Abstract
Background
Measurement of peptide/protein concentrations in biological samples for biomarker discovery commonly uses high-sensitivity mass spectrometers with a surface-processing procedure to concentrate the important peptides. These time-of-flight (TOF) instruments typically have low mass resolution and considerable electronic noise associated with their detectors. The net result is unnecessary overlapping of peaks, apparent mass jitter, and difficulty in distinguishing mass peaks from background noise. Many of these effects can be reduced by processing the signal using standard time-series background subtraction, calibration, and filtering techniques.
Methods
Surface-enhanced laser desorption/ionization (SELDI) spectra were acquired on a PBS II instrument from blank, hydrophobic, and IMAC-Cu ProteinChip® arrays (Ciphergen Biosystems, Inc.) incubated with calibration peptide mixtures or pooled serum. TOF data were recorded after single and multiple laser shots at different positions. Correlative analysis was used for time-series calibration. Target filters were used to suppress noise and enhance resolution after baseline removal and noise rescaling.
Results
The developed algorithms compensated for the electronic noise attributable to detector overload, removed the baseline caused by charge accumulation, detected and corrected mass peak jitter, enhanced signal amplitude at higher masses, and improved the resolution by using a deconvolution filter.
Conclusions
These time-series techniques, when applied to SELDI-TOF data before any peak identification procedure, can improve the data to make the peak identification process simpler and more robust. These improvements may be applicable to most TOF instrumentation that uses analog (rather than counting) detectors.
Current efforts in clinical research rely on the integration of proteomic technologies in the search for specific proteins or peptides (called biomarkers) that are associated with disease. Recent evidence suggests that single biomarkers may not be effective in improving detection, diagnosis, and prognosis. Thus, rather than focusing on the discovery of a single biomarker, protein profiling can maximize the use of samples collected from patients by mining larger segments of the proteome. When sequenced and identified (from databases or de novo), these protein biomarkers may also serve to elucidate potential new drug targets. Because diagnostics and drug design generally involve labor-intensive procedures, both for development and validation, highly parallel methods of preliminary screening are desirable. By shortening the preliminary research, these methods may allow for rapid development of tools for diagnosis and prognosis, which can be tailored for individual patients. This could facilitate the development of better strategies for treatment and offer higher recovery rates for patients. Measuring cancer-related changes in serum also may reduce unnecessary biopsies (1–4).
Laser desorption and ionization methods, including matrix-assisted laser desorption/ionization (MALDI)6 and surface-enhanced laser desorption/ionization (SELDI), can detect unfragmented singly charged “parent” ions with masses up to hundreds of kilodaltons in complex mixtures (5, 6) for concentrations below fmol/L. Although two-dimensional polyacrylamide gel electrophoresis (PAGE) imaging provides an alternative approach for proteomics to measure relative concentrations of proteins [see, e.g., Ref. (7)], PAGE has inferior mass resolution and sensitivity and does not provide information in the mass range <20 kDa.
SELDI time-of-flight mass spectrometry (TOF-MS) is performed with chromatographic (on-chip) purification of the samples (6). Like MALDI, this technique ionizes the sample by use of a light-absorbing matrix that is added to the spot surface after the purification step. Similar to MALDI, SELDI results depend on sample preparation and the protocols for laser irradiation (5, 8). Moreover, the large quantity of matrix typically produces multiple chemical adducts and neutral losses that also appear in the corresponding MS spectra in addition to parent peptides. SELDI does not deploy reflectron or quadrupole elements for mass focusing (9) and therefore provides far lower resolution than the highest resolution TOF instruments (5, 9, 10). Unlike two-dimensional PAGE, ion yields from SELDI are not easily related to the actual relative concentrations of individual peptides or proteins on the surface. This is because the relative intensities of SELDI peaks depend on interactions between proteins, between the proteins and the matrix, and between the proteins and the chip surface (5, 8, 11). However, when strict experimental protocols are followed, SELDI intensities are reproducible (4, 12, 13).
For diagnostic applications, the goal of MS is to find spectral patterns that indicate the presence or absence of a disease (2–4). The detection of patterns in the multitude of mass peaks that arise from complex clinical samples depends on the uniformity of instrumental response in both mass and intensity. Hence, the instrument must be thoroughly calibrated. The fundamental weaknesses of SELDI are excessive background noise, reduced signal-to-noise ratios at high masses, misassignment of peak masses, formation of multiple chemical adducts, and substantial overlap of peaks resulting from low resolution. All of these effects make it much more difficult to distinguish and identify peaks in the mass spectrum (2–4, 12–14). Furthermore, high concentrations of low-mass species (e.g., matrix or contaminants) frequently overload the detector and obscure the peptide peaks <2 kDa, which may be medically important. An improvement in the quality of current SELDI data and an understanding of its signal to noise are therefore essential steps for protein profile screening (12–14) and for the identification of biomarker peptides (2–4).
To become a viable (inexpensive, noninvasive, rapid) tool for clinical diagnostics, SELDI must provide ion signals that quantify the relative amounts of peptides or proteins that correlate with a specific medical condition. These operational requirements challenge MS electronics and data analysis methods. MS for heavy molecules was not originally targeted at measuring relative amounts of analytes over a broad concentration range (5). Instead, the engineering effort was initially focused on improving the ultimate sensitivity while maintaining as much as possible the assignment of a mass number (5, 9). Much of the recent research effort has been invested in the design of stable sample preparation protocols to ensure spectrum reproducibility (4, 6, 8, 11–14), but with the exception of careful isotopic labeling methods (15), even these more repeatable mass spectra have not yet shown a direct correlation between observed ion yields and the actual relative concentrations of the analytes. This is because maintaining constant instrument gain over several orders of magnitude in mass and concentration is still not possible with the current choices of detectors and electronics.
Here we describe a set of calibration and filtering procedures that correct for instrumental artifacts in SELDI spectra before analysis and interpretation of the data. Following these procedures should ensure that the data recorded from different instruments and in different laboratories with the same protocol for sample preparation can be compared directly. In these studies, we characterized the response of the PBS II instrument, using several ProteinChip® arrays with different surface chemistries. SELDI-TOF spectra were recorded after single and multiple laser shots for various hardware settings and several chip surfaces. We characterized the sources of SELDI baseline shifts, peak broadening, and apparent jitter in peak position. Using optimal smoothing and target-filtering methods that were developed by the time-series analysis and spectroscopy community (16–18), we created algorithms for subtracting baselines, for removing small jitters in peak timing, and for enhancing instrument resolution. We suggest that the improved practices described here for data acquisition, calibration, preprocessing, and filtering should become a part of the experimental routine for TOF-MS profiling of peptide expression. They suppress instrumental artifacts and automatically reduce the number of variables (through noise filtering) for classification of diseases from MS data. This is an important prerequisite for finding the most significant features (biomarkers) in a consistent statistical analysis of SELDI-MS data. Further improvements are possible and are easily incorporated into the methodology.
