Abstract
Studies of protein and organothiol interactions with silver nanoparticles (AgNPs) are important for understanding AgNP nanotoxicity, antimicrobial activity, and material fabrications. Reported herein is a systematic investigation of the effects of both reduced and oxidized protein cysteine residues on protein interactions with AgNPs. The model proteins included wild-type and mutated protein GB3 variants that contain 0, 1, or 2 reduced cysteine residues, respectively. Bovine serum albumin (BSA) that contains a total of 34 oxidized (disulfide-linked) cysteine residues and one reduced cysteine residue was also included. Protein cysteine content has no detectable effect on the kinetics of protein/AgNP binding. However, only proteins that contain reduced cysteine residues induce significant AgNP dissolution. Proteins can slow down, but do not prevent the AgNP dissolution induced by subsequently added organothiols. The insights provided in this work are important to the mechanistic understanding of AgNP stability in biofluids that are rich in proteins and amino acid thiols.
INTRODUCTION
Silver nanoparticles (AgNPs) have been widely used in biosensing, chemical catalysis, and solar energy harvesting.1–3 One key complication in understanding the AgNP structure and properties is the AgNP’s susceptibility to oxidation and dissolution. It is generally accepted that the surface silver atoms in as-synthesized AgNPs are oxidized and are likely silver oxide.4–6 Without silver chelating agents, the as-synthesized AgNPs covered in insoluble silver oxide can be stable in aqueous solution for up to several months under ambient conditions. However, organothiols can continuously react with AgNPs, converting the silver oxide and silver atoms into water-insoluble silver thiolate salts.7 The rate of such conversion depends strongly on the organothiol structure and conformation. For example, aromatic organothiols produce large silver-thiolate precipitates that can accumulate on the AgNP surface or settle elsewhere,7 while long-chain 1-alkanethiols on the AgNP are mostly adsorbed as a monolayer.8,9 The latter is due to the surface silver-alkanethiolate salts that are highly ordered on the AgNP surfaces, which impose a strong steric hindrance preventing further alkanethiol reaction with AgNPs. In contrast, alkanethiols on AuNPs are highly disordered regardless of their carbon-chain length.10
We recently investigated the effect of cysteine on protein binding to AuNPs.11 One key observation was that cysteine has no significant effect on the kinetics of the protein/AuNP binding, but it plays a critical role in stabilizing the AuNPs against organothiol displacement and organothiol-induced AuNP aggregation. This finding implies that protein and AuNP binding is initiated by forces including long-range electrostatic and van der Waals forces, but not the covalent cysteine/AuNP bonding that forms only after the protein is adsorbed and deformed onto the AuNPs.
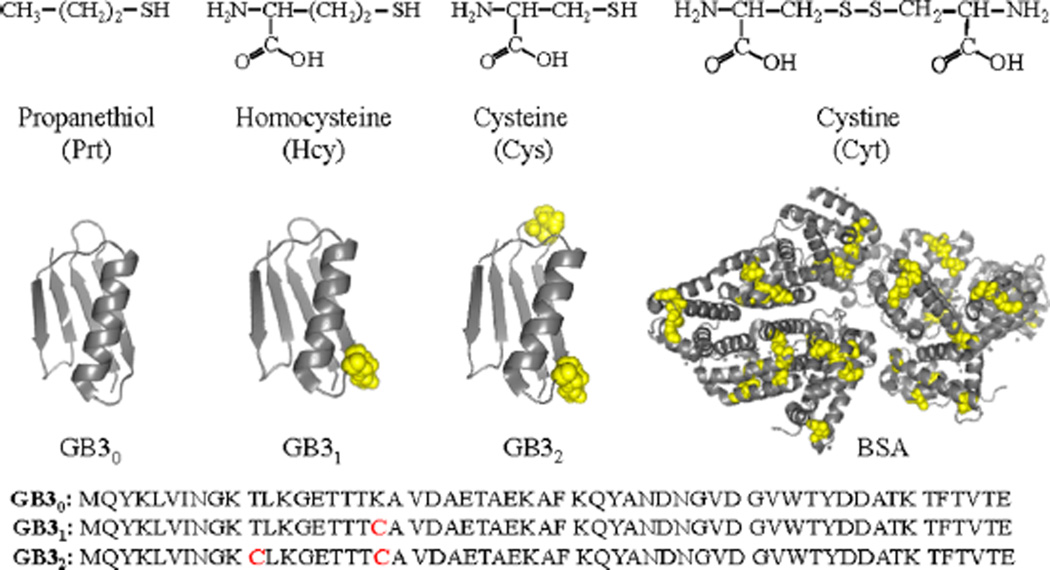
Reported herein is a systematic investigation of the effect of protein cysteine residues on protein interactions with AgNPs in water. The model proteins include bovine serum albumin (BSA), and wild-type and mutated third IgG-binding domain of protein G (GB3) (Figure 1). These proteins were also used in our study of protein binding with AuNPs,11–13 which enables us to compare and contrast the protein binding with AuNPs and AgNPs. The wild-type GB3 protein contains 56 amino acid residues with no cysteine (GB30).14,15 However, the mutated GB3 variants contain one (GB31) and two cysteine residues (GB32), respectively. The lysine residue in GB30 at the 19th position was replaced by a cysteine residue in GB31, while both the threonine and lysine at the 11th and 19th positions in GB30 were replaced by cysteines in GB32 (Figure 1). BSA has 17 interchain disulfide bonds formed by 34 oxidized cysteines and 1 free sulfhydryl group in one reduced cysteine.16
Figure 1.
(Top) Model organothiols used. (Bottom) Cartoon representation of GB3 (from PDB 2-OED) and BSA (from PDB 4OR0) proteins highlighting cysteine residues in yellow CPK spheres and amino acid sequence of GB3 variants. Image created using PyMOL software.
Organothiols have been used as probe molecules to study the protein structure and conformational modification when adsorbed onto AuNPs.12,13 The protein overlayer on AuNPs is highly permeable to small organothiol molecules that can trigger protein desorption or be adsorbed with protein onto the AuNP surface.11,13 In this work, a series of organothiols were employed to investigate the organothiols’ interaction with AgNPs that are pretreated with proteins. This study is critical for evaluating the effectiveness of protein in stabilization of AgNPs to organothiol-induced AgNP aggregation and dissolution. Such information is particularly relevant for AgNP biological applications because proteins and amino acid thiols are abundant in biofluids. For the sake of simplicity, we will use the notation of A/B to represent a two-component solution, and (A/B)/C a three-component solution in which the two components inside the parentheses are mixed first before the addition of the third component.
The model organothiols used in this study include the amino acid thiols cysteine (Cys) and homocysteine (Hcy), and the highly hydrophobic oragnothiol propanethiol (Prt). We also included the disulfide-linked organothiol cystine (Cyt) that is composed of two disulfide-linked (oxidized) Cys molecules. Investigation of both the Cys and Cyt interactions with AgNPs allows us to compare the reactivity of reduced (R-SH) and oxidized (R–S–S–R) organothiols with AgNPs.
