ABSTRACT
PpaA from Rhodobacter sphaeroides is a member of a family of proteins that are thought to function as antirepressors of PpsR, a widely disseminated repressor of photosystem genes in purple photosynthetic bacteria. PpaA family members exhibit sequence similarity to a previously defined SCHIC (sensor containing heme instead of cobalamin) domain; however, the tetrapyrrole-binding specificity of PpaA family members has been unclear, as R. sphaeroides PpaA has been reported to bind heme while the Rhodobacter capsulatus homolog has been reported to bind cobalamin. In this study, we reinvestigated tetrapyrrole binding of PpaA from R. sphaeroides and show that it is not a heme-binding protein but is instead a cobalamin-binding protein. We also use bacterial two-hybrid analysis to show that PpaA is able to interact with PpsR and activate the expression of photosynthesis genes in vivo. Mutations in PpaA that cause loss of cobalamin binding also disrupt PpaA antirepressor activity in vivo. We also tested a number of PpaA homologs from other purple bacterial species and found that cobalamin binding is a conserved feature among members of this family of proteins.
IMPORTANCE Cobalamin (vitamin B12) has only recently been recognized as a cofactor that affects gene expression by interacting in a light-dependent manner with transcription factors. A group of related antirepressors known as the AppA/PpaA/AerR family are known to control the expression of photosynthesis genes in part by interacting with either heme or cobalamin. The specificity of which tetrapyrroles that members of this family interact with has, however, remained cloudy. In this study, we address the tetrapyrrole-binding specificity of the PpaA/AerR subgroup and establish that it preferentially binds cobalamin over heme.
INTRODUCTION
In most species of anoxygenic phototrophic prokaryotes, photosynthesis gene expression is tightly regulated in response to changes in cellular redox poise and light intensity (1). Under high light intensity, pigment levels are reduced, presumably to balance the oxidation-reduction potential of the ubiquinone pool (2, 3). When grown under aerobic conditions, most purple nonsulfur bacteria repress the expression of photosynthesis genes and instead grow chemoheterotrophically or chemoautotrophically.
A number of regulatory elements have been identified that are involved in redox and light control of photosystem synthesis (reviewed in references 2 and 4). One well-studied redox- and light-regulated transcription factor that controls this process is PpsR (also called CrtJ in some species) (5–9). PpsR/CrtJ homologs are linked to a cluster of genes coding for enzymes involved in bacteriochlorophyll and carotenoid biosynthesis in almost all purple phototrophic bacteria. Several species contain two functional copies of PpsR, each with its own functionality (8, 10, 11). For example, in Bradyrhizobium ORS278, PpsR1 acts as a redox-responsive activator while a second homolog, PpsR2, acts as a light-regulated repressor in conjunction with the light-responding antirepressor/photoreceptor BphB2 (11).
Most PpsR-regulated genes are involved in photosynthesis, specifically, those for enzymes involved in the synthesis of the photopigments bacteriochlorophyll and carotenoids and structural proteins of the light-harvesting and reaction center photosystem (4–7). Other genes identified in the R. capsulatus and R. sphaeroides PpsR/CrtJ regulons include those for enzymes involved in heme biosynthesis and genes that code for cytochrome apoproteins (12–14). Each of the characterized PpsR/CrtJ-regulated promoters contains a variant of the DNA recognition sequence TGT-N12-ACA (5), which is present in tandem (typically 8 bp apart) or at distant sites up to 240 bp apart (15, 16). Mutational studies indicate that PpsR/CrtJ regulons bind cooperatively to these tandem sites and that subtle alterations in the spacing between these sites can have significant effects in promoter regulation (11, 15, 17, 18).
The DNA-binding properties of PpsR homologs are regulated, in part, by redox-dependent modifications of key cysteine residues present in the helix-turn-helix DNA-binding region (11, 17–21). It has also been reported that a PAS domain of PpsR binds heme and that heme binding affects the DNA-binding properties of PpsR (22). In addition to oxidation and heme, several antirepressors are known to interact with and disrupt the DNA-binding properties of PpsR. As mentioned above, the activity of PpsR2 from Bradyrhizobium sp. strain ORS278 is inhibited by a light-regulated interaction with the red-light-absorbing phytochrome-like photoreceptor BphB2 (11). In Rhodobacter sphaeroides, the activity of PpsR is inhibited by an interaction with the antirepressor AppA that converts PpsR from an active tetramer to an inactive dimer (17, 23–25). The antirepressor activity of AppA is regulated by both blue light absorption via a bound flavin and the presence of a bound heme (17, 26–28).
Recently, the DNA-binding properties of CrtJ from R. capsulatus was demonstrated to be inhibited by an interaction with another light-regulated antirepressor called AerR (29). AerR (PpaA in some species) has homologs in almost all purple nonsulfur bacteria and is typically located just upstream of the PpsR/CrtJ loci (29). These PpaA/AerR homologs are characterized by the presence of a B12-binding domain with an absence of enzymatic or other identifiable output domains. Phylogenetic analysis indicates that the B12-binding domain in PpaA/AerR homologs exhibit a notable evolutionary distance from that of B12-dependent enzymes, leading to the hypothesis that this family no longer binds cobalamin but instead may bind heme (30). Indeed, a study on a truncated B12-binding domain from R. sphaeroides PpaA showed a preference for the binding of heme over cobalamin like that of the related light-regulated antirepressor AppA (31). However, recently work by Cheng and coworkers showed that full-length AerR from R. capsulatus is, in fact, a bona fide cobalamin-binding protein that can be readily purified with tightly bound hydroxylcobalamin (29). Furthermore, a mutation of the strongly conserved histidine in the B12-binding motif that is thought to form an axial ligand with Co resulted in a loss of cobalamin binding in vitro (29). Analysis of the same His mutation in vivo also resulted in a loss of antirepressor activity, further demonstrating a role for B12 binding in the regulation of the antirepressor activity of AerR.
While cobalamin has long been recognized as a cofactor in a number of enzymatic reactions, it has only recently been shown to play a role as a sensing cofactor. In several bacteria, the mRNA for the cobalamin transporter BtuB was shown to contain a B12-binding riboswitch that downregulates translation at elevated cobalamin levels (32, 33). In Myxococcus xanthus, CarH regulates the light-dependent expression of carotenoid genes in a B12-dependent manner (34). In CarH, blue-light-driven photolysis of bound adenosylcobalamin to hydroxylcobalamin leads to a conformational change that subsequently releases CarH from target DNA (34). AerR also has a specificity for hydroxylcobalamin, which is a by-product of photohydrolyzed adenosylcobalamin, indicating that AerR also acts as a light antireceptor of CrtJ (29). Indeed, R. capsulatus strains with the gene for AerR deleted show an in vivo defect in light regulation of photosystem synthesis (29).
In this study, we reinvestigated the tetrapyrrole specificity of PpaA from R. sphaeroides. Our results show that a truncated B12-binding domain of PpaA binds substoichiometric amounts of heme but that isolated full-length PpaA preferentially binds hydroxylcobalamin. By overexpressing various mutant forms of PpaA in an AppA deletion strain, we also show that cobalamin binding is indeed the preferred mode of action of PpaA. Finally, we demonstrate that a number of PpaA/AerR homologs from other purple nonsulfur bacteria show that they preferentially bind cobalamin instead of heme.
MATERIALS AND METHODS
Strains and plasmids.
For the strains and plasmids used in this study, see Table S2 in the supplemental material. R. sphaeroides strain HR was grown in Sistrom's minimal medium (35) with succinate and Casamino Acids as a carbon source or in LB at 30°C. Escherichia coli was grown at 37°C in Luria broth (LB). Rhodospirillum centenum was grown in CENS medium (36) at 37°C. Rhodopseudomonas palustris and Rubrivivax gelatinosus were grown in PYVS medium (37) at 30°C.
