Abstract
We used literature searches and a database of all reported emerging infectious diseases (EIDs) to analyze the most important transmission pathways (e.g., vector-borne, aerosol droplet transmitted) for emerging zoonoses. Our results suggest that at the broad scale, the likelihood of transmission occurring through any one pathway is approximately equal. However, the major transmission pathways for zoonoses differ widely according to the specific underlying drivers of EID events (e.g., land-use change, agricultural intensification). These results can be used to develop better targeting of surveillance for, and more effective control of newly emerged zoonoses in regions under different underlying pressures that drive disease emergence.
Key Words: : Surveillance, Transmission routes, Pathway, Direct contact, Vector-borne, Virus, Zoonosis
Introduction
Emerging infectious diseases (EIDs) have significant public health and economic impacts and are increasing in frequency (Brahmbhatt 2005, Jones et al. 2008). Nearly two-thirds of EIDs are zoonotic and three-quarters of those originate in wildlife (Jones et al. 2008), thus targeted disease surveillance may be useful to optimize prevention and control measures and reduce the threat of future zoonotic EIDs (Karesh et al. 2005, Vrbova et al. 2010). Recent approaches to disease surveillance and control have largely been disease specific and reactive in nature, tackling pathogens after they have already emerged (Childs and Gordon 2009). Proactive approaches include pathogen discovery in wildlife to identify potential zoonoses (Anthony et al. 2013, Lipkin 2013), however the number of microbes in wildlife remaining to be discovered is likely large and the causes and dynamics of transmission from wildlife to human are poorly understood (Jones et al. 2008, Childs and Gordon 2009, Anthony et al. 2013). As pathogens continue to emerge from wildlife, a better understanding of the ways through which transmission could potentially occur is needed. Because pathogens with different transmission pathways may require very different prevention and control strategies, understanding the relative importance of each pathway for a given pathogen is essential.
Zoonoses can be transmitted from wildlife to humans by a range of routes (or pathways), yet research exploring the role of transmission pathways in past EID events has not been consolidated. Here, we analyze transmission pathways of all known previously emerging zoonoses. Our results suggest that the relative importance of different transmission pathways varies by EID driver (e.g., land use change, bushmeat consumption, climate, and weather). This suggests that targeting pathogen discovery and surveillance programs to different transmission pathways will increase our capacity to identifying important pathogens, either known or unknown.
Materials and Methods
We identified all unique zoonotic pathogens (n=183) from a published database of 335 emerging infectious disease “events” (the original case or cluster of cases representing an infectious disease emerging in human populations for the first time) from 1940 to 2004 (Jones et al. 2008). This database includes EID events caused by newly evolved strains of pathogens, novel pathogens that have entered human populations for the first time, and pathogens that have likely been established in humans historically, but that have recently increased in incidence (Jones et al. 2008). We excluded emergence events that arose through the evolution of antimicrobial drug resistance and those caused by newly evolved strains of known pathogens (e.g., multidrug-resistant tuberculosis and chloroquine-resistant malaria), as well as those attributed to human susceptibility to infection (disease outbreaks that are secondary to immunodeficiency diseases), leaving 148 records in our database.
We then conducted a systematic literature search to identify all documented animal-to-human transmission pathways for each pathogen and the relative contribution of each when more than one transmission route was described. Transmission pathways were identified through a comprehensive search in the ISI Web of Science online database, performing a topic search using the string “[pathogen name]? AND *transmission* AND (route* OR pathway*) from 1940 to present. The use of this combination of key words allowed for the identification of all documented pathways. Pathogen synonyms were included in the search. Supplementary references, including World Health Organization (WHO) and Centers for Disease Control and Prevention (CDC) fact sheets, were used to provide additional information about transmission routes from animals to humans. We identified five broad but distinct transmission pathway categories: (1) Direct contact (skin-to-skin contact; scratches; animal bites; contact with body fluids, organs, and tissues; direct large droplet exposure); (2) airborne transmission (via dust particles and airborne small droplets); (3) vector-borne (by biting or mechanical transfer by arthropods); (4) oral transmission (consumption of contaminated food or water); and (5) contaminated environment or fomite (indirect contact with soil or vegetation, contact with water, indirect transmission by contaminated inanimate objects). Where no transmission route was documented, this information was assumed to be unknown, and these pathogens were excluded from the analysis (n=3), leaving a total of 145 records in our database (Supplemental Table 1).
