Abstract
Halophilic microorganisms are able to grow in the presence of salt and are also excellent source of enzymes and biotechnological products, such as exopolysaccharides (EPSs) and polyhydroxyalkanoates (PHAs). Salt-tolerant bacteria were screened in the Organic Composting Production Unit (OCPU) of São Paulo Zoological Park Foundation, which processes 4 ton/day of organic residues including plant matter from the Atlantic Rain Forest, animal manure and carcasses and mud from water treatment. Among the screened microorganisms, eight halotolerant bacteria grew at NaCl concentrations up to 4 M. These cultures were classified based on phylogenetic characteristics and comparative partial 16S rRNA gene sequence analysis as belonging to the genera Staphylococcus, Bacillus and Brevibacterium. The results of this study describe the ability of these halotolerant bacteria to produce some classes of hydrolases, namely, lipases, proteases, amylases and cellulases, and biopolymers. The strain characterized as of Brevibacterium avium presented cellulase and amylase activities up to 4 M NaCl and also produced EPSs and PHAs. These results indicate the biotechnological potential of certain microorganisms recovered from the composting process, including halotolerant species, which have the ability to produce enzymes and biopolymers, offering new perspectives for environmental and industrial applications.
Keywords: halophilic, protease, lipase, amylase, cellulase
Introduction
The biocatalysts required in several industrial processes exhibit optimal activities at high ranges of salt concentration, pH and temperature. Halophiles are excellent sources of such enzymes and are found in nearly all major microbial clades, including prokaryotic (Bacteria and Archaea) and eukaryotic forms; two categories have been defined: halotolerant microorganisms that are adapted in live at high salinity, and halophiles that require salinity for growth. Halotolerant species tend to live in areas of salinity, such as hypersaline lakes, coastal dunes, saline deserts and salt seas (Ventosa and Nieto, 1995).
Halophilic enzymes perform the same enzyme function as their non-halophilic counterparts but require 1–4 M salt concentrations for their full activity and stability. In addition, these enzymes typically demonstrate a large excess of acidic amino acids compared to basic residues (Enache and Kamekura, 2010).
Proteases constitute approximately 66% of the total enzymes employed in biotechnological and commercial processes (Gupta et al., 2002), and the moderately halophilic aerobic bacteria of genera Bacillus, Pseudomonas, Halomonas and Serratia are important sources of proteases (Ventosa et al., 1998). Amylases are extensively studied due to their potential application in the food, detergent, paper and pharmaceutical industries, representing approximately 25% of the total enzymes in the industrial market. The extracellular production of β-amylase by halophilic Halobacillus sp. LY9 and of two α-amylases from Chromohalobacter sp. has been reported (Li and Yu, 2011; Prakash et al., 2009). Cellulases also have industrial application, including the generation of bioethanol and in the textile industry, and a halotolerant cellulase was characterized in a soil metagenome analysis (Voget et al., 2006). Lipolytic enzymes are of particular industrial interest, and their identification in halophilic bacteria has been reported and recently reviewed (Gomez et al., 2012). Exopolysaccharides (EPSs) and polyhydroxyalkanoates (PHAs) are biotechnological products that were identified and produced from halophilic/halotolerant microorganisms (Legat et al., 2010; Litchfield, 2012).
In this sense, the Organic Composting Production Unit (OCPU) of SPZPF is a potential source of microorganisms, as demonstrated by an OCPU metagenomic analysis, which revealed a diversity of biomass degradation functions and organisms (Martins et al., 2013). The composting process is predominantly aerobic, with organic residues being degraded by microorganisms, generating a humus-like material. In recent years, composting has attracted attention as a viable and environmentally adequate alternative for the treatment of organic waste. The initial phase of composting is thought to be the most dynamic part of the process and is characterized by a rapid increase in temperature, a large change in pH, and the degradation of simple organic compounds (Schloss et al., 2003). A detailed comparison of the bacterial diversity from different composting plants revealed a large difference at both the species and strain levels (Partanen et al., 2010).
This paper reports the screening of the composting process of OCPU at SPZPF for bacteria in the presence of a range of NaCl concentrations and also the evaluation of their potential for the production hydrolases and biopolymers. To date, the microbial diversity of this ecosystem has not been explored, particularly with regard to the screening of halotolerant microorganisms.
Material and Methods
Bacterial strains and isolation of DNA
Composting process
The composting process was conducted in the SPZPF OCPU in 2.5 × 2.0 × 1.6 m (length×width×height) cells, as shown in Figure 1. The piles were formed by organic residues including food, droppings and excreta, the beds of native and exotic wild animals, carcasses and wood chips from gardening. The pile has decomposition phases that were considered active degradation (before aeration) and mature compost (after aeration). Pile aeration was achieved by the mechanical turning of the material after 50 to 60 days of composting. The temperature of the pile was monitored at five different points (four sides and one center). The average temperature of the pile at the time of collection was 50 °C.