To make clean deletions, plasmid pAJV1 was used. Plasmid pAJV1 was constructed by fusing sacB from plasmid pZJD29a (38) to the puc promoter of R. sphaeroides. The puc promoter was amplified with primers Ppuc-sacB-FW1 and Ppuc-sacB-RV1, and the sacB reading frame was amplified with primers Ppuc-sacB-FW2 and Ppuc-sacB-RV2 (see Table S1 in the supplemental material). The products were then used in a crossover PCR. The resulting Ppuc-sacB cassette was then inserted into BstB1-digested and blunted pJP5603 (39). The resulting plasmid, pAJV1, confers both kanamycin resistance and sucrose sensitivity. Flanking 500-bp regions of ppaA were PCR amplified with primers appA-up-F, appA-up-R, appA-down-F, and appA-down-R (see Table S1). The products were then ligated into SmaI-digested pAJV1 by isothermal assembly. The resulting plasmid contains the flanking regions of appA linked by a 21-bp nonsense reading frame. The resulting plasmids were sequenced to ensure the sequence fidelity of inserts.
Deletion plasmids were transferred into R. sphaeroides by conjugal mating with E. coli S17-1(λpir). E. coli S17-1(λpir) was transformed with the deletion plasmid, grown to exponential phase, and washed twice to remove antibiotics. A 750-μl volume of washed E. coli cells was then mixed with 750 μl of an overnight culture of R. sphaeroides. Cells were pelleted and resuspended in 50 μl of LB. The resuspended cells were then spotted onto an LB plate in 50-μl aliquots and incubated for 24 to 48 h at 30°C. At that point, the cells were restreaked onto LB agar with 25 μg/ml kanamycin. The selective plates also contained 5 μg/ml gentamicin to inhibit the growth of E. coli, as R. sphaeroides has innate gentamicin resistance and its growth is not affected by gentamicin at low concentrations. The plates were incubated for 3 days at 30°C, after which several colonies were restreaked onto LB agar with 50 μg/ml kanamycin. To select for double recombinants, mutants were streaked onto LB without antibiotics and supplemented with 10% (wt/vol) sucrose. Colonies were selected for complete segregation by testing for kanamycin sensitivity. Deletion mutants were confirmed by colony input PCR.
To overexpress PpaA in R. sphaeroides, plasmid pSRKKm (40) was used. pSRKkm is a broad-host-range plasmid with a multiple cloning site with protein expression under the control of a T7 promoter. This plasmid allows induction of overexpression by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG). A PCR-amplified ppaA gene was cloned into NdeI- and NotI-digested pSRKkm. Several different mutations were subsequently introduced by using a quick-change protocol (Agilent Technologies). R. sphaeroides transformed with pSRKKm and its derivatives was grown in LB or Sistrom's minimal medium supplemented with 25 μg/ml kanamycin. Overexpression of ppaA was induced by the addition of IPTG to a final concentration of 1 mM.
Bacterial two-hybrid screening.
In vivo interaction between AppA and PpaA or PpsR was tested by BacterioMatch II bacterial two-hybrid screening (Agilent Technologies). The genes encoding PpaA, PpsR, and AppA were cloned into both plasmids pBT and pTRG. Interaction was tested by transforming BacterioMatch II validation cells with different combinations of bait and target plasmids. As a positive control, cells were transformed with plasmids pBT-LGF2 and pTRG-Gal11, which were supplied with the BacterioMatch II kit. As a negative control, cells were transformed with a bait plasmid in combination with an empty target plasmid. The transformants were then plated on selective medium (M9, His dropout) and incubated for 48 h at 30°C in the dark. Growth on selective medium indicated protein-protein interactions.
Protein purification.
Plasmid pET-MBP was used as a vector to overexpress various PpaA homologs. pET-MBP was constructed by PCR amplifying the maltose-binding protein (MBP) domain and tobacco etch virus (TEV) protease site from plasmid pMHT-delta238 (41) with primers MBP-mod-F and MBP-mod-R (see Table S1 in the supplemental material) and ligating the product into NcoI- and BamHI-digested plasmid pET28a(+) (Novagen). The resulting plasmid encodes a His6--tagged MBP domain that can be cleaved with TEV protease. Genes of interest were PCR amplified from genomic DNA (R. sphaeroides HR [primers Rsph-ppaA-F and Rsph-ppaA-R] PpA, Rubrivivax gelatinosus AerR [primers Rgel-aerR-F and Rgel-aerR-R], Rhodopseudomonas palustris CGA009 RPA1540 [primers Rpal-ppaA-F and Rpal-ppaA-R], and Rhodospirillum centenum AerR [primers Rcen-aerR-F or Rcen-aerR-FL-F and Rcen-aerR-R]) or synthesized as codon-optimized genes (Erythrobacter sp. strain NAP1 PpaA, Methylobacterium extorquens MA-1 PpaA, Jannaschia sp. strain CCS1 PpaA) (IDT, Coralville, IA, USA). The program JCAT was used to optimize sequences for overexpression in E. coli (42).
Proteins were overexpressed in E. coli BL21(DE3) in LB with 25 μg/ml kanamycin. Cultures were grown at 37°C to an optical density at 600 nm (OD600) of ∼0.3 and then incubated at 16°C for an additional 1.5 h. Overexpression was induced by adding IPTG to a final concentration of 0.4 mM. After 16 to 20 h of growth at 16°C, cells were harvested and pellets were stored at −80°C. For purification, cell pellets were thawed on ice, resuspended in lysis buffer (20 mM Tris-HCl, 150 mM NaCl, 10 mM imidazole), and lysed by three passages through a Microfluidizer. Except when apoprotein was needed, hydroxylcobalamin was added to a final concentration of 25 μM. Cell debris was removed by centrifugation for 30 min at 15,000 rpm (Sorvall SS-34 rotor) and filtered through a 0.45-μm syringe filter. The clarified lysate was then applied to a gravity nickel column. The column was washed with 30 ml of wash buffer (lysis buffer with 30 mM imidazole). Bound protein was eluted with elution buffer (lysis buffer with 250 mM imidazole). The buffer was exchanged for 20 mM Tris-HCl (pH 8)–150 mM NaCl with a desalting column (Bio-Rad EconoPac 10DG).
When needed, the His6-MBP tag was cleaved by adding TEV protease in a 1:20 (TEV protease to MBP-tagged protein) molar ratio and incubating the reaction mixture for 2.5 h at room temperature in 20 mM Tris-HCl (pH 8)–150 mM NaCl. TEV protease-cleaved tags and uncleaved protein were then removed by first applying the protein to a column with amylose resin (NEB). The flowthrough was then applied to a nickel column and eluted with wash buffer. Finally, the resulting protein was run over a Superose 12 column to remove aggregated protein. The final protein was concentrated with an Amicon Ultra-4 centrifugal filter (Millipore; 10-kDa molecular mass cutoff).
Cofactor binding.
To test for binding of different forms of cobalamin, protein was overexpressed in E. coli as described before. Cell lysate was then aliquoted in six equal amounts, and different forms of cobalamin were added to a final concentration of 25 μM. One aliquot was exposed to high-intensity light (∼1,000 μmol · m−2 · s−1) for 5 min after the addition of adenosylcobalamin. Other aliquots were kept in the dark. As a control, 1 aliquot was purified in the absence of cobalamin. The lysates were incubated on ice for 1 h. The lysate was clarified by centrifugation for 30 min at 15,000 rpm (Sorvall SS-34 rotor). Proteins were then purified with amylose resin (New England BioLabs, USA). All procedures were performed in the dark under a dim red safety light. A pink color and the presence of absorbance peaks in the 500- to 550-nm region were used as indicators of cobalamin binding.
Photosynthetic growth.