To determine whether zoonotic diseases are more likely to be transmitted through specific pathways, we first assigned all transmission pathways per pathogen equal weighting as per methods published in Taylor et al. (2001) and calculated the total proportion of zoonotic EIDs transmitted by each pathway at a broad scale. However, for many pathogens, some modes of transmission are more frequently implicated than others. To account for this, we then assigned all transmission pathways per pathogen different weightings based on how likely transmission is to occur through that particular pathway—either “likely” or “less likely” (Supplemental Table 1).
Each pathogen in the analysis received a total value of 1. For those pathogens that could only be transmitted through one pathway (e.g., West Nile), that pathway received the full value of 1 (assuming transmission would occur via that pathway 100% of the time). However, for organisms that could be transmitted by more than one pathway (e.g., rabies), any pathway that is documented in the literature as likely to occur (e.g., rabies infection via a bite or scratch) received a weight of 0.9, while the remaining value was split equally between the less common pathways (e.g., rabies infection via airborne transmission).
Likewise, if the literature indicated that transmission through a particular pathway was rare or unlikely to occur, this pathway received a weight of 0.1 (assuming that transmission via that pathway was likely to occur less than 10% of the time), and the remaining value of 0.90 was split equally among the other pathway(s). If a pathogen could be transmitted via multiple “unlikely” or “rare” pathways, the value of 0.1 was split equally between those pathways. In the literature, rare or unlikely routes of transmission generally refer to those that have been demonstrated in experimental laboratory studies and/or where only a handful of human cases have been documented (e.g., four cases of rabies attributed to the airborne pathway, two documented suspected cases of tick-borne Q fever; Supplemental Table 1).
For example, for the bacteria Coxiella burnetii (Q fever), the most common mode of transmission reported is airborne transmission via inhalation of aerosols from contaminated soil or animal waste. More rare modes of transmission to humans include tick bites and ingestion of unpasteurized milk or dairy products (Anderson et al. 2013). For this pathogen, we assigned the common pathway, the airborne pathway, a probability of 0.9, whereas the two less common pathways were assigned a probability of 0.05 each (Supplemental Table 1).
We then examined which transmission pathways were most likely to occur within various EID drivers, as defined in Smolinski et al. (2003) and Morse (1995) and modified by Jones et al. (2008). These drivers are largely environmental, ecological, political, economic, and social forces, functioning on a range of different scales, which facilitate the expansion and adaptation of a pathogen to a new niche (Smolinski et al. 2003). The first classification of these drivers was published in 1992 by the Institute of Medicine (IOM). This report identified six factors in the emergence of infectious diseases including: (1) Human demographics and behavior, (2) technology and industry, (3) economic development and land use, (4) international travel and commerce, (5) microbial adaptation and change, and (6) breakdown of public health measures. Seven additional drivers were added in a follow-up IOM report in 2003 (Smolinski et al. 2003) including: “human susceptibility to infection,” “climate and weather,” “changing ecosystems,” “poverty and social inequity,” “war and famine,” “lack of political will,” and “intent to harm.” To calculate which pathways were associated with each EID driver, we split the data into subsets by driver, summed the weights for each transmission category, and divided the sum by the total number of EID events per driver.
Last, to determine whether transmission routes differ significantly by disease driver, we used a permutation t-test (Hothorn et al. 2006, 2008) to compare all possible pairwise disease drivers (121 possible combinations). To minimize the probability of detecting false positives (i.e., decrease Type I error) we selected a rejection alpha of 0.10 (Quinn and Keough 2002). This level of significance was selected given the inherent reporting bias of EID data (e.g., developed countries tend to report more than other countries) and the low sample size for some of the drivers (e.g., Bushmeat n=4).