Materials and Methods
CALIBRATION SAMPLES AND m/z AXIS
The five-in-one protein (β-galactosidase, cytochrome C, myoglobin, glyceraldehyde-3-phosphate dehydrogenase, and albumin) calibration mixture was obtained from Ciphergen Biosystems, Inc. and was applied to a hydrophobic chip (NP20) according to a protocol specified by the manufacturer. Ciphergen’s proprietary ProteinChip software uses these spectra to automatically calibrate the instrument by a linear regression to fit time positions of the dominant peaks to their known mass values. The fitting function is a three-parameter quadratic function so that the mass eventually increases as the square of the measured TOF arrival time. Because the instrument samples a spectrum at a constant rate (every 4 ns for PBS II), this leads to a decreasing sample density per unit mass. Thus, the higher masses appear compressed in a spectrum viewed in the time domain.
SERUM PROCESSING AND STORAGE
The donor samples were collected at Eastern Virginia Medical School from properly consenting individuals according to a protocol approved by the Eastern Virginia Medical School Institutional Review Board. Venipuncture was performed, and blood was collected into a 10-mL Vacutainer® serum separator tube. The blood was clotted in a refrigerator at 3–6 °C for 30 min and then centrifuged at 1000g for 15 min. The serum was immediately decanted, aliquoted, and stored at −80 °C. These samples were stored for 5–8 months before SELDI analysis without freeze-thaw cycles. Before the SELDI analysis, the samples were thawed and divided into 40- to 50-µL aliquots. Serum samples for SELDI analysis were prepared as described previously (4).
SELDI ACQUISITION CONDITIONS
TOF spectra after single and multiple shots for blank chips (i.e., bare aluminum, no sample or matrix), calibration mixtures on hydrophobic chips (NP20), and pooled serum on IMAC-Cu chip were acquired. All chips had eight spots, labeled A-H. Each spot was subdivided into 100 sections, called subpositions, as illustrated in Fig. 1 of the Data Supplement that accompanies the online version of this article at http://www.clinchem.org/content/vol51/issue1/. For the blank chip, all shots were performed at subposition 20. For single shots with pooled serum, no warming shots were done and subpositions 20–80 were scanned. For average spectra with pooled serum, 30 shots per subposition with two warming shots (at a laser intensity of 240), were performed, scanning subpositions 20–80 with a 4-subposition step (16 steps and 192 shots overall). Multiple acquisitions were done with sensitivity settings of 7 and 3, detector voltages of 1600 and 1200 V, and laser intensity settings at 210 and 180. Raw data records and records processed by the instrument’s default variable width moving average filter were analyzed. The default moving average filter has a variable (mass-dependent) window width according to the manufacturer’s supplied table. This dependence is approximately linear on the m/z scale, and intermediate values for window width are calculated by linear interpolation between tabulated values.
STRATEGY OF CUSTOMIZED DATA PROCESSING
We recommend practices based on time-series analysis of the MS data in three basic areas: characterization and reduction of noise; calibration of peak timing; and resolution enhancement.
Noise characterization and reduction
For noise reduction, we present ways to remove the baseline, to remove some of the nonlinear effects attributable to detector saturation, and to rescale the signal to maintain a constant noise level. Most of the linear baseline arises from a constant offset and the accumulation of a slowly decaying charge by the detector electronics. The measured signal can thus be modeled as:
| (1) |
where yn is the incoming ion signal, and sn is the observed combination of incoming signal plus accumulated charge and constant offset, c. The constant a represents the fraction of signal that accumulates, and τ represents the number of time steps that this accumulated charge takes to decay. This model can be inverted by finding the baseline, bn, as follows:
| (2) |
In addition to this slow change, the baseline shifts as a step function after a large event saturates the detector. This step function has a constant value from the time starting at the first saturation event, t0, and ending after r · n time steps by returning to zero. The variable n describes the number of saturated data points in the peak, and r is a recovery rate, in time points per saturation, that depends on the gain settings of an individual instrument.
The random noise can be reduced at high masses by use of the default moving average filter, but we recommend rescaling the signal by the square root of the number of points in the moving average window to regain a constant level of random noise. The default settings for variable width moving average depend only weakly on the mass calibration; therefore, the same rescaling factor can be used for all records.
Calibration of peak timing
To improve the peak timing resolution, we recalibrated the mass transformation settings at each laser subposition by introducing small time shifts to maximize the cross-correlation between spectra near sharp spectral features before the average spectrum was computed. The same technique can be used for automatic alignment of the peaks in different spectra before classification.
Resolution enhancement
For resolution enhancement, we used a deconvolution filter to smooth and shape the signal so that individual mass peaks were easier to resolve. Our filter design, which simultaneously reduces the noise and narrows the incoming signal into our desired target shape, uses filter coefficients, ak, implicitly defined by the following equations (16):
| (3) |
where dk is the observed initial signal wavelet; bk is the desired target signal; gt is the cross-correlation between the desired and observed wavelets (defined in the above equation); rt − k is the autocorrelation function of the observed wavelet; qt − k is an estimation of the noise autocorrelation function; M is the number of points in the initial target region; and ν is a weighting factor that determines the relative importance of noise suppression to shaping. For each time point in the TOF record, this filter can be applied to all incoming signals, st, as:
| (4) |
The filtered signal, yt, at time t depends only on the incoming signal, st − k, from the earlier times (t − k, lower masses). One could equivalently create a filter that depends only on the signal at later times (higher masses) or one that is symmetric. The low mass edge of the mass peaks is sharper because of the absence of adducts and the natural skew of isotopic distributions (19), and we find that this choice of filters best uses the inherent instrument resolution. We used five filters of different widths and produced a net filtered signal by taking the fifth root of the product of the outputs of these five filters to dramatically reduce the noise and simultaneously increase the resolution. The choices of settings for artifact suppression will be optimized in future work.
