Abstract
The results of a previous study suggested that Cherrie's cane rat (Zygodontomys cherriei) is the principal host of Necoclí virus (family Bunyaviridae, genus Hantavirus) in Colombia. Bayesian analyses of complete nucleocapsid protein gene sequences and complete glycoprotein precursor gene sequences in this study confirmed that Necoclí virus is phylogenetically closely related to Maporal virus, which is principally associated with the delicate pygmy rice rat (Oligoryzomys delicatus) in western Venezuela. In pairwise comparisons, nonidentities between the complete amino acid sequence of the nucleocapsid protein of Necoclí virus and the complete amino acid sequences of the nucleocapsid proteins of other hantaviruses were ≥8.7%. Likewise, nonidentities between the complete amino acid sequence of the glycoprotein precursor of Necoclí virus and the complete amino acid sequences of the glycoprotein precursors of other hantaviruses were ≥11.7%. Collectively, the unique association of Necoclí virus with Z. cherriei in Colombia, results of the Bayesian analyses of complete nucleocapsid protein gene sequences and complete glycoprotein precursor gene sequences, and results of the pairwise comparisons of amino acid sequences strongly support the notion that Necoclí virus represents a novel species in the genus Hantavirus. Further work is needed to determine whether Calabazo virus (a hantavirus associated with Z. brevicauda cherriei in Panama) and Necoclí virus are conspecific.
Key Words: : Necoclí virus, Calabazo virus, Hantavirus, Hantavirus pulmonary syndrome, Zygodontomys cherriei, Zygodontomys brevicauda cherriei, Cherrie's cane rat, Colombia
Introduction
Hantavirus pulmonary syndrome (HPS) is a rodent-borne zoonosis caused by certain members of the virus family Bunyaviridae, genus Hantavirus. The viruses known to cause HPS include Sin Nombre virus (SNV) in Canada and the United States (Stephen et al. 1994, MacNeil et al. 2011); Choclo virus (CHOV) in Panama (Vincent et al. 2000); Rio Mamoré virus (RIOMV) in Brazil, French Guiana, and Peru (Casapía et al. 2012, Matheus et al. 2012, Carvalho de Oliveira et al. 2014); Laguna Negra virus (LANV) in Argentina, Bolivia, Brazil, and Paraguay (Johnson et al. 1997, Levis et al. 2004, Raboni et al. 2009); Andes virus (ANDV) in Argentina, Chile, and Uruguay (López et al. 1996, Toro et al. 1998, Padula et al. 2000); and 11 other named viruses (Table 1). Humans usually become infected with hantaviruses by inhalation of aerosolized droplets of urine, saliva, or respiratory secretions from infected rodents or inhalation of dust contaminated with secretions or excretions from infected rodents.
Table 1.
Rodent-Borne Hantaviruses Included in the Analyses of Nucleotide and Amino Acid Sequences
| GenBank accession numbersc | ||||||
|---|---|---|---|---|---|---|
| Virusa | Strain(s) | Hostb | Country | N protein | GPC | RdRp |
| DOBV | Ano-poroia/Afl9/1999 | Afla | Greece | AJ410615 | AJ410616 | AJ410617 |
| HTNV | 76–118 | Aagr | South Korea | M14626 | M14627 | X55901 |
| SAAV | Saaremaa–160v | Aagr | Estonia | AJ009773 | AJ009774 | AJ410618 |
| SEOV | 80–39 | Rnor | South Korea | AY273791 | S47716 | X56492 |
| THAIV | 741 (Thai749) | Bind | Thailand | AB186420 | (L08756) | (No data) |
| ISLAV | MC-SB-47 | Mcal | United States | U19302 | (No data) | (No data) |
| KHAV | MF-43 | Mfor | Russia | U35255 | AJ011648 | (No data) |
| PHV | PH-1 | Mpen | United States | M34011 | X55129 | EF646763 |
| PUUV | Sotkamo | Mgla | Finland | X61035 | X61034 | Z66548 |
| TOPV | Ls136V | Lsib | Russia | AJ011646 | AJ011647 | (no data) |
| TULV | Tula/Moravia/5302v | Marv | Czech Republic | Z69991 | Z69993 | AJ005637 |
| CARV | 26/2006 | Rsum | Mexico | AB620103 | AB620104 | AB620105 |
| ELMCV | RM-97 | Rmeg | USA | U11427 | U26828 | (no data) |
| HUIV | 200/2006 | Rmeg | Mexico | AB620106 | AB620107 | AB620108 |
| MTNV | 104/2006 | Pbea | Mexico | AB620100 | AB620101 | AB620102 |
| NYVd | Rhode Island-1 | Hsap | United States | U09488 | U36801 | (No data) |
| RIOSV | RMx-Costa-1 | Rmex | Costa Rica | U18100 | (No data) | (No data) |
| SNVd | NM R11 | Pman | United States | L37904 | L37903 | L37902 |
| ANAJVd | H759113/BRA270 | Hsap | Brazil | JX443690 | (No data) | (No data) |
| ANDVd | Chile-9717869 | Olon | Chile | AF291702 | AF291703 | AF291704 |
| ARAUVd | HPR/03–99 | Hsap | Brazil | AY740630 | (No data) | (No data) |
| ARAVd | P5/Cajuru | Hsap | Brazil | EF571895 | (No data) | (No data) |
| BAYVd | HV F0260003 | Opal | United States | GQ200820 | GQ244521 | GQ244526 |
| BCCVd | SPB 9408076 | Shis | United States | L39949 | L39950 | (No data) |
| BMJVd | Oc22531 | Ocha | Argentina | AF482713 | (No data) | (No data) |
| CADV | VHV-574 | Sals | Venezuela | DQ285566 | DQ284451 | GQ200821 |
