Abstract
Microbial infections are recognized by the innate immune system both to elicit immediate defense and to generate long-lasting adaptive immunity. To detect and respond to vastly different groups of pathogens, the innate immune system uses several recognition systems that rely on sensing common structural and functional features associated with different classes of microorganisms. These recognition systems determine microbial location, viability, replication and pathogenicity. Detection of these features by recognition pathways of the innate immune system is translated into different classes of effector responses though specialized populations of dendritic cells. Multiple mechanisms for the induction of immune responses are variations on a common design principle wherein the cells that sense infections produce one set of cytokines to induce lymphocytes to produce another set of cytokines, which in turn activate effector responses. Here we discuss these emerging principles of innate control of adaptive immunity.
Innate control of adaptive immunity is now a well-established paradigm. First introduced by Charles Janeway Jr. in 1989 (ref. 1), it states that recognition of conserved features of microbial pathogens by the innate immune system is mediated by pattern-recognition receptors (PRRs), which detect conserved pathogen-associated molecular patterns (PAMPs), such as bacterial and fungal cell-wall components and viral nucleic acids. Detection of PAMPs by PRRs leads to the induction of inflammatory responses and innate host defenses. In addition, the sensing of microbes by PRRs expressed on antigen-presenting cells, particularly dendritic cells (DCs), leads to the activation of adaptive immune responses. Several classes of PRRs have now been identified and characterized in some detail. These include Toll-like receptors (TLRs), nucleotide-binding oligomerization domain (Nod)-, leucine-rich repeat–containing receptors (NLRs), RIG-I-like receptors (RLRs), C-type lectin receptors (CLRs) and AIM-2 like receptors, as well as a family of enzymes that function as intracellular sensors of nucleic acids, including OAS proteins and cGAS2–4. The physiological role of different PRRs in the sensing of microbes and the induction of adaptive immune responses continues to be investigated, but in general, the available evidence unequivocally supports the view that PRR-mediated sensing instructs the adaptive immune responses: specifically, PRRs determine the origin of the antigens recognized by the antigen receptors expressed on T cells and B cells, as well as determine the type of infection encountered, and instruct lymphocytes to induce the appropriate effector class of the immune response.
Studies over the past decade have also revealed several important aspects of immune system’s function that require an expanded view of the innate control of adaptive immunity. For example, the type 2 immune response induced by parasitic worms and allergens appears to be largely independent of PRRs, perhaps because multicellular parasites lack molecular structures that are both conserved across different groups of parasites and distinct from the host organism. Similarly, allergens are not microbial in origin, lack conserved structural features and induce type 2 immune responses through mechanisms that remain largely unknown5. Another big puzzle is the apparent ability of the immune system to distinguish between beneficial commensal microorganisms and pathogenic microorganisms. Both types of microbes express PAMPs and are detectable by PRRs; however, the outcome of their recognition can depend on additional characteristics, such as invasiveness and production of toxins. Recent progress in understanding the complexity and diversity of commensal microbiota suggests that perhaps the classic notion of ‘pathogens’ is in fact applicable only to rare outliers of the entire spectrum of the microorganisms that can colonize a mammalian host. However, this does not mean that the immune system is concerned only with these few bad apples; even the normal commensal inhabitants need to be continuously monitored and somehow ‘managed’ by the immune system to prevent their outgrowth and mischief6–8.
Many discoveries made in the field over the past decade call for a more complete and nuanced picture of innate instruction of the adaptive immune responses. Here we review some of the recent developments in studies of innate control of adaptive immunity. We highlight some emerging concepts that expand the pattern-recognition paradigm. Finally, we discuss some of the major gaps and unknowns.
Recognition by the innate immune system
Microbial targets of recognition by PRRs are structurally diverse and include complex polysaccharides, glycolipids, lipoproteins, nucleotides and nucleic acids. Several families of PRRs detect these structures through the use of distinct ligand-recognition domains, including leucine-rich repeats, C-type lectin domains and various nucleic acid–binding domains. In addition to being characterized by their structure and specificity, PRRs are characterized by their tissue-specific expression, as well as by their localization in distinct cellular compartments, including the plasma membrane, endosomes, lysosomes and cytosol9. These differences in expression patterns are related to two general modes of recognition by the innate immune system and immune responses: cell-intrinsic recognition is mediated by intracellular cytosolic sensors that operate in infected cells. All cell types that can be infected by a given class of pathogens express the corresponding sensors (for example, sensors of viral nucleic acids). Cell-extrinsic recognition does not require that the cell expressing PRRs be infected. Instead, the PRRs involved in cell-extrinsic recognition (for example, TLRs and CLRs) are expressed in cells specialized in pathogen detection, such as surface epithelia and myeloid cells.
The detection of extracellular pathogens is mediated mainly by PRRs expressed on the plasma membranes of macrophages, DCs and other cell types. These PRRs, including TLR1, TLR2, TLR4, TLR5 and TLR6, and CLRs, including dectin-1, dectin-2 and mincle, are specialized in the recognition of common components of bacterial and fungal cell walls, such as lipopolysaccharides, bacterial lipopeptides, lipoteichoic acids, flagellin, glycolipids and β-glucans. Cell-extrinsic recognition of viruses is mediated by TLR3, TLR7, TLR8 and TLR9, which are expressed in the endosomes, where they detect viral nucleic acids. Recognition by TLRs and CLRs is generally not dependent on microbial viability, replication or invasiveness and thus it can detect a broad spectrum of microbial PAMPs regardless of their origin (that is, whether the PAMPs are produced by pathogenic or commensal microbes, dead or alive microbes, and replicating or quiescent microbes).
Cytosolic sensors of viral nucleic acids detect viral RNA (RLRs) and viral DNA (cytosolic DNA sensors) in infected cells10–12. Intracellular bacteria can be sensed by the endoplasmic reticulum membrane-resident adaptor STING through the detection of bacterial cyclic diguanylate monophosphate13. The same pathway is activated by the related dinucleotide cyclic AMP-GMP, which is produced by cGAS in response to the intracellular recognition of DNA3. Invasive bacteria are also recognized by the cytosolic PRRs Nod1 and Nod2, as well as by other NLRs that detect bacterial PAMPs (such as peptidoglycan fragments and flagellin) introduced into the cytosol14,15. Some NLRs detect intracellular bacterial and viral infections and induce the inflammasome complex, which leads to caspase-1-dependent processing and secretion of members of the interleukin 1 (IL-1) family of cytokines, such as IL-1β and IL-18. The recognition of microbes by the intracellular sensors generally depends on microbial invasiveness, viability and, in some cases, replication16–19. Thus, this class of PRRs is more restricted in what they can sense. Most PAMPs can be sensed both by cell-extrinsic pathways (mediated by TLR) and by cell-intrinsic pathways (mediated by RLRs, NLRs and cytosolic DNA sensors): this is the case for viral nucleic acids, lipopolysaccharide (LPS) and flagellin. Presumably the two modes of recognition have evolved to provide different signals to the immune system (discussed below).
In addition to the recognition of structural features (that is, direct PRR-mediated recognition of microbial ligands), the innate immune system can detect pathogens through the recognition of functional features, by sensing functional characteristics indicative of a pathogen’s presence, such as common effects of virulence factors20. The best available example of this mode of sensing is activation of the NLRP3 inflammasome by bacterial pore-forming exotoxins as well as by viroporins, which form ion channels in the membrane of host intracellular organelles21–23. Sensing of enzymatic activities of allergens is another example of functional feature recognition. It is likely that multicellular parasites are detected by the immune system mainly through this mode of sensing as well. This type of detection ‘by proxy’ is analogous to the function of Guard proteins in plant immunity24. There are several advantages of this mode of recognition. First, it eliminates the need for a very large number of specific receptors for each of the structurally diverse virulence factors, toxins or allergens. Instead, the common activities of these agents are detected and trigger the response. Second, the recognition of functional features, when combined with pattern recognition, can inform the immune system that the microbe is a pathogen. Finally, the recognition of functional features may be the only available mode of recognition of multicellular parasites that lack structurally invariant targets for direct pattern recognition. Instead, they may have some common activities, such as excretion of cysteine proteases and disruption of basement membranes. These types of effects on the host tissues may also be shared with certain classes of allergens and may be detectable through common sensing pathways. These pathways most probably overlap with pathways that sense tissue damage, which are designed to detect common consequences of tissue damage rather than the structure of the damaging agents25. Thus, PRR-mediated recognition in combination with the recognition of functional features leads to the type 1 immune response to microorganisms (viruses, bacteria, fungi and possibly protozoa), whereas the recognition of functional features in the absence of PRR signal leads to the type 2 immune response and tissue-repair response to macroparasites. Indeed, the case has been made that type 2 immunity to parasitic worms is in many ways a specialized form of tissue-repair response26,27. Finally, tissue damage induces a tissue-repair response that can involve the activation of specialized arms of innate and adaptive immunity, such as activation of regulatory T cells, group 2 innate lymphoid cells (ILC2 cells), eosinophils and M2 macrophages but does not lead to adaptive immunity28–31 (Fig. 1).
Figure 1.