In the present work, we chose a target region in the vicinity of a sodium atomic peak (mass range, 15–28 Da) for the filter construction. The full width of the skewed target peak in time was 19 points, whereas the broadened peak wavelet was 30 points wide and showed a signal-to-noise ratio of ~20. The lengths of the five filters were 150, 147, 143, 135, and 129 points; and the optimum value for ν (minimizing artifacts in the filtered signal) was 6.5.
COMPUTATIONAL RESOURCES
The algorithms for the data analysis were implemented in Matlab 6.12, and all calculations were performed on a Unix SunBlade workstation (500 MHz CPU, 384M of RAM). The target filter for TOF data is the subject of a patent application. The rights to scripts and implementation details belong to INCOGEN, Inc., but are available for academic users on request by sending an e-mail to filter@incogen.com.
Results and Discussion
A brief description of the techniques for sample preparation, data acquisition, and theoretical parameter estimation is given in the Materials and Methods. Below we begin our overview of results with a description of the sources of systematic noise in the PBS II data. This appears as a baseline signal (linear and nonlinear) or as random variations. We next suggest a method of dejittering between subpositions during acquisition from a single spot and between patient records from large data sets to correct peak timing and improve resolution of average records after multiple laser shots. We then follow with several experimental examples of resolution enhancement through deconvolution and noise suppression of experimental SELDI records using smoothing and shaping target filters.
CHARACTERIZATION AND REDUCTION OF NOISE
The SELDI-TOF instrument registers ion abundances as records of voltages induced in the detector when the ions arrive. For analog detectors, these voltages are sampled in time with a constant dwell period (4 ns for the PBS II instrument). The basic premise of a TOF measurement is that each arriving ion produces a signal at its arrival time but does not affect signals at other times. Moreover, because the signals are the sum of the charges arriving at a specified time, they should always be positive. Nevertheless, in most SELDI-TOF spectra, the peak signals usually ride on top of a large, relatively smooth background. The details of this background vary with the instrument settings and with the size and characteristics of the signal, but it is not unusual to observe background at 20% of the peak signal and to have that background persist for more than 15 000 data points. The instrument can attempt to subtract this baseline without a physical model by use of a segmented convex hull algorithm supplied in a proprietary software package from Ciphergen, Inc (20), but this method often subtracts signal for overlapping broad clusters at low and high masses. Such errors could compromise later comparisons of relative intensities.
The most common contribution to this background, and the easiest to correct, can be modeled as charge accumulation that decays on a much larger time scale than the typical ion pulse length, as in Eq. 1. If we assume that a small portion of the incoming signal accumulates and then decays with a decay time of τ, then the observed signal will have the form described by Eq. 1. This can be readily inverted by Eq. 2 and then subtracted from the original signal to produce a baseline-free signal, as in panels A and B of Fig. 1. Fig. 1 also includes a second horizontal axis showing the conversion from time point to m/z. The three model variables for this baseline will generally depend on the instrument settings, but they can be easily determined from a calibration experiment. In our studies, we found values of a = 0.05– 0.15%, τ = 800, and c = 3.8. We determined c from the average signal at very large masses (>30 kDa, above 100 µs acquisition time). We adjusted the parameter a to minimize the integrated signal under the constraint that the resulting signal never became appreciably negative. The slow decay of the signal in the intermediate mass region, where the incoming signal is small, determined the time constant, τ.
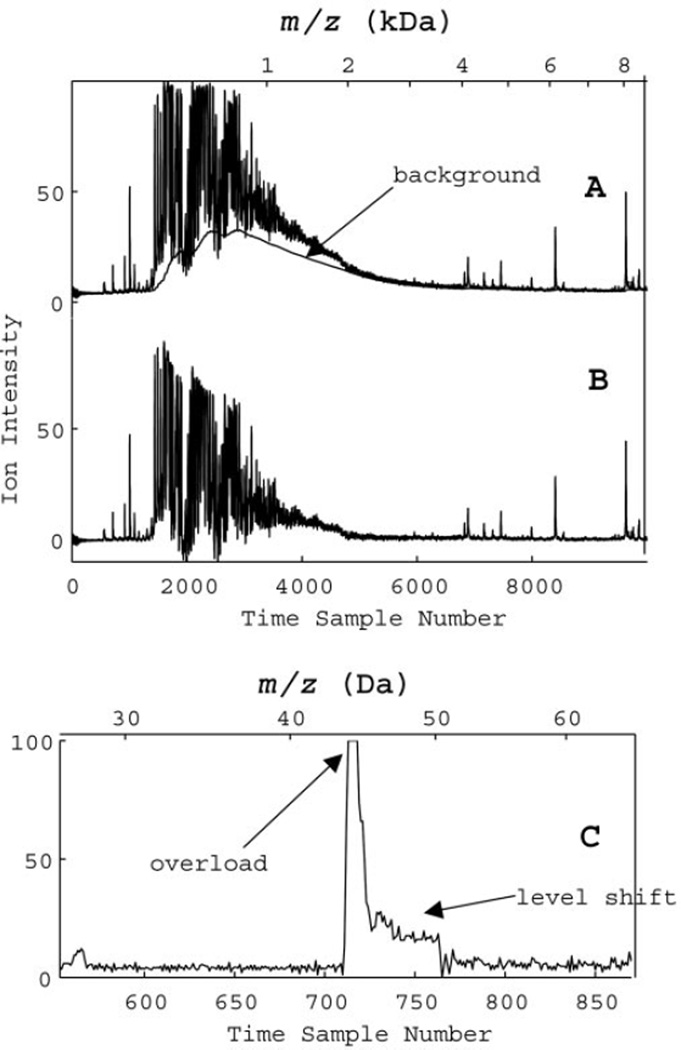
Fig. 1. Accumulated charge model and shift of baseline for SELDI data.
(A), original data (average of 192 laser shots at 16 subpositions) with a background representing 0.05% of the signal accumulating and lasting for 800 time steps. (B), corrected data after an accumulated charge subtraction. (C), detector output after an overload (after a single laser shot) shows a shift that changes suddenly back to the original value. The top axes are horizontal mass scales.
This linear background correction for charge accumulation can be applied to data collected after a single laser shot or to data averaged over many laser shots. We found very little variation in the time constant or in the offset variable in our experiments. The accumulation efficiency, a, apparently depends primarily on the sensitivity settings of the instrument and does not vary much as long as the experimental parameters are kept constant. Of course, the actual baseline magnitude will depend on the masses and concentrations of analytes on the SELDI surface because charge accumulation is an integrated effect of the incoming signals (Eq. 2). The charge accumulation baseline may show significant contributions to the observed intensity of species up to 20 kDa. This baseline subtraction technique has two major advantages over a default variable width convex hull fitting: it has very few adjustable parameters (physically related to instrumental settings); and it does not subtract signal attributable to slowly varying structure. Hence, very broad peaks will not be mistakenly eliminated as background.