It is important to note that there are extensive literature reports on single component protein or organothiol interactions with nanoparticles.17–27 However, cross-comparisons of the chemical reactivity of reduced and oxidized organothiols with AgNPs are to our knowledge very limited, so is the comparison of the effects of reduced and oxidized protein cysteine residues on the protein interactions with AgNPs. Filling this knowledge gap is critical for deepening our understanding of molecular-level interfacial interactions of AgNPs. As an example, the time-dependent surface enhanced Raman spectroscopic (SERS) measurements in this work revealed that Cyt initially binds to AgNP through its carboxylate group. Its disulfide bond is cleaved only after relatively long sample incubation of the Cyt/AgNP mixture. This result may explain the literature controversy on the structure of Cyt on AgNP surfaces.22,28
EXPERIMENTAL SECTION
Materials and Instruments
All chemicals including BSA were purchased from Sigma-Aldrich. The citrate-reduced AgNPs with nominal diameter of 10 nm (Figure S1, Supporting Information) were purchased from Nanocomposix Inc. AgNPs synthesized in house with the Lee and Meisel method have also been used in this work.29 Similar experimental observations were seen with both the in-house and commercial AgNPs. However, the data presented in this manuscript are all obtained with the commercial citrated reduced AgNPs because of their smaller particle size and more uniform particle size distribution. Indeed, because of their smaller size (10 nm versus ~65 nm for the inhouse AgNPs), the protein- and organothiol-induced AgNP structural modification are much more readily detectable with the commercial AgNPs. Nanopure water (Thermo Scientific) was used in sample preparation. An Olis HP 8452 A diode array spectrophotometer was used for the time-resolved UV–vis measurements. All solutions were stored in a refrigerator at ~4 °C. Normal Raman and SERS spectra were acquired using the Lab Ram HR800 confocal Raman microscope system with a 633 nm Raman excitation laser.
Synthesis of GB3 and GB3 Variants
A pET-11b plasmid encoding for GB3 was provided as a generous gift from Ad Bax (National Institutes of Health). After heat-shock transformation, Escherichia coli BI21*DE3 cells (Invitrogen) were incubated in 1 L of LB media at 37 °C. When the culture reached an OD600 of 0.5–0.7, expression was induced with 1 mM IPTG. After growing at 37 °C for 4 h the cells were harvested and then resuspended in lysis buffer (50mMNaCl, 20mMNaH2PO4, pH 7.5, 5 mM EDTA). For K19C and T11C K19C GB3 50 mM DTT was added to ensure all thiols were reduced. The resuspended cells were sonicated (Branson Sonifier 250) on ice at power level 6. Processed lysate was incubated at 85 °C for 15 min and was swirled every 3–4 min. After the mixture was cooled on ice, DNA was precipitated by adding 0.5% streptomycin sulfate and swirling an additional 10 min. The lysate was centrifuged (Beckman Coulter) at 18000g for 45 min, with GB3 remaining in the soluble fraction. Further DNA removal was performed using anion exchange column (GE Healthcare 5 mL HiTrap Q FF). The collected fractions were loaded onto HiLoad26/600 Superdex 75pg column (Amerisham biosciences/GE healthcare) and eluted with 50 mM NaCl, 20 mM NaH2PO4, and pH 7.5 (5 mM DTT for K19C and T11C K19C GB3). Pooled protein fractions were exchanged buffer to nanopure water by performing HiPrep26/10 desalting column (GE healthcare) and frozen at −80 °C. The protein was then lyophilized, and purity was estimated at >98% by SDS-PAGE (Biorad) analysis.
Protein Interactions with AgNPs
As-received colloidal AgNPs (7.8 nM, 0.5 mL) were mixed with GB30 protein (30 µM, 0.5 mL) and time-resolved UV–vis spectra were acquired immediately after the mixing. The same procedure was followed for GB31, GB32 and BSA proteins. Concentration dependence of protein interaction with AgNPs was studied by using 1, 10, 30, and 90 µM protein solutions. The time-resolved UV–vis spectra were taken immediately after the protein and AgNP mixing. The time-dependent UV–vis spectra were taken periodically during the entire time-course (up to 3 weeks) of the experiment period.
Effect of Ag+ on Protein Interactions with AgNPs
A 0.5 mL aliquot of 600 µM AgNO3 was mixed with an equal volume of each protein (60 µM), and then the solutions were refrigerated overnight to allow protein binding with Ag+. The AgNO3-treated proteins were then mixed with an equal volume of as-received AgNPs. Time-resolved UV spectra were acquired immediately after AgNP addition.
Sequential Protein and Organothiol Interactions with AgNPs
A 400 µL sample of as-received colloidal AgNPs was mixed with an equal volume of a 30 µM solution of each protein, respectively. The solutions were incubated overnight before the addition of 400 µL of 900 µM organothiol. Time-dependent UV–vis spectra were acquired immediately after the organothiol addition.
Transmission Electron Microscopy (TEM) Measurement
TEM measurement was acquired using a JEOL 2100 instrument. The washed AgNP samples were deposited onto Cu grids covered with a Formvar carbon film. The measurements were conducted at an accelerating voltage of 200 kV.
Raman and SERS Measurements
All Raman and SERS spectra were acquired with an Olympus 10× objective (NA = 0.25), a spectrograph grating of 300 grooves/mm, and a laser intensity before entering the sample of 1.3 mW. The spectral integration time varied from 10 to 200 s. The Raman shift was calibrated with a neon lamp, and the Raman shift accuracy was ~0.5 cm−1. When it was needed, 100 µL of 5% KCl was used as the aggregation reagent to induce AgNP aggregation before the SERS acquisition.