| CASVd | AN717313/BRA300 | Outi | Brazil | JX443692 | JX443702 | JX443698 |
| CATV | HV C1280001 | Ocou | Honduras | DQ256126 | DQ177347 | FJ858378 |
| CHOVd | 588 | Oful | Panama | DQ285046 | DQ285047 | EF397003 |
| JUQVd | LH_076_12 | Hsap | Brazil | JX173798 | (No data) | (No data) |
| LANVd | 510B | Clau | Paraguay | AF005727 | AF005728 | AF005729 |
| LECVd | 22819 | Ofla | Argentina | AF482714 | AF028022 | (No data) |
| MAPV | HV 97021050 | Odel | Venezuela | AY267347 | AY363179 | EU788002 |
| MCLV | 13796 | Nben | Argentina | AF482716 | (No data) | (No data) |
| MULV | SH-Tx-339 | Shis | United States | U54575 | (No data) | (No data) |
| NECV | HV O-0020003 | Zche | Colombia | KF481954 | KF494345 | KF735065 |
| ORNVd | 22996 | Olon | Argentina | AF482715 | AF028024 | (No data) |
| PRGV | 14403 | Aaza | Argentina | AF482717 | (No data) | (No data) |
| RIOMVd | HTN-007 | Omic | Peru | FJ532244 | FJ608550 | FJ809772 |
Hantaviruses principally associated with: Murine rodents (Muridae, Murinae)—DOBV, Dobrava-Belgrade virus; HTNV, Hantaan virus; SAAV, Saaremaa virus; SEOV, Seoul virus; THAIV, Thailand virus. Arvicoline rodents (Cricetidae, Arvicolinae)—ISLAV, Isla Vista virus; KHAV, Khabarovsk virus; PHV, Prospect Hill virus; PUUV, Puumala virus; TOPV, Topografov virus; TULV, Tula virus. Neotomine rodents (Cricetidae, Neotominae)—CARV, Carrizal virus; ELMCV, El Moro Canyon virus; HUIV, Huitzilac virus; MTNV, Montano virus; NYV, New York virus; RIOSV, Rio Segundo virus; SNV, Sin Nombre virus. Sigmodontine rodents (Cricetidae, Sigmodontinae)—ANAJV, Anajatuba virus; ANDV, Andes virus; ARAUV, Araucaria virus; ARAV, Araraquara virus; BAYV, Bayou virus; BCCV, Black Creek Canal virus; BMJV, Bermejo virus; CADV, Caño Delgadito virus; CASV, Castelo dos Sonhos virus; CATV, Catacamas virus; CHOV, Choclo virus; JUQV, Juquitiba virus; LANV, Laguna Negra virus; LECV, Lechiguanas virus; MAPV, Maporal virus; MCLV, Maciel virus; MULV, Muleshoe virus; NECV, Necoclí virus; ORNV, Orán virus; PRGV, Pergamino virus; RIOMV, Rio Mamoré virus. Italicized names indicate species recognized by the International Committee on Taxonomy for Viruses (Plyusnin et al. 2012).
Afla, Apodemus flavicollis; Aagr, Apodemus agrarius; Rnor, Rattus norvegicus; Bind, Bandicota indica; Mcal, Microtus californicus; Mfor, Microtus fortis; Mpen, Microtus pennsylvanicus; Mgla, Myodes glareolus; Lsib, Lemmus sibiricus; Marv, Microtus arvalis; Rsum, Reithrodontomys sumichrasti; Rmeg, Reithrodontomys megalotis; Pbea, Peromyscus beatae; Hsap, Homo sapiens; Rmex, Reithrodontomys mexicanus; Pman, Peromyscus maniculatus; Olon, Oligoryzomys longicaudatus; Opal, Oryzomys palustris; Shis, Sigmodon hispidus; Ocha, Oligoryzomys chacoensis; Sals, Sigmodon alstoni; Outi, Oligoryzomys utiaritensis; Ocou, Oryzomys couesi; Oful, Oligoryzomys fulvescens; Clau, Calomys laucha; Ofla, Oligoryzomys flavescens; Odel, Oligoryzomys delicatus; Nben, Necromys benefactus; Zche, Zygodontomys cherriei; Aaza, Akodon azarae; Omic, Oligoryzomys microtis.
N protein, nucleocapsid protein gene; GPC, glycoprotein precursor gene; RdRp, RNA-dependent RNA polymerase gene.
Etiologic agent of hantavirus pulmonary syndrome.
The genomes of hantaviruses consist of three single-stranded, negative-sense RNA segments, designated small (S, ∼1.9 kb), medium (M, ∼3.7 kb), and large (L, ∼6.6 kb). These segments encode the nucleocapsid (N) protein, glycoprotein precursor (GPC) to the envelope glycoproteins (Gn and Gc), and RNA-dependent RNA polymerase (RdRp), respectively (Plyusnin et al. 2012). The S genomic segments of some hantaviruses also encode a functional nonstructural (NSs) protein (Jääskeläinen et al. 2007).
Specific knowledge of the hantaviruses associated with rodents in Colombia is limited to a 427-nucleotide fragment of the N protein gene and 1465-nucleotide fragment of the GPC gene of Necoclí virus (NECV) strain 230 (GenBank acc. nos. JN717148 and JN717149, respectively). These sequences were determined from RNA isolated from a Cherrie's cane rat (Zygodontomys cherriei) captured in 2008 in the Municipality of Necoclí, Department of Antioquia, northwestern Colombia (Londoño et al. 2011). Bayesian analyses of chimeric nucleotide sequences (a 376-nucleotide fragment of the N protein gene concatenated to a 202-nucleotide fragment of the GPC gene) suggested that NECV is phylogenetically closely related to Maporal virus (MAPV) and Calabazo virus (CALV), which are naturally associated with the delicate pygmy rice rat (Oligoryzomys delicatus) in western Venezuela (Fulhorst et al. 2004, Hanson et al. 2011) and Cherrie's short-tailed cane rat (Z. brevicauda cherriei) on mainland Panama (Vincent et al. 2000), respectively. The objective of this study was to examine further the phylogenetic relationship of NECV to other hantaviruses.
