Highlight
Arabidopsis CyclinD3 genes are revealed as central regulators of cambial cell proliferation and vascular development, which constitutes part of a novel mechanism controlling secondary growth and radial organ size.
Key words: Arabidopsis, cambium, cell cycle, cell division, cell expansion, cyclin D3, inflorescence stem, organ size, secondary growth, vascular development, xylem.
Abstract
A major proportion of plant biomass is derived from the activity of the cambium, a lateral meristem responsible for vascular tissue formation and radial organ enlargement in a process termed secondary growth. In contrast to our relatively good understanding of the regulation of primary meristems, remarkably little is known concerning the mechanisms controlling secondary growth, particularly how cambial cell divisions are regulated and integrated with vascular differentiation. A genetic loss-of-function approach was used here to reveal a rate-limiting role for the Arabidopsis CYCLIN D3 (CYCD3) subgroup of cell-cycle genes in the control of cambial cell proliferation and secondary growth, providing conclusive evidence of a direct link between the cell cycle and vascular development. It is shown that all three CYCD3 genes are specifically expressed in the cambium throughout vascular development. Analysis of a triple loss-of-function CYCD3 mutant revealed a requirement for CYCD3 in promoting the cambial cell cycle since mutant stems and hypocotyls showed a marked reduction in diameter linked to reduced mitotic activity in the cambium. Conversely, loss of CYCD3 provoked an increase in xylem cell size and the expression of differentiation markers, showing that CYCD3 is required to restrain the differentiation of xylem precursor cells. Together, our data show that tight control of cambial cell division through developmental- and cell type-specific regulation of CYCD3 is required for normal vascular development, constituting part of a novel mechanism controlling organ growth in higher plants.
Introduction
Plant growth and development relies on the co-ordination of cell division and cell expansion, which occur in the stem-cell systems of the meristems (Gonzalez et al., 2012). The lateral meristem of the procambium is established during primary growth from the apical meristems and is responsible for forming the vascular (conductive) tissues of the primary xylem and primary phloem. In stems and roots of dicotyledonous plants, the procambium, as well as a subset of specialized parenchyma cells (as in stems), gives rise to a further lateral meristem, the cambium, which is responsible for the production of secondary xylem and secondary phloem, leading to the radial enlargement of the plant body in a process termed secondary growth. Vascular tissue formed through secondary growth constitutes the major proportion of plant biomass and is the basis of wood production in tree species. Vascular development in the Arabidopsis inflorescence stem is an attractive model system to study the co-ordination of cell division and cell expansion in organ growth as the two processes are spatially separated in radially expanding stems (Sehr et al., 2010; Sanchez et al., 2012). At the stem apex, the first of two specialized cambial cell populations, termed the fascicular cambium (FC), proliferates to form daughter cells that are displaced either inwards or outwards following a pathway towards terminal differentiation to adopt a xylem or phloem cell fate, respectively. Differentiation of cambial daughter cells is associated with cell expansion, lignification, and secondary cell wall biosynthesis (Schuetz et al., 2013). At the base of elongated stems, a second specialized cambial cell population termed the interfascicular cambium (IC) is established de novo from cells lying within interfascicular regions, culminating in a ring-like continuous meristematic zone constituting the vascular cambium (Sanchez et al., 2012). Once formed, the vascular cambium undergoes rapid proliferation marking the major phase of secondary growth and radial organ expansion.
Tight control of the rate of cambial cell division and the timing of cell expansion/differentiation in cambial daughter cells is critical for the maintenance and size of the cambium and, ultimately, for determining radial organ size. These processes are known to be under developmental, environmental, and hormonal control (Miyashima et al., 2013). However, as only a small number of genes have been identified as having roles in the control of cambial development and activity, remarkably little is known concerning the spatiotemporal regulation of cambial cell division and its integration with vascular differentiation.
