Significance
Circadian clocks are molecular machineries that allow organisms to anticipate daily cyclic challenges and to temporally modulate different processes. Thus, plant defense mechanisms against pathogens have been reported to vary daily in Arabidopsis thaliana. Although the plant–pathogen interaction is a two-sided story, nothing is known regarding circadian regulation of pathogenic traits. Herein we characterize a functional circadian clock in the necrotrophic fungal plant pathogen Botrytis cinerea, postulating additional roles for BcFRQ1, the Botrytis ortholog of the core clock component FREQUENCY of Neurospora crassa. By using different plant and Botrytis clock-null mutants, we demonstrate that the interaction between this pathogen and its host varies with the time of day, being the B. cinerea circadian clock key in regulating this outcome.
Keywords: circadian clock, virulence, plant–pathogen interaction, Arabidopsis thaliana, Botrytis cinerea
Abstract
The circadian clock of the plant model Arabidopsis thaliana modulates defense mechanisms impacting plant–pathogen interactions. Nevertheless, the effect of clock regulation on pathogenic traits has not been explored in detail. Moreover, molecular description of clocks in pathogenic fungi—or fungi in general other than the model ascomycete Neurospora crassa—has been neglected, leaving this type of question largely unaddressed. We sought to characterize, therefore, the circadian system of the plant pathogen Botrytis cinerea to assess if such oscillatory machinery can modulate its virulence potential. Herein, we show the existence of a functional clock in B. cinerea, which shares similar components and circuitry with the Neurospora circadian system, although we found that its core negative clock element FREQUENCY (BcFRQ1) serves additional roles, suggesting extracircadian functions for this protein. We observe that the lesions produced by this necrotrophic fungus on Arabidopsis leaves are smaller when the interaction between these two organisms occurs at dawn. Remarkably, this effect does not depend solely on the plant clock, but instead largely relies on the pathogen circadian system. Genetic disruption of the B. cinerea oscillator by mutation, overexpression of BcFRQ1, or by suppression of its rhythmicity by constant light, abrogates circadian regulation of fungal virulence. By conducting experiments with out-of-phase light:dark cycles, we confirm that indeed, it is the fungal clock that plays the main role in defining the outcome of the Arabidopsis–Botrytis interaction, providing to our knowledge the first evidence of a microbial clock modulating pathogenic traits at specific times of the day.
Circadian clocks are autonomous oscillators based on transcription–translation negative feedback loops (TTFLs), which temporally coordinate a series of processes, from gene expression to metabolism (1, 2). From an evolutionary perspective, these biological clocks have independently emerged at least three times throughout evolution and its molecular description is well documented in photosynthetic organisms like cyanobacteria, algae, and plants, as well as in insects, mammals, birds, and fungi (2). Recent evidence indicates that these machineries provide organisms with adaptive advantages (3–7), whereas the relevance of circadian regulation on the control of immune and defense responses in mammals (8, 9), flies (10, 11), and plants (12–16) has also been documented. Therefore, the time of the day at which a host–pathogen interaction first occurs has measurable consequences in the overall development of the pathogenesis process, at least in all cases where this paradigm has been assessed.
The idea that a circadian clock could modulate the outcome of a plant–pathogen interaction was postulated in recent years (17), and since then, its importance has been experimentally addressed in the plant (18). Nevertheless, the focus has been mostly centered on the circadian system of the host, partially due to the lack of molecular description of oscillatory machineries in the pathogens under study. Thus, no molecular evidence of a circadian clock modulating a pathogen’s behavior has been reported, with the exception of phenotypic evidence observed for insects (16) and malaria parasites (19).
The nonpathogenic fungus Neurospora crassa has been a premier model for chronobiology studies (1, 20, 21) but surprisingly, even after 25 y of the characterization of the central component of the N. crassa clock—the protein FREQUENCY (FRQ) (22)—no other fungal circadian system has been molecularly described. Thus, the work in Neurospora has unveiled that in its core oscillator the transcription factor (TF)/photoreceptor White Collar-1 (WC-1) associates with its partner White Collar-2 (WC-2) forming the White Collar Complex (WCC) to activate the expression of the frequency (frq) gene. FRQ is produced, associates with the FRQ-interacting RNA helicase (FRH), and acts as the negative element of the TTFL shutting down its own expression by inhibiting the WCC (21). As FRQ is progressively phosphorylated, it decreases its negative effect on WCC, and it is finally degraded around the time that a new cycle of frq expression is initiated (23). Thus, this process can be visualized as daily oscillations in frq mRNA and protein levels.
To unravel the role of a circadian clock on pathogenic traits, herein we characterize and describe the circadian oscillator of the plant pathogen Botrytis cinerea. This necrotrophic ascomycete is responsible for the gray mold disease on more than 200 different dicotyledonous plants (24) and is considered the second most important phytopathogenic fungus based on its scientific and economic importance (25). By generating arrhythmic mutants of the well-established B. cinerea B05.10 strain, we determine that in this necrotroph, a functional clock modulates its virulence with a minimal infection potential at dawn. Also, we present evidence that in B. cinerea the FRQ protein also serves noncircadian roles that have not been previously described for its homolog in N. crassa.
Results
B. cinerea Possesses a Functional Circadian Oscillator.
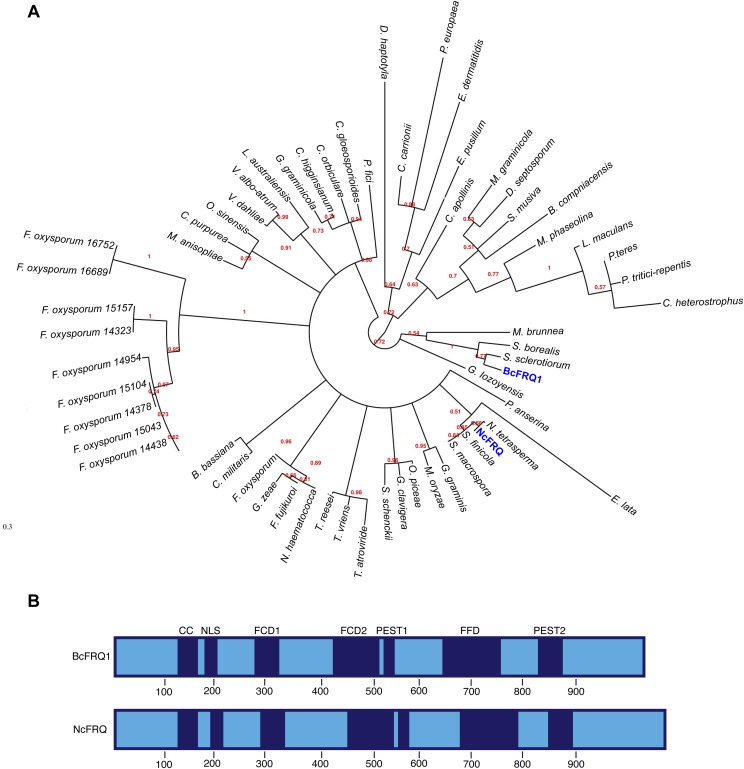
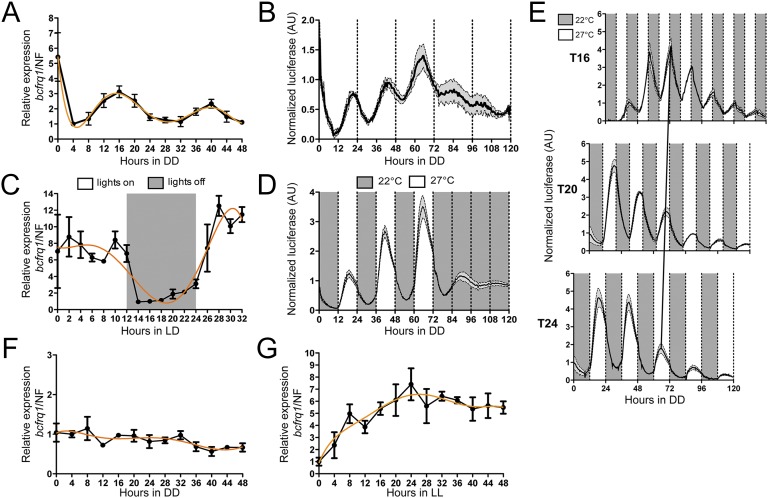
We have identified in the genome of B. cinerea (26) homologs for all main components of the N. crassa core oscillator. The proteins conforming the WCC in B. cinerea (BcWCL1 and BcWCL2) interact (27) and we have shown that BcWCL1 is a TF and photoreceptor, required for the light-activated expression of several transcriptional units, including the B. cinerea frequency gene (bcfrq1) (28). The latter encodes for a protein with 31.3% identity to its Neurospora homolog and contains all main domains described as key for FRQ function (21) (Fig. S1). Consistent with a functional TTFL oscillator, we observed that under free running conditions (FRCs; constant darkness, DD) bcfrq1 mRNA (Fig. 1A) and protein levels—measured as a translational bioluminescent reporter (BcFRQ1-LUC, Fig. S2 and Movie S1)—oscillate daily (Fig. 1B). Moreover, bcfrq1 mRNA levels increase and anticipate the lights-on transition when measured under light:dark cycling conditions (LD) (Fig. 1C), whereas levels of BcFRQ1-LUC exhibit an anticipatory behavior when evaluated under temperature cycles (Fig. 1D), demonstrating that these rhythms can be entrained by external cues, as expected (1).
