ABSTRACT
Gene transfer agents (GTAs) morphologically resemble small, double-stranded DNA (dsDNA) bacteriophages; however, their only known role is to package and transfer random pieces of the producing cell genome to recipient cells. The best understood GTA is that of Rhodobacter capsulatus, termed RcGTA. We discovered that homologues of three genes involved in natural transformation in other bacteria, comEC, comF, and comM, are essential for RcGTA-mediated gene acquisition. This paper gives genetic and biochemical evidence that RcGTA-borne DNA entry into cells requires the ComEC and ComF putative DNA transport proteins and genetic evidence that putative cytoplasmic ComM protein of unknown function is required for recipient capability. Furthermore, the master regulator of RcGTA production in <1% of a cell population, CtrA, which is also required for gene acquisition in recipient cells, is expressed in the vast majority of the population. Our results indicate that RcGTA-mediated gene transfer combines key aspects of two bacterial horizontal gene transfer mechanisms, where donor DNA is packaged in transducing phage-like particles and recipient cells take up DNA using natural transformation-related machinery. Both of these differentiated subsets of a culture population, donors and recipients, are dependent on the same response regulator, CtrA.
IMPORTANCE Horizontal gene transfer (HGT) is a major driver of bacterial evolution and adaptation to environmental stresses. Traits such as antibiotic resistance or metabolic properties can be transferred between bacteria via HGT; thus, HGT can have a tremendous effect on the fitness of a bacterial population. The three classically described HGT mechanisms are conjugation, transformation, and phage-mediated transduction. More recently, the HGT factor GTA was described, where random pieces of producing cell genome are packaged into phage-like particles that deliver DNA to recipient cells. In this report, we show that transport of DNA borne by the R. capsulatus RcGTA into recipient cells requires key genes previously thought to be specific to natural transformation pathways. These findings indicate that RcGTA combines central aspects of phage-mediated transduction and natural transformation in an efficient, regulated mode of HGT.
INTRODUCTION
The first evidence of prokaryotic genetic exchange was transformation, a term coined in 1928 by Griffith (1). Subsequently, conjugation was observed in 1946 (2), followed by transduction, which was discovered in 1952 (3). More recently, another prokaryotic mode of horizontal gene transfer, dependent on an extracellular particle called a gene transfer agent (GTA), was described (4), and GTAs subsequently have been discovered in diverse prokaryotes (5).
GTAs varying in morphology have been reported, although most resemble small, tailed double-stranded DNA (dsDNA) bacteriophages. The general criteria that define a GTA are (i) the DNA packaged within the head is insufficient to carry the GTA structural genes; (ii) all GTA particles package only random parts of the producing cell's genome (as opposed to packaging of host cell genomic DNA in generalized transducing phages, which is very infrequent); and (iii) production is controlled by cellular regulatory systems (5–7). As a consequence, the frequency of cellular gene transduction by GTAs is much greater than that by generalized transducing phages.
The best-understood GTA, found in the alphaproteobacterium Rhodobacter capsulatus, is called RcGTA. RcGTA particles morphologically resemble a small, siphoviridae-like bacteriophage (8), and RcGTA structural gene organization and sequence composition are similar to those of genuine bacteriophages (5). However, nucleic acid analyses revealed that essentially random ∼4-kb linear, dsDNA fragments of the producing cell genome with 3′ overhangs are packaged within particles (7, 8), so genetic markers are readily transferred from donor to recipient cells (5, 6). The RcGTA primary structural gene cluster consists of an ∼15-kb region of the chromosome, and additional essential factors, including a holin and endolysin, are encoded in distant genome regions (5, 7, 9). Production of RcGTA requires the CtrA response regulator, and transcription is modulated by the GtaI/R quorum-sensing system (10–12). The ability of cells to receive an RcGTA-carried genetic marker also is regulated by CtrA and quorum sensing, and in recipient cells DNA recombination occurs via a RecA/DprA recombination mechanism as in natural transformation systems (13).
The phrase “natural genetic competence” is defined as a physiological state in which bacteria actively take up exogenous DNA that may be recombined into the cell genome (14). The term “transformation” refers to the successful acquisition of a new genetic trait by recombination into the genome (14). It is now clear that natural competence systems are widespread in both Gram-positive and Gram-negative bacteria, and new functional systems are continually being discovered (14). Although some details differ, the general mechanisms of natural competence systems are the following. Exogenous DNA is bound by a pilus structure that brings DNA into proximity of the cytoplasmic membrane (the inner membrane in Gram-negative bacteria). DNA is thought to be transported through the membrane by the ComEC DNA transporter in a process also involving ComF, resulting in a single-stranded DNA (ssDNA) molecule entering the cytoplasm (14–16). The ssDNA is bound by the DprA protein, which facilitates the formation of RecA filaments on the ssDNA, and recombination with the recipient cell genome if sequence similarity exists (17, 18). In addition, the comM gene is required for maximal Haemophilus influenzae transformation, although the function is unknown (19).
In a previous report, it was shown that the RcGTA recipient capability (the capability of an R. capsulatus cell to receive an RcGTA-borne genetic marker) requires the CtrA response regulator, and that expression of homologues of natural competence genes dprA, comEC, comF, and comM is modulated by CtrA. Genetic analysis showed that one of these genes, dprA, is essential for RcGTA recipient capability, as is the RecA homologue (13). These findings indicated that RcGTA-carried DNA recombines into the chromosome of recipient cells via a natural transformation-like mechanism rather than one resembling temperate phages (20, 21). However, it was unknown whether three other homologues of natural competence genes downregulated in a ctrA mutant, namely, comEC, comF, and comM, were involved in RcGTA recipient capability (13).
Here, we provide genetic and biochemical evidence that comEC and comF facilitate uptake of RcGTA-delivered DNA and are required for recipient capability. Additionally, it was found that comM is required for recipient capability. We present a model in which RcGTA releases DNA into the periplasm of the recipient cell, which then is transported into the cytoplasm by the ComEC/ComF machinery in the same fashion as that in natural competence pathways.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The Escherichia coli strains DH5α λpir, S17-1 λpir (22), and TEC5 (23) were used for gene cloning and conjugation of plasmids into R. capsulatus. E. coli strains were grown at 37°C in LB medium (24) supplemented with the appropriate antibiotics at the following concentrations (in micrograms per milliliter): ampicillin, 150; gentamicin sulfate, 10; kanamycin sulfate, 50; tetracycline hydrochloride, 10.
R. capsulatus strains, listed in Table 1, were grown at 30°C in RCV defined medium (25) aerobically with shaking at 200 rpm for recipient capability, conjugation, and growth rate assays or were sealed in test tubes phototrophically for UV sensitivity and RcGTA tracking assays. For production of gentamicin-resistant (Gmr) RcGTA in the tracking assay, cultures of the RcGTA overproducer strain DE442(pd1080::Gm) were grown phototrophically in YPS complex medium (26). Cells were harvested in the stationary phase of growth. When appropriate, media were supplemented with the following (in micrograms per milliliter): gentamicin sulfate, 3; kanamycin sulfate, 10; rifampin, 80; and tetracycline hydrochloride, 1.
TABLE 1.
Plasmids and strains used and/or generated in this study
| Plasmid or strain | Feature(s)/inserted sequencea or description | Reference or source |
|---|---|---|
| Plasmids | ||
| pUC19Fup | comF upstream flanking region ligated into pUC19 | This study |
| pUC19Fdown | comF downstream flanking region ligated into pUC19 | This study |
| pUC19Mup | comM upstream flanking region ligated into pUC19 | This study |
| pUC19Mdown | comM downstream flanking region ligated into pUC19 | This study |
| pUC19EC | comEC gene and flanking regions ligated into pUC19 | This study |
| pComF | ∼500 bp 5′ of comF gene, the comF gene, and ∼200 bp downstream ligated into pCM62 | This study |
| pComM | ∼500 bp 5′ of comM gene, the comM gene, and ∼200 bp downstream ligated into pCM62 | This study |
| pComEC | ∼500 bp 5′ of comEC gene, the comEC gene, and ∼200 bp downstream ligated into pCM62 | This study |
| pZDJΔcomF | Upstream and downstream flanks of comF fused together in pZDJ | This study |
| pZDJΔcomM | Upstream and downstream flanks of comM fused together in pZDJ | This study |
| pZDJ | Gmr; Gram-negative suicide vector; contains sacB gene and tetR promoter | 27 |
| pUC19 | Ampr LacZα; gene cloning vector | Invitrogen |
| pCM62 | Tcr LacZα; broad-host-range vector | 29 |
| pCMmCherry | mCherry gene ligated into pCM62 | 13 |
| pZDJΔ1081 | rcc01081 knockout construct; used in HR expt | 27 |
| pCtrAmCherry | ctrA translationally fused to mCherry in pCMmCherry | This study |
| R. capsulatus strains | ||
| B10 | Wild-type isolate | 4 |
| DE442 | Rifr; RcGTA overproducer; believed to originate from Y262 | 8 |
| ΔcomF mutant | Markerless deletion of comF gene; B10 background | This study |
| ΔcomEC mutant | Kanr; KIXX cassette inserted 1,012 bp into the comEC gene in forward direction; B10 background | This study |
| ΔcomM | Markerless deletion of comM gene; B10 background | This study |
| DE442(pd1080Gm) | Rifr Gmr; RcGTA overproducer; harbors plasmid pd1080Gm | Westbye, unpublished |
Where applicable, the inserted sequence is described briefly.