RESULTS AND DISCUSSION
Protein Interactions with AgNPs
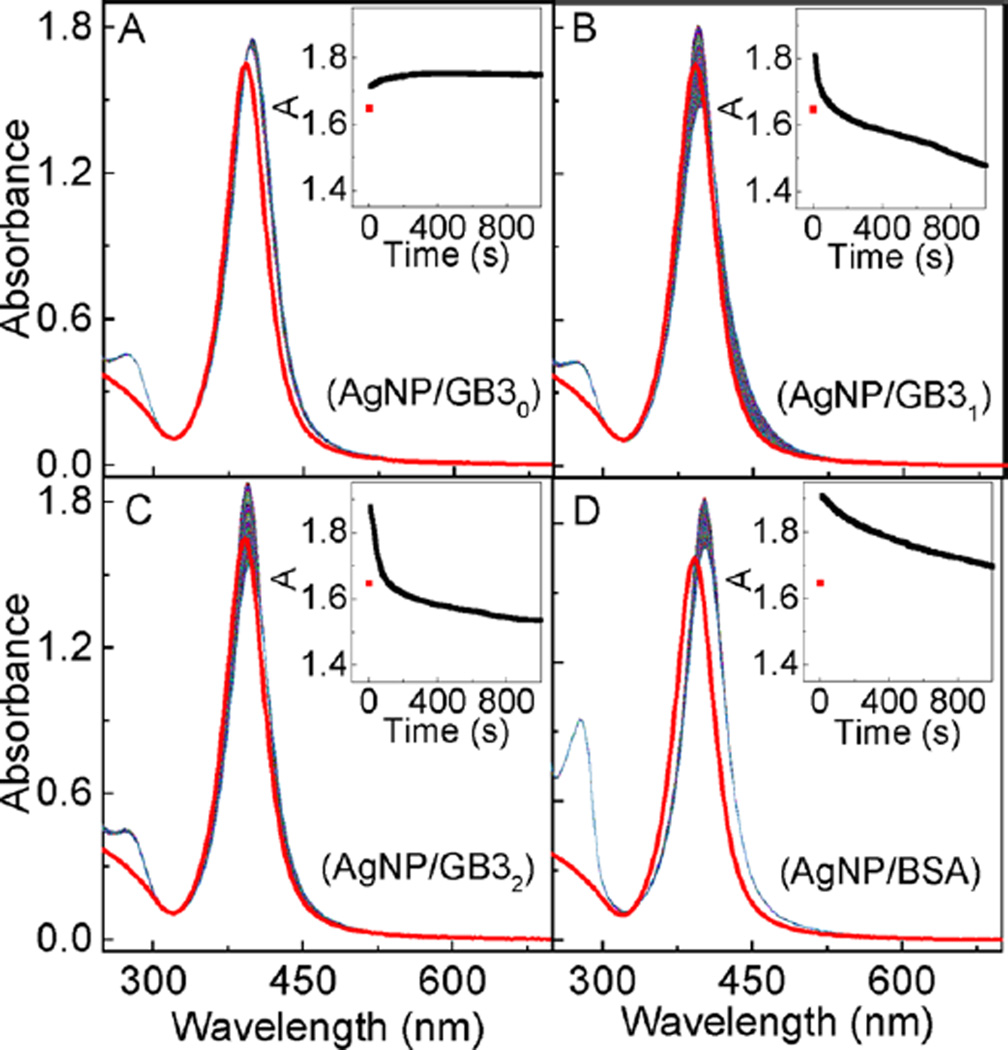
The effects of cysteine on the protein interactions with AgNPs were studied with time-resolved UV–vis spectroscopic method, which monitored the protein-adsorption-induced change in the AgNP localized surface plasmonic resonance (LSPR) (Figure 2). Upon protein addition, the AgNP LSPR peak absorbance increases immediately (within the first two seconds, the instrument dead time for time-resolved UV–vis). After reaching a maximum absorbance within the first few seconds, however, the AgNP LSPR peak absorbance monotonically decreases upon prolonged sample incubation for AgNPs treated with GB31, GB32, and BSA, while the LSPR peak of the GB30 treated AgNP is totally stable after the initial LSPR intensity change.
Figure 2.
Time-resolved UV–vis spectra of (A) AgNP/GB30, (B) AgNP/GB31, (C) AgNP/GB32, and (D) AgNP/BSA. The spectrum in red is the AgNP control. Insets are the time course of the peak AgNP UV–vis absorbance as a function of sample incubation time. The nominal concentrations of AgNPs and proteins are 3.9 nM and 10 µM, respectively.
It is instructive to compare the AgNP versus AuNP LSPR changes induced by protein adsorption. Immediate nanoparticle LSPR increase was also observed when the same sets of model proteins were added into AuNPs.11,12 This result indicates that protein adsorption onto AuNPs and AgNPs are both extremely rapid processes, and the protein cysteine content has no appreciable effect on either protein/AuNP or protein/AgNP binding kinetics. However, unlike the AuNPs for which their LSPR time-courses induced by GB3 protein binding are approximately identical for all three GB3 protein variants,11 the LSPR time-courses of the GB3 treated AgNPs are much more complicated (Figure 2). The increments in the AgNP LSPR peak absorbance induced by GB3 proteins increase from 0.11 for GB30, to 0.17 and 0.23 for GB31 and GB32, respectively. However, the LSPR peak absorbance of the AgNPs treated with GB30 remains constant after its initial increase, but the AgNP LSPR peak absorbance decreases significantly in the AgNP/GB31 and AgNP/GB32 from their respective maximum peak absorbance.
The increase of the AgNP LSPR peak absorbance is due to the protein binding to the AgNP surfaces, which increases the dielectric constant of the medium immediately surrounding the AgNP surfaces. However, the subsequent decrease of the AgNP LSPR peak absorbance is due to the protein-induced AgNP dissolution. The fact that GB30 induces no significant AgNP dissolution indicates that except for cysteine, the other 15 different amino acid residues in the GB3 proteins have no significant effect on AgNP dissolution. The fact that BSA induces significantly less AgNP LSPR attenuation than that for both GB31 and GB32 strongly indicates disulfide-linked cysteine is not effective in inducing AgNP dissolution. The latter was experimentally confirmed later in this work.
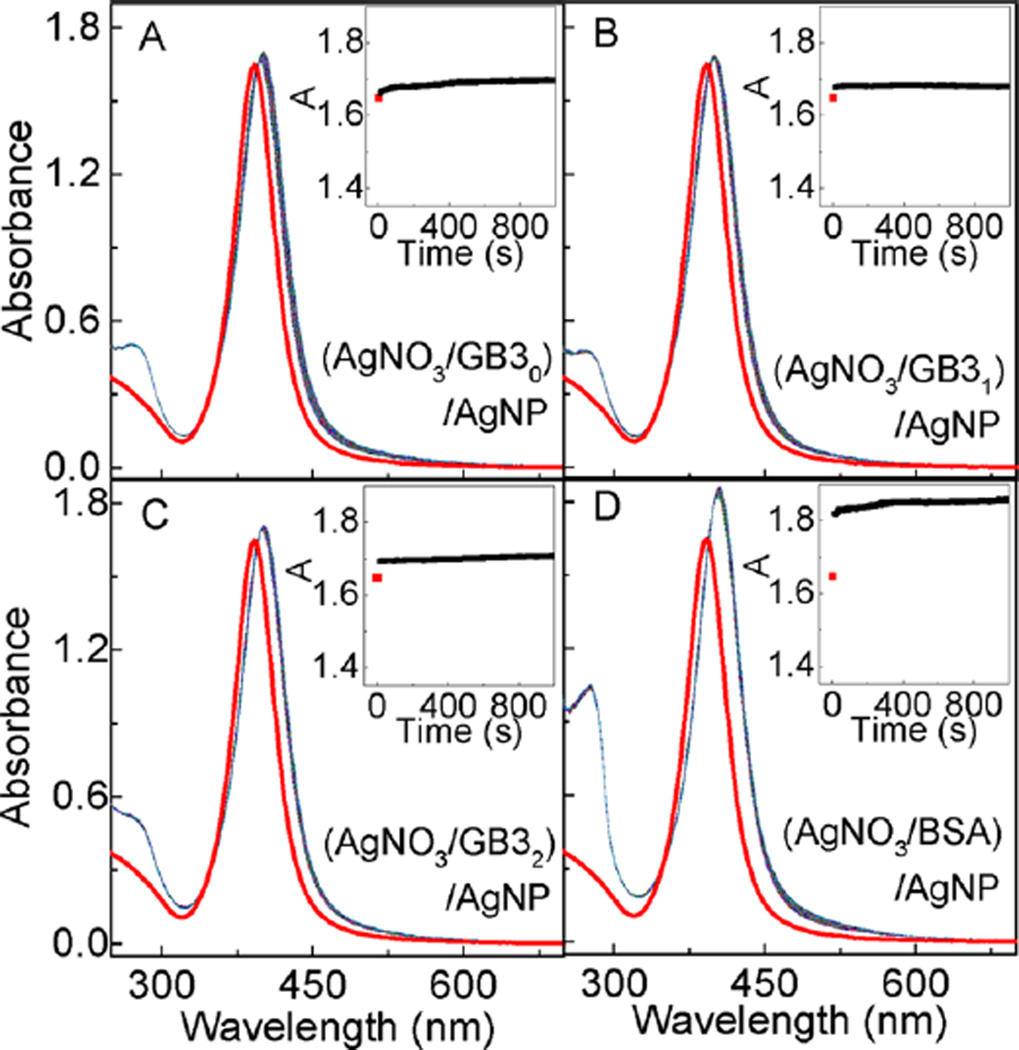
Experimental confirmation that protein-induced AgNP dissolution is caused by protein chelating Ag+ comes from the study of AgNP binding with proteins pretreated with excess AgNO3 (Figure 3). In this case, the AgNP LSPR peak absorbance increased immediately after the addition of the AgNO3-treated protein. This is similar to what has been observed with AgNPs mixed with intact proteins. However, subsequent aging of (protein/AgNO3)/AgNP solutions has no significant effect on the AgNP LSPR absorbance. It is important to note that the ratio of the Ag+/protein in these (protein/AgNO3)/AgNP samples are 10/1. The amount of Ag+ is in large excess to saturate all the reduced protein cysteine residues for all GB3 proteins and in BSA, but is not adequate for the 34 oxidized (disulfide-linked) protein cysteine residues in BSA. This design allows us to differentiate the effectiveness of oxidized and reduced protein cysteine residues in inducing AgNP dissolution. The data in Figure 3 have two notable implications. First, Ag+ chelated proteins can also rapidly adsorb onto the AgNP surface, which provides further evidence that the protein/AgNP binding is initiated by nonspecific forces. Second, oxidized (disulfide-linked) protein cysteine residues have no significant effect in inducing AgNP dissolution. Otherwise, there should be a significant AgNP LSPR decrease in the (BSA/AgNO3)/AgNP sample. The fact that no significant AgNP LSPR decrease is observed in the (protein/AgNO3)/AgNP samples also indicate that besides cysteine, other protein amino acid residues cannot induce significant AgNP dissolution. Otherwise, one should also observe significant AgNP dissolution at least in (BSA/AgNO3)/AgNP, considering that each BSA molecule contains 583 amino acids.
