Abstract
The Simbu serogroup currently consists of a highly diverse group of related arboviruses that infect both humans and economically important livestock species. Cat Que virus (CQV), a Simbu serogroup virus of the genus Orthobunyavirus (family Bunyaviridae), was first isolated in 2004 from mosquitoes during surveillance of arbovirus activity in acute pediatric encephalitis in northern Vietnam. We report here the complete genome sequence of SC0806 isolated from mosquitoes (Culex tritaeniorhynchus) in Sichuan Province, China. Consistent with the genomic organization of Simbu serogroup viruses, the SC0806 genome comprises three RNA segments—a large (L) segment (6928 nucleotides) that encodes the 2261-amino-acid RNA-dependent RNA polymerase, a medium (M) segment (4481 nucleotides) that encodes the 1433-amino-acid polyprotein, and a small (S) segment (984 nucleotides) that encodes a 234-amino-acid nucleocapsid protein and a 95-amino-acid nonstructural protein. The respective lengths of the 5′-untranslated region (UTR) and 3′-UTR of L, M, and S are 56 and 86, 43 and 136, and 44 and 238 nucleotides. Sequence (nucleotide and deduced amino acid) comparison and phylogenetic analysis revealed that SC0806 was closely related to the reported Vietnam isolate CQV. This is the first time that CQV has been isolated in Sichuan Province, China. Anti-SC0806 immunoglobulin M (IgM) and IgG antibodies were found in pigs reared locally, indicating that CQV has formed a natural cycle in the local area. Surveillance of the distribution and pathogenicity of SC0806 should be strengthened.
Key Words: : Cat Que virus, Whole genome, Phylogenetics, Seroprevalence, China
Introduction
Viruses with tripartite, negative-sense, single-stranded RNA (ssRNA) genomes are classified in the family Bunyaviridae (Elliott 1990, Bouloy 1991, Elliott et al. 1991). The L segment encodes a large polypeptide, RNA-dependent RNA polymerase (RdRp), which has replicase and transcriptase activities. The M segment encodes a polyprotein that undergoes posttranslational proteolytic cleavage to give rise to virion surface glycoproteins G1 and G2, and a nonstructural protein called NSm. The S segment encodes two proteins—the nucleocapsid (N) protein and a nonstructural protein (NSs). These proteins are encoded in overlapping reading frames of the same mRNA (Elliott 1990, Bouloy 1991, Elliott et al. 1991).
The genus Orthobunyavirus is the largest in the family Bunyaviridae, with over 170 named viruses classified into 18 serogroups and 48 species complexes (de Brito Magalhaes et al. 2011, Hoffmann et al. 2012, Briese et al. 2013, Rodrigues et al. 2014). In recent years, increasing numbers of bunyaviruses have been identified as important human pathogens. Among the 170 bunyaviruses, 60 can cause human diseases, including influenza-like symptoms (Tahyna virus), fever and joint pain (Oropouche virus), encephalitis (La Crosse virus), and hemorrhagic fever (Garissa virus). Some of them are fatal in humans (Anderson et al. 1961, 1960, Lambert and Lanciotti 2008, Aguilar et al. 2011, Vasconcelos et al. 2011).
The Simbu serogroup is the largest in the genus Orthobunyavirus, with 24 viruses isolated worldwide (Calisher et al. 1969, Qin et al. 2010, Kim et al. 2011, Oem et al. 2012, Briese et al. 2013, Ladner et al. 2014, Rodrigues et al. 2014), and have been grouped into the following seven species complexes: Akabane, Manzanilla, Oropouche, Sathuperi, Simbu, Shamonda, and Shuni (Saeed et al. 2001, Ladner et al. 2014). Cat Que virus (CQV) is a member of the Manzanilla species complex of Simbu serogroup, which was isolated in 2004 from mosquitoes during surveillance of arbovirus activity in acute pediatric encephalitis in northern Vietnam (Bryant et al. 2005). Pigs are the primary mammalian host of CQV. Here, we report a CQV strain (SC0806) isolated from mosquito samples collected in China. The SC0806 genome was sequenced, and the evolutionary relationships between SC0806 and other members of the family Bunyaviridae were analyzed. We also assessed the seroprevalence of SC0806 antibodies in the pig population.
Materials and Methods
Ethics Statement
The program for collection of mosquitoes and pig serum samples was approved by the Ethical Committee of the National Institute of Viral Disease Control and Prevention, China CDC. The mosquito specimens were collected from pigpens in Bazhong (31°15′–32°45′N, 106°21′–107°45′E) and Longchang (29°11′–29°32′N, 105°02′–105°26′E) counties. Pig serum was collected in Fuling (29°21′–30°01′N, 106°56′–107°43′E), Yubei (29°21′–30°01′N, 106°56′–107°43′E), and Wanzhou (30°24′–31°14′N, 107°55′–108°53′E) counties. The field study is part of the national surveillance of arboviruses and their infection. No specific permits were required for collection of mosquitoes and pig serum samples from the described locations. Field studies did not involve endangered or protected species.