The optical density at 650 nm (OD650) was used as a measure of the number of R. capsulatus colony-forming units per milliliter; an OD650 of 1 is ∼4.5 × 108 CFU ml−1.
Recombinant DNA techniques, plasmids, and PCR primers.
Standard methods of DNA purification, restriction enzyme digestion, and other modification techniques were used (24). All plasmids and primer sequences used in this study are listed in Tables 1 and 2, respectively. The plasmid pUC19 was used for subcloning and pCM62 as a complementation plasmid.
TABLE 2.
Primers used in this study
| Primer | 5′–3′ sequencea | Product | Restriction cut site |
|---|---|---|---|
| comF_up_for | GCTAGAGCTCGAACAATATCCTTGC | pUC19Fup | SacI |
| comF_up_rev | GCTAGAATTCCACCCGCAAGGCCGC | pUC19Fup | EcoRI |
| comF_down_for | GCATGAATTCTCGGTCGCGGTTCTG | pUC19Fdown | EcoRI |
| comF_down_rev | CTTATCTAGACGGTCCGAGGACAGA | pUC19Fdown | XbaI |
| comM_up_for | GTACGAGCTCAACCATTGCGAAACG | pUC19Mup | SacI |
| comM_up_rev | TACAGGATCCGAAAGCCACCGTATA | pUC19Mup | BamHI |
| comM_down_for | GCACGGATCCTCGCCGATCTGGACG | pUC19Mdown | BamHI |
| comM_down_rev | GACATCTAGACGGCACCAATGTCGC | pUC19Mdown | XbaI |
| JAB-rec-2-fwd | GTGATCCGGCTCACTCTAGATCAGGTGCG | pUC19EC | XbaI |
| JAB-rec-2-rev | GGCAGGCGTTTTCAAGCTTGTCCAGGG | pUC19EC | HindIII |
| comF_screen_for | GCGGCGTCGTCGTGC | NAb | NA |
| comF_screen_rev | TGCGGGTGGTGTAGATT | NA | NA |
| comM_screen_for | GGATCTAGGCTACTG | NA | NA |
| comM_screen_rev | TCCTGAGCGATGATGC | NA | NA |
| comF_comp_for | TACAGAGCTCCGCCATAGCTGACCC | pComF | SacI |
| comF_comp_rev | CAGTAAGCTTGGTGTAGATTTCGAC | pComF | HindIII |
| comM_comp_for | CGTAGAGCTCCGCTGGCTCGCTTCGG | pComM | SacI |
| comM_comp_rev | CGTAGGATCCCCGAGGTGCTGCTGC | pComM | BamHI |
| comEC_comp_for | GTACGAGCTCCTCCTGGGTGGCGA | pComEC | SacI |
| comEC_comp_rev | TAGTAAGCTTCTCGTCGAGGATCTG | pComEC | HindIII |
| gm_for | GTTATGGAGCAGCAACGA | NA | NA |
| gm_rev | GGAGTAGGTGGCTAC | NA | NA |
| pctrA_F | AGAGAAGCTTCTCTGGTCGCGCATCGG | pCtrAmCherry | HindIII |
| pctrA_R | AGAGGGATCCATCCTGGGTTCTCCGCA | pCtrAmCherry | BamHI |
| Gm_qPCR_for | GGTGGCTCAAGTATGGGCAT | NA | NA |
| Gm_qPCR_rev | CGAAAAGATCAAGAGCAGCCC | NA | NA |
| puhA_qPCR_for | AACGACGACGGCAAGCTCT | NA | NA |
| puhA_qPCR_rev | GCTTCTTCGTTCTCGACCGA | NA | NA |
Restriction cut sites (if applicable) are underlined.
NA, not applicable.
Creation of the ΔcomF (rcc197) and ΔcomM (rcc460) mutant strains.
The ΔcomF mutant we generated is a markerless in-frame deletion of ∼80% of the gene and was constructed from the wild-type (WT) strain B10. Approximately 900-bp (upstream) and 450-bp (downstream) flanking regions of the comF gene were PCR amplified as SacI-EcoRI (upstream) and EcoRI-XbaI (downstream) fragments and inserted into pUC19, generating the pUC19Fup and pUC19Fdown plasmids. The resultant plasmids were digested with the appropriate enzymes and SacI-EcoRI and EcoRI-XbaI fragments were inserted into the suicide plasmid pZDJ, generating pZDJΔcomF. This plasmid was conjugated into the WT strain B10 and allowed to recombine into the chromosome, selected for by acquisition of gentamicin resistance. Cells that underwent a second recombination event, resulting in the loss of the plasmid, were selected by aerobic growth on RCV agar medium containing 7% sucrose and screened for the loss of Gmr. Replacement of the WT comF with the ΔcomF allele was confirmed by PCR amplicon size and DNA sequencing using the primers comF_seq_for and comF_seq_rev. The ΔcomM mutant was constructed in the same fashion, with approximately 1,000-bp flanking regions (for both upstream and downstream) of the comM gene amplified and ultimately subcloned as SacI-BamHI (upstream) and BamHI-XbaI (downstream) fragments into the suicide plasmid pZDJ, generating pZDJΔcomM. The steps thereafter were identical to those described for the construction of the ΔcomF mutant, except the primers comM_seq_for and comM_seq_rev were used to evaluate comM allele size and for DNA sequencing. For a description of the pZDJ suicide plasmid, see reference 27.
Creation of the ΔcomEC (rcc02368) mutant strain.
The open reading frame (ORF) encoding ComEC (rcc02362) was amplified by PCR from the genome of WT strain B10 as an XbaI-to-HindIII fragment, using the primers JAB-rec-2-Fwd and JAB-rec-2-Rev. The amplified product was cloned into plasmid pUC19, and the gene was disrupted by insertion of a kanamycin resistance-encoding ∼1.4-kb SmaI-KIXX cartridge (28) into the unique NruI restriction site within the comEC coding region (1,012 bp 3′ of the start codon). Both orientations of the SmaI-KIXX cartridge were obtained and confirmed by DNA sequencing. Both constructs were transformed into E. coli TEC5 and conjugated into the RcGTA overproducer strain DE442. Mutant strains were generated by RcGTA-mediated transfer of the disrupted versions of the genes into the chromosome of WT strain B10 (4). PCR using the original amplification primers and restriction endonuclease analysis were used to confirm the resultant kanamycin-resistant strains. Strains containing the KIXX cartridge in both orientations had an identical phenotype (data not shown), and results from the mutant strain with KIXX inserted in the forward direction (the direction of transcription of the KIXX neo gene is the same as that of the disrupted comEC gene) are used in this report.
Complementation of comEC, comF, and comM mutants.
The complementation plasmids pComEC, pComF, and pComM were generated by amplifying the R. capsulatus comEC, comF, and comM genes and ∼450 bp 5′ of the start codons and ∼100 bp 3′ of the stop codons as SacI-HindIII, SacI-HindIII, and SacI-BamHI fragments, respectively. The resultant amplicons were ligated into the vector pCM62 (29) and verified by DNA sequencing. The resultant plasmids, pComEC, pComF, and pComM, were conjugated into the ΔcomEC, ΔcomF, and ΔcomM strains, respectively, to generate the native promoter-driven trans-complemented strains.
Fluorescence microscopy.
Approximately ∼1 kb of DNA sequence 5′ of the ctrA start codon was PCR amplified using primers pctrAF and pctrAR and cloned as a HindIII-BamHI fragment into pCM62::mCherry (13), resulting in an in-frame fusion of the ctrA start codon to the mCherry coding region. The resultant construct (pCtrAmCherry) was conjugated into R. capsulatus B10 WT cells and evaluated for fluorescence. Images of cells grown in RCV medium to the stationary phase at 30°C were taken using a Sony DSC-S75 digital camera mounted on a Zeiss Axioskop fluorescence microscope equipped with a ×1,000 magnification objective. A mercury lamp (HBO 50X) and Zeiss filter set 15 (excitation, 546 nm and 12 nm; emission, 590 nm) were used to observe mCherry fluorescence.
RcGTA-borne DNA tracking assay.