To test for in vivo functionality, an R. sphaeroides HR ΔappA mutant was transformed with plasmid pSRK-ppaA, pSRK-ppaA-H146A, or pSRK-ppaA-G149E. As a control, the ΔappA mutant was transformed with pSRKkm. Aerobic cultures were grown until log phase and inoculated into fresh Sistrom's medium with 4% (wt/vol) succinate and 0.2% (wt/vol) Casamino Acids at an OD660 of ∼0.3. The cultures were transferred to screw-cap tubes filled to the top. When needed, expression was induced with IPTG at a final concentration of 1 mM. The cultures were incubated under incandescent light for 48 h at 30°C. Ten-milliliter volumes of the cultures were harvested, resuspended in 20 mM Tris-HCl (pH 8)–150 mM NaCl, and lysed by sonication. The lysates were centrifuged at maximum speed for 10 min. The UV-visible (UV-vis) absorbance spectrum was measured from 300 to 900 nm and normalized to protein content.
RESULTS
R. sphaeroides PpaA binds hydroxylcobalamin.
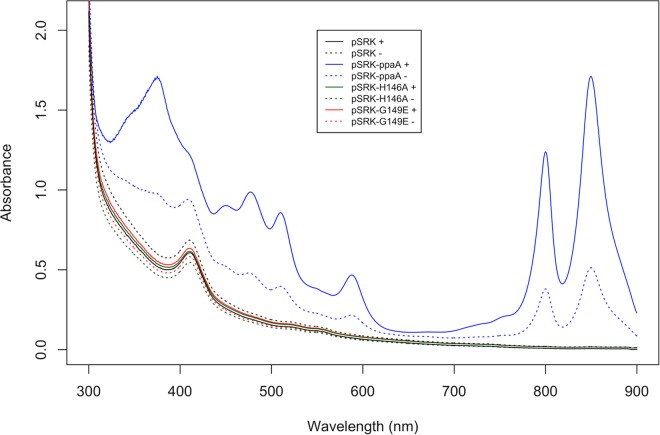
In a previous study, we showed that addition of hydroxycobalamin (OH-Cbl) to E. coli cell lysates that contained overexpressed AerR from R. capsulatus results in tight binding of this cobalamin derivative (29). Consequently, we tested whether full-length PpaA from R. sphaeroides is also able to bind cobalamin by adding various cobalamin derivatives to E. coli lysate that contained overexpressed PpaA. When OH-Cbl is added to the cell lysate, the protein elutes as a pink fraction during both affinity chromatography and size exclusion chromatography, with UV-vis spectral analysis indicating the presence of bound OH-Cbl (Fig. 1). The cobalamin remained associated with the protein in subsequent buffer exchange and gel filtration steps, showing that PpaA binds OH-Cbl tightly.
FIG 1.
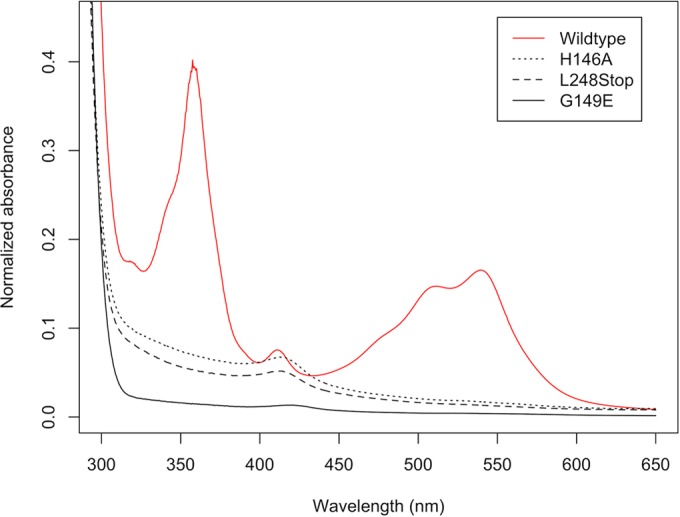
Absorption spectra of R. sphaeroides PpaA purified in the presence of hydroxycobalamin. Hydroxycobalamin was added to cell lysate at a final concentration of 25 μM. Wild-type PpaA copurifies with cobalamin, while truncated (L248Stop) or single-residue mutant proteins all lost the ability to bind hydroxycobalamin. Interestingly, a small peak around 412 nm is still visible in these mutant protein spectra, indicating substoichiometric heme binding. All spectra of His6-MBP tagged proteins were recorded after Ni affinity purification, followed by size exclusion chromatography. Spectra were normalized to A280.
We also tested the affinity of R. sphaeroides PpaA for different forms of cobalamin by the addition of cyano-, hydroxyl-, adenosyl-, or methylcobalamin to cell lysates, followed by purification of PpaA. The PpaA homolog from R. capsulatus, AerR, has a high degree of binding specificity for OH-Cbl over adenyl-, cyano-, and methylcobalamin (Ado-Cbl, CN-Cbl, and Met-Cbl, respectively) (Table 1) (29). The latter cobalamin derivatives have bulkier upper axial ligands to the centrally coordinated cobalt. As is the case with R. capsulatus AerR (29), none of these other cobalamin derivatives containing tighter upper ligands was able to significantly bind R. sphaeroides apo-PpaA, indicating that, like AerR, full-length PpaA is also selective for OH-Cbl (Table 1). Finally, light excitation of Ado-Cbl results in well-characterized photohydrolysis of the upper axial ligand to generate OH-Cbl as a product. Thus, addition of Ado-Cbl to apo-PpaA in cell lysates in the presence of high-intensity light led to PpaA with bound OH-Cbl, while addition of Ado-Cbl without light excitation did not (data not shown). Similar photohydrolysis-mediated cobalamin binding was also reported to occur with AerR (29).
TABLE 1.
Abilities of PpaA homologs to bind cobalamin
| Species | Protein | Binding of: |
|||
|---|---|---|---|---|---|
| OH-Cbl | CN-Cbl | Met-Cbl | Ado-Cbl | ||
| Rhodobacter sphaeroides HR | PpaA | + | − | − | − |
| Rubrivivax gelatinosus IL-144 | AerR | + | − | − | − |
| Methylobacterium extorquens PA1 | PpaA | + | − | − | − |
| Rhodopseudomonas palustris | RPA1540 | − | − | − | − |
| Jannaschia sp. strain CCS1 | PpaA | + | − | + | + |
| Rhodospirillum centenum | AerR | + | + | + | + |
| Erythrobacter sp. strain NAP1 | PpaA | + | + | + | + |
A previous study by Moskvin et al. (31) suggested that PpaA from R. sphaeroides is a heme-binding protein that is unable to bind cobalamin. However, in their experiments, they used a truncated variant of PpaA that contained just a region with homology to the AppA SCHIC (sensor containing heme instead of cobalamin) domain fused to an MBP domain. To test whether this truncation influences cofactor binding, we introduced a stop codon at Leu248, mimicking the C-terminal truncation that was used in their study. Interestingly, the L248Stop mutant was no longer able to bind cobalamin, which indicates that the carboxyl tail of PpaA (beyond codon 248) is important for OH-Cbl binding (Fig. 1). This result shows that an absence of cobalamin binding as reported before is mostly likely an artifact of the truncation introduced.
