at early times after the induction of DNA damage. Within 90 min following camptothecin (Fig. 1A) or γ-irradiation (Fig. 1B) treatment of MCF7 cells expressing a control NS shRNA, FBXO31 levels increased, as was consistent with previous results from our laboratory (9) and others (11). This increase in FBXO31 levels was accompanied by decreased MDM2 levels and increased p53 levels, as expected. Following camptothecin or γ-irradiation treatment of FBXO31 knockdown (KD) cells, there was a marked reduction in FBXO31 levels, as expected, but, notably, MDM2 and p53 levels remained constant. Similar results were obtained in p53-positive IMR90 cells (Fig. S1 A and B) and using a second, unrelated FBXO31 shRNA (Fig. S1 C–E).
Fig. 1.
FBXO31 is required for decreased MDM2 and increased p53 levels following DNA damage. (A and B) Immunoblot monitoring MDM2 [using a monoclonal (AB1) or polyclonal (N20) antibody], p53, FBXO31, phosphorylated ATM [p-ATM(1981)], and total ATM (t-ATM) in MCF7 cells expressing NS or FBXO31 shRNA and treated in the presence (45 or 90 min) or absence (0 min) of camptothecin (A) or γ-irradiation (IR) (B). α-tubulin (TUBA) was monitored as loading control. (C) Immunoblot monitoring Flag-MDM2, p53, and FBXO31 in MCF7 cells expressing Flag-MDM2 and NS or FBXO31 shRNA and treated in the presence or absence of camptothecin. GFP, expressed from a cotransfected plasmid, was used as a transfection and loading control. (D) Mitotic index analysis of HCT116 cells expressing NS or FBXO31 shRNA. Error bars indicate SD. *P ≤ 0.05, **P ≤ 0.01.
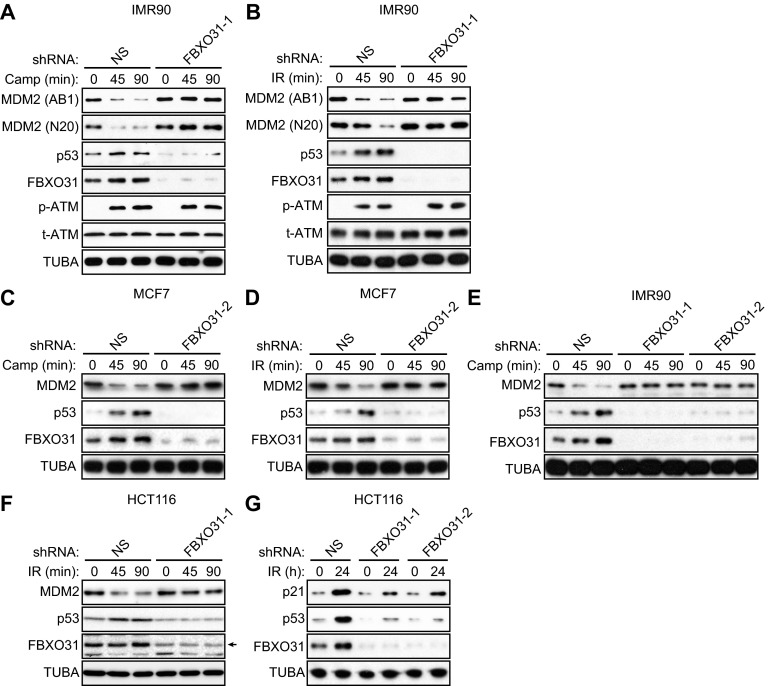
Fig. S1.
Confirmation of the results in Fig. 1 in other p53-positive cell lines and using a second FBXO31 shRNA. (A and B) Immunoblot analysis monitoring levels of MDM2 [using a monoclonal (AB1) or polyclonal (N20) antibody], p53, FBXO31, phosphorylated ATM [p-ATM(1981)], and total ATM (t-ATM) in IMR90 cells expressing NS or FBXO31 shRNA and treated in the presence or absence of camptothecin (A) or γ-irradiation (B). α-Tubulin (TUBA) was monitored as loading control. (C and D) Immunoblot analysis monitoring levels of MDM2, p53, and FBXO31 in MCF7 cells expressing NS or FBXO31 shRNA (unrelated to that used in Fig. 1 A and B) and treated in the presence (45 or 90 min) or absence (0 min) of camptothecin (C) or γ-irradiation (D). (E) Immunoblot analysis monitoring levels of MDM2, p53, and FBXO31 in IMR90 cells expressing NS or FBXO31 shRNA (unrelated to that used in A) and treated in the presence or absence of camptothecin. (F) Immunoblot analysis monitoring levels of MDM2, p53, and FBXO31 in HCT116 cells expressing NS or FBXO31 shRNA and treated in the presence or absence of γ-irradiation. (G) Immunoblot analysis monitoring levels of p21, p53, and FBXO31 in HCT116 cells expressing NS or one of two unrelated FBXO31 shRNAs and treated in the presence or absence of γ-irradiation.
A previous study suggested that following DNA damage the apparent decrease in MDM2 levels as monitored by immunoblotting in actuality is caused not by MDM2 degradation but rather by a conformational change in MDM2 that results in the masking of epitopes recognized by monoclonal anti-MDM2 antibodies (12) such as the AB1 antibody used in the experiments shown in Fig. 1 A and B and Fig. S1 A and B. Although this conclusion has been challenged by other studies (13), we performed two additional experiments to confirm that MDM2 is truly degraded following DNA damage in control but not in FBXO31 KD cells. First, in the experiments described above, MDM2 levels also were monitored using a polyclonal anti-MDM2 antibody (N20). Similar to the results with the AB1 monoclonal anti-MDM2 antibody, we found that after DNA damage MDM2 levels decreased in cells expressing the control NS shRNA but not in FBXO31 KD cells (Fig. 1 A and B and Fig. S1 A and B). Second, we ectopically expressed an N-terminal Flag-tagged MDM2 in control or FBXO31 KD MCF7 cells and, after DNA damage, monitored MDM2 levels using an anti-Flag antibody. The immunoblot results of Fig. 1C show that after camptothecin treatment in control MCF7 cells, the levels of ectopically expressed Flag-MDM2 decreased, and this decrease was accompanied by increased levels of endogenous p53. In contrast, after camptothecin treatment in FBXO31 KD cells, the levels of ectopically expressed Flag-MDM2 and endogenous p53 were unaffected.
The finding that in FBXO31 KD cells p53 levels failed to increase following DNA damage suggested that growth arrest would not occur efficiently. To test this prediction, we measured the mitotic index of control and FBXO31 KD cells in the presence of nocodazole to trap cells in mitosis. After DNA damage, cells harboring p53 arrest in G2 and G1, whereas cells lacking p53 will progress through the cell cycle and enter mitosis (14).