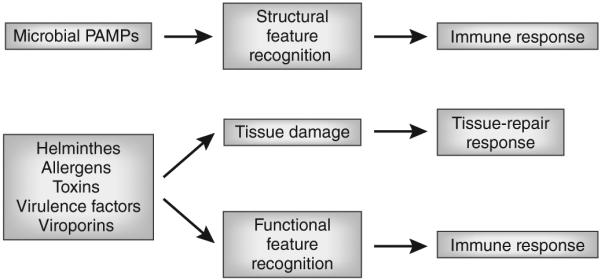
Recognition of structural and functional features. Sensors of the innate immune system recognize infectious agents through detection of unique molecular structures associated with the pathogens (structural feature recognition) or through detection of the characteristic functional activities (functional feature recognition) associated with the infection. The process of infection is often accompanied by tissue damage caused by the pathogens or by the immune response to pathogens. Tissue damage induces a tissue-repair program that can include some specialized immune responses.
In addition to pattern recognition, another strategy for recognizing structural features is through missing-self recognition32. This mode of discriminating self versus non-self is ancient; it is used, for example, by bacterial restriction-modification system to defend against parasitic DNA of phages and other bacteria. In this strategy, methylation of adenine nucleotides in specific DNA sequences protects self DNA from cleavage by restriction endonucleases, thereby permitting destruction of only foreign DNA. In mammalian immunity, missing-self recognition is important in multiple settings. This strategy is used by natural killer (NK) cells to detect infected or stressed cells32. Inhibitory receptors on NK cells containing an immunoreceptor tyrosine-based inhibitory motif detect major histocompatibility complex class I molecules on target cells to avoid killing healthy uninfected ‘self’ cells. Viral infection and various forms of stress reduce the surface expression of major histocompatibility complex class I, which flags these cells for NK cell– mediated lysis33. Other examples of this include macrophages that use inhibitory receptors based on immunoreceptor tyrosine-based inhibitory motifs to detect self ligands to avoid the phagocytosis and killing of normal cells34; the complement system uses a similar strategy to prevent lysis of cells that express complement inhibitory proteins, such as CD46 and CD55; factor H of the alternative complement pathway detects sialic acids present on host cells but absent on most bacteria and fungi and inhibits activation of the C3 convertase of the alternative pathway. Sialic acids are also detected by the Siglec family of inhibitory receptors, such as CD22 expressed on B cells35, which helps to determine the origin of the antigen recognized by the B cell antigen receptor. It is interesting to note that missing-self recognition is often used to inhibit effector responses, such as phagocytosis, cell killing and complement activation, which indicates that missing-self–recognition pathways generally operate along with the pathways that activate these effector responses. In some cases, the activating pathways are mediated by receptors containing an immunoreceptor tyrosine-based activation motif; in others, they are induced by PRR-based pathways (as is the case of the PRR properdin in the alternative complement pathway36).
In summary, different modes of recognition by the innate immune system form distinct layers of sensing by the immune system that provide requisite information to the adaptive immune system. Working in different combinations, the sensing pathways of the innate immune system instruct the activation of relevant effector responses of the adaptive immune system.
Recognition in different compartments
Microbial pathogens enter the host via mucosal surfaces, through a breach in the skin or via bites of insect vectors. To recognize and respond to invasion by vastly different classes of infectious agents, innate sensors are strategically located in distinct anatomical, tissue, cellular and subcellular compartments. The anatomical compartment in which the pathogen is recognized informs the host of the degree of threat it poses. Microorganisms in the lumen of the gut do not trigger inflammation, whereas those that have crossed the epithelial layer induce a local inflammatory response. Moreover, pathogens in the bloodstream signify a breach in barrier and are met with a full-blown systemic response. This compartment-specific outcome of sensing by the innate immune system is mediated by distinct sentinel cells that are strategically located to detect the location of pathogens.
The first surveillance system encountered by pathogens entering the host through mucosal surfaces is the supra-epithelial phagocytes. Specialized mononuclear phagocytes provide surveillance in certain mucosal surfaces above the epithelial layer. In the airways, alveolar macrophages are situated inside the lumen for access to airborne pathogens, whereas in the intestine and respiratory tracts, DCs form tight junctions with the epithelial cells and extend dendrites into the lumen37.
The epithelial cells constitute the next level of surveillance and are often the target of replication by a variety of pathogens. Even a non-epithelium-tropic pathogen must cross the epithelial barrier to access its replicative niche. Thus, the recognition of pathogens by epithelial cells informs the host of a breach in the first barrier. Epithelial cells are equipped with PRRs that induce chemokines able to recruit circulating leukocytes to initiate an innate defense. Colonic epithelial cells express small amounts of TLRs, and some TLRs, including TLR2, TLR3, TLR4 and TLR5, are selectively expressed on the basolateral surface of intestinal epithelial cells38. This expression pattern ensures that only invasive bacteria, but not luminal bacteria, trigger TLR stimulation in the intestinal mucosa. In addition, in response to damage or stress, epithelial cells release cytokines, including IL-25, IL-33, IL-1α and TSLP, to activate local ILCs and memory lymphocytes39.
Beneath the epithelial layer, in the lamina propria, reside DCs, macrophages and mast cells that are specialized in detecting pathogens that have crossed the epithelial barrier. These cells express PRRs that induce the secretion of inflammatory cytokines and chemokines that facilitate the recruitment of monocytes, neutrophils, eosinophils and basophils from the circulation. TLR5 expressed on the surface of DCs detects extracellular bacteria through flagellin, activating the release of cytokines, chemokines and antimicrobial peptides40. If the bacterium can invade the host cell successfully, flagellin in the cytosol is recognized by NAIP5 and NAIP6, which physically associate with NLRC4 to trigger activation of inflammasomes41,42. This signifies to the host that the bacteria has secretion systems capable of injecting effector molecules into the cytosol, and such infection is met with a greater degree of urgency, which causes the activation of inflammasomes and caspase-11 and the induction of pyroptosis43. The amplification in the number of leukocytes at the site of infection enables rapid containment of replicating pathogens through phagocytosis, secretion of toxic granules, and lysis of pathogens and infected cells.
When such local defenses are insufficient to contain the pathogen, the immune system engages a systemic-level acute-phase response to combat the spreading pathogen. Bacteria in the bloodstream are recognized immediately by monocytes and neutrophils through TLRs, NLRs and CLRs. Activation of these PRRs leads to enhanced phagocytosis, degranulation, respiratory burst and killing of bacterial, fungal and protozoan pathogens44. Engagement of TLRs also induces the formation of neutrophil extracellular traps consisting of extruded DNA, which entrap bacteria and mount the rapid induction of inflammatory cytokines and chemokines45. The inflammatory cytokines IL-1β, TNF and IL-6 act on the liver and the central nervous system. In the brain, these cytokines induce sickness behavior and fever46. In the liver, these cytokines induce the production of acute-phase response proteins, including C-reactive protein, serum amyloid protein and mannose-binding protein, to promote pathogen clearance through complement activation and phagocytosis47.
A parallel anatomical regulation of innate sensing exists for viruses. Viruses that infect the epithelial cells trigger cytosolic nucleic acid sensors to induce local type 1 interferon response. In addition, secreted cytokines enter the circulation and induce interferon-stimulated genes in distal tissues to increase hematopoiesis and prepare for systemic viral spread48. When this local response fails to restrict the virus replication, the viral load in the bloodstream, or ‘viremia’, triggers a robust systemic type 1 interferon response. The systemic virus infection is patrolled by plasmacytoid DCs. These plasmacytoid DCs are absent from peripheral tissues at steady state, but are found in the circulation, are constitutively localized in the red pulp of the spleen49,50 and are specialized in responding to blood-borne viruses51. Once TLRs are engaged in plasmacytoid DCs, they rapidly secrete large amounts of type 1 interferons, which are key to suppressing viral load throughout the body.
Although the mechanisms by which multicellular parasites are detected are not yet known, the defense strategies used also demonstrate clear compartment-specific effects. Thus, the detection of macroparasites in the intestinal and respiratory tracts promotes their expulsion through increased production of mucus and peristalsis, whereas the detection of parasitic eggs in the tissues can promote their sequestration through granuloma formation52. Finally, the detection of tick bites triggers basophil-mediated acquired tick resistance53,54.
The ‘choice’ of effector responses
The various pathways and modes of recognition by the innate immune system discussed above provide information about microbial presence, viability, replication, location and virulence. Each of these parameters of recognition by the immune system, as well as their combinations, are critical for the choice of the appropriate effector response. How the immune system interprets these signals is not yet fully understood, but here we review several possible mechanisms.