Plant cell division is regulated by cyclin-dependent kinases (CDK) which require the binding of a positive regulatory cyclin subunit for promoting the entry and progression of cells through the cell cycle (Gonzalez et al., 2012). In Arabidopsis, at least 110 genes have so far been recognized as belonging to a small number of gene families comprising the ‘core’ cell cycle machinery, including CDKs and cyclins, that act at different phases of the cell cycle to control division and the transition to differentiation (Vandepoele et al., 2002; Menges et al., 2005; Van Leene et al., 2010). One gene family, the D-type cyclins (CYCD), play a central role in promoting cell division through their activation of the CYCD-RETINOBLASTOMA-RELATED (RBR)-E2F pathway at the G1-S checkpoint in response to mitogenic signals such as auxin and cytokinin (Riou-Khamlichi et al., 1999; Oakenfull et al., 2002; Dewitte et al., 2007). The CYCD family is conserved between all plant species so far characterized including Arabidopsis, rice, and poplar, with individual gene members belonging to one of six subgroups (Menges et al., 2007). In Arabidopsis, the CYCD3 subgroup consists of three members (CYCD3;1, CYCD3;2, and CYCD3;3) which were shown to control the balance between cell division and cell expansion in shoot lateral organs by regulating the duration of mitotic activity, as well as mediating the response of meristematic cells to cytokinin (Dewitte et al., 2007). A partial expression analysis of CYCD3;1 using in situ hybridization revealed activity in the procambium of developing leaves and inflorescence stems (Dewitte et al., 2003). CYCD genes are thus prime candidates for playing roles in the regulation of cambial cell division and its integration with vascular differentiation.
The wider role of the cell cycle in vascular development and secondary growth has been investigated here. Based on public gene expression profiling data, it is revealed that a small set of core cell-cycle regulators are associated with both primary and secondary vascular development. The CYCD3 subgroup is identified as positive regulators of cambial cell proliferation and secondary growth based on a detailed gene expression and loss-of-function analysis of the CYCD3 genes during vascular development. Furthermore, an additional role for the CYCD3 subgroup in restraining cell expansion and differentiation of xylem precursor cells is uncovered. Thus, tight control of the cambial cell cycle and co-ordination with cell expansion and differentiation processes through developmental- and cell-type-specific regulation of CYCD3 is required for proper vascular development and radial organ growth.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana ecotype Columbia was used as the wild type in all experiments. The pCYCD3;1:GUS, pCYCD3;2:GUS, pCYCD3;3:GUS, and cycd3;1–3 lines were as described by Dewitte et al. (2007) and were obtained from James Murray (Cardiff, UK). The genotype of the cycd3;1–3 line was confirmed by the absence of transcripts for all three CYCD3 genes using RT-PCR (see Supplementary Fig. S1 at JXB online). Seeds were surface-sterilized with 70% ethanol (v/v) for 3min, followed by 20% hypochlorite (v/v) for 15min, and rinsed six times with sterile deionized water. Sterilized seed were sown onto square Petri plates containing sterile, solid, half-strength Murashige and Skoog (MS) medium. Plates were shifted to a cold room at 4 °C for 3 d to synchronize germination, then transferred to a growth chamber and grown at 22±2 °C under a 16/8h light/dark photoperiod. For the analysis of mature plants, 11-d-old seedlings were transplanted from the growth medium to soil (Fafard 4P Mix; Conrad Fafard, USA) contained in rectangular plastic tray inserts (6×3 compartments) inside black trays with humidity domes. Each seedling was placed in a separate compartment in each insert (to ensure uninhibited growth to maximum height). Trays were moved to a growth chamber and grown at 22±2 °C under a 16/8h light/dark photoperiod at 60% humidity. Humidity domes were removed after 3 d growth.