Fig. S1.
Characterization of BcFRQ1 from B. cinerea. (A) Phylogenetic reconstruction of 60 FRQ-like fungal proteins. FRQ proteins from B. cinerea (BcFRQ1) and N. crassa (NcFRQ) are indicated in blue. (B) Schematic representation of BcFRQ1 and NcFRQ showing the position of the translational start and stop codons, structural domains, and their respective positions. Previously defined functional domains (blue) include: CC, coiled-coil domain; NLS, nuclear localization signal; FCD1 and FCD2, FRQ-CKI interacting domains; FFD, FRQ-FRH interacting domain; PEST1 and PEST2, [proline (P), glutamate (E), serine (S), and threonine (T) rich sequence] domains. Protein domains were inferred from multiple sequence alignments of the FRQ-like proteins.
Fig. 1.
B. cinerea presents a functional circadian oscillator. (A) RT-qPCR showing bcfrq1 mRNA circadian oscillations under FRCs (DD). (B) BcFRQ1-LUC protein levels display circadian rhythms under DD. (C) bcfrq1 mRNA levels oscillate under LD conditions. (D) BcFRQ1-LUC levels reveal an anticipatory behavior under temperature cycles in DD. (E) BcFRQ1-LUC traces confirm entrainment to symmetric temperature cycles of different period length (T16, T20, and T24) under DD. The black diagonal line represents the phase of entrainment. (F) bcfrq1 mRNA levels depend on the bcwcl1 gene. (G) Under LL conditions, bcfrq1 mRNA levels fail to show rhythmic oscillations. For A, C, F, and G, each point, in black, represents mean ± SEM, whereas a trend line is depicted in orange. For B, D, and E, mean values are plotted as a black line, whereas SEM is represented as gray-filled area.
Fig. S2.
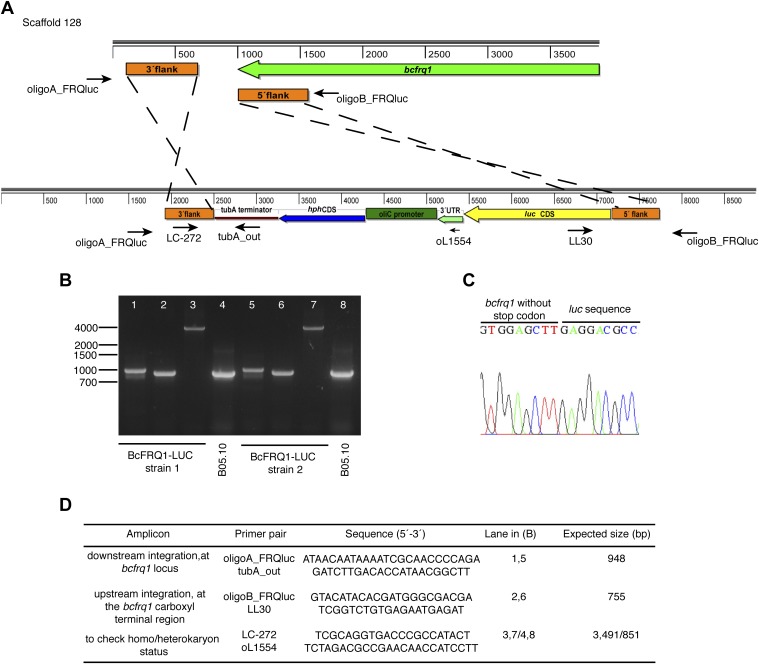
Generation of the BcFRQ1-LUC reporter strain. (A) Replacement strategy scheme showing the expected in-locus insertion of the knock-in cassette. The bcfrq1 gene and its transcriptional orientation (2,958 bp; BC1G_13940, genomic coordinates 37,815–40,772) are represented as a green arrow. The gene replacement cassette used to generate the BcFRQ1-LUC allele is shown bellow. The positions of the regions used for the homologous recombination (orange boxes) are shown. Black arrows show primers used for diagnostic PCRs, indicating their respective relative position and orientation. (B) Diagnostic PCRs showing in-locus integration. (C) Sequence chromatogram showing the junction of bcfrq1 and luc. (D) Primer pairs used in diagnostic PCRs.
To provide additional proof of the functionality of this oscillator, we confirmed the existence of changes in phase angle of the peaks of BcFRQ1-LUC expression when subjected to temperature cycles of different period length (T) (Fig. 1E) (29). Importantly, bcfrq1 levels cease to oscillate in the absence of BcWCL1 (Fig. 1F) or under conditions known to disrupt the circadian oscillator in Neurospora (21), such as in the presence of constant light (LL) (Fig. 1G). Ectopic overexpression of an additional copy of bcfrq1 under the control of a strong promoter (OE::bcfrq1, Fig. S3) abrogates rhythms of endogenous bcfrq1 expression by constantly closing the negative feedback loop and leading to arrhythmicity (Fig. S4). In toto, these results confirm that bcwcl1, bcfrq1, and BcFRQ1 are part of the circuitry of the B. cinerea TTFL circadian oscillator.
Fig. S3.
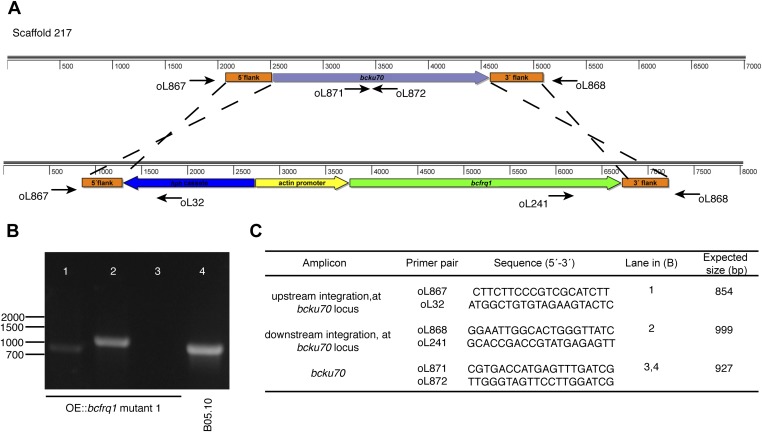
Generation of the OE::bcfrq1 strain. (A) Replacement strategy scheme showing the expected in-locus insertion of the OE::bcfrq1 construct, as well as the used replacement cassette (Bottom), at the bcku70 locus. The latter and its transcriptional orientation (2,063 bp; BC1G_16015, genomic coordinates 15,622–17,684) are represented as a purple arrow. To overexpress bcfrq1, the B. cinerea actin promoter (ActA, Bc1G_08198; 1,000 bp) was used. Genomic regions used for the homologous recombination (orange boxes) are shown. Black arrows show primers used for diagnostic PCRs, indicating their respective relative position and orientation. (B) Diagnostic PCRs showing in-locus integration (a representative mutant is shown). (C) Primer pairs used in diagnostic PCRs.
Fig. S4.
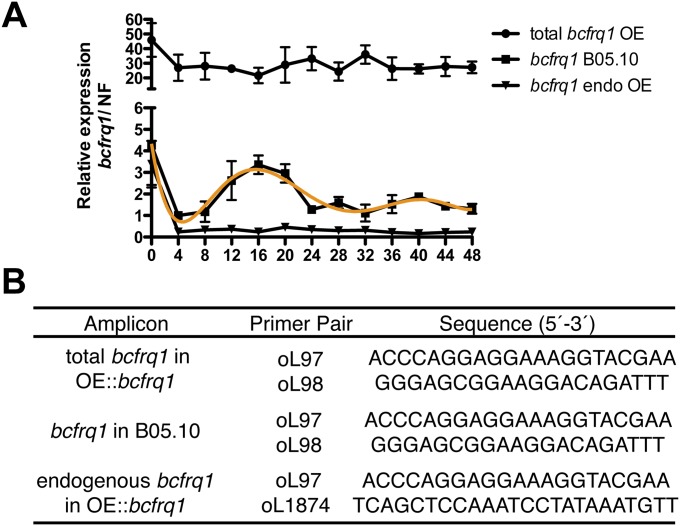
bcfrq1 transcript levels become elevated and arrhythmic in the OE::bcfrq1 genetic background. (A) Total and endogenous bcfrq1 transcript levels were determined by RT-qPCR in the OE::bcfrq1 mutant and compared with bcfrq1 transcript levels in the B05.10 strain. Each point indicates mean values ± SEM and a trend line is depicted in orange. (B) Primer pairs used in the RT-qPCR procedure.
