Highlight
Unlike in glycophytes, NaCl facilitates the nitrate uptake in the euhalophyte S. europaea, while calcium signalling plays important roles in this process.
Key words: [Ca2+]cyt, calcium signalling, 2D-DIGE, NaCl, nitrate uptake, plasma membrane, Salicornia europaea.
Abstract
Improving crop nitrogen (N) use efficiency under salinity is essential for the development of sustainable agriculture in marginal lands. Salicornia europaea is a succulent euhalophyte that can survive under high salinity and N-deficient habitat conditions, implying that a special N assimilation mechanism may exist in this plant. In this study, phenotypic and physiological changes of S. europaea were investigated under different nitrate and NaCl levels. The results showed that NaCl had a synergetic effect with nitrate on the growth of S. europaea. In addition, the shoot nitrate concentration and nitrate uptake rate of S. europaea were increased by NaCl treatment under both low N and high N conditions, suggesting that nitrate uptake in S. europaea was NaCl facilitated. Comparative proteomic analysis of root plasma membrane (PM) proteins revealed 81 proteins, whose abundance changed significantly in response to NaCl and nitrate. These proteins are involved in metabolism, cell signalling, transport, protein folding, membrane trafficking, and cell structure. Among them, eight proteins were calcium signalling components, and the accumulation of seven of the above-mentioned proteins was significantly elevated by NaCl treatment. Furthermore, cytosolic Ca2+ concentration ([Ca2+]cyt) was significantly elevated in S. europaea under NaCl treatment. The application of the Ca2+ channel blocker LaCl3 not only caused a decrease in nitrate uptake rate, but also attenuated the promoting effects of NaCl on nitrate uptake rates. Based on these results, a possible regulatory network of NaCl-facilitated nitrate uptake in S. europaea focusing on the involvement of Ca2+ signalling was proposed.
Introduction
Salinity is among the most severe abiotic stresses in agriculture that affects ~6% of the total land on earth (Munns and Tester, 2008). In saline soil, the salt concentration is high, whereas the nitrogen (N) content is usually deficient (Hamed et al., 2013); most crops could not survive on it. However, many halophytes can thrive in this type of infertile soil, suggesting that they may have specific N uptake and utilization mechanisms. Salicornia europaea is one of the most salt-tolerant plant species in the world (Ungar, 1987). A previous study demonstrated that S. europaea has a strong ability to utilize N; this plant can thrive under both N-limited and high N conditions (Nie et al., 2012). However, the effect of NaCl on N utilization has not been investigated in S. europaea. Elucidating the regulatory networks of efficient N uptake of S. europaea under salinity would be helpful for creating salt-tolerant crops with high N use efficiency, which is essential for the development of sustainable agriculture in marginal lands.
N is an essential macronutrient for plant growth. Among various N forms (nitrate, ammonium, amino acids, and peptides), nitrate (NO3 –) is the predominant form available in aerobic soils (Marschner and Rimmington, 1988). Salinity can severely influence nitrate assimilation in plants. Generally, a high Cl– concentration acts as an antagonist and represses nitrate uptake; thus, NaCl negatively affects nitrate uptake, assimilation, and protein synthesis which may be responsible, at least in part, for the depressed plant growth under saline conditions (Bar et al., 1997). In addition, the state of the membrane and/or membrane proteins also affects uptake of NO3 – by altering plasma membrane (PM) integrity under salinity (Frechilla et al., 2001). However, in halophytic Distichlis spicata (Pessarakli et al., 2012), NaCl has no negative effect on N uptake. Another report on the euhalophyte Suaeda physophora showed that NaCl application significantly increased leaf NO3 − concentration under N-sufficient conditions (Yuan et al., 2010). In fact, sodium-dependent NO3 − uptake has been reported in the marine diatom Phaeodactylum tricornutum (Rees et al. 1980), cyanobacteria (Lara et al. 1993), and the marine halophyte Zostera marina (Rubio et al., 2005). These results indicated that NaCl may have a promoting effect on nitrate uptake in some halophytes, which is different from glycophytes, whereas the underlying molecular networks are still unclear.
Nitrate uptake occurs in the outer cell layers of roots and relies on nitrate transporters, which are mainly localized at the PM together with the regulatory proteins. To date, four families of nitrate transporters have been identified: nitrate transporter 1/peptide transporter family (NRT1/PTR), nitrate transporter 2 family (NRT2), chloride channel family (CLC), and slow anion channel-associated homologues (SLAC1/SLAH) (Krapp et al., 2014). More than 60 nitrate transporters have been estimated in Arabidopsis, the regulation of which involves complex networks that are not fully understood. Investigations on root PM protein accumulation under different NaCl and nitrate levels would significantly enhance our knowledge of nitrate signalling under salinity. Proteomics is a powerful tool to study PM proteins depending on two main approaches, namely gel-free and two-dimensional electrophoresis (2-DE) (Nouri and Komatsu, 2010). The gel-free method has better coverage and higher solubility for hydrophobic membrane proteins than 2-DE. However, proteins containing a single transmembrane domain or peripheral membrane-associated proteins can be more readily resolved in 2-DE. In addition, 2-DE can give information on post-translational modifications (Tang, 2012) and it is also a valuable tool in deciphering N-terminal processing of proteins (Taylor et al., 2011). Nouri and Komastu (2010) used both gel-free and 2-DE methods to study soybean PM proteins, and concluded that the two techniques are complementary for comparative analysis. Meanwhile, Tang et al. (2008) used two-dimensional fluorescence difference gel electrophoresis (2D-DIGE) to study Arabidopsis PM proteins, and three homologous brasinosteroid-signalling kinases were successfully identified; thus, 2D-DIGE is demonstrated to be a powerful approach for studying signalling proteins localized in the PM.
Proteomic research on glycophytes has focused on PM proteins under salt stress (Cheng et al., 2009). Meanwhile, in a few studies, whole-plant proteomics have been applied in Arabidopsis (Wang et al., 2012), Zea mays (Prinsi et al., 2009), and barley (Moller et al., 2011) to detect their responses to different N conditions. However, no research has been performed to analyse the PM proteome of halophytes under different nitrate and NaCl levels. The aim of this study was to investigate the characteristics of nitrate uptake in S. europaea under NaCl treatment and to identify the underlying regulatory components. Through physiological analysis, it was demonstrated that in contrast to the situation in glycophytes, NaCl facilitates nitrate uptake in S. europaea. Comparative proteomics and cell biology studies further revealed that calcium signalling plays important roles in this process. These results open up exciting perspectives for further investigations on the specific regulatory pathways involved in efficient N uptake under NaCl conditions.
Materials and methods
Plant materials and growth conditions
Salicornia europaea seeds were collected from the coastal area of Dafeng City, Jiangsu Province, China. The plants were grown in a greenhouse with a day/night temperature regime of 25 °C/20 °C, photoperiod of 16h, and relative humidity of ~60%. At 20 d after sowing on perlite granules, seedlings were irrigated with four solutions containing different concentrations of NO3 – and NaCl in modified 1/2 Hoagland solution for 30 d. Then the plants were harvested and washed with distilled water to remove the surface ions and used for further analysis. The modified 1/2 Hoagland solution contained 0.5mM KH2PO4, 1mM MgSO4, 2.5mM CaCl2, 0.05mM Fe-EDTA, 2.5mM KCl, and micronutrients, with the pH adjusted to 6.5±0.1. The four treatments were 0.1mM NO3 – (LN), 0.1mM NO3 –+200mM NaCl (LN+S), 10mM NO3 – (HN), and 10mM NO3 –+200mM NaCl (HN+S).
Measurement of fresh weight (FW), dry weight (DW), root length, and root volume
FW was measured immediately after harvesting. DW was measured after drying for 48h in an oven at 80 °C. Roots were scanned with a digital scanner (Epson, Nagano, Japan) and then analysed with WinRHIZO software (Regent Instruments Inc., Quebec, Canada).
Quantification of total Na, total N contents, and NO3 – concentrations
Plants were dried for 72h in an oven at 80 °C and then weighted and subsequently ground into powder in a mixer mill (Retsch MM400, Hann, Germany). The powder was digested with a mixture of nitric acid and hydrogen peroxide using a microwave system (MARS; CEM Corporation, Matthews, NC, USA), which was used to determine total Na contents by atomic emission spectrometry (ICP-AES, Thermo, Waltham, MA, USA). N contents were determined by a Vario EL III CHNOS Elemental Analyzer (Elementar Analysensysteme GmbH, Germany). The NO3 – concentration was measured as previously described (Jampeetong and Brix, 2009). Dried material (5–10mg) was extracted with 10ml of Milli Q water at 80 °C in a water bath for 20min. Then, NO3 – concentrations were analysed by an AA3 continuous flow-analyser (Seal Analytical, Germany).
Analysis of nitrate reductase (NR) activities
NR activities were measured as previously described (Yaneva et al., 2002) with some modifications. The seedlings were homogenized in extraction buffer [50mM HEPES-KOH, pH 7.5, 1mM EDTA, 10mM glutathione (GSH), 0.1% (w/v) polyvinylpyrrolidone (PVP), 1mM dithiothreitol (DTT), and 1mM phenylmethylsulphonyl fluoride (PMSF)] and centrifuged for 20min at 12 000 g. The supernatant was incubated with reaction buffer (50mM HEPES-KOH, pH 7.5, 5mM KNO3, 0.2mM NADH) in the dark at 25 °C for 20min; the reaction was terminated by addition of 1% sulphanylamide in 1.5M HCl and 0.01% N-(1-naphthyl)-ethylenediammonium dichloride. Nitrite production was determined by reading the absorbance at 540nm.