Figure 1. Partial view of the Organic Composting Production Unit (OCPU) of São Paulo Zoological Park Foundation. (A) Aerobic composting process in the thermophilic phase; (B) grinding of plant matter from the Atlantic Rain Forest; (C) maturation phase of the composting process.
Microbial isolation
Compost samples (10 g) were collected from the piles and diluted in 90 mL of sterile water. Serial dilutions (−2, −4 and −6) were performed and spread on agar plates of two selective halophilic media: JCM nº 377 medium (10% [w/v] NaCl, 0.5% [w/v] casamino acids, 0.5% [w/v] yeast extract, 0.2% [w/v] KCl, 0.3% [w/v] sodium citrate, 2% [w/v] MgSO4•7H2O, 0.036% [w/v] FeCl2 4H2O, 0.00036% [w/v] MnCl2 4H2O and 2% [w/v] agar, pH 7.2) and YPC medium (0.5% [w/v] yeast extract, 0.1% [w/v] peptone and 0.1% [w/v] casamino acids with 60% [v/v] of salt water solution 24% [w/v] NaCl, 3% [w/v] MgCl2 6H2O, 3.5% [w/v] MgSO4•7H2O, 0.1% [w/v] KCl, 20 mM Tris HCl pH 7.5 and 3 mM CaCl2) and incubated at 30, 37 and 42 °C. After 24 or 48 h, the colonies were selected and transferred separately to obtain purified colonies.
Screening of secreted extracellular hydrolytic activities
Enzymatic agar plate assays were performed to detect the presence of extracellular hydrolases. All media were adjusted to pH 7.3, and NaCl was added to obtain a salt concentration in the range of 0–4 M. The composition of the media used is described below.
Determination of extracellular amylase activity
Amylolytic activity on plates was determined qualitatively using a previously described method (Pascon et al., 2011), which was modified for halophilic microorganisms by adding NaCl in the medium. After incubation at 37 °C for 5 days, the plates were exposed to iodine crystals for 5 min to reveal the starch degradation zone that indicates amylolytic activity.
Determination of extracellular protease activity
The cultures were screened in JCM nº 377 medium and YPC medium supplemented with 1% skim milk for the determination of protein hydrolytic activity. Clear zones around the colonies after 7 days were taken as evidence of proteolytic activity.
Determination of extracellular lipase activity
Lipase production by the isolated microorganisms was evaluated in nutrient agar tributyrin medium (NAT), which consisted of 1.3% nutrient broth, 1% tributyrin and 2% agar (Ben-Gigirey et al., 2000). After incubation at 37 °C for 7 days, the hydrolytic zones around the bacterial colonies were considered an indication of lipase production.
Determination of extracellular cellulase activity
Cellulase activity was screened on a solid medium containing carboxymethyl cellulose (CMC) (Rohban et al., 2009). After incubation at 37 °C for 7 days, the plates were flooded with 0.1% Congo red solution. The clear zone around colonies indicated cellulolytic activity.
Screening of polyhydroxyalkanoates and exopolysaccharides
Detection of polyhydroxyalkanoate (PHA)-producing microorganisms
The isolates were evaluated in mineral medium (Schlegel et al., 1970) with 2.5 M NaCl and containing glucose, xylose or octanoic acid as the carbon source. Glucose is known to be a carbon source for the production of short-chain-length PHAs, whereas octanoic acid produces medium-chain-length PHAs. Sugarcane bagasse contains xylose, and its excess is a promising substrate for producing by-products, such as second-generation bioethanol and PHAs (Lopes et al., 2009). After 24 h of incubation (30 °C), the isolated strains were evaluated for their ability to grow on these carbon sources; the isolates were stained with Sudan Black B after 72 h to verify their potential to produce PHAs.
Detection of exopolysaccharide (EPS) producers
The isolates were cultivated in Bushnell Haas Salt Medium (50 mL) containing 2.5 M NaCl with glycerol as sole the carbon source for the microbial growth. After incubation for 5 days at 30 °C in a rotary shaker (150 rpm), the cultures were centrifuged at 8,200 × g for 15 min (4 °C). The emulsification index (E24) of the supernatant was evaluated according to the method described by Fleck et al. (2000) using hexadecane as a hydrophobic model compound. The chemical composition of EPSs precipitated from the supernatant with ethanol up to 70% was dialyzed against pure water, and carbohydrates, proteins and uronic acids were quantified in the retained high molecular weight fraction, as reported (Tanasupawat et al., 2010).