The ∆bcfrq1 Mutant Presents Decreased Macroconidiation, Enhanced Sclerotia Formation, and Impaired Virulence That Is Dependent on the Composition of the Culture Media.
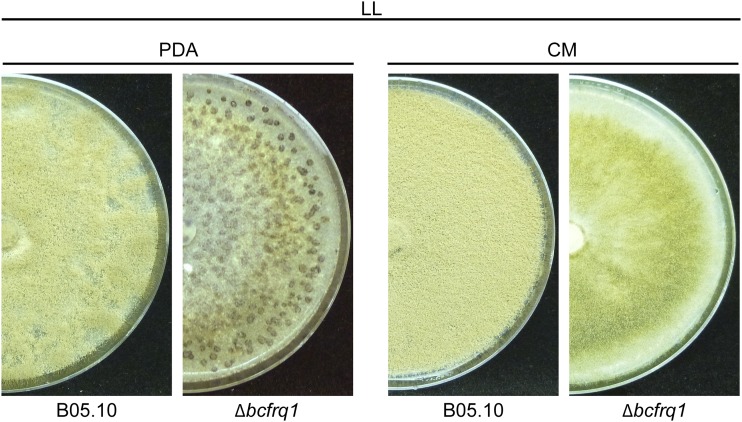
To assess the role of bcfrq1 in B. cinerea circadian regulation, we generated a bcfrq1 deletion strain (Fig. S5). Unexpectedly, we observed an altered developmental phenotype for the Δbcfrq1 mutant, which proved to be dependent on culture media composition. When cultivated on rich undefined potato dextrose agar (PDA) media, Δbcfrq1 forms sclerotia and produces microconidia under LD conditions, in contrast to the B. cinerea B05.10 wild-type (WT) strain (Fig. S6), which always develops sclerotia in the absence of light and produces macroconidia in its presence (28). On the other hand, when using a complete defined medium (CM), macroconidia production was restored in the Δbcfrq1 strain (Fig. S6). As previously described, microconidia are not able to infect (30), and therefore the Δbcfrq1 mutant grown on PDA is weakly pathogenic (Fig. S7). Nevertheless, Δbcfrq1 macroconidia were able to produce lesions of regular size (see below). Importantly, genetic complementation of bcfrq1 in the Δbcfrq1 genetic background restores macroconidia production and thereby full virulence (Fig. S8). These unanticipated phenotypes observed in the Δbcfrq1 strain, which have not been observed in the Neurospora Δfrq strain, suggest novel extra circadian functions for BcFRQ1 (Discussion).
Fig. S5.
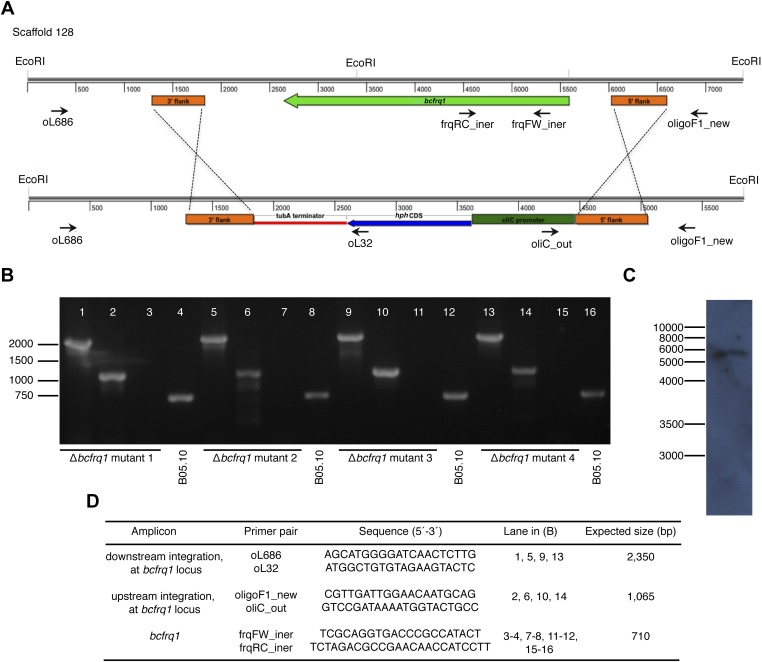
Generation of the Δbcfrq1 strain. (A) Schematic representation of the Δbcfrq1 locus depicting the in-locus insertion of the gene replacement cassette used for generation of the mutant (Bottom). Orange boxes denote the genomic regions used for homologous recombination of the replacement cassette, whereas black arrows show primers used for diagnostic PCRs, indicating their relative position and orientation. (B) Diagnostic PCRs showing in-locus integration. (C) Southern blot hybridization of a representative independent mutant (mutant 1). (D) Primer pairs used in diagnostic PCRs.
Fig. S6.
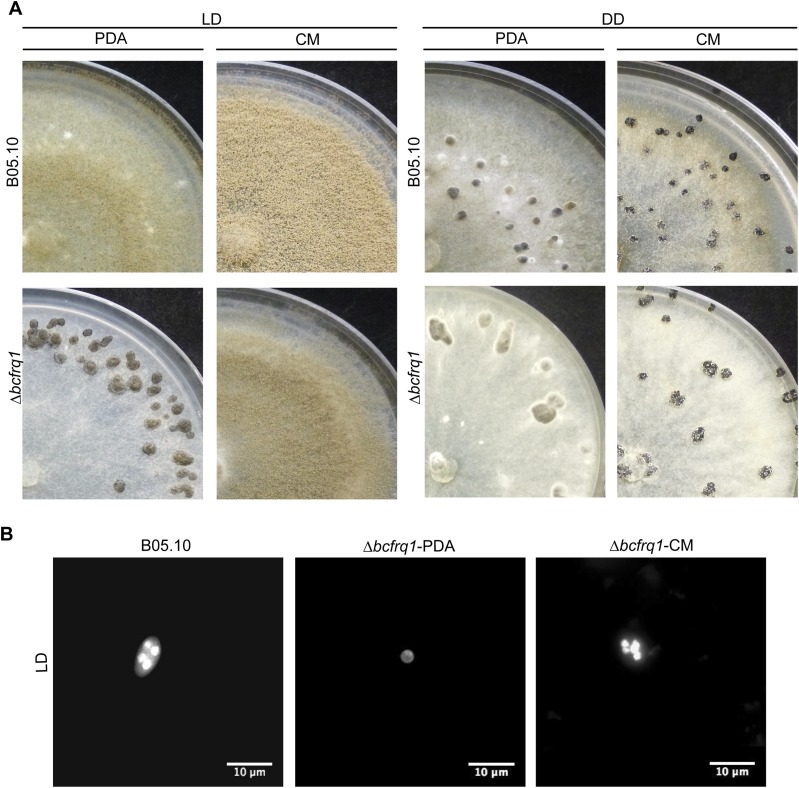
The Δbcfrq1 strain presents a culture media-dependent differentiation phenotype. (A) B05.10 and Δbcfrq1 strains were incubated on PDA and CM plates during 14 d under LD and DD conditions. (B) Hoechst-stained spores of B05.10 and Δbcfrq1 strains visualized by fluorescence microscopy.
Fig. S7.