An RcGTA stock was prepared from a DE442 overproducer mutant harboring a plasmid containing the rcc01080 gene (pd1080Gm) interrupted by a Gmr cassette (A. Westbye, unpublished data). This RcGTA stock was used because the Gmr gene is not present in the WT and mutant R. capsulatus strains studied in our work; thus, it can be tracked by PCR analysis from an extracellular location through periplasmic delivery to entry into the cytoplasm. For the tracking assay, WT, ΔcomEC, and ΔcomF strains were grown phototrophically in RCV medium to the stationary phase at 30°C. All harvesting and wash steps were done at room temperature (∼22°C). Cells were harvested by centrifugation and suspended at an OD650 of 3.0 in G-buffer (10 mM Tris-HCl [pH 7.8], 1 mM MgCl2, 1 mM CaCl2, 1 mM NaCl, 500 μg/ml bovine serum albumin [BSA]). A 500-μl sample of RcGTA stock and 500 μl of cells were mixed together and incubated at 30°C with gentle shaking for 60 min (for the no-GTA control, 500 μl of G-buffer and 500 μl of WT cells were mixed together). After the 60-min incubation, cells were harvested by centrifugation and washed three times in 1 ml of RCV medium. Cells then were suspended in 1 ml of RCV and incubated at 30°C with gentle shaking for an additional 120 min (for the 3-h time point and quantitative PCR [qPCR] measurements) or 23 h. At this point, 50 μl of each culture was plated onto RCV plus Gm to determine the Gmr colony-forming units per milliliter of each reaction, and the remaining cells were collected by centrifugation, washed 5 times with 1 ml of RCV medium, suspended in 400 μl of 50 mM EDTA, and incubated at 37°C for 60 min to disrupt the outer membrane (OM) and release periplasmic contents (30). Cells were pelleted by centrifugation to yield a cellular fraction, and the supernatant liquid (containing OM and periplasmic contents) was transferred to a fresh tube. DNA was extracted from the OM/periplasmic and cellular fractions by phenol-chloroform extraction, followed by isopropanol precipitation, and OM/periplasmic DNA was dissolved in 30 μl of distilled water (dH2O), whereas cellular (chromosomal) DNA preparations were dissolved in 500 μl of dH2O. Wash efficacy was evaluated by taking samples after each wash step and determining the presence of RcGTA-borne DNA by PCR using the primers Gm_qPCR_for and Gm_qPCR_rev. RcGTA-borne PCR amplification was undetectable after ∼5 washes (see Fig. S3 in the supplemental material).
To quantify the relative amount of RcGTA-borne DNA in the OM/periplasmic fraction, qPCR was used with primers specific for a donor allele, with cells taken after 3 h of incubation with RcGTA. The primers GmqPCR_for and GmqPCR_rev were used to detect the relative amounts of the Gmr gene in each periplasmic/OM fraction; these primers amplify an 85-bp region within the Gmr cartridge. Residual periplasm/OM DNA levels were normalized to the level of the chromosomal puhA gene (rcc00659) in the chromosomal DNA fraction of each sample. The puhA gene is an endogenous photosynthesis gene present in R. capsulatus; the primers puhA_qPCR_for and puhA_qPCR_rev were used for quantification. The SYBR select master mix (Applied Biosystems) was used per the manufacturer's instructions. Reaction mixtures of 20 μl containing SYBR select master mix, 400 nM specific primers, and the target template were used to quantify target DNA in an Applied Biosystems StepOne plus real-time PCR system, using the following program: 50°C for 2 min, followed by 40 cycles of 95°C for 15 s, 60°C for 15 s, and 72°C for 60 s (amplification), and then 95°C for 15 s, 60°C for 15 s, and 95°C for 15 s (melting curve). Amplicons were clearly distinguishable from primer dimers based on melting curve analysis. Amplification of targets occurred from 15 to 25 cycles. As negative controls, reaction mixtures containing no DNA template were always included. As positive controls, standard concentrations of DNA containing either the Gmr cartridge or puhA were included in all qPCR runs and were used to quantify the amount of DNA present in each test sample. One microliter of each DNA sample was used as the template in all qPCRs, and values are presented as the ratio of residual periplasmic/OM Gmr cartridge-containing DNA to the level of puhA DNA in the chromosomal fraction of cells. Additional control experiments, such as evaluation of wash stringency, are presented in Fig. S3 in the supplemental material. Standard curves for both the Gm cassette and puhA primer sets are given in Fig. S4 in the supplemental material, and the raw qPCR data are given in Table S6 in the supplemental material.
RcGTA recipient capability assay.
The rifampin-resistant (Rifr) RcGTA overproducer strain DE442 was the source of RcGTA for recipient capability and adsorption experiments. The titer of a sample of stationary-phase culture grown phototrophically in YPS complex medium, passed through a 0.2-μm-pore-diameter filter, was determined for RcGTA activity, and a diluted stock solution that produced ∼800 Rifr transductants per 100 μl of donor filtrate was used. All strains used as RcGTA recipients were derived from the WT strain B10. Recipient cultures were grown aerobically in RCV defined medium with shaking at 200 rpm to the stationary phase, harvested, and resuspended in an equal volume of G-buffer. A transduction assay then was performed in which 100 μl of RcGTA stock, 100 μl of recipient cells, and 400 μl of G-buffer were mixed together. This mixture was incubated for 90 min at 30°C with gentle agitation, after which 900 μl of RCV medium was added, followed by incubation under the same conditions for 3 h. Cells were spread on RCV plates containing rifampin and incubated aerobically for 3 days. The number of Rifr colonies was counted, and the average was corrected by subtracting the number of spontaneous Rifr colonies (no addition of RcGTA; usually less than 3%). Because of variability in total numbers of transductants between individual experiments, RcGTA recipient capability efficiencies are normalized to the WT control in each experiment.
RcGTA adsorption assay.
To quantitatively measure the ability of different strains to bind RcGTA, cultures were grown aerobically in RCV defined medium with shaking at 200 rpm to the stationary phase, and cells were harvested and resuspended at the same concentration in G-buffer. One hundred microliters of RcGTA solution (the same as the amount used in the RcGTA recipient capability assay), 100 μl of recipient cells, and 400 μl of G-buffer were mixed and incubated for 90 min at 30°C with gentle agitation, after which the mixture was passed through a 0.2-μm filter. The number of RcGTA particles in the filtrate (RcGTA that did not adsorb to cells) was quantified by using 100 μl of this filtrate as the RcGTA donor and WT strain B10 cells as the recipient. As controls, an assay that included no recipient cells for adsorption (no cells) and an assay that contained no RcGTA also were performed.
Plasmid conjugation and HR frequency.
Stationary-phase R. capsulatus cells were harvested by centrifugation and suspended at an OD650 of 1.0 in fresh RCV medium. E. coli strain S17-1 λpir, containing the suicide plasmid pZDJΔ1081, was grown overnight (to the stationary phase) and harvested by centrifugation. Cells were washed three times in RCV medium and suspended in RCV medium at an OD600 of 1.0. A 2:1 mixture of R. capsulatus-to-E. coli cells was spotted onto RCV agar plates and incubated at 30°C overnight. Mating spots then were suspended in 500 μl of RCV medium, and dilutions plated on RCV agar with or without gentamicin were incubated at 30°C for 3 days. The number of colonies was counted, and the homologous recombination (HR) frequency was calculated by dividing the number of antibiotic-resistant colonies by the total number of colony-forming units.
UV sensitivity assays.
R. capsulatus cultures were grown phototrophically in RCV defined medium at 30°C to late log phase and diluted in 10-ml portions of fresh RCV to ∼104 CFU ml−1. Diluted cultures were exposed to a 15-W germicidal lamp at a distance of 50 cm for 0, 5, 15, or 30 s as indicated, with gentle mixing. At each time point, 100-μl samples were taken, plated onto RCV agar in serial dilutions, and grown at 30°C for 4 days. The percent survival was calculated as the number of CFU arising from UV-treated samples divided by the number of CFU from the nonexposed cells.
Bioinformatic analyses.
The predicted amino acid sequence of R. capsulatus ComEC (encoded by rcc02362), ComF (encoded by rcc00197), ComM (encoded by rcc00460), and DprA (encoded by rcc03098) were used to probe bacterial genomes for the presence of homologues using BLASTP (31). Hits in other organisms then were verified to be reciprocal best hits to the R. capsulatus homologues, and full-length alignments of R. capsulatus proteins and homologues using EMBOSS Needle pairwise sequence alignment software (32) were done to verify that proteins are genuine homologues. Phylogenetic trees of ComM and DprA homologues were generated using MUSCLE MSA alignments and a neighbor-joining algorithm (33, 34). Domain analysis of proteins was performed using Pfam (35).
RESULTS
Bioinformatic analysis of comEC, comF and comM.
We began our study by analyzing the amino acid sequence of ComEC, ComF, and ComM predicted proteins. ComEC is essential for DNA transport through the cytoplasmic membrane (CM) in all natural transformation systems studied to date, including the well-studied Gram-positive Streptococcus pneumoniae and Bacillus subtilis and Gram-negative Haemophilus influenzae and Vibrio cholerae (14, 16). The ComEC protein family members are predicted to contain six transmembrane segments (TMS), which is supported by in vitro studies of the B. subtilis protein (16). Hydropathy analysis of R. capsulatus ComEC (rcc02362) predicts six TMS, that the protein is located in the CM, and that it contains the two conserved domains of ComEC proteins, pfam03772 and DUF4131 (Table 3 and Fig. 1A). Because these features are present in genuine ComEC proteins, we hypothesized that R. capsulatus rcc02362 encodes a ComEC.
