SUMMARY
The regulation of mRNA translation is of fundamental importance in biological mechanisms ranging from embryonic axis specification to formation of long-term memory. POS-1 is one of several CCCH zinc-finger RNA-binding proteins that regulate cell-fate specification during C. elegans embryogenesis. Paradoxically, pos-1 mutants exhibit striking defects in endo-mesoderm development, but have wildtype distributions of SKN-1, a key determinant of endo-mesoderm fates. RNAi screens for pos-1 suppressors identified genes encoding the cytoplasmic poly(A)-polymerase homolog GLD-2, the Bicaudal-C homolog GLD-3, and the protein NEG-1. We show that NEG-1 localizes in anterior nuclei where it negatively regulates endo-mesoderm fates. In posterior cells, POS-1 binds the neg-1 3′UTR to oppose GLD-2 and GLD-3 activities that promote NEG-1 expression and cytoplasmic lengthening of the neg-1 mRNA poly(A) tail. Our findings uncover an intricate series of post-transcriptional regulatory interactions that together achieve precise spatial expression of endo-mesoderm fates in C. elegans embryos.
INTRODUCTION
Eukaryotic cells exercise remarkable control over the post-transcriptional expression of mRNA. This is most notable in specialized cells such as large polarized embryonic cells and neurons, where a host of RNA-binding factors have been shown to regulate the spatial and temporal expression of mRNAs by controlling mRNA translation, stability, and localization within the cell (Darnell and Richter, 2012; Richter and Lasko, 2011). Translation efficiency for mRNAs is often positively correlated with poly(A) tail length, while tail shortening is frequently associated with mRNA turnover (Eckmann et al., 2011; Mangus et al., 2003). However, for some mRNAs tail shortening does not lead to turnover but instead correlates with storage. Cytoplasmic poly(A) tail lengthening can restore translation of these stored mRNAs (Weill et al., 2012). The conserved cytoplasmic poly(A) polymerase GLD-2 has been implicated in lengthening poly(A) tails and activating the translation of mRNAs in the C. elegans germline (Wang et al., 2002), mouse and frog oocytes (Barnard et al., 2004; Kwak et al., 2004; Nakanishi et al., 2006), and Drosophila embryos (Cui et al., 2008).
GLD-2 polyadenylation of its targets is thought to be regulated through RNA-binding cofactors (D’Ambrogio et al., 2013). For example, in C. elegans GLD-2 binds to the conserved KH-domain protein GLD-3, a homolog of Drosophila Bicaudal-C (Eckmann et al., 2004). GLD-3 stimulates GLD-2 activity in vitro (Wang et al., 2002). In addition, GLD-2 polyadenylation of gld-1 mRNA requires both GLD-3 and the RNA-binding-domain-containing protein RNP-8 (Kim et al., 2010). gld-2 mutants are sterile, and therefore the role of this gene in poly(A) tail synthesis has been examined in the germline alone. The role of GLD-2, if any, in controlling mRNA translation in C. elegans embryos has not been studied.
Genetic studies have identified several mRNA binding factors that control cell-fate specification during early C. elegans embryogenesis. These factors include the KH-domain protein MEX-3 (Draper et al., 1996) and several tandem CCCH zinc-finger proteins related to the vertebrate Tis11 gene including POS-1 and MEX-5 (Mello et al., 1992; Schubert et al., 2000; Tabara et al., 1999). Although a few target mRNAs for these factors have been identified, the targets, mechanism of regulation by RNA binding, and developmental outcomes remain largely unknown. For example, the tandem CCCH protein POS-1 is best studied for its role in restricting the translation of the GLP-1 mRNA to the anterior of the early embryo (Ogura et al., 2003). However, misregulation of GLP-1 (a Notch receptor homolog) cannot explain the nearly complete lack of endo-mesoderm specification observed in pos-1 mutant embryos.
Here we explore the role of POS-1 in early embryonic events that specify endo-mesoderm precursor cells that give rise to the majority of the C. elegans alimentary canal, including the pharynx and intestine. The endo-mesoderm components of the C. elegans alimentary canal are specified through both cell-intrinsic and inductive mechanisms (Goldstein, 1992; Mello et al., 1994; Priess and Thomson, 1987). A major endo-mesoderm precursor cell named EMS produces the entire intestine and the posterior portion of the pharynx. EMS is born through two asymmetric divisions that sequentially segregate the potential to express endo-mesoderm fates to the posterior sister cell during the first division of the egg, and then to the anterior sister cell during the second division (Figure 1A). The transcription factor SKN-1 is a major determinant of EMS development and accumulates asymmetrically in early 4-cell stage embryos, where its levels become high in posterior sister cells EMS and P2 (Bowerman et al., 1992). SKN-1 activity is further restricted to EMS through the activity of PIE-1, which localizes in the nucleus of P2 where it prevents SKN-1 from activating gene expression (Mello et al., 1992; Mello et al., 1996). In EMS, SKN-1 activates the expression of endo-mesoderm-promoting transcription factors, including MED-1, which promotes the expression of downstream genes required for EMS development (Maduro et al., 2007; Maduro et al., 2001). Although POS-1 is critical for EMS specification, the expression and localization of SKN-1 protein is wild-type in pos-1 mutant embryos, suggesting that POS-1 promotes EMS differentiation through other as yet unknown factors.
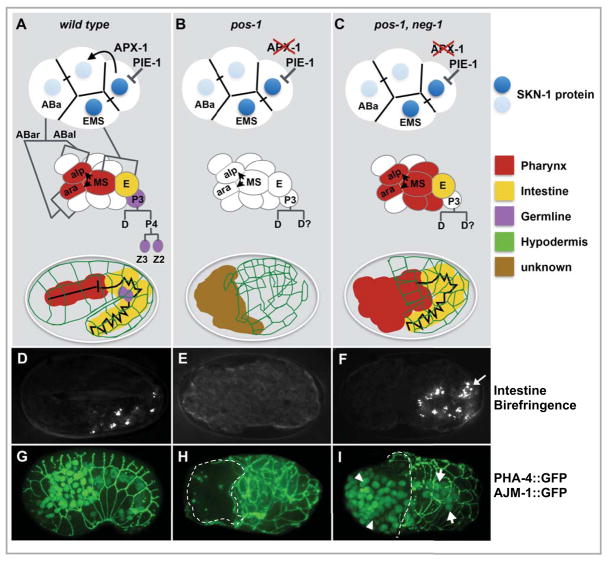
Figure 1. The pos-1 gutless phenotype is suppressed by neg-1 loss of function.
(A–C) Schematic diagrams showing key features in endo-mesoderm differentiation in wild-type and mutant backgrounds (as indicated). At the 4-cell stage SKN-1 localization and APX-1 signaling restrict endo-mesoderm potential to the blastomeres EMS and ABa, at the 12-cell stage MS signaling induces pharyngeal development in adjacent ABa descendants, finally body morphogenesis leads to enclosure of internal organs inside a network of hypodermal cells (diagrammed in green). The germ lineage P3 continues to divide asymmetrically producing the germline precursors Z2 and Z3 (purple). Defects in these events are indicated in the mutant contexts (B, C). (D–F) Polarized light micrographs showing gut differentiation as indicated by birefringent gut granule accumulation (D and F). (G–I) GFP fluorescence micrographs showing two differentiation markers: AJM-1::GFP which is expressed at cell-cell junctions of epidermal cells surrounding the embryo (McMahon et al., 2001), and PHA-4::GFP which is expressed in the nuclei of pharyngeal and intestinal precursors (Horner et al., 1998). In (H, I) failure of the AJM::GFP network to enclose the anterior region of the embryo is indicated by a dashed line, and PHA-4::GFP nuclear staining, absent in (H), is restored in pos-1, neg-1 double mutants (I, arrows).