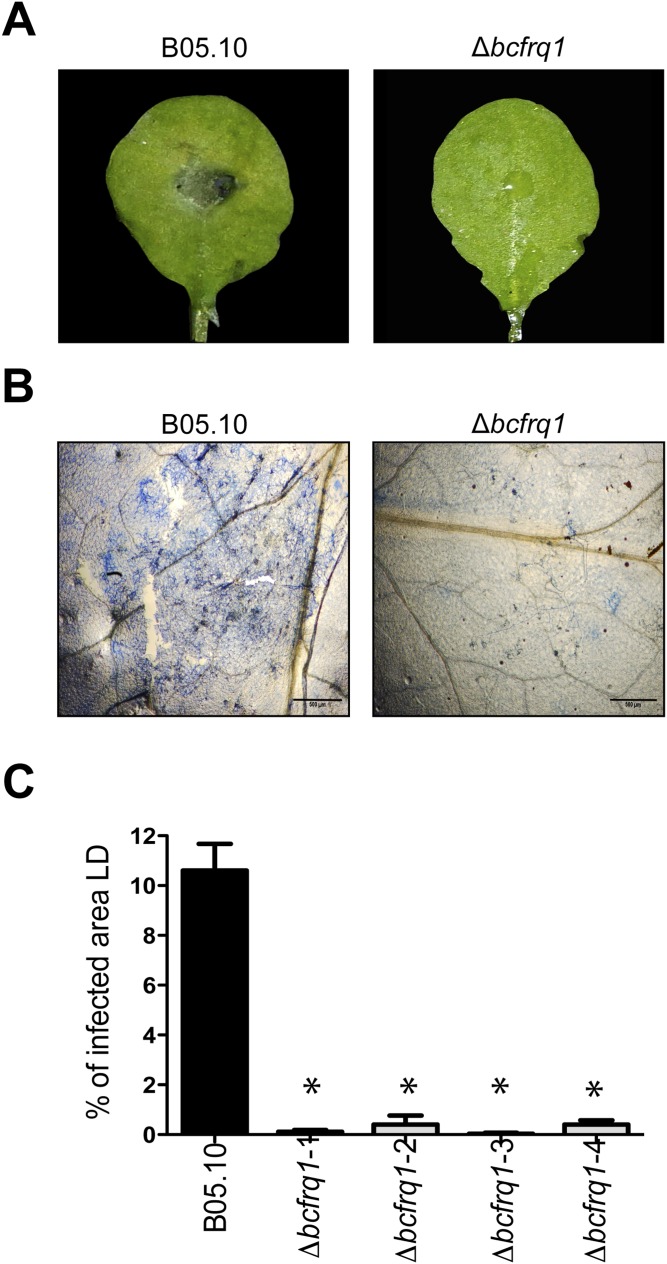
Δbcfrq1 strain presents an altered infection phenotype. (A) Conidia from the B05.10 and microconidia (obtained from PDA plates) of a representative Δbcfrq1 mutant were inoculated on A. thaliana Col-0 plants and kept under LD conditions for 3 d. Representative leaves for each infection condition are shown. (B) Trypan blue staining showing fungal growth on plants at 3 dpi. (Scale bars, 500 μm.) (C) Quantification of the lesion area. Bars represent mean values ± SEM. Significant differences in comparison with the lesion area observed for B05.10 are indicated with asterisks.
Fig. S8.
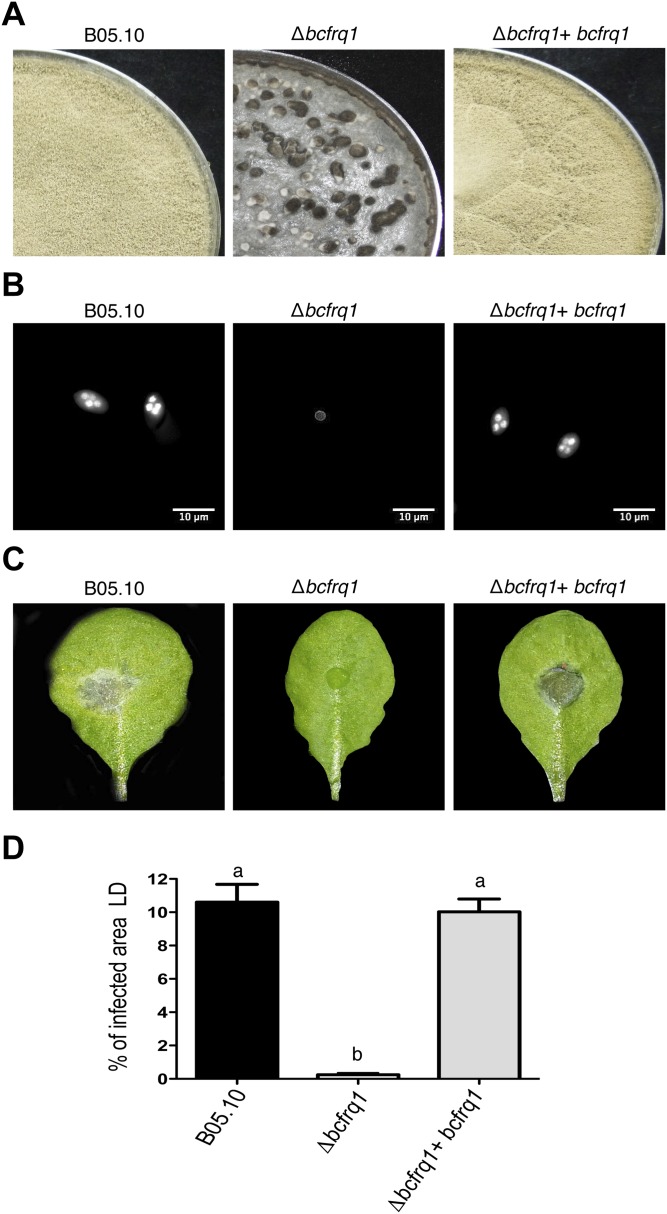
Genetic complementation of the Δbcfrq1 strain allows recovering wild-type phenotype. (A) B05.10, Δbcfrq1 and Δbcfrq1+bcfrq1 strains were incubated on PDA plates for 14 d under LD. (B) Hoechst-stained spores of B05.10 and one representative Δbcfrq1 and Δbcfrq1+bcfrq1 strain visualized by fluorescence microscopy. (C) Lesion spreading of B05.10, Δbcfrq1, and Δbcfrq1+bcfrq1 strains on A. thaliana Col-0 plants incubated under LD conditions for 3 d. (D) Quantification of the lesion area produced by B05.10 and one representative Δbcfrq1 and Δbcfrq1+bcfrq1 mutant at 3 dpi. Bars represent mean values ± SEM. Different letters indicate statistical differences.
The Botrytis–Arabidopsis Interaction Presents a Time-of-Infection–Dependent Outcome.
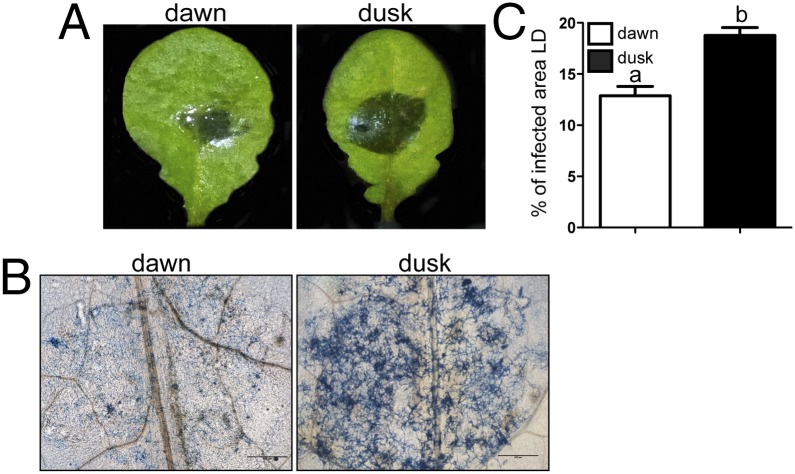
Recent studies (12–15) describing the ability of the plant defense mechanisms to anticipate infection processes, prompted us to test this paradigm when a clock-containing pathogen like B. cinerea is assessed as the attacking organism. The results depicted in Fig. 2 show that the outcome of the host–pathogen interaction, under LD environmental oscillations, clearly depends on the time of the day at which inoculation occurred. When the first physical contact between the fungus and the plant occurs at dawn, a significantly smaller percentage of infected area is observed, compared with when the challenge takes place at dusk (on average, 12.8% and 18.7% of the leaves surface, respectively). Nevertheless, from these results we cannot rule out if the differences are explained by a cyclical behavior—driven by the environment—in one of the organisms or in both, or if instead it is being endogenously (circadianly) controlled. Consequently, we proceeded to define the contribution of each circadian system in this time-of-day effect on the pathogenic outcome.
Fig. 2.
The outcome of the B. cinerea–A. thaliana interaction differs with the time of day. (A) Lesion spreading of B05.10 strain on A. thaliana Col-0 plants. Arabidopsis leaves were inoculated at dawn or dusk, and then plants were incubated in LD for 72 h. Representative pictures are shown. (B) Trypan blue staining showing fungal growth after 3 dpi. (Scale bars, 500 μm.) (C) Quantification of lesion spreading of three independent virulence assays. Bars represent mean ± SEM (different letters indicate statistical differences).
An Endogenous Circadian Clock Controls the Infection Process of B. cinerea on A. thaliana.
