Significance
Centromeres are crucial as they avoid genomic instability during mitosis, but the mechanisms involved in their assembly and maintenance are not yet fully elucidated in eukaryotes. Here, we describe a previously unidentified aspect of centromere regulation mediated by γ-tubulin complex protein 3-interacting proteins (GIPs). Our data correlate centromere assembly and cohesion through the recruitment of specific protein complexes in the nucleus. Due to the conservation of GIPs/mitotic spindle organizing protein 1 among fungi, mammals, and plants, our results open a new field of investigation for centromere regulation.
Keywords: centromere assembly, centromeric cohesion, ploidy stability, Arabidopsis, MZT1
Abstract
Centromeres play a pivotal role in maintaining genome integrity by facilitating the recruitment of kinetochore and sister-chromatid cohesion proteins, both required for correct chromosome segregation. Centromeres are epigenetically specified by the presence of the histone H3 variant (CENH3). In this study, we investigate the role of the highly conserved γ-tubulin complex protein 3-interacting proteins (GIPs) in Arabidopsis centromere regulation. We show that GIPs form a complex with CENH3 in cycling cells. GIP depletion in the gip1gip2 knockdown mutant leads to a decreased CENH3 level at centromeres, despite a higher level of Mis18BP1/KNL2 present at both centromeric and ectopic sites. We thus postulate that GIPs are required to ensure CENH3 deposition and/or maintenance at centromeres. In addition, the recruitment at the centromere of other proteins such as the CENP-C kinetochore component and the cohesin subunit SMC3 is impaired in gip1gip2. These defects in centromere architecture result in aneuploidy due to severely altered centromeric cohesion. Altogether, we ascribe a central function to GIPs for the proper recruitment and/or stabilization of centromeric proteins essential in the specification of the centromere identity, as well as for centromeric cohesion in somatic cells.
In eukaryotes, centromeres play a critical role in accurate chromosome segregation and in the maintenance of genome integrity through their regulated assembly and the maintenance of their cohesion until anaphase. Centromeres consist each of a central core (1) characterized epigenetically by the recruitment of the histone H3 variant CENH3 (CENP-A in animals). Extensive studies are still ongoing to identify the regulatory factors for loading and maintenance of CENH3 at centromeres. In yeast, suppressor of chromosome missegregation protein 3 was identified as a specific chaperone for CENP-A loading (2). In animals, the Mis18 complex, including the CENH3 assembly factor Kinetochore Null 2 (KNL2; also called Mis18BP1), recruits the cell cycle-dependent maintenance and deposition factor of CENP-A, HJURP (Holliday junction recognition protein), to centromeres (3). Recently, two Mis18-complex components, Eic1 and Eic2, were identified in fission yeast (4). Whereas Eic1 promotes CENH3 loading and maintenance, Eic2 is recruited at centromeres independently of its association with Mis18. Together with CENH3, the conserved kinetochore assembly protein CENP-C participates in pericentromeric cohesin recruitment (5). The CENH3 loading machinery changed rapidly during evolution, and a CENH3 chaperone has not been identified in plants thus far. Moreover, nothing is known about a possibly conserved interplay between CENH3 loading and sister chromatid cohesion at centromeres. Recently, the plant homolog of KNL2 was proposed as an upstream component for CENH3 deposition at centromeres (Table S1) (6). Finally, the regulation of centromeric complex positioning at the nuclear envelope environment is still elusive in plants.
Table S1.
Gene accession numbers
| Gene | Accession number |
| GIP1 | At4G09550 |
| GIP2 | At1g73790 |
| CENH3 | At1g01370 |
| CENP-C | At1g15660 |
| KNL2 | At5G02520 |
| CTF7 | At4g31400 |
| MAD3-1 | At2g33560 |
| MAD3-2 | At5g05510 |