These experiments were performed in p53-positive HCT116 cells, which previously have been shown to undergo p53-dependent growth arrest in a mitotic index assay (14). Similar to the other p53-positive cell lines analyzed above, in FBXO31 KD HCT116 cells, MDM2 levels did not decrease and p53 levels did not increase after DNA damage (Fig. S1F). The results shown in Fig. 1D demonstrate that at 18 and 24 h following γ-irradiation the mitotic index of FBXO31 KD HCT116 cells was markedly higher than that of control HCT116 cells expressing an NS shRNA. Notably, the difference in mitotic index between control and FBXO31 KD HCT116 cells correlated with levels of p53 and the p53 target p21 (Fig. S1G), which plays a critical role in p53-mediated growth arrest (15, 16).
FBXO31 Directs Degradation of MDM2.
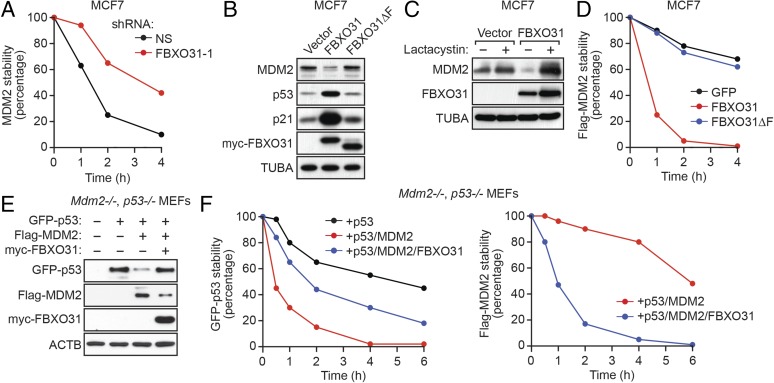
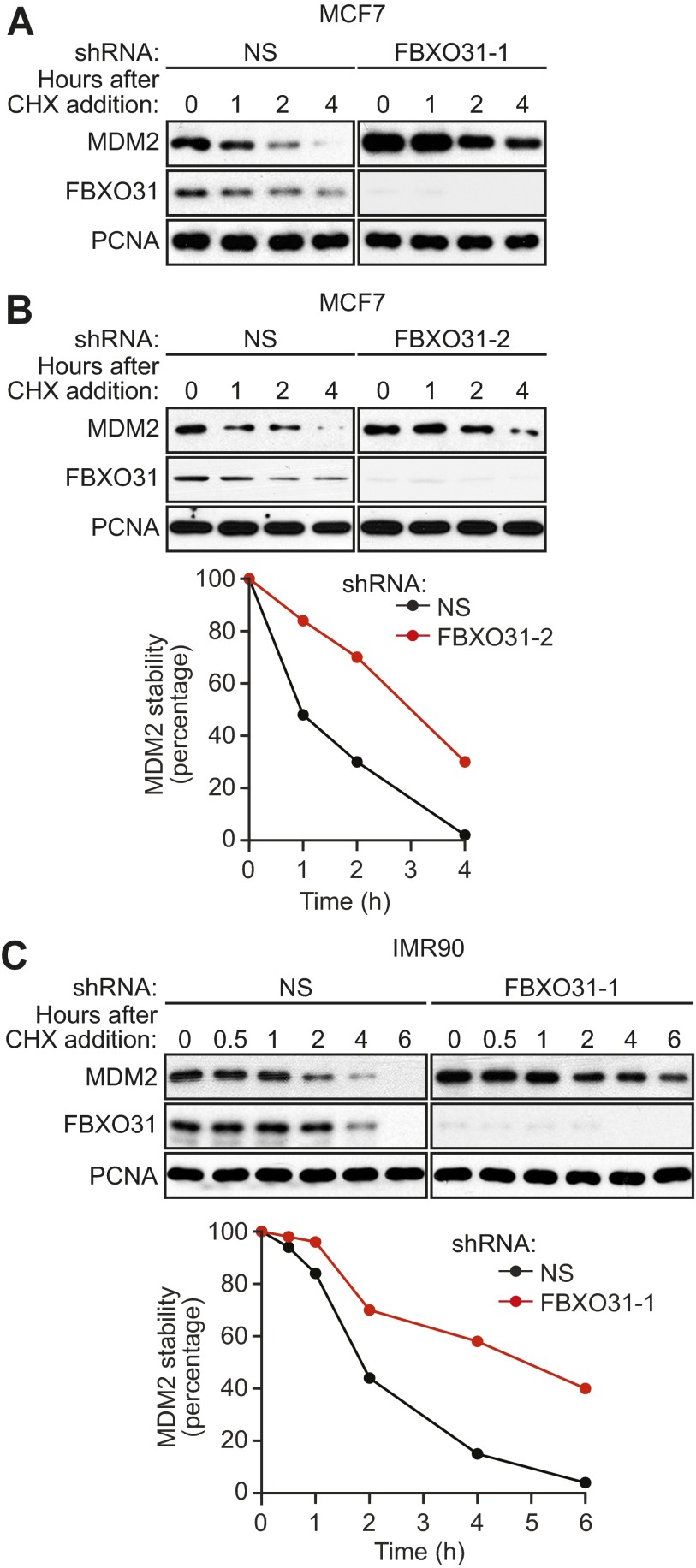
The results described above suggested that FBXO31 directly mediates degradation of MDM2, and we performed a series of experiments to confirm this possibility. First, we measured the half-life of endogenous MDM2 using a cycloheximide-chase/immunoblot assay. The results show that the half-life of MDM2 was substantially longer in FBXO31 KD MCF7 cells than in control cells (Fig. 2A and Fig. S2 A and B). Similar results were obtained in IMR90 cells (Fig. S2C).
Fig. 2.
FBXO31 directs degradation of MDM2. (A) Quantification of a cycloheximide-chase/immunoblot assay monitoring MDM2 stability in MCF7 cells expressing NS or FBXO31 shRNA following treatment with cycloheximide. The graph shows the ratio of the relative levels of MDM2 and PCNA (control) at each time point; time 0 was set to 100%. (B) Immunoblot monitoring MDM2, p53, and p21 in MCF7 cells expressing empty vector, FBXO31, or FBXO31∆F. (C) Immunoblot monitoring MDM2 in MCF7 cells expressing vector or FBXO31 and treated in the presence or absence of lactacystin. (D) Quantification of a cycloheximide-chase/immunoblot assay monitoring Flag-MDM2 stability in cycloheximide-treated MCF7 cells expressing GFP (control), FBXO31, or FBXO31∆F. (E) Immunoblot monitoring GFP-p53, Flag-MDM2, and myc-FBXO31 in Mdm2−/−, p53−/− MEFs coexpressing combinations of p53, MDM2, and FBXO31. β-Actin (ACTB) was monitored as a loading control. (F) Quantification of a cycloheximide-chase/immunoblot assay monitoring GFP-p53 and Flag-MDM2 stability in cycloheximide-treated Mdm2−/−, p53−/− MEFs coexpressing combinations of p53, MDM2, and FBXO31.
Fig. S2.