Figure 3.
AgNP interactions with AgNO3-treated proteins. Time-resolved UV–vis spectra of AgNPs mixed with AgNO3 treated (A) GB30, (B) GB31, (C) GB32, and (D) BSA, respectively. Insets are the time course of the AgNP peak UV–vis absorbance. The concentration of GB30, GB31, GB32 and BSA proteins are 60 µM. Ag+/protein ratio was 10/1. AgNO3 and protein were mixed and incubated overnight. Time-resolved UV–vis spectra were obtained immediately after the addition of AgNPs to the AgNO3-treated proteins.
Figure 4 illustrates the protein concentration dependence of the protein/AgNP interactions. When protein concentration is low the AgNP LSPR peak increases instantly following addition of protein. Further aging the protein/AgNP mixtures has no effect on the AgNP LSPR feature. This is true regardless of the proteins used. However, once the protein concentrations surpass the saturation packing density estimated on the basis of the AgNP sizes, concentrations, and the size of the proteins (Section S2, Supporting Information), AgNP/GB31 and AgNP/GB32 mixtures exhibit significant protein concentration dependence in the degree of the protein-induced AgNP dissolution. In contrast, the degree of the BSA-induced AgNP dissolution within the probed time period is independent of the BSA concentration. The fact that no GB30-induced AgNP dissolution occurs regardless of the protein concentration further confirms that only cysteine residues in the GB3 proteins can induce significant AgNP dissolution.
Figure 4.
Concentration dependence of protein interaction with AgNPs. Time–resolved UV–vis spectra of (A1–4) (AgNP/GB30), (B1–4) (AgNP/GB31), (C1–4) (AgNP/GB32), and (D1–4) (AgNP/BSA) as a function of protein concentration. The spectrum in red is the AgNP control. The nominal protein concentrations in 1–4 are 0.5, 5, 15, and 45 µM, respectively. The time course of the peak AgNP UV–vis absorbance of (A5) (AgNP/GB30), (B5) (AgNP/GB31), (C5) (AgNP/GB32), and (D5) (AgNP/BSA) mixtures as a function of sample incubation time. In A5–D5, 0.5, 5, 15, and 45 µM protein solutions are represented in black, green, blue, and magenta, respectively.
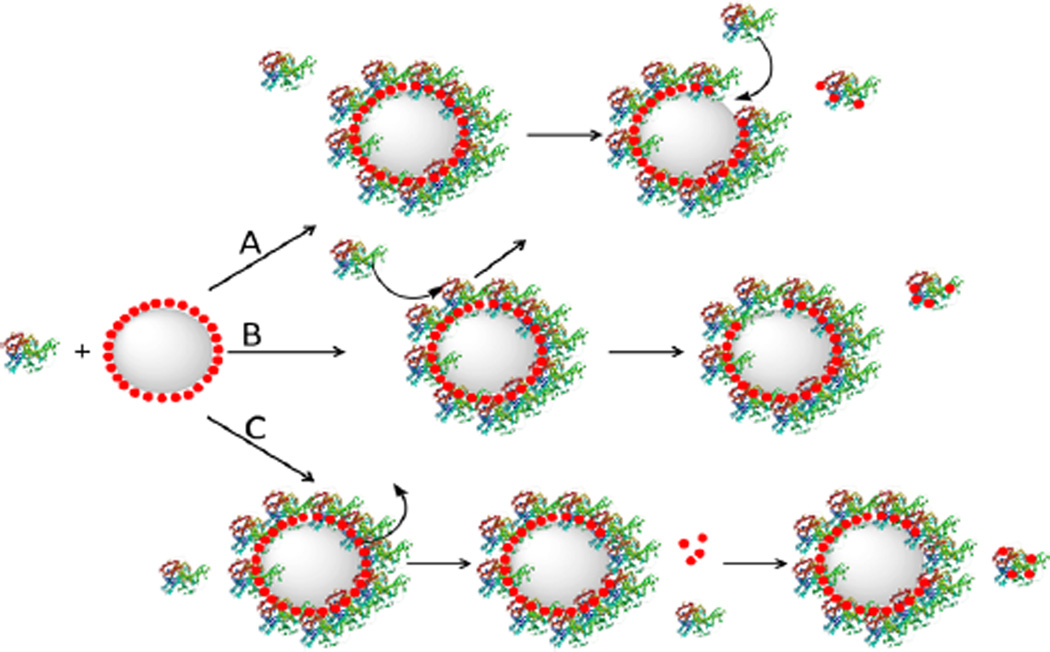
Three possible pathways can be proposed to explain the protein concentration dependence of protein-induced AgNP dissolution (Figure 5). The first is protein dissociative exchange on the AgNPs in which the protein initially is adsorbed onto the AgNPs, chelates with the surface silver ion, and then is dissociatively exchanged with free protein in solution. AgNP dissolution proceeds until all the reduced cysteine residues in the proteins are saturated with Ag+. The second pathway involves protein displacement exchange in which the incoming protein displaces the Ag+-loaded protein on the AgNP surfaces. The third pathway involves Ag+ leakage where the oxidized Ag+ or silver oxide diffuses through the protein overlayer into the surrounding medium, and reacts with protein free in solution. All three pathways could be concurrently in play in all protein/AgNP mixture solutions. However, the likely main reaction pathway for GB31 and GB32 induced AgNP dissolution is through the protein displacement exchange (pathway B in Figure 5) in which the rate of the AgNP dissolution depends critically on the protein concentration. In contrast, the BSA-induced AgNP dissolution likely follows the dissociative protein exchange and/or silver leakage pathway where the rate of protein-induced AgNP dissolution is mostly independent of protein concentration (pathways A and C in Figure 5). It is known that the rate of displacement exchange critically depends on the concentration of the incoming ligands,30,31 but the kinetics of the dissociative exchange depends on the amount of the protein adsorbed onto the AgNP surfaces. The silver leakage pathway is a two-step process. The surface silver atom is first oxidized and detached from the AuNP surface, and then diffuses through the protein overlayer. Subsequently, the diffuse silver ion chelates with a protein cysteine residue, leading to further AgNP oxidation and dissolution. The rate limiting step in this silver leakage pathway should be the AgNP oxidation and silver ion diffusion, not the Ag+ ion chelating with the protein cysteine residue.