TABLE 3.
Predicted and verified RcGTA recipient capability genese
| R. capsulatus genee | Annotation | Predicted/verified function(s) and/or characteristic(s) of gene product | WT/ΔctrA ratio |
Predicted TMS (n) | Phenotype of mutant(s) |
||
|---|---|---|---|---|---|---|---|
| Stationarya | Logb | Transformationc | RcGTA recipient capabilityd | ||||
| rcc00197 | comF | Unknown; required for DNA transport from periplasm to cytoplasm | 3.23 | 5.35 | 1 | Not detectable | Unknown |
| rcc00460 | comM | Unknown; putative ATPase and helicase-like domains | 2.91 | 1.89 | 0 | Variable; 0– to 300-fold reduction | Unknown |
| rcc02362 | comEC-rec2 | Integral membrane protein; transports DNA from periplasm to cytoplasm | 4.88 | 2.67 | 6 | Not detectable | Unknown |
| rcc03098 | dprA | Recombination mediator; loads RecA into ssDNA | 9.38 | 3.75 | 0 | Not detectable | Not detectable |
| rcc01751 | recA | Recombination protein A; RecA; DNA repair and homologous recombination | 1.47 | 1.22 | 0 | Not detectable | Not detectable |
| rcc01663 | ctrA | Cell cycle transcriptional regulator CtrA; regulates transcription of RcGTA and com genes | 0.94 | 1.04 | 0 | Unknown | Not detectable |
Ratio of WT to ΔctrA microarray expression values for cells grown to the exponential phase of growth.
Ratio of WT to ΔctrA microarray gene expression values for cells grown to the stationary phase of growth.
A 107-fold reduction in transformation was used as the limit of detection in other species.
A 106-fold reduction in recipient capability was used as the limit of detection in R. capsulatus.
Shown are predicted and verified RcGTA recipient capability genes, their annotations, predicted or verified functions, expression levels in the ΔctrA mutant versus the wild type in both stationary and log phases, the numbers of predicted transmembrane segments (TMS), the phenotypes in natural transformation systems, and the phenotypes resulting from mutations in these genes in natural transformation systems and in RcGTA recipient capability. Microarray expression values are derived from data reported in reference 61, and ΔdprA, ΔrecA, and ΔctrA mutant phenotypes were reported previously by Brimacombe et al. (13).
FIG 1.
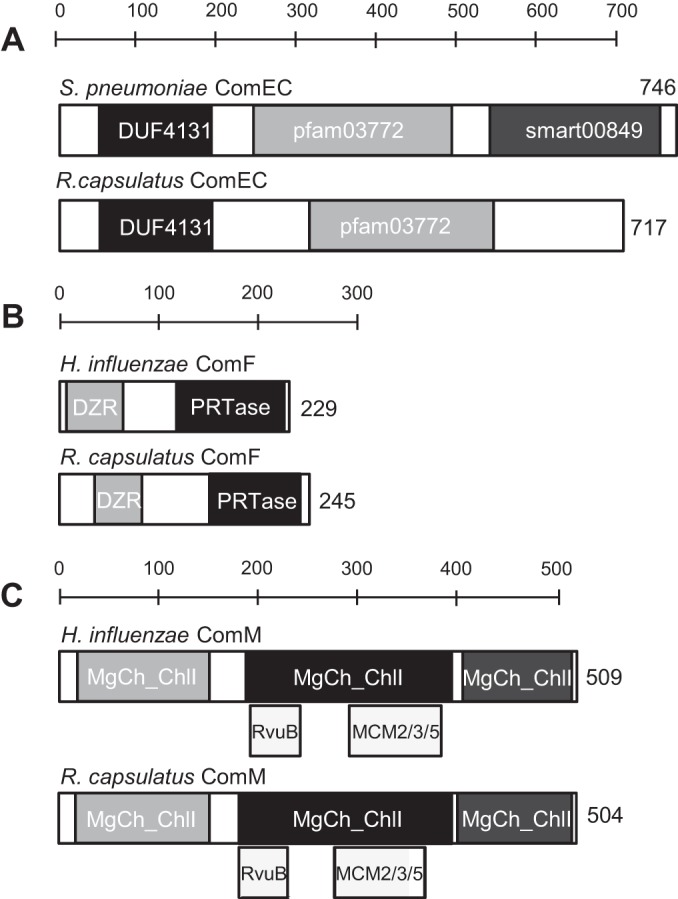
Summary of bioinformatic analyses of R. capsulatus ComEC, ComF, and ComM proteins. (A) Comparison of S. pneumoniae ComEC to R. capsulatus ComEC. (B) Comparison of H. influenzae ComF to R. capsulatus ComF. (C) Comparison of R. capsulatus ComM to H. influenzae ComM. Amino acid numbers and domains are indicated. DUF4131 is a functionally uncharacterized domain found frequently in the N-terminal section of ComEC homologues; pfam03772 is an uncharacterized domain ubiquitously present in ComEC homologues; smart00849, present in many ComEC homologues, may bind zinc ions as a cofactor (14); DZR stands for the double zinc ribbon domain most often involved in DNA binding in transcriptional regulators (38); PRTase stands for phosphoribosyltransferase, a domain often involved in nucleotide salvage pathways (37); MgCh-ChlI stands for magnesium chelatase subunit ChlI, an AAA+ ATPase that provides energy for insertion of Mg2+ into protoporphyrin IX (40); RvuB is a domain found in some Holliday junction DNA helicases (41); MCM2/3/5 stands for maintenance of minichromosomes and functions in the replication of genomes (42).
Although the ComF mechanism is unknown, it appears to be involved in DNA transport through the CM, because comF null mutants have the same phenotype as comEC null mutants (19, 36). Hydropathy analysis showed that the R. capsulatus ComF homologue has one putative TMS near the C terminus, with the bulk of the protein being cytoplasmic (Table 3). Of two putative cytoplasmic domains of ComF, one is homologous to a phosphoribosyltransferase domain often involved in nucleotide salvage pathways (37), and the other is homologous to a double zinc ribbon domain often involved in DNA binding (38) (Fig. 1B). Therefore, it is possible that ComF is involved in a DNA processing step associated with DNA import into the cell via ComEC (19, 36).
A comM gene has been studied in H. influenzae; its expression is induced during competence, and it is required for maximal transformation efficiency (19, 39). The function of ComM proteins is not known; however, our bioinformatics analysis yielded some clues. The R. capsulatus ComM is predicted to be cytoplasmic (Table 3), and several regions of the protein share homology with an AAA+ ATPase (subunit I of Mg2+ chelatase) (40). ComM also contains domains with weak similarity to RuvB, a Holliday junction DNA helicase (41), and an MCM2/3/5 (Fig. 1C), which are helicases in the initiation of DNA replication in archaea and eukaryotes (42). We also observed that ComM homologues are not ubiquitous in the genomes of naturally transformable bacteria (see Fig. S1 in the supplemental material). Instead, ComM homologues are detectable only in genomes in which the corresponding DprA homologue contains a DprA domain 3 (DD3) (see Fig. S1), which is predicted to confer dsDNA binding capability to DprA proteins (13). Based on these analyses, we speculate that ComM is involved in a recombination step within the cytoplasm of recipient cells, and that its function is related to the DD3 (13, 18).
comEC, comF, and comM are required for RcGTA recipient capability.
The hypothesis that comEC (rcc02362), comF (rcc00197), and comM (rcc00460) are involved in RcGTA recipient capability was addressed by generating knockouts of these genes.