Previously, we characterized the γ-tubulin complex protein 3-interacting proteins (GIPs), GIP1 and GIP2 (Table S1), as essential for the recruitment of γ-tubulin complexes at microtubule (MT) organizing centers in Arabidopsis (7, 8). This function seems conserved in the human and Schizosaccharomyces pombe GIP homologs named mitotic spindle organizing protein 1 (MZT1) (9–11). More recently, we localized GIPs at the nucleoplasm periphery, close to chromocenters, where they modulate the nuclear architecture (12, 13). Here, we exploit the various phenotype gradations of knockdown gip1gip2 mutants to investigate the role of GIPs at centromeres. We demonstrate that GIPs are required for CENH3 stabilization and centromere cohesion in Arabidopsis. We further show that these nuclear functions are not related to mitotic checkpoint controls and occur in addition to the previously established role of GIPs/MZT1 in spindle microtubule robustness. Our results highlight a previously unidentified aspect of centromere regulation mediated by GIPs/MZT1 to maintain genomic and ploidy stability.
Results and Discussion
GIPs Are Required for Proper Centromere Cohesion.
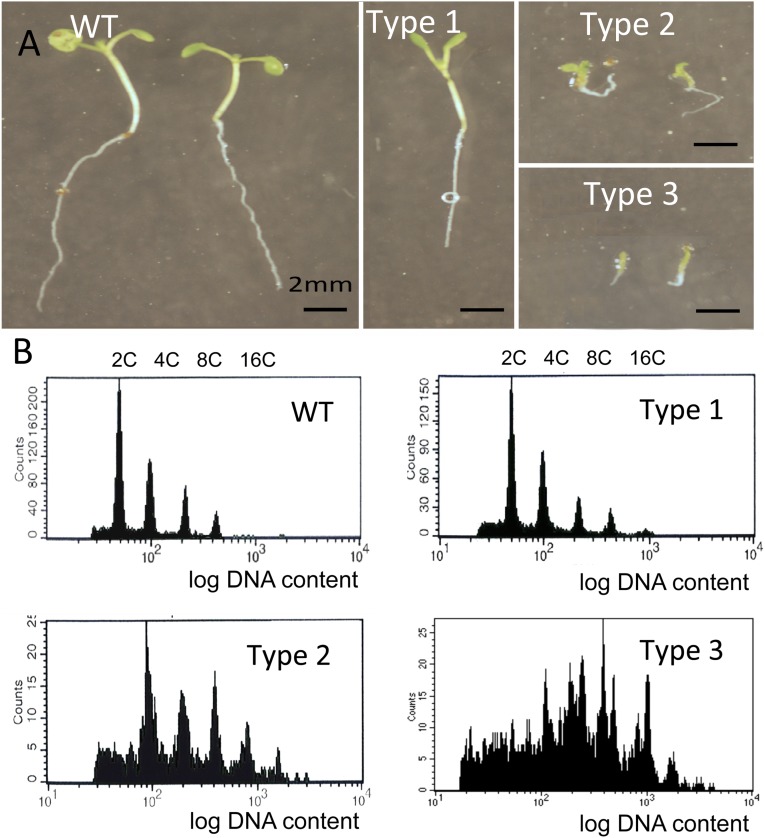
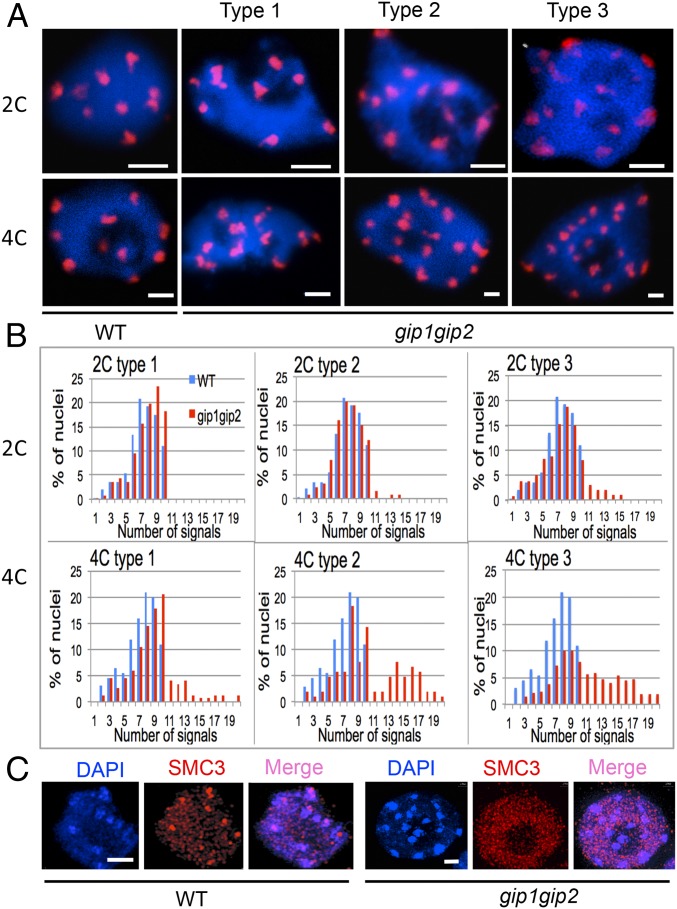
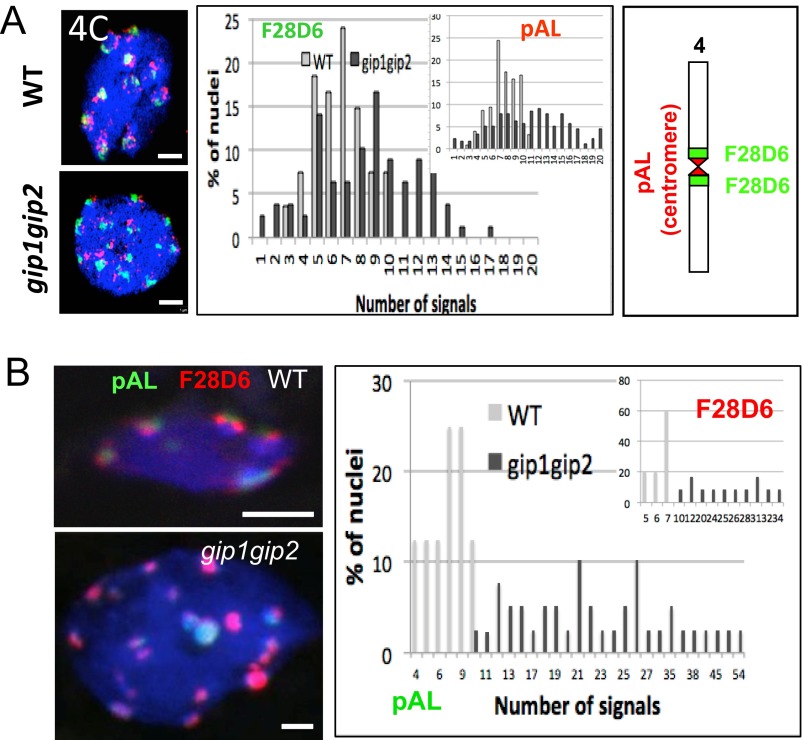
Previously, we observed aberrant chromosome segregation and micronuclei formation in meristematic gip1gip2 cells (8). As these may suggest defects in centromere/kinetochore functions, we analyzed centromere cohesion and subsequent ploidy maintenance in gip1gip2. Because these mutants—hereafter named gip—exhibit variability in growth and development, we used qualitative phenotypic traits to classify mutants into three categories (Fig. S1A, types 1–3). Then, using the 180-bp-centromeric (pAL) FISH probe, we compared the distribution of the centromeric signals between 2C and 4C nuclei flow sorted from WT and gip plantlets (Fig. 1 A and B). In type 1 mutants, the number of pAL signals in 2C nuclei was similar to WT and never exceeded ten (2n = 10 in Arabidopsis). In contrast, although 4C nuclei showed 10 pAL signals in WT, as previously described (14), 18% of 4C nuclei showed more than 10 signals in the type 1 gip mutants, highlighting centromere cohesion defects. Three percent to 9% of 2C nuclei in type 2 and 3 mutants, respectively, presented more than 10 signals, indicating ploidy instability as confirmed by FACS profile analyses (Fig. S1B). To support this hypothesis, we observed an increased number of pAL signals (>10) in up to 38% and 41% of the 4C fraction in type 2 and 3 mutants, respectively (Fig. 1B). Next, we focused our analyses on the most affected classes of gip mutants (types 2 and 3). Using the pericentromeric BAC probe F28D6, we observed a similar increase in the number of signals in 4C nuclei as with the pAL probe (Fig. S2A), which indicated that both centromeric and pericentromeric cohesions were affected in gip. These observations were in accordance with the whole-mount FISH analyses performed in mutant root tips (Fig. S2B).
Fig. S1.
Phenotypic characterization of gip1gip2 mutants. (A) Impaired gip1gip2 mutant phenotypes (types 1–3) compared with WT. Type 1 plantlets exhibit not quite fully expanded cotyledons, type 2 has a stunted phenotype, and type 3 is dwarf and presents severe morphological defects. (B) Corresponding FACS profiles.
Fig. 1.
gip1gip2 mutants exhibit centromeric cohesion defects. (A) FISH detection of centromeric pAL signals in 2C and 4C flow-sorted nuclei from WT and three seedling phenotypes (types 1–3) of gip1gip2 mutants. (B) Number of pAL signals in nuclei (2C and 4C WT, n = 120; gip1gip2 type 1, n = 116; type 2, n = 122; type 3, n = 125). (C) Immunolocalization of the SMC3 cohesin subunit in meristematic root nuclei. DAPI staining is shown in blue. (Scale bars, 2 µm.)
Fig. S2.
Centromeric and pericentromeric defects in gip1gip2. (A) FISH analyses of pAL (red) and pericentromeric BAC probes (green) in 4C leaf nuclei of type 2 (WT, n = 159; gip1gip2, n = 136; P < 0.01). The number and distribution of signals are indicated (Right). (B) Whole mount FISH pAL (green) and pericentromeric (red) signals in root nuclei were quantified (n = 40; P < 0.01). (Scale bars, 2 µm.)