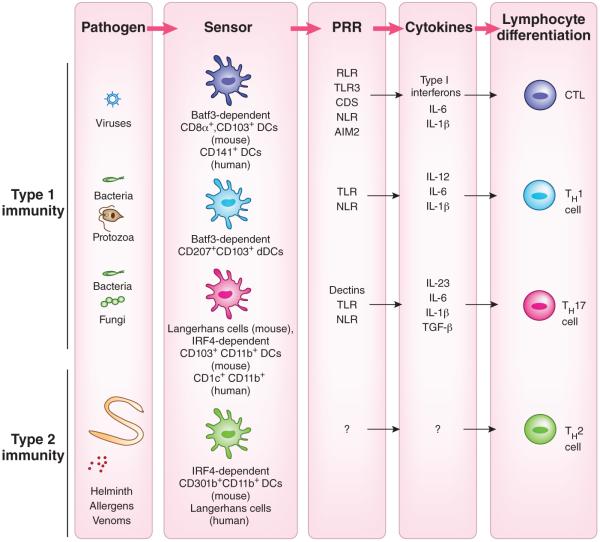
Because DCs have a critical role in the initiation of T cell–mediated responses, it has long been suspected that different DC populations might be specialized for the induction of different T cell effector responses. Much data have accumulated over the past decade on DC functional heterogeneity, including DC subset–specific expression of PRRs and production of signals, including cytokines, involved in differentiation of T cells into effector cells55–57. In addition, several transcriptional master regulators of DC lineages have been defined, which has provided the means for elucidating their functions in vivo58,59. Tissue-resident DCs consist largely of Langerhans cells, CD103+ DCs and CD11b+ DCs. Langerhans cells marked by CD207 reside in the stratified squamous epithelium of the skin and mucosal tissues. Within the dermis and submucosa, CD103+CD207+CD11b− DCs (developmentally dependent on the transcription factor Batf3) and CD301b+CD207−CD11b+ DCs (functionally dependent on the transcription factor IRF4) are present. In the intestine and lung, a third subset of DCs are found beneath the epithelial layer: CD103+CD11b+ DCs (IRF4 dependent)60,61. All four subsets of tissue-resident DCs migrate into the draining lymph nodes and contribute in a distinct manner to priming responses of the adaptive immune system. In addition to the migrant DCs, within lymphoid organs CD8α+ DCs (Batf3 dependent) and CD11b+ DCs are constitutively present. Studies of mice with specific deletions of subpopulations of DCs have revealed an emerging view of the unique capacity of DC subpopulations to generate different classes of T cell responses (Table 1).
Table 1.
In vivo requirement for DC subsets in T cell responses
| T cell class |
Tissue | Antigen pathogen |
Adjuvant | Route | DC subset studied |
TF tested | DC subset required? |
DC subset sufficient? | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| CTL | Lung | OVA-coated apoptotic cell |
± Poly(I:C) | i.n. | CD103+ | Batf3 | Yes | NA | 65 |
| Lung | Soluble OVA | None | i.n. | CD103+ | Batf3 | Yes | NA | 65 | |
| Lung | Soluble OVA | None | i.t. | CD103+ | NA | NA | Yes (AT) | 64 | |
| Lung | SeV | None | i.n. | CD103+ | Batf3 | Yes | NA | 63 | |
| Systemic | Lm-OVA | None | i.v. | CD8α+ | Batf3 | Yes | NA | 74 | |
| Intraperitoneal | T. gondii | None | i.p. | CD8α+CD103+ | Batf3 | Yes | NA | 73 | |
| Skin | OVA/Eα-expressing C. albicans |
None | s.s. | CD207+ dDC | Batf3 | Yes | NA | 69 | |
| Systemic or peritoneal cavity |
DEC205-OVA | Anti-CD40 | i.v. | CD8α+ | NA | NA | Yes (Ab targeting) | 133 | |
| Skin | OVA (self) | None | Skin (keratino- cyte-expressed) |
CD103+ | NA | Yes | Yes (coculture) | 134 | |
| Lung | OVA | PR8 influenza |
i.n. | CD103−
CD11b+ |
NA | NA | Yes (coculture) | 135 | |
| Lung | X-31 influenza | None | i.n. | CD207+ | NA | Yes (partial ↓) | Yes (coculture) | 136 | |
|
| |||||||||
| TH1 cells |
Skin | OVA/Eα-expressing C. albicans |
None | s.s. | CD207+ dDC | NA | Yes | NA | 69 |
| Skin | MOG | CFA/PTX | s.c. | CD103+CD207+ | GM-CSF | Yes | Yes (coculture) | 76 | |
| Skin | MOG | CFA/PTX | s.c. | CD103+CD11b− | Batf3 | No | NA | 75 | |
|
| |||||||||
| TH17 | Skin | MOG | CFA/PTX | s.c. | CD103+CD11b− | Batf3 | No | NA | 75 |
| cells | Skin | MOG | CFA/PTX | s.c. | CD103+CD207+ | GM-CSF | Yes (partial ↓) | NA | 76 |
| Intestine | NA | NA | TNBS-DSS colitis | CD103− | NA | NA | Yes (AT) | 137 | |
| Intestine | NA | NA | NA | CD103+CD11b+ | Notch2, Rbpj |
Yes (partial ↓) | NA | 84 | |
| Intestine | None | None | NA | CD103+CD11b+ | IRF4 | Yes (partial ↓) | NA | 60 | |
| Lung | A. fumigatus | None | i.n. | CD103−
CD11b+CD24+ |
IRF4 | Yes (partial ↓) | NA | 60 | |
| Intestine | NA | NA | NA | CD103+CD11b+ | NA | Yes (↓ in huLangDTA) |
NA | 83 | |
| Skin | OVA- and/or Eα-expressing C. albi- cans, anti-huLangerin- conjugated OVA and/or Eα |
None Ea | e.c., i.p. | LC | NA | Yes | Yes (Ab targeting) |
69 | |
| Intestine | OVA | LPS+ anti- CD40 |
i.p. | CD103+CD11b+ | IRF4 | Yes | Yes (coculture) | 61 | |
|
| |||||||||
| TH2 cells |
Lung | HDM (high dose) | HDM | i.t. | moDC CD103−
CD11b+ |
NA | NA | Yes (AT) | 138 |
| Skin | OVA, N. brasiliensis |
Papain none |
s.c. | CD301b+ | NA | Yes | No | 86 | |
| Skin | OVA, N. brasiliensis |
Papain none |
s.c. | PD-L2+ | IRF4 | Yes | No | 87 | |
| Skin | FITC | Acetone + dibu- tylphthalate |
Skin | CD301b+ | NA | NA | Yes (AT) | 88 | |
Abbreviations: OVA, ovalbumin; MOG, myelin oligodendrocyte glycoprotein; s.c., subcutaneous; i.t., intratracheal; i.p., intraperitoneal; s.s., skin scarification; NA, not addressed; ↓, reduction; Ab, antibody; AT, adoptive transfer of sorted DCs; CFA, complete Freund’s adjuvant; PTX, pertussis toxin; TF, transcription factor; HDM, house dust mite; T. gondii, Toxoplasma gondii; C. albicans, Candida albicans; A. fumigatus, Aspergillus fumigatus; N. brasiliensis, Nippostrongylus brasiliensis. Table prepared with help of Y. Kumamoto.
The clearance of viral infection relies on cytotoxic lymphocytes (CTLs), whose differentiation follows the engagement of nucleic acid sensors, which leads to the production of type 1 interferons and IL-12. Batf3-dependent CD103+ DCs found within or under various mucosal epithelial cells, as well as the Batf3-dependent CD8α+ DCs in lymphoid organs, are required for cross-presentation and the activation of CD8+ T cells62–64. These DCs express RLRs, NLRs and TLR3 but not TLR7 (refs. 65,66) and produce type 1 interferons and IL-12 in response to the recognition of viruses and virus-infected cells (Fig. 2). These cells express the C-type lectin DNGR-1 (CLECL9A), which detects F-actin exposed by necrotic cells for the uptake and cross-presentation of necrotic cells67,68. In addition to requiring CD103+ DCs, infection with fungal pathogens requires Batf3-dependent CD207+ dermal DCs for CTL responses69. The CD141hiCLEC9A+ DCs found in the dermis, lung and liver interstitia of humans share a transcriptome signature similar to that of mouse CD8α+ and CD103+ DCs and are also superior to other DCs in cross-priming CD8+ T cells ex vivo70. In addition, human CD1a+ Langerhans cells induce functional maturation of CD8+ T cells superior to that induced by other skin DC subsets ex vivo71,72.
Figure 2.
The instruction of various classes of T cell responses by DCs. The development and function of DCs in vivo requires transcription factors that are critical for each stage of their differentiation program. Studies probing the requirement or sufficiency for DC subsets have suggested a relationship between the DC lineage and T cell programming. CTL responses are induced by lymphoid organ–resident, Batf3-dependent CD8α+ DCs and tissue CD103+ DCs (mouse) and the human CD141+ DC counterpart. TH1 cell–mediated immunity requires stimulation, in a GM-CSF-dependent manner, by the CD207+CD103+ DC subset, a minor population of dermal DCs (mouse). TH17 responses are induced by mucosal IRF4-dependent CD103+CD11b+ DCs as well as by skin Langerhans cells, depending on the location of the pathogen (mouse). The human CD1c+ DCs prime TH17 cell responses. Finally, TH2 cell–mediated immunity requires IRF4-dependent CD301b+CD11b+ DCs (mouse). These DCs comprise the majority of dermal DC (dDC) population and are distinct from CD207+ CD103+ dermal DCs. In contrast, human Langerhans cells induce TH2 cytokine secretion ex vivo.
Defense by the immune system against intracellular bacteria and protozoa requires responses by CD8+ T cells and T helper type 1 cells (TH1 cell), which develop as a consequence of the engagement of TLRs by PAMPs that leads to the production of IL-12. CTL responses to bacterial and protozoan infection are ‘preferentially’ induced by Batf3-dependent CD103+ DCs that produce IL-12 (refs. 73,74). TH1 cell–mediated immunity to fungal pathogens depends on Batf3-dependent dermal DCs that express CD103 and CD207 (ref. 69). The GM-CSF-dependent DC population is required for the induction of TH1 cells after immunization with antigen and complete Freund’s adjuvant75,76, which demonstrates the key role of this DC subset in TH1 cell–mediated responses to bacterial and fungal pathogens (Fig. 2). However, the exact identity of the TH1-priming DCs remains controversial. While one report has shown a requirement for CD103+CD207+ DCs in TH1 priming76, another has shown that TH1 priming occurs normally in the absence of those DCs75 following immunization with myelin oligodendrocyte glycoprotein in complete Freund's adjuvant.