Materials and Methods
Rodent tissues
Frozen samples of lung (n=13), liver (n=13), and kidney (n=6) from 13 Cherrie's cane rats, all from the Department of Antioquia, were assayed for hantavirus. Rat #4 was captured in July of 2010 near Las Changas (8°32′52.5″N, 76°34′23.7″W), Municipality of Necoclí. Rat #84 was captured in February of 2011 near Alto de Mulatos (8°8′12.5″N, 76°33′1.7″W), Municipality of Turbo. Rat #340 and the 10 other rats were captured in 2007–2008 near Las Changas, Municipality of Necoclí. The species identity of each rodent was determined from external body measurements, morphological features of the skull, and pelage coloration; and the skins, skulls, and skeletons of rats #4 and #84 were archived in a mammal collection at Universidad de Antioquia under accession numbers CTUA 1713 and CTUA 1714, respectively (Dr. Sergio Solari, solari.udea@gmail.com). Previously, hantaviral N protein gene RNA was found in spleens from rats #4 and #84 (C.M.R., unpublished data) and lungs from the 11 rats captured in 2007–2008 (Londoño et al. 2011). The tissue samples in this study were shipped on dry ice to the University of Texas Medical Branch at Galveston in 2013. A majority of these samples had been thawed at least once prior to shipment.
Virus assay
The samples of lung, liver, and kidney were assayed for hantavirus by cultivation in monolayers of Vero E6 cells maintained under a fluid overlay (Milazzo et al. 2010). The weights of the samples tested for hantavirus ranged from 3.4 to 51.5 mg (median, 20.2 mg); Vero E6 cells from the monolayers inoculated with crude homogenates of the tissues were blindly passaged twice, with 12 or 13 days between each passage. Cells harvested on day 36 or 37 after inoculation were tested for hantaviral antigen, using an indirect fluorescent antibody test in which the primary antibody was a hyperimmune mouse ascitic fluid raised against SNV. The inoculation of cultured cells was done inside a class II biosafety cabinet in a biosafety level 2 laboratory; all other work with potentially infectious materials, including cultivation of the inoculated cells, was done in a biosafety level 3 laboratory.
Assay for hantaviral RNA
Samples of lung (n=3), liver (n=2), and kidney (n=2) from rats #4, #84, and #340 were tested for N protein gene RNA. Total RNA was isolated from 29.7–46.0 mg of tissue, using TRIzol® Reagent (Invitrogen Life Technologies, Inc., Carlsbad, CA). First-strand cDNA was synthesized by using Super Script II RNase H- Reverse Transcriptase (Invitrogen Life Technologies, Inc.) with oligonucleotide 5′-GGTGGTTGTGGTAGTAGTAGACTCC-3′ (Morzunov et al. 1995). The first-round PCR assay used MasterTaq (5 PRIME, Inc., Gaithersburg, MD) with oligonucleotides HTS90 (5′-TAGTAGTAGACTCCTTGAGAAGCTA-3′) and HTS17 (5′-GTGCCYACAGACTTTGATGCCAT-3′). The second-round (heminested) PCR assay used MasterTaq with HTS90 and HTS16 (5′-CATTGTRGATTGTGCWGTWGGCA-3′), which flank a 558-nucleotide fragment of the N protein gene of MAPV strain HV 97021050 (GenBank acc. no. AY267347).
Genetic characterization of hantavirus HV O-0020002
The nucleotide sequences of a 1907-nucleotide fragment of the S segment, 3615-nucleotide fragment of the M segment, and 403-nucleotide fragment of the L segment of HV O-0020002 were determined from the first-strand cDNA generated from lung from rat #84. The first-round and second-round PCR assays used MasterTaq. The sequences of the oligonucleotides that were used to prime the PCR assays are available from the first author (C.M.R.).
Genetic characterization of rat #84
The nucleotide sequence of a 1112-bp fragment of the cytochrome b (Cytb) gene of rat #84 was determined to confirm the genus-level identification of this rodent. DNA was isolated from liver by using the DNeasy® Blood and Tissue Kit (Qiagen, Valencia, CA). The PCR assay used REDTaq® (Sigma-Aldrich, St. Louis, MO) with oligonucleotides L14724 (Irwin et al. 1991) and MVZ14 (Smith and Patton 1993).
Sequencing reactions and analysis
Both strands of each purified amplicon were sequenced directly using the Big Dye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). The products of the sequencing reactions were analyzed on an ABI PRISM® 3100-Avant™ or 3130-Avant™ Genetic Analyzer (Applied Biosystems). The N protein gene sequences of hantaviruses HV O-0020001 (rat #4, lung) and HV O-0020011 (rat #340, lung); the nucleotide sequences of the S, M, and L segments of HV-0020002 (rat #84, lung); and the nucleotide sequence of the Cytb gene of rat #84 were deposited into the GenBank nucleotide sequence database under accession numbers KM196123, KM196124, KF481954, KF494345, KF735065, and KM111292, respectively.
Data analysis
The predicted amino acid sequences of the N proteins, GPC, and RdRp of HV O-0020002 and other hantaviruses (Table 1) were aligned by using ClustalW (Thompson et al. 1994). The nucleotide sequences of the N protein genes, GPC genes, and RdRp genes were aligned manually, guided by the computer-generated amino acid sequence alignments. Nonidentities between amino acid sequences were equivalent to uncorrected p distances (Tamura et al. 2011).
The analysis of Cytb gene sequences included four short-tailed cane rats (Z. brevicauda) from Isla Coiba, Panama (GenBank acc. nos. GU397414–GU397417), nine other cane rats (Zygodontomys sp.) from Isla Coiba (GU397410, GU397418–GU397425), 18 cane rats (Zygodontomys sp.) from Brazil (EU652749–EU652766), an Alfaro's rice rat (Handleyomys alfaroi) from Nicaragua (EU579489), and an Atlantic Forest oecomys (Oecomys catherinae) from Brazil (GU126525). The Cytb gene sequences were aligned manually.