Analysis of nitrate uptake rates
Nitrate uptake rates were measured as previously described (Muños et al., 2004). Salicornia europaea plants of ~30 d old irrigated with 1/2 Hoagland solution plus 200mM NaCl were transferred to 0.1mM CaSO4 for 1min and then to four solutions containing different NaCl and K15NO3 (99% atom excess 15N) concentrations, as follows: 0.1mM K15NO3, 0.1mM K15NO3+200mM NaCl, 10mM K15NO3, and 10mM K15NO3+200mM NaCl for 15min, and finally, to 0.1mM CaSO4 for 1min. Six roots for each treatment and three independent biological replications were dried at 70 °C for 48h and ground. The powder (2–3mg) was used for 15N determination by the Delta Plus-MS system (Thermo).
Thirty-day-old S. europaea plants irrigated with 1/2 Hoagland solution plus 200mM NaCl were pre-treated with 1mM of the non-specific Ca2+ channel blocker LaCl3 for 24h. The LaCl3-pre-treated and control plants were used for 15NO3 – uptake assay following the above-mentioned method.
Plasma membrane (PM) isolation
PMs were isolated as previously reported (Tang, 2012). Roots were homogenized in grinding buffer (pH 7.5) (25mM HEPES, 0.33M sucrose, 10% glycerol, 0.6% PVP, 5mM ascorbic acid, 5mM EDTA, 5mM DTT, and 1mM PMSF). The homogenate was filtered through Miracloth and centrifuged at 10 000 g for 15min. Total microsomal (TM) fractions were pelleted by centrifugation at 80 000 g for 1h and resuspended in suspension buffer [5mM KH2PO4/K2HPO4 buffer (pH 7.8), 0.33mM sucrose, 3mM KCl, 1mM DTT, and 1mM protease cocktail]. Two-phase partitioning was performed by using a solution containing 6.2% polyethylene glycol (PEG) 3350, 6.2% dextran T-500, 0.33M sucrose, 3mM KCl, and 5mM KH2PO4/K2HPO4 buffer (pH 7.8). The mixture was centrifuged at 1500 g for 10min. After partitioning, the upper phase fraction was diluted with 10 vols of dilution buffer (0.33M sucrose, 25mM HEPES, and 1mM DTT) and spun at 120 000 g for 1h to collect the PMs. Then, the PM vesicles were incubated with 0.02% Brij-58 detergent on ice for 10min to invert the vesicles and release the cytosolic contaminants. Samples were then diluted 20 times with double-distilled H2O and centrifuged at 120 000 g for 60min (Elmore et al., 2012). The pellets were resuspended in 100 μl of suspension buffer (5mM KH2PO4/K2HPO4 buffer, pH 7.8, 3mM KCl, 1mM DTT, 0.1mM EDTA, and 1 μM protease cocktail). For each sample, five independent biological replicates were prepared.
Immunodetection assay and H+-ATPase hydrolytic activity assay
Immunoblot analysis was performed according to standard methods (Zhang et al., 2011). Equal amounts of 5 μg of proteins from TM and PM fractions were separated by SDS–PAGE (Supplementary Fig. S1 availabe at JXB online) and transferred to nitrocellulose membranes (GE Amersham Biosciences, NY, USA) using a wet transblot system (Bio-Rad, Hercules, USA). The TM and PM proteins were probed with specific primary antibodies, and visualized using the enhanced chemiluminescence method. The primary antibodies used were anti-H+-ATPase (PM marker, Agrisera, Vännäs, Sweden), anti-Sar1 [secretion-associated and Ras-related protein 1, an endoplasmic reticulum (ER) marker, Agrisera], and anti-V-ATPase (tonoplast marker, Agrisera).
Hydrolytic activity of the PM H+-ATPase was measured according to the method of Nouri and Komastu (2010). The PM fraction was added to the reaction solution containing 30mM MES-TRIS (pH 6.5), 50mM KCl, 3mM MgSO4, and 3mM ATP in the presence or absence of the specific H+-ATPase inhibitor 0.1mM Na3VO4 and incubated at 30 °C for 15min. The reaction was stopped by the addition of 0.5% ammonium molybdate, 1% SDS, and 0.4M H2SO4. Ascorbate was then added at a final concentration of 0.3%, the mixture was placed at room temperature for 30min, and the absorbance was measured at 750nm.
Labelling of proteins with Cy dye
PM proteins were precipitated with the methanol/chloroform method as previously reported (Wessel and Flügge, 1984). The protein pellets were recovered in modified 2D-DIGE buffer (7M urea, 2M thiourea, 4% CHAPS, 25mM TRIS-HCl, and 1% n-dodecyl β-d-maltoside). The pH of protein samples was adjusted to 8.8 with HCl and NaOH, and the protein concentration was determined using the Bradford method. The internal standard was prepared by mixing equal amounts of all analysed samples. Protein samples were labelled using minimal fluorescent dyes (Cy3, Cy5, and Cy2) (GE Healthcare, Pittsburgh, PA, USA) following the manufacturer’s instruction.
2D-DIGE and image scanning
The labelled protein samples were mixed with the internal standard (Supplementary Table S1 at JXB online), adjusted to a total volume of 450 μl with rehydration buffer [7M urea, 2M thiourea, 4% CHAPS, 1% DTT, and 1% immobilized pH gradient (IPG) buffer, pH 4–7], and used for isoelectric focusing (IEF). IEF was performed on an IPG strip holder with 24cm, linear gradient IPG strips with pH 4–7 (GE Healthcare) and then on an Ettan DALT System (GE Healthcare) according to the manufacturer’s instructions. The images were analysed using DeCyder 6.5 (GE Healthcare). Spots reproducible in 24 of 30 images were used to identify protein abundance change by two-way analysis of variance (ANOVA) (P<0.05). The gels used for 2D-DIGE analyses were stained with Coomassie Brilliant Blue (CBB) to observe highly abundant spots. Then, additional gels with 1mg of internal standard proteins were stained with CBB for spots that could not be determined from 2D-DIGE gels.
Protein identification and database searching
The 117 most abundant spots were digested in-gel with bovine trypsin (Roche Molecular Biochemicals, Indianapolis, IN, USA) as described previously (Wang et al., 2007). The ultrafleXtreme matrix-assisted laser desorption ionization-time of flight/time of flight-mass spectrometer (MALDI-TOF/TOF-MS; Bruker Daltonics, Billerica, MA, USA) was used to reveal the MS/MS spectra. The peptides were suspended with 10 μl of 70% acetonitrile (ACN) containing 0.1% trifluoroacetic acid (TFA), and 1 μl was taken and spotted onto the AnchorChip™ MALDI target plate (Bruker Daltonics). Then 1 μl of matrix solution (1mg ml–1 α-cyano-4-hydroxycinnamic acid in 70% ACN containing 0.1% TFA) was spotted after the sample solution dried. All spectra were obtained in positive ion reflection mode under the control of FlexControl 3.3. The matrix suppression was set to deflection, 500Da. The spectra detection mass range was set at 700–4000 m/z. External calibration involved the use of the Bruker standard peptide calibration kit. For each spot, the top 30 strongest intensity peaks in the MS spectra and single-to-noise threshold >6 were automatically selected as precursor ions for MS/MS analysis. MS spectra were acquired with 400 laser shots per spectrum, whereas MS/MS spectra were obtained using 1500 laser shots per fragmentation spectrum. Then MS and MS/MS data were transferred to BioTools 3.2 (Bruker Daltonics) and searched against three databases with the Mascot engine 2.2.03 (http://www.matrixscience.com). The three databases were the NCBInr protein database (http://www.ncbi.nlm.nih.gov/; green plants, 1 669 695 sequences in NCBI 20131226) and two S. europaea transcriptome-translated protein databases, as follows: database 1 (162 969 sequences; Ma et al., 2013) and database 2 (35 219 sequences; Fan et al., 2013). Monoisotopic and [M+H]+ were selected for mass values. Peptide mass tolerance was set at 50 ppm, fragment mass tolerance was set as 0.5Da, and one missing cleavage was permitted. Carbamidomethyl (C) was set as fixed modification and Oxidation (M) was set as variable modification. All proteins were matched by Mascot with scores that exceeded their 95% confidence threshold and contained at least one peptide with a score at a significance threshold (P<0.05). If several database entries of homologous proteins matched these criteria, only the entry with the highest score is reported. For proteins having only one matching peptide with a score at a significant threshold (P<0.05), the MS/MS spectra were also presented (Supplementary Fig. S3; Supplementary Table S3 at JXB online).
Protein classification and bioinformatics analysis
The identified proteins were searched against the NCBI protein database (http://www.ncbi.nlm.nih.gov/) and Uniprot (http://www.uniprot.org/) and were divided into different groups based on their molecular and biological functions. The subcellular location of each protein was determined by SUBAIII (http://suba3.plantenergy.uwa.edu.au) and published information on the homologous proteins in Arabidopsis. Principal component analysis (PCA) was performed using the extended data analysis module within the DeCyder software (GE Healthcare). Heat map was performed using MultipleExperiment Viewer 4.8.1 software based on the Log2-transformed fold change.
RNA isolation and quantitative real-time PCR (qRT-PCR) analysis
Total RNA was extracted by Trizol reagent and treated with RNase-free DNase I. A 0.5 μg aliquot of RNA was used for the first-strand cDNA synthesis with the SuperScript III first-strand synthesis system (Invitrogen, Carlsbad, CA, USA). qRT-PCR was performed with an Mx3000P Real-Time PCR System (Agilent, USA) using SYBR qPCR Mix (Toyobo, Japan). The relative gene expression levels were calculated by the 2–ΔΔCT method, and the α-tubulin gene was used as internal control (Lv et al., 2012). qRT-PCR of each sample was repeated three times. The oligonucleotide primers used are listed in Supplementary Table S5 at JXB online.