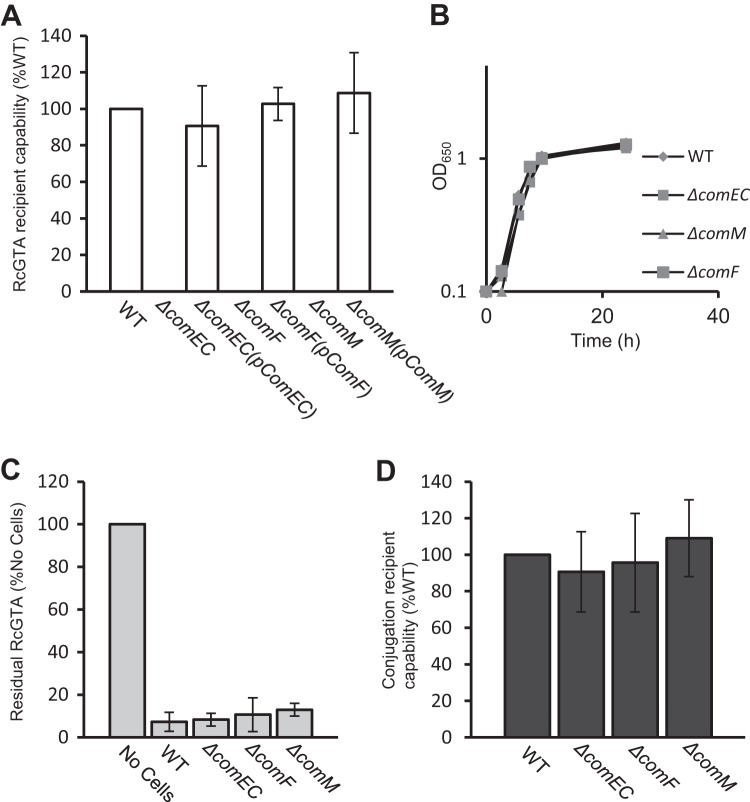
Null mutations of comEC, comF, or comM all resulted in an absolute loss in RcGTA recipient capability (<10−6 of WT levels), and these mutations were complemented to WT levels by the native gene in trans (Fig. 2A). In all three mutant strains, the growth kinetics, RcGTA adsorption capability, UV sensitivity, and frequency of RecA-dependent integration of a suicide plasmid containing an insert homologous to a region on the chromosome were found to be identical to those of the WT strain (Fig. 2; also see Fig. S2 in the supplemental material). Therefore, the R. capsulatus comEC, comF, and comM homologues are essential for RcGTA recipient capability, but these genes are not needed for RcGTA binding to cells, RecA-mediated homologous recombination, growth, or UV damage-induced DNA repair, indicating that the defect(s) in these mutants is specific to RcGTA recipient capability at some stage after particle adsorption to cells. Because ComEC and ComF are essential for DNA transport through the CM in natural competence systems (14, 19, 36), we hypothesized that RcGTA-borne DNA enters the recipient cell cytoplasm via the ComEC/ComF machinery. A corollary of this hypothesis is that there would be a build-up of RcGTA-borne DNA in the periplasm of recipient cells lacking either the ComEC or ComF protein, and that this DNA could be detected in biochemical experiments.
FIG 2.
(A) Relative RcGTA recipient capability of WT, ΔcomEC, ΔcomF, and ΔcomM strains and the trans-complemented ΔcomEC(pComEC), ΔcomF(pComF), and ΔcomM(pComM) strains. Error bars represent standard deviations from the means (n = 3). (B) Comparison of the growth rate of WT, ΔcomEC, ΔcomF, and ΔcomM strains grown in RCV liquid medium aerobically with shaking at 200 rpm. Culture turbidity (OD650) was measured for each strain at the indicated time points and plotted as a function of time. (C) Relative RcGTA adsorption capability of WT, ΔcomEC, ΔcomF, and ΔcomM strains. Error bars represent the standard deviations from the means (n = 3). (D) Relative homologous recombination frequency of the suicide plasmid pZDJ1081 into the chromosome of WT, ΔcomEC, ΔcomF, and ΔcomM strains, displayed as relative conjugation efficiency (%WT). Error bars represent the standard deviations from the means (n = 3).
RcGTA-borne DNA build-up in the periplasmic fraction of ΔcomEC and ΔcomF mutants.
To track the relative amount of RcGTA-borne DNA in the periplasm and cytoplasm of recipient cells, a modified RcGTA transduction assay of a Gmr cartridge was used to monitor incoming DNA entry into the cell (see Materials and Methods).
DNA was purified from the periplasmic and cytoplasmic fractions after 3 and 24 h of incubation with RcGTA. A nonquantitative PCR of each fraction revealed that RcGTA-borne DNA was present in the periplasmic but not the cytoplasmic fraction of WT, ΔcomEC, and ΔcomF strains 3 h after exposure to RcGTA (Fig. 3A). However, after 24 h, the RcGTA-borne Gmr marker was detectable within the cytoplasm of only WT and not mutant cells (Fig. 3B). These data indicate that the comEC and comF genes are needed to import DNA from RcGTA to the cytoplasm, and that at some point between 3 and 24 h, the DNA is degraded in the periplasm of the ΔcomEC and ΔcomF strains. To more directly evaluate whether ComEC and ComF play a role in DNA transport through the CM, we quantified the amount of RcGTA-borne DNA in the periplasmic fraction at the 3-h time point; if there were a greater amount of RcGTA-borne DNA in the periplasm of ΔcomEC and ΔcomF mutants than in the WT strain, then ComEC and ComF would facilitate transport of DNA through the CM.
FIG 3.
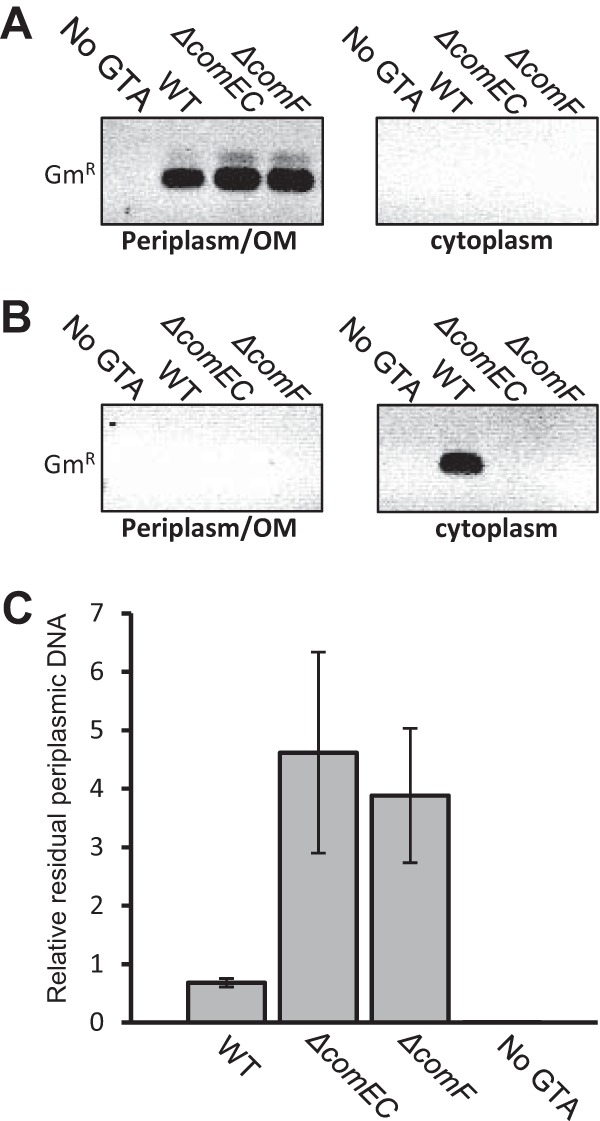
Tracking of RcGTA-borne DNA levels in the periplasm and cytoplasm of WT, ΔcomEC, and ΔcomF strains. (A) PCR products obtained from cell fractions after incubation with RcGTA for 3 h. (B) PCR products obtained from cell fractions after incubation with RcGTA for 24 h. (C) qPCR values, displayed as ratios of periplasmic RcGTA-borne DNA to chromosomal puhA levels. Error bars represent the standard deviations from the means (n ≥ 3). Statistical analysis was done by one-way ANOVA, with the results given in Table S6 in the supplemental material.
To quantify the residual periplasmic RcGTA-derived DNA at the 3-h time point, qPCR was performed using the Gmr cartridge as a target. Values obtained were normalized to the amounts of the puhA (photosynthetic reaction center) single-copy gene in the chromosomal fraction. It was found that the ΔcomEC and ΔcomF mutants contained 6.7-fold and 5.7-fold more incoming RcGTA DNA, respectively, in the periplasmic fraction than WT cells at this time point (Fig. 3C). These data indicate that the defect in RcGTA-borne gene acquisition in the ΔcomEC and ΔcomF mutants is due to a bottleneck in DNA transport from the periplasm to the cytoplasm.
Prevalence of ComEC and ComF in GTA-containing organisms.
The RcGTA major structural gene cluster and close homologues appear limited to and vertically inherited in the alphaproteobacteria (5, 43). In addition to RcGTA, several other functional GTAs have been observed, such as in Ruegeria mobilis, Ruegeria pomeroyi, and Roseovarious nubinhibens (5, 44, 45). Furthermore, RcGTA-like full gene clusters, or partial/rearranged clusters, were present in most of the alphaproteobacterial genomes (43). Although genes encoding comEC and comF homologues are widespread, they are not ubiquitous in prokaryotes (14). We hypothesized that if com genes are important for GTA-mediated genetic exchange, homologues would be present in all previously identified GTA gene-containing genomes (43). Therefore, we performed BLASTP searches using the R. capsulatus genes as a query against a number of such alphaproteobacterial genomes, as well as unrelated bacteria that produce a non-RcGTA-homologous GTA. We observed that of 23 organisms that contain a functional GTA (either homologous to RcGTA or not) or those that contain full or partial/rearranged RcGTA-like gene clusters, all contain a ComEC homologue and a ComF homologue, except for Sphingomonas alaskensis, which contains only a ComEC homologue (summarized in Table S5 in the supplemental material).
The RcGTA regulator CtrA is expressed throughout the population.