Conventional reverse transcription-PCR and reverse transcription quantitative real-time PCR
Tissue was obtained from the top (5mm under the shoot meristem), middle, and at the base (immediately above the rosette) of inflorescence stems from 30-cm-tall plants. Dissected tissue was collected from five individual plants for each stem position and pooled for RNA isolation. Total RNA was extracted using TRIzol Reagent (Invitrogen) and further purified using the DNA-free Kit (Ambion) according to the manufacturer’s instructions. RNA concentration and purity were determined using a NanoDrop 2000 Spectrophotometer (Thermo Scientific). RNA integrity was visualized by denaturing agarose gel electrophoresis and ethidium bromide staining. All reverse transcriptions were performed using 2 μg total RNA in a total reaction volume of 20 μl with the RETROscript kit (Ambion) using an oligo (dT)18 primer according to the manufacturer’s instructions. Briefly, samples were heated to 70 °C for 3min, held at 42 °C for 1h in the presence of 1 μl MMLV-RT, and inactivated at 92 °C for 10min. After cDNA synthesis, all samples were diluted 10 times in molecular biology grade water (Thermo Scientific) and stored at –20 °C.
All standard PCR reactions were performed using Taq DNA Polymerase (New England BioLabs) according to the manufacturer’s instructions. All quantitative real-time PCR (qPCR) reactions were prepared in twin.tec 96-well PCR plates (Eppendorf). Each 25 μl reaction consisted of 12.5 μl 2× QuantiTect SYBR Green PCR Master Mix (Qiagen), 1.5 μl forward and reverse primer (final concentration of 0.3 μM each primer), 3.0 μl diluted cDNA, and 8.0 μl molecular biology grade water (Thermo Scientific), typically prepared using master-mixes. Triplicate reactions from two biological replicates were performed on a Mastercycler ep realplex cycler (Eppendorf) using the following thermal cycling profile: 95 °C for 15min (to activate the polymerase), followed by 45 cycles of 94 °C for 15 s, 60 °C for 30 s, and 72 °C for 30 s. Finally, a dissociation (melting curve) analysis was performed to confirm the amplification of a single expected PCR product and the absence of primer dimer formation, according to the manufacturer’s instructions (Eppendorf). Minus reverse transcription and no template control reactions were included in all assays to confirm the absence of contaminating DNA in RNA preparations and PCR reagents, respectively. Normalized relative expression levels were calculated using the ΔΔCt method (Pfaffl, 2001) using the ACTIN2 gene as the endogenous control (Collins et al., 2012). Primers were designed using nucleotide sequences from The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org). The specificity of all primer sets for their respective target sequence was demonstrated by: (i) amplification of a single PCR product followed by sequencing of the resulting PCR fragment; (ii) generation of a single peak in the melting curve analysis; (iii) an in silico specificity screen using BLAST analysis (NCBI). The following primer pairs were used: ACTIN2, 5′-GAAGAACTATGAATTACCCGATGGGC-3′ and 5′-CCCGGGTTAGAAACATTTTCTGTGAACG-3′ (Collins et al., 2012); CYCD3;1, 5′-GCAAGTTGATCCCTTTGACC-3′ and 5′-CAGCTTGGACTGTTCAACGA-3′ (Collins et al., 2012); CYCB1;1, 5′-TAAGCAGATTCAGTTCCGGTCAAC-3′ and 5′-GGGAGCTTTACGAAAGAAATACTCC-3′ (Collins, 2008); IRX3, 5′-GCGTGTGCACCCATATCC-3′ and 5′-TCATCC ATTCTTTCCCGCC-3′ (Etchells and Turner, 2010); SND2, 5′-CAGACTCAACCACGTCAATGC-3′ and 5′-AGGGATAAAA GGTTGAGAGTC-3′ (Zhong et al., 2008).
