Abstract
In this study, we revealed that OA and UA significantly inhibited the expression of most genes related to peptidoglycan biosynthesis in S. mutans UA159. To the best of our knowledge, this is the first report to introduce the antimicrobial mechanism of OA and UA against S. mutans.
Keywords: ursolic acid, oleanolic acid, peptidoglycan biosynthesis, Streptococcus mutans UA159
Ursolic acid (UA, (3β)-hydroxy-urs-12-en-28-oic acid), oleanolic acid (OA, 3β-3-hydroxyolean-12-en-28-oic acid), and betulinic acid (BA, 3β-3-hydroxy-lup-20(29)-en-28-oic acid) are derivatives of triterpenoid saponins (Liu, 1995; Fontanay et al., 2008). These compounds are naturally found in a large variety of vegetables, medicinal herbs, and plants that have been investigated for antibacterial activity (Fontanay et al., 2008). The antimicrobial activity of OA and UA is stronger than that of BA (Fontanay et al., 2008). OA and UA inhibit growth of Gram-positive bacteria but not Gram-negative bacteria (Fontanay et al., 2008), but their antimicrobial mechanism is unknown. Previously, we reported that OA and UA have strong antimicrobial activity against Streptococcus mutans (Kim et al., 2010; Kim et al., 2011) but BA had no antimicrobial activity against S. mutans (data not shown). One of the characteristic differences between Gram-positive and Gram-negative bacteria is the thickness of the peptidoglycan layer. Considering this difference, it has been suggested that the mechanism of OA and UA antimicrobial activity may be related to inhibition of peptidoglycan biosynthesis (Horiuchi et al., 2007). Thus, the objective of this study was to investigate the effect of BA, OA, and UA on peptidoglycan biosynthesis in S. mutans UA 159 using the quantitative real-time polymerase chain reaction (qPCR) method to identify the antimicrobial mechanism against S. mutans.
S. mutans UA159 was a kind gift from Dr. Robert A. Burne, Department of Oral Biology, College of Dentistry, University of Florida. The strain was cultured in brain heart infusion (BHI) broth (Difco, Lab., Detroit, MI, USA) or on BHI agar plates in a 37 °C incubator.
Overnight cultures (1 mL) of S. mutans UA 159 were transferred to 9 mL BHI broth and grown at 37 °C to the mid-log phase (OD600 = 0.35). UA (Sigma, St. Louis, MO, USA), OA (Sigma), or BA (Sigma) solutions were added to each tube to a final concentration of 64 μg/mL. The bacterial culture solutions were incubated in a 37 °C incubator for 90 min and then harvested by centrifugation at 10,000 × g for 10 min at 4 °C. After discarding the supernatant, liquid nitrogen was immediately added to the tube. The frozen bacterial pellets were homogenized using a mortar and pestle (Smile Science, Seoul, Korea). Total RNAs were extracted from the homogenized bacteria following the manufacturer's instructions of the RNAqueous® kit (Ambion, Austin, TX, USA). DNase I treatment was conducted to completely remove the bacterial genomic DNAs using a TURBO DNA-free™ Kit (Ambion) according to the manufacturer's protocol. RNA concentration was determined at 260 nm 280 nm with a UV spectrophotometer (Ultrospec 2000, Pharmacia Biotech., UK).
We consulted the KEGG pathway database to determine the target genes related to S. mutans UA159 peptidoglycan biosynthesis (KEGG pathway). qPCR primers were designed based on the nucleotide sequences of the target genes (GenBank accession no. AE014133.2) using the MegAlign and PrimerSelect programs (Lasergene™ 8.0, DNAStar, Inc., Madison, WI, USA) (Table 1). The qPCR primers were synthesized by Bioneer Co. (Daejeon, Korea).