Bacterial identification
Mass spectrometry
The isolated microorganisms were treated with ethanol/formic acid for content extraction, following a previously described protocol (Pascon et al., 2011). Measurements were conducted with a Microflex LT mass spectrometer (Bruker Daltonics) using FlexControl software (version 3.0, Bruker Daltonics) in the positive linear mode (laser frequency, 20 Hz; ion source 1 voltage, 20 kV; ion source 2 voltage, 18.6 kV; lens voltage, 7.5 kV; mass range, 2000 to 20 000 Da). For each spectrum, 240 shots in 50-shot steps from different positions of the target spot (automatic mode) were collected and analyzed. The spectra were internally calibrated using Escherichia coli ribosomal proteins. The raw spectra were imported into the BioTyper software (version 2.0, Bruker Daltonics) and processed by standard pattern matching with default settings; the results were reported in a ranking table.
Amplification and sequencing of 16S rRNA gene fragment
DNA (30–50 ng) from each strain was incubated in a 50-μL reaction mixture containing 2 mM MgCl2, 200 μM dNTPs, 0.3 μM universal primer 27f (5-AGAGTTGATCCTGGCTCAG-3), 0.3 μM 1525r (5-AAGGAGGTGWTCCARCC-3) and 2 U Taq DNA polymerase (Invitrogen) in the recommended buffer. Amplification was performed in a Veriti 96 well Thermal Cycler (Applied Biosystems) with an initial temperature at 94 °C for 2 min, 30 cycles at 94 °C for 1 min, 55 °C for 1 min and 72 °C for 3 min. A final extension at 72 °C was included for 10 min. The PCR products were purified with a GFX PCR DNA and gel band purification kit (GE Healthcare), and the sequence analysis was performed using a 3500 Genetic Analyzer Sequencer (Applied Biosystems). Subsequently, 5.0 μL purified PCR product was mixed with 4.0 μL of BigDye v. 3.1 (Applied Biosystems) and 1.0 μL sequencing primer (0.5 μmol). The primers used in the sequencing reactions were 27f (Dojka et al., 1998), 782r (5ACCAGGGTATCTAATCCTGT3) (Chun and Goodfellow, 1995) and 1401r (5CGGTGTGTACAAGACC C3) (Nübel et al., 1996). The sequencing program consisted of 25 cycles at 95 °C for 20 s, 50 °C for 15 s and 60 °C for 60 s. The 16S rRNA gene sequence of all the analyzed strains was compared to bacterial sequences deposited in GenBank. Sequences with similarity were retrieved, and the consensus sequences were aligned using CLUSTALW with MEGA 5.05. EzTaxon tools (http://147.47.212.35:8080/) were further employed to confirm the similarities, and phylogenetic trees were constructed based on neighbor-joining, maximum-likelihood and maximum-parsimony methods. The resulting tree topologies were evaluated by a bootstrap analysis based on 1000 replicates.
Nucleotide sequence accession numbers
The sequenced strains SR5-6, SR5-7, SR5-12, YPC-6, YPC-8, YPC-11, YPC-13 and YPC-15 were deposited in GenBank under accession numbers JX154082, JX154083, JX154084, JX154085, JX154086, JX154087, JX154088 and JX154089, respectively.
Results and Discussion
Identification of halophilic strains
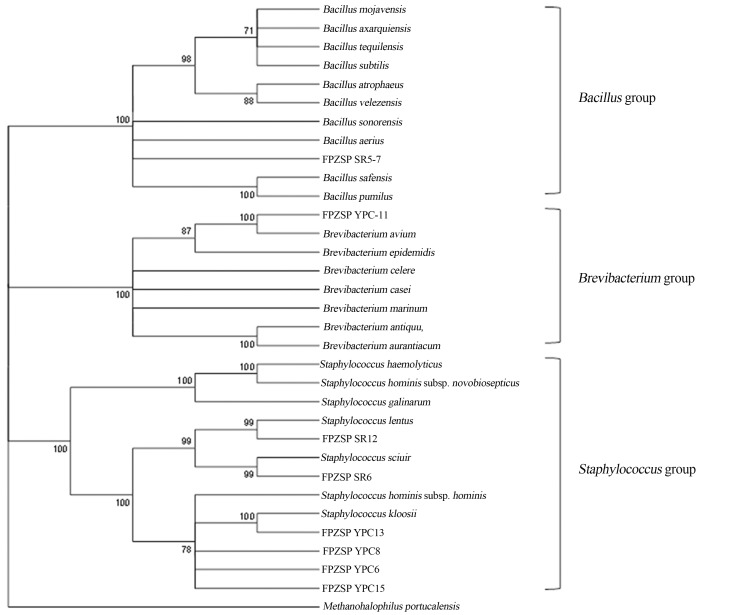
The isolated bacteria were obtained from the composting process during the turning stage (60th day). Eight out of eleven halophilic isolates in 2.5 M NaCl from the composting process were subjected to a MALDI-TOF mass spectrometry analysis, which indicated that the genera of all isolates were Gram positive, which was confirmed by Gram staining. These procedures ensured and confirmed the purity of the isolates. The 16S rRNA gene sequences of eight strains (> than 1300 bp) were compared with those previously deposited in GenBank. The neighbor-joining and maximum-likelihood trees showed the taxonomic position of these strains, which were affiliated with Bacillus, Staphylococcus and Brevibacterium genera (Figure 2).