The half-life of MDM2 is increased following FBXO31 knockdown. (A) Cycloheximide-chase/immunoblot assay monitoring MDM2 stability in MCF7 cells expressing NS or FBXO31 shRNA at the indicated time points following treatment with cycloheximide (CHX). PCNA was monitored as a control. (B, Upper) Cycloheximide-chase/immunoblot assay monitoring MDM2 stability in cycloheximide-treated MCF7 cells expressing NS or FBXO31 shRNA (unrelated to that used in A and Fig. 2A). (Lower) Quantification of MDM2 levels. The graph shows the ratio of the relative levels of MDM2 and PCNA (control) at each time point; time 0 was set to 100%. (C, Upper) Cycloheximide-chase/immunoblot assay monitoring MDM2 stability in cycloheximide-treated IMR90 cells expressing NS or FBXO31 shRNA. (Lower) Quantification.
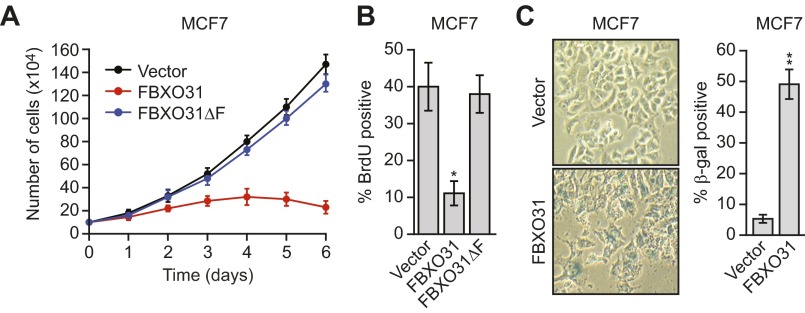
We next asked whether ectopic expression of FBXO31 would result in the degradation of endogenous MDM2. As was consistent with previous results (17), we found that ectopic expression of FBXO31 in MCF7 cells promoted growth arrest, as evidenced by reduced proliferation (Fig. S3A) and decreased DNA replication (BrdU incorporation) (Fig. S3B), and induced senescence, as evidenced by positive staining for senescence-associated β-gal (Fig. S3C). The immunoblot of Fig. 2B shows that ectopic expression of FBXO31 resulted in decreased levels of MDM2, which, as expected, were accompanied by increased levels of p53 and p21. Notably, previous studies have shown that increased p21 levels are sufficient to induce growth arrest and senescence (18, 19). In contrast to wild-type FBXO31, ectopic expression of an FBXO31 derivative in which the F-box had been deleted (FBXO31∆F) (17) failed to result in decreased levels of MDM2 or increased levels of p53 and p21.
Fig. S3.
Ectopic expression of FBXO31 promotes growth arrest, induces senescence, and reduces the half-life of MDM2. (A) Cell proliferation assay in MCF7 cells expressing vector, FBXO31, or FBXO31∆F. (B) DNA replication assay, monitored by BrdU incorporation, in MCF7 cells expressing empty vector, FBXO31, or FBXO31∆F. (C) Senescence-associated β-gal activity assay in MCF7 cells expressing empty vector or FBXO31. (Left) Representative images. (Right) Quantification of the percentage of β-gal+ cells.
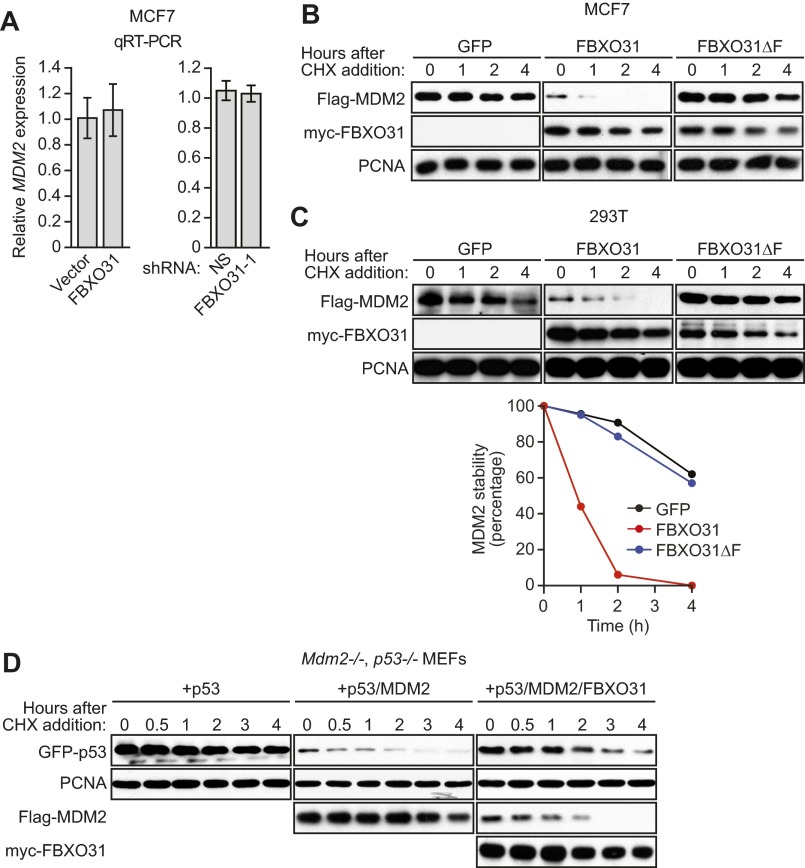
Consistent with our finding that FBXO31 affected MDM2 stability, the addition of the proteasome inhibitor lactacystin blocked the ability of ectopically expressed FBXO31 to decrease MDM2 levels (Fig. 2C). In addition, quantitative RT-PCR (qRT-PCR) analysis showed that MDM2 mRNA levels were unaffected by ectopic FBXO31 expression or after FBXO31 knockdown (Fig. S4A). Moreover, ectopic expression of FBXO31, but not FBXO31∆F, substantially reduced the half-life of MDM2 in MCF7 cells (Fig. 2D and Fig. S4B) and in 293T cells (Fig. S4C).
Fig. S4.
Ectopic expression of FBXO31, but not FBXO31∆F, substantially reduces the half-life of MDM2. (A) qRT-PCR analysis monitoring MDM2 mRNA levels in MCF7 cells expressing vector or FBXO31 (Left) or expressing NS or FBXO31 shRNA (Right). Error bars indicate SD. (B) Cycloheximide-chase/immunoblot assay monitoring Flag-MDM2 stability in cycloheximide-treated MCF7 cells expressing GFP (control), FBXO31, or FBXO31∆F. PCNA was monitored as a control. (C, Upper) Cycloheximide-chase/immunoblot assay monitoring Flag-MDM2 stability in 293T cells expressing GFP (control), FBXO31, or FBXO31∆F at the indicated time points following treatment with cycloheximide. (Lower) Quantification. (D) Cycloheximide-chase/immunoblot assay monitoring GFP-p53 and Flag-MDM2 stability in cycloheximide-treated Mdm2−/−, p53−/− MEFs coexpressing p53, MDM2, and FBXO31 in the indicated combinations.