CtrA-dependent transcription of RcGTA genes is limited to <1% of a WT population (7, 46). Because CtrA is essential for R. capsulatus RcGTA recipient capability and the natural competence gene homologues comEC, comF, comM, and dprA are downregulated in a ΔctrA mutant, we evaluated ctrA expression in single cells as an indicator of RcGTA recipient capability. For this purpose, an mCherry fluorescent protein coding sequence fused in frame with the ctrA start codon on plasmid pCtrAmCherry was conjugated into the WT strain B10. Cells were grown to the stationary phase and imaged by phase-contrast and fluorescence microscopy. It was found that ctrA, like the CtrA-induced competence gene dprA, is expressed in the vast majority of cells (Fig. 4A). Furthermore, a single peak of fluorescent cells was obtained when the culture was evaluated by flow cytometry (Fig. 4B).
FIG 4.
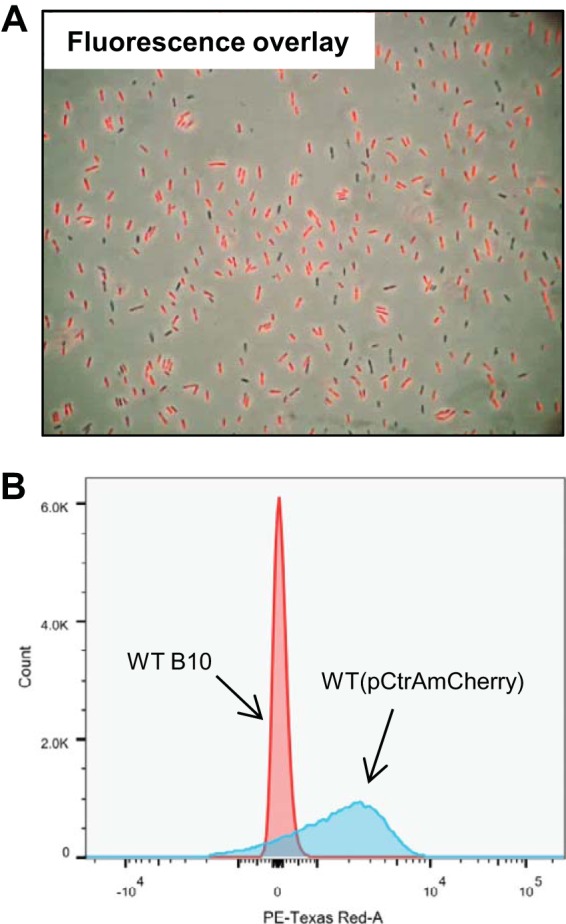
(A) Fluorescence image where WT(pCtrAmCherry) cells were excited by 561-nm light (fluorescence emission, 610 nm) overlain on a light microscopy image of the same cells. (B) Flow cytometry graphical representation of WT cells containing the plasmid pCtrAmCherry, on which the mCherry fluorescent protein gene is driven by the ctrA promoter, compared to WT cells lacking the plasmid. The horizontal axis represents arbitrary fluorescence units, and the vertical axis gives cell counts (the total number of cells in each sample, indicated by the integrated area under each curve, was 50,000). PE, phycoerythrin.
DISCUSSION
We provide genetic and biochemical evidence that ComEC/ComF natural competence-related machinery facilitates DNA carried within a phage-like RcGTA particle to enter the cytoplasm of recipient cells, and that the ComM protein of unknown function is involved in RcGTA recipient capability.
The RcGTA morphologically resembles a small, noncontractile tailed phage (8), and based on the data presented in this report, we suggest that RcGTA uses a mode of DNA entry into the cell different from that of phage model systems.
During infection by the contractile long-tailed phage T4, receptor binding triggers a conformational change resulting in the contraction of the tail sheath, causing the tail tube to puncture the outer membrane of the recipient cell. The tail tube then penetrates the peptidoglycan (PG), which is cleaved by the lysozyme domain on the tail tube (gp5), the tail tube crosses the cytoplasmic membrane, and subsequently it delivers DNA into the cytoplasm (47, 48).
The noncontractile long-tailed phage λ differs from T4. An initial binding to the LamB receptor (49) triggers a conformational change in the tail, bringing it into contact with the cell surface. This triggers release of the DNA, which is thought to be accompanied by movement of the tape measure protein (TMP) and central tail fiber protein from the tail into the cell envelope, forming a channel for DNA entry into the periplasm (50) and subsequently into the cytoplasm in a process involving the mannose permease complex, although the mechanism is not understood (50–52). The process of breaching the peptidoglycan also is unclear.
Infection by short-tailed phages, such as T7 or p22, begins with initial receptor binding via tail fibers. Often, interaction with a secondary receptor (such as an integral OM protein) also is required. Proteins present in the head then are injected into the periplasm via the tail prior to DNA entry, and these proteins may degrade the peptidoglycan. Subsequently, the short tail is lengthened, or a translocation tube is formed by injected proteins to form a channel into the cytoplasm, allowing DNA delivery (53, 54). In addition, it was recently shown that the apparently tail-less phage ϕX174 forms a tail-like structure during infection to span the periplasmic space for DNA transport (55).
Our data indicate that there is an accumulation of RcGTA-borne DNA in the periplasm of ΔcomEC and ΔcomF mutants. One possible explanation for the bottleneck is that the ComEC DNA transporter serves as a CM receptor for a lengthening tail or a TMP, analogous to the poorly understood function of ManY for phage λ (50, 51, 56). In this scenario, the CM receptor is not required for DNA ejection from the particle that has docked on an OM receptor but is required for injection into the cytoplasm, so the DNA comes to reside in the periplasm. Alternatively, RcGTA may inject DNA into the periplasm of WT cells and the ComEC/ComF proteins transport the DNA through the CM, as in natural transformation (14). Although our data do not definitively differentiate between these two possibilities, we favor the latter explanation for several reasons.
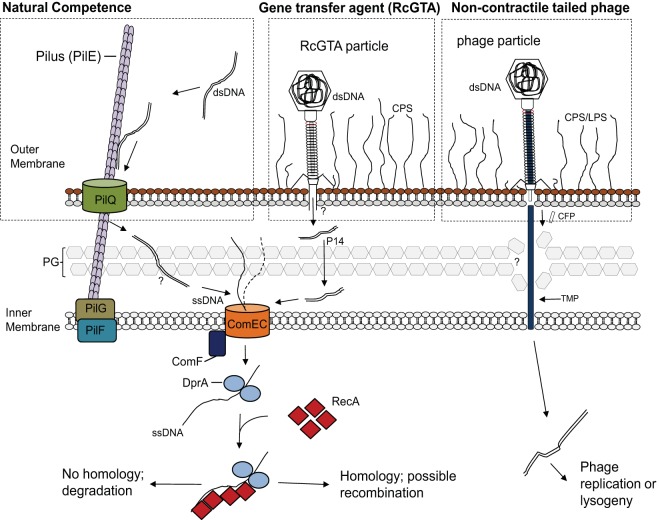
One reason is the fact that both comEC and comF expression are controlled by CtrA as part of a set of natural transformation genes, which also includes the RcGTA-borne DNA-specific cytoplasmic recombination mediator DprA (13). This indicates that all of these genes are part of a regulated system of DNA uptake and recombination, which contrasts with the use of ManY (which has a function unrelated to DNA transport) by phage λ (50). Another reason is the need for ComF in DNA passage through the CM. Because the bulk of ComF is predicted to reside in the cytoplasm, it appears unlikely to be used as an IM receptor. Lastly, we note that the RcGTA predicted TMP protein is only 219 amino acids in length, which is much shorter than typical noncontractile tailed phage TMPs (>600 amino acids) that are thought to bridge the periplasmic space (50). A model is shown in Fig. 5.
FIG 5.
Schematic diagram comparing an overview of natural transformation systems in Gram-negative bacteria to the proposed RcGTA recipient capability pathway and a noncontractile tailed phage pathway. The conserved steps in natural transformation are shown on the left. DNA is taken up by a transformation-dedicated pilus (Tfb), which binds dsDNA and brings it into the periplasm, where it is processed into ssDNA by an unknown process. DNA then enters the cytoplasm via the ComEC inner membrane protein in a pathway involving ComF. Upon cytoplasmic internalization, ssDNA is bound by DprA, which functions to recruit the RecA recombinase to polymerize onto ssDNA, promoting a homology search and subsequent recombination into the chromosome (14). The proposed model of RcGTA, shown in the center, involves the same steps for DNA import and recombination into the chromosome as those in natural transformation. It is unknown how RcGTA-injected DNA traverses the periplasm or whether DNA enters the cell as ssDNA or dsDNA. Passage through the peptidoglycan (PG) likely is facilitated by the RcGTA protein p14, which degrades PG (46). Shown on the right is a model of noncontractile tailed phage infection, with the tape measure protein (TMP) used to traverse the outer membrane, periplasm, and cytoplasmic membrane (50). CFP, central tail fiber protein; LPS, lipopolysaccharide; CPS, capsular polysaccharide. (The natural transformation pathway image was adapted from reference 14 with permission of the publisher.)