The Bayesian analyses were done with MrBayes (v. 3.2.2; Ronquist and Huelsenbeck 2003). The analyses of N protein, GPC, and RdRp gene sequences used the generalized time-reversible (GTR) model of nucleotide substitution, with a proportion of invariable sites and gamma distributed rate variation among sites; the analysis of Cytb gene sequences used the Hasegawa–Kishino–Yano (HKY) model of nucleotide substitution. The models of evolution were selected based on the Bayesian information criterion (BIC) implemented in MEGA (Tamura et al. 2011). The nucleotide sequences of Thottapalayam virus strain VRC 66412 (GenBank acc. nos. AY526097, DQ825771, and DQ825770) were the designated outgroups in the analyses of the N protein, GPC, and RdRp gene sequences, respectively; the nucleotide sequence of the Atlantic Forest Oecomys was the designated outgroup in the analysis of the Cytb gene sequences. Each analysis used the following options in MrBayes (v. 3.2.2)—two independent Markov chain Monte Carlo (MCMC) searches of four chains each, 2 million generations, sample frequency=every 100th generation. The first 200,000 (burn-in) trees were discarded, and the consensus tree (50% majority rule) was estimated from the remaining trees. Probability values in support of the clades were calculated a posteriori, and clades with probability values ≥0.95 were considered supported by the data (Erixon et al. 2003).
Results
The assays for hantavirus in the tissue samples from rats #4, #84, #340, and 10 other cane rats captured in the Department of Antioquia were negative; however, hantaviral N protein gene RNA was found in all seven samples from rats #4 (Las Changas), #84 (Alto de Mulatos), and #340 (Las Changas). In pairwise comparisons, identities among the N protein gene sequences of hantaviruses HV O-0020001 (rat #4), HV O-0020002 (rat #84), HV O-0020011 (rat #340), and NECV strain 230 (GenBank acc. no. JN717148) ranged from 99.6% to 100%, indicating that HV O-0020001, HV O-0020002, HV O-0020011, and strain 230 were conspecific.
The 1907-nucleotide fragment of the S segment and 3615-nucleotide fragment of the M segment of HV O-0020002 included the complete N protein gene (1287 nucleotides) and complete GPC gene (3417 nucleotides), respectively. The N protein gene sequence alignment (40 taxa), GPC gene sequence alignment (30 taxa), and RdRp gene sequence alignment (22 taxa) were 1332, 3477, and 408 characters in length, respectively.
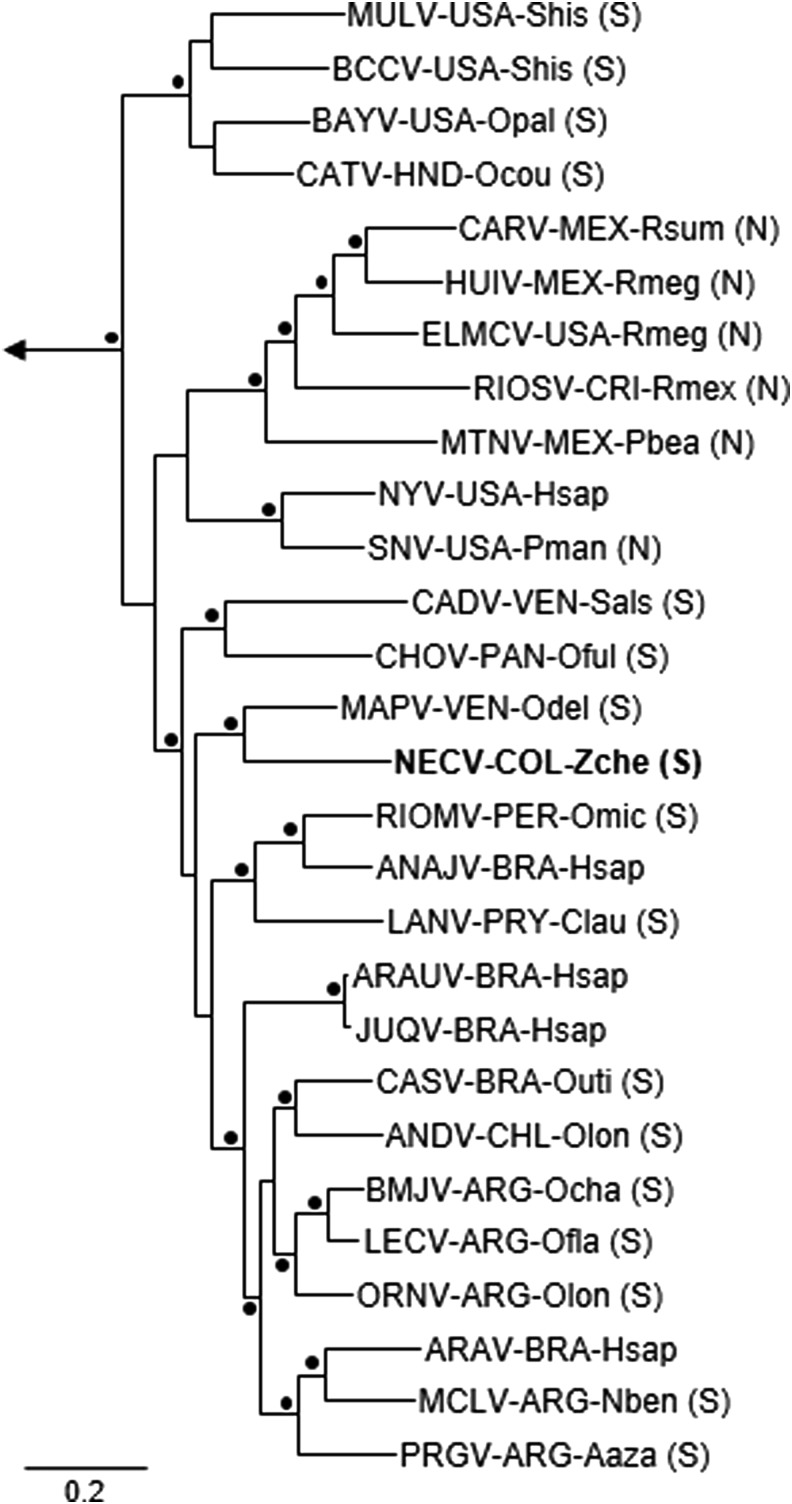
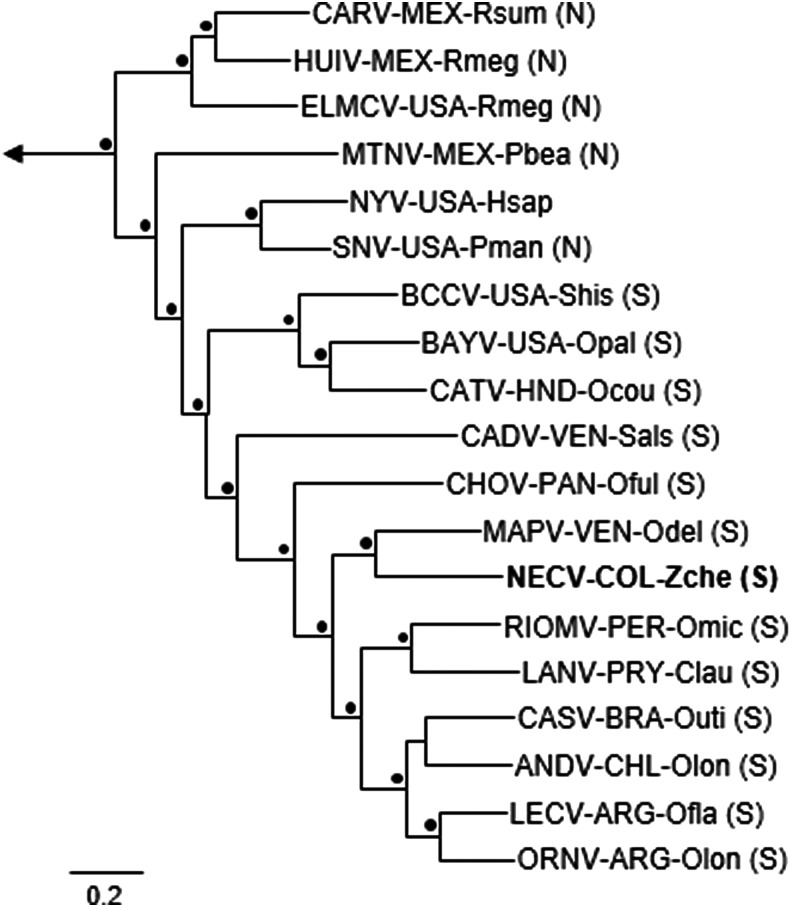
The Bayesian analyses of N protein gene sequences, GPC gene sequences, and RdRp gene sequences segregated the rodent-borne hantaviruses into three groups, in agreement with the taxonomic assignment of their principal hosts—(1) family Muridae, subfamily Murinae (Old World rats and mice), (2) family Cricetidae, subfamily Arvicolinae (voles and lemmings), and (3) family Cricetidae, subfamilies Neotominae and Sigmodontinae (New World rats and mice). Monophyly of the viruses in each group was supported by the results of analyses done a posteriori (clade probability values ≥0.95). The Bayesian analyses of