Table 1. Quantitative real-time polymerase chain reaction (qPCR) primers used in this study.
| Genes | Primer names and oligonucleotide sequences (5 – > 3) | Size of amplicon (bp) |
|---|---|---|
| glmU | UA159-glmU-F:
TGATCCTTTCGGTTATGGTCGTAT UA159-glmU-R: CGTTCCCGTGTTAATTTCTTTGA |
118 |
| murA | UA159-murA-F1:
CCTCCGGGCATAGAAACCTTAG UA159-murA-R1: ATTTTAGATGTGGCTCCTTATGAA |
110 |
| murB | UA159-murB-F1:
CGAGATATGCGCTTTGGTT UA159-murB-R1: AACGCTGCATTTCCTGACTG |
121 |
| murC | UA159-murC-F1:
GGTAACCATTTTCGAGAGCATAGG UA159-murC-R1: ACACGGATTGGAAAAAGCAGGAA |
140 |
| murC2 | UA159-murC2-F:
CAAATGGATCGTTACGGTGAAAT UA159-murC2-R: TTGGCGTTAAAGAGTGGGCTATC |
116 |
| murD | UA159-murD-F:
TCAAACGCAGGCAAAGATTATTC UA159-murD-R: TTTCAACATTATGGCTTCCTGGTA |
143 |
| murE | UA159-murE-F1:
CGTCCGGCCATGATTTCTACCA UA159-murE-R1: GCTCTCGGGGGTTGTTAGTTTTGA |
81 |
| alr | UA159-alr-F:
AGAAGGCGGGAGCGACTG UA159-alr-R: CAATTGACTGATAAGGGTGGTAAA |
116 |
| ddI | UA159- D-Ala-F:
AGATGTGGCTTTTTATGATTACGA UA159- D-Ala-R: AAAGTCCACAGCAGCCCAAAGT |
146 |
| murI | UA159-murI-F:
AATGGGCCGTAAAAGTGGATAATG UA159-murI-R: TGCCCCAAATTTGTTCCTAT |
147 |
| murF | UA159-murF-F1:
GGTGCTGGCAGATATGAAAGAAT UA159-murF-R1: AGTGGTCCAAAAAGAAAAATACGA |
111 |
| bacA | UA159-bacA-F:
AACGGCCTTTTTCATTGGTCTG UA159-bacA-R: GTTGCGACTTGCCGACTGG |
114 |
| mraY | UA159-mraY-F:
CTTTGCATTTGGGCGTGTTTTA UA159-mraY-R: GAAGCCAAGCCATCAATACCATC |
100 |
| murG | UA159-murG-F:
AAGGTGGGTTTGTTTCAGTTCC UA159-murG-R: GCAATTCGGTTAGCCAGTCC |
106 |
| murM | SM159-murM-F:
TATTTTTGGCAGTACGGATGGAT SM159-murM-R: ATGGCTGGGACAAACAAGTGA |
131 |
| murN | UA159-murN-F:
AAACAAAGAGACTGCCTGCTAAGA UA159-murN-R: AAAACCAATTGCGTGAACTGTC |
123 |
| pbp1a | UA159-pbp1a-F:
TTGATGCCGCTGGATTAGATACTG UA159-pbp1a-R: CACTGCTGGCACCATACTTTTGTT |
132 |
| pbp1b | UA159-pbp1b-F:
ATGGTGCTTCTCTTGATGATG UA159-pbp1b-R: CAAAGGCGTGATTATTCTGAT |
132 |
| pbp2a | UA159-pbp2a-F:
TATGTTGAAGGGTCCGGGTATCTA UA159-pbp2a-R: CGGTTCCCCATTCCCACTTT |
150 |
| pbp2b | UA159-pbp2b-F1:
ACCGCGTTGGAACATCTTATCT UA159-pbp2b-R1: TAGTCCCTTTGGCAGTGGTCTTA |
129 |
| pbp2X | UA159-pbp2X-F:
AAAGACGGACAAGTGACCTACCAA UA159-pbp2X-R: CCATCTGCGTTTCCAAATAAGTCT |
145 |
| dacA | UA159-dacA-F:
GTACCGCCAACTTTTTCAGC UA159-dacA-R: CAGCGCCAGCAATGTCC |
120 |
| 16S rDNA | SM159-16S -F:
CACACCGCCCGTCACACCAC SM159-16S -R: CCAGCCGCACCTTCCGATACG |
137 |
glmU, UDP-N-acetylglucosamine pyrophosphorylase gene; murA, UDP-N-acetylglucosamine 1-carboxyvinyltransferase gene; murB, UDP-N-acetylenolpyruvoylglucosamine reductase gene; murC, UDP-N-acetyl muramate-alanine ligase gene; murC2, UDP-N-acetylmuramyl tripeptide synthetase gene; murD, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate synthetase gene; murE, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-L-lysine ligase gene; alr, alanine racemase gene; ddI, D-alanine-D-alanine ligase gene; murI, glutamate racemase gene; murF, UDP-N-acetylmuramoyl-tripeptide—D-alanyl-D-alanine ligase gene; bacA, undecaprenyl pyrophosphate phosphatase gene; mraY, phospho-N-acetylmuramoyl-pentapeptide-transferase gene; murG, undecaprenyldiphospho-muramoylpentapeptide beta-N-acetylglucosaminyltransferase gene; murM, UDP-N-acetylmuramoylpentapeptide-lysine N6-alanyltransferase gene; murN, alanine adding enzyme gene; pbp1a, penicillin binding protein 1a gene; pbp1b, penicillin binding protein 1b gene; pbp2a, penicillin binding protein 2a gene; pbp2b, penicillin binding protein 2b gene; pbp2X, penicillin binding protein 2× gene; dacA, serine-type D-Ala-D-Ala carboxypeptidase (DD-transpeptidase, DacA) gene; 16S rDNA, 16S ribosomal RNA gene.