Sample collection
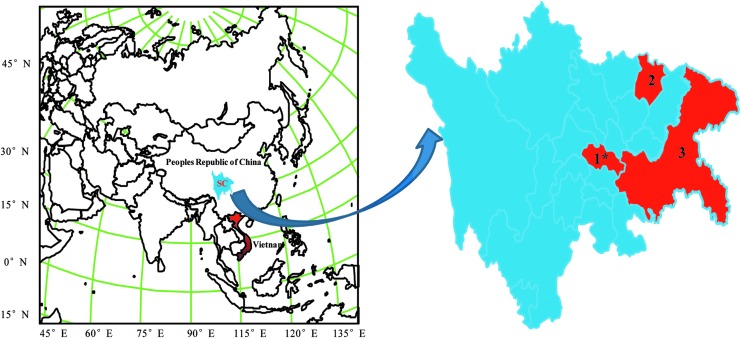
The mosquito specimens were collected from pigpens in Bazhong and Longchang counties, in eastern Sichuan, China, during July in 2006 and 2008 (Fig. 1). A mosquito-lure lamp (12 V, 300 mA; Wuhan Lucky Star Environmental Protection Technology, Hubei, China) was used to capture mosquitoes at night. The next morning, the cages with captured mosquitoes were placed at −20°C for 20 min. Following their identification, 50–100 mosquitoes of the same species were pooled, placed in a cryogenic vial, and cryopreserved rapidly in liquid nitrogen.
FIG. 1.
Collection locations of mosquito specimens and pig serum samples. Note: 1, Mosquito specimens collection site 1 (Longchang county); 2, mosquito specimens collection site 2 (Bazhong county); 3, pig serum samples collection site (eastern Sichuan Basin). (*) Where SC0806 was isolated. Color images available online at www.liebertpub.com/vbz
Pig serum was collected in Fuling, Yubei, and Wanzhou counties, in the eastern Sichuan Basin from May 21, 2008, to June 8, 2008 (Fig. 1). The samples were transported to the laboratory chilled and stored at −80°C until processed.
Virus isolation
All specimens were homogenized and centrifuged at 12,000×g for 30 min at 4°C. To isolate the virus, 150 mL of supernatant was added to monolayers of both C6/36 and BHK-21 cells and cultured at 28°C and 37°C, respectively, in a 5% CO2 incubator. The cells were monitored at 24-h intervals to identify cytopathic effects associated with infection (Li et al. 2011a, Li et al. 2011b).
Preliminary identification of the isolates using random PCR
The supernatant of SC0806-infected BHK-21 cells was filtered through a 0.22-μm filter (Millipore). Then, 200 U of DNase I (Sigma) was added to 200 μL of filtrate and incubated at 37°C for 1 h. A QIAamp Viral RNA Mini Kit (QIAGEN) was used to extract viral RNA according to the instructions. The specific primer was 20 μmol/L K2Sr (5′-GACCATCTAGCGACCTCCACNNNNNN-3′). SuperScript III™ reverse transcription reagent (Invitrogen) was used to synthesize the first cDNA strand according to the manufacturer's instructions. The enzyme was inactivated at 70°C for 10 min. Next, 2.5 U of Klenow enzyme (New England Biolabs) was incubated at 37°C for 1 h after adding 20 μL of the first-strand cDNA template pre-denatured at 94°C for 2 min; finally, the enzyme was inactivated at 75°C for 10 min. Each 3-μL aliquot of cDNA template synthesized by reverse transcription was amplified via random PCR with the specific primer K2S (5′-GACCATCTAGCGACCTCCAC-3′). The 50-μL reaction included 38.5-μL H2O for injection, 5 μL of 10×Ex-Taq buffer, 1.5 μL of potassium sulfide (K2S) (20 μmol/L), 1.5 μL of 10 mmol/L deoxynucleotides (dNTPs), 0.5 μL of Ex Taq (2.5 U), and 3 μL of template. The reaction consisted of a 94°C denaturation for 5 min, 40 cycles of 94°C for 1 min and 65°C for 3 min, and a final 5-min extension at 68°C. The products of random PCR amplification were subjected to 1% agarose gel electrophoresis, and fragments >500 bp were recovered.
The QIAamp Gel Purification Kit (QIAGEN) was used to purify the products and to connect with a pGEM-T vector (pGEM2T Easy Kit, Promega), transforming competent Escherichia coli DHα5. After blue–white screening, the bacteria were amplified using 2 μL of a bacterial suspension as template. Primer KS was used for PCR amplification (annealing at 58°C, 25 cycles) to detect the presence or absence of the inserted element in the carrier. Finally, the PCR products were subjected to 1% agarose gel electrophoresis, and the presence of amplified bands indicated an inserted sequence. The corresponding clones were sequenced. The sequencing results were subject to a BLAST online comparison with the National Center for Biotechnology Information (NCBI) database to determine the source of the inserted sequence.