Finally, to confirm the antagonistic roles of MDM2 and FBXO31 on p53 levels, we performed reconstitution experiments in homozygous knockout mouse embryo fibroblasts (MEFs) lacking MDM2 and p53 (Mdm2−/−, p53−/− MEFs). We ectopically expressed GFP-p53 alone, GFP-p53 and Flag-MDM2, or GFP-p53, Flag-MDM2, and myc-FBXO31 and measured p53 protein levels by immunoblotting. The results in Fig. 2E show, as expected, a reduction in p53 levels in the presence of MDM2. Notably, expression of FBXO31 led to decreased levels of MDM2 and a restoration of p53 levels. To confirm that these effects resulted from alterations of protein stability, the half-lives of p53 and MDM2 were measured by a cycloheximide-chase/immunoblot assay. The results in Fig. 2F show that the half-life of p53 was markedly reduced in the presence of MDM2 and that expression of FBXO31 led to a large decrease in the half-life of MDM2 and a concomitant increase in the half-life of p53 (see also Fig. S4D).
FBXO31 Interacts Directly with MDM2.
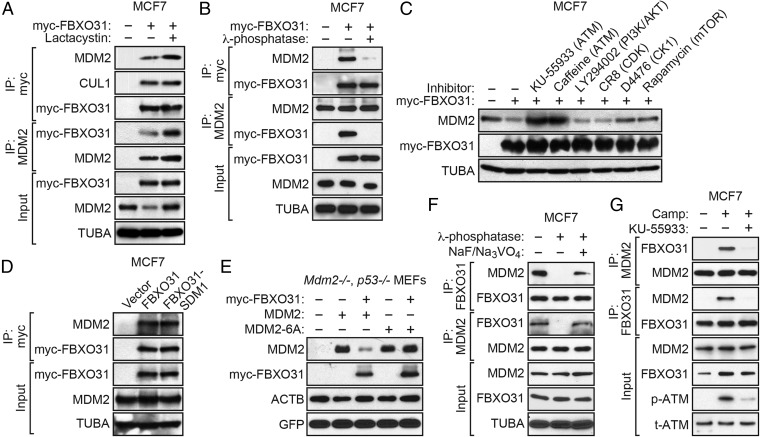
F-box proteins impart substrate specificity to the SCF ubiquitin ligase machinery by interacting directly with their protein targets (8). To test whether FBXO31 and MDM2 interact, we performed a series of coimmunoprecipitation experiments. First, MCF7 cells were stably transduced with a retrovirus expressing myc-tagged FBXO31, and FBXO31 was immunoprecipitated using an anti-myc antibody. Fig. 3A shows the presence of MDM2 in the FBXO31 immunoprecipitate, which was increased by lactacystin addition. The reciprocal coimmunoprecipitation experiment showed the presence of myc-FBXO31 in the MDM2 immunoprecipitate.
Fig. 3.
FBXO31 interacts with MDM2 directly in a manner that is dependent on phosphorylation of MDM2 by ATM. (A) Coimmunoprecipitation monitoring the interaction between ectopically expressed FBXO31 and endogenous MDM2 in MCF7 cells expressing vector or myc-FBXO31 and treated in the presence or absence of lactacystin. IP, immunoprecipitation. (B) Coimmunoprecipitation monitoring the FBXO31–MDM2 interaction in MCF7 cells treated in the presence or absence of λ-phosphatase. (C) Immunoblot monitoring MDM2 in MCF7 cells ectopically expressing FBXO31 and treated with kinase inhibitors. (D) Coimmunoprecipitation monitoring the FBXO31–MDM2 interaction in MCF7 cells expressing vector, myc-FBXO31, or myc-FBXO31-SDM1. (E) Immunoblot monitoring MDM2 in Mdm2−/−, p53−/− MEFs expressing vector or myc-FBXO31 and wild-type MDM2 or MDM2-6A. (F) Coimmunoprecipitation monitoring the endogenous FBXO31–MDM2 interaction in camptothecin-treated MCF7 cells in the presence or absence of λ-phosphatase or NaF/Na3VO4. (G) Coimmunoprecipitation monitoring the endogenous FBXO31–MDM2 interaction in MCF7 cells treated in the presence or absence of camptothecin or KU-55933.
The FBXO31–MDM2 Interaction Is Dependent on Phosphorylation of MDM2 by ATM.
Recognition by F-box proteins typically requires phosphorylation of the substrate, which serves as a signal for ubiquitin-dependent destruction (8). Fig. 3B shows that the FBXO31–MDM2 interaction was lost, as expected, after treatment of cell extracts with λ-phosphatase, which nonspecifically removes phosphoryl groups.
MDM2 is phosphorylated by several protein kinases including the DNA damage response-initiating kinase ATM (20, 21), AKT (22, 23), cyclin-dependent kinases (CDKs) (24), casein kinase 1 (CK1) (25), and mammalian target of rapamycin/S6K1 (mTOR) (26). We analyzed the effect of chemical inhibitors of these kinases on MDM2 levels in MCF7 cells after ectopic expression of FBXO31. Fig. 3C shows that treatment of cells with the two different ATM inhibitors, KU-55933 (27) and caffeine (28), resulted in a substantial increase in the levels of MDM2, indicating decreased FBXO31-directed degradation. In contrast, treatment of cells with inhibitors of the PI3K/AKT pathway, i.e., LY294002 (29), CDKs [CR8 (30)], CK1 [D4476 (31)], or mTOR [rapamycin (32)], had either no or only a modest effect on MDM2 levels. Consistent with the results of a previous study (25), treatment with the CK1 inhibitor D4476 also increased MDM2 levels, but the effect was much less than that observed following ATM inhibition.
We have shown previously that FBXO31 itself is phosphorylated by ATM (9). Therefore, it remained possible that the loss of the FBXO31–MDM2 interaction following ATM inhibition was caused, at least in part, by a failure to phosphorylate FBXO31. To rule out this possibility, we analyzed an FBXO31 derivative containing a mutation within the ATM phosphorylation site that prevents phosphorylation (FBXO31-SDM1) (9). Fig. 3D shows that wild-type FBXO31 and the FBXO31-SDM1 mutant interacted with MDM2 comparably.
Finally, we analyzed an MDM2 derivative in which all six ATM phosphorylation sites were mutated (MDM2-6A) (33). A plasmid expressing wild-type MDM2 or MDM2-6A was cotransfected with an FBXO31 expression plasmid into Mdm2−/−, p53−/− MEFs, and MDM2 levels were monitored by immunoblotting. As shown in Fig. 3E, FBXO31 failed to reduce MDM2-6A levels, confirming the essential role of ATM in FBXO31-directed degradation of MDM2.