First-strand cDNAs synthesis was performed with 4 μg of total RNA, random hexamers, and an AccuPower ® RocketScript™ RT PreMix (Bioneer Co., Deajeon, Korea) using the Exicycler™ 96 Real-Time Quantitative Thermal Block (Bioneer Co.) and following the manufacturer's protocol. qPCR was performed with the AccuPower® GreenStar™ qPCR PreMix (Bioneer Co.) using the Exicycler™ 96 Real-Time Quantitative Thermal Block. Each PCR reaction was performed in a total volume of 20 μL containing 1 μL each of the forward and reverse primers (final concentration, 500 nM each), 0.63 μL cDNA, and the appropriate dose of sterilized DNase-RNase-free water in PreMix PCR tubes. The qPCR conditions were initial denaturation at 95 °C for 3 min, 40 cycles of denaturation at 95 °C for 15 sec, primer annealing and extension at 55 °C for 15 sec, and final cooling at 25 °C for 1 min. Each qPCR reaction was performed in triplicate.
The expression levels of each target gene in the group were normalized with those of 16S rDNA. The normalized expression of each gene (N) was determined as:
Relative expression of the target genes between each experimental group (BA, OA, or UA) and the negative control group (1% DMSO) was calculated as [N of each experimental group (BA, OA, or UA)/N of the negative control group].
Data are expressed as mean ± standard deviation. Independent sample two-tailed t-tests were conducted to calculate the p-values using SPSS version 12 software (SPSS Inc., USA). A p-value ≤ 0.05 was considered statistically significant.
We determined the BA, OA, and UA concentration of 64 μg/mL by investigating the antimicrobial mechanism against S. mutans UA159 by calculating the growth curve at 16, 32, and 64 μg/mL BA, OA, and UA for 60, 90, 120, and 180 min in S. mutans UA159 at the mid-log phase (OD600 = 0.35). The growth of S. mutans UA159 was in a static (inhibited) state at concentrations of 64 and 32 μg/mL of OA and UA, but in the exponential state at concentrations of 64 and 32 μg/mL in the BA-treated and control groups (data not shown). We decided to further explore using 64 μg/mL BA, OA, and UA.