Although the phenotype of comEC and comF null mutants has been documented in several studies of natural transformation systems (19, 36), the role of comM homologues has not. Here, we establish that the R. capsulatus comM homologue is essential for detectable RcGTA recipient capability. This is in contrast to transformation of H. influenzae, which was reduced by only ∼6-fold (19) in a comM mutant, although the reason for this is not clear. One interesting observation we made was that genes encoding ComM homologues appear to be absent from genomes containing DprA homologues that lack a DD3, including S. pneumoniae and B. subtilis, in which DprA proteins have been extensively studied and bind ssDNA but not dsDNA. Because of the cooccurrence of ComM homologues and DprA proteins that contain a DD3, we speculate that ComM and DprA function in the same pathway. Because DprA is directly involved in recombination in natural transformation and ComM is predicted to be cytoplasmic and contains predicted domains similar to those of helicases, we speculate that ComM is involved in a recombination step in the cytoplasm of R. capsulatus cells and possibly in natural transformation systems as well.
CtrA is a widely conserved regulator among the alphaproteobacteria. In the prototypic example of Caulobacter crescentus, CtrA is a regulator of flagellar motility, the cell cycle, and asymmetric cell division, and it is essential for viability (57–59). In R. capsulatus, ctrA is not essential for viability but was found to control a regulon partially overlapping that of C. crescentus, including flagellum synthesis (60, 61). Although R. capsulatus CtrA is essential for both RcGTA production and recipient capability (13, 60, 62), the RcGTA promoter is activated in <1% of WT cells in the stationary phase (7, 46). In contrast, the essential recipient capability factor (dprA) promoter is expressed in essentially all cells in a population (13), as we found here for ctrA itself (Fig. 4). The mechanism for restricting RcGTA production to a subset of CtrA-induced cells remains to be determined.
Because RcGTA genetic exchange requires homologous recombination within the same species, coregulation of RcGTA production and recipient capability may be explained in terms of natural selection. Although RcGTA production requires lysis of the producing cell, only a small fraction of a population expresses these genes (7). Because the cost of producing RcGTA (i.e., death) is high, having the surrounding population primed as RcGTA recipients by transcription of the com genes would provide a selective advantage to such cells, because they could take up the DNA packaged by the producing cells, perhaps facilitating spread of beneficial mutations HR-mediated DNA repair during growth under mutation-prone conditions.
We suggest that RcGTA represents a mechanism of horizontal gene transfer that combines aspects of transduction and natural transformation, resulting in a novel genetic exchange mechanism that provides recipients with new genes or alleles. In a natural aquatic environment, where RcGTA-producing Rhodobacteraceae are prevalent (63), the encapsidation of DNA within RcGTA would protect the DNA from degradation by nucleases until the particle encountered a cell that had been rendered capable of acquisition of RcGTA-borne genes, via com gene induction, regulated by the same systems that induced the production of RcGTA particles.
Supplementary Material
ACKNOWLEDGMENTS
We thank L. Eltis for access to qPCR components and H. Otani for technical assistance. We thank the Canadian Institutes of Health Research for an Operating Grant (93779 to J.T.B.) and the University of British Columbia for a postgraduate scholarship (to C.A.B.).
C.A.B. designed research, performed experiments, analyzed data, and wrote the manuscript; H.D. performed fluorescence microscopy analysis, analyzed data, and wrote the manuscript; J.A.J. generated the ΔcomEC mutant and edited the manuscript; J.T.B. designed research, analyzed data, and wrote the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00332-15.
REFERENCES
- 1.Griffith F. 1928. The significance of Pneumococcal types. J Hyg 27:113–159. doi: 10.1017/S0022172400031879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lederberg J, Tatum EL. 1946. Gene recombination in Escherichia coli. Nature 158:558. [DOI] [PubMed] [Google Scholar]
- 3.Zinder ND, Lederberg J. 1952. Genetic exchange in Salmonella. J Bacteriol 64:679–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marrs B. 1974. Genetic recombination in Rhodopseudomonas capsulata. Proc Natl Acad Sci U S A 71:971–973. doi: 10.1073/pnas.71.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lang AS, Zhaxybayeva O, Beatty JT. 2012. Gene transfer agents: phage-like elements of genetic exchange. Nat Rev Microbiol 10:472–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lang AS, Beatty JT. 2001. The gene transfer agent of Rhodobacter capsulatus and “constitutive transduction” in prokaryotes. Arch Microbiol 175:241–249. doi: 10.1007/s002030100260. [DOI] [PubMed] [Google Scholar]
- 7.Hynes AP, Mercer RG, Watton DE, Buckley CB, Lang AS. 2012. DNA packaging bias and differential expression of gene transfer agent genes within a population during production and release of the Rhodobacter capsulatus gene transfer agent, RcGTA. Mol Microbiol 85:314–325. doi: 10.1111/j.1365-2958.2012.08113.x. [DOI] [PubMed] [Google Scholar]
- 8.Yen HC, Hu NT, Marrs BL. 1979. Characterization of the gene transfer agent made by an overproducer mutant of Rhodopseudomonas capsulata. J Mol Biol 131:157–168. doi: 10.1016/0022-2836(79)90071-8. [DOI] [PubMed] [Google Scholar]
- 9.Westbye AB, Leung MM, Florizone SM, Taylor TA, Johnson JA, Fogg PC, Beatty JT. 2013. Phosphate concentration and the putative sensor kinase protein CckA modulate cell lysis and release of the Rhodobacter capsulatus gene transfer agent. J Bacteriol 195:5025–5040. doi: 10.1128/JB.00669-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaefer AL, Taylor TA, Beatty JT, Greenberg EP. 2002. Long-chain acyl-homoserine lactone quorum-sensing regulation of Rhodobacter capsulatus gene transfer agent production. J Bacteriol 184:6515–6521. doi: 10.1128/JB.184.23.6515-6521.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lang AS, Beatty JT. 2002. A bacterial signal transduction system controls genetic exchange and motility. J Bacteriol 184:913–918. doi: 10.1128/jb.184.4.913-918.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leung MM, Brimacombe CA, Spiegelman GB, Beatty JT. 2012. The GtaR protein negatively regulates transcription of the gtaRI operon and modulates gene transfer agent (RcGTA) expression in Rhodobacter capsulatus. Mol Microbiol 83:759–774. doi: 10.1111/j.1365-2958.2011.07963.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brimacombe CA, Ding H, Beatty JT. 2014. Rhodobacter capsulatus DprA is essential for RecA-mediated gene transfer agent (RcGTA) recipient capability regulated by quorum-sensing and the CtrA response regulator. Mol Microbiol 92:1260–1278. doi: 10.1111/mmi.12628. [DOI] [PubMed] [Google Scholar]
- 14.Johnston C, Martin B, Fichant G, Polard P, Claverys JP. 2014. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol 12:181–196. doi: 10.1038/nrmicro3199. [DOI] [PubMed] [Google Scholar]
- 15.Mell JC, Hall IM, Redfield RJ. 2012. Defining the DNA uptake specificity of naturally competent Haemophilus influenzae cells. Nucleic Acids Res 40:8536–8549. doi: 10.1093/nar/gks640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Draskovic I, Dubnau D. 2005. Biogenesis of a putative channel protein, ComEC, required for DNA uptake: membrane topology, oligomerization and formation of disulphide bonds. Mol Microbiol 55:881–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mortier-Barriere I, Velten M, Dupaigne P, Mirouze N, Pietrement O, McGovern S, Fichant G, Martin B, Noirot P, Le Cam E, Polard P, Claverys JP. 2007. A key presynaptic role in transformation for a widespread bacterial protein: DprA conveys incoming ssDNA to RecA. Cell 130:824–836. doi: 10.1016/j.cell.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 18.Yadav T, Carrasco B, Hejna J, Suzuki Y, Takeyasu K, Alonso JC. 2013. Bacillus subtilis DprA recruits RecA onto single-stranded DNA and mediates annealing of complementary strands coated by SsbB and SsbA. J Biol Chem 288:22437–22450. doi: 10.1074/jbc.M113.478347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sinha S, Mell JC, Redfield RJ. 2012. Seventeen Sxy-dependent cyclic AMP receptor protein site-regulated genes are needed for natural transformation in Haemophilus influenzae. J Bacteriol 194:5245–5254. doi: 10.1128/JB.00671-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fogg PC, Colloms S, Rosser S, Stark M, Smith MC. 2014. New applications for phage integrases. J Mol Biol 426:2703–2716. doi: 10.1016/j.jmb.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groth AC, Calos MP. 2004. Phage integrases: biology and applications. J Mol Biol 335:667–678. doi: 10.1016/j.jmb.2003.09.082. [DOI] [PubMed] [Google Scholar]