Because the structural maintenance of chromosome 3 (SMC3) cohesin subunit is enriched at centromeres in root meristems (15), we further investigated whether its localization was modified in gip. SMC3 immuno-signals were severely decreased at centromeric chromocenters in gip nuclei compared with WT (Fig. 1C), thus supporting a role of GIPs in stabilizing and/or assembling the cohesin complex at centromeres during interphase.
To further characterize the centromere function of GIPs, we analyzed, in gip mutants, the spatial localization of CENH3 at centromeres in G2 cells and at kinetochores in mitotic cells (Fig. 2). Although the expected doublets of CENH3 signals were observed after replication in WT G2 cells (Fig. 2A), as previously described (16), additional single signals (i.e., from one to five per cell) were visible in gip mutants (Fig. 2B, arrows). Similar results were obtained in prometaphase and metaphase gip cells compared to WT (Fig. 2 E and F). Interestingly, more than 20 chromatids were observed in gip1gip2 metaphase cells (Fig. 2F), which are indicative of ploidy instability. Additionally, intercentromere and interkinetochore mean distances were found increased in gip compared with WT by 32% and 42%, respectively (Fig. 2 C–I). Such increases led to isolated chromatids (in 16.6% of gip cells; n = 30) or premature chromatid separation (Fig. 2F, arrowheads), indicating the occurrence of aneuploidy, as suggested by FACS profile analyses (Fig. S1B). Together, our results are in favor of defects in centromeric cohesion as a leading cause of aneuploidy in gip mutants.
Fig. 2.
Intercentromere/kinetochore distances between homologous chromosomes are increased in gip1gip2 mutants. CENH3 was immunolocalized at centromeres in G2 (A and B) and kinetochores during division (E–G) in WT (A and E), gip1gip2 (B and F), and mad3-1mad3-2 (G). Average (±SDs) intercentromere/kinetochore distance (C and H) and the relative frequency of centromere/kinetochore distances grouped into categories ranging from <0.4 to >1 (D and I) were determined in G2 and mitotic cells from WT, gip1gip2, and mad3-1mad3-2. A Student t test was used to calculate confidence values. ***P < 0.001; n = 50 chromosomes. Single CENH3 signals (arrows), sister chromatids (brackets), and early separated chromatids (arrowheads) are indicated. (Scale bars, 2 µm.)
To assess whether such reduced cohesion between centromeres is linked to the mitotic spindle assembly defects that we previously described in gip mutants (8), we quantified the intercentromere distance in the defective spindle assembly mitotic arrest deficient (mad) mutants (Table S1). Even though mad3.1mad3.2 show mitotic spindle defects (17), we did not observe any deviation from the interkinetochore distances measured in mitotic WT cells (Fig. 2 G and H). Altogether, our data strongly support a specific role of GIPs in the maintenance of centromere cohesion in addition to their role in γ-tubulin complex recruitment for mitotic spindle assembly (8).
GIPs Form a Protein Complex with the Centromeric Histone CENH3 in Cycling Cells.
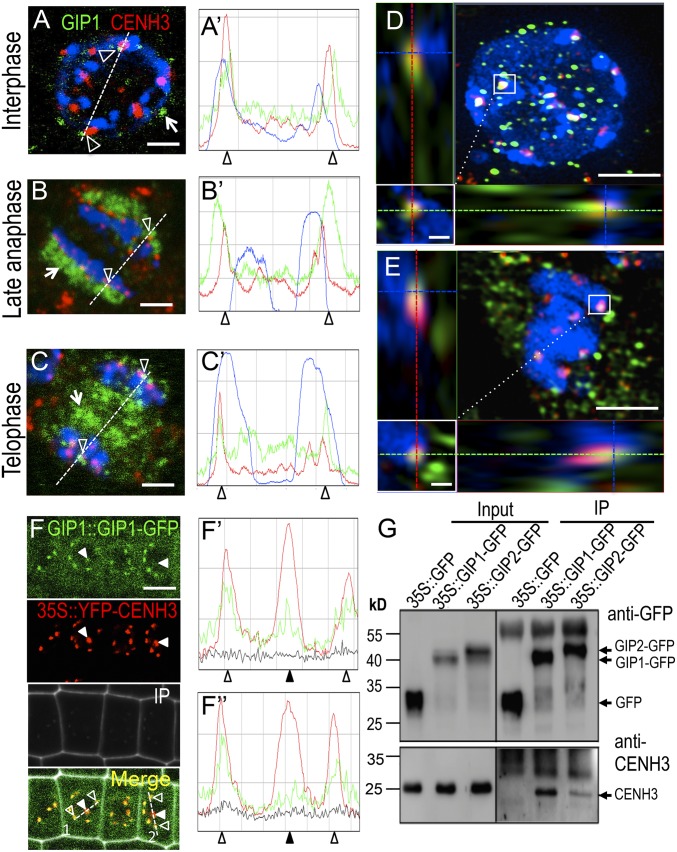
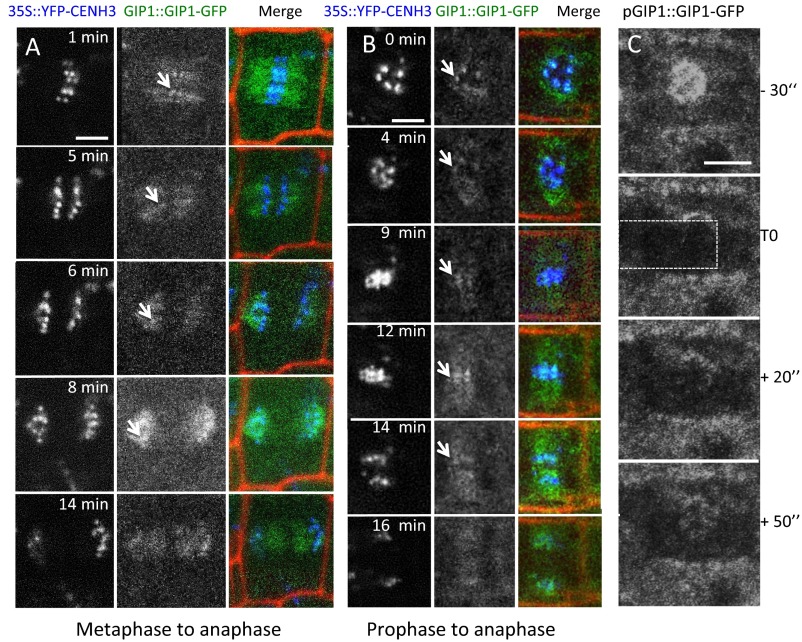
To address the spatial and temporal relationships between GIPs and centromeres/kinetochores, we labeled fixed root tip nuclei of gip lines expressing GIP1::GIP1-GFP with anti-CENH3 antibodies. During interphase, GIP1-GFP exhibited a punctuated distribution at the nuclear envelope, at the outer nuclear membrane where MTs are nucleated (Fig. 3A, arrows) and also close to the inner nuclear membrane (Fig. 3A, arrowheads) where centromeres are embedded into chromocenters (18). When present at centromeres, GIP1 colocalized with CENH3 as shown in fluorescence profile analyses (Fig. 3A′), as well as by superresolution structured illumination microscopy (SIM) (Fig. 3D). During mitosis, besides its localization on MT arrays, GIP1 was also present at the kinetochores where it colocalized with CENH3 and centromeric DNA (Fig. 3 B, B′, C, C′, and E). To precisely follow the distribution kinetic of GIP1-GFP, we introgressed a GIP1::GIP1-GFP construct into a gip1 line constitutively expressing EYFP-CENH3. Comparing the fluorescent signal intensities per centromere, we observed fluctuations in the amount of GIP1, whereas that of CENH3 remained stable (Fig. 3 F and F′, arrowheads). This variability in the intensity of GIP1-GFP signals was also observed during mitosis (Fig. S3 A and B), with a decrease at kinetochores from metaphase to midanaphase followed by an increase during late anaphase. These results were further confirmed by FRAP analyses in the nucleus (Fig. S3C and Movie S1) and suggest a dynamic localization of GIP1 at centromeres. In addition, the constitutive expression of EYFP-CENH3 appeared to enhance the GIP1-GFP recruitment at centromeres/kinetochores (Fig. 3F), thus pointing out a possible functional link between GIP1 and CENH3. To test whether GIP1 and CENH3 were structurally linked, we performed coimmunoprecipitation assays, using plantlets expressing functional GFP-tagged GIP1 or GIP2 (8). A significant amount of endogenous CENH3 was detected in GIP1 complexes, indicating that both GIP1 and CENH3 belong to the same protein complex in vivo (Fig. 3G). It is worth noting that CENH3 was also detected in GIP2 complexes albeit in a rather weak amount (Fig. 3G).