Here, we identify GLD-2, GLD-3, and NEG-1 as factors whose loss of function restore endoderm differentiation in pos-1 mutant embryos. We show that NEG-1::GFP accumulates asymmetrically to higher levels in the nuclei of anterior blastomeres of 4-cell stage wild-type C. elegans embryos. GLD-2 and GLD-3 are required for NEG-1::GFP expression, while POS-1 is required to restrict the accumulation of NEG-1 protein to anterior blastomeres. We show that POS-1 binds directly to two consensus binding sites in the neg-1 3′UTR and that POS-1 activity correlates with short neg-1 mRNA poly(A) tails, while GLD-2 and GLD-3 activities correlate with long neg-1 poly(A) tails. Moreover, we employ a deep-sequencing approach termed PAT-seq to analyze poly(A) tail length transcriptome-wide in early embryos. We identify numerous transcripts whose poly(A)-tail lengths depend reciprocally on POS-1 activity and the activities of GLD-2 and GLD-3. Finally, we show that NEG-1 activity is required to prevent anterior blastomeres from ectopically expressing endo-mesoderm fates, and that POS-1 restricts NEG-1 activity to ensure proper endo-mesoderm development in the posterior. Interestingly, the GLD-3 homolog Bicaudal-C antagonizes posterior development in the anterior of Drosophila embryos, suggesting that portions of this regulatory circuit are conserved across phyla.
RESULTS
Restoration of gut in pos-1 embryos
Gut specification requires the activity of SKN-1 and a downstream gene regulatory network including the GATA transcription factors MED-1 and END-1 (Bowerman et al., 1992; Maduro et al., 2007). SKN-1 protein levels and distribution are unaffected in the early blastomeres of pos-1 embryos (Tabara et al., 1999). Downstream transcription factors such as MED-1 and END-1, however, are not detected in the presumptive EMS lineage of pos-1 embryos (Data Not Shown). Moreover, analysis of the med-1 mRNA by in situ hybridization revealed reduced levels of med-1 mRNA relative to wild-type embryos (Figure S1A). Together these findings suggest that although SKN-1 is expressed and localized properly in pos-1 mutants, it fails to initiate the gut developmental program. Consistent with this idea, we found that overexpression of the downstream transcription factor MED-1 using a multicopy transgenic array could bypass the pos-1 gutless phenotype, restoring gut differentiation in 83% (618/762) of med-1(+++); pos-1 embryos (compared to 2% (9/437) in pos-1(zu148) embryos).
Analysis of genetic interactions between pos-1 and a panel of maternal RNA-binding proteins revealed that RNAi of the KH-domain gene gld-3 strongly suppresses the pos-1 gutless phenotype (70.3% ± 11.6, n=144 embryos, and Data Not Shown) (Figure S1B & S1C). This suppression resulted in embryos with well-differentiated endoderm and pharyngeal tissue, but did not correct other defects, including the failure in pos-1 mutants to properly specify the ABp fate through a GLP-1 mediated interaction, and the failure to properly specify the germline cells Z2 and Z3. To search for additional factors whose loss of function could restore gut development in pos-1 mutants, we used RNAi to systematically screen a set of 944 genes previously annotated as required for embryogenesis. Homozygous pos-1 hermaphrodites were exposed to RNAi and their embryos were then examined under the light microscope for intestinal birefringence (Experimental Procedures). We found that RNAi targeting 7 different genes restored gut differentiation in the pos-1 embryos (Table S1). In each case, as was observed for gld-3(RNAi), suppression of pos-1 was limited to the restoration of endo-mesoderm differentiation while proper ABp and P4 cell fates were not restored (See below and Data Not Shown).
Among the genes identified in our screen were gld-2, which encodes a cytoplasmic poly(A) polymerase required for germline development, and F32D1.6, which encodes a protein with no obvious functional domains. We have named this gene neg-1 due to its negative effect on gut differentiation [note that while this study was under preparation, another study identified a role for neg-1 in anterior morphogenesis (Nishimura et al., 2015)]. We found that knockdown of gld-2 by RNAi resulted in sterility, but that weak gld-2(RNAi) (see Experimental Procedures) resulted in partial suppression of pos-1 (41% n=38, Table S1. RNAi of neg-1 caused a robust suppression of the pos-1 endo-mesoderm differentiation defect (79%, n=152, Table S1). Similarly, the neg-1(tm6077) mutation restored endo-mesoderm specification in pos-1 mutant embryos to 74%, (n=293), suggesting that neg-1(tm6077) behaves like a loss of function mutation (Figure 1F, I). The 407 bp neg-1(tm6077) deletion removes approximately 60% of the of neg-1 open reading frame, including the C-terminal 97 amino acids and deletes a carboxy-terminal domain in the NEG-1 protein that is conserved in homologs found in related nematode species (Figure S5B).
POS-1 restricts NEG-1::GFP expression to anterior cell lineages
GLD-2 and GLD-3 have been proposed to activate translation of target mRNAs by lengthening their poly(A) tails (Crittenden et al., 2003; Eckmann et al., 2004; Wang et al., 2002). Therefore, an attractive model is that GLD-2 and GLD-3 promote the expression of one or more endo-mesoderm antagonists, and POS-1 functions in endo-mesoderm lineages to oppose GLD-2 and GLD-3 activities. NEG-1 could function with GLD-2 and GLD-3 to promote the expression of such an antagonist. Alternatively, NEG-1 itself might be the hypothetical endo-mesoderm antagonist.
To begin to explore these possibilities, we engineered a strain expressing a single-copy full genomic fusion neg-1::gfp transgene (Frokjaer-Jensen et al., 2008). Strikingly, we found that NEG-1::GFP is asymmetrically localized in the early embryo (Figure 2 and Supplemental Movie 1). NEG-1::GFP was detected in the zygotic nucleus and at equal levels in both nuclei of the two-cell embryo (23/23). However, at the four-cell stage, NEG-1::GFP expression was markedly higher in nuclei of the anterior AB blastomeres than in the nuclei of EMS and P2 (31/34). Following the four-cell stage, NEG-1::GFP remained high in the granddaughters of the AB blastomere and progressively diminished in subsequent divisions (data not shown). In the adult germline, we observed NEG-1::GFP in the nuclei of distal germ cells and the nuclei of growing oocytes, except for the most proximal oocyte (Figure S2A). Moreover, we observed intense sub-nuclear localization of NEG-1:GFP on condensed chromatin (11/11 NEG-1::GFP+ nuclei) (Figure S2B).
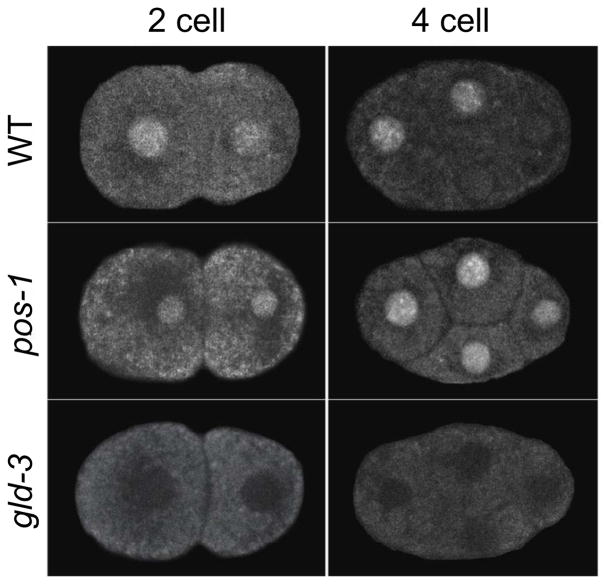
Figure 2. NEG-1::GFP is a asymmetrically expressed in the early embryo.
Representative confocal images of NEG-1::GFP rescuing neg-1(tm6077) in otherwise wild-type (WT), pos-1(zu148) and gld-3(RNAi) 2-cell, and 4-cell embryos.
Next we examined the effect of POS-1, GLD-2 and GLD-3 activities on NEG-1::GFP expression. We found that the asymmetry in NEG-1::GFP expression was abolished in pos-1 mutant embryos (Figure 2). Instead, NEG-1::GFP was expressed at high levels characteristic of AB descendants in all lineages of the pos-1(RNAi) (14/14) and pos-1(zu148) (6/6) embryos examined. In contrast, we found that NEG-1::GFP expression was absent or greatly reduced at all embryonic stages in gld-2 and gld-3 depleted embryos (Figure 2 and data not shown). As expected, we found that gld-3 is epistatic to pos-1. NEG-1::GFP expression, including its ectopic expression in EMS and P2, was abolished in pos-1; gld-3 double mutants (n=6). Taken together, these findings suggest that POS-1 represses the expression of NEG-1 in posterior blastomeres, while GLD-2 and GLD-3 are required to promote NEG-1 expression.