Complete genome sequencing, including the 5′- and 3′-untranslated regions
Viral RNA was extracted from 140-μL aliquots of virus-infected BHK-21 cell culture supernatant using a QIAamp Viral RNA Mini Kit (QIAGEN) according to the manufacturer's instructions. cDNA was produced with a Ready-To-Go Kit (GE Healthcare) using random hexanucleotide primers. Samples were amplified as described previously (Wang et al. 2002, 2003). The amplification products were pooled, ligated to an adaptor, and sequenced at the Washington University Genome Sequencing Center on a 454 GS FLX platform (454 Life Sciences, Branford, CT). The sequences were trimmed to remove the primer sequences before data analysis and assembly. Because the nucleic acids used for sequencing contained a mixture of host cell DNA and viral RNA, sequencing reads were filtered using the custom informatics pipeline VirusHunter to identify viral sequences (Zhao et al. 2013). Briefly, the default parameters in VirusHunter were set to cluster sequences that share ≥95% identity over 95% of the sequence length. The longest sequence from each cluster was retained as the representative sequence and used for downstream analysis. For filtering host sequences, the golden hamster Mesocricetus auratus genome was used as the reference (GenBank assembly ID, GCA_000349665.1) because the isolate was cultured in the BHK (hamster) cell line.
Sequences retained from the previous step were queried against the NCBI nucleotide database using BLASTn. Sequences with significant hits (expect [E] value cutoff 1E-10) are broadly classified as human, mouse, fungal, bacterial, phage, viral, or other based on the taxonomy identity of the best BLAST hit. Sequences identified as most similar to Orthobunyavirus were assembled with Newbler (454 Life Sciences) using the default parameters. Then, the 5′-rapid amplification of cDNA ends (5′-RACE) and 3′-RACE systems (v. 2.0, Invitrogen) were used to amplify the 5′- and 3′-untranslated regions (UTRs) from each of the three segments. 5′-RACE was performed according to the manufacturer's instructions. For 3′-RACE, a poly(A) tail was first added to the RNA using poly(A) polymerase. Then, the 3′-UTR sequences were generated by RT-PCR using sequence-specific and oligo(dT)-adapter primers.
Sequence analysis and phylogenetic comparisons
The nucleic acid sequences and deduced amino acid products were analyzed and assembled using DNAStar (Lasergene). BLAST searches of all available databases were carried out using the National Center for Biotechnology Information (NCBI) server (www.ncbi.nlm.nih.gov). An open reading frame (ORF) search was performed with the NCBI ORF Finder. The sequences of Simbu serogroup viruses and representative members of the California encephalitis, Bunyamwera, and Wyeomyia serogroups were downloaded from GenBank and used to build the phylogenetic trees. The sequences were aligned using ClustalX (v. 2.0). The nucleic acid and amino acid sequence identities were calculated using MEGA (v. 5.1) with the default settings (www.megasoftware.net) (Tamura et al. 2011). Neighbor-joining phylogenetic trees of the L, M, and S segments were also generated in MEGA, using the p-distance and Poisson correction algorithms. The robustness of the branching was evaluated by bootstrapping using 1000 replications.
Enzyme-linked immunosorbent assay test of anti-virus antibodies in pig sera
BHK21 cells were inoculated with SC0806 virus. The cell supernatant was collected when >75% of the cells were diseased and centrifuged at 5000 rpm for 10 min. The supernatant was filtered through a 0.22-μm filter, and the filtrate was concentrated using a 100K Millipore concentration tube (Millipore, Billerica, MA). The concentrated virus was inactivated at 56°C for 30 min; the inactivated virus was used to detect immunoglobulin M (IgM) and IgG enzyme-linked immunosorbent assay (ELISA) antigen of the newly isolated virus and stored at −70°C.
Serum samples were tested for SC0806 virus-specific IgM antibodies by ELISA, as described previously (Wang et al. 2011). Carbonate buffer at pH 9.6 was used to dilute each virus antigen 1:400, which was left overnight at 4°C, incubated with 5% skimmed milk at 37°C for 1 h, and washed with phosphate-buffered saline Tween (PBS-T) four times for 1 min each time. Then 100 μL of serum to be tested at 1:100 dilution was incubated at 37°C for 1 h and washed four times with PBS-T. Next, 100 μL of goat anti-pig IgM (1:500) and IgG antibody (1:50,000) (Sigma) labeled with horseradish peroxidase (HRP) were added to each well at 37°C for 1 h and washed four times with PBS-T. Finally, 100 μL of an equal mixture of substrate color liquids A and B were added to each well at room temperature for developing at 10 min. NH2SO4 was used to terminate the reaction, and a microplate reader was used to read the optical density (OD). The result was considered positive when P/N was >2.1.
Results
Isolation and preliminary identification of SC0806
Isolation from mosquito samples was attempted in BHK-21 and C6/36 cells. One virus isolate (named SC0806) was obtained from Culex (Cx.) tritaeniorhynchus collected in Longchang County, Sichuan Province. SC0806 caused syncytial cytopathogenic effects in BHK-21 cells 48 h after infection, including granulating, shrinking, rounding, seining, and falling off. In comparison, no apparent cytopathic effects were observed in C6/36 cells. Random PCR was performed to amplify the SC0806 gene sequences. In total, three positive clones were obtained and 731 nucleotides were sequenced. The sequence obtained had high identities with the S segments of Simbu serogroup viruses, including CQV and Oya virus (OYAV) (Kono et al. 2002, Ladner et al. 2014). Phylogenetic analysis based on a partial sequence of the S segment showed that SC0806 was closely related to CQV and grouped in the Manzanilla species complex branch (date not shown) of the Simbu serogroup in the genus Bunyavirus.