Detection of [Ca2+]cyt
Thirty-day-old S. europaea plants irrigated with 1/2 Hoagland solution plus 200mM NaCl were used. In the 2h NaCl exposure experiment, the intact roots were incubated in loading solution containing 20 μΜ Fluo-4/AM, 50mM sorbitol, 0.2mM CaCl2, and 0.05% Pluronic F127 (pH 4.2) at 4 °C for 4h in the dark, followed by a 2h incubation with 0.2mM CaCl2 with or without 200mM NaCl at 20 °C in the dark. In the 24h, 3 d, and 30 d NaCl exposure experiments, the plants were first pre-treated with 200mM NaCl for 18h, 66h, and 30 d. Then, the intact roots of treated and control plants were incubated in loading solution with or without 200mM NaCl at 4 °C for 4h in the dark, after which the roots were incubated for 2h with 0.2mM CaCl2 with or without 200mM NaCl at 20 °C in the dark. After washing with buffer, [Ca2+]cyt was determined under a laser scanning confocal microscope (Leica TCS SP5, Wetzlar, Germany) at excitation and emission wavelengths of 488nm and 505–530nm, respectively. Observations were focused on the primary root tips of plants. The same area of root tips of each sample was analysed using Image J software (National Institutes of Health, Bethesda, MD, USA) to determine the fluorescence intensity (mean pixel intensity).
Statistical analysis
Statistical analysis was carried out using SPSS 18.0 software. Data were evaluated by two-way ANOVA. To determine the type of interaction between saline and nitrogen treatment, taking the FW for example, the effect of saline–high nitrogen treatment (FWHN+S/FWLN) was compared with the effect of each factor when applied individually (i.e. FWHN/FWLN×FWLN+S/FWLN). Interaction was synergistic if FWHN+S/FWLN was substantially more than FWHN/FWLN×FWLN+S/FWLN; and it was additive if FWHN+S/FWLN was approximately equal to FWHN/FWLN×FWLN+S/FWLN, whereas it was antagonistic if FWHN+S/FWLN was substantially less than FWHN/FWLN×FWLN+S/FWLN (Striker et al., 2015). For each experiment, at least three replications were used for analysis. The methods of significance testing are described in the figure legends.
Results
NaCl had a synergetic effect with nitrate on the growth of S. europaea
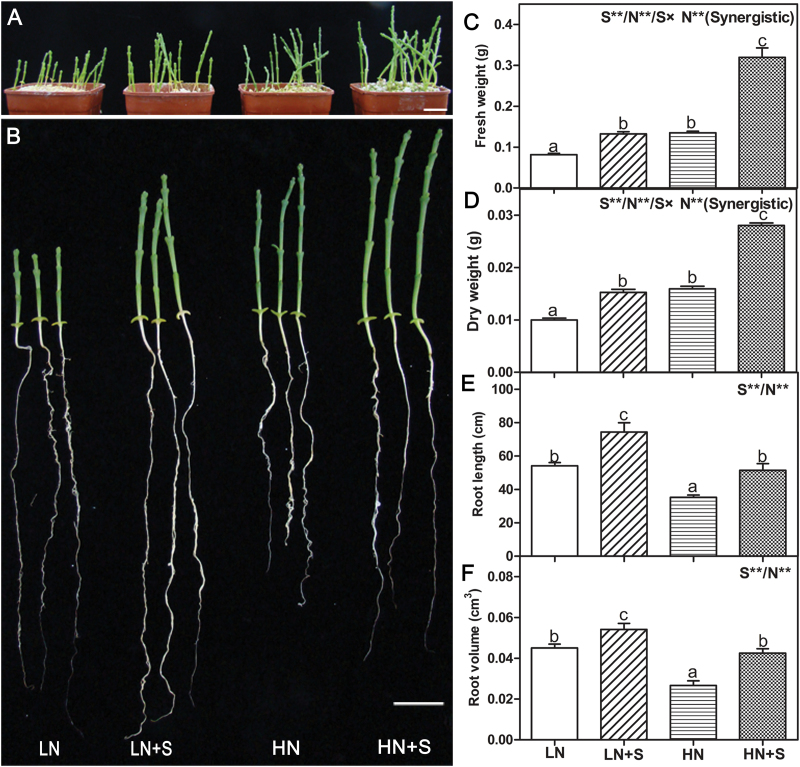
In this study, the effect of NaCl and nitrate on the growth of S. europaea was first investigated. Seedlings were treated with four different solutions, LN, LN+S, HN, and HN+S. Generally, NaCl has a positive influence on the growth of euhalophytes, for which zero NaCl is an abnormal condition whereas 200mM NaCl is in the range of optimal condition (Flowers and Colmer, 2008). Consistently, in this study, it was found that S. europaea grows better in the presence of 200mM NaCl under both LN and HN conditions. In addition, high N also promotes the growth of S. europaea (Fig. 1A, B). The FW, DW, root length, and root volume were measured, and two-way ANOVA (P<0.05) was used to reveal the effects of S, N, and S×N on these parameters. All the four traits showed significant S and N effects. The FW and DW increased under NaCl and NO3 – treatments, and seedlings treated with HN+S exhibited the highest values, whereas LN showed the lowest (Fig. 1C, D). Moreover, a significant S×N interaction was observed for FW and DW, and the type of interaction was synergistic (Fig. 1C, D), suggesting that NaCl and nitrate had synergistic effects on the growth of S. europaea. NaCl facilitated the root growth of S. europaea, whereas high nitrate inhibited it. Both the root length and volume were largest under LN+S treatment, the values of which were 110.8% and 103.1% higher than those treated with HN, respectively (Fig. 1E, F).
Fig. 1.
Phenotypic and physiological changes of S. europaea grown under different NaCl and NO3 – concentrations. S. europaea seedlings were treated with different nitrate and NaCl concentrations for 30 d, and the phenotypes (A and B), fresh weight (C), dry weight (D), root length (E), and root volume (F) were determined. Scale bars=2cm. Values were means ±SE (n=12). Different letters above the bars indicated significant differences (P<0.05). For C, D, E, and F, two-way ANOVA was performed to indicate significant S, N, and S×N effects. Double asterisks indicate significant differences at P<0.01. Combined effects are symbolized by a slash (/). Synergistic indicates the type of interaction.
The effects of NaCl on nitrate uptake and assimilation
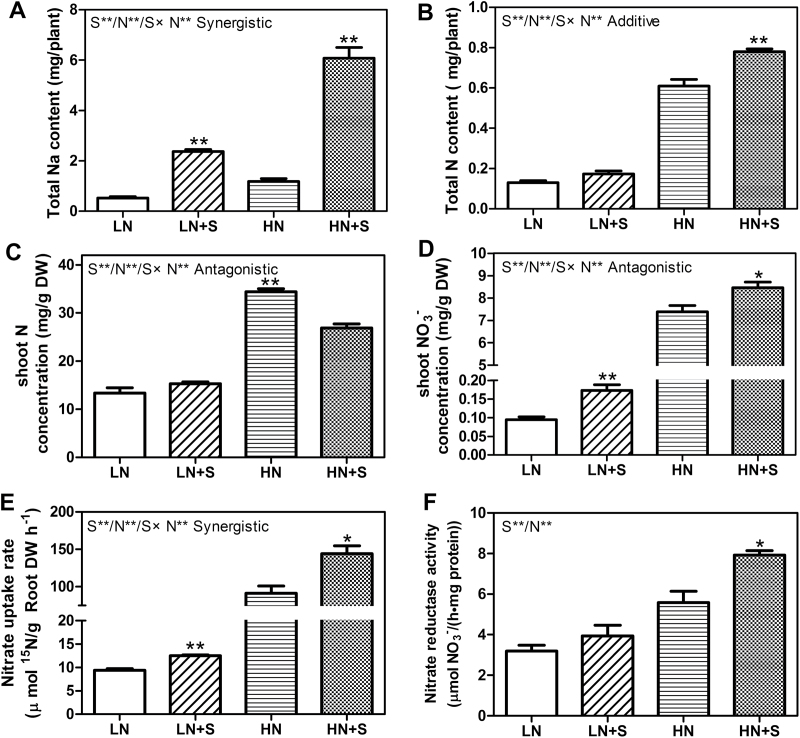
In order to investigate the effects of NaCl on the nitrate uptake and assimilation, the total Na and total N contents per plant, the N and NO3 – concentrations in shoot of S. europaea, the nitrate uptake rates, and nitrate reductase activities were measured (Fig. 2). All of the traits except NR activities showed significant S, N, and S×N effects. As expected, total Na contents were significantly increased under NaCl treatments, which were also promoted by N treatment (Fig. 2A). Under low N conditions, no significant effect of NaCl on total N content and shoot N concentrations was observed (Fig. 2B, C), but the shoot NO3 – concentrations and the nitrate uptake rates increased significantly by NaCl treatment. The values increased by 83.4% and 32.8% (Fig. 2D, E), respectively. Compared with the LN condition, NR activity under LN+S increased by 23.5%, but the values were very low and the difference was not statistically significant (Fig. 2F). Under high N conditions, the shoot N concentrations were significantly decreased by NaCl treatment (Fig. 2C), but the total N contents, shoot NO3 – concentration, the nitrate uptake rates, and the NR activities were all significantly increased by NaCl treatment. The values increased by 27.8, 14.6, 57.6, and 41.7%, respectively (Fig. 2B–F). Notably, NaCl and NO3 – also had a synergistic interaction effect on nitrate uptake rate. These results showed that NaCl could promote nitrate uptake and assimilation in S. europaea.
Fig. 2.