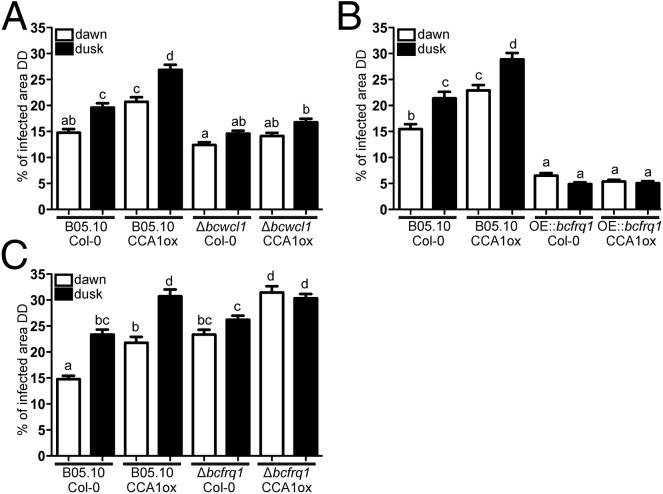
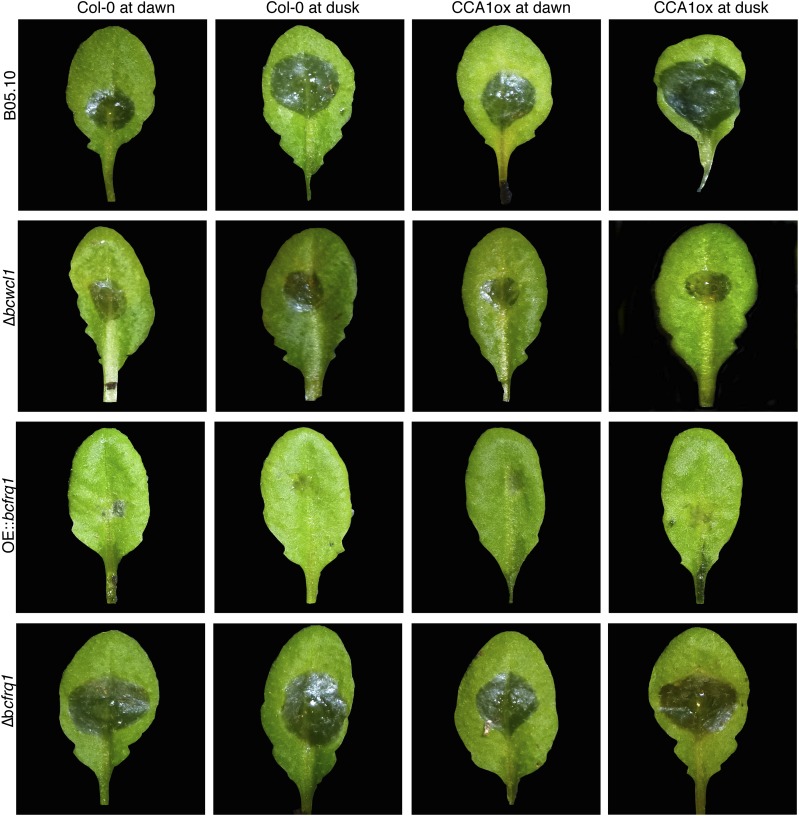
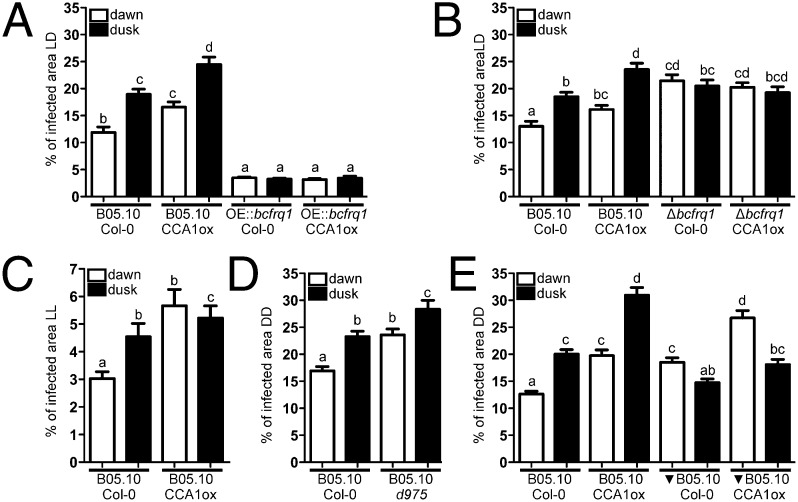
To confirm that the observed effects were not just driven responses to environment cycles (LD), additional analyses were conducted under constant (circadian) conditions. First, to assess the role of the plant circadian clock we used both WT and arrhythmic A. thaliana ecotypes [Columbia 0 (Col-0) and constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1ox), respectively], keeping them for 72 h under DD after their inoculation at dawn or dusk. The results show that, under FRCs, significant differences in the percentage of the leaves infected areas were once again observed when B05.10 WT B. cinerea infected Col-0 plants, with the smaller lesions being produced when inoculation occurred at dawn (14.8% and 19.6% at dawn and dusk, respectively) (Fig. 3A and Fig. S9). Remarkably, when CCA1ox arrhythmic plants were challenged with the B05.10 strain, significant differences were still detected, with a bigger percentage of lesion area at dusk (20.7% and 26.8% at dawn and dusk, respectively; Fig. 3A and Fig. S9). Importantly, the basal level of infection was higher in the CCA1ox genetic background, indicative of an increased susceptibility. The fact that there is a time-of-day difference even when an A. thaliana clock-deficient mutant is used, strongly suggests that the fungal clock is playing a key role in the outcome of this plant–pathogen interaction. In agreement with this observation, the arrhythmic ∆bcwcl1 strain (Fig. 1F) (28) is unable to produce a statistically significant different lesion area when inoculated at dusk compared with dawn, in the Col-0 plant (Fig. 3A and Fig. S9). To confirm this result, arrhythmic CCA1ox plants were challenged with the ∆bcwcl1 mutant, revealing the same behavior (Fig. 3A and Fig. S9).
Fig. 3.
A fungal circadian clock controls the infection process of B. cinerea on A. thaliana. (A) Measurements of lesion spreading of B05.10 and Δbcwcl1 strains. (B) Comparison of lesion spreading between B05.10 and OE::bcfrq1 strains. (C) Lesion spreading of B05.10 and Δbcfrq1 strains. Conidia were inoculated at dawn or dusk on A. thaliana Col-0 and CCA1ox plants, as indicated. Lesions were quantified 3 dpi under DD. Bars represent mean ± SEM (different letters indicate statistical differences).
Fig. S9.
B. cinerea has increased ability to infect at night in rhythmic and clock-null Arabidopsis plants. Representative pictures of lesion spreading of B05.10, Δbcwcl1, OE::bcfrq1, and Δbcfrq1 strains on A. thaliana Col-0 and CCA1ox plants, inoculated with a conidial suspension at dawn or dusk under FRCs (DD).
To exclude the possibility that the loss of temporal variation seen for the infections with ∆bcwcl1 in DD resulted from defects in the blue light signaling pathway (28) rather than by the absence of a functional circadian clock, the arrhythmic OE::bcfrq1 mutant (Fig. S4) was used. Notably, the OE::bcfrq1 strain also lacks the ability to achieve a better infection at night when infecting Col-0 or CCA1ox plants (Fig. 3B and Fig. S9). Surprisingly, this strain not only has lost the circadian modulation of virulence, but moreover it displays smaller lesions and shows the inability to form secondary spreading lesions (Fig. S9, third row).
Finally, using the macroconidia from Δbcfrq1 we further confirmed the influence of the fungal clock in the plant–pathogen circadian phenotype under study. Consistent with the previous observations, infection with this clock-deficient strain resulted in no significant difference in the percentage of infected leaf area when inoculation was performed at dawn or at dusk (Fig. 3C and Fig. S9). In aggregate, these results (Fig. 3) indicate that the outcome of the B. cinerea–A. thaliana interaction is strongly influenced by the fungal circadian clock leading the pathogen to achieve differential infection levels at night versus at day.
Light:Dark Cycles Confirm the Importance of the B. cinerea Circadian Clock in Regulating Virulence Under Environmentally Relevant Conditions.
To obtain further data defining the importance of fungal versus plant clocks in mediating the described interaction, additional infection assays were performed under LD cycling conditions. Fig. 4 A and B depict that, whereas the B. cinerea WT strain presents differences in its infection capability between inoculations at dawn and dusk on both Col-0 and CCA1ox plants, neither of the B. cinerea's clock mutants (OE::bcfrq1 or Δbcfrq1) displayed time-differential infection capabilities. This finding reflects that the differences in virulence observed between dawn and dusk for the B. cinerea WT strain under both DD and LD conditions are indeed due to the control of its circadian clock. Thus, these results are consistent with a scenario in which the fungal clock, rather than an environmentally driven behavior or the plant clock, is largely responsible for the outcome of the interaction.
Fig. 4.
The B. cinerea circadian clock influences the outcome of the interaction under 12:12 photocycles as well as under constant darkness conditions. (A) Disruption of the clock by bcfrq1 overexpression (OE::bcfrq1) affects temporal variation of virulence in LD. (B) There are no differences in the infection behavior observed between dawn and dusk for the Δbcfrq1 strain in LD. (C) Under LL, the Arabidopsis clock—but not the Botrytis clock—provides a time-of-day effect on susceptibility. (D) The B05.10 strain has the ability to produce bigger lesions at dusk in both Col-0 and d975 arrhythmic mutant plants. (E) The relative magnitude of lesions achieved at dawn or dusk follows the time sensed by the fungus and not the plant. WT B05.10 strain was grown under the same LD cycle or 12 h out of phase (represented as a black inverted triangle) than the plants. Arabidopsis were inoculated at dawn or dusk with B. cinerea grown in the same phase or having opposite times. In all graphs, after macroconidia inoculations, WT (Col-0) and mutant plants were kept under the specified LD, LL, or DD regimes. White and black bars indicate conidia inoculation at dawn and dusk, respectively. Lesion area measurements were performed 3 dpi. Bars represent mean ± SEM (different letters indicate statistical differences).