Fig. 3.
GIPs and CENH3 colocalize and associate in the same protein complex. (A–E) Immunodetection of GIP1-GFP and CENH3 in gip1gip2 meristematic root cells. Confocal microscopy (A–C) shows that GIP1-GFP is present at the nuclear periphery (A) and at the spindle (B) and phragmoplast (C) during mitosis (arrows) as well as at the centromeres/kinetochores (arrowheads). Fluorescent profiles (A'–C') indicate the colocalization of both proteins at centromeres in interphase nuclei and mitotic cells. (D and E) Superresolution microscopy (SIM) reveals the colocalization of GIP1-GFP and CENH3 in both interphase nuclei (D) and prometaphase cells (E). (Insets) Magnification of the GIP1-CENH3 colocalization. Green, red, and blue axes indicate x, y, and z axes, respectively. (F) GIP1::GIP1-GFP is expressed in a gip1 line overexpressing YFP-CENH3. GIP1-GFP is present at centromeres and colocalizes with CENH3 but with at different relative intensities (filled arrowheads). Corresponding fluorescent profiles (empty arrowheads in F' and F″) indicate protein colocalization. (G) Coimmunoprecipitation of GIP1- or GIP2-GFP with endogenous CENH3 using anti-GFP antibodies. (Scale bars, 2 µm; Insets, 0.2 µm.)
Fig. S3.
Dynamics of GIP proteins. (A and B) Time-lapse imaging of cells coexpressing GIP1-GFP and YFP-CENH3. Propidium iodide marked cell walls. Arrows indicate GIP1 and CENH3 localization at centromeres. (Scale bars, 5 µm.) (C) GIP1-GFP in interphase cells. See Movie S1. The white rectangle indicates the FRAP bleached zone.
GIPs Are Essential for CENH3 Loading and/or Maintenance in Cycling Cells.
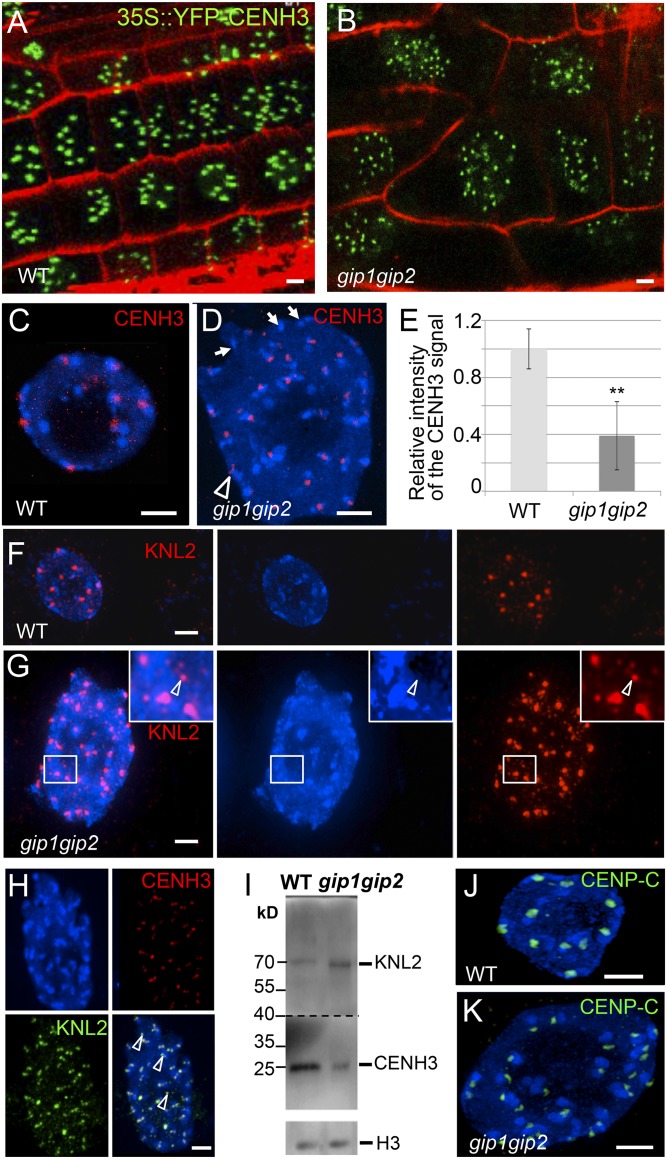
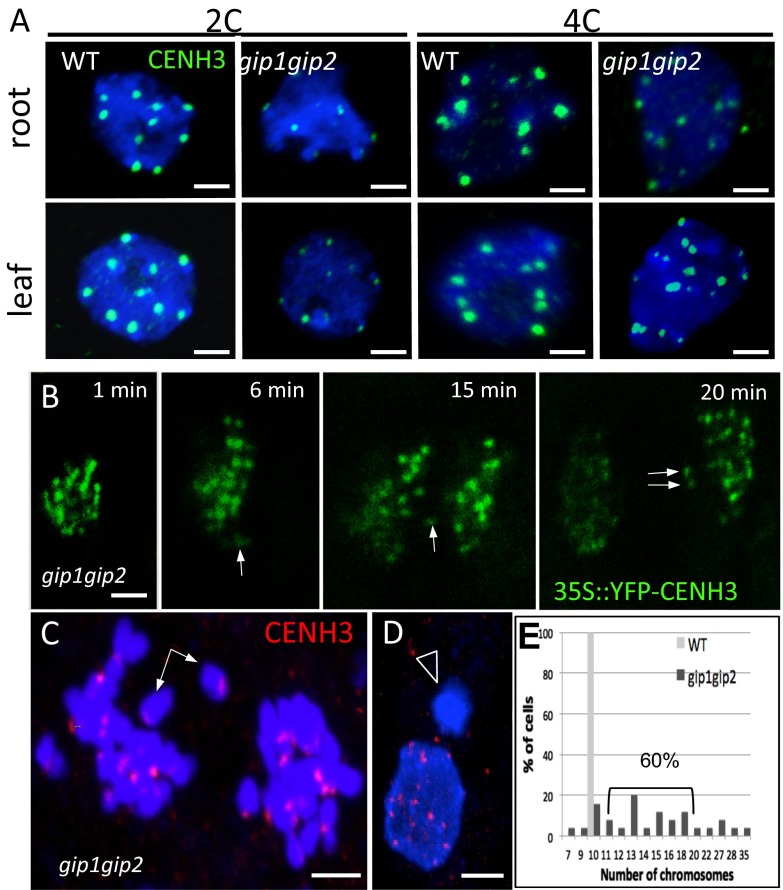
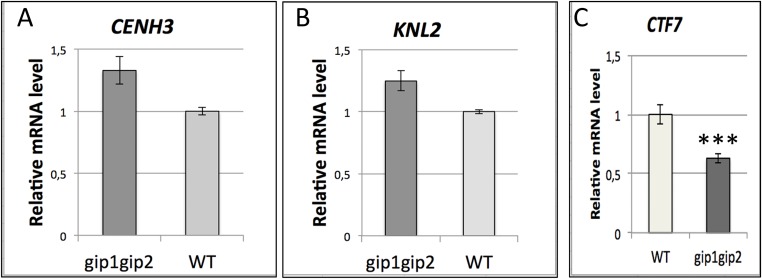
To further delineate the molecular functions of GIPs at centromeres, we introgressed the 35S::EYFP-CENH3 construct (16) into gip mutants. In root meristematic nuclei, the number of CENH3 signals was severely increased compared with WT (Fig. 4 A and B). This increase and the dispersion of CENH3 signals in seedlings confirmed centromere cohesion defects that may occur early during embryogenesis, leading to impaired embryo development (8). In addition, the decreased intensity of CENH3 signals in meristematic nuclei (Fig. 4 C–E) may also reflect defects in CENH3 loading/maintenance at centromeres. This decreased intensity was confirmed using 2C and 4C nuclei sorted from gip young leaves and roots (Fig. S4A). We observed 49% of chromocenters associated with irregular CENH3 signals in gip mutants (Fig. 4D; n = 200), suggesting impeded loading and/or maintenance of CENH3 at centromeres. To support this hypothesis and reflect impaired chromosome segregation, lagging centromeres (47.6% of anaphase cells, n = 42) and micronuclei formation (7% of the interphase cells with at least one micronucleus, n = 426), which