Genome organization and characteristics of SC0806
To obtain the entire SC0806 genome, we performed shotgun 454 Life Sciences pyrosequencing of total nucleic acids extracted from a plaque-purified isolate of SC0806. Following random PCR amplification, samples were pooled (with barcodes), and sequenced using the Roche/454 FLX Titanium platform, producing a total of 34,930 reads. A total of 3454 (200–2500 bp) unique sequences were obtained after VirusHunter classification. After filtering, 1450 of these reads mapped to mammalian genomes. Following BLAST and mapping analyses, 1845 viral reads (300–2500 bp) shared above 80% sequence identity to viruses in the Manzanilla species complex in the Simbu serogroup of genus Orthobunyavirus. De novo assembly of these sequence reads generated the complete sequences of the L, M, and S segments of SC0806. The degree of enrichment times sequencing coverage was >20. There was no region within the segments that had notable lower coverage. To validate the viral genome, we designed primer pairs that generated seven, five, and two overlapping amplicons, respectively, for the L, M, and S gene segments (the primer sequences are available on request). In addition, the 5′ and 3′ ends of the segments were confirmed by 5′- and 3′-RACE. The L, M, and S sequences determined in this study were deposited in GenBank (JX983192–JX983194).
Consistent with the genome composition of Simbu serogroup viruses, the SC0806 genome includes three RNA segments (L, M, and S); both ends of each gene segment contained 5′- and 3′-UTRs of different lengths (Table 1). The complete SC0806 L segment was 6928 nucleotides long. A single 6783-nucleotide ORF encodes the 2261-amino-acid RdRp. The ORF is flanked by a 56-nucleotide 5′-UTR and an 86-nucleotide 3′-UTR. The complete SC0806 M segment was 4481 nucleotides long, and included a single 4481-nucleotide ORF encoding the 1433-amino acid polyprotein. The M segment ORF is flanked by a 43-nucleotide 5′-UTR and 136-nucleotide 3′-UTR. The 984-nucleotide-long SC0806 S segment contains two overlapping ORFs (699 and 285 nucleotides), encoding a 233-amino-acid N protein and 95-amino-acid Ns protein, respectively. The S gene 5′-UTR is 44 nucleotides long and the 3′-UTR is 238 nucleotides long. The predicted protein-coding ORFs of SC0806 were similar in length and had start and stop positions similar to the reported Vietnam CQV isolate (Table 1).
Table 1.
Genomic Organization and Biological Character Comparison of SC0806 and Representatives of Recognized Orthobunyavirus Serogroups
| L segment | M segment | S segment | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Serogroups | Species | Virus | 5′-UTR (nt) | ORF (nt)/RdRp (aa) | 3′-UTR (nt) | 5′-UTR (nt) | ORF (nt)/Polyprotein (aa) | 3′-UTR (nt) | 5′-UTR (nt) | ORF (nt)/NP (aa) | ORF (nt)/NSs (aa) | 3′-UTR (nt) | Natural Host Range | Human disease | Distribution |
| Simbu | Manzanilla | SC0806 | 56 | 6783/2261 | 86 | 43 | 4299/1433 | 136 | 44 | 699/233 | 285/95 | 238 | Mosquitoes | Febrile illness? | China |
| Manzanilla | Cat Que virus | 56 | 6783/2261 | 84 | 43 | 4299/1433 | 136 | 44 | 699/233 | 285/95 | 237 | Mosquitoes | No human pathology has been documented | Vietnam | |
| Manzanilla | TRVL 3586 | 59 | 6783/2261 | 71 | 55 | 4299/1433 | 159 | 44 | 699/233 | 285/95 | 236 | Mosquitoes, alouatta seniculus | Febrile illness | Trinidad | |
| Akabane | OBE-1 | 30 | 6753/2251 | 82 | 22 | 4203/1401 | 81 | 33 | 699/233 | 273/91 | 123 | Mosquitoes, cattle | Febrile illness | Japan, Australia, Kenya, Taiwan, Viet Nam, Bandung, Thailand | |
| Oropouche | TRVL-9760 | 31 | 6756/2252 | 21 | 17 | 4260/1420 | 77 | 44 | 693/231 | 273/91 | 9 | Mosquitoes, human, livestock, bradypus tridactylus, culicoides | Febrile illness with central nervous system signs (including encephalitis) | Trinidad, Brazil, Colombia | |
| Sathuperi | Sathuperi virus | 18 | 6762/2254 | 78 | 6 | 4209/1403 | 112 | 43 | 699/233 | 273/91 | 98 | Mosquitoes, human, cattle, culicoides, | Febrile illness | India, Nigeria | |
| Simbu | SA Ar 53 | 21 | 6759/2253 | 112 | 13 | 4227/1409 | 174 | 24 | 699/233 | 273/91 | 134 | Mosquitoes, human, | Febrile illness | South Africa, Senegal, Cameroun, Central African Republic, Botswana | |