Figure 2. Phylogenetic tree showing the position of the halotolerant isolates, as based on a partial 16S rRNA gene sequence comparison obtained by neighbor-joining and maximum-likelihood trees. The nucleotide sequence accession numbers were deposited in GenBank, as described in Material and Methods.
Strain SR5-7 showed high 16S rRNA gene sequence similarity to Bacillus when compared with the 184 different species of this genus. However, based on the similarity matrix of the 16S rRNA gene, this isolate did not show 100% similarity with any of the species already reported. The species of Bacillus described as halophilic to date are as follows: B. hemicentroti (Chen et al., 2011); B. humanensis (Chen et al., 2011); B. xianensis (Sanchez-Porro et al., 2003; Schlegel et al., 1970); B. alkaliphilic (Zhang et al., 2012); B. halochares (Pappa et al., 2010); B. chungangensis (Cho et al., 2010) and B. subtilis (Takenaka et al., 2011). Thus, the possibility that a new species of halophilic Bacillus was isolated from a compost process is noteworthy.
The YPC-11 strain was identified as Brevibacterium avium (100% similarity). An EzTaxon analysis confirmed that this strain shared 100% 16S rRNA gene sequence similarity with B. avium and 99.97% with Brevibacterium epidermidis, the only halotolerant bacterium (Nagata and Wang, 2005) described in the genus Brevibacterium.
Strains SR5-12, SR5-6, YPC-6, YPC-8, YPC-13 and YPC-15 were classified as members of the Staphylococcus genus. The isolates SR5-12 and SR5-6 showed 100% 16S rRNA gene sequence similarity with S. lentus and S. sciuri, respectively, a result that was confirmed by EzTaxon. However, to date, these species have not been described as high salt concentration-tolerant bacteria, with the only species of Staphylococcus known as halophilic being Staphylococcus equorum (Essghaier et al., 2009).
All the selected isolates were deposited at the São Paulo Zoo Park Culture Collection (SPZSP-CCol).
Salt tolerance and growth of halophilic isolates
All the bacteria isolated in 2.5 M NaCl were tested for their ability to grow at different salt concentrations. A slowing of bacterial growth was observed in the presence of high salt concentrations, as indicated by the time (in days) required for detecting the presence of bacteria in the culture medium (Table 1). Staphylococcus strains SR5-12, YPC-6, SR5-6 and YPC-8 showed similar growth behavior from 0 to 4.0 M NaCl. Strain YPC-13 had the slowest growth at high salinity, and strain YPC-15 grew preferentially at 2.5 M NaCl or higher. Bacillus strains SR5-7 and YPC-11 (affiliated with B. avium) exhibited a preference for growing in a culture medium containing 0.5 to 2.0 M NaCl but failed to grow in 4.0 M NaCl. It is important to note that although all of these bacteria tolerated high salinities (2.5 M NaCl or higher), they are not strictly halophilic bacteria. According to Kushner (1978), bacteria that are able to grow in the absence of salt as well as in the presence of relatively high salt concentrations are designated halotolerant or extremely halotolerant if growth extends above 2.5 M. Based on this classification, seven out of the eight isolated microorganisms isolated from composting process were halotolerant or extremely halotolerant. It should be noted that the salt requirement and tolerance of many species vary according to the growth conditions, such as temperature and medium composition.
Table 1. Different types of hydrolytic activities, amylase (A), cellulase (C), lipase (L) and protease (P), found in the eight strains isolated from OCPU at the 0 – 4.0 M NaCl concentration range.