- 22.Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat Biotech 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 23.Taylor DP, Cohen SN, Clark WG, Marrs BL. 1983. Alignment of genetic and restriction maps of the photosynthesis region of the Rhodopseudomonas capsulata chromosome by a conjugation-mediated marker rescue technique. J Bacteriol 154:580–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- 25.Beatty JT, Gest H. 1981. Biosynthetic and bioenergetic functions of citric acid cycle reactions in Rhodopseudomonas capsulata. J Bacteriol 148:584–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wall JD, Weaver PF, Gest H. 1975. Gene transfer agents, bacteriophages, and bacteriocins of Rhodopseudomonas capsulata. Arch Microbiol 105:217–224. doi: 10.1007/BF00447140. [DOI] [PubMed] [Google Scholar]
- 27.Brimacombe CA, Stevens A, Jun D, Mercer R, Lang AS, Beatty JT. 2013. Quorum-sensing regulation of a capsular polysaccharide receptor for the Rhodobacter capsulatus gene transfer agent (RcGTA). Mol Microbiol 87:802–817. doi: 10.1111/mmi.12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barany F. 1985. Single-stranded hexameric linkers: a system for in-phase insertion mutagenesis and protein engineering. Gene 37:111–123. doi: 10.1016/0378-1119(85)90263-X. [DOI] [PubMed] [Google Scholar]
- 29.Marx CJ, Lidstrom ME. 2001. Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology 147:2065–2075. [DOI] [PubMed] [Google Scholar]
- 30.Hancock RE. 1984. Alterations in outer membrane permeability. Annu Rev Microbiol 38:237–264. doi: 10.1146/annurev.mi.38.100184.001321. [DOI] [PubMed] [Google Scholar]
- 31.Dereeper A, Audic S, Claverie JM, Blanc G. 2010. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol 10:8. doi: 10.1186/1471-2148-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R. 2013. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res 41:W597–W600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 35.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. 2014. Pfam: the protein families database. Nucleic Acids Res 42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seitz P, Blokesch M. 2013. DNA-uptake machinery of naturally competent Vibrio cholerae. Proc Natl Acad Sci U S A 110:17987–17992. doi: 10.1073/pnas.1315647110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murray AW. 1971. The biological significance of purine salvage. Annu Rev Biochem 40:811–826. doi: 10.1146/annurev.bi.40.070171.004115. [DOI] [PubMed] [Google Scholar]
- 38.Klug A, Rhodes D. 1987. Zinc fingers: a novel protein fold for nucleic acid recognition. Cold Spring Harbor Symp Quant Biol 52:473–482. doi: 10.1101/SQB.1987.052.01.054. [DOI] [PubMed] [Google Scholar]
- 39.Gwinn ML, Ramanathan R, Smith HO, Tomb JF. 1998. A new transformation-deficient mutant of Haemophilus influenzae Rd with normal DNA uptake. J Bacteriol 180:746–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reid JD, Siebert CA, Bullough PA, Hunter CN. 2003. The ATPase activity of the ChlI subunit of magnesium chelatase and formation of a heptameric AAA+ ring. Biochemistry 42:6912–6920. doi: 10.1021/bi034082q. [DOI] [PubMed] [Google Scholar]
- 41.Yamada K, Miyata T, Tsuchiya D, Oyama T, Fujiwara Y, Ohnishi T, Iwasaki H, Shinagawa H, Ariyoshi M, Mayanagi K, Morikawa K. 2002. Crystal structure of the RuvA-RuvB complex: a structural basis for the Holliday junction migrating motor machinery. Mol Cell 10:671–681. doi: 10.1016/S1097-2765(02)00641-X. [DOI] [PubMed] [Google Scholar]
- 42.Maiorano D, Lutzmann M, Mechali M. 2006. MCM proteins and DNA replication. Curr Opin Cell Biol 18:130–136. doi: 10.1016/j.ceb.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 43.Lang AS, Beatty JT. 2007. Importance of widespread gene transfer agent genes in alpha-proteobacteria. Trends Microbiol 15:54–62. doi: 10.1016/j.tim.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 44.Biers EJ, Wang K, Pennington C, Belas R, Chen F, Moran MA. 2008. Occurrence and expression of gene transfer agent genes in marine bacterioplankton. Appl Environ Microbiol 74:2933–2939. doi: 10.1128/AEM.02129-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McDaniel LD, Young E, Delaney J, Ruhnau F, Ritchie KB, Paul JH. 2010. High frequency of horizontal gene transfer in the oceans. Science 330:50. doi: 10.1126/science.1192243. [DOI] [PubMed] [Google Scholar]
- 46.Fogg PCM, Westbye AB, Beatty JT. 2012. One for all or all for one: heterogeneous expression and host cell lysis are key to gene transfer agent activity in Rhodobacter capsulatus. PLoS One 7:e43772. doi: 10.1371/journal.pone.0043772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rossmann MG, Mesyanzhinov VV, Arisaka F, Leiman PG. 2004. The bacteriophage T4 DNA injection machine. Curr Opin Struct Biol 14:171–180. doi: 10.1016/j.sbi.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 48.Leiman PG, Shneider MM. 2012. Contractile tail machines of bacteriophages. Adv Exp Med Biol 726:93–114. [DOI] [PubMed] [Google Scholar]
- 49.Schwartz M. 1975. Reversible interaction between coliphage lambda and its receptor protein. J Mol Biol 99:185–201. doi: 10.1016/S0022-2836(75)80167-7. [DOI] [PubMed] [Google Scholar]
- 50.Davidson AR, Cardarelli L, Pell LG, Radford DR, Maxwell KL. 2012. Long noncontractile tail machines of bacteriophages. Adv Exp Med Biol 726:115–142. [DOI] [PubMed] [Google Scholar]
- 51.Williams N, Fox DK, Shea C, Roseman S. 1986. Pel, the protein that permits lambda DNA penetration of Escherichia coli, is encoded by a gene in ptsM and is required for mannose utilization by the phosphotransferase system. Proc Natl Acad Sci U S A 83:8934–8938. doi: 10.1073/pnas.83.23.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scandella D, Arber W. 1976. Phage lambda DNA injection into Escherichia coli pel- mutants is restored by mutations in phage genes V or H. Virology 69:206–215. doi: 10.1016/0042-6822(76)90207-5. [DOI] [PubMed] [Google Scholar]
- 53.Casjens SR, Molineux IJ. 2012. Short noncontractile tail machines: adsorption and DNA delivery by podoviruses. Adv Exp Med Biol 726:143–179. [DOI] [PubMed] [Google Scholar]
- 54.Hu B, Margolin W, Molineux IJ, Liu J. 2013. The bacteriophage t7 virion undergoes extensive structural remodeling during infection. Science 339:576–579. doi: 10.1126/science.1231887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun L, Young LN, Zhang X, Boudko SP, Fokine A, Zbornik E, Roznowski AP, Molineux IJ, Rossmann MG, Fane BA. 2014. Icosahedral bacteriophage PhiX174 forms a tail for DNA transport during infection. Nature 505:432–435. [DOI] [PubMed] [Google Scholar]
- 56.Boulanger P, Jacquot P, Plancon L, Chami M, Engel A, Parquet C, Herbeuval C, Letellier L. 2008. Phage T5 straight tail fiber is a multifunctional protein acting as a tape measure and carrying fusogenic and muralytic activities. J Biol Chem 283:13556–13564. doi: 10.1074/jbc.M800052200. [DOI] [PubMed] [Google Scholar]
- 57.Quon KC, Marczynski GT, Shapiro L. 1996. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84:83–93. doi: 10.1016/S0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 58.Wu J, Ohta N, Newton A. 1998. An essential, multicomponent signal transduction pathway required for cell cycle regulation in Caulobacter. Proc Natl Acad Sci U S A 95:1443–1448. doi: 10.1073/pnas.95.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reisenauer A, Quon K, Shapiro L. 1999. The CtrA response regulator mediates temporal control of gene expression during the Caulobacter cell cycle. J Bacteriol 181:2430–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lang AS, Beatty JT. 2000. Genetic analysis of a bacterial genetic exchange element: the gene transfer agent of Rhodobacter capsulatus. Proc Natl Acad Sci U S A 97:859–864. doi: 10.1073/pnas.97.2.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mercer RG, Callister SJ, Lipton MS, Pasa-Tolic L, Strnad H, Paces V, Beatty JT, Lang AS. 2010. Loss of the response regulator CtrA causes pleiotropic effects on gene expression but does not affect growth phase regulation in Rhodobacter capsulatus. J Bacteriol 192:2701–2710. doi: 10.1128/JB.00160-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mercer RG, Quinlan M, Rose AR, Noll S, Beatty JT, Lang AS. 2012. Regulatory systems controlling motility and gene transfer agent production and release in Rhodobacter capsulatus. FEMS Microbiol Lett 331:53–62. doi: 10.1111/j.1574-6968.2012.02553.x. [DOI] [PubMed] [Google Scholar]
- 63.Yu F, MacLeod DM, Rivkin RB, Chen F, Buchan A, Lang AS. 2010. High diversity of Rhodobacterales in the subarctic north Atlantic Ocean and gene transfer agent protein expression in isolated strains. Aquatic Microb Ecol 59:283–293. doi: 10.3354/ame01398. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.