Consensus POS-1 binding elements are required for NEG-1 asymmetry
Early C. elegans embryos are transcriptionally quiescent (Guven-Ozkan et al., 2008; Seydoux and Dunn, 1997), and therefore regulation of NEG-1::GFP likely occurs at the level of mRNA translation or protein stability. To ask if NEG-1 expression is regulated through the 3′ untranslated region of the neg-1 mRNA, we fused gfp containing a nuclear localization sequence to the 3′UTR of neg-1 (hereafter gfp::3′UTRneg-1) and placed this reporter under the promoter of maternally-expressed gene oma-1 (Experimental Procedures). We found that the pattern of GFP expression from this reporter, which entirely lacks neg-1 coding sequences, was identical to that of the full-length neg-1::gfp fusion gene (Figure S3A and Supplemental Movie 2). We therefore conclude that the 3′UTR of neg-1 is sufficient to confer asymmetric expression in the early embryo.
The 3′UTR of neg-1 contains 3 overlapping, predicted RNA binding protein (RBP) elements, referred to here as the RBP cluster (Figure 3A). This RBP cluster begins with a uracil-rich sequence that represents a consensus MEX-5 binding region, which is adjacent to overlapping MEX-3 and POS-1 predicted binding elements (Farley et al., 2008; Pagano et al., 2009; Pagano et al., 2007). To address whether MEX-3, MEX-5, and POS-1 physically bind to the RBP cluster, we carried out electrophoretic mobility shift assays (EMSA) and fluorescence polarization (FP) assays (Experimental Procedures). We found that POS-1, MEX-3, and MEX-5 bind the RBP cluster with affinities comparable to those previously determined between each protein and confirmed biological targets (Figure 3B and 3C), and that MEX-5 binding to the RBP is favored over POS-1 binding (Figure S3B). Moreover, POS-1 binding was reduced when its putative binding site was mutated (Figure 3B, 3C and S3B). In addition to binding within the RBP cluster, we found that MEX-5 binds a second region (M5B), (Figure 3A & 3C). We also tested an additional region 3′ of the RBP cluster (P1M3B) that contains a consensus POS-1 binding element and an overlapping MEX-3 consensus sequence (Figure 3A), and found that both recombinant proteins bind this site with high affinity in vitro (Figure S3C and S3D).
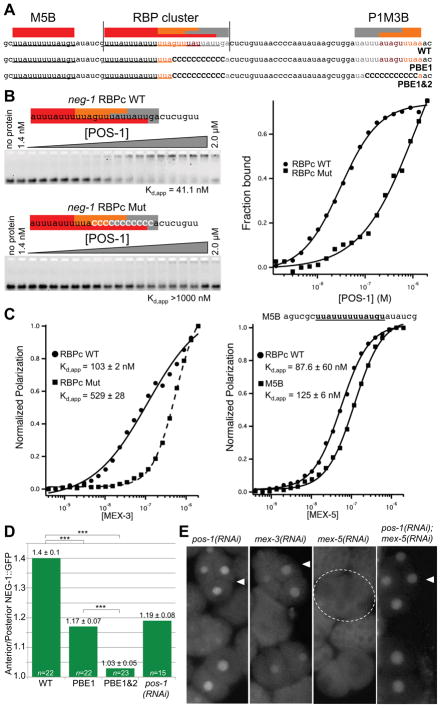
Figure 3. POS-1 and MEX-5 bind and regulate the expression of neg-1 mRNA.
(A) Nucleotide sequence of a portion of the neg-1 mRNA 3′UTR indicating predicted binding sites for POS-1 (grey), MEX-3 (orange) and MEX-5 (red). Wild-type (WT) and two sequences with mutations in the POS-1 consensus elements (PBE1 and PBE1&2) are shown.
(B) Fluorescence electrophoretic mobility shift assays with recombinant POS-1 and fluorescently labeled wild-type (RBPc WT) and mutant (RBPc Mut) fragments of the neg-1 RBP cluster (as indicated). The mobility of each labeled probe is shown after incubation with increasing amounts of POS-1 protein as indicated at top of the gel images. The apparent dissociation constant, Kd,app indicated below the gel images, represents the average of three independent replicates ± the standard deviation, SD. A graphical quantification of the data is shown at right.
(C) Graphical representations of fluorescence polarization data obtained from incubating the fluorescent probes (as indicated) with increasing amounts of MEX-3 (Left graph) or MEX-5 (Right Graph). The RBP cluster probes were the same as those used in (B). The sequence of the MEX-5 B probe (M5B) is shown above the right graph. The Kd,app values were calculated as described in (B).
(D) Graphical representation of relative Anterior/Posterior GFP intensity observed in 4-cell stage embryos transgenic for neg-1 promoter-driven GFP under the control of the neg-1 wild-type (WT) or mutated the neg-1 3′UTR. The mutations in the PBE1 and PBE1&2 UTRs are shown in (A). Relative Anterior/Posterior GFP intensity after pos-1(RNAi) is also indicated for comparison. The y-axis indicates the average ratio of GFP intensity in the “Anterior” (ABa + ABp) divided by intensity in the “Posterior” (EMS + P2) (see Experimental Procedures). Data are represented as mean ± standard deviation and *** indicates a t-test p-value < 0.0001. Sample size (n) is indicated for each column.
(E) GFP fluorescence micrographs showing the expression of a wild-type NEG-1::GFP reporter in representative wild-type embryos (WT) and embryos depleted by RNAi of various RBPs (as indicated).
To determine whether these consensus binding elements direct the in vivo regulation of NEG-1 expression, we generated gfp::3′UTRneg-1 reporters containing the same mutations used for the in vitro binding assays. We found that mutation of the predicted POS-1 binding element within the RBP cluster caused a more symmetric distribution of NEG-1::GFP between anterior and posterior blastomeres (Figure 3D, column PBE1). Combining this lesion with a lesion in the second POS-1 binding element P1M3B completely abolished the anterior-posterior asymmetry in NEG-1::GFP levels (column PBE1&2). Interestingly, this double binding-site mutant consistently exhibited NEG-1::GFP expression that although equal between anterior and posterior blastomeres was also lower in intensity than levels observed in wild-type anterior nuclei (Data Not Shown), suggesting that one or both of these 3′UTR elements may also contribute positively to NEG-1 expression.
mex-5 positively regulates neg-1 expression
MEX-5 and MEX-3 are enriched in anterior blastomeres (Draper et al., 1996; Schubert et al., 2000) where NEG-1 protein levels are high. Moreover, as shown above, these factors bind in vitro to regions in the NEG-1 3′UTR that are also bound by POS-1 protein, raising the possibility that MEX-5 and MEX-3 are positive regulators of NEG-1 expression that directly compete with POS-1 for binding. Consistent with this possibility, we found that RNAi of mex-5 and mex-3 reduced the levels of NEG::GFP expression. In the case of mex-3(RNAi) the asymmetry of NEG-1::GFP expression was not affected, but the overall levels appeared slightly reduced (Figure 2E and Data Not Shown). Strikingly, however, RNAi knockdown of mex-5 completely abolished NEG-1::GFP expression (0/30) (Figure 3E). RNAi of mex-6, a partially redundant paralog of mex-5, did not affect NEG-1::GFP expression (data not shown). The complete absence of NEG-1 expression in mex-5(RNAi) early embryos is opposite to the consequence of pos-1 loss of function. We therefore examined the consequences of mex-5 knockdown in a pos-1(zu148); neg-1::gfp strain. We found that all pos-1(zu148); mex-5(RNAi) early embryos exhibited bright NEG-1::GFP expression characteristic of pos-1 mutants (n=14), including equal and high levels of NEG-1::GFP in all blastomeres at the 4-cell stage (n=4) (Figure 3E). Similar results were obtained from pos-1(RNAi); mex-5(RNAi) double knockdown embryos (Figure 3E). These findings indicate that pos-1 is epistatic to and genetically downstream of mex-5 for the regulation of neg-1 expression and suggest that MEX-5 protein promotes neg-1 expression in the anterior by countering POS-1 repression. A previous study has shown that the POS-1 protein exhibits wild-type localization in mex-5 mutant embryos (Tenlen et al., 2006), suggesting that the absence of neg-1 in mex-5(RNAi) embryos is not caused by ectopic anterior accumulation of POS-1 protein. The finding that in vitro binding sites for MEX-3, MEX-5 and POS-1 overlap, suggests that direct competition between these positive and negative factors may contribute to NEG-1 regulation and may also explain why mutating these elements abolishes both the asymmetry and the overall level of NEG-1::GFP expression.