Total Na and N contents, shoot N and NO3 – concentrations, nitrate uptake rates, and NR activities of S. europaea grown under different NaCl and NO3 – concentrations. S. europaea seedlings were treated with different nitrate and NaCl concentrations for 30 d, total Na (A) and total N (B) content of the whole plant, shoot N (C) and NO3 – (D) concentration, and NR activity (F) were measured. (E) S. europaea seedlings were treated with four different NaCl and 15N labelled K15NO3 concentrations for 15min, and the content of 15N in roots was measured. Values were means ±SE (n=4 for A, B, C, D, and F; and n=3 for E). Asterisks indicate significant differences (P<0.05) between salt-treated and untreated samples. In addition, two-way ANOVA was performed to indicate significant S, N, and S×N effects. Double asterisks indicate significant differences at P<0.01, and combined effects are symbolized by a slash (/). Synergistic, additive, and antagonistic represent different types of interaction.
Identification of NaCl- and nitrate-regulated PM proteins by 2D-DIGE
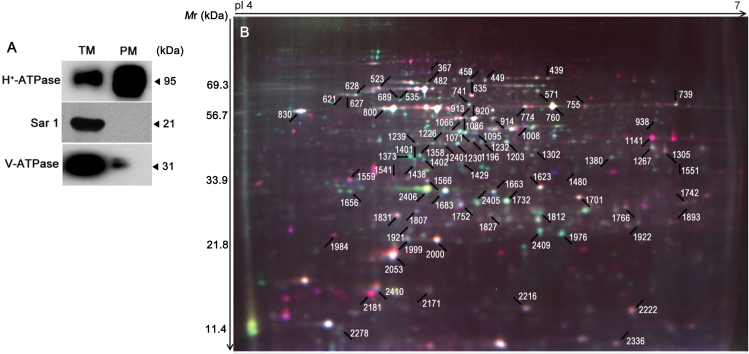
To determine the proteins regulated by NaCl and nitrate in S. europaea, a comparative proteomic analysis of root PM proteins under the four treatments was conducted. Root PM fractions were purified by the two-phase partitioning method (Tang, 2012), and their purity was evaluated by western blot analysis. Compared with the TM fraction, H+-ATPase was strongly enriched in the PM fraction. Conversely, V-ATPase, a tonoplast membrane protein, was more strongly enriched in the TM fraction than in the PM fraction. Sar1, an integral ER membrane protein, was only detected in the TM fraction and was absent in the PM fraction (Fig. 3A). Furthermore, the hydrolytic activity of PM H+-ATPase in the presence or absence of 0.1mM of the specific PM H+-ATPase inhibitor Na3VO4 was determined, and the purity of PM vesicles was 84.76±2.80%. These results demonstrated that PM vesicles of relatively high purity were obtained.
Fig. 3.
Purity assessment and a representative 2D-DIGE image. (A) Western blot analysis of extracted PM proteins with antibodies against proteins from different cell compartments. (B) PM proteins were separated by 2D-DIGE and detected with a Typhoon 9400 Scanner. The differentially accumulated protein spots under different NaCl and nitrate concentrations were determined using two-way ANOVA, and those positively identified by MALDI-TOF-TOF are marked by arrows.
2D-DIGE with pH 4–7 strips was used to separate root PM proteins, and ~2400 protein spots were found in each image (Fig. 3B; Supplementary Fig. S2 at JXB online). The 20 protein samples representing five biological repeats of each of the four treatments were assigned to 10 DIGE gels, which were labelled with Cy2, Cy3, and Cy5. A total of 30 images were acquired (Supplementary Table S1; Supplementary Fig. S2). After matching the 30 spot maps, 717 protein spots were found in 80% of them (Supplementary Table S2). PCA based on these 717 protein spots was performed to determine the distribution and similarities of different experimental groups. The five spot maps of each treatment were exactly clustered in the score plot (Fig. 4A), indicating high reproducibility among the replicate gels. Principal component 1 (PC1) explained 47.7% of the variance and separated the experimental groups according to NaCl treatment, whereas the axis PC2 accounted for 13.7% of the variance and separated the experimental groups according to nitrate treatment.
Fig. 4.
PCA, spot significant analysis, and functional classification of the identified proteins. (A) PCA of the different experimental groups. Samples plotted on the first two principal components (PCs) are shown. (B) Venn diagram showing the distribution of significant spots following two-way ANOVA. The numbers outside the parentheses are the total numbers of significant spots and at least one absolute variation was obtained above 1.5-fold by performing comparisons among the different treatments. The numbers in parentheses are the number of protein spots identified in the MALDI-TOF/TOF-MS. (C) Functional classification of 81 identified proteins. The percentage of proteins from each category is shown. The two most abundant categories, metabolism and cell signalling, were subsequently divided into subgroups.
Two-way ANOVA (P<0.05) with false discovery rate (FDR) correction for multiple testing revealed that a total of 406 spots displayed significant S, N, and S×N effects and at least one absolute variation >1.5-fold by comparison within the different treatments. Overall, among the differentially accumulated proteins, 166, 18, and 14 spots displayed pure S, N, and S×N effects, respectively, showing that NaCl treatment had a greater impact on protein accumulation than nitrate. In addition, 69 spots displayed pure combined S and N effects, which indicated that both NaCl and nitrate affected the accumulation of these proteins, but the two factors were independent. Finally, 64 spots displayed significant S×N effect in combination with S and N effects, indicating that NaCl and nitrate had interactive effects on the accumulation of these proteins (Fig. 4B).
Functional classification of NaCl- and nitrate-regulated proteins
After removing the dim spots and noise, the 117 most abundant spots displaying significant S, N, and/or S×N effects were manually excised from the gels, and further analysed by MS/MS spectra. Since there is a lack of information on the genome and comprehensive protein sequence of S. europaea, these spectra were searched against the NCBInr green plant database and two S. europaea transcriptome-translated protein databases (Fan et al., 2013; Ma et al., 2013). Most proteins blasted by the three databases were homologous proteins from different species, and the ones with the highest scores were finally selected for annotation (Supplementary Fig. S3; Supplementary Table S3 at JXB online). In total, 81 out of the 117 spots were annotated, among which 65 proteins have been previously shown or predicted to be associated with the PM. The localization information determined by the SUBAIII database and the literature is listed in Supplementary Table S3.
Furthermore, the identified proteins were grouped into six categories based on their molecular and biological functions, namely metabolism (46%), cell signalling (27%), transport (10%), protein folding (7%), membrane trafficking (5%), and cell structure (5%). The two major categories were further classified into subcategories. Specifically, proteins related to metabolism were divided into six subcategories, as follows: N metabolism, energetic metabolism, secondary metabolism, acid phosphates, cell wall-associated proteins, and proteolysis. Proteins involved in cell signalling were assigned to three subcategories, as follows: calcium signalling components, stress response, and detoxication (Fig. 4C; Supplementary Table S3 at JXB online).
The accumulation pattern of the identified proteins
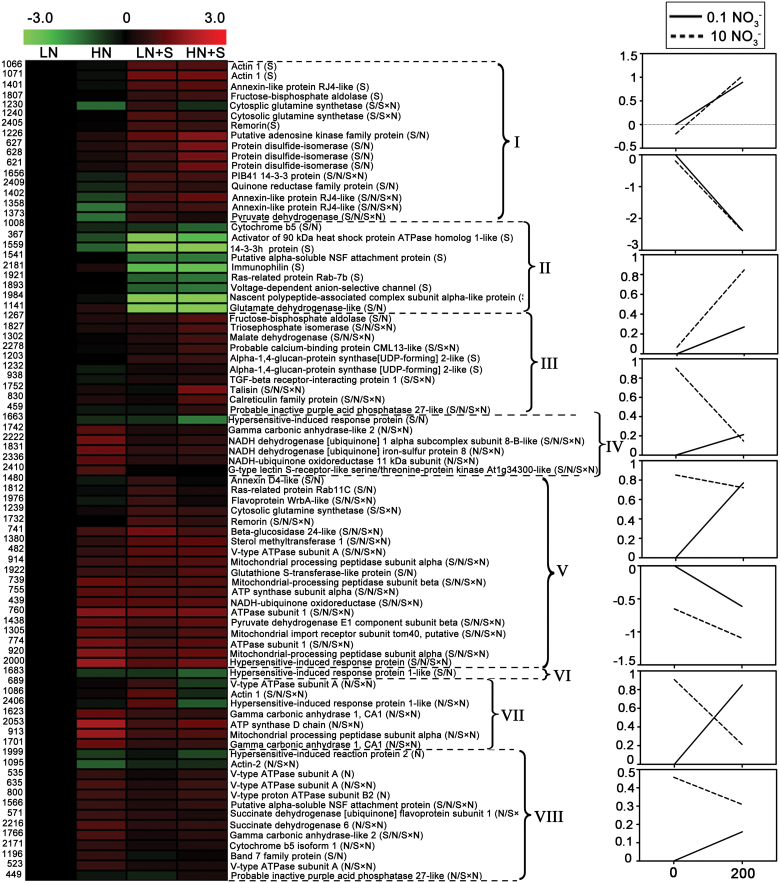
The 81 annotated, differentially accumulated proteins were further grouped into eight distinct clusters (Clusters I–VIII, Fig. 5; Supplementary Table S4 at JXB online) according to their change in abundance pattern. These clusters were illustrated by reaction norm diagrams consisting of a graphical representation highlighting the contribution of NaCl treatment (0mM and 200mM) to the observed protein abundance for different levels of nitrate treatments (0.1mM and 10mM). Cluster I contained 16 proteins whose accumulation increased significantly by NaCl treatment under both LN and HN conditions. Seven of these proteins were also significantly changed under nitrate treatments. Cluster II comprised nine proteins that were significantly repressed by the NaCl treatment under both LN and HN conditions. Four of these nine proteins changed significantly under nitrate treatments. Clusters III and IV comprised proteins whose abundance was increased or decreased significantly by NaCl treatment, but only under HN conditions. Cluster III included 10 proteins, three of which were significantly changed under nitrate treatments. Cluster IV contained six proteins, all of which were significantly changed under nitrate treatments. The largest cluster (Cluster V) comprised 19 proteins, which showed increased accumulation by NaCl treatment under LN conditions. Sixteen of these 19 proteins were also significantly changed under nitrate treatments. Only one protein in Cluster VI was repressed by NaCl treatment under LN conditions, and this protein was also down-regulated under nitrate supplement without NaCl. Cluster VII comprised seven proteins showing opposite responses to NaCl under LN and HN conditions. All of these proteins displayed a clear S×N interaction effect. Cluster VIII comprised 13 proteins whose abundance was not changed significantly by NaCl treatment whether under LN or HN conditions, but the abundance of such proteins changed significantly in response to nitrate.