Figure 5.
Schematics of possible pathways for protein-induced AgNP dissolution. (A) Dissociative protein exchange in which the dissociated protein carries the dissolved silver atoms, (B) displacement protein exchange in which the displaced protein carries the dissolved silver atoms, and (C) silver ion leakage in which surface silver is oxidized and diffused out of the protein overlayer. Large gray spheres and small red spheres represent AgNPs and Ag+ ions, respectively.
The protein concentration dependence of the AgNP LSPR feature is in sharp contrast to the AuNP LSPR feature induced by protein binding (Figure S2, Supporting Information). The AuNP LSPR peak absorbance reaches a constant once the protein concentration is beyond a threshold value in the AuNP/protein mixtures. Further addition of protein has no effect on the AuNP LSPR features. This result indicates that protein can only adopt a monolayer adsorption onto AuNPs. It also highlights the difference between AgNPs and AuNPs in their molecular-level interfacial interactions.
The reason that the AgNP dissolution rate induced by BSA is lower than that of the same amount of GB31 and GB32 (Figure 4) is likely due to the former’s much larger size. The molecular weight of BSA is about 10 times higher than that of GB3 proteins. Accordingly, the rates of protein/protein exchange on AgNP surface, protein dissociation from the AgNP, and the silver leakage from the AgNP surface in AgNP/BSA mixtures should be drastically slower than their respective counterpart process in the AgNP/GB3 protein mixtures.
Sequential Protein and Organothiol Interactions with AgNPs
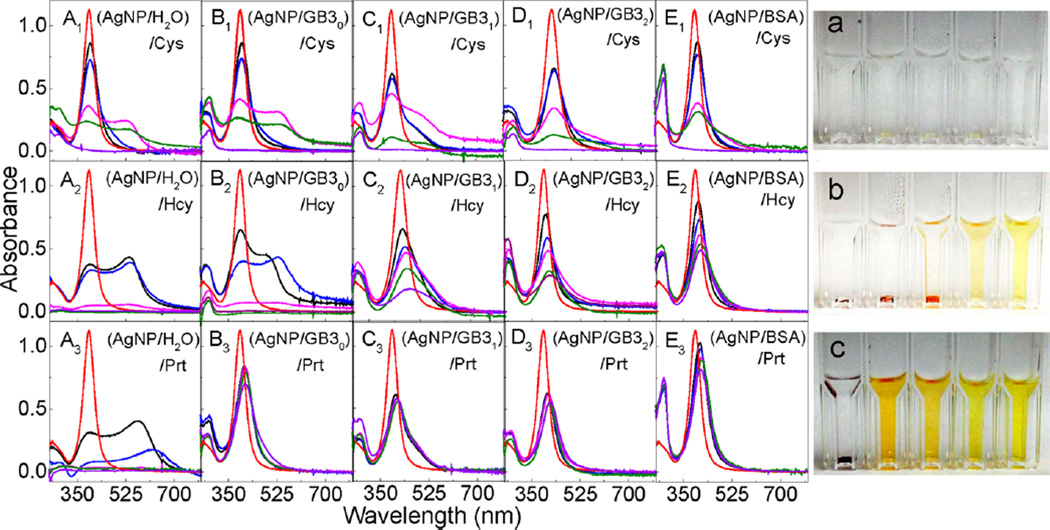
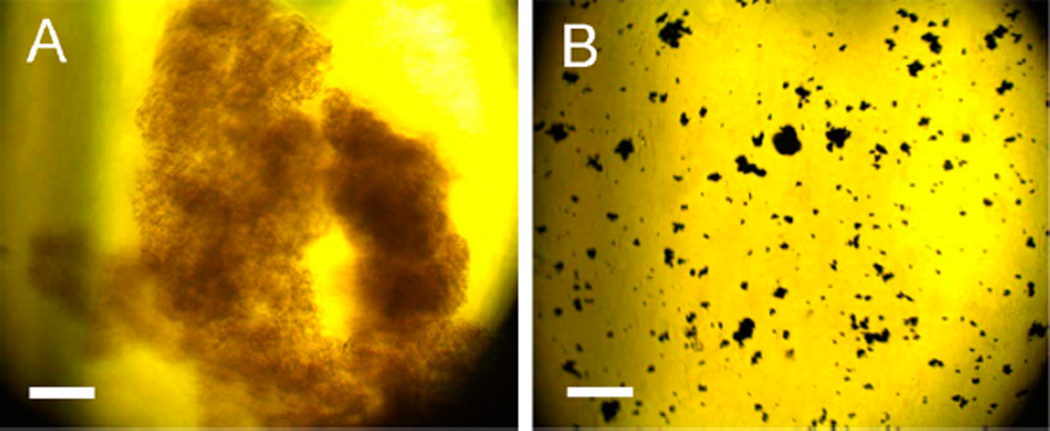
The sequential protein and organothiol interactions with AgNPs are enormously complicated (Figure 6). AgNPs mixed with the GB3 proteins and BSA maintain excellent dispersion stability (no aggregation) in water (Figure 2). However, organothiols can induce both significant AgNP dissolution and AgNP aggregation (Figures 6, parts A1–A3). It is critical to note that AgNP dissolution and aggregation is not mutually exclusive even though AgNP aggregation should invariably reduce the rate of AgNP dissolution. The AgNP dissolution rate in the AgNP/organothiol mixture should also increase with increasing water solubility of the silver–thiolate salts. After prolonged (~3 weeks) sample incubation, no AgNPs can be observed in the AgNPs mixed with excess Cys (Figure 6). The AgNPs were converted into amorphous white precipitates that can be seen under microscopy examinations (Figure 7). In contrast, the AgNP dissolution rate in AgNP/Hcy and AgNP/Prt are much slower. The precipitate in the AgNP/Prt mixture is mostly the aggregated AgNPs, the amorphous silver-propylthiolate salt can only be observed in the TEM image of the prolonged incubated AgNP/Prt mixture (Figure S3, Supporting Information), but not in the optical image shown in Figure 7.
Figure 6.