the N protein gene sequences (Fig. 1) and GPC gene sequences (Fig. 2) grouped HV O-0020002 with the other South American hantaviruses and CHOV, and placed HV O-0020002 in a close sister relationship to MAPV. Similarly, the Bayesian analysis of the RdRp gene sequences (tree not shown) grouped HV O-0020002 with ANDV, Castelo dos Sonhos virus (CASV), LANV, MAPV, RIOMV, and CHOV (clade probability value, 1.00), but did not solve the relationships between HV O-0020002 and the six other viruses in this group.
FIG. 1.
Phylogenetic relationships among Necoclí virus strain HV O-0020002 and other hantaviruses naturally associated with neotomine or sigmodontine rodents, on the basis of Bayesian analysis of complete nucleocapsid protein gene sequences. The analysis included five hantaviruses associated with murine rodents (not shown) and six hantaviruses associated with arvicoline rodents (not shown). The branch labels include (in the following order) virus, country, and host species (N, Neotominae; S, Sigmodontinae). ANAJV, Anajatuba virus; ARAUV, Araucaria virus; ARAV, Araraquara virus; ANDV, Andes virus; BAYV, Bayou virus; BCCV, Black Creek Canal virus; BMJV, Bermejo virus; CADV, Caño Delgadito virus; CARV, Carrizal virus; CASV, Castelo dos Sonhos virus; CATV, Catacamas virus; CHOV, Choclo virus; ELMCV, El Moro Canyon virus; HUIV, Huitzilac virus; JUQV, Juquitiba virus; LANV, Laguna Negra virus; LECV, Lechiguanas virus; MAPV, Maporal virus; MCLV, Maciel virus; MTNV, Montano virus; MULV, Muleshoe virus; NECV, Necoclí virus; NYV, New York virus; ORNV, Orán virus; PRGV, Pergamino virus; RIOMV, Rio Mamoré virus; RIOSV, Rio Segundo virus; SNV, Sin Nombre virus. ARG, Argentina; BRA, Brazil; CHL, Chile; COL, Colombia; CRI, Costa Rica; HND, Honduras; MEX, Mexico; PAN, Panama; PER, Peru; PRY, Paraguay; USA, United States of America; VEN, Venezuela. Aaza, Akodon azarae; Clau, Calomys laucha; Hsap, Homo sapiens; Nben, Necromys benefactus; Ocha, Oligoryzomys chacoensis; Ocou, Oryzomys couesi; Odel, Oligoryzomys delicatus; Ofla, Oligoryzomys flavescens; Oful, Oligoryzomys fulvescens; Olon, Oligoryzomys longicaudatus; Omic, Oligoryzomys microtis; Opal, Oryzomys palustris; Outi, Oligoryzomys utiaritensis; Pbea, Peromyscus beatae; Pman, Peromyscus maniculatus; Rmeg, Reithrodontomys megalotis; Rmex, Reithrodontomys mexicanus; Rsum, Reithrodontomys sumichrasti; Sals, Sigmodon alstoni; Shis, Sigmodon hispidus; Zche, Zygodontomys cherriei. ANAJ, ARAV, and JUQV are naturally associated with the sigmodontine species Oligoryzomys fornesi, Bolomys lasiurus, and O. fornesi, respectively (Chu et al. 2009, Firth et al. 2012, Suzuki et al. 2004); NYV is principally associated with the neotomine species Peromyscus leucopus (Huang et al. 1996); and ARAUV is naturally associated with the sigmodontine species Akodon montensis, Akodon paranaensis, Oligoryzomys nigripes, and Oxymycterus judex (Raboni et al. 2012). The length of the scale bar is equivalent to 0.2 substitution per site. A black dot (•) at a node indicates that the clade probability values were ≥95.0. Thottapalayam virus strain VRC 66412 was the designated outgroup.