We also observed a major nonlinear contribution to the baseline after detector overload events. After an overload event, the baseline shifts for a short time, forming a shelf-like structure. Fig. 1C shows an example of this baseline shift during a single laser shot. The nominal background shifts by a large amount and stays shifted for a time that depends on the duration (and perhaps the height) of the overload. It then suddenly changes back to its original level. These sudden changes often appear as sharp features, peaks or dips, in the averaged signal from many laser shots, making them very difficult to remove from the baseline. They can, however, be corrected on a shot-by-shot basis by subtracting the constant shelve over the detector recovery period (see Materials and Methods). According to the specifications and to our calibration measurements, the recovery rate for the PBS II detector is ~7.5 time points (30 ns) per overload event; therefore, as an example, a 4-point-long detector saturation would shift the baseline for 30 time points after the first overload event (26 points after the last one).
Decreasing the laser fluence or the detector sensitivity can eliminate this nonlinear shift associated with overload events, but that may also reduce the high mass signal below an acceptable value. A shot-by-shot removal of the shifts before averaging of multiple laser shots would eliminate this entirely, although that is not currently an option in the instrument’s standard data acquisition software.
Once the charge accumulation background and nonlinear effects have been eliminated, the remaining noise in the Ciphergen PBS II system appears to be random fluctuations in the detector. These can be reduced considerably by averaging: either over many laser shots or over many time points. Of course, averaging over sample points may decrease the instrument’s resolution. Most analog TOF devices sample at constant time intervals so that the time measurement precision, T/Δt, grows without bound. Once this precision is sufficiently high, any individual mass peak will be distributed over many dwell times. This decreases the signal-to-noise ratio because the inherent random fluctuations per dwell time are constant. Thus, for the high masses, integrating or averaging over many dwell times is a natural correction for random noise that may not decrease the resolution. The default Ciphergen PBS II signal-processing routine (20) uses a variable width moving average filter in the mass domain to gain this increase in the high-mass signal to noise.
The results of a single laser shot on a nominally blank chip as a function of sample time are shown in panels A through C of Fig. 2. The left pane of Fig. 2 also shows a second horizontal axis illustrating the sample to m/z conversion. The unprocessed data (Fig. 2A) have nearly constant amplitude variations of ~3 bits in the analog-to-digital converter. Because this is a single laser shot, Fig. 2A clearly shows that the recorded signal has only a few discrete values as measured by the analog-to-digital converter. Some of these fluctuations in this time sequence are certainly not random. We have distinguished a major component of the noise at the clock frequency, which is presumably attributable to parasitic coupling of the clock to the detector amplifier. This will be substantially reduced for averaging windows greater than a few points, although it would be better eliminated before the average by subtracting of the coherent, period-two component. Other groups (21) have also observed additional sharp features at 4096 clock cycles, which are presumably attributable to switching in a memory bank. Again, these features can best be removed before the moving average, although the moving average will also significantly reduce the effects of these single time step events at high masses (Fig. 2B).
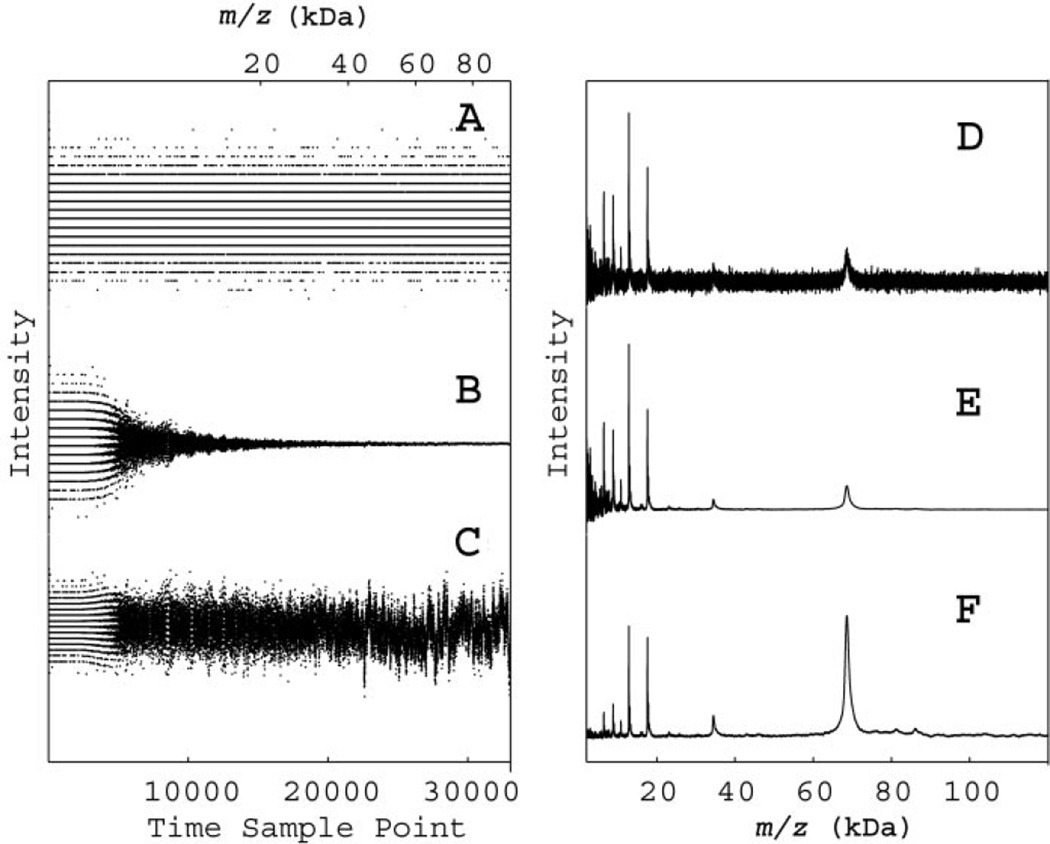
Fig. 2. Data recorded after a single laser shot (A–C), and SELDI signals from a pooled serum sample (D–F).