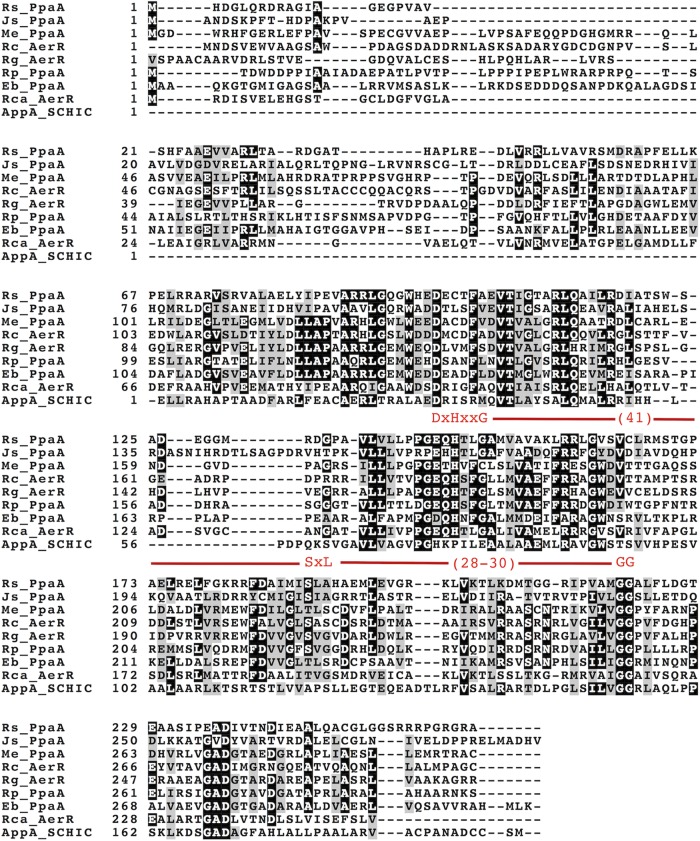
We tested the importance of the strongly conserved signature cobalamin-binding motif E(D)xHxxG-(41)-S(T)xL-(26-28)-GG (Fig. 2) (43) by introducing a histidine-to-alanine mutation into it and then assaying for cobalamin binding. Our reasoning for choosing this residue is that the His in this motif, specifically, His145 in AerR (His146 in PpaA), is known to form a lower axial ligand to OH-Cbl (29). When we mutated His146 to Ala, PpaA was no longer able to bind OH-Cbl (Fig. 1), which is similar to the result reported for the AerR H145A mutant (29). We also introduced mutations into the strongly conserved glycine located three residues downstream of the conserved histidine that is also part of the conserved E(D)xHxxG sequence. In the heme-binding SCHIC domain of AppA, this glycine is replaced by a glutamate (27, 28), which presumably introduces a negative charge repelling the phosphate group of the tail of cobalamin. This substitution also provides steric hindrance preventing the 5,6-dimethylbenzimidazole (DMBI) tail from inserting itself into the Rossman fold of the protein, which presumably allows AppA to bind heme over cobalamin (Fig. 3). Consequently, we mutated this glycine to a glutamate in PpaA (G149E) and observed that it also abrogated OH-Cbl binding (Fig. 1).
FIG 2.
Alignment of PpaA/AerR peptide sequences that were characterized for cobalamin binding in this study with that of AppA. The conserved cobalamin-binding motif is highlighted in red. Rs, R. sphaeroides; Js, Jannaschia sp. strain CCS1; Me, M. extorquens; Rc, R. centenum; Rg, R. gelatinosus; Rp, R. palustris; Eb, Erythrobacter sp. strain NAP1; Rca, R. capsulatus.
FIG 3.
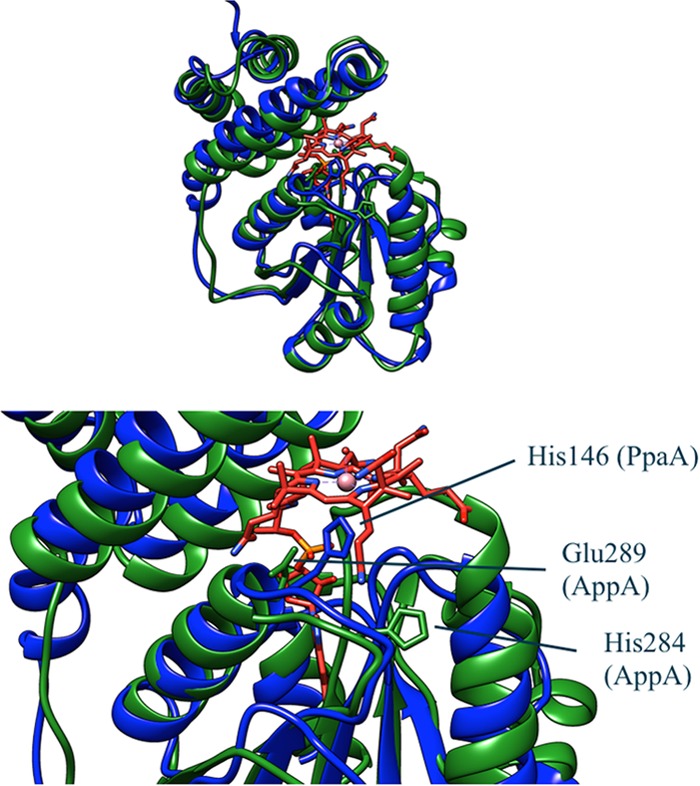
Comparison of PpaA with the structure of the AppA SCHIC domain (Protein Data Bank accession no. 4HEH). PpaA is blue, AppA is green, and cobalamin is red. The structure of PpaA was predicted by the Phyre homology modeler. The strongly conserved histidine is shown in stick representation in both structures. The glutamate that replaces a strongly conserved glycine in AppA is also shown in stick representation. This glutamate is in close proximity to the phosphate group of the DMBI tail of cobalamin and may explain why AppA binds heme instead of cobalamin.
PpaA purified in the absence of added OH-Cbl shows a small UV-vis absorption peak at 412 nm, suggesting substoichiometric heme binding. The same peak is also present in the spectra of the H146A and G149E mutant forms of PpaA (Fig. 1). We tried to increase the amount of bound heme by incubating purified wild-type or mutant apo-PpaA with free hemin at a 1:1 molar ratio. All forms of PpaA showed slight substoichiometric heme binding, as indicated by a red shift of the Soret peak from 385 nm for free hemin to 412 nm for bound hemin (Fig. 4). This red shift was less obvious in the H146A mutant (Fig. 3), indicating that a fraction of heme may be coordinating with His146. This histidine, however, is not essential for heme binding, as was hypothesized by Moskvin et al. (31), as some heme binding still occurs in the H146A mutant form. The G149E mutant form, which mimics the sequence found in the heme-binding AppA SCHIC domain, exhibits more pronounced heme binding, as evidence by an increase in the 412-nm heme peak over that observed with wild-type PpaA (Fig. 4).
FIG 4.
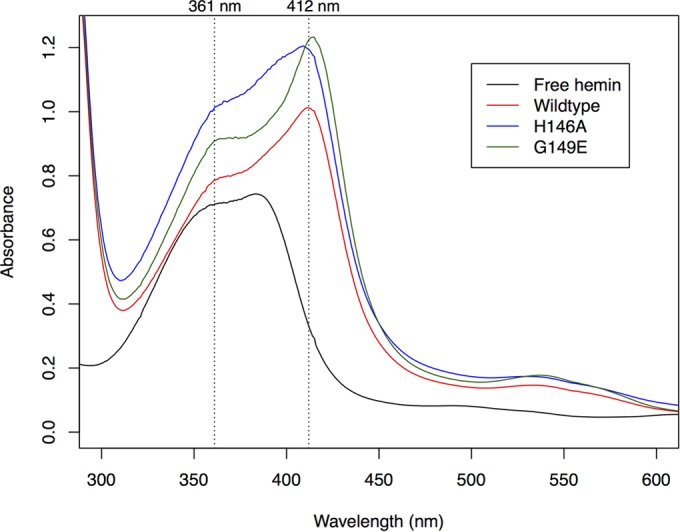
Reconstitution of nickel affinity-purified His6-MBP-PpaA with hemin (vertical dotted lines at 361 and 412 nm). Purified protein was mixed with hemin in a 1:1 molar ratio and incubated overnight at 4°C. The spectrum of heme is red shifted, suggesting that PpaA does have some heme-binding capacity. Heme binding by the H146A mutant is less apparent, while the G149E mutant shows a more pronounced spectral change.
In vivo analysis of PpaA antirepressor activity.