lead to strong aneuploidy (60% of cells with 11–19 chromosomes, n = 50), were detected in gip mutants (Fig. S4 B–E). As KNL2 participates in CENH3 loading in Arabidopsis (6), we investigated its location in gip mutants. In addition to the classical centromere localization observed in WT (Fig. 4F), KNL2 also appeared as speckles throughout the nucleoplasm in gip (Fig. 4 G and H and Fig. S5). This pattern may explain the increased KNL2 protein level detected in gip nuclear protein extracts (Fig. 4I). However, such an overaccumulation seems insufficient to maintain appropriate CENH3 level at centromeres. Although ectopic deposition of KNL2 was observed, ectopic loading of CENH3 may be prevented by the induction of CENH3 degradation as was already described in yeast (19). This functional hypothesis is reinforced by the severe decrease in the CENH3 protein level detected in gip (Fig. 2H) compared with its stable mRNA level (Fig. S6A). Altogether, our data strongly support the essential role played by GIPs in the loading/maintenance of CENH3 at centromeres. Interestingly, as the level of KNL2 transcripts was not significantly affected in gip (Fig. S6B), the increased protein level of KNL2 may result from its reduced proteasome-mediated degradation (6).
Fig. 4.
gip1gip2 mutants exhibit defects of centromeric components in cycling cells. 35S::YFP-CENH3 expression in root tips from WT (A) and gip1gip2 (B) using identical imaging settings. (C–H) Immunolocalization of CENH3 and/or KNL2 in WT (C and F) and gip nuclei (D, G, and H). DAPI staining is shown in blue. (D) Arrowhead(s) and arrows indicate elongated signal(s) or absence of signal, respectively. (E) Mean intensity of the CENH3 signals: WT, n = 50; gip mutants, n = 156. **P < 0.01. (G) The arrowhead shows a KNL2 signal outside of a chromocenter. (H) Arrowheads in the merged image indicate KNL2 signals not colocalized with CENH3 in the nucleoplasm. (I) Western blot analysis performed on the same blot of endogenous CENH3 and KNL2 protein amounts in nuclear extracts from WT and gip1gip2. Histone H3 was used as a loading control. This experiment was reproduced four times. (J and K) Immunolocalization of CENP-C in WT (J) and gip nuclei (K). (Scale bars, 2 µm.)
Fig. S4.
Centromere defects in gip1gip2. (A) 2C and 4C sorted nuclei from roots and leaves of gip1gip2 and WT plantlets. Immunolabeling was performed using anti-CENH3 antibodies. (B) Live cell imaging showing mitotic defects in gip1gip2 overexpressing CENH3. Arrows indicate mis-segregated chromatids. (C and D) Immunostaining of gip1gip2 mitotic cells using anti-CENH3 antibodies. Arrows indicate lagging chromosomes and one micronucleus is indicated (arrowhead). (E) Quantification of the number of chromosomes per mitotic cell (n = 50). (Scale bars, 2 µm.)
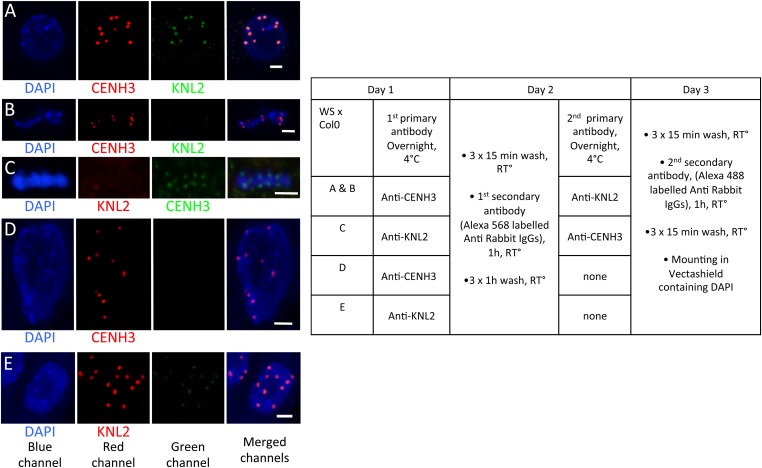
Fig. S5.