The TH17 subset of helper T cells is specialized in eliminating extracellular bacteria and fungal pathogens77,78. The priming of TH17 cells begins with the engagement of CLRs such as dectin-1 and dectin-2 expressed by DCs, Langerhans cells and macrophages79–81. In addition to inducing dectins, the phagocytosis of bacteria-infected apoptotic cells induces the production of TGF-β and IL-6 by DCs, which promotes TH17 cell differentiation82. Several studies support the notion that CD11b+CD103+ DCs that develop under the control of IRF4 have a key role in TH17 cell induction in vivo60,61,83,84. The cytokines critical for TH17 responses, IL-6 and IL-23, are both selectively secreted by the CD11b+CD103+ DCs60,61. In addition to the CD11b+CD103+ DCs, Langerhans cells are necessary and sufficient for induction of the TH17 cell response to fungal pathogens in the skin69. We are tempted to speculate that these DCs co-engage TLRs and CLRs to selectively induce IL-23 and suppress IL-12 for optimal TH17 cell induction in vivo85. Human CD1c+ DCs, the counterpart of mouse CD11b+ DCs, also express IRF4, secrete IL-23 and promote TH17 cells in response to Aspergillus fumigatus60 (Fig. 2).
Although sensors in the innate immune system that detect protease allergens and helminthes are not yet known, studies have demonstrated that induction of the TH2 cell response to both requires the PD-L2+CD301b+ DC population86–88. The CD301b+ DCs comprise the majority of dermal DCs that are CD207−, and these cells require IRF4 for their ability to promote TH2 cell–mediated immunity. Of note, in humans, Langerhans cells induce the secretion of TH2 cell cytokines in mixed-leukocyte reactions more efficiently than other skin DC subsets do71,72, which suggests that different skin DC subsets may carry out the priming of TH2 cells in mice and humans (Fig. 2).
Recognition by the innate immune system is also critical for antibody responses. T cell–independent B cell responses are induced by signals from both TLRs and B cell antigen receptors (BCRs). B-1a and B-1b cells produce natural immunoglobulin M (IgM) antibodies that confer protection against infection with certain bacteria, such as Streptococcus pneumonia90 and Borrelia hermsii91, as well as the early IgM response to viruses such as influenza virus92. B-1a cells express TLR9 and TLR7 and respond robustly to their ligands93. The BCR induces noncanonical signaling via the transcription factor NF-κB that complements TLR4-induced canonical signaling via NF-κB in response to LPS to induce activation-induced cytidine deaminase expression needed for immunoglobulin class switching94.
T cell–dependent antibody response requires at least three signals, consisting of engagement of the BCR by an antigen, and signals from follicular helper T cells (TFHcells) in the form of CD40L (the ligand for the costimulatory receptor CD40) and cytokines that induce class switch to a particular heavy-chain isotype. In addition, T cell–dependent antibody responses are controlled by the recognition of B cells by the intrinsic innate immune system. First, co-engagement of BCR with the complement receptor CD21 determines whether the antigen has been flagged by the complement system in the form of attachment of the complement component C3d. Attachment of the C3d fragment to a protein antigen has been shown to substantially increase the sensitivity of B cells95. CD21 then signals through the signal-transduction receptor CD19 complex to induce signaling via the kinase PI(3)K that co-stimulates the BCR signaling96. Second, another co-receptor, CD22, determines whether the antigen bound by BCR contains α2,6 sialic acids. Because α2,6 sialic acid is present on host glycoconjugates but not on most bacterial carbohydrates97, this co-engagement helps to determine the origin of the antigen. When CD22 is engaged by its sialic acid ligand, it activates the lipid phosphatase SHIP, which converts phosphatidylinositol-(3,4,5)-trisphosphate (generated by PI(3)K) into phosphatidylinositol-(3,4)-bisphosphate and thus inhibits downstream steps in the PI(3)K signaling pathway96. Finally, antibody responses, particularly those of IgG2 isotype (mouse) or IgG1 and IgG3 isotypes (human), are regulated by B cell–intrinsic TLRs signaling. In this context, TLRs probably function to determine whether the antigen bound by the BCR is a PAMP or is associated with a PAMP. The importance of TLR signaling in B cells has been demonstrated in response to pathogens in infection models98–102, in response to self antigens in lupus models103,104, and in LPS-stimulated and CpG-stimulated responses98,105. The systemic IgA response to virus-like particles containing RNA also requires B cell–intrinsic, but not DC-intrinsic, TLR7 expression106.
In summary, different sensing pathways of the innate immune system are designed to determine the class of infecting pathogens on the basis of their localization, viability, replication and virulence. These parameters are sensed by distinct sensory pathways, often operating in combination, and are translated into the signals that initiate the appropriate types of effector responses (Fig. 2).
Costs associated with effector responses
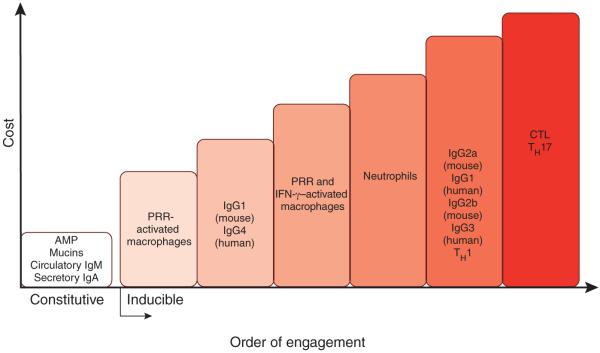
The ‘choice’ of effector response is determined not only by the class of infecting pathogen (for example, a virus versus a fungus) but also by the cost of the immune response107. The symptoms of infectious disease are caused by both direct pathogen-inflicted damage, as well as by the immune response itself, or ‘immunopathology’. Immunopathology is generally considered to be a consequence of excessive immune response, and therefore the magnitude and duration of the immune response are thought to be optimized so as to provide maximal protection from infection with minimal immunopathology. However, in addition to considering the quantitative characteristic of the immune response, it may be important to consider the fact that different effector responses have different immunopathological potential; for example, defensins and IgA secreted into intestinal lumen have lower costs to host fitness than do cytotoxic responses. In general, all effector responses can be viewed on a spectrum defined by the costs (immunopathology) associated with their maximal deployment. This difference in the cost of effector responses appears to correspond to the temporal order of their induction: low-cost responses are induced first; if they are insufficient to prevent or clear the infection, the response of next lowest cost is induced second and so on (Fig. 3). The responses of highest cost are induced as a last resort. Some of the responses that have negligible costs (such as secretory IgA and antimicrobial peptides produced by surface epithelia) are expressed constitutively, whereas the costly responses are inducible. It is interesting to note that the ‘choice’ of effector response is probably determined by the innate sensing pathway activated by a given challenge—for example, PRR sensing of microbial PAMPs by surface epithelia alone can induce only low-cost responses; the same PRR activated in macrophages or DCs behind epithelial barriers may induce responses of higher cost; the sensing of pathogen invasiveness, viability, replication and virulence activities induce responses of progressively higher cost. Thus, macrophages activated by PRRs alone induce effector response of lower cost (secretion of cytokines and antimicrobial peptides) than those induced by macrophages activated by PRRs and interferon-γ (IFN-γ) (enhanced microbicidal activities and collateral damage) (Fig. 3). This strategy should optimize the immune response so as to minimize its cost while providing sufficient protection. In addition to immunopathology, immune responses can have other costs, most notably metabolic costs. Therefore, immune responses are probably calibrated on the basis of weighted contribution of different costs.
Figure 3.
The order of effector function engagement, from lowest cost to highest cost. Defense mechanisms of the immune system have different costs to host fitness. The order of engagement of effector functions corresponds to their increasing costs (here, their potential to cause immunopathology). The low-cost effector mechanisms, such as antimicrobial peptides (AMPs), secretory IgA and circulatory IgM, are expressed constitutively. When they are insufficient to contain pathogens, the response of next lowest cost is induced (here, tissue-resident macrophages); when this response is insufficient, neutrophils are recruited, and so on. The examples of effector responses presented here and their costs are for illustration purposes only; they are neither comprehensive nor quantitative.
Design principle of immune responses
Over the past decade, many exciting discoveries have been made about the functions and regulation of ILCs108–110, tissue-resident memory T cells (TRM cells)111–114 and epithelial cells, as well as the signals that activate these cell types and the cytokines they produce. These discoveries paint a complex picture of multiple cell types connected by cytokines that perform distinct functions. However, behind this complexity, there is a simpler pattern that reveals a common design principle of the immune response, with many details being a variation on a common theme.