| Shamonda | An5550 | 19 | 6762/2254 | 79 | 17 | 4209/1403 | 85 | 30 | 699/233 | 273/91 | 195 | Cattle | Febrile illness | Nigeria | |
| Shuni | An10107 | 12 | 6759/2253 | 106 | 11 | 4212/1404 | 100 | 34 | 699/233 | 273/91 | 114 | Mosquitoes, human, cattle, sheep, culicoides | Febrile illness | Nigeria, South Africa | |
| Bunyamwera | Bunyamwera | Bunyamwera virus | 50 | 6714/2238 | 108 | 56 | 4299/1433 | 100 | 85 | 699/233 | 303/101 | 174 | Human, chimpanzee, monkeys, domestic animals, rodents, birds | Febrile illness with rash or central nervous system signs (including encephalitis) | South Africa, Uganda, Nigeria, Cameroun, Central African Republic, Kenya, Senegal, Congo, Senegal, Mozambique, Egypt, Tunisia, Tanzania, Angola |
| California encephalitis | California encephalitis | La Crosse virus | 61 | 6789/2263 | 127 | 61 | 4323/1441 | 138 | 79 | 705/235 | 276/92 | 195 | Mosquitoes | Febrile illness with central nervous system signs (including encephalitis) | California, Utah, New Mexico, Texas, USA, Manitoba, Canada |
| Wyeomyia | Wyeomyia | Wyeomyia virus | 36 | 6708/2236 | 107 | 20 | 4251/1417 | 266 | 71 | 699/233 | 78/26* | 294 | Mosquitoes, human | Febrile illness | Colombia, Panama, Trinidad, Brazil, French Guiana |
Note: GenBank accession numbers used: SC0806, JX983192–JX983194; Cat Que virus, JQ675598–JQ675600; TRVL 3586, KF697148–KF697150; OBE-1, NC009894–NC009896; TRVL-9760, KC759122–KC759124; Sathuperi virus, HE795102–HE795104; SA Ar 53, NC018476–NC018478; An5550, HE795105–HE795107; An10107, HE800141–HE800143; Bunyamwera virus, NC001925–NC001927; La Crosse virus, GU206144–GU206146; Wyeomyia virus, JN572080–JN572082.
Truncated open reading frame (ORF) that may not be expressed.
The distribution, natural host range, and human disease of each serogroup are based on the documentation in ArboCAT (wwwn.cdc.gov/arbocat).
L segment, large RNA segment; M segment, medium RNA segment; S segment, small RNA segment; UTR, untranslated region; ORF, open reading frame; nt, nucleotides; aa, amino acids.
The SC0806 sequences were compared with published Orthobunyavirus sequences for members of Simbu, Bunyamwera, California encephalitis, and Wyeomyia serogroups. Comparison of the nucleotide and deduced amino acid sequences revealed that SC0806 was closely related to the Manzanilla species complex of the Simbu serogroup in the genus Bunyavirus (Table 2). The nucleotide (amino acid,) identity of the L, M, and S gene segments between SC0806 and CQV was 92.3% (99.1%), 97.3% (99.1%), and 85% (100% for Np and NSs), respectively. By contrast, the identity between SC0806 and other orthobunyaviruses was very low (less than 28.8%) (Table 2). This implies that SC0806 is a CQV strain of the Manzanilla species complex in the Simbu serogroup.
Table 2.
Nucleotide and Amino Acid Sequence Identity (%) Between SC0806 and Representatives of Recognized Orthobunyavirus Serogroups
| L segment | M segment | S segment | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Serogroups | Species | Virus | nt | aa | nt | aa | nt | aa (Np) | aa (NSs) |
| Simbu | Manzanilla | Cat Que virus | 92.3 | 99.1 | 97.3 | 99.1 | 85 | 100 | 100 |
| Manzanilla | TRVL 3586 | 26.9 | 89.6 | 29.2 | 78.5 | 78.6 | 96.1 | 88.4 | |
| Akabane | OBE-1 | 27.1 | 7.7 | 26.7 | 6.4 | 23.3 | 62.2 | 44.2 | |
| Oropouche | TRVL-9760 | 27.1 | 9.5 | 27.2 | 14.5 | 16.2 | 7.7 | 53.6 | |
| Sathuperi | Sathuperi virus | 27.2 | 7.6 | 26.3 | 5.5 | 22.5 | 61.8 | 42.1 | |
| Simbu | SA Ar 53 | 27.4 | 7.3 | 27 | 5.8 | 23.7 | 64.3 | 48.4 | |
| Shamonda | An5550 | 26.4 | 7.6 | 27.8 | 6.4 | 23.5 | 61.8 | 44.2 | |
| Shuni | An10107 | 26 | 26 | 26.7 | 6.5 | 22 | 64.8 | 46.3 | |
| Bunyamwera | Bunyamwera | Bunyamwera virus | 28.4 | 7.8 | 27.6 | 8.7 | 25.7 | 23.6 | 9.9 |
| California encephalitis | California encephalitis | La Crosse virus | 26.8 | 8.1 | 27.5 | 6.6 | 28.8 | 10.2 | 10.3 |
| Wyeomyia | Wyeomyia | Wyeomyia virus | 28.3 | 7.3 | 27.7 | 6.4 | 26.7 | 23.8 | 10.5 |
Note: GenBank accession numbers used: SC0806, JX983192–JX983194; Cat Que virus, JQ675598–JQ675600; TRVL 3586, KF697148-KF697150; OBE-1, NC009894–NC009896; TRVL-9760, KC759122–KC759124; Sathuperi virus, HE795102–HE795104; SA Ar 53, NC018476–NC018478; An5550, HE795105–HE795107; An10107, HE800141–HE800143; Bunyamwera virus, NC001925–NC001927; La Crosse virus, GU206144–GU206146; Wyeomyia virus, JN572080-JN572082.