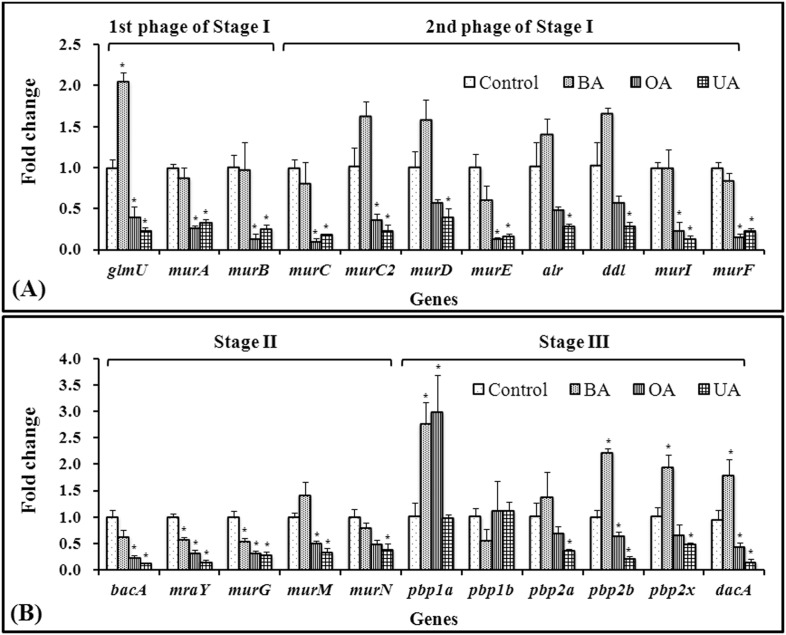
The synthesis of UDP-N-acetylglucosamine (UDP-GlcNAc) and UDP-N-actetylmuramic acid (UDP-MurNAc), the building blocks of peptidoglycan, is the first phase of first stage (stage I) of intracellular peptidoglycan assembly (Bugg and Walsh, 1992). UDP-GlcNAc is synthesized from GlcNAc-1-P by UDP-N-acetylglucosamine pyrophosphorylase (GlmU). UDP-MurNAc is synthesized from UDP-GlcNAc by UDP-N-acetylglucosamine 1-carboxyvinyltransferase (MurA) and UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) (Bugg and Walsh, 1992). The data showed that UA and OA, but not BA, significantly downregulated glmU, murA, and murB expression (Figure 1A). Interestingly, glmU was overexpressed by a factor of two in the BA-treated group compared to that in the control group. The second phase of stage I of peptidoglycan biosynthesis is the assembly of the UDP-MurNAc-pentapeptide (Bugg and Walsh, 1992). The D-amino acids, D-alanine, D-glutamate, and DL-diaminopimelate, occur in peptidoglycan (Bugg and Walsh, 1992). For S. mutans UA159, D-alanine and D-glutamate are catalyzed by alanine racemase (Alr) and glutamate racemase (MurI), respectively (the KEEG pathway). Biosynthesis of DL-diaminopimelate (DL-DAP) is catalyzed by DAP epimerase or meso-DAP D-dehydrogenase (Bugg and Walsh, 1992). These genes are missing in S. mutans UA159, even though UDP-N-acetylmuramyl tripeptide synthetase (MurC2) which catalyzes adding DL-DAP to UDP-MurNAc-L-alanine-D-glutamate occurs (KEEG Pathway). The reason for the discrepancy is unclear, but it is possible that DL-DAP is synthesized through an unknown pathway or that DL-DAP is not used in peptidoglycan synthesis in S. mutans UA159. Generally, DL-DAP is added as the third residue in the pentapeptide of Gram-negative bacteria and some Gram-positive bacteria, whereas lysine is added in other Gram-positive bacteria (http://www.enzyme-database.org/reaction/polysacc/PepGly1.html). Using these D-amino acids and D-Ala-D-Ala formed by D-alanine-D-alanine ligase (DdI), two types of UDP-MurNAc-pentapeptide, UDP-MurNAc-L-Ala-γ-D-Glu-DL-DAP-D-Ala-D-Ala (UDP-MurNAc-pentapeptide I) and UDP-MurNAc-L-Ala-γ-D-Glu-DL-DAP-D-Ala-D-Ala (UDP-MurNAc-pentapeptide II) are synthesized by UDP-N-acetyl muramate-alanine ligase (MurC), UDP-N-acetylmuramoyl-L-alanyl-D-glutamate synthetase (MurD), MurC2 or UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-L-lysine ligase (MurE), and UDP-N-acetylmuramoyl-tripeptide-D-alanyl-D-alanine ligase (MurF) (the KEGG Pathway). The data showed that second stage-related genes were downregulated by OA and UA, but murE was downregulated only by BA (Figure 1A). These results reveal that OA and UA inhibited expression from the first stages of S. mutans UA159 peptidoglycan biosynthesis, whereas BA induced overexpression of UDP-GlcNAc, the first substrate for peptidoglycan synthesis.