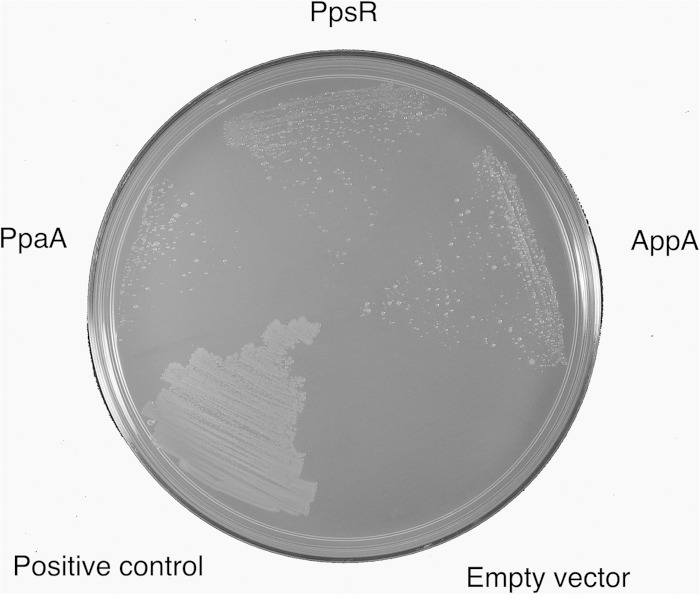
In most purple bacterial species, the ppaA gene is located just upstream of the ppsR gene that codes for a well-characterized repressor of photosystem gene expression (29). In R. capsulatus, the PpaA homolog AerR was shown to physically interact with the PpsR homolog CrtJ in vitro by coelution during gel filtration (29). In this study, we extended this analysis by addressing whether PpaA can interact with PpsR in vivo by bacterial-two-hybrid screening with PpsR as bait. The results of this analysis indicate that PpaA indeed interacts with PpsR in vivo, as evidence by growth on selective minimal medium (Fig. 5). PpaA as a target shows a level of interaction with PpsR that is similar in regard to growth on minimal medium to positive controls such as PpsR interacting with itself (PpsR is known to form a stable tetramer [17]) or with AppA, which is also known to interact with PpsR (Fig. 5). Thus, R. sphaeroides PpsR appears to have two structurally related regulators, PpaA and AppA, the former of which binds cobalamin and the latter of which binds heme.
FIG 5.
Bacterial two-hybrid screening with PpsR as bait. The different targets are indicated. The screening shows interaction between PpsR and PpaA. This interaction may be weaker than that between PpsR and AppA and the interaction of PpsR with itself, as this strain shows less vigorous growth. The positive control is a strain transformed with pBT-LGF2 and pTRG-Gal11 as supplied in the BacterioMatch II kit.
Previous studies have suggested that PpaA plays only a minor role in regulating the expression of photosynthetic genes in R. sphaeroides, as the amount of photopigment synthesis by a PpaA null mutant strain is nearly the same as that observed with wild-type cells (Fig. 6) (44). The PpaA mutant also retains nearly normal photosynthetic growth capabilities, indicating a minor role in affecting PpsR regulation of photosynthesis gene expression. This is contrasted by the major role that AppA plays in regulating the DNA-binding activity of PpsR, as shown by the AppA deletion, which causes a complete loss of pigmentation (Fig. 6 and 7). Prior analysis indicates that the loss of pigmentation in the AppA null mutant is a consequence of constitutive repression of photosynthesis gene expression by PpsR (24). To further address the in vivo activity of PpaA, we overexpressed PpaA in vivo by using a broad-host-range overexpression plasmid (pSRK-ppaA) that utilized the lac promoter to express PpaA. The vector also codes for the LacI repressor, allowing tunable expression based on IPTG induction. When overexpressing PpaA in the AppA deletion mutant, we observed complete restoration of photopigment production (Fig. 7) and photosynthetic growth. This indicates that overexpression of PpaA relieves constitutive repression of bacteriochlorophyll gene expression that occurs by PpsR when AppA is absent. It also indicates that PpaA can indeed function as a PpsR antirepressor but that its activity is overshadowed by AppA in regard to photosystem synthesis.
FIG 6.
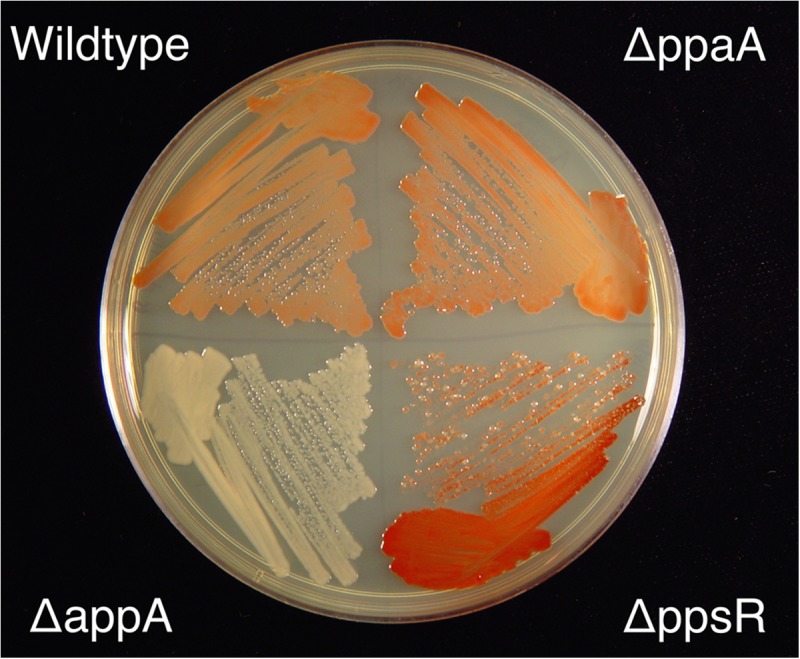
Pigmentation levels exhibited by R. sphaeroides colonies grown under aerobic conditions in the dark on LB agar solidified growth medium. Relevant deletion constructs are indicated.
FIG 7.
Extracted pigments from a ΔappA::pSRK-PpaA mutant grown photosynthetically in the presence (solid line) or absence (dotted line) of IPTG. Overexpression of PpaA leads to restoration of pigment production and allows photosynthetic growth.
Using this PpaA overexpression vector, we also tested the abilities of several mutant PpaA proteins to function as antirepressors of PpsR in vivo. The PpaA H146A mutant protein, which did not exhibit cobalamin binding in vitro, was also not able to restore growth under photosynthetic conditions when overexpressed by IPTG induction (Fig. 7). This indicates that coordination of cobalamin by the conserved histidine is indeed essential for the in vivo antirepressor activity of PpaA. Similarly, the G149E mutant protein, which also is defective in cobalamin binding, also did not restore photosynthetic growth (Fig. 7).
PpaA/AerR homologs from six other species bind cobalamin.
We next asked whether binding of cobalamin is widespread among PpaA/AerR homologs from various species, each of which contains a signature cobalamin-binding motif (Fig. 2). For this analysis, we cloned homologs of PpaA/AerR from six additional photosynthetic species, expressed them in E. coli, and then purified them in the presence of hydroxycobalamin (OH-Cbl), cyanocobalamin (CN-Cbl), methylcobalamin (Met-Cbl), and adenosylcobalamin (Ado-Cbl). All of the homologs tested were able to bind one or more forms of cobalamin, with the exception of RPA1540 from Rhodopseudomonas palustris (Fig. 8; Table 1). The R. palustris PpaA homolog is an outlier, as it is not located upstream of the PprR-encoding gene and is instead located outside the photosynthesis gene cluster upstream of a heme oxygenase. Given that we did not observe binding of cobalamin by this protein, we also tested for R. palustris PpaA for binding of biliverdin, the end product of the reaction catalyzed by heme oxygenase. However, binding of biliverdin was also not detected (data not shown).