L segment, large RNA segment; M, segment, medium RNA segment; S segment, small RNA segment; nt, nucleotides; aa, amino acids; Np, nucleoprotein; NSs, nonstructural protein.
Phylogenetic classification of SC0806
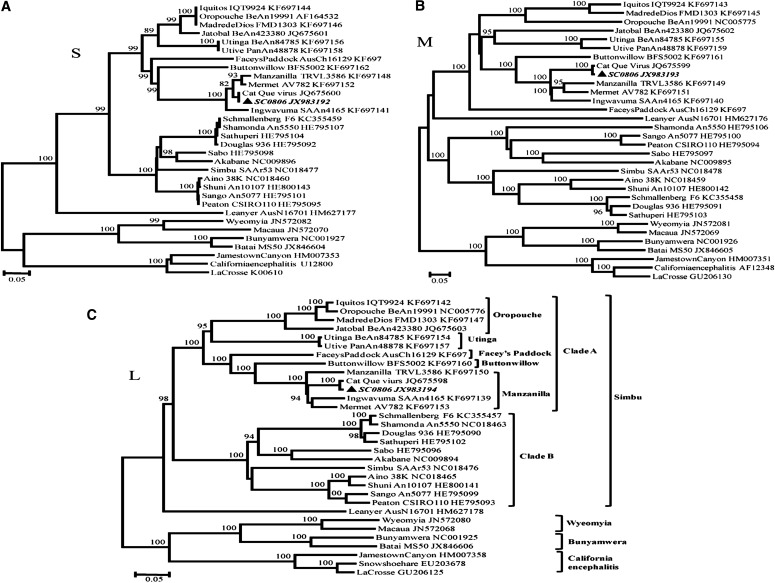
To further examine the phylogeny of SC0806, available sequences of Simbu serogroup viruses and of representative members of the California encephalitis, Bunyamwera, and Wyeomyia serogroups were aligned and phylogenetic analyses were carried out. The phylogenetic analyses of the L, M, and NP ORFs indicated that SC0806 was distantly related to members of the Simbu serogroup (Fig. 2). The members of the Simbu serogroup were divided into two clades, clades A and B (Ladner et al. 2014). Clade A was subdivided into five species complexes—Manzanilla, Oropouche, Utinga, Facey's Paddock, and Buttonwillow. In all three amino acid trees, SC0806 grouped with the Manzanilla species and was on the same evolutionary branch as CQV (Fig. 2). The nucleotide trees exhibited similar topology (data not shown).
FIG. 2.
Phylogenetic tree of members of the genus Orthobunyavirus on the basis of the protein-coding portion of the small (S) (A), medium (M) (B), and large (L) (C) segments. The tree was built in MEGA version 5.1 (www.megasoftware.net) using the neighbor-joining algorithm and a p-distance matrix. The tree is unrooted, and the node labels represent percentage bootstrap support values after 1000 resampling events. The sequence of SC0806 from this study is in bold italic.
Detection of anti-SC0806 antibodies in local pigs
To investigate the infection of pigs by SC0806, 91 pig serum samples were collected from Fuling, Yubei, and Wanzhou in the eastern Sichuan Basin in 2008 (Fig. 1). IgM and IgG to SC0806 in serum samples were detected using an ELISA: 21.98% (20/91) were SC0806 IgM antibody positive, 60.43% (55/91) were SC0806 IgG antibody positive, with 7.53% (7/91) positive for both (Table 3). Therefore, SC0806 infection is common in the pigs in the area. Further analysis showed that the positive rate of the SC0806 IgM antibody was highest in piglets (younger than 4 months) and decreased gradually with increasing age, whereas the positive rate of the SC0806 IgG antibody was lowest in piglets and increased gradually with age, reaching 100% at 8 months of age (Table 3).
Table 3.