Under Constant Light the A. thaliana Clock Acquires a Dominant Role.
Additional assays were performed under constant light (LL) because this setting allows the plant clock to “free run” (31), whereas the B. cinerea clock (as the one in Neurospora) fails to do so (Fig. 1G). Thus, inoculation of Col-0 plants at dusk with the B05.10 strain showed larger lesions, but notably, no significant differences were observed between dawn and dusk when the CCA1ox plants were challenged with the same fungal strain (Fig. 4C), demonstrating that under LL conditions—and in contrast to what is seen in LD and DD—the A. thaliana circadian clock acquires a preponderant role.
Additional Plant Arrhythmic Mutants and Inverted-Cycle Infection Further Confirm the Relevance of the Fungal Clock Under FRCs.
To provide additional lines of evidence supporting the importance of the fungal versus the plant clock under FRCs (DD), we tested the behavior of WT B. cinerea on the triple mutant of PSEUDO-RESPONSE REGULATORS prr9 prr7 prr5 (d975) arrhythmic Arabidopsis mutant (32). Once again, we observed that despite the absence of a functional clock in the plant, the WT B. cinerea strain is capable of producing differential lesions with larger ones at night, and that moreover, this arrhythmic triple Arabidopsis mutant is—as also observed for CCA1ox—more sensitive to this pathogen (Fig. 4D).
Finally, we proceeded to work with inverted cycles to determine in a different manner the relevance of both clocks. Thus, the fungus was entrained in an inverted light:dark cycle (12 h out of phase) to the one under which the plants were grown. Then we proceeded to infect the plants at dawn or dusk with B. cinerea that had the same internal time or that had the opposite time of the plants, maintaining them thereafter in FRCs for 72 h. Plants inoculated with the fungus that had been under the same LD entrainment recapitulated the type of results already shown (Fig. 3). Notably, in the inverted cycle experiment, we observed that plants challenged at dawn showed larger lesions as the fungus that was inoculated on them had dusk time, whereas plants inoculated at dusk with fungus that had dawn time displayed smaller lesions (Fig. 4E). In aggregate, these experiments provide additional and compelling evidence supporting the importance of the B. cinerea clock in controlling its pathogenic potential.
Discussion
In B. cinerea B05.10, rhythms in macroconidiation can be observed under light:dark cycles, but not under constant darkness (28). This result differs from what is known in Neurospora, where conidiation rhythms can also be detected in the latter condition, but only in strains containing a mutant allele of the ras-1 gene (ras-1bd), which leads to enhanced circadian output (33). The absence of overt rhythms in B. cinerea could be due to a similar reason, to the divergence of output pathways, or to the absence of a functional clock. Therefore, motivated by the interest in understanding the impact of circadian regulation on fungal virulence, we were able to establish the existence of an endogenous circadian clock in this ascomycete by using several molecular assays. bcfrq1 mRNA levels oscillate in LD as well as under free running conditions, whereas rhythms are lost under LL stimulation, or upon deletion of bcwcl1 (Fig. 1 A, C, F, and G). These results are consistent with the notion that in N. crassa frq transcript levels oscillate in a cyclical environment (1) or in constant darkness (34). Likewise, in Neurospora a dysfunctional circadian clock is evidenced under LL—due to high frq expression levels (35)—and also in a ∆wc-1 strain (36). The latter observations are also consistent with the fast light responsiveness of bcfrq1 mRNA upon light stimulation, which depends on bcwcl1 (28).
In addition, by overexpressing an extra copy of bcfrq1 under a constitutive promoter, we demonstrated that bcfrq1 expression levels at the endogenous locus become low and arrhythmic (Fig. S4), recapitulating the iconic experiment performed by Aronson et al. (34) in N. crassa, which provided solid evidence for the central role of negative feedback loops in circadian core mechanisms. Additionally, we showed that BcFRQ1 protein levels not only oscillate under DD culture conditions, but also anticipate changes in temperature cycles and can be entrained by cycles of different T (Fig. 1 B, D, and E). Together, these data demonstrate the presence of a functional circadian oscillator in B. cinerea, closing the gap in knowledge regarding clock mechanisms in fungi other than Neurospora. Whereas circadian rhythms (output) have been described in Aspergillus (37) and yeast (38), and more recently in bioluminescence in a mushroom (7), the molecular bases of the underlying oscillators remain obscure.
Remarkably, the absence of bcfrq1 has a strong impact on sexual/asexual reproduction and virulence, which is further modulated by modifying media culture conditions (Fig. S6). Such a widespread effect of bcfrq1 in B. cinerea biology clearly differs with what has been reported for the frq knockout strain (frq10) of N. crassa, which shows no reported phenotypic alterations related to the production of conidia (39). This result suggests novel extra circadian functions for BcFRQ1, because the developmental phenotype of ∆bcfrq1 is observed even under conditions where the fungal clock is already disrupted by a constant environmental cue (LL; Fig. S10), and also because in the absence of BcWCL1 (where the clock is also broken) multinucleated conidia capable of infection are easily produced (28). With respect to the latter, we noticed that the phenotypes of Δbcwcl1 and Δbcfrq1 strains are quite different, even though one of the molecular consequences of BcWCL1 absence is low level of bcfrq1 transcripts (28). Therefore, although part of the phenotypes observed in the Δbcwcl1 strain should overlap with the ones seen in Δbcfrq1, this overlap does not occur. A plausible explanation resides in the known ability of FRQ to modify WCC activity. Thus, in Neurospora, the absence of FRQ leads to hyperactivity of WC-1 and its destabilization due to transcription-associated turnover (40). So, defects in signaling connected with BcWCL1 and other associated proteins involved in light signaling and development (i.e., BcVEL1) (41) could be impacted by the absence of BcFRQ1. Importantly, media composition can revert the sexually related phenotype of B. cinerea, suggesting that nutritional cues can partially override some of the developmental alterations observed in this mutant. The mechanisms behind these extracircadian functions of BcFRQ1 will be the subject of a different study.
Fig. S10.
The Δbcfrq1 strain develops sclerotia under LL culture conditions. B05.10 and Δbcfrq1 strains were incubated on PDA and CM plates during 21 d under LL conditions.
Previous studies have highlighted the ways by which circadian clocks regulate plant defense mechanisms against bacteria, insects, and oomycetes (18). However, this paradigm had not been tested for fungi in general and for necrotrophic fungi in particular. Based on a typical type of assay conducted for these experiments (infection of WT and clock-deficient Arabidopsis plants at different circadian times followed by the development of infection under constant light conditions) the conclusion would have been that the plant clock solely regulates this interaction. Indeed, as we demonstrated herein, in LL, a condition known to reduce B. cinerea basal infection levels (28), the environmental disruption of the fungal clock does not dramatically change the time-dependent outcome of the interaction, whereas in return the plant clock seems to adopt an important role as revealed by the data in the CCA1ox mutant (Fig. 4C), results that are aligned to what has been previously described for bacterial and oomycete pathogens (12, 13, 15). Nevertheless, by generating and evaluating B. cinerea clock mutants, we have demonstrated that in this fungus, a functional clock modulates its virulence under DD (Figs. 3 and 4D) and also under conditions that better reflect the real world (LD cycling conditions) (Fig. 4 A and B). In addition, the experiments conducted with inverted cycles (Fig. 4E) are key in ruling out potentially strange explanations of the results, further supporting the importance of the fungal clock in the outcome of the Botrytis–Arabidopsis interaction.
Intriguingly, once we compensated Δbcfrq1’s failure to produce microconida—by manipulating media composition—we observed that its macroconidia have a bigger infection capability than the B05.10 WT strain. However, the temporal difference to infect better at night is lost (Figs. 3C and 4B). It is intriguing that when removing a central clock component, such as BcFRQ1 (but not BcWCL1), the infection is significantly enhanced. Nonetheless, similar results have been described for A. thaliana: Zhang et al. (15) reported that arrhythmic CCA1ox plants have enhanced resistance to the oomycete pathogen Hpa, which is in agreement with the results published by Wang et al. (13). Nevertheless, Bhardwaj et al. (12) and Zhang et al. (15) showed that the CCA1ox plants have enhanced susceptibility and loss of temporal regulation in defense against the virulent bacteria Pseudomonas syringae. Thus, although one would intuitively predict that an arrhythmic plant is always more prone to infection (due to the loss of the adaptive advantage of the clock), some of the results in A. thaliana reveal the opposite for the CCA1ox strain. Thus, as it was described for the plant clock protein CCA1, BcFRQ1 could be acting on other output pathways not regulated by the circadian clock (42), further complicating the molecular dissection of clock regulation on virulence mechanisms.