Experimental control for double immunolabeling of centromeres using both rabbit anti-CENH3 and rabbit anti-KNL2 antibodies, on WT roots. The table indicates the followed protocol. The three (blue, red, and green) splitted channels (using ImageJ) are merged on the fourth panel (Right). (A) Colocalization of CENH3 and KNL2 in interphase. When KNL2 antibodies are incubated after (B) or before (C) CENH3 antibodies in metaphase, the absence of KNL2 labeling attests for the absence of artefactual fluorescence cross reactions. No fluorescence is observed in interphase nuclei, in the green channel, after either anti-CENH3 (D) or anti-KNL2 (E) preincubation, confirming the specificity of the antibodies. DAPI staining in blue. (Scale bars, 2 μm.)
Fig. S6.
Relative transcript levels of CENH3 (A), KNL2 (B), and CTF7 (C) in gip1gip2 plantlets. Four independent experiments were performed: SD are indicated; P < 0.001.
It was previously established in Arabidopsis that CENH3 and another centromeric protein, CENP-C (Table S1), colocalize at centromeres throughout the cell cycle (20). In gip meristematic root nuclei, CENP-C showed the same altered distribution as CENH3 (Fig. 4 J and K). Therefore, our results suggest that in addition to their involvement in the recruitment and/or maintenance of CENH3, GIP proteins may also be involved, directly or indirectly, in the recruitment and maintenance of CENP-C at the centromeres/kinetochores.
GIPs as a Cornerstone of Centromere Regulation at the Nuclear Envelope.
Little is known about the centromeric regulation at the nuclear envelope. As described above, we present evidence for a central role of GIPs in centromere cohesion and CENH3 loading and/or maintenance. These functions are distinct from the previously described role of GIPs in the recruitment of γ-tubulin complexes (8). Interestingly, the recently characterized GIP homolog, MZT1 in S. pombe, was shown to have two different functions: one for the stabilization of mitotic spindle MTs and the other for the proper segregation of chromosomes without affecting spindle MTs (11). Our data are consistent with these observations because centromeric cohesion was strongly perturbed in gip mutants, whereas centromeres remained cohesive in mad mutants affected in the spindle assembly checkpoint.
Similarly to GIPs (13), the recently characterized nuclear matrix constituent protein CRoWded Nuclei 4 (CRWN4) controls the sizing and shaping of the nucleus, as well as the ploidy level (21). In addition, both gip and crwn4 mutants showed an increase in the number of centromere signals, and both GIPs and CRWN4 are located at the nuclear periphery close to the inner nuclear membrane (13, 22). Although CRWN4 is a component of the nuclear matrix, our results indicate that GIPs are key players at the nuclear envelope in the recruitment of the γ-tubulin complexes at the outer nuclear membrane (8) and in the regulation of the centromere architecture close to the inner nuclear membrane. Cohesin loading at centromeres may depend on CENH3 and CENP-C levels at centromeres/kinetochores, as previously observed in Saccharomyces cerevisiae (5). Moreover, as the establishment of sister chromatid cohesion is mainly ensured through the acetylation of SMC3 by Chromosome Transmission Fidelity 7/Establishment of Cohesion 1 (CTF7/ECO1) during DNA replication in eukaryotes (23), the reduced level of CTF7 mRNA (Table S1) in gip mutants (Fig. S6C) may impair SMC3 stabilization at centromeres.
Centromere dysfunction, due to both impaired centromere cohesion and decreased CENH3 and CENP-C recruitment in gip, may lead to kinetochore instability and subsequent mis-segregation of chromosomes, resulting in aneuploidy and genomic instability. Failure to segregate chromosomes was already reported in human and Drosophila cell lines affected in CENH3 deposition, in which HJURP and Chromosome Alignment defect 1 (CAL1) were depleted (3, 24), respectively. Such defects were recently described in the Arabidopsis knl2 mutant (6). However, even though KNL2 was proposed as an upstream factor for CENH3 loading, its overaccumulation in gip mutants does not improve the CENH3 loading at centromeres, highlighting GIPs as central actors in CENH3 loading and/or stabilization at centromeres. The CENH3 loading machinery has changed rapidly during evolution, resulting in no sequence conservation between CENH3 chaperones identified thus far in Drosophila, yeast, or humans (2, 3, 25). Based on this variability, GIPs may be the cornerstone of an alternative pathway involved in the centromere regulation at the nuclear envelope. Potentially operating as a multifunctional hub at the nuclear envelope, GIPs may coordinate centromere functions essential for proper chromosome segregation.
Materials and Methods
Plant Materials and Growth Conditions.
gip1, gip1gip2, GIP1::AtGIP1-GFP, 35S::AtGIP1-GFP, and 35S::AtGIP2-GFP Arabidopsis lines were described previously (8, 13). A 35S::EYFP-AtCENH3 line (16) was introgressed into either gip1 or sesquimutant gip1gip2 lines. The GIP1::GIP1-GFP construct (13) was introduced by agro-transformation to produce gip1 lines expressing GIP1-GFP. Arabidopsis transformation was performed as described previously (8). The Arabidopsis lines were grown in vitro on Murashige and Skoog medium (SERVA Electrophoresis) at 20 °C with a 16-h photoperiod (70 µmol/m2 per second of fluorescent lighting).
RT-PCR.
Total RNA was extracted from 10-day-old Arabidopsis seedlings with the Nucleospin RNA plant kit (Macherey-Nagel) following the manufacturer’s instructions. Quantitative RT-PCR was performed as previously described (26). Forward and reverse gene-specific primers were used (Table S2) in the experiments, and results were normalized relative to three standard genes (SI Materials and Methods).
Table S2.
List of primers used in RTqPCR
| Gene | Fw | Rv |
| CENH3 | AGGCGGCAGGTCCAACTAC | GGTGAAGTTGTAGGATTTGTTTGTTG |
| KNL2 | CCAATGCGGCGTCTACTGAT | CGCCAAAACTCTAGTGATGACACA |
| CTF7 | CGATCTGCGATTACCGAAAG | CGCTTCTTTTTGTGATTTTTAGG |
| St UbiCred | ACAAGCCAATTTTTGCTGAGC | ACAACAGTCCGAGTGTCATGGT |
| St HexoK | GGCGTTTTCTGATAGCGAAAA | ATGGATCAGGCATTGGAGCT |
| St Actin2 | CATCCAAGCTGTTCTCTCCTTG | TGGCATGAGGAAGAGAAACC |
Standard (St) primers were used for internal gene references.
Coimmunoprecipitation and Immunoblotting.
Twelve-day-old transgenic Arabidopsis seedlings (500 mg fresh weight) expressing GFP, GIP1-, or GIP2-GFP (8) were frozen in liquid nitrogen and ground to powder. The extraction buffer [50 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, 1% Nonidet P-40, and 5% (vol/vol) glycerol], supplemented with protease inhibitors (Roche), was added to the powder. The supernatants were filtered through a 50-μm nylon mesh after centrifugation at 5,000 × g. Protein complexes containing CENH3 associated with GIP1-GFP or GIP2-GFP were enriched with polyclonal anti-GFP antibodies bound to the Dynabeads protein A (Invitrogen), following the manufacturer’s instructions. Protein fractions were separated with SDS/PAGE and transferred to Immobilon membranes (Millipore) for immunoblotting. GFP, GIP1-GFP, and GIP2-GFP recombinant proteins were detected using polyclonal rabbit anti-GFP antibodies (1:10,000 dilution). Anti-CENH3 antibodies (Novus Biologicals; 1:1,000) were used and revealed with Lumi-Light Plus (Roche).