SC0806-Specific IgM and IgG Antibodies in Pig Serum Samples Tested by ELISA
| <4 month old | 4 month–8 month old | ≥8 month old | Total | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Location | IgM | IgG | IgM+IgG+ | IgM | IgG | IgM+IgG+ | IgM | IgG | IgM+IgG+ | IgM | IgG | IgM+IgG+ |
| Fuling | 44.44% (12/27) | 7.40% 2/27 | 7.40% (2/27) | 33.33% (1/3) | 0% (0/3) | 0% (0/3) | — | — | — | 43.33% (13/30) | 6.67% (2/30) | 6.67% (2/30) |
| Yibei | 40% (4/10) | 50% (5/10) | 40% (4/10) | 17.65% (3/17) | 82.35% (14/17) | 5.88% (1/17) | 0% (0/3) | 100% (3/3) | 0% (0/3) | 23.33% (7/30) | 73.33% (22/30) | 16.67% (5/30) |
| Wanzhou | — | — | — | — | — | — | 0% (0/31) | 100% (31/31) | 0% (0/31) | 0% (0/34) | 100% (34/34) | 0% (0/34) |
| Total | 43.24% (16/37) | 18.92% (7/37) | 16.22% (6/37) | 20% (4/20) | 70% (14/20) | 5% (1/20) | 0% (0/34) | 100% (34/34) | 0% (0/34) | 21.98% (20/91) | 60.43% (55/91) | 7.53% (7/91) |
IgM, immunoglobulin M; IgG, immunoglobulin G; ELISA, enzyme-linked immunosorbent assay.
Discussion
We sequenced the entire SC0806 genome, and the sequence analysis showed that the SC0806 genome was consistent with those of the Simbu serogroup viruses. The number and length of ORFs, the positions of the start and stop codons, and the lengths of the terminal sequence in three gene fragments of SC0806 were similar to those of the Vietnamese isolate CQV (Table 1). In the whole-genome sequence alignment, SC0806 had the highest identity with CQV (nucleotide identity >85%, amino acid identity >99.1%). The analysis based on the three gene fragments showed that SC0806 and CQV were on the same evolutionary branch. CQV has not been officially assigned to a species complex by the International Committee on Taxonomy of Viruses (ICTV). However, using a combination of genetic and serological data, it was clear that CQV belonged to the Manzanilla species complex (Ladner et al. 2014). Therefore, on the basis of these results, SC0806 is a CQV strain that belongs to the Manzanilla species complex in the Simbu serogroup.
Previous studies have shown that pigs are the primary mammalian host of CQV (Bryant et al. 2005, Ladner et al. 2014). To understand the natural spread of SC0806 in the area, SC0806 infection in pigs was investigated. SC0806 IgM and IgG antibodies were found in the serum of pigs. The results obtained by ELISA assays are consistent with the results obtained by indirect immunofluorescence assay (IFA) (data not shown). We also found that the proportion of pigs with SC0806 IgM antibodies was highest in piglets younger than 4 months old, and the proportion decreased gradually with increasing age, whereas the opposite relationship was true for SC0806 IgG antibodies, indicating that the piglets had been infected by SC0806. In addition, we performed PCR to detect CQV nucleic acid in CQV IgM-positive serum samples of piglets and local unknown fever patients. The positive results (data not shown) indicate that CQV has formed a natural cycle in mosquitoes and pigs in the local area and may be associated with human and animal diseases.
We are the first to isolate CQV from Cx. tritaeniorhynchus in the Sichuan Province of China. Cx. tritaeniorhynchus is widely distributed in mainland China and Southeast Asia and is the most important vector of Japanese encephalitis virus (JEV) in this region (Sun et al. 2009, Wang et al. 2011). The isolation of CQV from Cx. tritaeniorhynchus implies that CQV can replicate in Cx. tritaeniorhynchus and use it as a vector. In addition, CQV was first isolated in Vietnam at 8–23°N latitude. We isolated CQV in Sichuan Province, central China at 26–34°N latitude, indicating that CQV is now distributed widely.
Conclusions
This article reports the isolation of CQV, a Simbu serogroup virus in the genus Bunyavirus (family Bunyaviridae), from Cx. tritaeniorhynchus collected in Sichuan Province, China. In addition, the study detected SC0806 antibody in the serum of local pigs, suggesting the existence of a natural cycle of SC0806 in mosquitoes and pigs in Sichuan Province, China. In view of the ability of the virus to spread via mosquitoes and cause disease in pigs and other animal populations, it is important to continue to study and monitor the transmission range and pathogenicity of SC0806.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (grant no. 81290342), the Ministry of Science and Technology of China (grant no. 2011CB504702), the National Key Technology R&D Program of the Ministry of Science and Technology (grant no. 2014BAI13B04), and State Key Laboratory for Infectious Disease Prevention and Control (grant no. 2014SKLID03). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Disclosure Statement
No competing financial interests exist.