FIG 8.
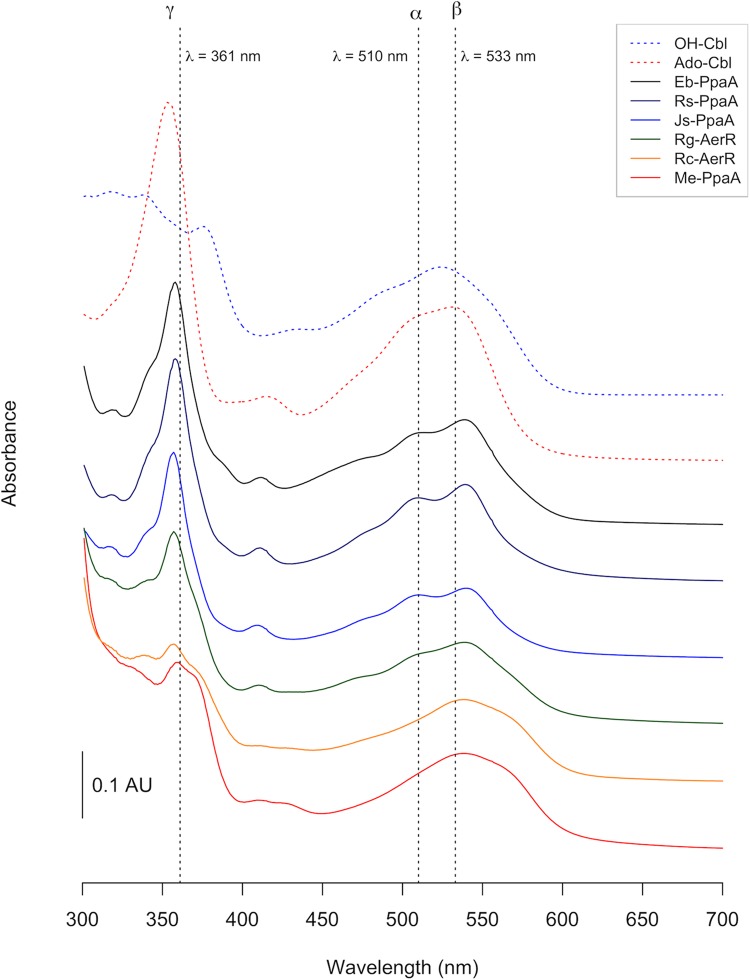
Absorption spectra of purified PpaA homologs. All proteins were purified in the presence of light-excited adenosylcobalamin. Spectra were recorded after removal of the His6-MBP tag. Erythrobacter sp. strain NAP1 PpaA (Eb-PpaA), R. sphaeroides PpaA (Rs-PpaA), and Jannaschia sp. strain CCS1 PpaA (Js-PpaA) all show spectra similar to that of free hydroxycobalamin. The spectrum of R. gelatinosus AerR (Rg-AerR) appears to be more red shifted. The α and β peaks (around 500 to 550 nm) of R. centenum AerR (Rc-AerR) and M. extorquens PpaA (Me-PpaA) are more strongly red shifted, while the γ peak (∼350 nm) is strongly reduced. AU, arbitrary unit.
We observed that PpaA homologs from Methylobacterium extorquens PA1 and Rubrivivax gelatinosus IL-144 bound only OH-Cbl, which is similar to AerR from R. capsulatus and PpaA from R. sphaeroides (Table 1). The PpaA homologs from Rhodospirillum centenum and Erythrobacter sp. strain NAP1 showed binding to all forms of cobalamin, while the PpaA homolog from Jannaschia sp. strain CCS1 bound OH-Cbl, Met-Cbl, and Ado-Cbl but not CN-Cbl. Overall, the spectra of the various Cbl homologs are similar to the spectrum of free OH-Cbl, with the addition of ∼4- and ∼8-nm red spectral shifts of the γ and β peaks, respectfully (Fig. 8). One notable difference is that the α and β peaks of cobalamin bound to M. extorquens PpaA and R. centenum AerR are red shifted by about 25 nm compared to the spectra of the other PpaA homologs (Fig. 8).
DISCUSSION
PpaA from R. sphaeroides was previously reported to be a heme-binding protein (31). However, our analysis shows that R. sphaeroides PpaA selectively and effectively binds OH-Cbl over both heme and other forms of cobalamin that contain a more tightly bound upper ligand. Full-length PpaA does have some heme-binding capacity, but it cannot be readily reconstituted to stoichiometric amounts by the addition of exogenous heme, indicating that PpaA has a lower affinity for heme than for cobalamin. There are several differences between our study and that of Moskvin et al. (31). The first is that we added excess hydroxylcobalamin to the PpaA-overexpressing E. coli cell lysate while Moskvin et al. used cyanocobalamin added to the growth medium (31). Another explanation for the observed differences is that Moskvin et al. used a PpaA truncation that mimics the AppA SCHIC domain. Our results show that an arginine-rich C terminus that was deleted in the study of Moskvin et al. is necessary for incorporation of OH-Cbl.
The well-characterized PpsR antirepressor AppA is known to bind heme at its SCHIC domain (27, 28). This sequence of this domain resembles the B12-binding domain of PpaA and other well-characterized B12-binding proteins. However, the E/DxHxxG region of the cobalamin-binding motif that is present in the PpaA/AerR homologs has been replaced in AppA with the sequence HxPxxE, such that the Glu/Asp (E/D) residue in PpsR/AerR is now a heme-binding His (Fig. 2) (28). The location where His binds Co in PpsR/AerR has also been replaced with a Pro in AppA, and the canonical Gly in PpsR/AerR has been replaced with a bulky charged Glu in AppA. Recent crystal structures of the AppA SCHIC domain show that the presence of a Glu at the terminal position of this motif may be providing electrostatic repulsion to the phosphate group in cobalamin, thereby affecting binding to this tetrapyrrole (23, 28). When we introduced a Gly-to-Glu mutation at the terminal position of this motif in PpaA, it indeed resulted in loss of cobalamin binding and a slight increase in heme binding. Interestingly, the PpaA homolog from M. extorquens has a cysteine instead of a glycine at this position of the B12-binding motif without disrupting OH-Cbl binding. This indicates that Gly at this location is not absolutely necessary for cobalamin binding. Clearly, the heme-binding site present in AppA is quite divergent from that of the more typical cobalamin-binding sites in the PpaA/AerR homologs. This presumably allows selective binding of heme by AppA over that of cobalamin by PpsR/AerR.
A chromosomal deletion of PpaA did not lead to an appreciable difference in pigmentation under photosynthetic conditions, indicating that the role of PpaA in the control of photopigment synthesis is minimal. A similar phenotype for a PpaA disruption was also reported by Gomelsky et al. (44). This is contrasted by a much stronger reduction in photopigment synthesis observed upon deletion of the PpaA homolog AerR in R. capsulatus (29). Indeed, the phenotype produced by the aerR deletion in R. capsulatus is more like that of an appA deletion in R. sphaeroides, which also exhibits a severe reduction in pigment synthesis. AppA and AerR are both known to function as antirepressors of PpsR/CrtJ, respectively (25, 29), so it seems likely that PpaA may also function as an antirepressor on PpsR in R. sphaeroides. In support of this conclusion, we observed that overexpression of PpaA in a ΔappA mutant background does, indeed, lead to restoration of photopigment synthesis and subsequent photosynthetic growth. The latter result indicates either that AppA has taken over the antirepressor role of PpaA and that PpaA is a cryptic antirepressor or that under some growth conditions, AppA is the main PpsR antirepressor, while under other growth conditions, PpaA controls PpsR activity.