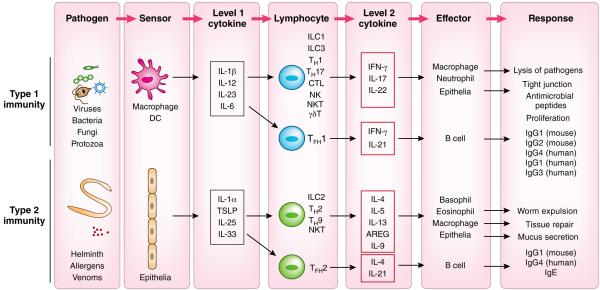
The commonalities become apparent with the consideration that all the cell types involved in the initiation and execution of the immune response fall into three categories (Fig. 4). The first category is made up of cell types that function as sensors of infection or damage. These are the cell types that express various PRRs and other sensing machinery to detect microorganisms, macroparasites, allergens, toxins and venoms and any other stimuli that elicit immune responses. The cell types that function as sensors include DCs and macrophages for the type 1 immune response, and epithelial cells and mast cells for the type 2 immune response. The different modes of recognition by the innate immune system described above operate in these cell types to induce production of the first level of cytokines. The ‘level 1’ cytokines include IL-12, IL-23, IL-6 and IL-1β for the type 1 immune responses, and TSLP, IL-25, IL-33 and IL-1α for the type 2 immune responses115,116. These cytokines act on the second category of cells, which is made up of various populations of lymphocytes, including ILCs, innate-like lymphocytes (ILLs; including NKT cells, MAIT cells and epithelial γδ T cells), TFH cells and TRM cells. In response to ‘level 1’ cytokines, these lymphocytes produce ‘level 2’ cytokines. These include IFN-γ, IL-17, IL-22 for type 1 immune response, and IL-4, IL-5, IL-9, IL-13 and AREG for type 2 immune response. IL-21 produced by TFH cells is common to both type 1 immune responses and type 2 immune responses. The ‘level 2’ cytokines act on the third category of cells, which are effectors of the immune response. These include macrophages, neutrophils, epithelial cells, eosinophils and basophils, and B cells (specifically B-2 cells), as well as sensory neurons, endothelium and smooth muscle cells. These cells perform diverse effector functions, including barrier defense, killing and expulsion of pathogens, antibody production, and tissue repair.
Figure 4.
The two-tiered design of the immune responses. Immune responses are orchestrated by three categories of cells that function as ‘sensors’ that detect pathogens and secrete ‘level 1’ cytokines, the tissue-resident lymphocytes that respond to ‘level 1’ cytokines to secrete ‘level 2’ cytokines, and the effector cells that respond to ‘level 2’ cytokines to carry out effector functions to eliminate the pathogen. DCs and macrophages serve as sensors for the type 1 immune response and epithelial cells and mast cells for the type 2 immune response. ‘Level 1’ cytokines act on terminally differentiated lymphocytes, including ILCs, ILLs, TFH cells and TRM cells. In response to ‘level 1’ cytokines, these lymphocytes produce ‘level 2’ cytokines. The ‘level 2’ cytokines act on the effector cells of the immune response. These include macrophages, neutrophils, epithelial cells, eosinophils and basophils, B cells as well as sensory neurons, endothelium and smooth muscle cells. These cells perform diverse effector functions, including barrier defenses, killing and expulsion of pathogens, antibody production and tissue repair.
Some cell types can function both as sensors and as effectors (Fig. 4). Macrophages function as sensors when activated by the PRRs and as effectors when activated by ‘level 2’ cytokines produced by lymphocytes. However, as discussed above, the responses elicited by these stimuli are distinct: as sensors, macrophages secrete ‘level 1’ cytokines to elicit responses from local lymphocytes. In contrast, macrophages stimulated as effectors by a ‘level 2’ cytokine (for example, IFN-γ) become highly activated effectors, induce robust antimicrobial and phagocytic responses, including the production of reactive oxygen and nitrogen species. Similarly, epithelial cells serving as sensors of helminth infection produce the ‘level 1’ cytokines IL-33, IL-25 and TSLP, whereas epithelial cells responding to the ‘level 2’ cytokine IL-13 become activated and differentiate into goblet cells to increase mucus secretion. The reason the same cell types can function as both sensors and effectors presumably reflects their evolutionary history when they operated in simpler pathways, performing both sensing functions and effector functions without the involvement of lymphocytes.
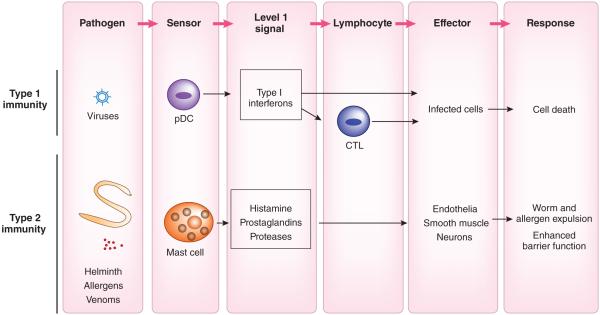
There are some variations on the common theme of two-tiered design of the immune response. For example, in some cases, the single-tiered cytokine system consists of sensor cell types that mediate direct effector response through secretion of ‘level 1’ signals (Fig. 5). For antiviral programs, plasmacytoid DCs serve as a key sensor for the detection viruses through endosomal TLRs to produce a burst of type 1 interferons, which act systemically to block viral replication117. Infected cells themselves produce type 1 interferons to mediate similar effects locally. Similarly, mast cells that reside in the mucosa sense the presence of the noxious substances and parasitic worms, and release histamines and prostaglandins, which act on local vasculature to induce vasodilation and leakage, and act on intestinal and airway smooth muscles to induce peristalsis, contraction and expulsion of parasites. Histamine produced by mast cells and TSLP produced by epithelial cells can also stimulate a subset of C-fiber neurons to induce the itching sensation, which helps rid of macroparasites and ectoparasites118. The single-tiered design probably reflects a more evolutionarily ancient strategy in which the sensor and effector functions were carried out by the same cells. Such a strategy may have been optimal in invertebrate deuterostomes that have numerous PRRs but have limited cell types119.
Figure 5.
Single-tiered immune responses. Some immune responses involve only the ‘level 1’ signals that act directly on the effector cells. For antiviral responses, plasmacytoid DCs serve as a key sensor for the detection of viruses through endosomal TLRs to induce type 1 interferons, which block viral replication. Mast cells that reside in the mucosa sense the presence of the noxious substances and parasitic worms and release histamines and prostaglandins, which act on local vasculature to induce vasodilation and leakage, and intestinal and airway smooth muscles to induce peristalsis, contraction and expulsion of parasites. Histamine produced by mast cells and TSLP produced by epithelial cells can also stimulate a subset of C-fiber neurons to induce the itching sensation.
The two-tiered design uses ILCs, ILLs and TRM cells to mediate the effector responses. Unlike secretion by ILCs or ILLs, the secretion of ‘level 2’ cytokines by TRM cells usually requires specific antigen, although not necessarily120. The ‘level 1’ cytokines usually work in combinations consisting of a cytokine of the IL-1 family (IL-1, IL-18 and IL-33) and a cytokine that activated the kinase Jak and transcription factors of the STAT family (IL-12, IL-23, IL-6, IL-15, IL-2, IL-7 and TSLP) to induce the secretion of distinct sets of ‘level 2’ cytokines from lymphocytes120. Macrophages and DCs detect pathogens through PRRs and secrete IL-12, IL-23 and IL-6. They also sense pathogens through inflammasomes and release IL-1β and IL-18. These ‘level 1’ cytokines act on differentiated lymphocytes of both innate classes and adaptive classes. IL-12 (an activator of STAT4), and IL-18 and IL-1β (activators of the adaptor MyD88) act on ILC1 cells, NK cells, γδT cells and memory CD4+ T cells and CD8+ T cells in the tissue to secrete IFN-γ. IL-1β (an activator of MyD88) together with IL-23 or IL-21 (activators of STAT3) to induce ILC3 cells and memory TH17 cells to secrete IL-17 and IL-22 (ref. 121). Similarly, the ‘level 1’ cytokine TSLP (STAT5 activator), together with IL-33 or IL-1α (activators of MyD88) released from epithelial cells, act on ILC2 cells and TH2 memory cells to secrete IL-5, IL-13 and AREG. Signaling via the receptor for IL-25 induces activation of NF-κB and, together with IL-33, induces the production of IL-5 and IL-13 by ILC2 cells122. In addition, the same ‘level 1’ cytokines also induce secretion of IL-9 from tissue-resident TH9 memory cells. Of note, secretion of IL-4 is induced only upon the engagement of T cell antigen receptors (TCRs) on memory TH2 cells123 or the treatment of ILC2 cells with the phorbol ester PMA and ionomycin124 and, unlike other TH2 cytokines, combinations of ‘level 1’ cytokines are not sufficient to induce IL-4 secretion120. ‘Level 1’ cytokines can also program antibody responses through their action on TFH cells. IL-12 induces TFH cells to secrete IFN-γ125, while an as-yet-unknown signal induces the secretion of IL-4 from TFH cells (Fig. 4). In mice, TFH cells that secrete IL-21 drive IgG1 responses, whereas IL-4-secreting and IFN-γ-secreting TFH cells drive IgE responses and IgG2 responses, respectively126.
It is important to note that naive T cells are not part of the common design of the immune response presented here. Indeed, naive T cells can become the source of ‘level 2’ cytokines only after their activation and differentiation into effector or memory cells. This is a reflection of the clonal distribution of their TCRs and the requirement for extra steps of proliferation and differentiation before they can become competent to function as cells denoted as the lymphocyte category. In contrast, ILCs that lack TCRs, ILLs that have invariant TCRs, and TRM cells that have many features in common with ILCs and ILLs, either do not need to undergo proliferation and differentiation steps (ILCs and ILLs), or have already done so in the past (TRM cells).