Figure 1. Relative expression of genes related to peptidoglycan biosynthesis, (A) stage I and (B) stage II and III, in Streptococcus mutans UA159 following treatment with betulinic acid (BA), oleanolic acid (OA), or ursolic acid (UA). glmU, UDP-N-acetylglucosamine pyrophosphorylase; murA, UDP-N-acetylglucosamine 1-carboxyvinyltransferase; murB, UDP-N-acetylenolpyruvoylglucosamine reductase; murC, UDP-N-acetyl muramate-alanine ligase; murC2, UDP-N-acetylmuramyl tripeptide synthetase; murD, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate synthetase; murE, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-L-lysine ligase; alr, alanine racemase; ddI, D-alanine-D-alanine ligase; murI, glutamate racemase; murF, UDP-N-acetylmuramoyl-tripeptide—D-alanyl-D-alanine ligase; bacA, undecaprenyl pyrophosphate phosphatase; mraY, phospho-N-acetylmuramoyl-pentapeptide-transferase; murG, undecaprenyldiphospho-muramoylpentapeptide beta-N-acetylglucosaminyltransferase; murM, UDP-N-acetylmuramoylpentapeptide-lysine N6-alanyltransferase; murN, alanine adding enzyme; pbp1a, penicillin binding protein 1a; pbp1b, penicillin binding protein 1b; pbp2a, penicillin binding protein 2a; pbp2b, penicillin binding protein 2b; pbp2x, penicillin binding protein 2x, dacA, serine-type D-Ala-D-Ala carboxypeptidase (DD-transpeptidase, DacA). *p ≤ 0.05, significantly downregulated or upregulated compared with the control group.
The second stage (stage II) of peptidoglycan biosynthesis is transport of the MurNAc-pentapeptide portion of the UDP-MurNAc-pentapeptide across the cytoplasmic membrane by a undecaprenyl lipid carrier, undecaprenyl phosphate (UP), which is synthesized from undecaprenyl pyrophosphate (UPP) by undecaprenyl pyrophosphate phosphatase (BacA) (Higashi et al., 1970; Bouhss et al., 2008). The coupling of UDP-MurNAc-pentapeptide to UP to produce UPP-MurNAc-pentapeptide (Lipid I) and UMP is catalyzed by phospho-N-acetylmuramoyl-pentapeptide-transferase (MraY). Then, another glucosamine residue, GlcNAc, is attached to Lipid I by undecaprenyldiphospho-muramoylpentapeptide beta-N-acetylglucosaminyltransferase (MurG) and forms UPP-MurNAc-(GlcNAc)-pentapeptide (Lipid II). 2 L-Alanine (L-Ala) is added to UDP-MurNAc-(GlcNAc)-L-Ala-γ-D-Glu-L-Lys-D-Ala-D-Ala by UDP-N-acetylmuramoylpentapeptide-lysine N6-alanyltransferase (MurM) and another alanine-adding enzyme (MurN) to form UDP-MurNAc-(GlcNAc)-L-Ala-γ-D-Glu-L-Lys-(L-Ala)2-D-Ala-D-Ala (the KEGG pathway). The data showed that OA and UA significantly inhibited expression of bacA, mraY, murG, murM, and murN and that BA also downregulated these genes, except murM and murN (Figure 1B). These findings reveal that OA, UA, and BA inhibited translocation of the MurNAc-pentapeptide, an intermediate or monomer of peptidoglycan in the cell wall.
The last stage (stage III) of peptidoglycan biosynthesis is transglycosylation, transpeptidation, and trimming of the peptidoglycan, which are catalyzed by penicillin-binding proteins (PBPs) and transpeptidase (Pinho et al., 2001; Lee et al., 2003; Mainardi et al., 2005; Fan et al., 2007). S. mutans UA159 has five PBPs such as PBP1a, PBP1b, PBP2a, PBP2b, and PBP2X, as well as serine-type D-Ala-D-Ala carboxypeptidase (DacA), which is involved in the last stage of peptidoglycan biosynthesis (the KEEG pathway). The data showed that these genes, except PBP1a and PBP1b, were downregulated 52–85% and 31–56% in the OA and UA groups compared to those in the control group. Our qPCR data showed that OA and UA significantly downregulated the PBP genes, except pbp1a and pbp1b, whereas BA significantly upregulated these genes, except pbp1b and pbp2a (Figure 1B). These results indicate that OA and UA inhibit polymerization between a newly synthesized peptidoglycan monomer and the existing peptidoglycan, and that BA increased polymerization in S. mutans UA159.