POS-1, GLD-2, and GLD-3 regulate neg-1 poly(A)-tail length
In the germline, GLD-2 and GLD-3 are thought to positively regulate gene expression by promoting the cytoplasmic poly-adenylation of target mRNAs (Crittenden et al., 2003; Eckmann et al., 2004; Wang et al., 2002). Indeed, several C. elegans transcripts expressed in the germline have been shown to be dependent on GLD-2 for their polyadenylation (Janicke et al., 2012; Kim et al., 2010). Moreover, the neg-1 transcript is one of hundreds of transcripts that have shortened poly(A)-tails in the sterile gld-2 adult compared to wild-type adults (Beilharz TH; in preparation). We therefore asked if GLD-2 and GLD-3 might exert their positive effects on NEG-1 expression by promoting the poly-adenylation of neg-1 mRNA in the embryo.
To measure the length of neg-1 poly(A)-tails in early embryos, we used a deep-sequencing approach termed Poly(A)-Tail cDNA-sequencing (PAT-seq; see Experimental Procedures; Harrison et al, MS ID#:RNA/2014/048355 MS TITLE: PAT-seq: A method to study the integration of 3′ UTR dynamics with gene-expression in the eukaryotic transcriptome.). If NEG-1 regulation is achieved through poly-adenylation, then NEG-1 would be expected to have long poly(A)-tails in the anterior blastomeres and short tails in posterior blastomeres. If this difference in poly(A)-tail length between blastomeres is significantly large, we would detect two different populations of tail lengths when analyzing whole embryos. Consistent with this idea, we observed a bimodal distribution of neg-1 poly(A)-tail lengths in wild-type embryos (Figure 4A), with a clustering of longer tails centered at around 80 A residues, and a second clustering of shorter tails centered at approximately 20 A residues in length (Figure 4B) (n=318 tails). In pos-1 mutants, the density of reads with approximately 80 A residues was increased and was more tightly clustered relative to wild type (Figure 4A arrow, and Figure 4B). Furthermore, the median poly(A) read length was slightly increased (red bars in Figure 4A), as was the average from 44.4 bases in WT to 48.9 bases in pos-1 (n=276 tails, p = 0.05). In contrast, read lengths were dramatically shifted toward shorter tails in gld-3 and gld-2 depleted embryos (Figure 4A and B), with an average length in gld-3(RNAi) embryos of 25.3 (n=250 tails, p = 0.00005), and in gld-2(RNAi) embryos of 36.3, (n=210 tails, p = 0.005). Taken together, these findings suggest that POS-1 and GLD-2/GLD-3 regulate the expression of NEG-1 by controlling the polyadenylation status of neg-1 mRNA.
Figure 4. POS-1, GLD-3 and GLD-2 regulate neg-1 mRNA poly(A)-tail length.
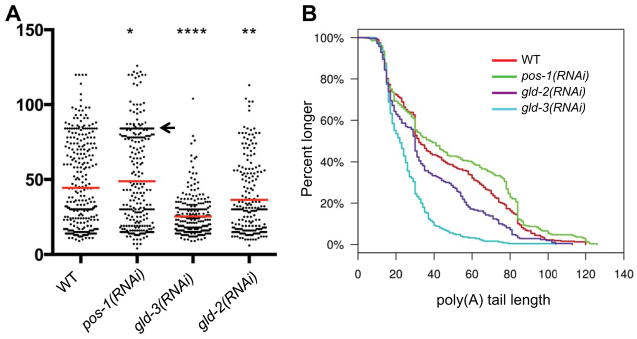
(A) Graphical representation of neg-1 mRNA Poly(A) tail lengths as detected by PAT-seq analysis of RNA prepared from WT and mutant early embryos (as indicated). Each dot represents a cDNA sequence and the number A residues in its poly(A) tail corresponding to its placement on the y-axis. A high density of dots may appear as a black line. The Red bar reflects the median according to Kolmagorov-Smirnov test. Asterisks indicate statistical significance (*) P ≤ 0.05, (**) P ≤ 0.01 and (****) P ≤ 0.0001. Arrow notes a range of overrepresented poly(A) tail lengths in WT, which increases in pos-1(RNAi) early embryos.
(B) A continuous histograms demonstrating the percentage of poly(A) tails (y-axis) longer than a given length (x-axis) in each genotype tested.
neg-1(+) activity represses anterior endo-mesoderm differentiation
To further analyze the function of neg-1, we characterized the loss-of-function phenotypes associated with neg-1(RNAi) or the deletion allele neg-1(tm6077). Interestingly, we found that neg-1, like pos-1, is required maternally for proper embryonic development. Approximately 75% of embryos produced by homozygous neg-1 mutant (or RNAi) mothers die before hatching. In most cases the dead embryos exhibit defective morphogenesis, characterized by a failure of hypodermal cells to properly enclose the anterior portion of the embryo (Figure 5A). These findings suggests that neg-1(+) activity promotes anterior cell fate specification and/or morphogenesis.
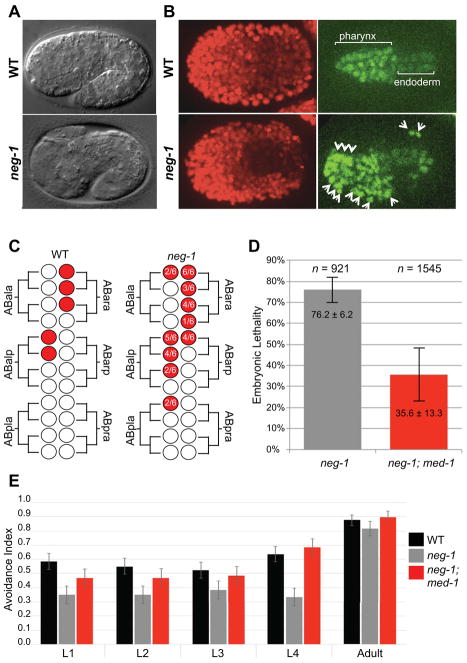
Figure 5. neg-1 prevents ectopic endo-mesoderm differentiation in the AB lineage.
(A) Nomarski light micrographs showing wild-type and neg-1 embryos approximately midway through body morphogenesis. Defective anterior enclosure with resulting external pharyngeal tissue (white arrows) is indicated in the neg-1 embryo.
(B, C) Ectopic PHA-4::GFP expression in neg-1(−) embryos. (B) Fluorescence micrographs of a wild-type (WT) and neg-1(tm6077) embryo expressing histone::mCherry (marking all nuclei), and pha-4::gfp (marking pharyngeal and endodermal cells). The white arrowheads indicate ectopic pharyngeal cells in the neg-1 embryo.
(C) Lineage diagrams constructed from 4D microscopy studies on 6 independently analyzed neg-1(tm6077) embryos. Wild-type lineages that produce pharynx are shown by red circles in the diagram at left. The neg-1 lineages that were positive for PHA-4::GFP are shown at right with the frequency of GFP detection indicated inside each red circle. Diagram is a summary of lineages in (Figure S4).