In this fencing contest between these two organisms, the dominance of each clock is judge and the results presented here indicate that under DD and LD conditions, the B. cinerea clock plays a fundamental role in the interaction, thus explaining the major part of the temporal variation of the infection process. In this regard, the most important plant defense mechanism against necrotrophic infection is the jasmonic acid (JA) pathway (43). Interestingly, a circadian gating effect allows higher JA-signaling activity response during the morning (14, 16). Although it is reasonable to hypothesize that the circadian clock of B. cinerea allows the fungus to anticipate the JA plant defense response, optimizing and generating its maximal virulence potential at dusk, the experiments using out-of-phase light:dark cycles (Fig. 4E) argue against this possible explanation because they show that the fungus can achieve maximum virulence even at dawn, as long as its internal clock indicates dusk time. A likely explanation, therefore, is that fungal virulence potential can override plant defense mechanisms. Further studies will help us to elucidate the mechanisms by which the circadian clock is modulating the pathogenic potential of this hostile organism at different times of the day.
Materials and Methods
B. cinerea Strains and Culture Conditions.
Strain B05.10 (26) of B. cinerea Pers.: Fr. [Botryotinia fuckeliana (de Bary) Whetzel], originally isolated from Vitis vinifera (Germany), was used as the recipient strain for genetic modifications. B. cinerea strains were cultivated in Petri dishes containing synthetic CM (44), PDA (Applichem), or Gamborg B5 (Duchefa Biochemie) supplemented with 2% (wt/vol) glucose. The strains were grown at 20 °C using Percival incubators, equipped with cool white light fluorescent tubes (light intensity up to 100 μM/m2/s; wavelength 400–720 nm) in a 12:12-h LD regime.
Time Course Experiments, RNA Extraction, and Real-Time Quantitative PCR.
All experiments were conducted in Percival incubators kept at 20 °C. For circadian experiments under DD conditions, B05.10, Δbcwcl1, and OE::bcfrq1 strains were grown on PDA plates covered with cellophane under LL conditions for 24 h and then transferred to DD every 4 h. For LL circadian experiments, B05.10 was grown on PDA plates covered with cellophane under DD for 24 h and then transferred to LL every 4 h. After 48 h, mycelia samples (obtained from independent PDA plates) were harvested. For the LD time course experiment, the B05.10 strain was inoculated on cellophane-covered PDA Petri dishes under LD culture conditions, during 96 h. Thereafter, cultures were harvested every 2 h, during 32 h. RNA isolation and real-time quantitative PCR (RT-qPCR) procedures were conducted as previously described (28). Expression values refer to the culture grown 4 h in the dark (1 = DD4) for Fig. 1 A, C, and F (1 = DD16) and for Fig. 1G expression levels refer to the culture obtained in the dark (1 = LL0). The plot for Fig. 1A represents four biological with three technical replicates each and for Fig. 1 C, F, and G two biological with two technical replicates each. In the case of Fig. S4A, transcript levels refer to the culture grown 4 h in the dark determined for the B05.10 strain (1 = DD4) and all of the plots represent three biological replicates. Each point, in black, represents mean values ± SEM, whereas a trend line is depicted in orange.
Cloning of Replacements Cassettes.
All of the replacement cassettes were assembled using a previously described strategy (28). The fully codon optimized (oluc) N. crassa luciferase gene (45) was used to generate a BcFRQ1-LUC reporter, as described for Neurospora frq (46).
Transformation of B. cinerea.
B. cinerea transformations were carried out using protoplast-mediated transformation as previously described (27).
Southern Blot.
PCR-verified mutants for Δbcfrq1 were checked by Southern blot hybridization using the DIG Easy Hyb Hybridization solution (Roche) and the PCR DIG Probe Synthesis Kit (Roche) following manufacturer’s instructions.
Assessment of Rhythmic Luminescence.
The luciferase reporter BcFRQ1-LUC (two biological replicates) was analyzed using a PIXIS-CCD camera (Princeton Instruments) at 20 °C, using Petri plates filled with Gamborg B5, as described (45). Before being transferred to DD, plates were entrained for 3 d in LD. The reporter strain was also monitored under temperature entrainment conditions using steps from 22 °C to 27 °C under DD conditions (12:12) and by using 22 °C to 27 °C temperature cycles of varying period length (16, 20, and 24 T). Bioluminescence traces were acquired using the WinView software and analyzed in more detail using a custom-made ImageJ macro. For Fig. 1 B, D, and E, the plots represent the mean of two biological with two technical replicates each.
Fluorescence Microscopy.
Conidia from 2-wk-old culture were collected by glass wool filtration, diluted in water to 108 conidia per milliliter and then pelleted by centrifugation (1,000 × g for 20 min). Thereafter, conidia were stained with a dilution of 1 mg/mL of Hoechst 33342 (Life Technologies) for 15 min in darkness and then rinsed twice with 1× phosphate-buffered saline. Conidia were mounted in 30% (vol/vol) glycerol. Fluorescence microscopy was performed with Cytation 3 Cell Imaging Multi-Mode Reader (Biotek). Photos were obtained using Gen5 Data Analysis Software (Biotek).
A. thaliana Growth.
Accession Col-0, constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1ox), and the triple mutant of PRR9, PRR7, and PRR5 (d975) were grown at 22 °C for 14 d under LL and then for another 2 wk at 20 °C under LD using Percival incubators. CCA1ox and d975 plants are arrhythmic with respect to circadian outputs in LL, DD, and LD (15, 32, 47, 48).
Virulence Assays.
Before infection, A. thaliana and B. cinerea were grown under LD conditions for 2 wk at 20 °C. Thereafter, leaves of approximately 1-mo-old plants were inoculated with B. cinerea conidia, which were obtained essentially as described previously (28). Briefly, spores were harvested from the same B. cinerea plate at dawn or dusk, and 7 µL of conidial suspensions of 2 × 105 spores per milliliter were used to inoculate unwounded A. thaliana leaves. Infections were done at dawn or dusk, according to the photoperiod of the plant growth chamber. All inoculated plants were kept inside plastic boxes at 20 °C under a humid environment, in LL, LD, or DD conditions for 72 h and then immediately analyzed (3 d postinoculation, dpi). Lesions on A. thaliana leaves—at least 180 infected leaves per B. cinerea strain and condition—were measured semiautomatically with ImageJ software using an external calibration scale. Because the different A. thaliana ecotypes have different leaf sizes, the lesion area was calculated by measuring the total and infected area of the leaf, being the total area of the leaf 100%. Therefore, measurements in the graphs are expressed as percentage. For Fig. 2B, the quantification was obtained from three independent virulence assays; the Mann–Whitney test was performed and P values < 0.05 were used as cutoff values for showing significance. For Figs. 3 and 4, the results were obtained from four and three independent virulence assays, respectively. For Figs. 3 and 4 A, B, and E, three-way analysis of variance and Tukey's HSD (P < 0.05) were used. The statistical analysis for Fig. 4 C and D was done by using two-way analysis of variance and Tukey's honest significant difference test (P < 0.05). For Figs. S7 and S8, two independent virulence assays were done; the Kruskal–Wallis test resulted in P < 0.05.
Trypan Blue Staining.
Detection of B. cinerea hyphae in infected plant tissues was done as previously described (28).
Supplementary Material
Acknowledgments
We thank Julia Schumacher and Paul Tudzynski (Westfälische Wilhelms-Universität) for advice and guidance. This work was supported by Millennium Nucleus for Fungal Integrative and Synthetic Biology Grant NC120043 (to L.F.L.), Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT) Grant 1131030 (to L.F.L.), FONDECYT Inicio 11140678 (to P.C.), and International Centre for Genetic Engineering and Biotechnology CHI09-02 (to L.F.L.). M.A.H. is a predoctoral fellow supported by CONICYT (Comisión Nacional de Investigación Científica y Tecnológica) Chile (AT-24121100).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1508432112/-/DCSupplemental.