Results
In a previous study, Taylor et al. (2001) included transmission route as a potential risk factor for human disease emergence. In their analysis, if an organism could be transmitted by more than one transmission pathway, all were included with equal weighting. They found that zoonotic diseases were more likely to be transmitted by vectors (by biting or mechanical transfer by arthropods) or indirect contact (via food or an environmental reservoir). In accordance with their results, we found that when all pathways in our analysis received an equal weighting, zoonotic diseases were more likely to be transmitted by the same two pathways that Taylor identified—oral transmission and by vectors (Supplemental Table 1; Fig. 1). Because some pathogens could be transmitted by more than one pathway, we found that, at a broad scale, 42% of all zoonotic pathogens were transmitted through oral transmission, 42% via vector-borne, 36% by airborne transmission, 29% by direct contact, and 24% via contact with a contaminated environment or fomite.
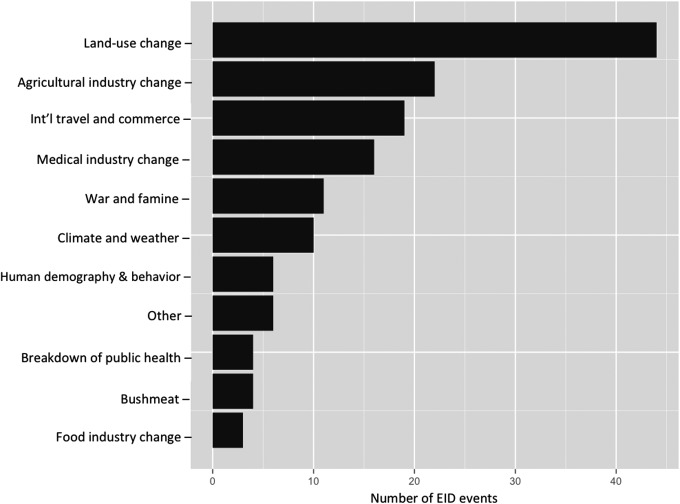
FIG. 1.
Number of previous emergence events by primary drivers of disease as defined by Jones et al. (2008).
When broken down by pathogen type, the majority of zoonotic EIDs were viral and bacterial in origin, with smaller proportions of helminth, fungi, and prion-origin diseases. For viruses, the vector-borne route of transmission was the most common, followed by airborne transmission and direct animal contact. Very few viral EIDs were transmitted through the foodborne pathway through exposure to a contaminated environment, or via fomites. For bacteria, transmission was most likely to occur through the foodborne, contaminated environment, and direct-contact pathways. Fewer bacterial EIDs were transmitted through the airborne and vector-borne pathways. The majority of rickettsial and protozoal infections were likely transmitted through vectors, whereas the direct contact and airborne pathways were most relevant for fungal diseases. Last, the oral transmission pathway was most relevant for helminth and prion-driven diseases.
When ranked by primary EID driver; land-use change, agricultural industry change, and international travel and commerce are globally the top three drivers of zoonoses (Fig. 1). At the broad scale, our results indicate that all pathways are approximately equally common, yet when stratified by EID driver, the relevant transmission pathways vary greatly (Fig. 2).
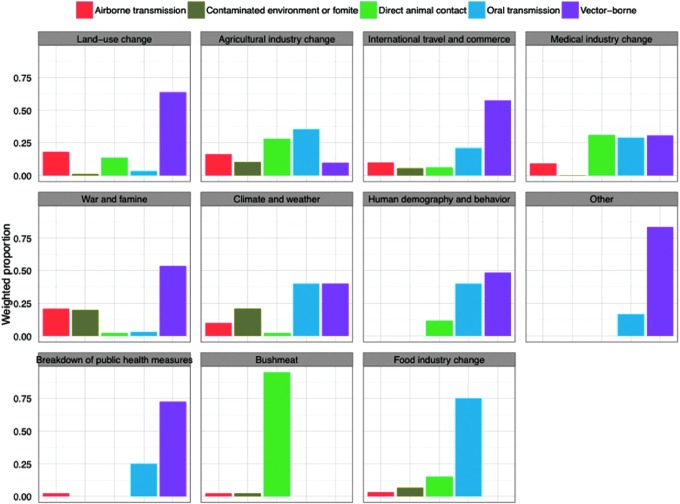
FIG. 2.
Scaled number of zoonotic emerging infectious diseases (EID) events (n=145) per transmission route categorized by the primary driver of disease emergence for each pathogen.