Sequential protein and organothiol interaction with AgNPs. (Left) Time-dependent UV–vis spectra of (A1–A3) (AgNP/H2O)/organothiol, (B1–B3) (AgNP/GB30)/organothiol, (C1–C3) (AgNP/GB31)/organothiol, (D1–D3) (AgNP/GB32)/OT, and (E1–E3) (AgNP/BSA)/OT. Spectra were obtained (red) 0 min, and (black) 2 min, (blue) 1 h, (magenta) 1day, (green) 3 days, and (purple) 3 weeks after the sample preparation. The concentrations of AgNP, protein, and organothiols were 2.6 nM, 10 µM, and 300 µM, respectively. (Right) Photographs of the AgNPs treated with Cys, Hcy, and Prt, respectively. Cuvettes form left to right corresponding to samples A–E in the same row. Photographs were taken after the samples were left in refrigerator for 3 weeks.
Figure 7.
Optical images of the precipitate formed in three-weeks aged (A) AgNP/Cys and (B) AgNP/Prt. Scale bar =100 µm.
All the model proteins including GB30 slow organothiol-induced AgNP aggregation in the (AgNP/protein)/organothiol samples, and BSA is the most effective protein in preventing AgNP aggregation triggered by the subsequently added organothiols. However, none of the proteins can prevent AgNP dissolution induced by the subsequently added organothiols, which is evident from the complete absence of AgNPs in the (AgNP/protein)/Cys sample and from the large AgNP LSPR intensity drop in the (AgNP/protein)/Hcy samples. Importantly, the effect of the protein binding to AgNP on AgNP dissolution induced by the subsequently added organothiol is likely highly complicated. On one hand, the protein enhances the AgNP dispersion stability in solution, which should enhance organothiol-induced AgNP dissolution. On the other hand, the protein overlayer on the AgNP should reduce the mass transfer rate of the organothiol in and out of the protein overlayer, which can reduce the rate of AgNP dissolution. Consequently, the dissolution rate of AgNPs sequentially treated with protein/organothiol mixture can be slower or faster than AgNPs mixed with organothiol alone depending on the structure and conformation of protein and organothiol on the AgNPs, and the solubility of the silver–thiolate salt.
All three pathways proposed in Figure 5 for the protein concentration dependence of protein-induced AgNP dissolution could be in play for the sequential protein and organothiol interactions with AgNPs. Besides possible silver ion leakage, the dissociative and displacement exchanges between protein/protein, organothiol/organothiol, and protein/organothiol can also contribute to the AgNP dissolution. Presumably, the organothiol/organothiol exchange is likely the predominant pathway for the AgNP dissolution in the (AgNP/protein)/organothiol mixtures. This hypothesis is consistent with the fact that Cys induces significantly faster AgNP dissolution than Hcy and Prt in their corresponding (AgNP/Protein)/organothiol mixture solutions (Figure 6).
Cyt Binding with AgNPs
Oxidized (disulfide-linked) cysteines play critical roles in protein structure and properties. Most cysteine residues in wild-type proteins are disulfide-linked to maintain the globular structure of the proteins. Possible AgNP interaction with oxidized protein cysteine residues has been studied with Cyt as the model molecule. However, Cyt differs significantly from the oxidized protein cysteine residues because the latter do not usually contain free carboxylate groups. In contrast, each Cyt has two free carboxylate groups for possible AgNP binding. Indeed, Lee et al. believe that Cyt binds to the AgNPs as a thiolate in which the S–S bond is cleaved by the formation of S–Ag bonds,28 while Lopez-Tobar et al. proposed that Cyt is adsorbed onto AgNPs as carboxylate.22 Resolving this controversy is critical for understanding AgNP interactions with oxidized protein cysteine residues. It would be impossible for disulfide-linked cysteine residues in protein to induce any significant AgNP dissolution if Cyt indeed can only bind to AgNP as carboxylate. This is because the carboxyl group in cysteine is mostly utilized in peptide bonds. Furthermore, the absence of significant AgNP dissolution in the GB30 containing samples regardless of the GB30 concentrations strongly suggests that the protein carboxyl groups are inadequate for inducing significant AgNPs dissolution under the investigated experimental conditions.
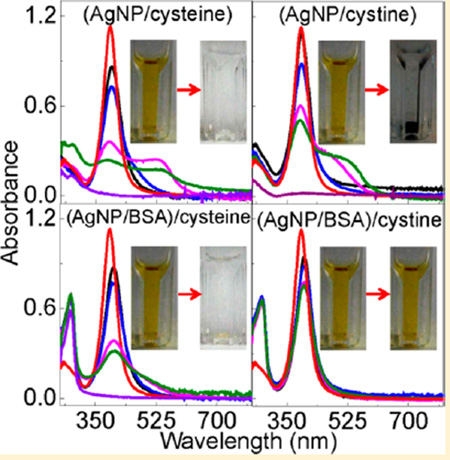
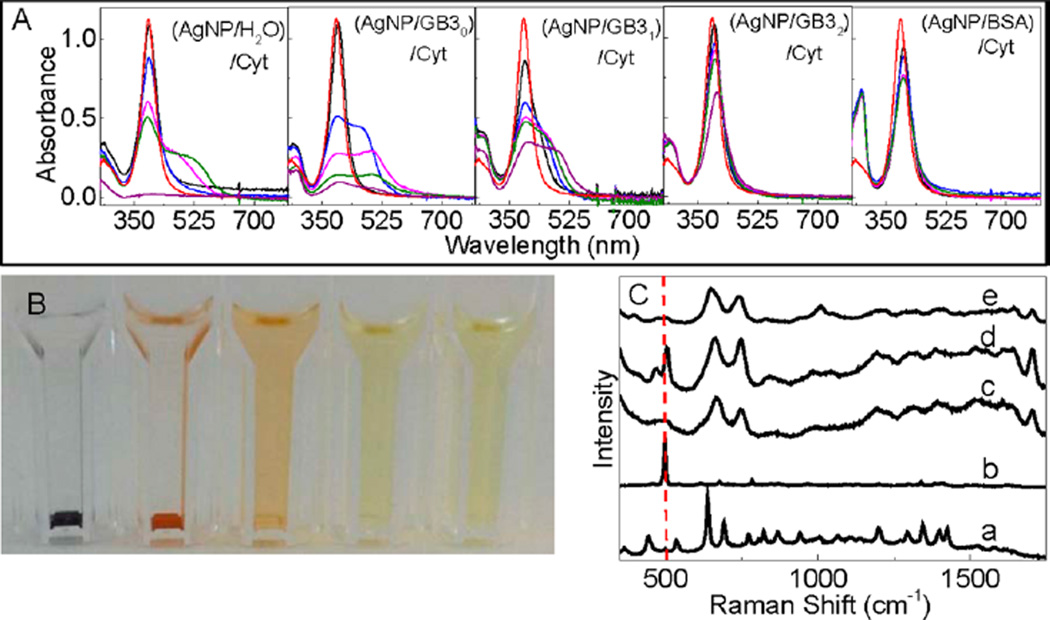
Figure 8 showed the photographs and UV–vis spectra of AgNP/Cyt and (AgNP/protein)/Cyt mixtures, and the SERS spectra of the Cys and Cyt on AgNPs. Cyt induces gradual AgNP aggregation and settlement in the AgNP/Cyt, (AgNP/GB30)/Cyt, and (AgNP/GB31)/Cyt mixtures, but no significant AgNP aggregation was observed in (AgNP/GB32)/Cyt and (AgNP/BSA)/Cyt mixtures. In stark contrast to Cys binding to AgNPs that leads to complete AgNP dissolution in all Cys-containing samples, there are still undissolved AgNPs in all the Cytcontaining AgNP samples even after 2 months of sample incubation. It is important to note that the amount of Cys and Cyt added into the AgNP/protein mixtures are all the same in their sulfur content. The fact that Cyt can induce AgNP aggregation or dissolution confirms that Cyt can bind to AgNPs as previously reported. However, the sharp difference in AgNP dissolution rates between the Cys- and Cyt-containing samples indicates that Cyt is much less effective than Cys for inducing AgNP dissolution.