In panels A–C, the signal intensity is plotted vs time (bottom axis) or m/z (top axis). In panel A, raw data show the bit conversion errors at times when little ion signal was recorded. In panel B, the default variable-width moving average has reduced the bit conversion noise at long times by averaging over many samples. In panel C, the signal has been rescaled by the square root of the number of averaged samples to recover a constant noise amplitude. In panels D–F, the SELDI signal (average of 192 laser shots) from a pooled serum sample are plotted vs m/z. In panel D, the raw data show a rapidly decreasing signal-to-noise ratio at high mass. In panel E, the default variable-width moving average has integrated the signal to reduce the noise at high mass. In panel F, rescaling by the square root of the number of averaged samples, as in panel C, has produced a constant noise value and has considerably enhanced the high mass features.
The effects of the manufacturer’s default moving average on the noise (Fig. 2A) are shown in Fig. 2B. Note that past 6000 time points, the averaging window includes enough points to completely obscure the digitized nature of the input and to rapidly decrease the random fluctuations. According to the default software settings, the window width of the moving average filter changes linearly with mass or quadratically with time sample. Fig. 2C shows a renormalization of the averaged noise from Fig. 2B, where we multiplied the signal by the square root of the window size in time points. In the case of truly random noise, this would produce constant amplitude noise and, as shown in Fig. 2C, is a good approximation. However, there is clearly some structure evident in the averaged noise because the moving average has more effectively removed the high-frequency components. Nevertheless, this rescaling does provide a nearly constant noise amplitude, making it easier to apply peak-picking routines with the global noise threshold, unlike those described by Fung and Enderwick (20).
The effects of this moving average and rescaling of intensity patterns for the mass spectrum of pooled serum sample are illustrated in panels D through F of Fig. 2. Similar to the left panes of Fig. 2, A and B, Fig. 2D shows the unprocessed data and Fig. 2E shows the result after application of the variable width moving average, which enhances the high mass peaks but attenuates the noise nonlinearly. Fig. 2F shows the result of rescaling done similar to Fig. 2C. Although the noise is visible only in the low mass region of Fig. 2E, it is relatively constant (but very small) throughout Fig. 2F. In Fig. 2F, the singly charged albumin peak (68 kDa) and doubly charged albumin (34 kDa) are clearly visible well above the noise. Apparent differences in mass numbers from the expected for albumin are attributable to mass axis calibration errors outside the mass-focusing range. It is important to realize that noise rescaling does not change the local signal-to-noise ratio because both noise and signal are multiplied by the same factor. However, when the constant noise is recovered in the broad mass range by rescaling, the peak magnitudes are proportional to the signal-to-noise ratio for that feature.
The default window width of the moving average filter grows with increasing mass so that it is always ~20% of the typical feature width expected by the manufacturer. We found the default settings for moving average to be consistent with our own measurements of typical increasing edges of mass features, as shown in Fig. 2 of the online Data Supplement. Because instrumental broadening and isotopic broadening cause peaks to spread out more to the high mass side (5, 8, 22), we measured the low mass edge of a variety of peaks in the spectra from the pooled sera by measuring the width from half height on the low mass side to the maximum height. As shown in Fig. 2 of the online Data Supplement, the instrument resolution in this mass-focusing region is nearly constant, although the peaks are broader than predicted by isotopic low-mass half-width, calculated using Protein Prospector® (19). We can therefore expect that application of time series analysis techniques relying on the constant instrumental resolution, such as correlation and convolution filters, would be warranted in the mass-focusing range.
CALIBRATION OF PEAK TIMING
Because TOF measurements relate the mass of an ion to the time that ion takes to reach the detector, the calibration that relates the arrival time to the actual ionic mass is a crucial characterization step. The Ciphergen software readily calculates the appropriate calibration constants based on the designation of specific known peptide peaks. The calibration model assumes a quadratic relationship between the arrival time and the ionic mass. However, these calibration constants are very sensitive to changes in the electronic timing or in the flight path distance. SELDI and MALDI measurements are particularly vulnerable to these types of errors because the ions are created in a fast-moving plume that may change from laser shot to laser shot (5, 11, 18). The effect of the dispersion of velocities in the plume has been minimized in the Ciphergen instrument by use of time-lag mass focusing, but variations in the average plume velocity can still introduce a calibration error. Moreover, the laser beam moves over a relatively large distance (see Fig. 1 in the online Data Supplement), altering the net flight time and introducing additional calibration shifts. Some SELDI instruments now ship with calibration software that corrects for small changes among the different sample spots. Even within a particular sample spot, however, one expects some variation. For a typical MALDI plume velocity of 500 m/s, the average time shift attributable to local height variations on the sample would be 2 ns/µm. Because SELDI samples consist of a collection of randomly oriented crystals tens of micrometers thick, one anticipates time shifts larger than the dwell time. In our single-lasers-hot measurements, we found that shot-to-shot variations at a single subposition were small, typically much less than the line width of the narrowest features in the time-lag-focused mass region. However, when we moved the laser between subpositions, we observed significant apparent shifts in the calibration timing.
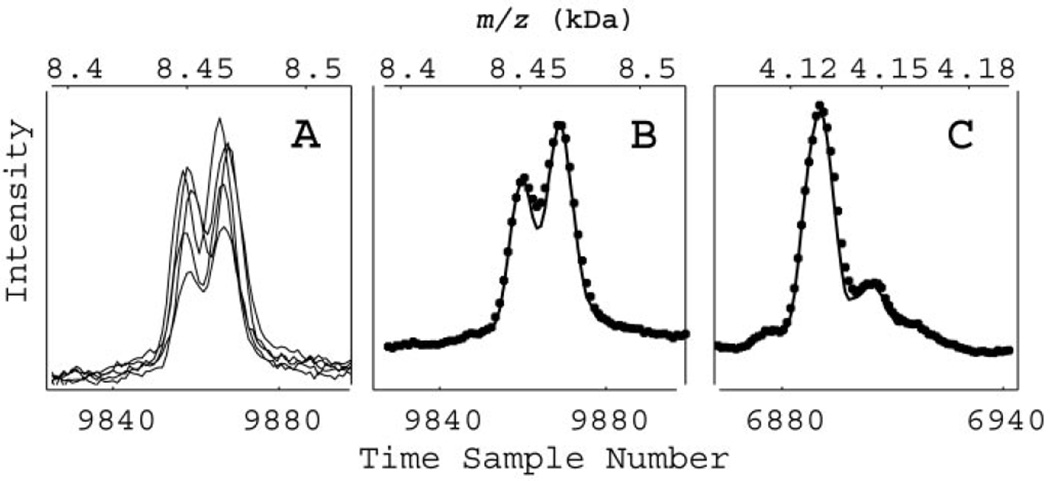
The collection of traces in Fig. 3A, taken at different subpositions on the same sample spot, illustrates what appears to be erratic time shifts. The amplitudes also vary because each of these traces comes from a different subposition on a spot (see Fig. 1 in the online Data Supplement) with apparently different surface coverage by the sample. Adding ~20 of these traces would typically generate a complete spectrum. The small shifts would unnecessarily broaden any features and decrease their peak amplitude. We corrected these shifts by introducing a subposition-dependent time shift derived by maximizing the cross-correlation between each individual trace and the average trace over the narrow doublet near the time point 9860 (8464 Da). When 12 of these shifted traces were then averaged, they produced a spectrum with improved resolution, as shown in Fig. 3B. In addition, the same time shifts can change the mass location of the peak at 4130 Da (time point 6895), as shown in Fig. 3C, and simultaneously improve the mass resolution there. The real benefit of using this correlative procedure (18) to correct for calibration changes between subpositions is that it does not require a known mass; it needs only a single feature with relatively narrow structure and can be easily automated (18). This autocalibration procedure provides an obvious improvement in correcting peak location and intensity. In addition to autocalibration during acquisition, a similar approach can be used for detecting and correcting shifts among different spots and chips, and hence for proper alignment of variables in the data matrix before classification.