Future perspectives
The focus of this Review has been the emerging principles of recognition by the innate immune system and control of adaptive immunity. Future discoveries will no doubt lead to additional revisions and modifications of the current paradigms. Some exciting emerging new directions include the sensing pathways of the immune system that detect specific dietary and microbial metabolites, including short-chain fatty acids and vitamins127–129. In addition, the detection of bacterial pigments by the aryl hydrocarbon receptor has been found to regulate an antibacterial response130. The sensing of xenobiotic substances by this receptor has been found to have a role in the enhancement of barrier immunity through IL-22 production and other mechanisms131. Another sensor of xenobiotic substances, PXR, has been found to control intestinal inflammation by sensing bacterial metabolites132. How all these pathways fit in with the current models of immune recognition is an exciting area of future investigation.
In addition to expanding the knowledge of sensing pathways of the innate immune system, a major challenge for the future studies is to define the rules of engagement that determine the functions of these pathways in the context of infectious diseases. This will be essential for advancing the ability to design effective vaccines. A major hurdle in this area has been the lack of understanding of the requirements for the induction of protective immunity. Instead, immunogenicity is often used as a surrogate for protective immunity in vaccine design and trials. However, immunogenicity, a property related to the ability to induce an immune response, is not equivalent to protective immunity, which is conferred by immune responses able to eliminate the pathogen without killing the host in the process. The immunogenicity of vaccines often leads to protective immunity when the latter is based on neutralizing antibodies. In most other cases, vaccines that are immunogenic often do not provide protection. A given immune response may or may not be protective for any one of several reasons; it could be of wrong specificity, effector class, location, magnitude or duration. Thus, just as the rules of immunogenicity have been defined over the past 15 years, a major challenge for the future will be to define the rules of protective immunity.
ACKNOWLEDGMENTS
We thank Y. Kumamoto for help with Table 1. Supported by the US National Institutes of Health (AI081884, AI062428 and AI064705 to A.I., and AI046688, AI089771 and DK071754 to R.M.), Else Kröner-Fresenius-Stiftung (R.M.) and the Howard Hughes Medical Institute (A.I. and R.M.).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Janeway CA., Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 1989;54:1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 3.Wu J, Chen ZJ. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 2014;32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 4.Rathinam VA, Vanaja SK, Fitzgerald KA. Regulation of inflammasome signaling. Nat. Immunol. 2012;13:333–342. doi: 10.1038/ni.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palm NW, Rosenstein RK, Medzhitov R. Allergic host defences. Nature. 2012;484:465–472. doi: 10.1038/nature11047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belkaid Y, Naik S. Compartmentalized and systemic control of tissue immunity by commensals. Nat. Immunol. 2013;14:646–653. doi: 10.1038/ni.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat. Immunol. 2013;14:676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chow J, Lee SM, Shen Y, Khosravi A, Mazmanian SK. Host-bacterial symbiosis in health and disease. Adv. Immunol. 2010;107:243–274. doi: 10.1016/B978-0-12-381300-8.00008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barton GM, Kagan JC. A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat. Rev. Immunol. 2009;9:535–542. doi: 10.1038/nri2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoneyama M, Onomoto K, Jogi M, Akaboshi T, Fujita T. Viral RNA detection by RIG-I-like receptors. Curr. Opin. Immunol. 2015;32C:48–53. doi: 10.1016/j.coi.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 11.Gürtler C, Bowie AG. Innate immune detection of microbial nucleic acids. Trends Microbiol. 2013;21:413–420. doi: 10.1016/j.tim.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rathinam VA, Fitzgerald KA. Cytosolic surveillance and antiviral immunity. Curr. Opin. Virol. 2011;1:455–462. doi: 10.1016/j.coviro.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burdette DL, Vance RE. STING and the innate immune response to nucleic acids in the cytosol. Nat. Immunol. 2013;14:19–26. doi: 10.1038/ni.2491. [DOI] [PubMed] [Google Scholar]
- 14.von Moltke J, Ayres JS, Kofoed EM, Chavarria-Smith J, Vance RE. Recognition of bacteria by inflammasomes. Annu. Rev. Immunol. 2013;31:73–106. doi: 10.1146/annurev-immunol-032712-095944. [DOI] [PubMed] [Google Scholar]
- 15.Lamkanfi M, Dixit VM. Inflammasomes and their roles in health and disease. Annu. Rev. Cell Dev. Biol. 2012;28:137–161. doi: 10.1146/annurev-cellbio-101011-155745. [DOI] [PubMed] [Google Scholar]
- 16.Blander JM, Sander LE. Beyond pattern recognition: five immune checkpoints for scaling the microbial threat. Nat. Rev. Immunol. 2012;12:215–225. doi: 10.1038/nri3167. [DOI] [PubMed] [Google Scholar]
- 17.Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stuart LM, Paquette N, Boyer L. Effector-triggered versus pattern-triggered immunity: how animals sense pathogens. Nat. Rev. Immunol. 2013;13:199–206. doi: 10.1038/nri3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sander LE, et al. Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature. 2011;474:385–389. doi: 10.1038/nature10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chovatiya R, Medzhitov R. Stress, inflammation, and defense of homeostasis. Mol. Cell. 2014;54:281–288. doi: 10.1016/j.molcel.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu. Rev. Immunol. 2009;27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- 22.Mariathasan S, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 23.Ichinohe T, Pang IK, Iwasaki A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat. Immunol. 2010;11:404–410. doi: 10.1038/ni.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dangl JL, Jones JD. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 25.Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454:428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- 26.Gause WC, Wynn TA, Allen JE. Type 2 immunity and wound healing: evolutionary refinement of adaptive immunity by helminths. Nat. Rev. Immunol. 2013;13:607–614. doi: 10.1038/nri3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allen JE, Wynn TA. Evolution of Th2 immunity: a rapid repair response to tissue destructive pathogens. PLoS Pathog. 2011;7:e1002003. doi: 10.1371/journal.ppat.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monticelli LA, et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat. Immunol. 2011;12:1045–1054. doi: 10.1031/ni.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heredia JE, et al. Type 2 innate signals stimulate fibro/adipogenic progenitors to facilitate muscle regeneration. Cell. 2013;153:376–388. doi: 10.1016/j.cell.2013.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burzyn D, et al. A special population of regulatory T cells potentiates muscle repair. Cell. 2013;155:1282–1295. doi: 10.1016/j.cell.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kärre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319:675–678. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 33.Raulet DH. Missing self recognition and self tolerance of natural killer (NK) cells. Semin. Immunol. 2006;18:145–150. doi: 10.1016/j.smim.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 34.Karlhofer FM, Ribaudo RK, Yokoyama WM. MHC class I alloantigen specificity of Ly-49+ IL-2-activated natural killer cells. Nature. 1992;358:66–70. doi: 10.1038/358066a0. [DOI] [PubMed] [Google Scholar]
- 35.Doody GM, et al. A role in B cell activation for CD22 and the protein tyrosine phosphatase SHP. Science. 1995;269:242–244. doi: 10.1126/science.7618087. [DOI] [PubMed] [Google Scholar]
- 36.Kemper C, Atkinson JP, Hourcade DE. Properdin: emerging roles of a pattern-recognition molecule. Annu. Rev. Immunol. 2010;28:131–155. doi: 10.1146/annurev-immunol-030409-101250. [DOI] [PubMed] [Google Scholar]
- 37.Rescigno M, et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2001;2:361–367. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- 38.Abreu MT. Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat. Rev. Immunol. 2010;10:131–144. doi: 10.1038/nri2707. [DOI] [PubMed] [Google Scholar]
- 39.Saenz SA, Taylor BC, Artis D. Welcome to the neighborhood: epithelial cell-derived cytokines license innate and adaptive immune responses at mucosal sites. Immunol. Rev. 2008;226:172–190. doi: 10.1111/j.1600-065X.2008.00713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uematsu S, et al. Detection of pathogenic intestinal bacteria by Toll-like receptor 5 on intestinal CD11c+ lamina propria cells. Nat. Immunol. 2006;7:868–874. doi: 10.1038/ni1362. [DOI] [PubMed] [Google Scholar]
- 41.Lightfield KL, et al. Differential requirements for NAIP5 in activation of the NLRC4 inflammasome. Infect. Immun. 2011;79:1606–1614. doi: 10.1128/IAI.01187-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao Y, et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477:596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- 43.Casson CN, et al. Caspase-11 activation in response to bacterial secretion systems that access the host cytosol. PLoS Pathog. 2013;9:e1003400. doi: 10.1371/journal.ppat.1003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas CJ, Schroder K. Pattern recognition receptor function in neutrophils. Trends Immunol. 2013;34:317–328. doi: 10.1016/j.it.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 45.Brinkmann V, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 46.Pecchi E, Dallaporta M, Jean A, Thirion S, Troadec JD. Prostaglandins and sickness behavior: old story, new insights. Physiol. Behav. 2009;97:279–292. doi: 10.1016/j.physbeh.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 47.Gabay C, Kushner I. Acute-phase proteins and other systemic responses to inflammation. N. Engl. J. Med. 1999;340:448–454. doi: 10.1056/NEJM199902113400607. [DOI] [PubMed] [Google Scholar]