| SR5-6 (Staphylococcus sp.) | SR5-7 (Bacillus sp.) | SR5-12 (S. lentus) | YPC-6 (Staphylococcus sp.) | YPC-8 (Staphylococcus sp.) | YPC-11 (B. avium) | YPC-13 (Staphylococcus sp.) | YPC-15 (Staphylococcus sp.) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|
|
|
|||||||||
| [NaCl] | Days | Hydrolase | Days | Hydrolase | Days | Hydrolase | Days | Hydrolase | Days | Hydrolase | Days | Hydrolase | Days | Hydrolase | Days | Hydrolase |
| 0 M | 1 | Nd | 1 | L, P | 1 | L, P | 1 | Nd | 2 | L | 2 | A, C | 2 | Nd | 4 | A, C, L |
| 0.5 M | 1 | L | 1 | L, P | 1 | L | 1 | Nd | 2 | L | 2 | A, C | 2 | Nd | 0 | Nd |
| 1.0 M | 1 | Nd | 1 | L | 1 | L | 1 | Nd | 2 | L | 2 | A, C | 2 | Nd | 0 | Nd |
| 1.5 M | 1 | Nd | 1 | L | 1 | Nd | 1 | Nd | 2 | L | 2 | A, C | 2 | Nd | 0 | Nd |
| 2.0 M | 1 | Nd | 1 | L | 1 | Nd | 1 | Nd | 2 | L | 3 | A, C | 3 | Nd | 1 | A, C |
| 2.5 M | 1 | Nd | 2 | Nd | 2 | Nd | 2 | Nd | 2 | L | 4 | A, C | 7 | Nd | 2 | A, C |
| 3.0 M | 2 | Nd | 2 | Nd | 2 | Nd | 2 | Nd | 2 | Nd | 7 | A, C | 7 | Nd | 2 | A, C |
| 3.5 M | 4 | Nd | 2 | Nd | 2 | Nd | 3 | Nd | 3 | Nd | 21 | A, C | 21 | Nd | 3 | A, C |
| 4.0 M | 7 | Nd | - | Nd | 4 | Nd | 4 | Nd | 4 | Nd | 21 | A, C | 21 | Nd | 3 | A, C |
The assays were performed as described in Material and Methods. Days: Time in days that visible growth was observed, considering zero to be the day of inoculation. (Nd): Hydrolytic activity not detected.
Several bacteria of Bacillus, Halobacillus and Staphylococcus have been found in saline environments, such as Salt Plains National Wildlife Refuge, Great Salt Plains of Oklahoma, a Bolivian hypersaline lake, deep-sea sediments and tropical marine sediments (Ventosa et al., 1998). Some species of Bacillus sp. are salt tolerant and are important degraders of organic pollutants. Examples include Bacillus cereus, which degrades 1,3-dichlorobenzene derivatives from town-gas industrial influent (Wang et al., 2003), and Bacillus subtilis, which degrades p-aminobenzene from textile industry wastewater (Zissi et al., 1997).
Hydrolytic activities of halotolerant isolates
The SR5-6, SR5-7, SR5-12, YPC-8, YPC-11 and YPC-15 strains were found to be moderate halophilic microorganisms and showed combined cellulolytic, amylolytic, lipolytic and proteolytic activities (Table 1). These strains have potential biotechnological applications with respect to their ability to produce different hydrolases (Rohban et al., 2009). In contrast, no hydrolytic activity was observed for YPC-6 and YPC-13.
Only YPC-11 (affiliated with B. avium) presented amylase and cellulase hydrolytic activities from 0 to 4 M NaCl. Members of the genus Bacillus are well known enzyme producers, and many industrial processes utilize species belonging to this genus for the commercial production of enzymes (Vasconcellos et al., 2011). The strain SR5-7 (affiliated with Bacillus) produced lipase and protease in 2.0 M and 0.5 M NaCl, respectively. It is interesting to note that the lipase producers reported thus far are limited to representatives of the genera Salinivibrio, Halomonas and Bacillus-Salibacillus (Sanchez-Porro et al., 2003).
Polyhydroxyalkanoate (PHA) producers
All the isolates were evaluated using a medium with nitrogen limitation and different carbon sources (Table 2). The isolates grew better with glucose as the sole carbon source compared to xylose and octanoic acid. The isolates YPC-13 (affiliated with Bacillus sp.), SR5-7 (affiliated with Bacillus sp.) and YPC-15 (affiliated with Staphylococcus sp.) accumulated PHAs in presence of octanoic acid, xylose and glucose, respectively. The genus Bacillus is known as a producer of PHAs (Lopes et al., 2009), and Staphylococcus epidermidis, which was isolated from sesame oil, presented the ability to produce poly-3-hydroxybutyrate (Wong et al., 2000). The strain YPC-11 (affiliated with B. avium) was detected as a potential producer of biopolymers using octanoic acid and xylose. This result is in accordance with the previous observation that Brevibacterium casei (SRKP2 strain) could produce PHAs in a medium containing dairy industrial waste, yeast extract and sea water (Pandian et al., 2009). Halotolerant microbes are important for the biotechnology industry due to their advantages for use in sterilization processes and the control of contaminants; the PHA-producing halophilic microorganisms have recently been reviewed (Poli et al., 2011). The production of PHAs using xylose is an alternative strategy to produce economically competitive PHAs using agro-industrial products such as sugarcane molasses and bagasse (Gomez et al., 2012).