Results from the permutation t-tests indicate that transmission pathways differ significantly between many disease drivers (Table 1). For example, zoonotic diseases attributed to land-use change were more likely to be transmitted via the vector-borne pathway (52.5%), followed by direct animal contact (23.8%), the airborne pathway (19.8%), and a smaller proportion from the contaminated environment (2%) and oral transmission pathways (2%; Fig. 2). For food industry change, the most relevant pathway is oral transmission (58%) (Fig. 2).
Table 1.
Pairwise Permutation t-Test Results Comparing the Difference in Documented Pathways Between All Possible Pairwise Disease Drivers (121 Possible Combinations)
| Driver comparison | Z score | p value |
|---|---|---|
| Agricultural industry change vs. food industry change | 2.243 | 0.016 |
| Agricultural industry change vs. breakdown of public health | 2.142 | 0.024 |
| Bushmeat vs. agricultural industry change | 2.062 | 0.032 |
| Human demography and behavior versus agricultural industry change | 1.993 | 0.048 |
| Bushmeat vs. land-use change | −1.466 | 0.056 |
| Food industry change vs. land-use change | −1.502 | 0.056 |
| Breakdown of public health vs. land-use change | −1.471 | 0.063 |
| Food industry change versus international travel and trade | −1.535 | 0.063 |
| Land-use change vs. other | 1.401 | 0.063 |
| Land-use change vs. breakdown of public health | −1.471 | 0.063 |
| Med industry change vs. breakdown of public health | −1.765 | 0.071 |
| Bushmeat vs. medical industry changes | −1.667 | 0.071 |
| Agricultural industry change vs. other | 1.822 | 0.079 |
| Food industry change vs. medical industry change | −1.917 | 0.079 |
| Breakdown of public health vs. international travel and trade | −1.442 | 0.087 |
This table shows only statistically significant combinations, sorted from highly significant (0.016) to less significant (0.087). We selected an alpha value of rejection of 0.10 (90% level of confidence). Analysis was performed using the coin library in the R statistics program (Hothorn et al. 2006, 2008).
Discussion
An important challenge to developing effective prevention and control strategies for zoonotic EIDs is identifying the relevant transmission routes between reservoir hosts and humans. In many cases, primary pathways for transmission are identified during outbreak investigations after a disease has already emerged. However, it is likely that many more novel transmission pathways remain undescribed for pathogens that pose a potential human health risk. The recent emergence of a number of pandemic zoonoses (e.g., severe acute respiratory syndrome [SARS], pandemic influenza H1N1), zoonotic viruses of pandemic potential (e.g., Middle East respiratory syndrome [MERS] coronavirus), and those of regional concern (e.g., Ebola virus), in addition to the increasing frequency of EID events (Jones et al. 2008), make the targeting of surveillance programs to early stages of emergence a crucial tool for combating pandemics (Morse et al. 2012). Our analysis also provides us with insights into which transmission pathways are important in regions where different drivers of EIDs predominate, thus allowing for more targeted prevention measures and surveillance approaches. This approach may have value for targeted surveillance of pathogens that are known to emerge through different pathways. For instance, although Nipah virus (NiV) has been attributed to preferential feeding by Pteropus bats on human-cultivated fruit or other plant products, as was seen with NiV in Malaysia (transmitted via mangoes fed to pigs) and Bangladesh (via date palm sap) (Chua et al. 2000, Luby et al. 2006), the mechanism through which the disease emerged differed between countries.
In Malaysia, agricultural intensification led to the planting of mango trees directly adjacent to intensively managed pig populations, attracting fruit bats to the area. This activity resulted in the initial spillover of NiV from Pteropus bats into intensively managed pig populations, leading to subsequent outbreaks to human via direct contact with live pigs (Chua 2003, Epstein et al. 2006, Pulliam et al. 2012). In Bangladesh, the most frequently implicated transmission pathway from animals to people is via the oral transmission pathway, specifically ingestion of fresh date palm sap contaminated by bat excreta (Hughes et al. 2009, Olival et al. 2013). By targeting this and other potential routes of food contamination, surveillance efforts and control measures may better minimize the risk of zoonotic disease emergence.