- 48.Hermesh T, Moltedo B, Moran TM, Lopez CB. Antiviral instruction of bone marrow leukocytes during respiratory viral infections. Cell Host Microbe. 2010;7:343–353. doi: 10.1016/j.chom.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Asselin-Paturel C, et al. Mouse type I IFN–producing cells are immature APCs with plasmacytoid morphology. Nat. Immunol. 2001;2:1144–1150. doi: 10.1038/ni736. [DOI] [PubMed] [Google Scholar]
- 50.Asselin-Paturel C, et al. Type I interferon dependence of plasmacytoid dendritic cell activation and migration. J. Exp. Med. 2005;201:1157–1167. doi: 10.1084/jem.20041930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barchet W, et al. Virus-induced interferon alpha production by a dendritic cell subset in the absence of feedback signaling in vivo. J. Exp. Med. 2002;195:507–516. doi: 10.1084/jem.20011666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anthony RM, Rutitzky LI, Urban JF, Jr., Stadecker MJ, Gause WC. Protective immune mechanisms in helminth infection. Nat. Rev. Immunol. 2007;7:975–987. doi: 10.1038/nri2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brown SJ, Askenase PW. Cutaneous basophil responses and immune resistance of guinea pigs to ticks: passive transfer with peritoneal exudate cells or serum. J. Immunol. 1981;127:2163–2167. [PubMed] [Google Scholar]
- 54.Wada T, et al. Selective ablation of basophils in mice reveals their nonredundant role in acquired immunity against ticks. J. Clin. Invest. 2010;120:2867–2875. doi: 10.1172/JCI42680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Merad M, Sathe P, Helft J, Miller J, Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 2013;31:563–604. doi: 10.1146/annurev-immunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ginhoux F, Jung S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat. Rev. Immunol. 2014;14:392–404. doi: 10.1038/nri3671. [DOI] [PubMed] [Google Scholar]
- 57.Geissmann F, et al. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Satpathy AT, Wu X, Albring JC, Murphy KM. Re(de)fining the dendritic cell lineage. Nat. Immunol. 2012;13:1145–1154. doi: 10.1038/ni.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Briseño CG, Murphy TL, Murphy KM. Complementary diversification of dendritic cells and innate lymphoid cells. Curr. Opin. Immunol. 2014;29:69–78. doi: 10.1016/j.coi.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schlitzer A, et al. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity. 2013;38:970–983. doi: 10.1016/j.immuni.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Persson EK, et al. IRF4 transcription-factor-dependent CD103+CD11b+ dendritic cells drive mucosal T helper 17 cell differentiation. Immunity. 2013;38:958–969. doi: 10.1016/j.immuni.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 62.Hildner K, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Edelson BT, et al. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J. Exp. Med. 2010;207:823–836. doi: 10.1084/jem.20091627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.del Rio ML, Rodriguez-Barbosa JI, Kremmer E, Forster R. CD103− and CD103+ bronchial lymph node dendritic cells are specialized in presenting and cross-presenting innocuous antigen to CD4+ and CD8+ T cells. J. Immunol. 2007;178:6861–6866. doi: 10.4049/jimmunol.178.11.6861. [DOI] [PubMed] [Google Scholar]
- 65.Desch AN, et al. CD103+ pulmonary dendritic cells preferentially acquire and present apoptotic cell-associated antigen. J. Exp. Med. 2011;208:1789–1797. doi: 10.1084/jem.20110538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Desch AN, et al. Dendritic cell subsets require cis-activation for cytotoxic CD8 T-cell induction. Nat. Commun. 2014;5:4674. doi: 10.1038/ncomms5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sancho D, et al. Identification of a dendritic cell receptor that couples sensing of necrosis to immunity. Nature. 2009;458:899–903. doi: 10.1038/nature07750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zelenay S, et al. The dendritic cell receptor DNGR-1 controls endocytic handling of necrotic cell antigens to favor cross-priming of CTLs in virus-infected mice. J. Clin. Invest. 2012;122:1615–1627. doi: 10.1172/JCI60644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Igyártó BZ, et al. Skin-resident murine dendritic cell subsets promote distinct and opposing antigen-specific T helper cell responses. Immunity. 2011;35:260–272. doi: 10.1016/j.immuni.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haniffa M, et al. Human tissues contain CD141hi cross-presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells. Immunity. 2012;37:60–73. doi: 10.1016/j.immuni.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Klechevsky E, et al. Functional specializations of human epidermal Langerhans cells and CD14+ dermal dendritic cells. Immunity. 2008;29:497–510. doi: 10.1016/j.immuni.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Segura E, et al. Characterization of resident and migratory dendritic cells in human lymph nodes. J. Exp. Med. 2012;209:653–660. doi: 10.1084/jem.20111457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mashayekhi M, et al. CD8alpha+ dendritic cells are the critical source of interleukin-12 that controls acute infection by Toxoplasma gondii tachyzoites. Immunity. 2011;35:249–259. doi: 10.1016/j.immuni.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Edelson BT, et al. CD8alpha+ dendritic cells are an obligate cellular entry point for productive infection by Listeria monocytogenes. Immunity. 2011;35:236–248. doi: 10.1016/j.immuni.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Edelson BT, et al. Batf3-dependent CD11blow/− peripheral dendritic cells are GM-CSF-independent and are not required for Th cell priming after subcutaneous immunization. PLoS ONE. 2011;6:e25660. doi: 10.1371/journal.pone.0025660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.King IL, Kroenke MA, Segal BM. GM-CSF-dependent, CD103+ dermal dendritic cells play a critical role in Th effector cell differentiation after subcutaneous immunization. J. Exp. Med. 2010;207:953–961. doi: 10.1084/jem.20091844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hernández-Santos N, Gaffen SL. Th17 cells in immunity to Candida albicans. Cell Host Microbe. 2012;11:425–435. doi: 10.1016/j.chom.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Romani L. Cell mediated immunity to fungi: a reassessment. Med. Mycol. 2008;46:515–529. doi: 10.1080/13693780801971450. [DOI] [PubMed] [Google Scholar]
- 79.Brown GD, Netea MG. Exciting developments in the immunology of fungal infections. Cell Host Microbe. 2012;11:422–424. doi: 10.1016/j.chom.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 80.Reid DM, Gow NA, Brown GD. Pattern recognition: recent insights from Dectin-1. Curr. Opin. Immunol. 2009;21:30–37. doi: 10.1016/j.coi.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Robinson MJ, et al. Dectin-2 is a Syk-coupled pattern recognition receptor crucial for Th17 responses to fungal infection. J. Exp. Med. 2009;206:2037–2051. doi: 10.1084/jem.20082818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Torchinsky MB, Garaude J, Martin AP, Blander JM. Innate immune recognition of infected apoptotic cells directs TH17 cell differentiation. Nature. 2009;458:78–82. doi: 10.1038/nature07781. [DOI] [PubMed] [Google Scholar]
- 83.Welty NE, et al. Intestinal lamina propria dendritic cells maintain T cell homeostasis but do not affect commensalism. J. Exp. Med. 2013;210:2011–2024. doi: 10.1084/jem.20130728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lewis KL, et al. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity. 2011;35:780–791. doi: 10.1016/j.immuni.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dennehy KM, Willment JA, Williams DL, Brown GD. Reciprocal regulation of IL-23 and IL-12 following co-activation of Dectin-1 and TLR signaling pathways. Eur. J. Immunol. 2009;39:1379–1386. doi: 10.1002/eji.200838543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kumamoto Y, et al. CD301b+ dermal dendritic cells drive T helper 2 cell-mediated immunity. Immunity. 2013;39:733–743. doi: 10.1016/j.immuni.2013.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gao Y, et al. Control of T helper 2 responses by transcription factor IRF4-dependent dendritic cells. Immunity. 2013;39:722–732. doi: 10.1016/j.immuni.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Murakami R, et al. A unique dermal dendritic cell subset that skews the immune response toward Th2. PLoS ONE. 2013;8:e73270. doi: 10.1371/journal.pone.0073270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tang H, et al. The T helper type 2 response to cysteine proteases requires dendritic cell–basophil cooperation via ROS-mediated signaling. Nat. Immunol. 2010;11:608–617. doi: 10.1038/ni.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Haas KM, Poe JC, Steeber DA, Tedder TF. B-1a and B-1b cells exhibit distinct developmental requirements and have unique functional roles in innate and adaptive immunity to S. pneumoniae. Immunity. 2005;23:7–18. doi: 10.1016/j.immuni.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 91.Alugupalli KR, et al. B1b lymphocytes confer T cell-independent long-lasting immunity. Immunity. 2004;21:379–390. doi: 10.1016/j.immuni.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 92.Baumgarth N, et al. B-1 and B-2 cell-derived immunoglobulin M antibodies are nonredundant components of the protective response to influenza virus infection. J. Exp. Med. 2000;192:271–280. doi: 10.1084/jem.192.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gururajan M, Jacob J, Pulendran B. Toll-like receptor expression and responsiveness of distinct murine splenic and mucosal B-cell subsets. PLoS ONE. 2007;2:e863. doi: 10.1371/journal.pone.0000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pone EJ, et al. BCR-signalling synergizes with TLR-signalling for induction of AID and immunoglobulin class-switching through the non-canonical NF-kappaB pathway. Nat. Commun. 2012;3:767. doi: 10.1038/ncomms1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dempsey PW, Allison ME, Akkaraju S, Goodnow CC, Fearon DT. C3d of complement as a molecular adjuvant: bridging innate and acquired immunity. Science. 1996;271:348–350. doi: 10.1126/science.271.5247.348. [DOI] [PubMed] [Google Scholar]