Table 2. Analysis of isolates for the ability to produce EPSs in mineral medium and PHAs using different carbon sources in the presence of 2.5 M NaCl.
| SR-6 (Staphylococcus sp.) | SR-7 (Bacillus sp.) | SR-12 (S. lentus) | YPC-6 (Staphylococcus sp.) | YPC-8 (Staphylococcus sp.) | YPC-11 (B. avium) | YPC-13 (Staphylococcus sp.) | YPC-15 (Staphylococcus sp.) | |
|---|---|---|---|---|---|---|---|---|
| EPS (E24) | 58% | 50% | 51% | 53% | 50% | 55% | 55% | 55% |
| PHA | Nd | Xylose (+) | Nd | Nd | Nd | Xylose (+) Octanoic acid (+) | Octanoic acid (+) | Glucose (+) |
(E24): Emulsification indexes of the isolates in a mineral medium containing 2.5 M NaCl, as described in Materials and Methods.
(+): indicates that PHAs were detected when using the indicated carbon source.
(Nd): PHA production not detected.
EPS production and emulsification potential
Microbial exopolymers (EPSs) correspond to compounds produced by microorganisms to solubilize essential nutrients for their survival or to promote their adherence onto surfaces (Ron and Rosenber, 2002). The use of glycerol as a sole carbon source and 2.5 M NaCl resulted in EPS values up to 60% of the emulsification index (E24) of hexadecane (Table 2). A colorimetric analysis showed that the biosurfactant produced by the evaluated halotolerant strains were mainly composed of carbohydrates (95%) but also contained proteins (0.5%) and uronic acids (4.5%) in their composition. A similar EPS composition was also reported in halophilic Archaea strains (Poli et al., 2011).
Conclusion
Screens for halotolerant or halophilic microorganisms in non-saline environments are scarce as is the detection of extracellular enzymes. This study found eight isolates from an organic residue composting process that showed the ability to tolerate a wide range of salinity. Some of these strains presented combined hydrolytic ability in the presence of NaCl. The possibility of these microorganisms, particularly YPC-11 (affiliated with B. avium), to produce EPSs and PHAs in the presence of 2.5 M NaCl can offer new biotechnological and bioremediation perspectives for the treatment of oilfield wastes as well as in MEOR (microbial-enhanced oil recovery) processes. The performance of the halotolerant isolates in the present work were not compared to other already known and classic halophilic microorganisms, and this should be performed in future work.
Acknowledgments
This study was supported by the Brazilian research agencies Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP - 2009/52030-5) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq - 471340/2011-1). The authors would like to express their gratitude to Fundação Parque Zoológico de São Paulo for providing the material from its composting facility and to recognize the relevant participation and support of Mr. Robson Carlos Santos and Mr. Carlos Augusto Magalhães Batista, at Fundação Parque Zoológico de São Paulo, in the composting unit processes.
References
- Ben-Gigirey B, de Sousa JMVB, Villa TG, et al. Characterization of biogenic amine-producing Stenotrophomonas maltophilia strains isolated from white muscle of fresh and frozen albacore tuna. Int J Food Microbiol. 2000;57:19–31. [Google Scholar]
- Chen YG, Hao DF, Chen QH, et al. Bacillus hunanensis sp nov., a slightly halophilic bacterium isolated from non-saline forest soil. Antonie Van Leeuwenhoek Int J Genet Mol Microbiol. 2011;99:481–488. doi: 10.1007/s10482-010-9512-7. [DOI] [PubMed] [Google Scholar]
- Chen YG, Zhang YQ, He JW, et al. Bacillus hemicentroti sp. nov., a moderate halophile isolated from a sea urchin. Int J Syst Evol Microbiol. 2011;61:2950–2955. doi: 10.1099/ijs.0.026732-0. [DOI] [PubMed] [Google Scholar]
- Cho SL, Jung MY, Park MH, et al. Bacillus chungangensis sp. nov., a halophilic species isolated from sea sand. Int J Syst Evol Microbiol. 2010;60:1349–1352. doi: 10.1099/ijs.0.013607-0. [DOI] [PubMed] [Google Scholar]
- Chun J, Goodfellow M. A phylogenetic analysis of the genus Norcadia with 16S rDNA gene sequences. Int J Syst Bacteriol. 1995;45:240–245. doi: 10.1099/00207713-45-2-240. [DOI] [PubMed] [Google Scholar]