FIG. 2.
Phylogenetic relationships among Necoclí virus strain HV O-0020002 and other hantaviruses naturally associated with neotomine or sigmodontine rodents, on the basis of Bayesian analysis of complete glycoprotein precursor gene sequences. The analysis included five hantaviruses associated with murine rodents (not shown) and five hantaviruses associated with arvicoline rodents (not shown). The labels are the same as those in Fig. 1. The length of the scale bar is equivalent to 0.2 substitution per site. A black dot (•) at a node indicates that the clade probability values were ≥95.0. Thottapalayam virus strain VRC 66412 was the designated outgroup.
Nonidentities among the amino acid sequences of the N proteins of the hantaviruses associated with neotomine or sigmodontine rodents ranged from 0.0% (Araucaria virus [ARAUV] and Juquitiba virus [JUQV]) to 18.6% (LANV and Rio Segundo virus [RIOSV]), and nonidentities between the amino acid sequence of the N protein of HV O-0020002 and the amino acid sequences of the N proteins of other hantaviruses associated with sigmodontine rodents ranged from 8.7% (Table 2) to 15.1% (Muleshoe virus [MULV]). Similarly, nonidentities among the amino acid sequences of the GPC of the hantaviruses associated with neotomine or sigmodontine rodents ranged from 4.3% (Lechiguanas virus [LECV] and Orán virus [ORNV]) to 26.3% (LANV and Montano virus [MTNV]), and nonidentities between the amino acid sequence of the N protein of HV O-0020002 and the amino acid sequences of the N proteins of other hantaviruses associated with sigmodontine rodents ranged from 11.7% to 24.2% (Table 2).
Table 2.
Nonidentities among the Complete Amino Acid Sequences of the Nucleocapsid Proteins and among the Complete Amino Acid Sequences of the Glycoprotein Precursors of Hantaviruses Associated with Sigmodontine Rodents
| Nucleocapsid protein (% sequence nonidentity) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Virusa | NECV | ANDV | CADV | CHOV | LANV | MAPV | RIOMV | Other |
| NECV | — | 10.8 | 14.6 | 11.1 | 12.0 | 8.7 | 10.8 | ≥10.4 |
| ANDV | 14.3 | — | 14.2 | 9.7 | 9.4 | 8.0 | 8.7 | ≥2.8 |
| CADV | 24.2 | 25.3 | — | 14.4 | 15.8 | 13.4 | 15.1 | ≥13.7 |
| CHOV | 17.4 | 16.8 | 23.0 | — | 12.0 | 10.1 | 10.1 | ≥9.0 |
| LANV | 15.1 | 13.0 | 25.5 | 18.0 | — | 10.4 | 7.3 | ≥7.3 |
| MAPV | 11.7 | 14.6 | 24.5 | 16.7 | 17.8 | — | 8.3 | ≥8.0 |
| RIOMV | 14.9 | 11.7 | 24.8 | 16.5 | 8.8 | 15.1 | — | ≥2.8 |
| Other | ≥14.0 | ≥6.0 | ≥23.6 | ≥16.7 | ≥12.9 | ≥14.2 | ≥10.5 | — |
| Glycoprotein precursor (% sequence nonidentity) | ||||||||
NECV, Necoclí virus; ANDV, Andes virus; CADV, Caño Delgadito virus; CHOV, Choclo virus; LANV, Laguna Negra virus; MAPV, Maporal virus; RIOMV, Rio Mamoré virus; Other—Anajatuba, Araucaria, Araraquara, Bayou, Black Creek Canal, Bermejo, Castelo dos Sonhos, Catacamas, Juquitiba, Lechiguanas, Maciel, Muleshoe, Orán, and Pergamino viruses. Nonidentities among the complete amino acid sequences of the nucleocapsid proteins and complete amino acid sequences of the glycoprotein precursors of the 14 “other” hantaviruses ranged from 0.0–15.1% and 4.3–24.3%, respectively.
Examination of the N protein gene sequence alignment revealed a 189-nucleotide open reading frame (ORF) in the N protein gene of HV O-0020002, 156-nucleotide ORF in the N protein gene of RIOSV, and 189-nucleotide ORF in the N protein gene of each of the 26 other hantaviruses associated with neotomine or sigmodontine rodents. The first nucleotide position in each ORF was 79 nucleotides downstream from the first nucleotide position in the corresponding N protein gene; as such, the reading frame of the ORF was +1 relative to the N protein gene. In pairwise comparisons, nonidentities among the amino acid sequences of the putative NSs proteins of the hantaviruses associated with neotomine or sigmodontine rodents ranged from 1.6% (ARAUV and JUQV, Bermejo virus [BMJV] and LECV) to 46.0% (Carrizal virus [CARV] and Maciel virus [MCLV]); nonidentities between the amino acid sequence of the NSs protein of HV O-0020002 and the amino acid sequences of the NSs proteins of the other South American hantaviruses ranged from 15.9% (LANV) to 42.9% (Caño Delgadito virus [CADV]); and the amino acid sequence of the NSs protein of HV O-0020002 was 20.6% different from the amino acid sequence of the NSs protein of MAPV.