References
- 1.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96(2):271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 2.Bell-Pedersen D, et al. Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat Rev Genet. 2005;6(7):544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dodd AN, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309(5734):630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 4.Lowrey PL, Takahashi JS. Genetics of circadian rhythms in mammalian model organisms. Adv Genet. 2011;74:175–230. doi: 10.1016/B978-0-12-387690-4.00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA. 1998;95(15):8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yerushalmi S, Green RM. Evidence for the adaptive significance of circadian rhythms. Ecol Lett. 2009;12(9):970–981. doi: 10.1111/j.1461-0248.2009.01343.x. [DOI] [PubMed] [Google Scholar]
- 7.Oliveira AG, et al. Circadian control sheds light on fungal bioluminescence. Curr Biol. 2015;25(7):964–968. doi: 10.1016/j.cub.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheiermann C, Kunisaki Y, Frenette PS. Circadian control of the immune system. Nat Rev Immunol. 2013;13(3):190–198. doi: 10.1038/nri3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curtis AM, Bellet MM, Sassone-Corsi P, O'Neill LA. Circadian clock proteins and immunity. Immunity. 2014;40(2):178–186. doi: 10.1016/j.immuni.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Lee JE, Edery I. Circadian regulation in the ability of Drosophila to combat pathogenic infections. Curr Biol. 2008;18(3):195–199. doi: 10.1016/j.cub.2007.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kahrstrom CT. Host response: Phagocytosis runs like clockwork. Nat Rev Microbiol. 2012;10(3):162. doi: 10.1038/nrmicro2751. [DOI] [PubMed] [Google Scholar]
- 12.Bhardwaj V, Meier S, Petersen LN, Ingle RA, Roden LC. Defence responses of Arabidopsis thaliana to infection by Pseudomonas syringae are regulated by the circadian clock. PLoS ONE. 2011;6(10):e26968. doi: 10.1371/journal.pone.0026968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W, et al. Timing of plant immune responses by a central circadian regulator. Nature. 2011;470(7332):110–114. doi: 10.1038/nature09766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin J, Heidrich K, Sanchez-Villarreal A, Parker JE, Davis SJ. TIME FOR COFFEE represses accumulation of the MYC2 transcription factor to provide time-of-day regulation of jasmonate signaling in Arabidopsis. Plant Cell. 2012;24(6):2470–2482. doi: 10.1105/tpc.111.095430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang C, et al. Crosstalk between the circadian clock and innate immunity in Arabidopsis. PLoS Pathog. 2013;9(6):e1003370. doi: 10.1371/journal.ppat.1003370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodspeed D, Chehab EW, Min-Venditti A, Braam J, Covington MF. Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc Natl Acad Sci USA. 2012;109(12):4674–4677. doi: 10.1073/pnas.1116368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roden LC, Ingle RA. Lights, rhythms, infection: The role of light and the circadian clock in determining the outcome of plant-pathogen interactions. Plant Cell. 2009;21(9):2546–2552. doi: 10.1105/tpc.109.069922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma M, Bhatt D. The circadian clock and defence signalling in plants. Mol Plant Pathol. 2014;16(2):210–218. doi: 10.1111/mpp.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mideo N, Reece SE, Smith AL, Metcalf CJ. The Cinderella syndrome: Why do malaria-infected cells burst at midnight? Trends Parasitol. 2013;29(1):10–16. doi: 10.1016/j.pt.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunlap JC, et al. A circadian clock in Neurospora: How genes and proteins cooperate to produce a sustained, entrainable, and compensated biological oscillator with a period of about a day. Cold Spring Harb Symp Quant Biol. 2007;72:57–68. doi: 10.1101/sqb.2007.72.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montenegro-Montero A, Larrondo LF. The Neurospora circadian system: From genes to proteins and back in less than 24 hours. In: Kasbekar D, McCluskey K, editors. Neurospora. Genomics and Molecular Biology. Caister Academic Press; Norfolk, UK: 2013. [Google Scholar]
- 22.McClung CR, Fox BA, Dunlap JC. The Neurospora clock gene frequency shares a sequence element with the Drosophila clock gene period. Nature. 1989;339(6225):558–562. doi: 10.1038/339558a0. [DOI] [PubMed] [Google Scholar]
- 23.Larrondo LF, Olivares-Yanez C, Baker CL, Loros JJ, Dunlap JC. Circadian rhythms. Decoupling circadian clock protein turnover from circadian period determination. Science. 2015;347(6221):1257277. doi: 10.1126/science.1257277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Staats M, van Baarlen P, van Kan JA. Molecular phylogeny of the plant pathogenic genus Botrytis and the evolution of host specificity. Mol Biol Evol. 2005;22(2):333–346. doi: 10.1093/molbev/msi020. [DOI] [PubMed] [Google Scholar]
- 25.Dean R, et al. The top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol. 2012;13(4):414–430. doi: 10.1111/j.1364-3703.2011.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amselem J, et al. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 2011;7(8):e1002230. doi: 10.1371/journal.pgen.1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schumacher J. Tools for Botrytis cinerea: New expression vectors make the gray mold fungus more accessible to cell biology approaches. Fungal Genet Biol. 2012;49(6):483–497. doi: 10.1016/j.fgb.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 28.Canessa P, Schumacher J, Hevia MA, Tudzynski P, Larrondo LF. Assessing the effects of light on differentiation and virulence of the plant pathogen Botrytis cinerea: Characterization of the White Collar Complex. PLoS ONE. 2013;8(12):e84223. doi: 10.1371/journal.pone.0084223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pregueiro AM, et al. Assignment of an essential role for the Neurospora frequency gene in circadian entrainment to temperature cycles. Proc Natl Acad Sci USA. 2005;102(6):2210–2215. doi: 10.1073/pnas.0406506102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jarvis WR. Botryotinia and Botrytis Species. Taxonomy and Pathogenicity. Ottawa: Research Branch, Canada Department of Agriculture; Ottawa, Canada: 1977. [Google Scholar]
- 31.McClung CR. CIRCADIAN RHYTHMS IN PLANTS. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:139–162. doi: 10.1146/annurev.arplant.52.1.139. [DOI] [PubMed] [Google Scholar]
- 32.Nakamichi N, Kita M, Ito S, Yamashino T, Mizuno T. PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 2005;46(5):686–698. doi: 10.1093/pcp/pci086. [DOI] [PubMed] [Google Scholar]
- 33.Belden WJ, et al. The band mutation in Neurospora crassa is a dominant allele of ras-1 implicating RAS signaling in circadian output. Genes Dev. 2007;21(12):1494–1505. doi: 10.1101/gad.1551707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aronson BD, Johnson KA, Loros JJ, Dunlap JC. Negative feedback defining a circadian clock: Autoregulation of the clock gene-frequency. Science. 1994;263(5153):1578–1584. doi: 10.1126/science.8128244. [DOI] [PubMed] [Google Scholar]
- 35.Crosthwaite SK, Loros JJ, Dunlap JC. Light-induced resetting of a circadian clock is mediated by a rapid increase in frequency transcript. Cell. 1995;81(7):1003–1012. doi: 10.1016/s0092-8674(05)80005-4. [DOI] [PubMed] [Google Scholar]
- 36.Lee K, Dunlap JC, Loros JJ. Roles for WHITE COLLAR-1 in circadian and general photoperception in Neurospora crassa. Genetics. 2003;163(1):103–114. doi: 10.1093/genetics/163.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greene AV, Keller N, Haas H, Bell-Pedersen D. A circadian oscillator in Aspergillus spp. regulates daily development and gene expression. Eukaryot Cell. 2003;2(2):231–237. doi: 10.1128/EC.2.2.231-237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eelderink-Chen Z, et al. A circadian clock in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2010;107(5):2043–2047. doi: 10.1073/pnas.0907902107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aronson BD, Johnson KA, Dunlap JC. Circadian clock locus frequency: Protein encoded by a single open reading frame defines period length and temperature compensation. Proc Natl Acad Sci USA. 1994;91(16):7683–7687. doi: 10.1073/pnas.91.16.7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi M, Collett M, Loros JJ, Dunlap JC. FRQ-interacting RNA helicase mediates negative and positive feedback in the Neurospora circadian clock. Genetics. 2010;184(2):351–361. doi: 10.1534/genetics.109.111393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schumacher J, et al. Natural variation in the VELVET gene bcvel1 affects virulence and light-dependent differentiation in Botrytis cinerea. PLoS ONE. 2012;7(10):e47840. doi: 10.1371/journal.pone.0047840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McClung CR. Plant biology: Defence at dawn. Nature. 2011;570:44–45. doi: 10.1038/470044a. [DOI] [PubMed] [Google Scholar]
- 43.Browse J. Jasmonate passes muster: A receptor and targets for the defense hormone. Annu Rev Plant Biol. 2009;60:183–205. doi: 10.1146/annurev.arplant.043008.092007. [DOI] [PubMed] [Google Scholar]
- 44.Pontecorvo G, Roper JA, Hemmons LM, Macdonald KD, Bufton AW. The genetics of Aspergillus nidulans. Adv Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- 45.Gooch VD, et al. Fully codon-optimized luciferase uncovers novel temperature characteristics of the Neurospora clock. Eukaryot Cell. 2008;7(1):28–37. doi: 10.1128/EC.00257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larrondo LF, Loros JJ, Dunlap JC. High-resolution spatiotemporal analysis of gene expression in real time: in vivo analysis of circadian rhythms in Neurospora crassa using a FREQUENCY-luciferase translational reporter. Fungal Genet Biol. 2012;49(9):681–683. doi: 10.1016/j.fgb.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakamichi N, et al. Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 2009;50(3):447–462. doi: 10.1093/pcp/pcp004. [DOI] [PubMed] [Google Scholar]
- 48.Wang ZY, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93(7):1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.