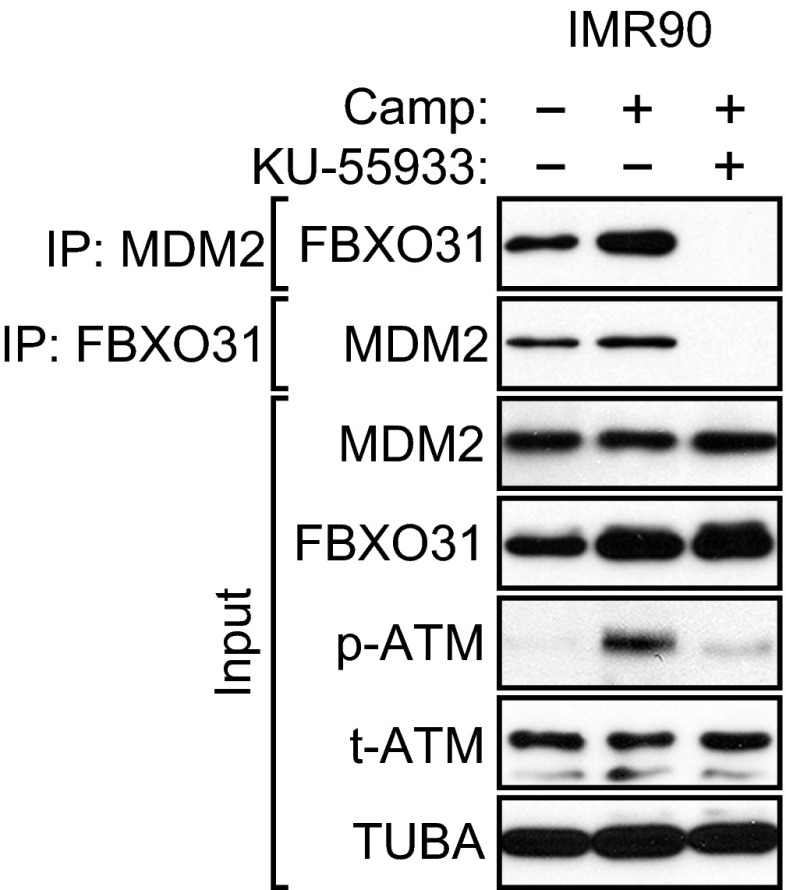
The experiments described above were performed with ectopically expressed FBXO31 and/or MDM2. We next performed coimmunoprecipitation experiments in camptothecin-treated MCF7 cells to detect an interaction between endogenous FBXO31 and MDM2. The coimmunoprecipitation experiment in Fig. 3F shows that an interaction could be detected between endogenous FBXO31 and MDM2. The FBXO31–MDM2 interaction was lost following treatment of cell extracts with λ-phosphatase but was restored upon the addition of phosphatase inhibitors. Also, as is consistent with the results described above, Fig. 3G shows that the interaction between endogenous FBXO31 and MDM2 was lost following treatment of MCF7 cells with the ATM inhibitor KU-55933. Finally, the coimmunoprecipitation experiment in Fig. S5 shows that an ATM-dependent interaction between endogenous FBXO31 and MDM2 also could be detected in IMR90 cells.
Fig. S5.
The endogenous FBXO31–MDM2 interaction is lost following treatment of IMR90 cells with the ATM inhibitor KU-55933. Coimmunoprecipitation analysis monitoring the endogenous FBXO31–MDM2 interaction in IMR90 cells treated in the presence or absence of camptothecin or KU-55933. Extracts were immunoprecipitated with an FBXO31 or MDM2 antibody.
FBXO31 Directs Polyubiquitination of MDM2.
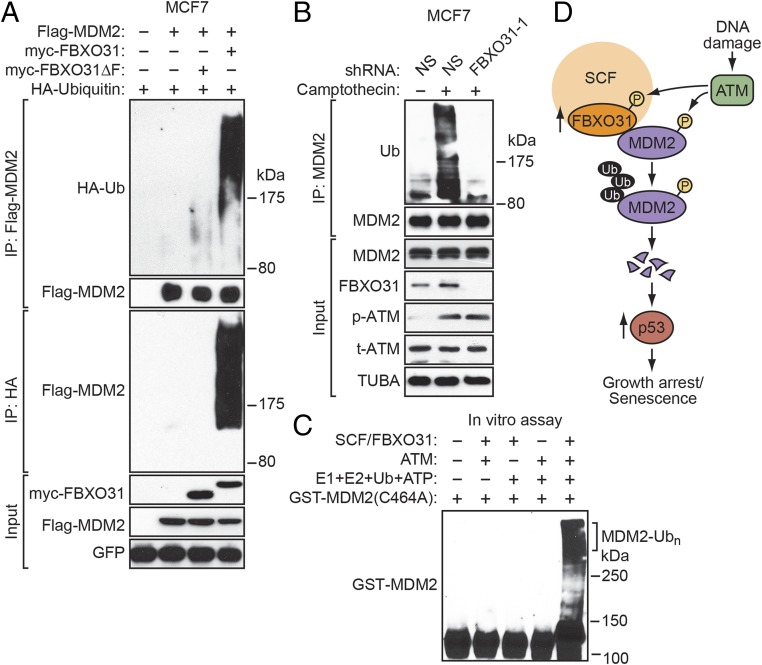
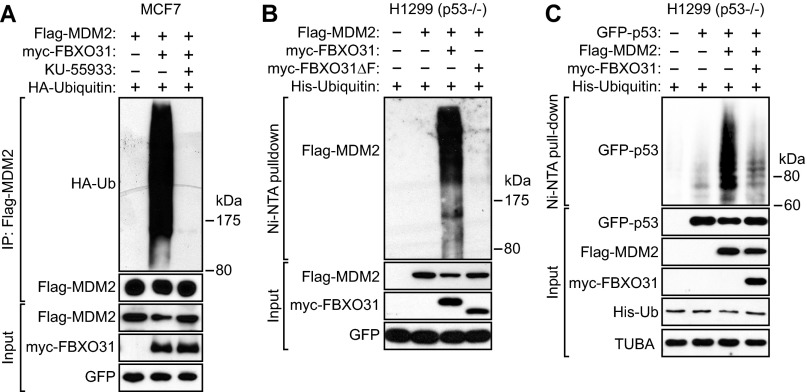
Typically, F-box proteins direct polyubiquitination of their substrates, resulting in proteasome-mediated degradation (7, 34). We performed a series of experiments to determine whether FBXO31 can direct polyubiquitination of MDM2. In these experiments, cells were treated with the proteasome inhibitor MG132 to minimize degradation of polyubiquitinated proteins. In the first experiment, MCF7 cells were cotransfected with plasmids expressing Flag-MDM2, HA-tagged ubiquitin, and either myc-FBXO31 or myc-FBXO31ΔF. Polyubiquitination of MDM2 was assessed by immunoprecipitation of Flag-MDM2 followed by immunoblotting for HA-ubiquitin. The results of Fig. 4A show that ectopic expression of FBXO31, but not FBXO31∆F, resulted in polyubiquitination of MDM2. Similar results were obtained in a reciprocal coimmunoprecipitation experiment. Notably, FBXO31-directed polyubiquitination of MDM2 was lost after treatment of cells with the ATM inhibitor KU-55933 (Fig. S6A).
Fig. 4.
FBXO31 directs polyubiquitination of MDM2. (A) In vivo assay monitoring ubiquitination of Flag-MDM2 in MCF7 cells expressing Flag-MDM2, HA-ubiquitin, and either myc-FBXO31 or myc-FBXO31∆F. GFP expressed from a cotransfected plasmid was used as transfection/loading control. (B) In vivo assay monitoring the ubiquitination of endogenous MDM2 in MCF7 cells expressing NS or FBXO31 shRNA and treated in the presence or absence of camptothecin. (C) In vitro ubiquitination assay monitoring the ability of a purified FBXO31/CUL1/SKP1/ROC1 complex to ubiquitinate GST-MDM2(C464A). (D) Model. Ub, ubiquitin.
Fig. S6.