- 96.Doody GM, Dempsey PW, Fearon DT. Activation of B lymphocytes: integrating signals from CD19, CD22 and Fc gamma RIIb1. Curr. Opin. Immunol. 1996;8:378–382. doi: 10.1016/s0952-7915(96)80128-2. [DOI] [PubMed] [Google Scholar]
- 97.Varki A, Gagneux P. Multifarious roles of sialic acids in immunity. Ann. NY Acad. Sci. 2012;1253:16–36. doi: 10.1111/j.1749-6632.2012.06517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hou B, et al. Selective utilization of Toll-like receptor and MyD88 signaling in B cells for enhancement of the antiviral germinal center response. Immunity. 2011;34:375–384. doi: 10.1016/j.immuni.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Browne EP. Toll-like receptor 7 controls the anti-retroviral germinal center response. PLoS Pathog. 2011;7:e1002293. doi: 10.1371/journal.ppat.1002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Barr TA, Brown S, Mastroeni P, Gray D. B cell intrinsic MyD88 signals drive IFN-gamma production from T cells and control switching to IgG2c. J. Immunol. 2009;183:1005–1012. doi: 10.4049/jimmunol.0803706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Guay HM, Andreyeva TA, Garcea RL, Welsh RM, Szomolanyi-Tsuda E. MyD88 is required for the formation of long-term humoral immunity to virus infection. J. Immunol. 2007;178:5124–5131. doi: 10.4049/jimmunol.178.8.5124. [DOI] [PubMed] [Google Scholar]
- 102.Heer AK, et al. TLR signaling fine-tunes anti-influenza B cell responses without regulating effector T cell responses. J. Immunol. 2007;178:2182–2191. doi: 10.4049/jimmunol.178.4.2182. [DOI] [PubMed] [Google Scholar]
- 103.Leadbetter EA, et al. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416:603–607. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- 104.Teichmann LL, Schenten D, Medzhitov R, Kashgarian M, Shlomchik MJ. Signals via the adaptor MyD88 in B cells and DCs make distinct and synergistic contributions to immune activation and tissue damage in lupus. Immunity. 2013;38:528–540. doi: 10.1016/j.immuni.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pasare C, Medzhitov R. Control of B-cell responses by Toll-like receptors. Nature. 2005;438:364–368. doi: 10.1038/nature04267. [DOI] [PubMed] [Google Scholar]
- 106.Bessa J, et al. Alveolar macrophages and lung dendritic cells sense RNA and drive mucosal IgA responses. J. Immunol. 2009;183:3788–3799. doi: 10.4049/jimmunol.0804004. [DOI] [PubMed] [Google Scholar]
- 107.Nish S, Medzhitov R. Host defense pathways: role of redundancy and compensation in infectious disease phenotypes. Immunity. 2011;34:629–636. doi: 10.1016/j.immuni.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Diefenbach A, Colonna M, Koyasu S. Development, differentiation, and diversity of innate lymphoid cells. Immunity. 2014;41:354–365. doi: 10.1016/j.immuni.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.McKenzie AN, Spits H, Eberl G. Innate lymphoid cells in inflammation and immunity. Immunity. 2014;41:366–374. doi: 10.1016/j.immuni.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 110.Cortez VS, Robinette ML, Colonna M. Innate lymphoid cells: new insights into function and development. Curr. Opin. Immunol. 2015;32C:71–77. doi: 10.1016/j.coi.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mueller SN, Gebhardt T, Carbone FR, Heath WR. Memory T cell subsets, migration patterns, and tissue residence. Annu. Rev. Immunol. 2013;31:137–161. doi: 10.1146/annurev-immunol-032712-095954. [DOI] [PubMed] [Google Scholar]
- 112.Shin H, Iwasaki A. Tissue-resident memory T cells. Immunol. Rev. 2013;255:165–181. doi: 10.1111/imr.12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cauley LS, Lefrancois L. Guarding the perimeter: protection of the mucosa by tissue-resident memory T cells. Mucosal Immunol. 2013;6:14–23. doi: 10.1038/mi.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schenkel JM, Masopust D. Tissue-resident memory T cells. Immunity. 2014;41:886–897. doi: 10.1016/j.immuni.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tait Wojno ED, Artis D. Innate lymphoid cells: balancing immunity, inflammation, and tissue repair in the intestine. Cell Host Microbe. 2012;12:445–457. doi: 10.1016/j.chom.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ahern PP, Izcue A, Maloy KJ, Powrie F. The interleukin-23 axis in intestinal inflammation. Immunol. Rev. 2008;226:147–159. doi: 10.1111/j.1600-065X.2008.00705.x. [DOI] [PubMed] [Google Scholar]
- 117.Liu YJ. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu. Rev. Immunol. 2005;23:275–306. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- 118.Wilson SR, et al. The epithelial cell-derived atopic dermatitis cytokine TSLP activates neurons to induce itch. Cell. 2013;155:285–295. doi: 10.1016/j.cell.2013.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Buckley KM, Rast JP. Diversity of animal immune receptors and the origins of recognition complexity in the deuterostomes. Dev. Comp. Immunol. 2015;49:179–189. doi: 10.1016/j.dci.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 120.Guo L, Junttila IS, Paul WE. Cytokine-induced cytokine production by conventional and innate lymphoid cells. Trends Immunol. 2012;33:598–606. doi: 10.1016/j.it.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu. Rev. Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 122.Salimi M, et al. A role for IL-25 and IL-33-driven type-2 innate lymphoid cells in atopic dermatitis. J. Exp. Med. 2013;210:2939–2950. doi: 10.1084/jem.20130351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Guo L, et al. IL-1 family members and STAT activators induce cytokine production by Th2, Th17, and Th1 cells. Proc. Natl. Acad. Sci. USA. 2009;106:13463–13468. doi: 10.1073/pnas.0906988106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Moro K, et al. Innate production of T(H)2 cytokines by adipose tissue–associated c-Kit+Sca-1+ lymphoid cells. Nature. 2010;463:540–544. doi: 10.1038/nature08636. [DOI] [PubMed] [Google Scholar]
- 125.Nakayamada S, et al. Early Th1 cell differentiation is marked by a TFH cell-like transition. Immunity. 2011;35:919–931. doi: 10.1016/j.immuni.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Reinhardt RL, Liang HE, Locksley RM. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat. Immunol. 2009;10:385–393. doi: 10.1038/ni.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tan J, et al. The role of short-chain fatty acids in health and disease. Adv. Immunol. 2014;121:91–119. doi: 10.1016/B978-0-12-800100-4.00003-9. [DOI] [PubMed] [Google Scholar]
- 128.Spencer SP, et al. Adaptation of innate lymphoid cells to a micronutrient deficiency promotes type 2 barrier immunity. Science. 2014;343:432–437. doi: 10.1126/science.1247606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Spencer SP, Belkaid Y. Dietary and commensal derived nutrients: shaping mucosal and systemic immunity. Curr. Opin. Immunol. 2012;24:379–384. doi: 10.1016/j.coi.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Moura-Alves P, et al. AhR sensing of bacterial pigments regulates antibacterial defence. Nature. 2014;512:387–392. doi: 10.1038/nature13684. [DOI] [PubMed] [Google Scholar]
- 131.Stockinger B, Di Meglio P, Gialitakis M, Duarte JH. The aryl hydrocarbon receptor: multitasking in the immune system. Annu. Rev. Immunol. 2014;32:403–432. doi: 10.1146/annurev-immunol-032713-120245. [DOI] [PubMed] [Google Scholar]
- 132.Venkatesh M, et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity. 2014;41:296–310. doi: 10.1016/j.immuni.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dudziak D, et al. Differential antigen processing by dendritic cell subsets in vivo. Science. 2007;315:107–111. doi: 10.1126/science.1136080. [DOI] [PubMed] [Google Scholar]
- 134.Henri S, et al. CD207+ CD103+ dermal dendritic cells cross-present keratinocyte-derived antigens irrespective of the presence of Langerhans cells. J. Exp. Med. 2010;207:189–206. doi: 10.1084/jem.20091964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ballesteros-Tato A, Leon B, Lund FE, Randall TD. Temporal changes in dendritic cell subsets, cross-priming and costimulation via CD70 control CD8+ T cell responses to influenza. Nat. Immunol. 2010;11:216–224. doi: 10.1038/ni.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.GeurtsvanKessel CH, et al. Clearance of influenza virus from the lung depends on migratory langerin+CD11b− but not plasmacytoid dendritic cells. J. Exp. Med. 2008;205:1621–1634. doi: 10.1084/jem.20071365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kourepini E, et al. Osteopontin expression by CD103− dendritic cells drives intestinal inflammation. Proc. Natl. Acad. Sci. USA. 2014;111:E856–E865. doi: 10.1073/pnas.1316447111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Plantinga M, et al. Conventional and monocyte-derived CD11b+ dendritic cells initiate and maintain T helper 2 cell-mediated immunity to house dust mite allergen. Immunity. 2013;38:322–335. doi: 10.1016/j.immuni.2012.10.016. [DOI] [PubMed] [Google Scholar]