Fig. 5.
Clustering analysis of differentially accumulated proteins. The 81 differentially accumulated proteins were grouped into eight clusters according to their change patterns, which are illustrated by reaction norm diagrams consisting of a graphical representation highlighting the contribution of NaCl treatment to the observed protein abundance under nitrate treatment. Each row in the heat map indicates a single protein, whose expression values were generated using Log2 ( treatments/ LN). For each protein, the spot number and the protein name with its ANOVA effect (S, N, and/or S×N) are shown. Reaction norms were generated using the means of Log2 ( treatments/ LN) of spots for each defined cluster. , means of normal volume/normal standard volume.
The involvement of Ca2+ signalling in NaCl-facilitated nitrate uptake in S. europaea
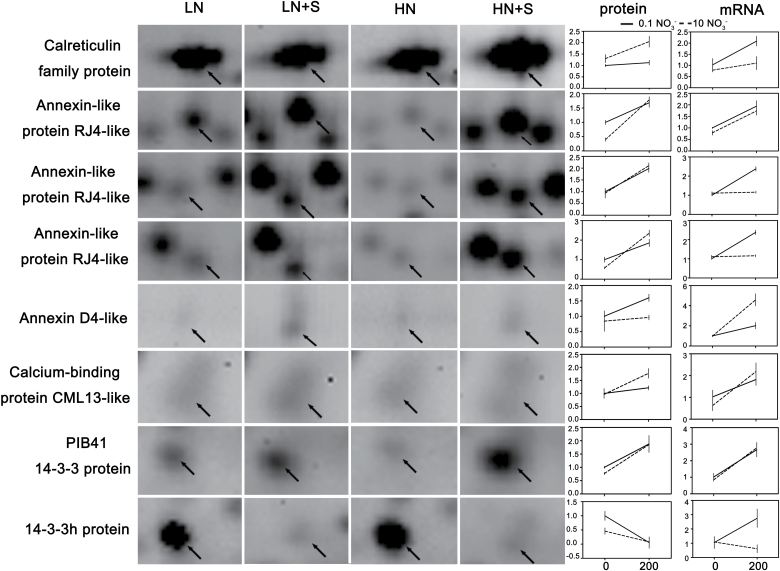
Eight calcium signalling components were identified by 2D-DIGE, and seven of these components were up-regulated, whereas the other one was down-regulated by NaCl treatment under HN and/or LN conditions (Fig. 6). qRT-PCR was performed to examine whether the eight components were also regulated at the transcription level. As shown in Fig. 6, the mRNA transcripts of the eight proteins were all up-regulated by NaCl treatment under HN and/or LN conditions. Two of them, annexin (ANN)-like protein RJ4-like and PBI41 14-3-3, displayed consistency between mRNA and protein variation trends, whereas the other six proteins showed some discrepancies (Fig. 6).
Fig. 6.
Expression profiles of calcium signalling components at protein and mRNA levels in response to NaCl and nitrate. The images on the left are magnified views of 2D-DIGE gel sections containing the spot of the calcium signalling components. The corresponding expression files at protein and mRNA levels are listed on the right. Expression levels of mRNAs were evaluated using qRT-PCR, n=3. The protein expression values were generated using ( treatments/ LN). , means of normal volume/normal standard volume, n=5. Values are means ±SD.
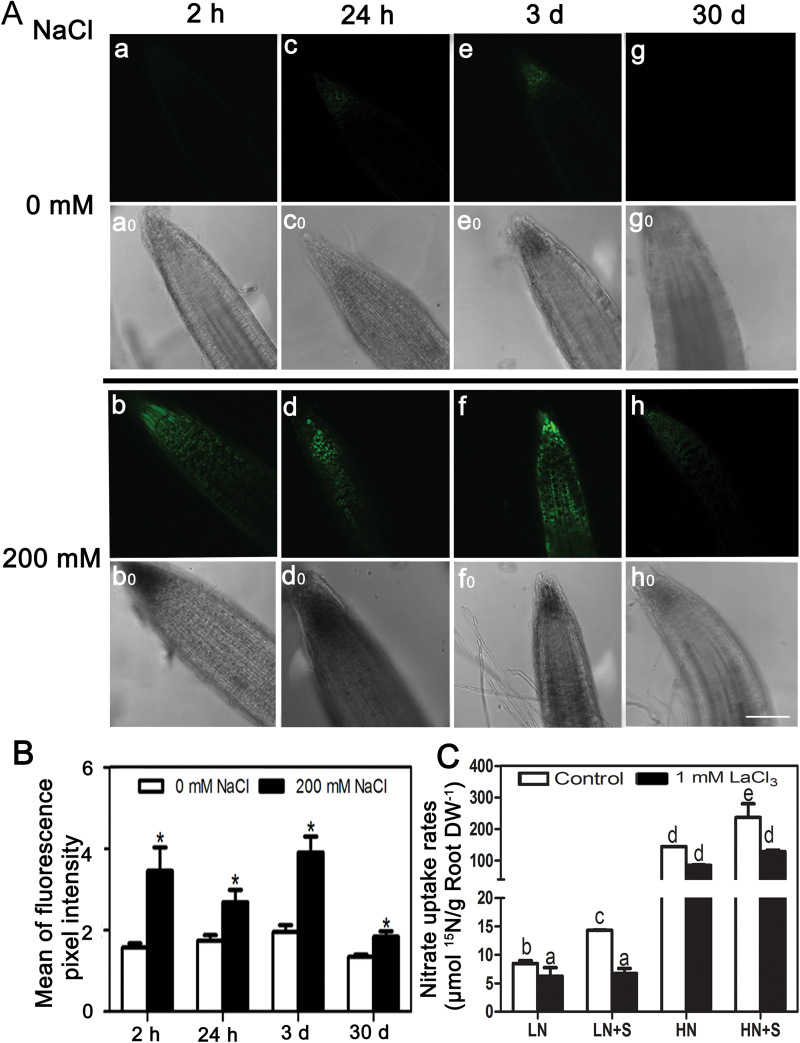
Previous studies have shown that [Ca2+]cyt fluctuations and the downstream calcium signalling proteins play pivotal roles in stress responses (Plieth, 2005). Thus, the dynamic changes of [Ca2+]cyt under 200mM NaCl treatment were determined in S. europaea roots using the Ca2+-sensitive fluorescent probe, Fluo-4/AM. In contrast to weak fluorescence detected in the control root tips, strong fluorescence was observed in roots treated with NaCl for 2h, 24h, and 3 d (Fig. 7A, B). Although the fluorescence from 30 d NaCl-treated root tips was weak, it was still significantly higher than that of the control. Statistical analysis of the mean intensity of fluorescence showed that [Ca2+]cyt was significantly elevated under NaCl treatments (Fig. 7B).
Fig. 7.
The involvement of Ca2+ signalling in NaCl-facilitated nitrate uptake in S. europaea. (A) The effects of NaCl on relative [Ca2+]cyt in S. europaea root tips. S. europaea seedlings treated with 200mM NaCl for 2h, 24h, 3 d, and 30 d were incubated with Fluo-4/AM, and root tips were observed by confocal microscopy (b, d, f, and h). The root tips without NaCl treatments served as controls (a, c, e, and g), a0 to h0 are light field images of the corresponding root tip. Scale bar=100 μm. (B) Quantification of average fluorescence intensities of root tips. Values are means ±SE (n=10). Asterisks indicate significant differences (P<0.05). (C) The effect of LaCl3 on nitrate uptake rates of S. europaea roots. LaCl3-pre-treated S. europaea seedlings were immersed in modified 1/2 Hoagland solutions with different NaCl and 15N-labelled KNO3 concentrations for 15min, and the content of 15N in roots was measured. Values are means ±SE (n=3). Different letters above the bars indicate significant differences at P<0.05, which were calculated separately under LN and HN conditions.
To investigate the effects of [Ca2+]cyt on nitrate uptake under NaCl treatments in S. europaea, LaCl3 (a Ca2+ channel blocker) was applied to S. europaea roots, and the nitrate influx rates were detected. Pre-treatment with LaCl3 resulted in decreased [Ca2+]cyt (Supplementary Fig. S4 at JXB online), and the nitrate uptake rates of roots were significantly reduced under the three treatments besides the HN conditions compared with the control. Moreover, the reduction was much more pronounced under NaCl treatments than without NaCl treatments. After LaCl3 pre-treatment, no significant difference in the nitrate uptake rate was observed between LN and LN+S, or between HN and HN+S (Fig. 7C). This result indicated that the decrease in [Ca2+]cyt not only caused a decrease in nitrate uptake rate, but also attenuated the promotion effects of NaCl on this parameter.