Figure 8.
Characterization of Cyt interactions with AgNPs. (A) Time-dependent UV–vis spectra of AgNP/Cyt and (AgNP/protein)/Cyt samples and (B) photograph of the AgNP/Cyt and (AgNP/protein)/Cyt samples used for the UV–vis measurement in part A. The photograph was taken after the samples were kept inside the refrigerator for 3 weeks. (C) Time-dependent SERS spectra of Cyt adsorbed onto AgNPs. Spectra a and b are the normal Raman spectra of Cys and Cyt, respectively, (c) SERS spectra of Cys on AgNPs, and spectra d and e were acquired with Cyt adsorbed onto AgNPs. The sample incubation time before the SERS spectral acquisition is ~2 h and ~12 h for spectrum (d) and (e), respectively. The dash line indicates the S–S stretch feature at 520 cm−1 region.
Time-dependent SERS studies revealed that the Cyt disulfide bond remains mostly intact in Cyt initially adsorbed onto AgNPs, but is cleaved after prolonged sample incubation (Figure 8). This is evident from the presence of the S–S stretch feature in the SERS spectra of the freshly prepared Cyt/AgNP sample and absence of the S–S peak in the aged sample. This aging effect on the Cyt structure on AgNPs may be due to both kinetic and thermodynamic reasons. It is possible that the thiolate formation with AgNPs is thermodynamically more favorable, but kinetically slower, than the reaction of the carboxylate group with AgNPs. Therefore, the Cyt initially binds as a Ag–carboxylate salt, but is subsequently converted to the more stable Ag-thiolate salt. It is also possible that the disulfide binding to AgNP is both kinetically and thermodynamically more unfavorable than that for carboxylate. In the latter case, the S–S cleavage only occurs when the carboxyl groups in Cyt are fully reacted with silver ions derived from AgNPs.
The fact that Cyt can induce AgNP dissolution in (AgNP/GB32)/Cyt and (AgNP/BSA)/Cyt does not necessarily imply that oxidized cysteine residues in protein can induce AgNP dissolution. This is because oxidized protein cysteine residues are commonly located in the interior core of globular proteins.32 Unlike the Cyt in (AgNP/GB32)/Cyt and (AgNP/BSA)/Cyt that can diffuse to the AgNP surfaces to initiate the Cyt/AgNP interactions, it is unlikely for the oxidized protein cysteine residues to directly bind to AgNPs. This steric hindrance combined with the low disulfide reactivity with AgNPs explains why BSA is drastically less effective than the thiol-containing GB3 variant in inducing AgNP dissolution. Indeed, the experimental data obtained from AgNP binding with protein pretreated with AgNO3 (Figure 3) strongly indicates that oxidized protein cysteine has no effect on protein-induced AgNP dissolution.
CONCLUSIONS
The effects of both reduced and oxidized protein cysteine residues on protein binding to AgNPs were investigated with a series of model protein and organosulfur molecules. The protein cysteine content has no effect on the kinetics of protein/AgNP binding. Only reduced protein cysteine induces significant AgNP dissolution when the protein concentration is as high as 45 µM. Other protein amino-acid residues including oxidized protein cysteine residues have no significant effect on AgNP dissolution. Pretreatment of AgNPs with protein can inhibit AgNP aggregation induced by subsequently added organothiols, but can not prevent the organothiol-induced AgNP dissolution. The insights provided in this work are important for enhancing the understanding of AgNP interfacial interactions with proteins and organothiols.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by an NIH Award (R15GM113152) to N.C.F. as well as an NSF CAREER Award (CHE 1151057) and NSF funds (EPS-0903787) provided to D.Z.
Footnotes
ASSOCIATED CONTENT
UV–vis absorption spectrum and TEM image of commercial silver nanoparticles, threshold protein concentration for saturation silver nanoparticle binding, protein concentration dependence of protein interactions with gold nanoparticles, and TEM image of propanethiol-containing AgNPs. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Ahamed M, AlSalhi MS, Siddiqui MKJ. Silver Nanoparticle Applications and Human Health. Clin. Chim. Acta. 2010;411:1841–1848. doi: 10.1016/j.cca.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 2.Choi O, Deng KK, Kim N-J, Ross L, Jr, Surampalli RY, Hu Z. The Inhibitory Effects of Silver Nanoparticles, Silver Ions, and Silver Chloride Colloids on Microbial Growth. Water Res. 2008;42:3066–3074. doi: 10.1016/j.watres.2008.02.021. [DOI] [PubMed] [Google Scholar]
- 3.Jana NR, Sau TK, Pal T. Growing Small Silver Particle as Redox Catalyst. J. Phys. Chem. B. 1998;103:115–121. [Google Scholar]
- 4.Li X, Lenhart JJ, Walker HW. Dissolution-Accompanied Aggregation Kinetics of Silver Nanoparticles. Langmuir. 2010;26:16690–16698. doi: 10.1021/la101768n. [DOI] [PubMed] [Google Scholar]
- 5.Yin Y, Li Z-Y, Zhong Z, Gates B, Xia Y, Venkateswaran S. Synthesis and Characterization of Stable Aqueous Dispersions of Silver Nanoparticles Through the Tollens Process. J. Mater. Chem. 2002;12:522–527. [Google Scholar]
- 6.Chen M, Wang L-Y, Han J-T, Zhang J-Y, Li Z-Y, Qian D-J. Preparation and Study of Polyacryamide-Stabilized Silver Nanoparticles through a One-Pot Process. J. Phys. Chem. B. 2006;110:11224–11231. doi: 10.1021/jp061134n. [DOI] [PubMed] [Google Scholar]
- 7.Ansar SM, Perera GS, Gomez P, Salomon G, Vasquez ES, Chu IW, Zou S, Pittman CU, Walters KB, Zhang D. Mechanistic Study of Continuous Reactive Aromatic Organothiol Adsorption onto Silver Nanoparticles. J. Phys. Chem. C. 2013;117:27146–27154. [Google Scholar]
- 8.Fenter P, Eisenberger P, Li J, Camillone N, Bernasek S, Scoles G, Ramanarayanan TA, Liang KS. Structure of Octadecyl Thiol Self-Assembled on the SilveR111) Surface: An Incommensurate Monolayer. Langmuir. 1991;7:2013–2016. [Google Scholar]