Another possibility is that PpaA functions as both a light and a redox sensor. In regard to light sensing, PpaA selectively binds OH-Cbl, which is known to be generated as a photohydrolysis product of light excitation of Ado-Cbl. Thus, hydroxylcobalamin selectivity may be a means to allow PpaA to indirectly sense the presence of light by interacting with a photolysis product of Ado-Cbl. In regard to redox sensing, cobalt is redox active with the ability to easily transition from Co(III) in the oxidized state to Co(II) and then to Co(I) in a series of one-electron transfer events. The stability of the axial ligands to Co is weakened as the Co is reduced from Co(III) to Co(II). Specifically, Co(III) has both upper and lower axial ligands, as well as the four ligands from the pyrrole ring (termed 6 coordinate) while Co(II) is 5 coordinate, as it has a loss of either the upper or lower axial ligand. Further reduction to Co(I) produces 4 coordinate cobalamin that has lost both upper and lower axial ligands with retention of just the four pyrrole ligands. These Co redox events are known to occur under physiologically relevant potentials (−350 mV to >150 mV) (45) and, in the case of methionine synthase, involve both molecular oxygen and flavodoxin as oxidants and reductants, respectively (46). One could therefore envision that PpaA may function as a redox sensor, as changes in the oxidation state of cobalamin could alter the binding of relevant axial amino acid ligands to Co, thus changing the structure and activity of PpaA.
Finally, with the exception of the outlier PpaA from R. palustris, which is not linked to PpsR, all of the PpaA/AerR homologs from other species bound OH-Cbl, with several (Jannaschia sp. strain CCS1, R. centenum, and Erythrobacter sp. strain NAP1) also capable of binding other cobalamin derivatives with tighter upper axial ligands. The set of homologs tested here is limited, but it is curious to see that two of the three more promiscuous homologs are from aerobic anoxygenic phototrophs, a group of purple nonsulfur bacteria that are obligately aerobic and use photosynthesis under aerobic conditions (47). The third promiscuous species is AerR from R. centenum, with R. centenum also capable of synthesizing significant amounts of photopigments under aerobic conditions (48, 49).
Collectively, our results suggest that the clade of proteins previously designated SCHIC proteins actually preferentially bind cobalamin over heme. The one outlier appears to be AppA, which has undergone distinct mutational changes in the cobalamin-binding domain (Fig. 2) that allow AppA to uniquely bind heme over cobalamin. The challenges going forward will be to gain a better understanding of the complementing roles of PpaA and AppA in controlling the DNA-binding activity of PpsR.
Supplementary Material
ACKNOWLEDGMENTS
We thank Zhuo Cheng for helpful discussions regarding the purification of AerR.
This study was supported by National Institutes of Health grant GM040941 awarded to C.E.B.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00374-15.
REFERENCES
- 1.Cohen-Bazire G, Sistrom WR, Stanier RY. 1957. Kinetic studies of pigment synthesis by non-sulfur purple bacteria. J Cell Comp Physiol 49:25–68. doi: 10.1002/jcp.1030490104. [DOI] [PubMed] [Google Scholar]
- 2.Bauer CE, Elsen S, Swem LR, Swem DL, Masuda S. 2003. Redox and light regulation of gene expression in photosynthetic prokaryotes. Philos Trans Biol Sci 358:147–154. doi: 10.1098/rstb.2002.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu J, Bauer CE. 2010. RegB kinase activity is controlled in part by monitoring the ratio of oxidized to reduced ubiquinones in the ubiquinone pool. mBio 1:e00272-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer CE, Setterdahl A, Wu J, Robinson BR. 2008. Regulation of gene expression in response to oxygen tension, p 707–725. In Hunter CN, Daldal F, Thurnauer MC, Beatty JT (ed), Purple photosynthetic bacteria. Kluwer Academic Press, Dordrecht, The Netherlands. [Google Scholar]
- 5.Penfold RJ, Pemberton JM. 1991. A gene from the photosynthetic gene cluster of Rhodobacter sphaeroides induces trans-suppression of bacteriochlorophyll and carotenoid levels in R. sphaeroides and R. capsulatus. Curr Microbiol 23:259–263. doi: 10.1007/BF02092027. [DOI] [Google Scholar]
- 6.Penfold RJ, Pemberton JM. 1994. Sequencing, chromosomal inactivation, and functional expression in Escherichia coli of ppsR, a gene which represses carotenoid and bacteriochlorophyll synthesis in Rhodobacter sphaeroides. J Bacteriol 176:2869–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ponnampalam SN, Buggy JJ, Bauer CE. 1995. Characterization of an aerobic repressor that coordinately regulates bacteriochlorophyll, carotenoid, and light harvesting-II expression in Rhodobacter capsulatus. J Bacteriol 177:2990–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elsen S, Jaubert M, Pignol D, Giraud E. 2005. PpsR: a multifaceted regulator of photosynthesis gene expression in purple bacteria. Mol Microbiol 57:17–26. doi: 10.1111/j.1365-2958.2005.04655.x. [DOI] [PubMed] [Google Scholar]
- 9.Yin L, Bauer CE. 2013. Controlling the delicate balance of tetrapyrrole biosynthesis. Philos Trans R Soc Lond B Biol Sci 368:20120262. doi: 10.1098/rstb.2012.0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braatsch S, Bernstein JR, Lessner F, Morgan J, Liao JC, Harwood CS, Beatty JT. 2006. Rhodopseudomonas palustris CGA009 has two functional ppsR genes, each of which encodes a repressor of photosynthesis gene expression. Biochemistry 45:14441–14451. doi: 10.1021/bi061074b. [DOI] [PubMed] [Google Scholar]
- 11.Jaubert M, Zappa S, Fardoux J, Adriano JM, Hannibal L, Elsen S, Lavergne J, Verméglio A, Giraud E, Pignol D. 2004. Light and redox control of photosynthesis gene expression in Bradyrhizobium: dual roles of two PpsR. J Biol Chem 279:44407–44416. doi: 10.1074/jbc.M408039200. [DOI] [PubMed] [Google Scholar]
- 12.Moskvin OV, Gomelsky L, Gomelsky M. 2005. Transcriptome analysis of the Rhodobacter sphaeroides ppsr regulon: PpsR as a master regulator of photosystem development. J Bacteriol 187:2148–2156. doi: 10.1128/JB.187.6.2148-2156.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smart JL, Willett JW, Bauer CE. 2004. Regulation of hem gene expression in Rhodobacter capsulatus by redox and photosystem regulators RegA, CrtJ, FnrL, and AerR. J Mol Biol 342:1171–1186. doi: 10.1016/j.jmb.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 14.Swem D, Bauer CE. 2002. Coordination of ubiquinol oxidase and cytochrome cbb3 oxidase expression by multiple regulators in Rhodobacter capsulatus. J Bacteriol 184:2815–2820. doi: 10.1128/JB.184.10.2815-2820.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elsen S, Ponnampalam SN, Bauer CE. 1998. CrJ bound to distant binding sites interacts cooperatively to aerobically repress photopigment biosynthesis and light harvesting II gene expression in Rhodobacter capsulatus. J Biol Chem 273:30762–30769. doi: 10.1074/jbc.273.46.30762. [DOI] [PubMed] [Google Scholar]
- 16.Ponnampalam SN, Bauer CE. 1997. DNA binding characteristics of CrtJ. J Biol Chem 272:18391–18396. doi: 10.1074/jbc.272.29.18391. [DOI] [PubMed] [Google Scholar]
- 17.Masuda S, Bauer CE. 2002. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell 110:613–623. doi: 10.1016/S0092-8674(02)00876-0. [DOI] [PubMed] [Google Scholar]