Additional in vivo ubiquitination experiments related to Fig. 4. (A) In vivo ubiquitination assay. Extracts from MCF7 cells treated in the presence or absence of KU-55933 and expressing Flag-MDM2, myc-FBXO31, and HA-ubiquitin (HA-Ub) were immunoprecipitated with a Flag antibody and probed for HA-ubiquitin. GFP expressed from a cotransfected plasmid was used as transfection/loading control. (B and C) In vivo ubiquitination assays. H1299 cells were transfected with plasmids expressing His-ubiquitin, Flag-MDM2, either myc-FBXO31 or myc-FBXO31∆F, and either GFP alone (B) or GFP-p53 (C). Proteins bound to His-ubiquitin were purified by Ni-NTA pull-down, washed, and eluted in imidazole. Ubiquitinated Flag-MDM2 and GFP-p53 were detected using an anti-Flag or anti-GFP antibody, respectively.
To confirm these results, we performed another in vivo ubiquitination experiment involving cotransfection of plasmids expressing Flag-MDM2, His-tagged ubiquitin, and either myc-FBXO31 or myc-FBXO31ΔF. His-ubiquitin–conjugated proteins were purified under stringent, denaturing conditions, followed by immunoblotting for Flag-MDM2. The results confirm that ectopic expression of FBXO31, but not FBXO31∆F, resulted in polyubiquitination of MDM2 (Fig. S6B).
We also used this His-ubiquitin pull-down assay to confirm the antagonistic relationship of MDM2 and FBXO31 on polyubiquitination of p53. In p53-negative H1299 cells we ectopically expressed His-ubiquitin with GFP-p53 alone, GFP-p53 and Flag-MDM2, or GFP-p53, Flag-MDM2, and myc-FBXO31. Polyubiquitinated p53 was detected by purifying His-ubiquitin–conjugated proteins followed by immunoblotting for GFP-p53. The results show that ectopic expression of MDM2 resulted in a substantial increase in polyubiquitination of p53, which was counteracted by the coexpression of FBXO31 (Fig. S6C).
The ubiquitination assays described above were performed with ectopically expressed proteins. We next performed an additional in vivo ubiquitination experiment to confirm that endogenous FBXO31 could polyubiquitinate endogenous MDM2. Extracts from untreated or camptothecin-treated MCF7 cells expressing either an NS or FBXO31 shRNA were immunoprecipitated with an anti-MDM2 antibody, and the immunoprecipitate was analyzed by immunoblotting with an anti-ubiquitin antibody. As expected, the results of Fig. 4B show that camptothecin treatment led to a large increase in polyubiquitinated MDM2. Notably, knockdown of FBXO31 substantially reduced the amount of polyubiquitinated MDM2 in camptothecin-treated cells. These results indicate that FBXO31 is required for polyubiquitination of MDM2 following DNA damage.
Finally, we performed an in vitro ubiquitination assay. Previous studies have shown that MDM2 has auto-ubiquitinating activity in vitro in the presence of an E1 ubiquitin-activating enzyme and E2 ubiquitin-conjugating enzyme (35, 36). Therefore, we used a previously described catalytically inactive MDM2 mutant, MDM2(C464A) (35). In addition to MDM2(C464A), the reaction mixtures contained or lacked the known cofactors (E1, E2, ubiquitin, and ATP), ATM, and the SCF/myc-FBXO31 complex purified from transfected 293T cells. The results in Fig. 4C show that in vitro polyubiquitination of MDM2(C464A) was dependent on the addition of the known cofactors, ATM, and the myc- SCF/FBXO31 complex.
Discussion
In this report we show an essential role for FBXO31 in MDM2 degradation following genotoxic stress that is summarized in the schematic model shown in Fig. 4D and discussed below. Following DNA damage, FBXO31 is phosphorylated by ATM, resulting in increased levels of FBXO31. FBXO31, as part of the SCF complex, then interacts directly with and mediates the degradation of MDM2, which is dependent on the phosphorylation of MDM2 by ATM. The decreased MDM2 results in increased levels of p53, which promotes growth arrest and senescence through transcriptional activation of p21 and other p53 target genes. Thus, FBXO31 is essential for the classic robust increase in p53 levels following DNA damage.
We have shown previously that in p53-deficient SK-MEL-28 cells FBXO31 also can induce G1 arrest following DNA damage through interaction with and degradation of cyclin D1 (9). Thus, following genotoxic stress, FBXO31 can induce growth arrest through two independent pathways that differ with regard to both substrates and p53 dependence.
Previous studies have shown that in response to DNA damage, MDM2 degradation also is mediated by the F-box protein beta-transducin repeat containing E3 ubiquitin ligase protein (b-TRCP; also called BTRC) and requires phosphorylation of MDM2 by CKIδ (25). Although our results do not rule out the possibility that β-TRCP contributes to MDM2 degradation, FBXO31 appears to have the predominant role, at least in the cell types we analyzed (see, for example, Fig. 3C).
ATM is considered the main transducer of the DNA damage response that is activated by double-strand breaks (37). Therefore it is notable that after DNA damage ATM phosphorylates both FBXO31, leading to its stabilization, and MDM2, enabling interaction with and degradation by FBXO31. Collectively, the results presented here highlight the critical function of FBXO31 in the DNA damage response and provide further support for the role of FBXO31 as a tumor suppressor.
Materials and Methods
DNA replication and senescence assays are described in SI Materials and Methods.
Cell Lines and Culture.
MCF7, IMR90, H1299, and 293T cells were obtained from ATCC and were grown as recommended. Cell lines stably expressing empty vector, myc-FBXO31, or myc-FBXO31∆F were generated by retroviral transduction as described (9). Cells were treated with camptothecin (20 µM; Sigma) or γ-irradiation (20 Gy) for 45 or 90 min, MG132 (10 µM; Sigma) for 4 h, lactacystin (5 µM; Calbiochem) for 8 h; or KU-55933 (10 µM; Tocris Bioscience), caffeine (1 mM; Sigma), LY294002 (10 µM; Cayman Chemical), CR8 (10 µM; Tocris Bioscience), D4476 (1 µM; Cayman), or rapamycin (5 µM; Calbiochem) for 24 h. FBXO31 KD cells were generated using lentiviral shRNAs (FBXO31-1, TGCTGTTGACAGTGAGCGAGCAAAGGTTCCTAAATGGTAATAGTGAAGCCACAGATGTATTACCATTTAGGAACCTTTGCCTGCCTACTGCCTCGGA and FBXO31-2, TGCTGTTGACAGTGAGCGAGCAAAGGTTCCTAAATGGTAATAGTGAAGCCACAGATGTATTACATTTAGGAACCTTTGCCTGCCTACGCCTCGGA; Open Biosystems/GE Dharmacon) as described (9). Mdm2−/−, p53−/− MEFS (provided by Stephen Jones, University of Massachusetts Medical School, Worcester, MA) were transfected with pCMV-MDM2-WT and pCMV-MDM2-6A plasmids (provided by Jiandong Chen, Moffitt Cancer Center, Tampa, FL). HCT116 cells (14) were provided by B. Vogelstein of Johns Hopkins University, Baltimore, and were grown in RPMI1640 medium containing 10% (vol/vol) FBS.