(D, E) med-1 loss of function suppresses neg-1 mutant phenotypes. (D) Bar graph showing the percent embryonic lethality in neg-1(tm6077) single mutant and neg-1(tm6077); med-1(ok804) mutant embryos (as indicated). n = number of embryos assayed. The error bars indicate the mean ± SD. (E) Bar graph showing the glycerol avoidance index for wild-type (n=80) and the neg-1(tm6077) (n=60) and neg-1(tm6077); med-1(ok804) (n=60) double mutant strains (as indicated). The larval stages analyzed L1 through Adult are indicated. The avoidance index is calculated as the number of positive responses divided by the total number of trials (see text) ± SEM.
Because neg-1 is an antagonist of gut specification, we wondered if NEG-1 might function during wild-type development to represses mesoderm or endoderm differentiation in lineages that normally specify portions of the anterior hypodermis. To explore this possibility we examined the expression pattern of PHA-4 in neg-1(tm6077). pha-4 encodes a FoxA transcription factor expressed in mesodermal and endodermal precursor cells (Du et al., 2014; Horner et al., 1998). Using 4D-microscopy to trace lineages (Hardin, 2011), we found that PHA-4::GFP was ectopically expressed in anterior AB sub-lineages of neg-1(tm6077) embryos, suggesting that NEG-1 prevents PHA-4 expression in lineages that are normally destined to become ectoderm (Figure 5B & 5C, Figure S4).
In addition to hypodermal lineages, neg-1 loss-of-function also affects neuronal lineages. We therefore analyzed neg-1 mutants that survived embryogenesis for behavioral deficits that might reflect impaired neuronal function. We found that neg-1 worms exhibit reduced osmotic avoidance. Wild-type L4 worms avoided 2 M glycerol with an avoidance index of 0.63 ± 0.05 (n = 80), whereas neg-1 worms of the same stage scored 0.33 ± 0.06 (n = 60) (Figure 5E).
As described above, we found that over-expression of the endo-mesoderm promoting transcription factor med-1 can partially suppress the pos-1 gutless phenotype. We reasoned therefore that med-1 loss-of-function might suppress ectopic mesoderm differentiation and thus restore anterior development in neg-1 embryos. Indeed, we found that med-1(ok804) dramatically reduced embryonic lethality of neg-1(tm6077) from ~75% to 35.6% ± 13.3 (n=1545 embryos; Figure 5D). Notably, the osmotic avoidance defect of neg-1 was also suppressed (0.68 ± 0.06, n = 60) (Figure 5E), suggesting that ectopic mesoderm differentiation also causes the neg-1 behavioral phenotype. Taken together these findings suggest that NEG-1 functions in the anterior to ensure robust ectoderm specification, at least in part by preventing endo-mesoderm development, and that POS-1 promotes posterior endo-mesoderm differentiation by restricting NEG-1 protein accumulation to the anterior.
DISCUSSION
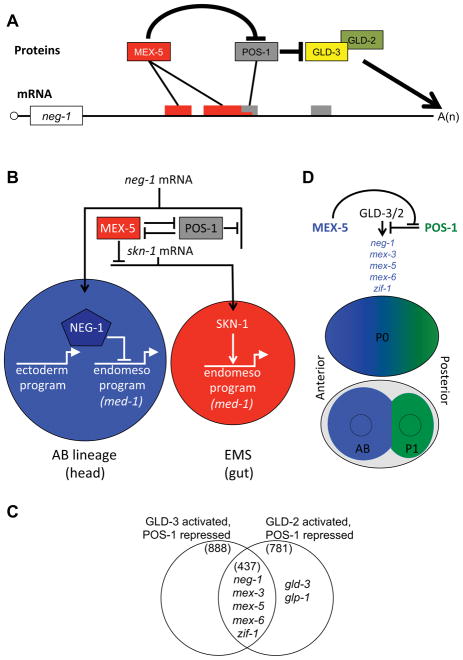
Early embryonic patterning in C. elegans involves the precise spatial and temporal control of mRNA translation (Begasse and Hyman, 2011). Maternal mRNAs encoding transcription factors, cell-signaling components, and other developmental regulators become differentially expressed in early embryonic cells, contributing to the rapid diversification of cell fates (Hwang and Rose, 2010). Here we have explored the role of the RNA-binding protein POS-1 in this process. We have shown that a maternal mRNA encoding an endo-mesoderm repressor NEG-1 is a key target for POS-1 regulation. Taken together, our genetic, molecular, and localization studies support a model in which POS-1 binds to the neg-1 mRNA and regulates its cytoplasmic poly adenylation (Figure 6A). Our findings suggest that POS-1 and its homolog MEX-5 bind adjacent sequences in the neg-1 mRNA 3′ UTR and have opposing effects on the recruitment of the cytoplasmic poly(A) polymerase GLD-2 and its co-factor GLD-3. In anterior blastomeres, where MEX-5 levels are high (Schubert et al., 2000), MEX-5 overcomes POS-1 inhibition leading to GLD-2/GLD-3 recruitment, extension of neg-1 poly(A) tails, and accumulation of NEG-1 protein which, in turn, represses endo-mesoderm fates, ensuring the proper expression of ectodermal (skin and neuronal) cell fates. Conversely, in posterior blastomeres, high POS-1 levels prevent GLD-2/GLD-3 mediated activation of neg-1 mRNA translation and thus protect endo-mesoderm precursors from NEG-1 protein accumulation (Figure 6B).
Figure 6. RBPs control cytoplasmic poly adenylation of mRNAs to establish anterior posterior asymmetries.
(A) Molecular regulation of neg-1 expression. MEX-5 and POS-1 bind the 3′UTR of neg-1. POS-1 prevents GLD-3/2 polyadenylation of neg-1 in posterior lineages and MEX-5 counters POS-1 repression in anterior lineages.
(B) Model for the regulation of cell fate specification in anterior and posterior lineages (blue and red circles respectively). The diagram indicates how regulation of mRNA translation by the RBPs POS-1 and MEX-5 ensures asymmetric accumulation of the endo-mesoderm activator (SKN-1) and of the endo-mesoderm antagonist (NEG-1), and how the downstream activities of these factors ensure proper expression of ectodermal and endo-mesodermal transcriptional programs. MEX-5 negative regulation of SKN-1 and POS-1 is based on Schubert et al., (2000).
(C) GLD-2/GLD-3 activated, POS-1 repressed genes are those that had shorter tails upon gld-3 RNAi or gld-2 RNAi and longer poly(A)-tails after pos-1 RNAi (overlap between both circles). This common group consists of 436 genes that include the anterior expressed neg-1, mex-3, mex-5, mex-6 and zif-1. gld-3 and glp-1 qualify as GLD-2 activated, POS-1 repressed genes but not as GLD-3 activated, POS-1 repressed.
(D) Model indicating how the regulatory circuit that controls neg-1 mRNA expression may also explain the regulation of many other mRNAs including those of several previously studied genes (indicated).
GLD-2 and GLD-3 as regulators of early embryogenesis
Previous work demonstrated that GLD-2 and GLD-3 function together during germline development to promote the transition from mitosis to meiosis (Crittenden et al., 2003; Eckmann et al., 2004; Wang et al., 2002). Here we have shown that GLD-2 and GLD-3 also promote embryogenesis by positively regulating the expression of the endo-mesoderm inhibitor NEG-1. Our findings suggest that GLD-2 and GLD-3 promote NEG-1 expression by lengthening the poly(A) tail of neg-1 mRNA. Similar regulatory interactions may control the expression of other proteins that accumulate in anterior blastomeres. Of more than 5000 genes represented in our early embryo PAT-seq data sets, we identified 436 with “GLD-2/GLD-3-activated, POS-1-repressed” poly(A) tail lengths (Figure 6C). These included mex-3, mex-5, mex-6, and zif-1, whose protein products are enriched in the anterior in a pattern similar to that of NEG-1 (DeRenzo et al., 2003; Draper et al., 1996; Schubert et al., 2000). A recent study used oligo-dT selection to recover mRNAs from isolated anterior and posterior blastomeres of 2-cell-stage C. elegans embryos (Osborne Nishimura et al., 2015). Interestingly, comparing these data to our PAT-seq data revealed that 40.5% (17/42) of the AB enriched mRNAs from the single-blastomere study, but only 1.8% (2/112) of P1 enriched mRNAs, were among the GLD-2/GLD-3-activated, POS-1-repressed group of mRNAs identified by PAT-seq (Table S2 and S3). These findings suggest that the POS-1/GLD-2/GLD-3 interactions that regulate neg-1 poly(A)-tail length may also function more generally to regulate the asymmetric expression of dozens of target mRNAs including neg-1, zif-1, and mex-3/5/6 (Figure 6C).