- 18.Shimizu T, Cheng Z, Matsuura K, Masuda S, Bauer CE. 2015. Evidence that altered cis element spacing affects PpsR mediated redox control of photosynthesis gene expression in Rubrivivax gelatinosus. PLoS One 10:e0128446. doi: 10.1371/journal.pone.0128446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng Z, Wu J, Setterdahl A, Reddie K, Carroll K, Hammad LA, Karty JA, Bauer CE. 2012. Activity of the tetrapyrrole regulator CrtJ is controlled by oxidation of a redox active cysteine located in the DNA binding domain. Mol Microbiol 85:734–746. doi: 10.1111/j.1365-2958.2012.08135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho SH, Youn SH, Lee SR, Yim HS, Kang SO. 2004. Redox property and regulation of PpsR, a transcriptional repressor of photosystem gene expression in Rhodobacter sphaeroides. Microbiology 150:697–706. doi: 10.1099/mic.0.26777-0. [DOI] [PubMed] [Google Scholar]
- 21.Masuda S, Dong C, Swem D, Setterdahl AT, Knaff DB, Bauer CE. 2002. Repression of photosynthesis gene expression by formation of an intramolecular disulfide bond in CrtJ. Proc Natl Acad Sci U S A 99:7078–7083. doi: 10.1073/pnas.102013099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin L, Dragnea V, Bauer CE. 2012. PpsR, a regulator of heme and bacteriochlorophyll biosynthesis, is a heme-sensing protein. J Biol Chem 287:13850–13858. doi: 10.1074/jbc.M112.346494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winkler A, Heintz U, Lindner R, Reinstein J, Shoeman RL, Schlichting I. 2013. A ternary AppA-PpsR-DNA complex mediates light regulation of photosynthesis-related gene expression. Nat Struct Mol Biol 20:859–867. doi: 10.1038/nsmb.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gomelsky M, Kaplan SS. 1995. AppA, a novel gene encoding a trans-acting factor involved in the regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J Bacteriol 177:4609–4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomelsky M, Kaplan S. 1997. Molecular genetic analysis suggesting interactions between AppA and PpsR in regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J Bacteriol 179:128–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han Y, Braatsch S, Osterloh L, Klug G. 2004. A eukaryotic BLUF domain mediates light-dependent gene expression in the purple bacterium Rhodobacter sphaeroides 2.4.1. Proc Natl Acad Sci U S A 101:12306–12311. doi: 10.1073/pnas.0403547101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han Y, Meyer MH, Keusgen M, Klug G. 2007. A haem cofactor is required for redox and light signalling by the AppA protein of Rhodobacter sphaeroides. Mol Microbiol, 64:1090–1104. doi: 10.1111/j.1365-2958.2007.05724.x. [DOI] [PubMed] [Google Scholar]
- 28.Yin L, Dragnea V, Feldman G, Hammad LA, Karty JA, Dann CE III, Bauer CE. 2013. Redox and light control the heme-sensing activity of AppA. mBio 4:e00563-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng Z, Li K, Hammad LA, Karty JA, Bauer CE. 2014. Vitamin B12 regulates photosystem gene expression via the CrtJ antirepressor AerR in Rhodobacter capsulatus. Mol Microbiol 91:649–664. doi: 10.1111/mmi.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moskvin OV, Kaplan S, Gilles-Gonzalez MA, Gomelsky M. 2007. Novel heme-based oxygen sensor with a revealing evolutionary history. J Biol Chem 282:28740–28748. doi: 10.1074/jbc.M703261200. [DOI] [PubMed] [Google Scholar]
- 31.Moskvin OV, Gilles-Gonzalez M-A, Gomelsky M. 2010. The PpaA/AerR regulators of photosynthesis gene expression from anoxygenic phototrophic proteobacteria contain heme-binding SCHIC domains. J Bacteriol 192:5253–5256. doi: 10.1128/JB.00736-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nahvi A, Barrick JE, Breaker RR. 2004. Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res 32:143–150. doi: 10.1093/nar/gkh167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. 2002. Genetic control by a metabolite binding mRNA. Chem Biol 9:1043–1049. doi: 10.1016/S1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 34.Ortiz-Guerrero JM, Polanco MC, Murillo FJ, Padmanabhan S, Elías-Arnanz M. 2011. Light-dependent gene regulation by a coenzyme B12-based photoreceptor. Proc Natl Acad Sci U S A 108:7565–7570. doi: 10.1073/pnas.1018972108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sistrom WR. 1960. A requirement for sodium in the growth of Rhodopseudomonas spheroides. J Gen Microbiol 22:778–785. doi: 10.1099/00221287-22-3-778. [DOI] [PubMed] [Google Scholar]
- 36.Favinger J, Stadtwald R, Gest H. 1989. Rhodospirillum centenum, sp. nov., a thermotolerant cyst-forming anoxygenic photosynthetic bacterium. Antonie Van Leeuwenhoek 55:291–296. doi: 10.1007/BF00393857. [DOI] [PubMed] [Google Scholar]
- 37.Ragatz L, Jiang Z-Y, Bauer CE, Gest H. 1995. Macroscopic phototactic behavior of the purple photosynthetic bacterium Rhodospirillum centenum. Arch Microbiol 163:1–6. doi: 10.1007/BF00262196. [DOI] [PubMed] [Google Scholar]
- 38.Masuda S, Bauer CE. 2004. Null mutation of HvrA compensates for loss of an essential RelA/SpoT-like gene in Rhodobacter capsulatus. J Bacteriol 186:235–239. doi: 10.1128/JB.186.1.235-239.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Penfold RJ, Pemberton JM. 1992. An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene 118:145–146. doi: 10.1016/0378-1119(92)90263-O. [DOI] [PubMed] [Google Scholar]
- 40.Khan SR, Gaines J, Roop RM, Farrand SK. 2008. Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of traR and traM expression on Ti plasmid quorum sensing. Appl Environ Microbiol 74:5053–5062. doi: 10.1128/AEM.01098-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blommel PG, Fox BG. 2007. A combined approach to improving large-scale production of tobacco etch virus protease. Protein Expr Purif 55:53–68. doi: 10.1016/j.pep.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grote A, Hiller K, Scheer M, Münch R, Nörtemann B, Hempel DC, Jahn D. 2005. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res 33(Web Server Issue):W526–W531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ludwig ML, Matthews RG. 1997. Structure-based perspectives on B12-dependent enzymes. Annu Rev Biochem 66:269–313. doi: 10.1146/annurev.biochem.66.1.269. [DOI] [PubMed] [Google Scholar]
- 44.Gomelsky L, Sram J, Moskvin OV, Horne IM, Dodd HN, Pemberton JM, McEwan AG, Kaplan S, Gomelsky M. 2003. Identification and in vivo characterization of PpaA, a regulator of photosystem formation in Rhodobacter sphaeroides. Microbiology 149:377–388. doi: 10.1099/mic.0.25972-0. [DOI] [PubMed] [Google Scholar]
- 45.Schumacher W, Holliger C, Zehnder AJ, Hagen WR. 1997. Redox chemistry of cobalamin and iron-sulfur cofactors in the tetrachloroethene reductase of Dehalobacter restrictus. FEBS Lett 409:421–425. doi: 10.1016/S0014-5793(97)00520-6. [DOI] [PubMed] [Google Scholar]
- 46.Koutmos M, Datta S, Pattridge KA, Smith JL, Matthews RG. 2009. Insights into the reactivation of cobalamin-dependent methionine synthase. Proc Natl Acad Sci U S A 106:18527–18532. doi: 10.1073/pnas.0906132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yurkov VV, Beatty JT. 1998. Aerobic anoxygenic phototrophic bacteria. Microbiol Mol Biol Rev 62:695–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yildiz FH, Gest H, Bauer CE. 1991. Attenuated effect of oxygen on photopigment synthesis in Rhodospirillum centenum. J Bacteriol 173:5502–5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masuda S, Berleman J, Hasselbring BM, Bauer CE. 2008. Regulation of aerobic photosystem synthesis in the purple bacterium Rhodospirillum centenum by CrtJ and AerR. Photochem Photobiol Sci 7:1267–1272. doi: 10.1039/b802365b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.