Discussion
NaCl facilitates nitrate uptake in S. europaea
Generally, NaCl has inhibitory effects on nitrate assimilation in glycophytes. For example, in Arabidopsis, NO3 – concentrations and NR activity in both leaves and roots were markedly decreased by NaCl during the second week of treatment (Debouba et al., 2013). However, in this study, it was found that NaCl can promote nitrate uptake in the euhalophyte S. europaea. The shoot NO3 – concentration and nitrate uptake rates of S. europaea were significantly promoted by NaCl under both low N and high N conditions (Fig. 2D, E). These results are consistent with the findings for the euhalophyte S. physophora which showed that NaCl application significantly increased the leaf NO3 – concentration under N-sufficient conditions (Yuan et al., 2010). The current results and previous studies together show that the effect of NaCl on nitrate uptake and assimilation may be significantly different between glycophytes and halophytes. NaCl generally has a positive influence on the growth of euhalophytes, for which zero NaCl is an abnormal condition whereas 200mM NaCl is in the range of the optimal condition (Flowers and Colmer, 2008). Previous studies showed that the growth of S. europaea is stimulated by 200–400mM NaCl (Wang et al., 2009; Lv et al., 2012). Consistently, in this study, it was found that supply of 200mM NaCl improved the growth of S. europaea under both LN and HN conditions (Fig. 1A, B). Better growth of S. europaea under salt conditions increased the demand for N, which could at least partly explain the stimulation of nitrate uptake and increased plant N content by salt treatment (Fig. 2B, E). Furthermore, there might be an NaCl-facilitated nitrate uptake mechanism in S. europaea, which is directly supported by the findings that nitrate uptake rates and shoot NO3 – concentrations of S. europaea were significantly promoted by NaCl treatment under both high N and low N conditions (Fig. 2D, E). In addition, the NR activities were significantly increased by salt under the high N condition (Fig. 2F). Consistent with this study, a sodium-dependent nitrate transport at the PM of leaf and root cells was also reported in Zostera marina, which is a seagrass that grows submerged in seawater where the NaCl concentration is ~500mM (Rubio et al., 2005).
The possible roles of Ca2+ signalling proteins identified by comparative proteomics in S. europaea
Ca2+ plays vital roles in plant development and in response to environmental stimuli (DeFalco et al., 2010). Developmental and environmental cues are perceived at the cell surface, thereby eliciting [Ca2+]cyt changes through concerted action of channels, pumps, and carriers (Kudla et al., 2010). Two protein families, ANN and calreticulin (CRT), involved in this process were identified in this proteomic study. ANN encodes unconventional calcium-permeable channels, which is a key component of root cell adaptation to salinity (Davies, 2014). Four ANN proteins (spots 1358, 1401, 1402, and 1480) were significantly increased in response to NaCl (Fig. 6), indicating that they may act as NaCl-regulated Ca2+-permeable channels in S. europaea. Interestingly, spots 1401 and 1402 represented the same protein, which have the same molecular mass but different pI values (Fig. 3B). It is speculated that the isoforms may be generated by phosphorylation of ANN. CRT is a highly conserved Ca2+-binding protein functioning in intracellular Ca2+ homeostasis and signalling and is predominantly located in the ER; CRT is reportedly associated with the extracellular membrane surface (Jia et al., 2009). CRT is an NaCl-responsive protein in potato (Aghaei et al., 2008), as well as in Arabidopsis (Jiang et al., 2007). In S. europaea, the abundance of CRT (spot 830) was up-regulated by NaCl only under HN conditions, and it was also induced by HN under NaCl application (Fig. 6), indicating that CRT may act as an NaCl- and HN-responsive protein in S. europaea regulating calcium homeostasis and signalling transduction.
The spatio-temporal Ca2+ signals are then decoded and transmitted by Ca2+-binding proteins (DeFalco et al., 2010). Calmodulin (CaM)-like (CML) and 14-3-3 proteins involved in this process were identified. CMLs are involved in Ca2+ signal transduction by associating with numerous downstream targets, such as protein kinase, phosphatases, transcription factors, transporters, and channels (Popescu et al., 2007). CMLs have been implicated in plant development processes and abiotic stress responses (Magnan et al., 2008). In this study, one CML13-like protein was up-regulated by NaCl under HN conditions (Fig. 6), and this protein may function in Ca2+ signalling as a part of the response of S. europaea to NaCl under N-sufficient conditions. In contrast, the accumulation of CML was significantly down-regulated after 48h salt treatment in roots of barely, which was accompanied by significant reduction of root Ca2+ content (Wu et al., 2014). These different trends may reflect diverse mechanisms of CML regulation in glycophytes and halophytes. 14-3-3 proteins are also involved in Ca2+ signalling, which can interact with Ser/Thr-phosphorylated proteins (Pauly et al., 2007). They have been reported to regulate plant N assimilation by tuning the activities of NRT2, NR, and glutamine synthase (GS). Two phosphorylation sites of NRT2.1 were identified as being regulated by NaCl treatment (Vialaret et al., 2014), and sequence analysis of NRT2s in tobacco and Arabidopsis has identified some possible 14-3-3-binding sites (Guo et al., 2011). The activities of cytosolic NR and GS, the key enzymes in nitrate assimilation, are regulated through phosphorylation by 14-3-3s, calcium-independent protein kinases (CIPKs), and calcium-dependent protein kinases (CDPKs) (Masclaux-Daubresse et al., 2010). NR is the target of CPK17 and 14-3-3, which is regulated by salt stress (Lambeck et al., 2010). Notably, the two 14-3-3 proteins identified in this study changed oppositely in response to NaCl treatment (Fig. 6). Similarly, in salt-tolerant rice strains, the abundance of two 14-3-3 proteins increased, whereas one 14-3-3 protein decreased under salt stress (Liu et al., 2014). These findings indicate that NaCl may regulate 14-3-3 proteins through multiple mechanisms.
Changes in [Ca2+]cyt and downstream Ca2+ signalling are essential for NaCl-facilitated nitrate uptake in S. europaea
Salt stress is first perceived at the cell membrane level, and then an intracellular signalling cascade is triggered via secondary messengers (Kader and Lindberg, 2010). The present results suggested that Ca2+ acted as an essential second messenger in NaCl-facilitated nitrate uptake in S. europaea and functioned through downstream signalling components. First, [Ca2+]cyt was significantly elevated in S. europaea under both short- and long-term NaCl treatment (Fig. 7A, B). Many studies have revealed an increase in [Ca2+]cyt after salt stress (Kader et al., 2007; D’Onofrio and Lindberg, 2009). NaCl induces a higher increase in [Ca2+]cyt in salt-tolerant rice cultivars than in salt-sensitive cultivars (Kader et al., 2007). However, in Arabidopsis root cells or corn root protoplast, NaCl induced a decrease in [Ca2+]cyt within minutes (Lynch and Läuchli, 1988; Halperin et al., 2003). Thus, the regulation of [Ca2+]cyt by NaCl varies with the tested species, cells, and tissues. It is likely that the amplitude, frequency, and duration of changes in [Ca2+]cyt are important for decoding the specific downstream responses for salt stress response in plants. Secondly, a decline in [Ca2+]cyt not only caused a decrease in nitrate uptake rate, but also eliminated the promotion effects of NaCl on this parameter (Fig. 7C), suggesting that the NaCl-induced [Ca2+]cyt elevation was essential for NaCl-facilitated nitrate uptake in S. europaea. Evidence on the direct relationship between [Ca2+]cyt and nitrate uptake is still lacking, but some research has shown links between them. Application of Ca2+ could increase nitrate uptake under salt stress, and this effect may be ascribed to its involvement in preserving the structural and functional integrity of cell membranes or in increasing the activities of nitrate transporters (Mansour, 2000). Meanwhile, plants treated with EGTA (a specific Ca2+ chelator) or La3+ (a Ca2+ channel blocker) showed decreased induction of the key genes involved in the nitrate assimilatory pathway, such as NR and nitrite reductase (NiR) (Vidal et al., 2010). Thirdly, eight calcium signalling proteins were identified in this proteomic study, seven of which were significantly up-regulated under NaCl treatments (Fig. 6); it is speculated that these may play important roles in NaCl-facilitated nitrate uptake in S. europaea. The important roles of Ca2+ signalling proteins in salt adaption have been reported in other plant species. In Arabidopsis, the phosphorylation levels of CPKs changed significantly under salt stress (Vialaret et al., 2014). Mehlmer et al. (2010) also reported that CPK3-mediated Ca2+ signalling is required for salt stress acclimation, and 28 potential CPK3 targets were identified by their studies. Among these, remorin, CRT, VDAC (voltage-dependent anion channel), 14-3-3 proteins, and RAB GTPase, all of which play roles in cellular ion homeostasis and signal transduction under salt stress (Mehlmer et al., 2010), were also identified in the present study. In addition, Latz et al. (2013) reported that the vacuolar two-pore K+ channel 1 (TPK1) was a target of CPK3 and 14-3-3 protein, and played an essential role in the regulation of the cytosolic K+/Na+ ratio under salt stress adaptations.
Based on the present results and previous studies, a possible regulatory network of NaCl-facilitated nitrate uptake in S. europaea focusing on the involvement of Ca2+ signalling is proposed (Fig. 8). In S. europaea roots, the NaCl stimuli induced Ca2+ entry across the PM by activating ANN protein and some unknown Ca2+ channels (Davies, 2014), thereby causing [Ca2+]cyt elevation. CRT is involved in cytosolic Ca2+ homeostasis (Jia et al., 2009). The [Ca2+]cyt elevation is sensed by calcium signalling proteins including CaM/CML, CBL/CIPKs, as well as CDPKs/CPKs (DeFalco et al., 2010). 14-3-3 protein can be activated by CPK3 or some unknown Ca2+ signalling components (Mehlmer et al., 2010). The calcium signalling proteins can transmit the signal into phosphorylation cascades capable of modulating gene expression, and target protein activity, which may function in ion transport/homeostasis, membrane trafficking, redox homeostasis, and so on (DeFalco et al., 2010). Thus, the elevated nitrate uptake rates and NR activity under NaCl treatment in S. europaea may be ascribed to calcium-dependent phosphorylation of proteins such as those described in the following sentences. CBL1/9 and CIPK8/23 are involved in the regulation of NRT1.1 by phosphorylation (Ho et al., 2009; Hu et al., 2009), while 14-3-3 is involved in the regulation of NRT2s (Guo et al., 2011). NRs in the cytosol are targets of CPK17 and 14-3-3, which are also regulated by salt stress (Lambeck et al., 2010). The subsequent steps of N assimilation in roots take place in plastids. GS participates in this processs and can be regulated by CIPK and 14-3-3 through phosphorylation (Masclaux-Daubresse et al., 2010).