A more apparent example highlighting the differences in transmission routes between drivers is land-use change versus climate and weather. Land-use change is the leading driver for emerging zoonoses (Fig. 1) and is likely to increase in the future (Smolinski et al. 2003, Patz et al. 2004, Murray and Daszak 2013). Our results suggest that disease emergence in regions under pressure from land use change has most often occurred through the vector-borne pathway and direct animal contact. Thus, effective control measures in regions of active land use change could be focused on ensuring or enhancing vector control (e.g., larval control through environmental management, use of mosquito nets, etc.), preventing transmission through direct animal contact (e.g., use of personal protective equipment, hand washing), and reducing risk of airborne transmission (e.g., use of mask or light cloth to prevent inhalation of infectious agents in high-risk occupations or in areas where land disruption activities are occurring).
Infectious diseases driven by climate and weather are more likely to be transmitted via the oral transmission and vector-borne pathways. The relevance of these particular pathways to this driver can be explained by examining the events in our database linked to climate and weather. For instance, there are 10 events/pathogens that are associated with climate and weather including five Vibro spp., Coccidioides immitis (Valley fever), and several vector-borne diseases, including Murray Valley encephalitis virus, Sindbis virus, St. Louis encephalitis virus, and Zika virus. Previous work has shown climate-related increases in sea surface temperature and sea level can lead to a higher incidence of food-borne (shellfish poisoning) and water-borne infectious diseases, highlighting the importance of the oral transmission pathway. In fact, recent studies examining climate change impacts on human and animal health found a positive correlation between the prevalence of Vibro spp., including V. cholerae, and increasing sea surface temperature. Similarly, changes in climate have also been shown to affect disease transmission of many vector-borne diseases by shifting the vector's geographic range, increasing reproductive and biting rates and by shortening the incubation period (Vezzulli et al. 2013, Patz et al. 2005). Although this finding is not surprising, it does allow us to identify strategies to reduce oral transmission (e.g., food safety measures) and vector-borne transmission in regions with changing climates.
In summary, these findings provide us with a novel approach to identifying and understanding all the ways by which a pathogen might spill over from nature into human hosts. These results may be particularly useful to target the growing number of efforts to discover and characterize new pathogens before they emerge in human populations, coupled with efforts that identify potential zoonotic pathogen reservoirs and specific high-risk behavior in a region. Our results could be compiled into a catalogue of relevant transmission pathways and reservoirs. Targeting pathogen discovery through this approach could tell us new information about how likely a newly discovered potentially zoonotic pathogen is to emerge. Public health efforts could develop control strategies that focus on behavioral change in high-risk populations to minimize exposure to potential reservoirs and target relevant transmission pathways (e.g., personal protective behaviors for direct animal contact pathway, vector control targeted at larval mosquitos, use of bed nets, etc., for the vector-borne pathway). This approach will allow zoonotic disease surveillance to shift toward a more preemptive strategy and use more targeted public health interventions to prevent zoonotic disease spillover and emergence.
Supplementary Material
Acknowledgments
This study was made possible by the generous support of the American people through the United States Agency for International Development (USAID) Emerging Pandemic Threats PREDICT (cooperative agreement no., GHN-A-OO-09-00010-00). The contents are the responsibility of the authors and do not necessarily reflect the views of USAID or the United States Government. This work was also supported by a National Institutes of Health, NIAID non-biodefense EID Research Opportunities Award (grant no. 1 R01 AI079231-01) and by the International Development Research Centre (project no. 106150-001).
Author Disclosure Statement
No competing financial interests exist.