References
- Aguilar PV, Barrett AD, Saeed MF, Watts DM, et al. Iquitos virus: A novel reassortant Orthobunyavirus associated with human illness in Peru. PLoS Negl Trop Dis 2011; 5:e1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CR, Spence L, Downs WG, Aitken TH. Oropouche virus: A new human disease agent from Trinidad, West Indies. Am J Trop Med Hyg 1961; 10:574–578 [DOI] [PubMed] [Google Scholar]
- Anderson CR, Spence LP, Downs WG, Aitken TH. Manzanilla virus: A new virus isolated from the blood of a howler monkey in Trinidad, W.I. Am J Trop Med Hyg 1960; 9:78–80 [DOI] [PubMed] [Google Scholar]
- Bouloy M. Bunyaviridae: Genome organization and replication strategies. Adv Virus Res 1991; 40:235–275 [DOI] [PubMed] [Google Scholar]
- Briese T, Calisher CH, Higgs S. Viruses of the family Bunyaviridae: Are all available isolates reassortants? Virology 2013; 446:207–216 [DOI] [PubMed] [Google Scholar]
- Bryant JE, Crabtree MB, Nam VS, Yen NT, et al. Isolation of arboviruses from mosquitoes collected in northern Vietnam. Am J Trop Med Hyg 2005; 73:470–473 [PubMed] [Google Scholar]
- Calisher CH, Kokernot RH, De Moore JF, Boyd KR, et al. Arbovirus studies in the Ohio–Mississippi Basin, 1964–1967. VI. Mermet: A Simbu-group arbovirus. Am J Trop Med Hyg 1969; 18:779–788 [DOI] [PubMed] [Google Scholar]
- de Brito Magalhaes CL, Drumond BP, Novaes RF, Quinan BR, et al. Identification of a phylogenetically distinct orthobunyavirus from group C. Arch Virol 2011; 156:1173–1184 [DOI] [PubMed] [Google Scholar]
- Elliott RM. Molecular biology of the Bunyaviridae. J Gen Virol 1990; 71(Pt 3):501–522 [DOI] [PubMed] [Google Scholar]
- Elliott RM, Schmaljohn CS, Collett MS. Bunyaviridae genome structure and gene expression. Curr Top Microbiol Immunol 1991; 169:91–141 [DOI] [PubMed] [Google Scholar]
- Hoffmann B, Scheuch M, Hoper D, Jungblut R, et al. Novel orthobunyavirus in Cattle, Europe, 2011. Emerg Infect Dis 2012; 18:469–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YH, Kweon CH, Tark DS, Lim SI, et al. Development of inactivated trivalent vaccine for the teratogenic Aino, Akabane and Chuzan viruses. Biologicals 2011; 39:152–157 [DOI] [PubMed] [Google Scholar]
- Kono Y, Yusnita Y, Mohd Ali AR, Maizan M, et al. Characterization and identification of Oya virus, a Simbu serogroup virus of the genus Bunyavirus, isolated from a pig suspected of Nipah virus infection. Arch Virol 2002; 147:1623–1630 [DOI] [PubMed] [Google Scholar]
- Ladner JT, Savji N, Lofts L, Travassos da Rosa A, et al. Genomic and phylogenetic characterization of viruses included in the Manzanilla and Oropouche species complexes of the genus Orthobunyavirus, family Bunyaviridae. J Gen Virol 2014; 95:1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert AJ, Lanciotti RS. Molecular characterization of medically important viruses of the genus Orthobunyavirus. J Gen Virol 2008; 89:2580–2585 [DOI] [PubMed] [Google Scholar]
- Li MH, Fu SH, Chen WX, Wang HY, et al. Genotype V Japanese encephalitis virus is emerging. PLoS Negl Trop Dis 2011a; 5:e1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YX, Li MH, Fu SH, Chen WX, et al. Japanese encephalitis, Tibet, China. Emerg Infect Dis 2011b; 17:934–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oem JK, Lee KH, Kim HR, Bae YC, et al. Bovine epizootic encephalomyelitis caused by Akabane virus infection in Korea. J Comp Pathol 2012; 147:101–105 [DOI] [PubMed] [Google Scholar]
- Qin XC, Zhou DJ, Chen XP, Xu JG, et al. [Viruses of the family Bunyaviridae and their associated-diseases]. Zhonghua Liu Xing Bing Xue Za Zhi 2010; 31:1111–1114 [PubMed] [Google Scholar]
- Rodrigues DS, Medeiros DB, Rodrigues SG, Martins LC, et al. Pacui Virus, Rio Preto da Eva Virus, and Tapirape Virus, three distinct viruses within the family Bunyaviridae. Genome Announc 2014; 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed MF, Li L, Wang H, Weaver SC, et al. Phylogeny of the Simbu serogroup of the genus Bunyavirus. J Gen Virol 2001; 82:2173–2181 [DOI] [PubMed] [Google Scholar]
- Sun X, Fu S, Gong Z, Ge J, et al. Distribution of arboviruses and mosquitoes in northwestern Yunnan Province, China. Vector Borne Zoonotic Dis 2009; 9:623–630 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasconcelos HB, Nunes MR, Casseb LM, Carvalho VL, et al. Molecular epidemiology of Oropouche virus, Brazil. Emerg Infect Dis 2011; 17:800–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Coscoy L, Zylberberg M, Avila PC, et al. Microarray-based detection and genotyping of viral pathogens. Proc Natl Acad Sci USA 2002; 99:15687–25692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Urisman A, Liu YT, Springer M, et al. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol 2003; 1:E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhang H, Sun X, Fu S, et al. Distribution of mosquitoes and mosquito-borne arboviruses in Yunnan Province near the China–Myanmar–Laos border. Am J Trop Med Hyg 2011; 84:738–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Krishnamurthy S, Cai Z, Popov VL, et al. Identification of novel viruses using VirusHunter—an automated data analysis pipeline. PLoS One 2013; 8:e78470. [DOI] [PMC free article] [PubMed] [Google Scholar]