- 9.Laibinis PE, Whitesides GM, Allara DL, Tao YT, Parikh AN, Nuzzo RG. Comparison of the Structures and Wetting Properties of Self-Assembled Monolayers of n-Alkanethiols on the Coinage Metal Surfaces, Copper, Silver, and Gold. J. Am. Chem. Soc. 1991;113:7152–7167. [Google Scholar]
- 10.Ansar SM, Gadogbe M, Siriwardana K, Howe JY, Dogel S, Hosseinkhannazer H, Collier WE, Rodriguez J, Zou S, Zhang D. Dispersion Stability, Ligand Structure and Conformation, and SERS Activities of 1-Alkanethiol Functionalized Gold and Silver Nanoparticles. J. Phys. Chem. C. 2014;118:24925–24934. [Google Scholar]
- 11.Siriwardana K, Wang A, Vangala K, Fitzkee N, Zhang D. Probing the Effects of Cysteine Residues on Protein Adsorption onto Gold Nanoparticles Using Wild-Type and Mutated GB3 Proteins. Langmuir. 2013;29:10990–10996. doi: 10.1021/la402239h. [DOI] [PubMed] [Google Scholar]
- 12.Vangala K, Ameer F, Salomon G, Le V, Lewis E, Yu L, Liu D, Zhang D. Studying Protein and Gold Nanoparticle Interaction Using Organothiols as Molecular Probes. J. Phys. Chem. C. 2012;116:3645–3652. [Google Scholar]
- 13.Vangala K, Siriwardana K, Vasquez ES, Xin Y, Pittman CU, Walters KB, Zhang D. Simultaneous and Sequential Protein and Organothiol Interactions with Gold Nanoparticles. J. Phys. Chem. C. 2012;117:1366–1374. [Google Scholar]
- 14.Ulmer TS, Ramirez BE, Delaglio F, Bax A. Evaluation of Backbone Proton Positions and Dynamics in a Small Protein by Liquid Crystal NMR Spectroscopy. J. Am. Chem. Soc. 2003;125:9179–9191. doi: 10.1021/ja0350684. [DOI] [PubMed] [Google Scholar]
- 15.Derrick JP, Wigley DB. The Third IgG-Binding Domain from Streptococcal Protein G: An Analysis by X-ray Crystallography of the Structure Alone and in a Complex with Fab. J. Mol. Biol. 1994;243:906–918. doi: 10.1006/jmbi.1994.1691. [DOI] [PubMed] [Google Scholar]
- 16.Kang YN, Kim H, Shin WS, Woo G, Moon TW. Effect of Disulfide Bond Reduction on Bovine Serum Albumin-Stabilized Emulsion Gel Formed by Microbial Transglutaminase. J. Food Sci. 2003;68:2215–2220. [Google Scholar]
- 17.MacCuspie R. Colloidal Stability of Silver Nanoparticles in Biologically Relevant Conditions. J. Nanopart. Res. 2011;13:2893–2908. [Google Scholar]
- 18.Tai J-T, Lai C-S, Ho H-C, Yeh Y-S, Wang H-F, Ho R-M, Tsai D-H. Protein–Silver Nanoparticle Interactions to Colloidal Stability in Acidic Environments. Langmuir. 2014;30:12755–12764. doi: 10.1021/la5033465. [DOI] [PubMed] [Google Scholar]
- 19.Martin MN, Allen AJ, MacCuspie RI, Hackley VA. Dissolution, Agglomerate Morphology, and Stability Limits of Protein-Coated Silver Nanoparticles. Langmuir. 2014;30:11442–11452. doi: 10.1021/la502973z. [DOI] [PubMed] [Google Scholar]
- 20.Ostermeyer A-K, Kostigen Mumuper C, Semprini L, Radniecki T. Influence of Bovine Serum Albumin and Alginate on Silver Nanoparticle Dissolution and Toxicity to Nitrosomonas Europaea. Environ. Sci. Technol. 2013;47:14403–14410. doi: 10.1021/es4033106. [DOI] [PubMed] [Google Scholar]
- 21.Gondikas AP, Morris A, Reinsch BC, Marinakos SM, Lowry GV, Hsu-Kim H. Cysteine-Induced Modifications of Zerovalent Silver Nanomaterials: Implications for Particle Surface Chemistry, Aggregation, Dissolution, and Silver Speciation. Environ. Sci. Technol. 2012;46:7037–7045. doi: 10.1021/es3001757. [DOI] [PubMed] [Google Scholar]
- 22.López-Tobar E, Hernández B, Ghomi M, Sanchez-Cortes S. Stability of the Disulfide Bond in Cystine Adsorbed on Silver and Gold Nanoparticles As Evidenced by SERS Data. J. Phys. Chem. C. 2012;117:1531–1537. [Google Scholar]
- 23.Pokhrel LR, Dubey B, Scheuerman PR. Impacts of Select Organic Ligands on the Colloidal Stability, Dissolution Dynamics, and Toxicity of Silver Nanoparticles. Environ. Sci. Technol. 2013;47:12877–12885. doi: 10.1021/es403462j. [DOI] [PubMed] [Google Scholar]
- 24.Toh H, Batchelor-McAuley C, Tschulik K, Compton R. Chemical Interactions between Silver Nanoparticles and Thiols: A Comparison of Mercaptohexanol against Cysteine. Sci. China Chem. 2014;57:1199–1210. [Google Scholar]
- 25.Navarro E, Piccapietra F, Wagner B, Marconi F, Kaegi R, Odzak N, Sigg L, Behra R. Toxicity of Silver Nanoparticles to Chlamydomonas Reinhardtii. Environ. Sci. Technol. 2008;42:8959–8964. doi: 10.1021/es801785m. [DOI] [PubMed] [Google Scholar]
- 26.Eigenheer R, Castellanos ER, Nakamoto MY, Gerner KT, Lampe AM, Wheeler KE. Silver Nanoparticle Protein Corona Composition Compared Across Engineered Particle Properties and Environmentally Relevant Reaction Conditions. Environ. Sci.: Nano. 2014;1:238–247. [Google Scholar]
- 27.Kim JA, Salvati A, Aberg C, Dawson KA. Suppression of Nanoparticle Cytotoxicity Approaching In Vivo Serum Concentrations: Limitations of In Vitro Testing for Nanosafety. Nanoscale. 2014;6:14180–14184. doi: 10.1039/c4nr04970e. [DOI] [PubMed] [Google Scholar]
- 28.Lee H, Kim MS, Suh SW. Raman Spectroscopy of Sulphur-Containing Amino Acids and Their Derivatives Adsorbed on Silver. J. Raman Spectrosc. 1991;22:91–96. [Google Scholar]
- 29.Lee PC, Meisel D. Adsorption and Surface-Enhanced Raman of Dyes on Silver and Gold Sols. J. Phys. Chem. 1982;86:3391–3395. [Google Scholar]
- 30.Maiolo D, Bergese P, Mahon E, Dawson KA, Monopoli MP. Surfactant Titration of Nanoparticle–Protein Corona. Anal. Chem. 2014;86:12055–12063. doi: 10.1021/ac5027176. [DOI] [PubMed] [Google Scholar]
- 31.Monopoli MP, Åberg C, Salvati A, Dawson KA. Biomolecular Coronas Provide the Biological Identity of Nanosized Materials. Nat. Nanotechnol. 2012;7:779–786. doi: 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- 32.Requejo R, Hurd TR, Costa NJ, Murphy MP. Cysteine Residues Exposed on Protein Surfaces Are The Dominant Intra-mitochondrial Thiol and May Protect Against Oxidative Damage. FEBS J. 2010;277:1465–1480. doi: 10.1111/j.1742-4658.2010.07576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.