Fig. 8.
A putative regulatory network of NaCl-facilitated nitrate uptake in S. europaea. In S. europaea roots, the NaCl stimuli induced Ca2+ entry across the PM by activating ANN protein and some unknown Ca2+ channels, thereby causing [Ca2+]cyt elevation. CRT is involved in cytosolic Ca2+ homeostasis. The [Ca2+]cyt elevation is sensed by calcium signalling proteins including CaM/CML, CBL/CIPKs, and CDPKs/CPKs. 14-3-3 protein can be activated by CPK3 or some unknown Ca2+ signalling components. The calcium signalling proteins can transmit the signal into phosphorylation cascades capable of modulating gene expression and target protein activity, which may function in ion transport/homeostasis, membrane trafficking, redox homeostasis, and so on. CBL1/9 and CIPK8/23 are involved in the regulation of NRT1.1 by phosphorylation while 14-3-3 is involved in the regulation of NRT2s. NRs in the cytosol are targets of CPK17 and 14-3-3, which are also regulated by salt stress. The subsequent steps of N assimilation take place in plastids. GS participates in this process and can be regulated by CIPK and 14-3-3 through phosphorylation. The components in red indicate those identified in the present study.
Conclusions
In this study, it was first found that, unlike in glycophytes, NaCl facilitates nitrate uptake in S. europaea, which may be a unique feature in halophytes. Further comparative proteomics of root PM proteins revealed 81 differentially accumulated proteins in response to salt and nitrate. Among them, there are eight calcium signalling components, and the accumulations of seven was increased in response to salinity. [Ca2+]cyt was significantly elevated in S. europaea under both short- and long-term NaCl treatment. In addition, application of the Ca2+ channel blocker LaCl3 not only caused a decrease in nitrate uptake rate but also reduced the promotion effects of NaCl on the nitrate uptake rates. It is proposed that NaCl induced [Ca2+]cyt elevation and the downstream calcium signalling are essential for NaCl-facilitated nitrate uptake in S. europaea. The calcium signalling proteins may function through regulating the expression or activity of nitrate transporters and some key enzymes in N assimilation, which can consequently affect nitrate assimilation.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Coomassie blue-stained gel image of TM and PM samples.
Figure S2. Ten images of 2D-DIGE.
Figure S3. Scores and matched peptides of the identified proteins based on ultrafleXtreme MALDI-TOF/TOF-MS.
Figure S4. The effect of LaCl3 on the relative [Ca2+]cyt in S. europaea root tips.
Table S1. Protein samples assigned for Cy dye label and DIGE.
Table S2. Expression profile data of the 717 spots present in at least 24 of the 30 images.
Table S3. The differentially accumulated S. europaea PM proteins identified by MALDI-TOF/TOF.
Table S4. Protein distribution in the eight clusters.
Table S5. Specific primers used for real-time PCR of genes encoding calcium signalling components of S. europaea.
Acknowledgements
This work was supported by the Research Programs from the Chinese Ministry of Agriculture (grant no. 2014ZX08009-003-002), the National Natural Science Foundation of China (grant no. 31200201), and the Science and Technology Service Network Initiative of the Chinese Academy of Sciences (grant no. KFJ-EW-STS-061-4). The authors thank Dafeng Jinglong Marine Industrialized Development Corporation, Ltd, Jiangsu Province for providing seeds of S. europaea. All authors have declared no conflict of interest.
Glossary
Abbreviations:
- ANN
annexin
- ANOVA
analysis of variance
- [Ca2+]cyt
cytosolic Ca2+ concentration
- CaM
calmodulin
- CBL
calcineurin B-like
- CDPK
calcium-dependent protein kinase
- CIPK
calcium-independent protein kinase
- CML
calmodulin-like
- CRT
calreticulin
- 2D-DIGE
two-dimensional fluorescence difference gel electrophoresis
- DW
dry weight
- FW
fresh weight
- GDH
glutamate dehydrogenase
- GS
glutamine synthase
- NiR
nitrite reductase
- NR
nitrate reductase
- NRT
nitrate transporter
- PM
plasma membrane
- TM
total microsomal
- VDAC
voltage-dependent anion-selective channel.
References
- Aghaei K, Ehsanpour AA, Komatsu S. 2008. Proteome analysis of potato under salt stress. Journal of Proteome Research 7, 4858–4868. [DOI] [PubMed] [Google Scholar]
- Bar Y, Apelbaum A, Kafkafi U, Goren R. 1997. Relationship between chloride and nitrate and its effect on growth and mineral composition of avocado and citrus plants. Journal of Plant Nutrition 20, 715–731. [Google Scholar]
- Cheng Y, Qi Y, Zhu Q, Chen X, Wang N, Zhao X, Chen H, Cui X, Xu L, Zhang W. 2009. New changes in the plasma-membrane-associated proteome of rice roots under salt stress. Proteomics 9, 3100–3114. [DOI] [PubMed] [Google Scholar]
- Davies JM. 2014. Annexin-mediated calcium signalling in plants. Plants 3, 128–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debouba M, Dguimi HM, Ghorbel M, Gouia H, Suzuki A. 2013. Expression pattern of genes encoding nitrate and ammonium assimilating enzymes in Arabidopsis thaliana exposed to short term NaCl stress. Journal of Plant Physiology 170, 155–160. [DOI] [PubMed] [Google Scholar]
- DeFalco TA, Bender KW, Snedden WA. 2010. Breaking the code: Ca2+ sensors in plant signalling. Biochemical Journal 425, 27–40. [DOI] [PubMed] [Google Scholar]
- D’Onofrio C, Lindberg S. 2009. Sodium induces simultaneous changes in cytosolic calcium and pH in salt-tolerant quince protoplasts. Journal of Plant Physiology 166, 1755–1763. [DOI] [PubMed] [Google Scholar]
- Elmore JM, Liu J, Smith B, Phinney B, Coaker G. 2012. Quantitative proteomics reveals dynamic changes in the plasma membrane during Arabidopsis immune signaling. Molecular and Cellular Proteomics 11, M111.014555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan PX, Nie LL, Jiang P, Feng JJ, Lv SL, Chen XY, Bao HXG, Guo J, Tai F, Wang JH. 2013. Transcriptome analysis of Salicornia europaea under saline conditions revealed the adaptive primary metabolic pathways as early events to facilitate salt adaptation. PLoS One 8, e80595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers TJ, Colmer TD. 2008. Salinity tolerance in halophytes. New Phytologist 179, 945–963. [DOI] [PubMed] [Google Scholar]
- Frechilla S, Lasa B, Ibarretxe L, Lamsfus C, Aparicio-Tejo P. 2001. Pea responses to saline stress is affected by the source of nitrogen nutrition (ammonium or nitrate). Plant Growth Regulation 35, 171–179. [Google Scholar]
- Guo C, Chang W, Gu J, Li X, Lu W, Xiao K. 2011. Molecular characterization, transcriptional regulation and function analysis of nitrate transporters in plants. Frontiers of Agriculture in China 5, 291–298. [Google Scholar]
- Halperin SJ, Gilroy S, Lynch JP. 2003. Sodium chloride reduces growth and cytosolic calcium, but does not affect cytosolic pH, in root hairs of Arabidopsis thaliana L. Journal of Experimental Botany 54, 1269–1280. [DOI] [PubMed] [Google Scholar]
- Hamed KB, Ellouzi H, Talbi OZ, Hessini K, Slama I, Ghnaya T, Bosch SM, Savouré A, Abdelly C. 2013. Physiological response of halophytes to multiple stresses. Functional Plant Biology 40, 883–896. [DOI] [PubMed] [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF. 2009. CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194. [DOI] [PubMed] [Google Scholar]
- Hu HC, Wang YY, Tsay YF. 2009. AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. The Plant Journal 57, 264–278. [DOI] [PubMed] [Google Scholar]
- Jampeetong A, Brix H. 2009. Nitrogen nutrition of Salvinia natans: effects of inorganic nitrogen form on growth, morphology, nitrate reductase activity and uptake kinetics of ammonium and nitrate. Aquatic Botany 90, 67–73. [Google Scholar]
- Jia XY, He LH, Jing RL, Li RZ. 2009. Calreticulin: conserved protein and diverse functions in plants. Physiologia Plantarum 136, 127–138. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Yang B, Harris NS, Deyholos MK. 2007. Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. Journal of Experimental Botany 58, 3591–3607. [DOI] [PubMed] [Google Scholar]
- Kader MA, Lindberg S. 2010. Cytosolic calcium and pH signaling in plants under salinity stress. Plant Signaling and Behavior 5, 233–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kader MA, Lindberg S, Seidel T, Golldack D, Yemelyanov V. 2007. Sodium sensing induces different changes in free cytosolic calcium concentration and pH in salt-tolerant and -sensitive rice (Oryza sativa) cultivars. Physiologia Plantarum 130, 99–111. [Google Scholar]
- Krapp A, David LC, Chardin C, Girin T, Marmagne A, Leprince A-S, Chaillou S, Ferrario-Méry S, Meyer C, Daniel-Vedele F. 2014. Nitrate transport and signalling in Arabidopsis . Journal of Experimental Botany 65, 789–798. [DOI] [PubMed] [Google Scholar]