In summary, OA and UA inhibited the expression of peptidoglycan biosynthesis-related genes of S. mutans UA159 at the transcriptional level which might be the one of the antimicrobial mechanism of OA and UA against S. mutans.
Acknowledgments
This study was supported by a grant from the Korea Healthcare Technology R&D Project, Ministry for Health, Welfare and Family Affairs, Republic of Korea (A091074).
References
- Bouhss A, Trunkfield AE, Bugg TD, et al. The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS Microbiol Rev. 2008;32:208–233. doi: 10.1111/j.1574-6976.2007.00089.x. [DOI] [PubMed] [Google Scholar]
- Bugg TD, Walsh CT. Intracellular steps of bacterial cell wall peptidoglycan biosynthesis: enzymology, antibiotics, and antibiotic resistance. Nat Prod Rep. 1992;9:199–215. doi: 10.1039/np9920900199. [DOI] [PubMed] [Google Scholar]
- Fan X, Liu Y, Smith D, et al. Diversity of penicillin-binding proteins. Resistance factor FmtA of Staphylococcus aureus . J Biol Chem. 2007;282:35143–35152. doi: 10.1074/jbc.M706296200. [DOI] [PubMed] [Google Scholar]
- Fontanay S, Grare M, Mayer J, et al. Ursolic, oleanolic and betulinic acids: antibacterial spectra and selectivity indexes. J Ethnopharmacol. 2008;120:272–276. doi: 10.1016/j.jep.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Higashi Y, Strominger JL, Sweeley CC. Biosynthesis of the peptidoglycan of bacterial cell walls. XXI. Isolation of free C55-isoprenoid alcohol and of lipid intermediates in peptidoglycan synthesis from Staphylococcus aureus . J Biol Chem. 1970;245:3697–3702. [PubMed] [Google Scholar]
- Horiuchi K, Shiota S, Hatano T, et al. Antimicrobial activity of oleanolic acid from Salvia officinalis and related compounds on vancomycin-resistant enterococci (VRE) Biol Pharm Bull. 2007;30:1147–1149. doi: 10.1248/bpb.30.1147. [DOI] [PubMed] [Google Scholar]
- KEGG Pathway Peptidoglycan biosynthesis - Streptococcus mutans UA159. 2013. [Accessed 5 March 2014]. Available at: http://www.genome.jp/kegg-bin/show_pathway?smu00550.
- Kim MJ, Kim CS, Ha WH, et al. Antimicrobial effects of oleanolic acid against Streptococcus mutans and Streptococcus sobrinus isolated from a Korean population. Int J Oral Biol. 2010;35:191–195. [Google Scholar]
- Kim MJ, Kim CS, Park JY, et al. Antimicrobial effects of ursolic acid against mutans streptococci isolated from Koreans. Int J Oral Biol. 2011;36:7–11. [Google Scholar]
- Lee M, Hesek D, Suvorov M, et al. A mechanism-based inhibitor targeting the DD-transpeptidase activity of bacterial penicillin-binding proteins. J Am Chem Soc. 2003;125:16322–16326. doi: 10.1021/ja038445l. [DOI] [PubMed] [Google Scholar]
- Liu J. Pharmacology of oleanolic acid and ursolic acid. J Ethnopharmacol. 1995;49:57–68. doi: 10.1016/0378-8741(95)90032-2. [DOI] [PubMed] [Google Scholar]
- Mainardi JL, Fourgeaud M, Hugonnet JE, et al. A novel peptidoglycan cross-linking enzyme for a beta-lactam-resistant transpeptidation pathway. J Biol Chem. 2005;280:38146–38152. doi: 10.1074/jbc.M507384200. [DOI] [PubMed] [Google Scholar]
- Pinho MG, Filipe SR, de Lencastre H, et al. Complementation of the essential peptidoglycan transpeptidase function of penicillin-binding protein 2 (PBP2) by the drug resistance protein PBP2A in Staphylococcus aureus . J Bacteriol. 2001;183:6525–6531. doi: 10.1128/JB.183.22.6525-6531.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]