Fig. 3. Example of dejittering.
(A), several spectra obtained at various subpositions within the same sample spot showing jitter in peak position. The variation in peak amplitude is typical for single subposition spectra resulting from irregular sample coverage of the chip spot. An autocorrelation dejittering technique enhances the resolution of a high mass doublet (B) and simultaneously improves the lower mass feature (C). ● represent data before autocalibration.
TARGET FILTER DECONVOLUTION
We found that target filter deconvolution can dramatically enhance the resolution in SELDI-TOF data, unmasking nearly obscured features to identify them clearly as peaks. As described in the Materials and Methods, the idea behind this filtering technique is to fit a part of the record that has a single peak on a noisy background (16). If this peak has a shape characteristic of the instrument function and also has a high signal-to-noise ratio, we can use it to create a filter that maps the instrument function into a wavelet of arbitrary shape. Typically, the desired shape is a symmetric peak with a narrower line width. The best filter also takes the spectrum of the noise into account implicitly weighted by the factor ν (Eq. 3). The physical meaning of ν is the ratio of the filtered noise within the bandwidth of the signal wavelet to the integrated signal. For large values of ν, the filter only suppresses noise and does not reshape the signal. After designing the best-fit filter, we then apply it to the entire mass-focusing region, assuming a constant instrumental function and stationary noise. This type of approach may seem to be unwarranted in a TOF spectrum because the resolution changes so rapidly with mass, but most of this variation is simply attributable to the transformation from time to mass. In fact, the instrumental resolution of peaks from masses as small as sodium (23 Da) up to ~12 kDa is nearly constant in time (see Fig. 2 in the online Data Supplement). As also shown Fig. 2 of the online Data Supplement, this filter would not be appropriate outside the mass-focusing range because the instrumental function changes rapidly. However, in a future report, we will show that we can extend the range of a best-fit filter by use of appropriate rescaling of the coefficients.
Even in the best cases, the deconvolution process introduces some artifacts, or spurious peaks, into the filtered signal. The fraction of the signal that is transferred to the artifacts increases for small length filters, for large decreases in linewidth, or for small weighting parameters, ν. Artifacts are distinct from the filtered signal in that their position, amplitude, and width change when the filter length is altered. The phase of the artifacts depends on the filter length; we therefore have geometrically averaged the output of multiple filters with different lengths to detect and eliminate these artifacts.
With the conditions listed in the Materials and Methods, we increased the resolution of the filtered signal by a factor of 2 from the raw signal and simultaneously increased its signal-to-noise ratio by a factor of 2. We used atomic sodium peaks from the very low mass region to create our filters because the observed shape of this atomic ion represents the instrument line shape function; we then successfully applied these filters to peptides as heavy as 9 kDa. This process is successful because the Ciphergen TOF mass spectrometer uses mass focusing that produces a nearly constant time resolution over a wide range of masses, from simple atomic ions of mass 23 Da up to nearly 12 kDa (see Fig. 2 in the online Data Supplement). Thus, when viewed as a time series rather than as a mass spectrum, standard time-series techniques for noise reduction and signal deconvolution become extremely valuable.
The net effect of resolution enhancement via target filtering in the mass-focusing range is that some spectra that appeared to have slight shoulders on large peaks can be deconvoluted to identify clear satellite peaks. Examples of how this filter deconvolutes data from a pooled sera sample are shown in Fig. 4. The sodium ion target region is shown in Fig. 4A. The SELDI instrument assigns this ion a mass of 19, although its mass determination is calibrated by use of much heavier peptides from a calibration mixture (see Materials and Methods). Because the instrument is not well calibrated for low masses, we have identified this monoatomic peak as sodium by use of an independent compositional analysis of the SELDI chips. Sodium occurs only as a single isotope; therefore, this low-mass peak should accurately reproduce the instrument function. The crosses in Fig. 4A show the SELDI data, whereas the diamonds show the desired wavelet form and the solid line shows the filtered result.