It is not yet known how GLD-2 and GLD-3 are recruited to their targets. A structural study indicates that domains KH2-KH5 of GLD-3 assemble a thumb-like structure that may present KH1 for direct contact with GLD-2 (Nakel et al., 2010; Eckmann et al., 2004). These findings raise the question of whether GLD-3 can bind both mRNA and GLD-2 simultaneously, and suggest that, rather than directly binding RNA to recruit GLD-2, GLD-3 may serve as an adaptor protein for GLD-2 recruitment by other RNA-binding factors. One attractive possibility is that CCCH-finger proteins contribute RNA-binding specificity that determines which mRNAs are targeted for poly(A) tail lengthening by GLD-2. With at least seven CCCH proteins (MEX-1, POS-1, MEX-5, MEX-6, OMA-1, OMA-2, and PIE-1) implicated in cell-fate determination in early embryos, the combinatorial complexity available for mRNA regulation is substantial. Hinting at additional complexity, our findings reveal a subset of mRNAs whose poly(A) tail lengths appear to depend on GLD-2, but not GLD-3 activity (Figure 6D). Among these, the glp-1 mRNA was previously shown to be a target of POS-1 negative regulation in the posterior of the embryo (Ogura et al., 2003). It will therefore be interesting to determine if a distinct GLD-2 complex, that does not include GLD-3, activates GLP-1 mRNA translation. It is also worth noting that the mRNA of the GLP-1 ligand APX-1, whose protein expression in the P2 blastomere depends on POS-1(+) activity, is among a subset of mRNAs whose poly(A) tail lengths were positively correlated with GLD-2 and POS-1 (but not GLD-3 activity) (Table S3). Thus, in addition to its role as a negative regulator of neg-1 mRNA, POS-1 may function as a positive regulator that recruits a distinct GLD-2 complex to promote the lengthening of apx-1 mRNA poly(A) tails.
A conserved mechanism for regulating anterior-posterior asymmetries?
How NEG-1 represses endo-mesoderm genes remains unclear. NEG-1 does not contain recognizable sequence domains that resemble other known functional elements, and only two homologs have been identified in related nematodes (Figure S5A, S5B). However, the nuclear localization of NEG-1 protein and its association with chromatin suggest that NEG-1 may prevent the activation of endo-mesoderm differentiation by acting as a transcriptional repressor.
Despite the absence of sequence homologs in other organisms, our findings suggest that NEG-1 partakes in a conserved mechanism that controls embryonic asymmetry in metazoans. Drosophila Bicaudal-C is required to prevent posterior (caudal/abdominal) fates from appearing ectopically in the anterior of the Drosophila embryo, a developmental function very similar to that described here for C. elegans gld-3. Interestingly, Drosophila Bicaudal-C appears to bind its own mRNA and repress translation by recruiting components of a conserved mRNA deadenylase complex (Chicoine et al., 2007). A recent study suggests that the Xenopus homolog Bic-C represses the expression of specific mRNAs in the vegetal hemisphere of early embryos by directly binding translational control element present in their 3′UTRs (Zhang et al., 2014). How these repressive functions of Bic-C proteins can be reconciled with our findings that GLD-3 functions to promote mRNA poly(A) tail length and gene expression remain to be seen. However, a dynamic interplay between activities that shorten and lengthen poly(A) tails in the cytoplasm is thought to be central to the regulation of mRNA translation during fly oogenesis and Xenopus oocyte maturation (Ivshina et al., 2014). It is possible, therefore, that Bic-C proteins mediate repression by shortening poly(A) tails to promote mRNA turnover or to promote the storage of certain mRNAs for later activation. Conversely, by engaging GLD-2 protein, Bic-C proteins may have the ability to reverse this repression, selectively re-activating specific mRNAs. Currently, we do not know how neg-1 mRNA becomes de-adenylated. Therefore it will be interesting to determine if GLD-3 plays a role in recruiting a deadenylase complex to the neg-1 mRNA, perhaps during oogenesis, to set the stage for re-adenylation later during embryogenesis.
In both fly and frog oocytes, conserved RNA binding factors called cytoplasmic polyadenylation-element-binding proteins (CPEBs) play a key role in recruiting the cytoplasmic poly(A) polymerase GLD-2 to specific targets (Barnard et al., 2004; Benoit et al., 2008; Cui et al., 2008). Although CPEB proteins are conserved in C. elegans and have been shown to function with GLD-2 to regulate germline development, we have not found a role for CPEBs in regulating neg-1 mRNA polyadenylation or other embryonic events. Perhaps POS-1 and its CCCH zinc-finger homologs provide RNA-binding and regulatory activities in C. elegans that are analogous to those provided by CPEB proteins in other organisms. There is clearly a rich complexity of mRNA regulation in metazoan embryos, especially in embryos where important early developmental events occur prior to the onset of zygotic transcription. It will be interesting in the future to determine the extent to which cytoplasmic polyadenylation and the rich combinatorial complexity of RNA-binding factors contribute to the remarkable unfolding of early embryonic patterning, organogenesis, and neurogenesis across phyla.
EXPERIMENTAL PROCEDURES
Strains
JJ462: +/nT1 IV; pos-1(zu148) unc-42(e270)/nT1 V.
WM317: neg-1(tm6077). The deletion was obtained by the National Bioresource Project (Mitani Laboratory) and removed a region encoding the last 97 amino acids as well as the terminating stop codon of the gene thus fusing the coding region with the 3′UTR and adding 11 codons to the predicted gene product before encountering an in-frame stop codon.
RW10425: stIs10116 [his-72(promoter)::his-24::mCherry + unc-119(+)]. stIs37 [pie-1(promoter)::mCherry::H2B + unc-119(+)]. stIs10389 [pha-4::GFP::TY1::3xFLAG].
WM310: gfp::neg-1; neg-1(tm6077) A region of cosmid F32D1 flanked by AflII and NotI digestion sites was subcloned and an ORF encoding GFP was introduced upstream of F32D1.6 (gfp::neg-1). The engineered transgene preserved neg-1 in its predicted operon and its downstream neighbor fipp-1 and was introduced using MosSCI transgenesis (Frokjaer-Jensen et al., 2008). gfp::neg-1 was crossed into the neg-1(tm6077) background.
WM311: oma-1 promoter::NLS::gfp::neg-1 3′UTR (J608.6)
WM312: oma-1 promoter::NLS::gfp::neg-1 3′UTR with mutated POS-1 binding site (See RBPc Mut in Table 2)
WM316: neg-1(tm6077); med-1(ok804)
WM326: pos-1(zu148) unc-42 (e230)/hT1 [med-1::gfp, rol-6(su1006)]
WM319: oma-1 promoter::NLS::gfp::neg-1 3′UTR with two mutated POS-1 binding elements
med-1 in situ hybridization
In situ hybridization was preformed as described in (Tabara et al., 1999). Probes were designed against gfp mRNA and used to identify med-1::gfp in the strain med-1::gfp, rol-6(su1006) (Maduro et al., 2001), before and after pos-1(RNAi).
pos-1 suppressor screen
pos-1 suppressor screen was performed by placing 5 to 10 L2 – L4 pos-1(zu148) unc-42(e270) worms on RNAi food and allowing them to reach adulthood and lay eggs. Embryos were then scored for gut granules. Positive hits were then retested by scoring the embryos (E) of 4 to 6 individual worms (n) fed the RNAi food in order to assess overall gut development and obtain average pos-1 suppression and standard deviation. The same protocol was followed for gld-3 and gld-2 suppression. For the effect of med-1 upregulation, a med-1::gfp multicopy transgenic array was crossed with pos-1(zu148) unc-42(e270) and embryos of homozygous hermaphrodites were scored for gut granules.