The Bayesian analysis of Cytb gene sequences indicated that rat #84 was more closely related to the four short-tailed cane rats (Z. brevicauda) from Isla de Coiba than to the nine other cane rats (Zygodontomys sp.) from Isla Coiba or 18 cane rats from Brazil. The monophyly of rat #84 and the four short-tailed cane rats from Isla de Coiba was supported by analyses done a posteriori (clade probability values, 1.00).
Discussion
Presently, there are no isolates of NECV, CALV, or 16 of the 27 other hantaviruses naturally associated with neotomine or sigmodontine rodents (Table 1), even though a majority of hantaviral isolates seem to grow readily in monolayers of Vero E6 cells. The failure to isolate hantavirus from the rodents in this study may be related to the small sizes or suboptimal quality of the tissue samples.
The results of the analyses of complete N protein gene sequences and complete GPC gene sequences confirm the close phylogenetic relationship between NECV and MAPV (Londoño et al. 2011). Presently, our knowledge of the genome of CALV is limited to the nucleotide sequences of a 376-nucleotide fragment of the N protein gene, 292-nucleotide fragment of the GPC gene that encodes the Gn, and 205-nucleotide fragment of the GPC gene that encodes the Gc (GenBank acc. nos. AF395443, AF402331, and AF402333, respectively). In pairwise comparisons, nonidentities between these sequences and the homologous sequences of NECV strain HV O-0020002 and MAPV strain HV 97021050 were 14.9% (amino acid sequence nonidentity, 0.0%) and 19.5% (4.8%), 14.0% (2.1%) and 24.7% (10.4%), and 14.6% (4.5%) and 23.4% (7.5%), respectively, We anticipate that analyses of complete N protein gene sequences and complete GPC gene sequences in a future study will determine whether CALV is phylogenetically more closely related to NECV or MAPV.
Specific members of the Muridae or Cricetidae are the principal hosts of many of the hantaviruses for which natural host relationships have been well characterized. The results of a previous study (Londoño et al. 2011) strongly suggested that Z. cherriei is the principal host of NECV in northwestern Colombia. Presently, our knowledge of the natural host relationships of CALV is limited to a single CALV-infected cane rat (Z. b. cherriei) captured on the Azuero Peninsula, mainland Panama (Vincent et al. 2000). The results of the Bayesian analysis of Cytb gene sequences in this study indicated that rat #84 from Alto de Mulatos was a zygodont (Zygodontomys sp.) and more closely related to the zygodonts from Isla de Coiba, Panama, than to the zygodonts from Brazil. Whether Z. b. cherriei is the principal host of CALV in Panama and whether Z. b. cherriei on the Azuero Peninsula and Z. cherriei in northwestern Colombia are synonymous remain to be determined.
The Ninth Report of the International Committee on Taxonomy of Viruses (Plyusnin et al. 2012) set forth criteria for species demarcation in the genus Hantavirus, including the virus must (1) exhibit at least a 7% difference in amino acid sequence identity from other hantaviruses in comparisons of complete N protein sequences and comparisons of complete GPC sequences, (2) occupy a unique ecological niche, and (3) represent a unique serotype (as defined by results of two-way cross-neutralization tests). The results of the pairwise comparisons of complete N protein sequences and complete GPC sequences in this study support the notion that the hantavirus associated with Z. cherriei in northwestern Colombia represents a novel hantaviral species, previously named “Necoclí virus” (Londoño et al. 2011). Whether NECV and CALV are conspecific may be decided by comparisons of complete N protein sequences and comparisons of complete GPC sequences in a future study.
The exact conservation of the location and length of the NSs ORF in the N protein genes of 27 of the 28 hantaviruses associated with neotomine or sigmodontine rodents suggests that the ORF is functional and that the NSs protein plays a critical role in the biology of these viruses in their respective principal hosts. Hypothetically, the NSs protein also affects the course and outcome of hantaviral infections in humans.
Presently, our knowledge of HPS in Colombia is limited to a nonlethal case that was treated in the Department of Córdoba, northwestern Colombia (Mattar et al. 2014). We note that the identity of the etiologic agent in this case was not determined. Whether NECV is an agent of HPS in the Department of Antioquia or elsewhere in Colombia remains to be investigated.
Acknowledgments
The rodents were captured by students and mammalogists from Universidad de Antioquia in the course of a project entitled “Ecological study of rickettsia endemicity in Colombia.” Sergio Solari (Insituto de Biología, Universidad de Antioquia) assisted with determination of the species identities of the rodents. Leidy Y. Acevedo-Gutiérrez (Universidad de Antioquia) helped prepare the manuscript.
This study was supported financially by grant 111554531578 and doctoral student scholarship grant 567 from Colciencias (Bogotá, D.C., Department of Cundinamarca, Colombia), and funds from the sustainability program 2013–2014, Universidad de Antioquia. The laboratory work at University of Texas Medical Branch (UTMB) was financially supported by grant AI-067947 from the US Department of Health and Human Services, National Institutes of Health.
Author Disclosure Statement
No competing financial interests exist.