Extraction of Arabidopsis Nuclear Proteins.
Twelve-day-old seedlings were ground in liquid nitrogen and incubated in lysis buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, and 1 mM EDTA) supplemented with 1% Triton X-100, 10% (vol/vol) glycerol, 0.1 mM PMSF, and 5 mM β-mercaptoethanol for 20 min at 4 °C. After centrifugation (3,300 × g for 20 min at 4 °C) and washing with lysis buffer, the pellet was resuspended in SDS/PAGE loading buffer. For immunoblotting, anti-H3 polyclonal antibodies (1/25,000; Millipore), anti-CENH3 (Novus Biologicals; 1:5,000), and anti-KNL2 (1:5,000) (6) antibodies were used.
Flow Sorting of Nuclei.
Nuclei of 10-day-old plantlets were isolated and flow-sorted according to their polyploidy level after formaldehyde fixation using a FACS Aria (BD Biosciences), as described previously (27).
Immunostaining and FISH.
Fixation and labeling protocols are provided in SI Materials and Methods. The antibodies used were rabbit polyclonal anti-CENH3 (Novus Biologicals; 1/500), rabbit polyclonal anti-KNL2 (1/500) (6), rabbit polyclonal anti-SMC3 (1/250) (28), mouse monoclonal anti-GFP (Molecular Probes; 1/500), and rabbit polyclonal anti-CENPC (1/100) (29) antibodies. Signals were detected using Alexa Fluor dyes-conjugated secondary antibodies (Alexa 568, 1:300; Alexa 488 1:200; Life Technologies) and counterstained with 2 µg/mL DAPI. FISH on slides with sorted and squashed nuclei was performed according to ref. 30. The pAL centromeric signal was scored using the two-sided Fisher exact test. Intercentromeric distances were measured using ImageJ software.
Confocal and Superresolution Microscopy.
Confocal images were acquired with a Zeiss LSM 780 microscope equipped with 20×/0.8 and 63×/1.4 oil objectives. Superresolution images were obtained using SIM on an Elyra PS.1 microscope system equipped with a C-Apo 63×/1.2 W Korr objective and by applying ZEN software (Carl Zeiss). Interkinetochore distances and signal intensity were measured with the ImageJ software (31). For whole-mount FISH analyses, images were acquired with an A1 Nikon confocal microscope (van Leeuwenhoek Center for Advanced Microscopy).
SI Materials and Methods
Labeling Probe.
The pAL plasmid (32) was used for the detection of the 180-bp centromeric tandem repeats and the BAC F28D6 (DDBJ/EMBL/GenBank accession no. AF147262 obtained from Nottingham Arabidopsis Stock Center) for the pericentromeric repeats. The BAC was isolated using the Purelink Hipure plasmid midiprep kit (Invitrogen). Probes were labeled with Cy3-dUTP or Cy5-dUTP (Invitrogen) using a nick translation kit (Roche).
Whole-Mount FISH.
Ten-day-old seedlings were fixed in 10 mL fixation buffer [1× PBS, 0.067 M EGTA, pH 7.5, 1% formaldehyde, and 10% (vol/vol) DMSO] for 25 min at room temperature and 5 min under vacuum. Then, the material was rinsed in methanol before storage in ethanol at 4 °C. The fixed material was incubated in 5 mL ethanol:xylene (1:1) for 30 min at room temperature. Rosettes were rinsed in ethanol, methanol, and PBT (1× PBS and 1% Tween-20) and treated with RNase A (0.1 mg/mL in 2× SSC) for 1 h at 37 °C. After washing in PBT, postfixation was performed in PBT with 1% formaldehyde for 25 min and washed in 2× SSC before storage at 4 °C. The samples were incubated in 2× SCC-HB50 buffer [50% formamide, 0.5 M sodium phosphate, 2× SSC, and 10% (vol/vol) dextran sulfate] for 30 min and then with HB50 buffer alone for 30 min. The seedlings were immersed in the hybridization solution containing 500–800 ng of probe in HB50 for 1 h at 37 °C. Seedlings and probes were then denatured at 90 °C for 4 min and placed on ice for 3 min and then 4 d at 37 °C. Successive washing was performed at 42 °C in 2× SSC, in 50% formamide in 2× SSC, and then 2× SCC. The material was mounted in Vectashield containing DAPI (2 µg/mL).
Immunostaining and FISH.
Ten-day-old Arabidopsis seedlings were placed for 20 min on ice in 4% paraformaldehyde in PEM buffer (50 mM Pipes, 5 mM EGTA, and 5 mM MgSO4, pH 6.9). Root tips were digested 10 min at 37 °C in PEM buffer supplemented with 2.5% (wt/vol) pectinase, 2.5% (wt/vol) cellulase, and 2.5% (wt/vol) pectolyase (Yakult Pharmaceutical). Unspecific binding was blocked by 8% (wt/vol) BSA and 0.1% Triton X-100 in PEM buffer for 1 h at room temperature. Primary (overnight at 4 °C) and secondary antibodies (1 h at room temperature) were diluted in PEM buffer supplemented with 1% BSA. Slides with flow-sorted nuclei were placed in 4% (vol/vol) paraformaldehyde and immunostained as previously described (33). For FISH, slides were baked for 30 min at 60 °C, washed in 2× SSC, postfixed in 4% (vol/vol) formaldehyde for 10 min, and dehydrated in an ethanol series [70%, 90%, and 96% (vol/vol) successively]. Probes were denatured in 50% formamide, 2× SCC, and 10% (vol/vol) dextran sulfate for 5 min at 95 °C and put on ice. Twenty microliters of probes per slide were added and submitted to denaturation at 80 °C for 2 min, and hybridization was performed overnight at 37 °C. Washes were performed as described above.
Supplementary Material
Acknowledgments
We thank J. Bruder, A. Kunze, M. Kühne, S. Mangold, D. Schatz, E. Stroh, and N. Pitzalis for technical help. We also thank B. Favery for providing mad mutants and L. Blech for valuable advice. This work was supported by the Centre National de la Recherche Scientifique (CNRS), the Ministère de l’Enseignement Supérieur et de la Recherche (MESR), the Partenariats Hubert Curien programs, 31504VJ and ALW2PJ/09053, the Netherlands’ Organization for Scientific Research, and the Deutscher Akademischer Austausch Dienst. M.B.’s PhD was funded by the MESR. Microscopy was carried out at the Strasbourg-Esplanade cellular imaging facilities (CNRS, Université de Strasbourg, Région Alsace, Association de la Recherche sur le Cancer, and Ligue Nationale contre le Cancer).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1506351112/-/DCSupplemental.