Binding Assays
Fluorescence anisotropy and electrophoretic mobility shift assays (EMSA) using purified recombinant POS-1 (80–180), MEX-3 (45–205) and MEX-5 (236–350) were done as described in Farley et al. 2008, Pagano et al. 2009 and Pagano et al. 2007, respectively. All RNA oligonucleotides used in this study were chemically synthesized and fluorescently labeled at the 3′ end with fluorescein amidite (FAM) by Integrated DNA Technologies (IDT).
Competition assays are set up similar to the EMSA assays. 550 nM of POS-1 (80–180) or 450 nM MEX-5 (236–350) was added to the RNA equilibration buffer to obtain a 70% RNA bound complex. Then the corresponding competing protein was titrated to the reaction mixture at varying concentrations. After 3 hours of equilibration, the reaction mixture was run on a 5% native polyacrylamide gel in 1X TB for 3 hours, at 120V. Quantifications were done by determining the pixel intensity of the RNA species bound by protein relative to the pixel intensity of total RNA species to give the fraction bound of RNA. The pixel intensities of each band were determined and background corrected by using Image Gauge (Fujifilm, Tokyo, Japan).
NEG-1::GFP quantification
Time-lapse imaging of NEG-1::GFP was conducting by capturing GFP fluorescence (exposure time: 40 milliseconds) at 3 different planes (z-stacks) every 1 minute from the 2 to 6-cell stages. Images spanning the four-cell stage were analyzed by measuring GFP intensity in nuclei (mean gray value) using ImageJ. The 3 time points preceding the division of ABa and ABp were selected to represent NEG-1::GFP intensity in the 4-cell stage. GFP intensity of ABa and ABp were summed for “Anterior GFP” and that of EMS and P2 were summed for “Posterior GFP”. NEG-1::GFP Asymmetry = Anterior GFP – Posterior GFP.
Embryonic cell lineaging and pha-4::gfp expression
The EMS lineage was performed using the pha-4::gfp lineaging strain RW10425 for wildtype. The strain was fed pos-1 RNAi food to lineage EMS in pos-1(−) embryos. The strain was crossed with pos-1(zu148) unc-42(e270) and fed gld-3 RNAi food to lineage EMS in pos-1(−); gld-3(−) embryos. Lineaging was performed according to Du et al. 2014. Images of pha-4::gfp expression generated during lineaging were used for Figure 6C.
PAT-seq
The Poly(A)-Tail focused RNA-seq, or PAT-seq approach was utilized to determine the gene expression, poly(A)-site and polyadenylation state of the transcriptome of early C. elegans embryos having been depleted for a series of RNA binding proteins. pos-1 was knocked down using dsRNA expressed E. coli fed to ~100,000 starved/synchronized L1 larvae. When half of the population reached adulthood and half were still in the L4 stage, worms were bleached and embryos harvested and stored in Trizol. This ensured an enrichment of early embryos between 1 and 24 cell stage. Since prolonged gld-3 and gld-2 RNAi causes sterility, starved/synchronized L1 larvae were fed diluted OP50 (200uL of concentrated OP50 diluted in 2mL of M9 and starved L1s) for 16 hours. After this initial step, the OP50 was mostly consumed by the larvae and concentrated gld-3 or gld-2 dsRNA expressing E. coli was added to the plates. When half of the population reached adulthood and half were still in the L4 stage, worms were bleached and embryos harvested and stored in Trizol. Total RNA was isolated using standard procedures. For wildtype samples, synchronized L1s were fed OP50 until the aforementioned stage before being processed as stated above. >90% knockdown of pos-1 and gld-3 was confirmed by qPCR, whereas the gld-2 knockdown was approximately 50% (data not shown).
Osmotic avoidance test
A drop of glycerol (2 M) was delivered near the tail of a worm as it moves forward. The glycerol drop instantly surrounds the worm and reaches the anterior sensory organs. Wildtype worms immediately sense the glycerol as a repellent and move backwards. Backward movement was scored.
Supplementary Material
Acknowledgments
We thank the Mello and Ambros labs for input and discussion, Darryl Conte and Myriam Aouadi for comments on the text and figures and James Mello for critical reading of the manuscript. neg-1(tm6077) was generated by Shohei Mitani. This work was supported by NIH Grants HD36247 and HD33769 to C.C.M. C.C.M is a Howard Hughes Medical Institute Investigator.
Footnotes
ACCESSION NUMBERS
GEO accession number: GSE57993
AUTHOR CONTRIBUTIONS
A.E. and C.C.M designed the experiments and wrote the paper. A.E., M.S., H.W. and D.Y. developed the transgenic worms. M.S. performed the med-1 ISH. A.E. E.K and S.R. performed, analyzed and interpreted the in vitro gel-shift assays. A.E., P.F.H., D.R.P and T.H.B. performed, analyzed and interpreted the PAT-seq experiments. A.E., Z.D. and Z.B. performed the cell-lineaging study. A.E., C.D.C and J.S. performed, analyzed and interpreted the behavior assay. T.I. performed additional imaging trials to quantify NEG-1::GFP intensity.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barnard DC, Ryan K, Manley JL, Richter JD. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Begasse ML, Hyman AA. The first cell cycle of the Caenorhabditis elegans embryo: spatial and temporal control of an asymmetric cell division. Results Probl Cell Differ. 2011;53:109–133. doi: 10.1007/978-3-642-19065-0_6. [DOI] [PubMed] [Google Scholar]
- Benoit P, Papin C, Kwak JE, Wickens M, Simonelig M. PAP- and GLD-2-type poly(A) polymerases are required sequentially in cytoplasmic polyadenylation and oogenesis in Drosophila. Development. 2008;135:1969–1979. doi: 10.1242/dev.021444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman B, Eaton BA, Priess JR. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell. 1992;68:1061–1075. doi: 10.1016/0092-8674(92)90078-q. [DOI] [PubMed] [Google Scholar]
- Chicoine J, Benoit P, Gamberi C, Paliouras M, Simonelig M, Lasko P. Bicaudal-C recruits CCR4-NOT deadenylase to target mRNAs and regulates oogenesis, cytoskeletal organization, and its own expression. Developmental cell. 2007;13:691–704. doi: 10.1016/j.devcel.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Crittenden SL, Eckmann CR, Wang L, Bernstein DS, Wickens M, Kimble J. Regulation of the mitosis/meiosis decision in the Caenorhabditis elegans germline. Philos Trans R Soc Lond B Biol Sci. 2003;358:1359–1362. doi: 10.1098/rstb.2003.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Sackton KL, Horner VL, Kumar KE, Wolfner MF. Wispy, the Drosophila homolog of GLD-2, is required during oogenesis and egg activation. Genetics. 2008;178:2017–2029. doi: 10.1534/genetics.107.084558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ambrogio A, Nagaoka K, Richter JD. Translational control of cell growth and malignancy by the CPEBs. Nat Rev Cancer. 2013;13:283–290. doi: 10.1038/nrc3485. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Richter JD. Cytoplasmic RNA-binding proteins and the control of complex brain function. Cold Spring Harb Perspect Biol. 2012;4:a012344. doi: 10.1101/cshperspect.a012344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRenzo C, Reese KJ, Seydoux G. Exclusion of germ plasm proteins from somatic lineages by cullin-dependent degradation. Nature. 2003;424:685–689. doi: 10.1038/nature01887.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper BW, Mello CC, Bowerman B, Hardin J, Priess JR. MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell. 1996;87:205–216. doi: 10.1016/s0092-8674(00)81339-2. [DOI] [PubMed] [Google Scholar]