References
- Carvalho de Oliveira R, Cordeiro-Santos M, Guterres A, Fernandes J, et al. Rio Mamore virus and hantavirus pulmonary syndrome, Brazil. Emerg Infect Dis 2014; 20:1568–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casapía M, Mamani E, García MP, Miraval ML, et al. Síndrome pulmonar por hantavirus (virus Río Mamoré) en la Amazonía Peruana. Rev Peru Med Exp Salud Publica 2012; 29:390–395 [DOI] [PubMed] [Google Scholar]
- Chu YK, Goodin D, Owen RD, Koch D, et al. Sympatry of 2 hantavirus strains, Paraguay, 2003–2007. Emerg Infect Dis 2009; 15:1977–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erixon P, Svennblad B, Britton T, Oxelman B. Reliability of Bayesian posterior probabilities and bootstrap frequencies in phylogenetics. Syst Biol 2003; 52:665–673 [DOI] [PubMed] [Google Scholar]
- Firth C, Tokarz R, Simith DB, Nunes MRT, et al. Diversity and distribution of hantaviruses in South America. J Virol 2012; 86:13756–13766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulhorst CF, Cajimat MNB, Utrera A, Milazzo ML, et al. Maporal virus, a hantavirus associated with the fulvous pygmy rice rat (Oligoryzomys fulvescens) in western Venezuela. Virus Res 2004; 104:139–144 [DOI] [PubMed] [Google Scholar]
- Hanson JD, Utrera A, Fulhorst CF. The delicate pygmy rice rat (Oligoryzomys delicatus) is the principal host of Maporal virus (family Bunyaviridae, genus Hantavirus). Vector Borne Zoonotic Dis 2011; 11:691–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Campbell WP, Means R, Ackman DM. Hantavirus S RNA sequence from a fatal case of HPS in New York. J Med Virol 1996; 50:5–8 [DOI] [PubMed] [Google Scholar]
- Irwin DM, Kocher TM, Wilson AC. Evolution of the cytochrome b genes of mammals. J Mol Evol 1991; 32:128–144 [DOI] [PubMed] [Google Scholar]
- Jääskeläinen KM, Kaukinen P, Minskaya ES, Plyusnina A, et al. Tula and Puumala hantavirus NSs ORFs are functional and the products inhibit activation of the interferon-beta promoter. J Med Virol 2007; 79:1527–1536 [DOI] [PubMed] [Google Scholar]
- Johnson AM, Bowen MD, Ksiazek TG, Williams RJ, et al. Laguna Negra virus associated with HPS in western Paraguay and Bolivia. Virology 1997; 238:115–127 [DOI] [PubMed] [Google Scholar]
- Levis S, Garcia J, Pini N, Calderón G, et al. Hantavirus pulmonary syndrome in northwestern Argentina: Circulation of Laguna Negra virus associated with Calomys callosus. Am J Trop Med Hyg 2004; 71:658–663 [PubMed] [Google Scholar]
- Londoño AF, Díaz FJ, Agudelo-Flórez P, Levis S, et al. Genetic evidence of hantavirus infections in wild rodents from northwestern Colombia. Vector Borne Zoonotic Dis 2011; 11:701–708 [DOI] [PubMed] [Google Scholar]
- López N, Padula P, Rossi C, Lázaro ME, et al. Genetic identification of a new hantavirus causing severe pulmonary syndrome in Argentina. Virology 1996; 220:223–226 [DOI] [PubMed] [Google Scholar]
- MacNeil A, Ksiazek TG, Rollin PE. Hantavirus pulmonary syndrome, United States, 1993–2009. Emerg Infect Dis 2011; 17:1195–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheus S, Lavergne A, de Thoisy B, Dussart P, et al. Complete genome sequence of a novel hantavirus variant of Rio Mamoré virus, Maripa virus, from French Guiana. J Virol 2012; 86:5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattar S, Garzon D, Tadeu L, Faccini-Martinez AA, et al. Serological diagnosis of hantavirus pulmonary syndrome in a febrile patient in Colombia. Int J Infect Dis 2014; 5:201–203 [DOI] [PubMed] [Google Scholar]
- Milazzo ML, Duno G, Utrera A, Richter MH, et al. Natural host relationships of hantaviruses native to western Venezuela. Vector Borne Zoonotic Dis 2010; 10:605–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morzunov SP, Feldman H, Spiropoulou CF, Semenova VA, et al. A newly recognized virus associated with a fatal case of hantavirus pulmonary syndrome in Louisiana. J Virol 1995; 69:1980–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padula PJ, Colavecchia SB, Martinez VP, Gonzalez Della Valle MO, et al. Genetic diversity, distribution, and serological features of hantavirus infection in five countries in South America. J Clin Microbiol 2000; 38:3029–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plyusnin A, Beaty BJ, Elliott RM, Goldbach R, et al. Family Bunyaviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. California: Elsevier, 2012:725–774 [Google Scholar]
- Raboni SM, de Borba L, Hoffmann FG, de Noronha L. Evidence of circulation of Laguna Negra-like hantavirus in the Central West of Brazil: Case report. J Clin Virol 2009; 45:153–156 [DOI] [PubMed] [Google Scholar]
- Raboni SM, Delfraro A, de Borba L, Teixeira BR, et al. Hantavirus infection prevalence in wild rodents and human anti-hantavirus serological profiles from different geographic areas of South America. Am J Trop Med Hyg 2012; 87:371–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003; 19:1572–1574 [DOI] [PubMed] [Google Scholar]
- Smith MF, Patton JL. The diversification of South American muroid rodents: Evidence from mitochondrial DNA sequence data for the Akodontine tribe. Biol J Linn Soc 1993; 50:149–177 [Google Scholar]
- Stephen C, Johnson M, Bell A. First reported cases of hantavirus pulmonary syndrome in Canada. Can Commun Dis Rep 1994; 20:121–125 [PubMed] [Google Scholar]
- Suzuki A, Bisordi I, Levis S, Garcia J, et al. Identifying rodent hantavirus reservoirs, Brazil. Emerg Infect Dis 2004; 10:2127–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W (1.7): Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 1994; 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro J, Vega DJ, Khan AS, Mills JN, et al. An outbreak of hantavirus pulmonary syndrome, Chile, 1997. Emerg Infect Dis 1998; 4:687–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent MJ, Quiroz E, Gracia F, Sanchez AJ, et al. Hantavirus pulmonary syndrome in Panama: Identification of novel hantaviruses and their likely reservoirs. Virology 2000; 277:14–19 [DOI] [PubMed] [Google Scholar]