References
- 1.Nagaki K, et al. Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics. 2003;163(3):1221–1225. doi: 10.1093/genetics/163.3.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Camahort R, et al. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell. 2007;26(6):853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 3.Dunleavy EM, et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137(3):485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 4.Subramanian L, Toda NR, Rappsilber J, Allshire RC. Eic1 links Mis18 with the CCAN/Mis6/Ctf19 complex to promote CENP-A assembly. Open Biol. 2014;4:140043. doi: 10.1098/rsob.140043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eckert CA, Gravdahl DJ, Megee PC. The enhancement of pericentromeric cohesin association by conserved kinetochore components promotes high-fidelity chromosome segregation and is sensitive to microtubule-based tension. Genes Dev. 2007;21(3):278–291. doi: 10.1101/gad.1498707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lermontova I, et al. Arabidopsis kinetochore null2 is an upstream component for centromeric histone H3 variant cenH3 deposition at centromeres. Plant Cell. 2013;25(9):3389–3404. doi: 10.1105/tpc.113.114736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janski N, Herzog E, Schmit AC. Identification of a novel small Arabidopsis protein interacting with gamma-tubulin complex protein 3. Cell Biol Int. 2008;32(5):546–548. doi: 10.1016/j.cellbi.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 8.Janski N, et al. The GCP3-interacting proteins GIP1 and GIP2 are required for γ-tubulin complex protein localization, spindle integrity, and chromosomal stability. Plant Cell. 2012;24(3):1171–1187. doi: 10.1105/tpc.111.094904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hutchins JR, et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328(5978):593–599. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhani DK, et al. Mzt1/Tam4, a fission yeast MOZART1 homologue, is an essential component of the γ-tubulin complex and directly interacts with GCP3(Alp6) Mol Biol Cell. 2013;24(21):3337–3349. doi: 10.1091/mbc.E13-05-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masuda H, Mori R, Yukawa M, Toda T. Fission yeast MOZART1/Mzt1 is an essential γ-tubulin complex component required for complex recruitment to the microtubule organizing center, but not its assembly. Mol Biol Cell. 2013;24(18):2894–2906. doi: 10.1091/mbc.E13-05-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Batzenschlager M, Herzog E, Houlné G, Schmit AC, Chabouté ME. GIP/MZT1 proteins orchestrate nuclear shaping. Front Plant Sci. 2014;5:29. doi: 10.3389/fpls.2014.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Batzenschlager M, et al. The GIP gamma-tubulin complex-associated proteins are involved in nuclear architecture in Arabidopsis thaliana. Front Plant Sci. 2013;4:480. doi: 10.3389/fpls.2013.00480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schubert V, et al. Sister chromatids are often incompletely aligned in meristematic and endopolyploid interphase nuclei of Arabidopsis thaliana. Genetics. 2006;172(1):467–475. doi: 10.1534/genetics.105.048363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lam WS, Yang X, Makaroff CA. Characterization of Arabidopsis thaliana SMC1 and SMC3: Evidence that AtSMC3 may function beyond chromosome cohesion. J Cell Sci. 2005;118(Pt 14):3037–3048. doi: 10.1242/jcs.02443. [DOI] [PubMed] [Google Scholar]
- 16.Lermontova I, et al. Loading of Arabidopsis centromeric histone CENH3 occurs mainly during G2 and requires the presence of the histone fold domain. Plant Cell. 2006;18(10):2443–2451. doi: 10.1105/tpc.106.043174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paganelli L, et al. Three BUB1 and BUBR1/MAD3-related spindle assembly checkpoint proteins are required for accurate mitosis in Arabidopsis. New Phytol. 2015;205(1):202–215. doi: 10.1111/nph.13073. [DOI] [PubMed] [Google Scholar]
- 18.Fransz P, De Jong JH, Lysak M, Castiglione MR, Schubert I. Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc Natl Acad Sci USA. 2002;99(22):14584–14589. doi: 10.1073/pnas.212325299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ranjitkar P, et al. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol Cell. 2010;40(3):455–464. doi: 10.1016/j.molcel.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shibata F, Murata M. Differential localization of the centromere-specific proteins in the major centromeric satellite of Arabidopsis thaliana. J Cell Sci. 2004;117(Pt 14):2963–2970. doi: 10.1242/jcs.01144. [DOI] [PubMed] [Google Scholar]
- 21.Wang H, Dittmer TA, Richards EJ. Arabidopsis CROWDED NUCLEI (CRWN) proteins are required for nuclear size control and heterochromatin organization. BMC Plant Biol. 2013;13:200. doi: 10.1186/1471-2229-13-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakamoto Y, Takagi S. LITTLE NUCLEI 1 and 4 regulate nuclear morphology in Arabidopsis thaliana. Plant Cell Physiol. 2013;54(4):622–633. doi: 10.1093/pcp/pct031. [DOI] [PubMed] [Google Scholar]
- 23.Ivanov D, et al. Eco1 is a novel acetyltransferase that can acetylate proteins involved in cohesion. Curr Biol. 2002;12(4):323–328. doi: 10.1016/s0960-9822(02)00681-4. [DOI] [PubMed] [Google Scholar]
- 24.Goshima G, et al. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316(5823):417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erhardt S, et al. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J Cell Biol. 2008;183(5):805–818. doi: 10.1083/jcb.200806038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roa H, et al. Ribonucleotide reductase regulation in response to genotoxic stress in Arabidopsis. Plant Physiol. 2009;151(1):461–471. doi: 10.1104/pp.109.140053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pecinka A, et al. Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma. 2004;113(5):258–269. doi: 10.1007/s00412-004-0316-2. [DOI] [PubMed] [Google Scholar]
- 28.Schubert V, Lermontova I, Schubert I. The Arabidopsis CAP-D proteins are required for correct chromatin organisation, growth and fertility. Chromosoma. 2013;122(6):517–533. doi: 10.1007/s00412-013-0424-y. [DOI] [PubMed] [Google Scholar]
- 29.Lermontova I, et al. Knockdown of CENH3 in Arabidopsis reduces mitotic divisions and causes sterility by disturbed meiotic chromosome segregation. Plant J. 2011;68(1):40–50. doi: 10.1111/j.1365-313X.2011.04664.x. [DOI] [PubMed] [Google Scholar]
- 30.Lysak M, Fransz P, Schubert I. Cytogenetic analyses of Arabidopsis. Methods Mol Biol. 2006;323:173–186. doi: 10.1385/1-59745-003-0:173. [DOI] [PubMed] [Google Scholar]
- 31.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez-Zapater JM, Estelle MA, Somerville CR. A highly repeated DNA sequence in Arabidopsis thaliana. Mol Gen Genet. 1986;204(3):417–423. [Google Scholar]
- 33.Jasencakova Z, Meister A, Walter J, Turner BM, Schubert I. Histone H4 acetylation of euchromatin and heterochromatin is cell cycle dependent and correlated with replication rather than with transcription. Plant Cell. 2000;12(11):2087–2100. doi: 10.1105/tpc.12.11.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.