- Kudla J, Batistic O, Hashimoto K. 2010. Calcium signals: the lead currency of plant information processing. The Plant Cell 22, 541–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambeck I, Chi JC, Krizowski S, Mueller S, Mehlmer N, Teige M, Fischer K, Schwarz G. 2010. Kinetic analysis of 14-3-3-inhibited Arabidopsis thaliana nitrate reductase. Biochemistry 49, 8177–8186. [DOI] [PubMed] [Google Scholar]
- Lara C, Rodríguez R, Guerrero MG. 1993. Sodium dependent nitrate transport and energetics in cyanobacteria. Journal of Phycology 29, 389–395. [Google Scholar]
- Latz A, Mehlmer N, Zapf S, Mueller TD, Wurzinger B, Pfister B, Csaszar E, Hedrich R, Teige M, Becker D. 2013. Salt stress triggers phosphorylation of the Arabidopsis vacuolar K+ channel TPK1 by calcium-dependent protein kinases (CDPKs). Molecular Plant 6, 1274–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CW, Chang TS, Hsu YK, Wang AZ, Yen HC, Wu YP, Wang CS, Lai CC. 2014. Comparative proteomic analysis of early salt stress responsive proteins in roots and leaves of rice. Proteomics 14, 1759–1775. [DOI] [PubMed] [Google Scholar]
- Lv SL, Jiang P, Chen XY, Fan PX, Wang XC, Li YX. 2012. Multiple compartmentalization of sodium conferred salt tolerance in Salicornia europaea . Plant Physiology and Biochemistry 51, 47–52. [DOI] [PubMed] [Google Scholar]
- Lynch J, Läuchli A. 1988. Salinity affects intracellular calcium in corn root protoplasts. Plant Physiology 87, 351–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Zhang M, Xiao X, You J, Wang J, Wang T, Yao Y, Tian C. 2013. Global transcriptome profiling of Salicornia europaea L. shoots under NaCl treatment. PLoS One 8, e65877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnan F, Ranty B, Charpenteau M, Sotta B, Galaud JP, Aldon D. 2008. Mutations in AtCML9, a calmodulin-like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid. The Plant Journal 56, 575–589. [DOI] [PubMed] [Google Scholar]
- Mansour M. 2000. Nitrogen containing compounds and adaptation of plants to salinity stress. Biologia Plantarum 43, 491–500. [Google Scholar]
- Marschner H, Rimmington G. 1988. Mineral nutrition of higher plants. Plant, Cell and Environment 11, 147–148. [Google Scholar]
- Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A. 2010. Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Annals of Botany 105, 1141–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlmer N, Wurzinger B, Stael S, Hofmann-Rodrigues D, Csaszar E, Pfister B, Bayer R, Teige M. 2010. The Ca2+-dependent protein kinase CPK3 is required for MAPK-independent salt-stress acclimation in Arabidopsis . The Plant Journal 63, 484–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller AL, Pedas P, Andersen B, Svensson B, Schjoerring JK, Finnie C. 2011. Responses of barley root and shoot proteomes to long-term nitrogen deficiency, short-term nitrogen starvation and ammonium. Plant, Cell and Environment 34, 2024–2037. [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. 2008. Mechanisms of salinity tolerance. Annual Review of Plant Biology 59, 651–681. [DOI] [PubMed] [Google Scholar]
- Muños S, Cazettes C, Fizames C, Gaymard F, Tillard P, Lepetit M, Lejay L, Gojon A. 2004. Transcript profiling in the chl1-5 mutant of Arabidopsis reveals a role of the nitrate transporter NRT1.1 in the regulation of another nitrate transporter, NRT2.1. The Plant Cell 16, 2433–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie LL, Feng JJ, Lv SL, Jiang P, Fan PX, Tai F, Li YX. 2012. The response of euhalophyte Salicornia europaea L. to different nitrogen forms. Acta Ecologica Sinica 32, 5703–5712. [Google Scholar]
- Nouri MZ, Komatsu S. 2010. Comparative analysis of soybean plasma membrane proteins under osmotic stress using gel-based and LC MS/MS-based proteomics approaches. Proteomics 10, 1930–1945. [DOI] [PubMed] [Google Scholar]
- Pauly B, Lasi M, MacKintosh C, Morrice N, Imhof A, Regula J, Rudd S, David CN, Bottger A. 2007. Proteomic screen in the simple metazoan Hydra identifies 14-3-3 binding proteins implicated in cellular metabolism, cytoskeletal organisation and Ca2+ signalling. BMC Cell Biology 8, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessarakli M, Harivandi M, Kopec DM, Ray DT. 2012. Growth responses and nitrogen uptake by saltgrass (Distichlis spicata L.), a halophytic plant species, under salt stress, using the 15N technique. International Journal of Agronomy 2012, 896971. [Google Scholar]
- Plieth C. 2005. Calcium: just another regulator in the machinery of life? Annals of Botany 96, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu SC, Popescu GV, Bachan S, Zhang Z, Seay M, Gerstein M, Snyder M, Dinesh-Kumar S. 2007. Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proceedings of the National Academy of Sciences, USA 104, 4730–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinsi B, Negri AS, Pesaresi P, Cocucci M, Espen L. 2009. Evaluation of protein pattern changes in roots and leaves of Zea mays plants in response to nitrate availability by two-dimensional gel electrophoresis analysis. BMC Plant Biology 9, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees TAV, Cresswell RC, Syrett PJ. 1980. Sodium-dependent uptake of nitrate and urea by a marine diatom. Biochimica et Biophysica Acta 596, 141–144. [DOI] [PubMed] [Google Scholar]
- Rubio L, Linares-Rueda A, García-Sánchez M, Fernández J. 2005. Physiological evidence for a sodium-dependent high-affinity phosphate and nitrate transport at the plasma membrane of leaf and root cells of Zostera marina L. Journal of Experimental Botany 56, 613–622. [DOI] [PubMed] [Google Scholar]
- Striker GG, Teakle NL, Colmer TD, Barrett-Lennard ED. 2015. Growth responses of Melilotus siculus accessions to combined salinity and root-zone hypoxia are correlated with differences in tissue ion concentrations and not differences in root aeration. Environmental and Experimental Botany 109, 89–98. [Google Scholar]
- Tang W. 2012. Quantitative analysis of plasma membrane proteome using two-dimensional difference gel electrophoresis. Methods in Molecular Biology 876, 67–82. [DOI] [PubMed] [Google Scholar]
- Tang W, Kim TW, Oses-Prieto JA, Sun Y, Deng Z, Zhu S, Wang R, Burlingame AL, Wang ZY. 2008. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis . Science 321, 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NL, Heazlewood JL, Millar AH. 2011. The Arabidopsis thaliana 2-D gel mitochondrial proteome: refining the value of reference maps for assessing protein abundance, contaminants and post-translational modifications. Proteomics 11, 1720–1733. [DOI] [PubMed] [Google Scholar]
- Ungar IA. 1987. Population characteristics, growth and survival of the halophyte Salicornia europaea . Ecology 68, 569–575. [Google Scholar]
- Vialaret J, Di Pietro M, Hem S, Maurel C, Rossignol M, Santoni V. 2014. Phosphorylation dynamics of membrane proteins from Arabidopsis roots submitted to salt stress. Proteomics 14, 1058–1070. [DOI] [PubMed] [Google Scholar]
- Vidal EA, Tamayo KP, Gutierrez RA. 2010. Gene networks for nitrogen sensing, signaling, and response in Arabidopsis thaliana . Wiley Interdisciplinary Reviews: Systems Biology and Medicine 2, 683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Bian YY, Cheng K, Zou HF, Sun SSM, He JX. 2012. A comprehensive differential proteomic study of nitrate deprivation in Arabidopsis reveals complex regulatory networks of plant nitrogen responses. Journal of Proteome Research 11, 2301–2315. [DOI] [PubMed] [Google Scholar]
- Wang XC, Li XF, Deng X, Han HP, Shi WL, Li YX. 2007. A protein extraction method compatible with proteomic analysis for the euhalophyte Salicornia europaea . Electrophoresis 28, 3976–3987. [DOI] [PubMed] [Google Scholar]
- Wang XC, Fan PX, Song HM, Chen XY, Li XF, Li YX. 2009. Comparative proteomic analysis of differentially expressed proteins in shoots of Salicornia europaea under different salinity. Journal of Proteome Research 8, 3331–3345. [DOI] [PubMed] [Google Scholar]
- Wessel D, Flügge UI. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Analytical Biochemistry 138, 141–143. [DOI] [PubMed] [Google Scholar]
- Wu D, Shen Q, Qiu L, Han Y, Ye L, Jabeen Z, Shu Q, Zhang G. 2014. Identification of proteins associated with ion homeostasis and salt tolerance in barley. Proteomics 14, 1381–1392. [DOI] [PubMed] [Google Scholar]
- Yaneva IA, Hoffmann GW, Tischner R. 2002. Nitrate reductase from winter wheat leaves is activated at low temperature via protein dephosphorylation. Physiologia Plantarum 114, 65–72. [DOI] [PubMed] [Google Scholar]
- Yuan JF, Tian CY, Feng G. 2010. Effects of sodium on nitrate uptake and osmotic adjustment of Suaeda physophora . Journal of Arid Land 2, 190–196. [Google Scholar]
- Zhang Y, Ding S, Lu Q, Yang Z, Wen X, Zhang L, Lu C. 2011. Characterization of photosystem II in transgenic tobacco plants with decreased iron superoxide dismutase. Biochimica et Biophysica Acta 1807, 391–403. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.