- Dojka MA, Hugenholtz P, Haack SK, et al. Microbial diversity in a hydrocarbon-and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl Environ Microbiol. 1998;64:3869–3877. doi: 10.1128/aem.64.10.3869-3877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enache M, Kamekura M. Hydrolytic enzymes of halophilic microorganisms and their economic values. Rom J Biochem. 2010;47:47–59. [Google Scholar]
- Essghaier B, Fardeau ML, Cayol JL, et al. Biological control of grey mould in strawberry fruits by halophilic bacteria. J Appl Microbiol. 2009;106:833–846. doi: 10.1111/j.1365-2672.2008.04053.x. [DOI] [PubMed] [Google Scholar]
- Fleck LC, Bicca FC, Ayub MAZ. Physiological aspects of hydrocarbon emulsification, metal resistance and DNA profile of biodegrading bacteria isolated from oil polluted sites. Biotechnol Lett. 2000;22:285–289. [Google Scholar]
- Fuciños P, Gonzalez R, Atanes E, et al. Lipases and esterases from extremophiles: overview and case example of the production and purification of an esterase from Thermus thermophilus HB27. Methods Mol Biol. 2012;861:239–266. doi: 10.1007/978-1-61779-600-5_15. [DOI] [PubMed] [Google Scholar]
- Gomez J, Mendez B, Nikel P, et al. Making Green polymers even greener: Towards sustainable production of polyhydroxyalkanoates from agroindustrial by-products. In: Petre M, editor. Adv Appl Biotechnol. InTech Publisher; 2012. pp. 41–62. [Google Scholar]
- Gupta R, Beg QK, Lorenz P. Bacterial alkaline proteases: molecular approaches and industrial applications. Appl Microbiol Biotechnol. 2002;59:15–32. doi: 10.1007/s00253-002-0975-y. [DOI] [PubMed] [Google Scholar]
- Kushner DJ. Life in High Salt and Solute Concentrations: Halophilic Bacteria. Academic Press; London: 1978. [Google Scholar]
- Legat A, Gruber C, Zangger K, et al. Identification of polyhydroxyalkanoates in Halococcus and other haloarchaeal species. Appl Microbiol Biotechnol. 2010;87:1119–1127. doi: 10.1007/s00253-010-2611-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yu HY. Extracellular production of beta-amylase by a halophilic isolate, Halobacillus sp. LY9. J Ind Microbiol Biotechnol. 2011;38:1837–1843. doi: 10.1007/s10295-011-0972-1. [DOI] [PubMed] [Google Scholar]
- Litchfield CD. Potential for industrial products from the halophilic Archaea. J Ind Microbiol Biotechnol. 2012;38:1635–1647. doi: 10.1007/s10295-011-1021-9. [DOI] [PubMed] [Google Scholar]
- Lopes MSG, Rocha RCS, Zanotto SP, et al. Screening of bacteria to produce polyhydroxyalkanoates from xylose. World J Microbiol Biotechnol. 2009;25:1751–1756. [Google Scholar]
- Martins LF, Antunes LP, Pascon RC, et al. Metagenomic analysis of a tropical composting operation at the São Paulo zoo park reveals diversity of biomass degradation functions and organisms. PLoS One. 2013;8:e61928. doi: 10.1371/journal.pone.0061928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata S, Wang CX. Efficient utilization of ectoine by halophilic Brevibacterium species and Escherichia coli subjected to osmotic down shock. J Biosci Bioeng. 2005;99:61–67. doi: 10.1263/jbb.99.61. [DOI] [PubMed] [Google Scholar]
- Nübel U, Engelen B, Felske A, et al. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxadetected by temperature gradient gel electrophoresis. J Bacteriol. 1996;178:5636–5643. doi: 10.1128/jb.178.19.5636-5643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandian SRK, Deepak V, Kalishwaralal K, et al. Synthesis of PHB nanoparticles from optimized medium utilizing dairy industrial waste using Brevibacterium casei SRKP2: A green chemistry approach. Colloids Surf B-Biointerfaces. 2009;74:266–273. doi: 10.1016/j.colsurfb.2009.07.029. [DOI] [PubMed] [Google Scholar]
- Pappa A, Sanchez-Porro C, Lazoura P, et al. Bacillus halochares sp. nov., a halophilic bacterium isolated from a solar saltern. Int J Syst Evol Microbiol. 2010;60:1432–1436. doi: 10.1099/ijs.0.014233-0. [DOI] [PubMed] [Google Scholar]