References
- Anderson A, Bijlmer H, Fournier P-E, Graves S, Hartzell J, Kersh GJ, Limonard G, Marrie TJ, Massung RF, McQuiston JH. Diagnosis and Management of Q Fever—United States, 2013: Recommendations from CDC and the Q Fever Working Group. US Department of Health and Human Services, Public Health Service, Centers for Disease Control and Prevention, 2013 [PubMed] [Google Scholar]
- Anthony SJ, Epstein JH, Murray KA, Navarrete-Macias I, et al. . A strategy to estimate unknown viral diversity in mammals. MBio 2013; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmbhatt M. Avian and Human Pandemic Influenza—Economic and Social Impacts. The World Bank, 2005 [Google Scholar]
- Childs JE, Gordon ER. Surveillance and control of zoonotic agents prior to disease detection in humans. Mt Sinai J Med 2009; 76:421–428 [DOI] [PubMed] [Google Scholar]
- Chua K, Bellini W, Rota P, Harcourt B, et al. . Nipah virus: A recently emergent deadly paramyxovirus. Science 2000; 288:1432–1435 [DOI] [PubMed] [Google Scholar]
- Chua KB. Nipah virus outbreak in Malaysia. J Clin Virol 2003; 26:265–275 [DOI] [PubMed] [Google Scholar]
- Epstein JH, Field HE, Luby S, Pulliam JR, et al. . Nipah virus: Impact, origins, and causes of emergence. Curr Infect Dis Rep 2006; 8:59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn T, Hornik K, van de Wiel MA, Zeileis A. A Lego system for conditional inference. Am Statistician 2006; 60:257–263 [Google Scholar]
- Hothorn T, Hornik K, van de Wiel MA, Zeileis A. Implementing a class of permutation tests: The coin package. J Statist Software 2008; 28:1–23 [Google Scholar]
- Hughes JM, Wilson ME, Luby SP, Gurley ES, et al. . Transmission of human infection with Nipah virus. Clin Infect Dis 2009; 49:1743–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KE, Patel NG, Levy MA, Storeygard A, et al. . Global trends in emerging infectious diseases. Nature 2008; 451:990–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karesh WB, Cook RA, Bennett EL, Newcomb J. Wildlife trade and global disease emergence. Emerg Infect Dis 2005; 11:1000–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipkin WI. The changing face of pathogen discovery and surveillance. Nat Rev Microbiol 2013; 11:133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby SP, Rahman M, Hossain MJ, Blum LS, et al. . Foodborne transmission of Nipah virus, Bangladesh. Emerg Infect Dis 2006; 12:1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse SS. Factors in the emergence of infectious-diseases. Emerg Infect Dis 1995; 1:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse SS, Mazet JAK, Woolhouse M, Parrish CR, et al. . Prediction and prevention of the next pandemic zoonosis. Lancet 2012; 380:1956–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray KA, Daszak P. Human ecology in pathogenic landscapes: Two hypotheses on how land use change drives viral emergence. Curr Opin Virol 2013; 3:79–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olival KJ, Islam A, Yu M, Anthony SJ, et al. . Ebola virus antibodies in fruit bats, Bangladesh. Emerg Infect Dis 2013; 19:270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patz JA, Daszak P, Tabor GM, Aguirre AA, et al. . Unhealthy landscapes: Policy recommendations on land use change and infectious disease emergence. Environ Health Persp 2004; 112:1092–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patz JA, Campbell-Lendrum D, Holloway T, Foley JA. Impact of regional climate change on human health. Nature 2005; 438:310–317 [DOI] [PubMed] [Google Scholar]
- Pulliam JRC, Epstein JH, Dushoff J, Rahman SA, et al. . Agricultural intensification, priming for persistence and the emergence of Nipah virus: A lethal bat-borne zoonosis. J R Soc Interface 2012; 9:89–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn GP, Keough MJ. Experimental Design and Data Analysis for Biologists. Cambridge, UK: Cambridge University Press, 2002 [Google Scholar]
- Smolinski MS, Hamburg MA, Lederberg J. Microbial Threats to Health: Emergence, Detection, and Response. Washington DC: Institute of Medicine, National Academies Press, 2003 [PubMed] [Google Scholar]
- Taylor LH, Latham SM, Mark E. Risk factors for human disease emergence. Philos Trans R Soc Lond B Biol Sci 2001; 356:983–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezzulli L, Colwell RR, Pruzzo C. Ocean warming and spread of pathogenic vibrios in the aquatic environment. Microb Ecol 2013; 65:817–825 [DOI] [PubMed] [Google Scholar]
- Vrbova L, Stephen C, Kasman N, Boehnke R, et al. . Systematic review of surveillance systems for emerging zoonoses. Transbound Emerg Dis 2010; 57:154–161 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.