- Du Z, Santella A, He F, Tiongson M, Bao Z. De novo inference of systems-level mechanistic models of development from live-imaging-based phenotype analysis. Cell. 2014;156:359–372. doi: 10.1016/j.cell.2013.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann CR, Crittenden SL, Suh N, Kimble J. GLD-3 and control of the mitosis/meiosis decision in the germline of Caenorhabditis elegans. Genetics. 2004;168:147–160. doi: 10.1534/genetics.104.029264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann CR, Rammelt C, Wahle E. Control of poly(A) tail length. Wiley Interdiscip Rev RNA. 2011;2:348–361. doi: 10.1002/wrna.56. [DOI] [PubMed] [Google Scholar]
- Farley BM, Pagano JM, Ryder SP. RNA target specificity of the embryonic cell fate determinant POS-1. RNA. 2008;14:2685–2697. doi: 10.1261/rna.1256708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B. Induction of gut in Caenorhabditis elegans embryos. Nature. 1992;357:255–257. doi: 10.1038/357255a0. [DOI] [PubMed] [Google Scholar]
- Guven-Ozkan T, Nishi Y, Robertson SM, Lin R. Global transcriptional repression in C. elegans germline precursors by regulated sequestration of TAF-4. Cell. 2008;135:149–160. doi: 10.1016/j.cell.2008.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin J. Imaging embryonic morphogenesis in C. elegans. Methods Cell Biol. 2011;106:377–412. doi: 10.1016/B978-0-12-544172-8.00014-1. [DOI] [PubMed] [Google Scholar]
- Horner MA, Quintin S, Domeier ME, Kimble J, Labouesse M, Mango SE. pha-4, an HNF-3 homolog, specifies pharyngeal organ identity in Caenorhabditis elegans. Genes Dev. 1998;12:1947–1952. doi: 10.1101/gad.12.13.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SY, Rose LS. Control of asymmetric cell division in early C. elegans embryogenesis: teaming-up translational repression and protein degradation. BMB Rep. 2010;43:69–78. doi: 10.5483/bmbrep.2010.43.2.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivshina M, Lasko P, Richter JD. Cytoplasmic polyadenylation element binding proteins in development, health, and disease. Annual review of cell and developmental biology. 2014;30:393–415. doi: 10.1146/annurev-cellbio-101011-155831. [DOI] [PubMed] [Google Scholar]
- Janicke A, Vancuylenberg J, Boag PR, Traven A, Beilharz TH. ePAT: a simple method to tag adenylated RNA to measure poly(A)-tail length and other 3′ RACE applications. RNA. 2012;18:1289–1295. doi: 10.1261/rna.031898.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KW, Wilson TL, Kimble J. GLD-2/RNP-8 cytoplasmic poly(A) polymerase is a broad-spectrum regulator of the oogenesis program. Proc Natl Acad Sci U S A. 2010;107:17445–17450. doi: 10.1073/pnas.1012611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JE, Wang L, Ballantyne S, Kimble J, Wickens M. Mammalian GLD-2 homologs are poly(A) polymerases. Proc Natl Acad Sci U S A. 2004;101:4407–4412. doi: 10.1073/pnas.0400779101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro MF, Broitman-Maduro G, Mengarelli I, Rothman JH. Maternal deployment of the embryonic SKN-1-->MED-1,2 cell specification pathway in C. elegans. Dev Biol. 2007;301:590–601. doi: 10.1016/j.ydbio.2006.08.029. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Meneghini MD, Bowerman B, Broitman-Maduro G, Rothman JH. Restriction of mesendoderm to a single blastomere by the combined action of SKN-1 and a GSK-3beta homolog is mediated by MED-1 and -2 in C. elegans. Mol Cell. 2001;7:475–485. doi: 10.1016/s1097-2765(01)00195-2. [DOI] [PubMed] [Google Scholar]
- Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon L, Legouis R, Vonesch JL, Labouesse M. Assembly of C. elegans apical junctions involves positioning and compaction by LET-413 and protein aggregation by the MAGUK protein DLG-1. J Cell Sci. 2001;114:2265–2277. doi: 10.1242/jcs.114.12.2265. [DOI] [PubMed] [Google Scholar]
- Mello CC, Draper BW, Krause M, Weintraub H, Priess JR. The pie-1 and mex-1 genes and maternal control of blastomere identity in early C. elegans embryos. Cell. 1992;70:163–176. doi: 10.1016/0092-8674(92)90542-k. [DOI] [PubMed] [Google Scholar]
- Mello CC, Draper BW, Priess JR. The maternal genes apx-1 and glp-1 and establishment of dorsal-ventral polarity in the early C. elegans embryo. Cell. 1994;77:95–106. doi: 10.1016/0092-8674(94)90238-0. [DOI] [PubMed] [Google Scholar]
- Mello CC, Schubert C, Draper B, Zhang W, Lobel R, Priess JR. The PIE-1 protein and germline specification in C. elegans embryos. Nature. 1996;382:710–712. doi: 10.1038/382710a0. [DOI] [PubMed] [Google Scholar]
- Nakanishi T, Kubota H, Ishibashi N, Kumagai S, Watanabe H, Yamashita M, Kashiwabara S, Miyado K, Baba T. Possible role of mouse poly(A) polymerase mGLD-2 during oocyte maturation. Dev Biol. 2006;289:115–126. doi: 10.1016/j.ydbio.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Ogura K, Kishimoto N, Mitani S, Gengyo-Ando K, Kohara Y. Translational control of maternal glp-1 mRNA by POS-1 and its interacting protein SPN-4 in Caenorhabditis elegans. Development. 2003;130:2495–2503. doi: 10.1242/dev.00469. [DOI] [PubMed] [Google Scholar]
- Osborne Nishimura E, Zhang JC, Werts AD, Goldstein B, Lieb JD. Asymmetric Transcript Discovery by RNA-seq in C. elegans Blastomeres Identifies neg-1, a Gene Important for Anterior Morphogenesis. PLoS genetics. 2015;11:e1005117. doi: 10.1371/journal.pgen.1005117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano JM, Farley BM, Essien KI, Ryder SP. RNA recognition by the embryonic cell fate determinant and germline totipotency factor MEX-3. Proc Natl Acad Sci U S A. 2009;106:20252–20257. doi: 10.1073/pnas.0907916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano JM, Farley BM, McCoig LM, Ryder SP. Molecular basis of RNA recognition by the embryonic polarity determinant MEX-5. J Biol Chem. 2007;282:8883–8894. doi: 10.1074/jbc.M700079200. [DOI] [PubMed] [Google Scholar]
- Priess JR, Thomson JN. Cellular interactions in early C. elegans embryos. Cell. 1987;48:241–250. doi: 10.1016/0092-8674(87)90427-2. [DOI] [PubMed] [Google Scholar]
- Richter JD, Lasko P. Translational control in oocyte development. Cold Spring Harb Perspect Biol. 2011;3:a002758. doi: 10.1101/cshperspect.a002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert CM, Lin R, de Vries CJ, Plasterk RH, Priess JR. MEX-5 and MEX-6 function to establish soma/germline asymmetry in early C. elegans embryos. Mol Cell. 2000;5:671–682. doi: 10.1016/s1097-2765(00)80246-4. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Dunn MA. Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development. 1997;124:2191–2201. doi: 10.1242/dev.124.11.2191. [DOI] [PubMed] [Google Scholar]
- Tabara H, Hill RJ, Mello CC, Priess JR, Kohara Y. pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development. 1999;126:1–11. doi: 10.1242/dev.126.1.1. [DOI] [PubMed] [Google Scholar]
- Tenlen JR, Schisa JA, Diede SJ, Page BD. Reduced dosage of pos-1 suppresses Mex mutants and reveals complex interactions among CCCH zinc-finger proteins during Caenorhabditis elegans embryogenesis. Genetics. 2006;174:1933–1945. doi: 10.1534/genetics.105.052621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Eckmann CR, Kadyk LC, Wickens M, Kimble J. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature. 2002;419:312–316. doi: 10.1038/nature01039. [DOI] [PubMed] [Google Scholar]
- Weill L, Belloc E, Bava FA, Mendez R. Translational control by changes in poly(A) tail length: recycling mRNAs. Nat Struct Mol Biol. 2012;19:577–585. doi: 10.1038/nsmb.2311. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Park S, Blaser S, Sheets MD. Determinants of RNA binding and translational repression by the Bicaudal-C regulatory protein. J Biol Chem. 2014;289:7497–7504. doi: 10.1074/jbc.M113.526426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.