Immunoblotting.
Protein extracts were prepared and immunoblotting was performed as described (9) using the following antibodies: mouse monoclonal MDM2 (AB1; Calbiochem), rabbit polyclonal MDM2 (N20; Santa Cruz), mouse monoclonal p53 (DO1; Santa Cruz), FBXO31 (Bethyl Laboratories, Inc.), phospho-ATM Ser1981 (Cell Signaling), ATM (Cell Signaling), p21 (Cell Signaling), myc (Roche), Flag (Sigma), GFP (Santa Cruz), Proliferating cell nuclear antigen (PCNA; (Santa Cruz), 6×-His (Abcam), β-actin (Sigma), and α-tubulin (Sigma).
Mitotic Index.
HCT116 cells were seeded on a coverslip (5 × 104 cells per 12-well plate) and were exposed on the following day to 10-Gy ionizing radiation (Cobalt-60 irradiator). Thirty minutes later, nocodazole (200 nM; Sigma) was added to the cells for 18 or 24 h, or, as a control, cells were left untreated (0 h time point). Cells then were collected, fixed with 3.7% (vol/vol) formaldehyde in PBS for 30 min at 37 °C, and washed three times in PBS. When all time points were collected, cells were permeabilized with chilled methanol, stained with Hoechst 33258 (10 mg/mL) in PBS, and washed three times in PBS. Nuclei were visualized by fluorescence microscopy (Olympus). Nuclei with condensed, evenly staining chromosomes were scored as mitotic. At least 200 cells were counted for each sample.
Cycloheximide Chase.
For the experiments monitoring endogenous MDM2, MCF7 or IMR90 cells were infected with a lentivirus expressing an NS or FBXO31 shRNA and puromycin selected for 5 d. Cells then were treated with cycloheximide (100 µg/mL; Sigma). For the experiments monitoring Flag-MDM2, MCF7 cells or 293T cells were cotransfected with a plasmid expressing Flag-MDM2 (provided by Ze'ev Ronai, Sanford-Burnham Medical Research Institute, La Jolla, CA) and a plasmid expressing GFP (pEGFPC1; Clontech), myc-FBXO31, or myc-FBXO31∆F. Thirty hours later, cells were treated with cycloheximide (100 μg/mL). For experiments in Mdm2−/−, p53−/− MEFS, cells were transfected with plasmids expressing GFP-p53 (Addgene), Flag-MDM2, and myc-FBXO31 in various combinations, and 30 h later, cells were treated with cycloheximide (50 µg/mL). Total cell extracts were prepared as described above and subjected to immunoblotting with Flag (Sigma), myc (Roche), GFP (FL Santa Cruz), and PCNA (Cell Signaling) antibodies. Band intensities were quantified using Image J software version 1.47v (NIH).
qRT–PCR.
Total RNA was isolated and reverse transcription was performed as described (9), followed by qRT-PCR using MDM2 forward (5′-CATTGTCCATGGCAAAACAG-3′) and reverse (5′-GGCAGGGCTTATTCCTTTTC-3′) primers.
Coimmunoprecipitation.
Coimmunoprecipitation assays were performed as described (9). Lactacystin (5 µM; Calbiochem) was added for 8 h before preparation of the protein extract. For λ-phosphatase treatment (Fig. 3F), whole-cell extract prepared from MCF7 cells treated with camptothecin (20 µM for 45 min) was either mock treated or treated with λ-phosphatase (2,000 U) for 30 min at 30 °C in the presence or absence of 10 mM NaF (Sigma) and 1 mM Na3VO4 (Sigma). FBXO31 coimmunoprecipitations were performed using a previously described polyclonal antibody (9). Blots were probed with the antibodies described above, including Cullin 1 (CUL1; Cell Signaling). For the assays shown in Fig. 3G and Fig. S5, cells were treated with KU-55933 for 12 h before extract preparation.
Ubiquitination Assays.
For in vivo assays, MCF7 cells were cotransfected with plasmids expressing Flag-MDM2, HA-tagged ubiquitin (Addgene), and either myc-FBXO31 or myc-FBXO31ΔF; 36 h later, cells were treated with 10 µM MG132 for 4 h and were lysed in cell lysis buffer containing 10 mM N-ethylmaleimide (Sigma). For the His-ubiquitin pull-down assays, H1299 cells were transfected with plasmids expressing His-ubiquitin (Addgene), Flag-MDM2, myc-FBXO31, myc-FBXO31∆F, and either GFP alone or GFP-p53 in various combinations. Thirty hours posttransfection, cells were treated with MG132 (20 µm for 4 h) and then were harvested and used for purification of His6-tagged proteins by Ni-NTA beads. The cell pellet was lysed in buffer A [6 M guanidinium⋅HCl, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris⋅Cl (pH 8.0), 5 mM imidazole, 10 mM β-mercaptoethanol] and incubated with Ni-NTA beads (Qiagen) for 4 h at room temperature. The beads were washed with buffers A, B [8 M urea, 0.1 M Na2PO4/NaH2PO4, 0.01 M Tris⋅Cl (pH 8.0), 10 mM β-mercaptoethanol], and C [8 M urea, 0.1 M Na2PO4/NaH2PO4, 0.01 M Tris⋅Cl (pH 6.3), 10 mM β-mercaptoethanol], and bound proteins were eluted with buffer D [200 mM imidazole, 0.15 M Tris⋅Cl (pH 6.7), 30% (vol/vol) glycerol, 0.72 M β-mercaptoethanol, 5% (wt/vol) SDS]. The eluted proteins were analyzed by immunoblotting.
For the in vivo assay, extracts from MCF7 cells were treated with DMSO or camptothecin (25 µM for 45 min) and MG132 (10 µM for 6 h) before immunoprecipitation.
The in vitro ubiquitination assay was performed as described (9). Briefly, 293T cells were cotransfected with plasmids encoding myc-CUL1, myc-SKP1, myc-ROC1, and myc-FBXO31. Complexes containing FBXO31/CUL1/SKP1/ROC were immunopurified from the cell lysate using anti-myc beads and were incubated with recombinant GST-MDM2(C464A) (provided by Allan Weissman, National Cancer Institute, Bethesda), purified recombinant active ATM (provided by Tanya Paull, University of Texas at Austin, Austin, TX), 0.1 mm E1 (Boston Biochem), 0.25 mm E2 (UBCH5A; Boston Biochem), and 2.5 µg/mL ubiquitin (Boston Biochem) in ubiquitin assay buffer [10 mM Tris (pH 7.5), 100 mM NaCl, 2.5 mM MgCl2, 2.5 mM ATP, 1 mM DTT, and 10 mM MnCl2] for 2 h at 30 °C, and were analyzed by immunoblotting.
Statistical Analysis.
All quantitative data were collected from experiments performed in at least triplicate and are expressed as mean ± SD. Differences between groups were assayed using two-tailed Student t test using Microsoft Excel.
SI Materials and Methods
For DNA replication and senescence assays, cells were incubated with 20 µM BrdU (Sigma) for 4 h and stained as described (38). Cells were stained for β-gal activity as described (39) and were visualized on a Zeiss Axiovert 40 CFL microscope. Images were captured using QCapture Pro version 5 software (QImaging Corporation), and ∼500 cells were scored for each sample.