- Partanen P, Hultman J, Paulin L, et al. Bacterial diversity at different stages of the composting process. BMC Microbiol. 2010;10:1–11. doi: 10.1186/1471-2180-10-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascon RC, Bergamo RF, Spinelli RX, et al. Amylolytic Microorganism from São Paulo Zoo Composting: Isolation, Identification, and Amylase Production. Enzym Res. 2011;2011:679624. doi: 10.4061/2011/679624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli A, Di Donato P, Abbamondi GR, et al. Synthesis, Production, and Biotechnological Applications of Exopolysaccharides and Polyhydroxyalkanoates by Archaea. Archaea. 2011;2011:693253. doi: 10.1155/2011/693253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash B, Vidyasagar M, Madhukumar MS, et al. Production, purification, and characterization of two extremely halotolerant, thermostable, and alkali-stable alpha-amylases from Chromohalobacter sp TVSP 101. Process Biochem. 2009;44:210–215. [Google Scholar]
- Rohban R, Amoozegar MA, Ventosa A. Screening and isolation of halophilic bacteria producing extracellular hydrolases from Howz Soltan Lake, Iran. J Ind Microbiol Biotechnol. 2009;36:333–340. doi: 10.1007/s10295-008-0500-0. [DOI] [PubMed] [Google Scholar]
- Ron EZ, Rosenberg E. Biosurfactants and oil bioremediation. Curr Opin Biotechnol. 2002;13:249–252. doi: 10.1016/s0958-1669(02)00316-6. [DOI] [PubMed] [Google Scholar]
- Sanchez-Porro C, Martin S, Mellado E, Ventosa A. Diversity of moderately halophilic bacteria producing extracellular hydrolytic enzymes. J Appl Microbiol. 2003;94:295–300. doi: 10.1046/j.1365-2672.2003.01834.x. [DOI] [PubMed] [Google Scholar]
- Schlegel HG, Lafferty R, Krauss I. Isolation of mutants not accumulating poly-beta-hydroxybutyric acid. Arch Für Mikrobiol. 1970;71:283–294. doi: 10.1007/BF00410161. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Hay AG, Wilson DB, et al. Tracking temporal changes of bacterial community fingerprints during the initial stages of composting. FEMS Microbiol Ecol. 2003;46:1–9. doi: 10.1016/S0168-6496(03)00153-3. [DOI] [PubMed] [Google Scholar]
- Takenaka S, Yoshida N, Yoshida K, et al. Molecular cloning and sequence analysis of two distinct halotolerant extracellular proteases from Bacillus subtilis FP-133. Biosci Biotechnol Biochem. 2011;75:148–151. doi: 10.1271/bbb.100588. [DOI] [PubMed] [Google Scholar]
- Tanasupawat S, Chamroensaksri N, Kudo T, et al. Identification of moderately halophilic bacteria from Thai fermented fish (pla-ra) and proposal of Virgibacillus siamensis sp. nov. J Gen Appl Microbiol. 2010;56:369–379. doi: 10.2323/jgam.56.369. [DOI] [PubMed] [Google Scholar]
- Vasconcellos SP, Dellagnezze BM, Wieland A, et al. The potential for hydrocarbon biodegradation and production of extracellular polymeric substances by aerobic bacteria isolated from a Brazilian petroleum reservoir. World J Microbiol Biotechnol. 2011;27:1513–1518. doi: 10.1007/s11274-010-0581-6. [DOI] [PubMed] [Google Scholar]
- Ventosa A, Nieto JJ. Biotecnological applications and potentialities of halophilic microrganisms. World J Microbiol Biotechnol. 1995;11:85–94. doi: 10.1007/BF00339138. [DOI] [PubMed] [Google Scholar]
- Ventosa A, Nieto JJ, Oren A. Biology of moderately halophilic aerobic bacteria. Microbiol Mol Biol Rev. 1998;62:504–544. doi: 10.1128/mmbr.62.2.504-544.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voget S, Steele HL, Streit WR. Characterization of a metagenome-derived halotolerant cellulase. J Biotechnol. 2006;126:26–36. doi: 10.1016/j.jbiotec.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Wang L, Zhou Q, Zhang BS, et al. The biodegradation of 1,3-dichlorobenzene by an adapted strain Bacillus cereus PF-11 derived from town-gas industrial effluent. J Environ Sci Health Part A. 2003;38:1837–1848. doi: 10.1081/ese-120022882. [DOI] [PubMed] [Google Scholar]
- Wong AL, Chua H, Yu pH. Microbial production of polyhydroxyalkanoates by bacteria isolated from oil wastes. Appl Biochem Biotechnol. 2000;84–86:843–857. doi: 10.1385/abab:84-86:1-9:843. [DOI] [PubMed] [Google Scholar]
- Zhang GM, Li SY, Xue YF, et al. Effects of salts on activity of halophilic cellulase with glucomannanase activity isolated from alkaliphilic and halophilic Bacillus sp. BG-CS10. Extremophiles. 2012;16:35–43. doi: 10.1007/s00792-011-0403-2. [DOI] [PubMed] [Google Scholar]
- Zissi U, Lyberatos G, Pavlou S. Biodegradation of p-aminoazobenzene by Bacillus subtilis under aerobic conditions. J Ind Microbiol Biotechnol. 1997;19:49–55. [Google Scholar]