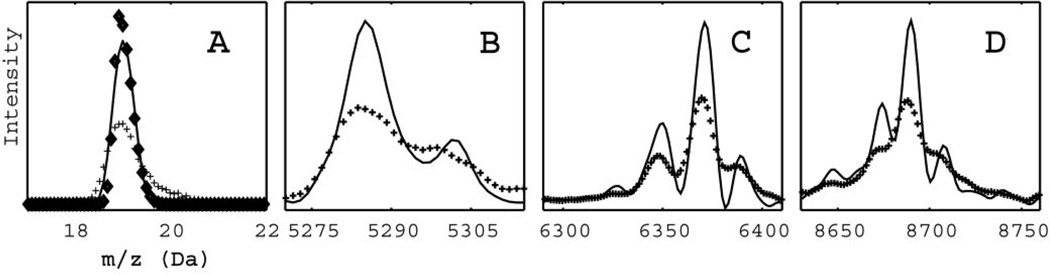
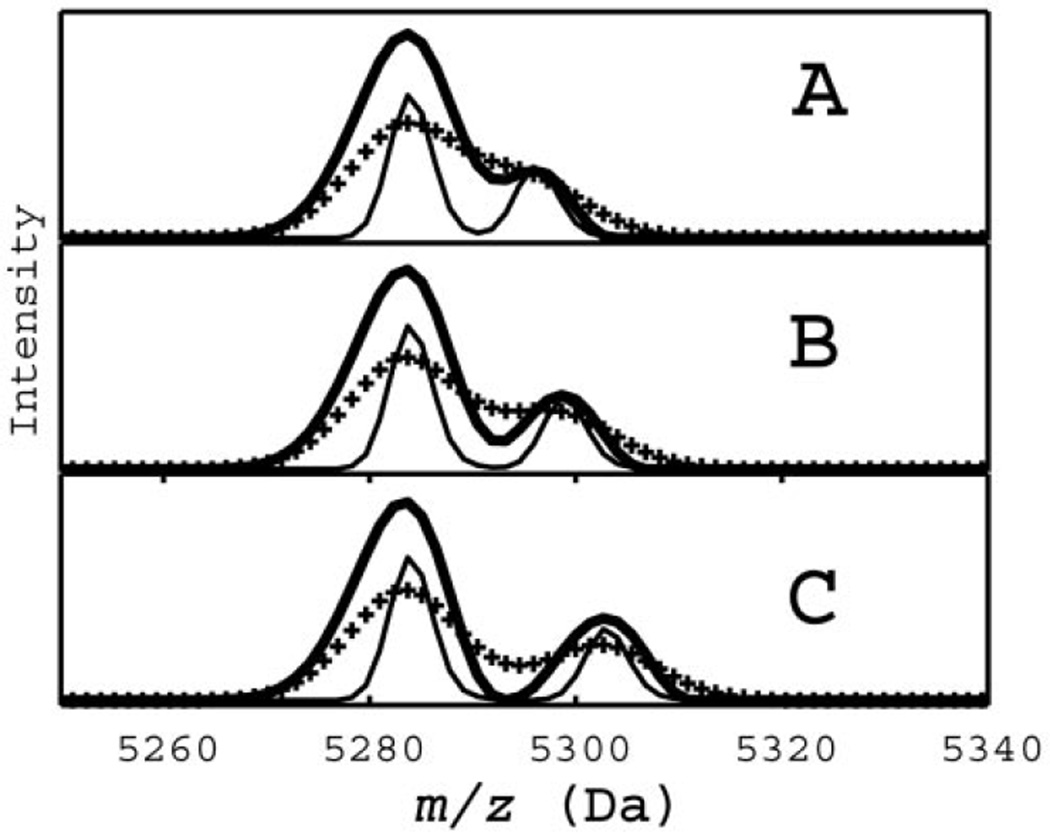
Fig. 4. Use of the atomic sodium target to construct a filter.
In (A), the SELDI data (✚) for a pooled serum are filtered (solid line) to a shape close to the target (♦). Applying this filter to data later in the time series reveals the doublet structure at 5300 Da (B) and quartets at 6370 Da (C) and 8700 Da (D).
In panels B, C, and D of Fig. 4, the raw data are represented by crosses and the signals deconvoluted by the filter by solid lines. In Fig. 4C, the small structure near 6400 Da actually consists of four distinct peaks. Similarly, the structure near 8700 Da in Fig. 4D also consists of four distinct peaks. The peaks in Fig. 4D correspond to small sinapinic acid adducts of large intensity peptide peaks preceding them on the left by 223 mass units (not shown). The increase in resolution suggests a clear chemical interpretation of deconvoluted features simply from a visual inspection. The masses of the peaks to the right of the highest intensity parent ion are separated by multiples of the sodium mass (22 Da) and thus represent sodium adducts. The two peaks to the left of a parent are shifted by −18 Da, suggesting neutral losses of water or ammonia, or by −44 Da, suggesting neutral loss of carbon dioxide. Interestingly, the majority of small peaks in the sera spectrum can be identified as adducts or neutral losses of a relatively small number of distinct peptides.
One potential danger with deconvolution filters is that they can introduce artifacts in the vicinity of large peaks, which might be mistaken for real satellite features. Consequently, small peaks in the vicinity of larger structures in a deconvoluted signal should be questioned. However, because our filter uses only data from lower masses to generate the deconvoluted spectrum (Eq. 4), most artifacts should occur primarily on the high mass side of large peaks. One easy check for the legitimacy of a small structure is to create a reversed filter that uses only signals from higher masses. These filters have inherently less resolution because of the tendency of various broadening mechanisms to skew peaks toward higher masses (5, 8, 22), but their artifacts will occur at different masses. Fig. 3 in the online Data Supplement shows a deconvoluted structure obtained with use of both high and low mass filters and has only one small feature that is an artifact. The small peak at 9820 Da in the filtered signal in Fig. 3 of the online Data Supplement (solid line) is apparently an artifact attributable to the preceding peptide parent peak at 9650 Da, which has an amplitude that is 100 times higher. This feature disappears when the data are filtered with the filter that uses only higher masses (dashed line in Fig. 3 of the online Data Supplement). In general, peaks identified by use of both filtering directions are valid; in those cases, the preferred spectrum will be the output of the low-mass filter only.
As a second test of the validity of our deconvolution filter, we constructed simulated data similar to the doublet in Fig. 4B. The unbroadened simulated peaks are shown in Fig. 5 as a thin line, the broadened data as crosses, and the filtered data as a thick solid line. In the three cases (panels A–C), we varied the separation between the two simulated peaks by approximately one third of a linewidth from the case where the smaller peak appears as a slight bump (Fig. 5A) to where it is clearly visible (Fig. 5C). In all three cases, the filter output accurately located the known position of the underlying simulated data. Note that filter coefficients used on the simulated data were calculated from the experimental atomic shape in Fig. 4A.
Fig. 5. A doublet, similar to that observed experimentally in Fig. 4B, modeled as the sum of two gaussian lines (thin line) with increasing separation (A–C).
The filter constructed for the data in Fig. 4A successfully deconvoluted the simulated broadened data (✚) to produce a filtered spectrum (thick solid line). This correctly reproduces the time positions of the original model peaks (thin solid line).
In summary, we have shown that processing of SELDI-TOF data in the time domain substantially improves the data reproducibility to enhance data interpretation and to ease comparisons among data sets. These time-series analysis techniques, as tailored for SELDI data, can provide automatic, real-time correction of artifacts introduced by SELDI hardware and can suppress instrumental noise, correct peak timing errors, enhance high mass signals, and deconvolute overlapped peaks. Further improvements, such as extrapolating these techniques to data outside the mass-focusing region and automating the optimization of the filtering parameters, will be addressed in future work.