Acknowledgments
We thank Jiandong Chen, Stephen Jones, Tanya Paull, Ze’ev Ronai, Allan Weissman, and Bert Vogelstein for providing reagents and Sara Deibler for editorial assistance. This work was supported in part by a Ramalingaswami Fellowship from the Department of Biotechnology, Ministry of Science and Technology, Government of India (to M.K.S.). M.R.G. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1510929112/-/DCSupplemental.
References
- 1.Kumari R, Kohli S, Das S. p53 regulation upon genotoxic stress: Intricacies and complexities. Mol Cell Oncol. 2014;1(3):e969653. doi: 10.4161/23723548.2014.969653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petitjean A, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: Lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28(6):622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408(6810):307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 4.Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13(2):83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Itahana K, et al. Targeted inactivation of Mdm2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell. 2007;12(4):355–366. doi: 10.1016/j.ccr.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 6.Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell. 2008;132(3):363–374. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol. 2013;14(6):369–381. doi: 10.1038/nrm3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho MS, Tsai PI, Chien CT. F-box proteins: The key to protein degradation. J Biomed Sci. 2006;13(2):181–191. doi: 10.1007/s11373-005-9058-2. [DOI] [PubMed] [Google Scholar]
- 9.Santra MK, Wajapeyee N, Green MR. F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature. 2009;459(7247):722–725. doi: 10.1038/nature08011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zilfou JT, Lowe SW. Tumor suppressive functions of p53. Cold Spring Harb Perspect Biol. 2009;1(5):a001883. doi: 10.1101/cshperspect.a001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, et al. F-box only protein 31 (FBXO31) negatively regulates p38 mitogen-activated protein kinase (MAPK) signaling by mediating lysine 48-linked ubiquitination and degradation of mitogen-activated protein kinase kinase 6 (MKK6) J Biol Chem. 2014;289(31):21508–21518. doi: 10.1074/jbc.M114.560342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng Q, Chen J. Mechanism of p53 stabilization by ATM after DNA damage. Cell Cycle. 2010;9(3):472–478. doi: 10.4161/cc.9.3.10556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, et al. DNA damage-induced activation of ATM promotes β-TRCP-mediated Mdm2 ubiquitination and destruction. Oncotarget. 2012;3(9):1026–1035. doi: 10.18632/oncotarget.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bunz F, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282(5393):1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 15.el-Deiry WS, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75(4):817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 16.Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55(22):5187–5190. [PubMed] [Google Scholar]
- 17.Kumar R, et al. FBXO31 is the chromosome 16q24.3 senescence gene, a candidate breast tumor suppressor, and a component of an SCF complex. Cancer Res. 2005;65(24):11304–11313. doi: 10.1158/0008-5472.CAN-05-0936. [DOI] [PubMed] [Google Scholar]
- 18.Fang L, et al. p21Waf1/Cip1/Sdi1 induces permanent growth arrest with markers of replicative senescence in human tumor cells lacking functional p53. Oncogene. 1999;18(18):2789–2797. doi: 10.1038/sj.onc.1202615. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Blandino G, Givol D. Induced p21waf expression in H1299 cell line promotes cell senescence and protects against cytotoxic effect of radiation and doxorubicin. Oncogene. 1999;18(16):2643–2649. doi: 10.1038/sj.onc.1202632. [DOI] [PubMed] [Google Scholar]
- 20.Khosravi R, et al. Rapid ATM-dependent phosphorylation of MDM2 precedes p53 accumulation in response to DNA damage. Proc Natl Acad Sci USA. 1999;96(26):14973–14977. doi: 10.1073/pnas.96.26.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maya R, et al. ATM-dependent phosphorylation of Mdm2 on serine 395: Role in p53 activation by DNA damage. Genes Dev. 2001;15(9):1067–1077. doi: 10.1101/gad.886901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogawara Y, et al. Akt enhances Mdm2-mediated ubiquitination and degradation of p53. J Biol Chem. 2002;277(24):21843–21850. doi: 10.1074/jbc.M109745200. [DOI] [PubMed] [Google Scholar]
- 23.Mayo LD, Donner DB. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc Natl Acad Sci USA. 2001;98(20):11598–11603. doi: 10.1073/pnas.181181198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang T, Prives C. Cyclin a-CDK phosphorylation regulates MDM2 protein interactions. J Biol Chem. 2001;276(32):29702–29710. doi: 10.1074/jbc.M011326200. [DOI] [PubMed] [Google Scholar]
- 25.Inuzuka H, et al. Phosphorylation by casein kinase I promotes the turnover of the Mdm2 oncoprotein via the SCF(beta-TRCP) ubiquitin ligase. Cancer Cell. 2010;18(2):147–159. doi: 10.1016/j.ccr.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai KP, et al. S6K1 is a multifaceted regulator of Mdm2 that connects nutrient status and DNA damage response. EMBO J. 2010;29(17):2994–3006. doi: 10.1038/emboj.2010.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hickson I, et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64(24):9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 28.Blasina A, Price BD, Turenne GA, McGowan CH. Caffeine inhibits the checkpoint kinase ATM. Curr Biol. 1999;9(19):1135–1138. doi: 10.1016/s0960-9822(99)80486-2. [DOI] [PubMed] [Google Scholar]
- 29.Vlahos CJ, Matter WF, Hui KY, Brown RF. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002) J Biol Chem. 1994;269(7):5241–5248. [PubMed] [Google Scholar]
- 30.Bettayeb K, et al. CR8, a potent and selective, roscovitine-derived inhibitor of cyclin-dependent kinases. Oncogene. 2008;27(44):5797–5807. doi: 10.1038/onc.2008.191. [DOI] [PubMed] [Google Scholar]
- 31.Rena G, Bain J, Elliott M, Cohen P. D4476, a cell-permeant inhibitor of CK1, suppresses the site-specific phosphorylation and nuclear exclusion of FOXO1a. EMBO Rep. 2004;5(1):60–65. doi: 10.1038/sj.embor.7400048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown EJ, et al. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369(6483):756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 33.Cheng Q, Chen L, Li Z, Lane WS, Chen J. ATM activates p53 by regulating MDM2 oligomerization and E3 processivity. EMBO J. 2009;28(24):3857–3867. doi: 10.1038/emboj.2009.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z, Liu P, Inuzuka H, Wei W. Roles of F-box proteins in cancer. Nat Rev Cancer. 2014;14(4):233–247. doi: 10.1038/nrc3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275(12):8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 36.Honda R, Yasuda H. Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 1999;18(1):22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shiloh Y, Ziv Y. The ATM protein kinase: Regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14(4):197–210. [PubMed] [Google Scholar]
- 38.Zeng YX, Somasundaram K, el-Deiry WS. AP2 inhibits cancer cell growth and activates p21WAF1/CIP1 expression. Nat Genet. 1997;15(1):78–82. doi: 10.1038/ng0197-78. [DOI] [PubMed] [Google Scholar]
- 39.Dimri GP, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92(20):9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]