Supplementary Material
Acknowledgments
This work was supported by the Virginia Commonwealth Research Technology Fund (IN2002-03), an SBIR award from the National Cancer Institutes (CA101479), and a grant from the National Cancer Institutes Early Detection Research Network (CA85087). We are grateful to staff members at INCOGEN, Inc., for organizing the data-sharing site and the raw SELDI data archives.
Footnotes
Nonstandard abbreviations: MALDI, matrix-assisted laser desorption/ionization; SELDI, surface-enhanced laser desorption/ionization; PAGE, polyacrylamide gel electrophoresis; TOF, time-of-flight; and MS, mass spectrometry.
References
- 1.Wright GL, Jr, Cazares LH, Leung S-M, Nasim S, Adam B-L, Yip T-T, et al. Surface enhanced laser desorption/ionization (SELDI) mass spectrometry: a novel protein biochip technology for detection of prostate cancer biomarkers in complex protein mixtures. Prost Cancer Prost Dis. 1999;2:264–276. doi: 10.1038/sj.pcan.4500384. [DOI] [PubMed] [Google Scholar]
- 2.Ball G, Mian S, Holding F, Allibone RO, Lowe J, Ali S, et al. An integrated approach utilizing artificial neural networks and SELDI mass spectrometry for the classification of human tumors and rapid identification of potential biomarkers. Bioinformatics. 2002;18:395–404. doi: 10.1093/bioinformatics/18.3.395. [DOI] [PubMed] [Google Scholar]
- 3.Petricoin EF, III, Ardekani AM, Hitt BA, Levine PJ, Fusaro VA, Steinberg SM, et al. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;359:572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 4.Adam B-L, Qu Y, Davis JW, Ward MD, Clements MA, Cazares LH, et al. Serum protein fingreprinting coupled with a pattern-matching algorithm distinguishes prostate cancer from benign prostate hyperplasia and healthy men. Cancer Res. 2002;62:1609–1614. [PubMed] [Google Scholar]
- 5.Cotter RJ. Time-of-flight mass spectrometry. Washington, DC: ACS; 1997. p. 326. [Google Scholar]
- 6.Merchant M, Weinberger SR. Recent advancements in surface-enhanced laser desorption/ionization-time of flight-mass spectrometry. Electrophoresis. 2000;21:1164–1167. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1164::AID-ELPS1164>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 7.Emmert-Buck MR, Gillespie JW, Paweletz CP, Ornstein DK, Basrur V, Apella E, et al. An approach to proteomic analysis of human tumors. Mol Carcinog. 2000;27:158–165. [PubMed] [Google Scholar]
- 8.Schiller J, Arnold K. Mass spectrometry in structural biology. In: Meyers RA, editor. Encyclopedia of analytical chemistry. New York: Wiley; 2000. pp. 1–26. [Google Scholar]
- 9.Jayaram R. Mass spectrometry, theory and applications. New York: Plenum Press; 1966. pp. 43–118. [Google Scholar]
- 10.Loboda AV, Krutchisky AN, Bromorski M, Ens W, Standing KG. A tandem quadrupole/time-of-flight mass spectrometer with a matrix-assisted laser desorption/ionization source: design and performance. Rapid Commun Mass Spectrom. 2000;14:1047–1057. doi: 10.1002/1097-0231(20000630)14:12<1047::AID-RCM990>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 11.Gusev AI, Wilkinson WR, Proctor A, Hercules DM. Improvement of signal reproducibility and matrix/comatrix effects in MALDI analysis. Anal Chem. 1995;65:1034–1041. [Google Scholar]
- 12.Cordingley HC, Roberts SLL, Tooke P, Armitage JR, Lane PW, Wu W, et al. Multifactorial screening design and analysis of SELDI-TOF ProteinChip® array optimization experiments. Biotechniques. 2003;34:364–373. doi: 10.2144/03342rr07. [DOI] [PubMed] [Google Scholar]
- 13.Baggerly KA, Morris JS, Coombes KR. Reproducibility of SELDI-TOF protein patterns in serum: comparing data sets from different experiments. Bioinformatics. 2004;20:777–785. doi: 10.1093/bioinformatics/btg484. [DOI] [PubMed] [Google Scholar]
- 14.Coombes KR, Fritsche HA, Clarke C, Chen J-N, Baggerly KA, Morris JS, et al. Quality control and peak finding for proteomics data collected from nipple aspirate fluid by surface-enhanced laser desorption and ionization. Clin Chem. 2003;49:1615–1623. doi: 10.1373/49.10.1615. [DOI] [PubMed] [Google Scholar]
- 15.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 16.Robinson EA, Treitel S. Statistical communication and detection. London: Griffin; 1967. pp. 249–283. [Google Scholar]
- 17.Marshall AG. Fourier transform in NMR, optical and mass spectrometry. New York: Elsevier; 1990. p. 450. [Google Scholar]
- 18.Nicola AJ, Gusev AI, Proctor A, Hercules DM. Automation of data collection for matrix-assisted laser desorption /ionization mass spectrometry using a correlative analysis algorithm. Anal Chem. 1998;70:3213–3219. doi: 10.1021/ac980143s. [DOI] [PubMed] [Google Scholar]
- 19.Senko MW, Beu SC, McLafferty FW. Determination of monoisotopic masses and ion populations for large biomolecules from resolved isotopic distributions. [accessed October 1, 2004];J Am Soc Mass Spectrom. 1995 6:229–233. doi: 10.1016/1044-0305(95)00017-8. http://prospector.ucsf.edu/ucsfhtml4.0/msiso.htm. [DOI] [PubMed] [Google Scholar]
- 20.Fung ET, Enderwick C. ProteinChip clinical proteomics: computational challenges and solutions. Biotechniques. 2002;32(Suppl):34–41. [PubMed] [Google Scholar]
- 21.Baggerly KA, Morris JS, Wang J, Gold D, Xiao L-C, Coombes KR. A comprehensive approach to the analysis of matrix-assited laser desorption/ionization-time of flight proteomics spectra from serum samples. Proteomics. 2003;3:1667–1672. doi: 10.1002/pmic.200300522. [DOI] [PubMed] [Google Scholar]
- 22.Zenobi R, Knochenmuss R. Ion formation in MALDI mass spectrometry. Mass Spectrom Rev. 1999;17:337–366. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.