Histology and microscopy
Histochemical staining for GUS activity in seedling and adult tissues was performed as described by Jefferson et al. (1987). GUS-stained and unstained tissue was cleared in 70% ethanol and mounted in 10% glycerol prior to microscopic analysis. Stem samples for sectioning were taken from (i) the base of inflorescences immediately above the rosette, and (ii) from 5mm below the shoot apex, of 30-cm-long plants in which the first internode was at least 3.5cm long. Samples were fixed in FAA (formalin/acetic acid/alcohol) and then dehydrated and embedded in paraffin, and microtome-cut sections 5 μm thick were stained with 0.05% toluidine blue. Whole plants were examined and photographed using a Leica S6 D dissecting microscope equipped with a Nikon coolpix 4500 digital zoom camera. Sectioned material was examined and photographed using a Zeiss Axioskop microscope equipped with a Jenoptik ProgRes digital zoom camera and ProgRes CapturePro (2.8.8) camera control software (Jenoptik). Adobe Photoshop CS5 (v12.1 x64) and Adobe Illustrator CS5 (v15.1.0) were used for image preparation.
Phenotypic analyses
In all quantitative measurements described, sampled plants were taken from a total population of 108 plants. Stem diameter was measured at the base of inflorescences immediately above the rosette when the inflorescence was 1, 10, 20, and 30cm long. Hypocotyl diameter was measured at the base of the hypocotyl adjacent to the rosette in 30-cm-tall plants. Stem lengths were measured at 21, 28, 35, 42, 49, 56, and 63 d from the date of sowing. In the analyses of stem and hypocotyl diameter, and stem length, ten individual randomly-selected plants were evaluated at each time point. Measurement of the width:length (tangential:radial) ratio of vascular bundles at the stem base was as described previously (Fisher and Turner, 2007), by evaluating three randomly selected vascular bundles each from tissue sections from 15 individual randomly selected plants. Measurement of the lateral extension of the FC-derived (FCD) and IC-derived (ICD) tissues at the stem base was as described earlier (Sehr et al., 2010), by evaluating both FCD and ICD extension at five randomly selected positions each from tissue sections from 15 individual randomly-selected plants. Cell numbers at the stem base were calculated from tissue sections from 15 individual randomly selected plants. Xylem cell sizes at the stem base were measured using ImageJ software v1.46r (http://rsbweb.nih.gov/ij/) from tissue sections from ten individual randomly selected plants. Briefly, images were adjusted (brightness/contrast) to reduce background noise using Adobe Photoshop CS5 (v12.1 x64). In ImageJ, images were converted to greyscale, and then to binary (threshold), followed by manual adjustments to remove further background noise. Cells were highlighted using the Outlines tool, and the Analyze Particles tool was used to obtain cell sizes (Calculated from cell area in μm2). Data were exported as a spreadsheet for further analysis. All statistical analyses were performed using JMP v10.0.2 software package (SAS Institute). A two-tailed independent Student’s t test was applied to data using significance levels of P <0.05, P <0.01, and P <0.001 as indicated by single, double, and triple asterisks, respectively.
Extraction and analysis of data from gene expression databases
The Arabidopsis Gene Expression Database (AREX Lite; Brady et al., 2007; http://www.arexdb.org/download.html) was used to extract expression levels of core cell-cycle and cambium marker genes in different cells and tissues of the root tip (Brady et al., 2007). In this dataset, a mixed-model analysis was used to normalize all arrays globally (Brady et al., 2007). Briefly, expression values were mean-normalized (centring to zero) and log2-transformed to yield relative expression indices. Therefore, high expression and low expression for any given gene (probe) is relative to zero (lowest expression). Supplementary Table S1 from Zhao et al. (2005) was the source of data for expression levels of core cell-cycle genes in phloem/cambium, xylem, and non-vascular tissues of the mature hypocotyl. Hierarchical clustering was performed using Cluster software v3.0 (Eisen et al., 1998; De Hoon et al., 2004; http://bonsai.hgc.jp/~mdehoon/software/cluster/) and visualized using Java TreeView v1.1.6r4 (Saldanha, 2004; http://jtreeview.sourceforge.net/).
Results
Global expression profiling of core cell-cycle genes identifies CYCD3 D-type cyclins as potential regulators of vascular development
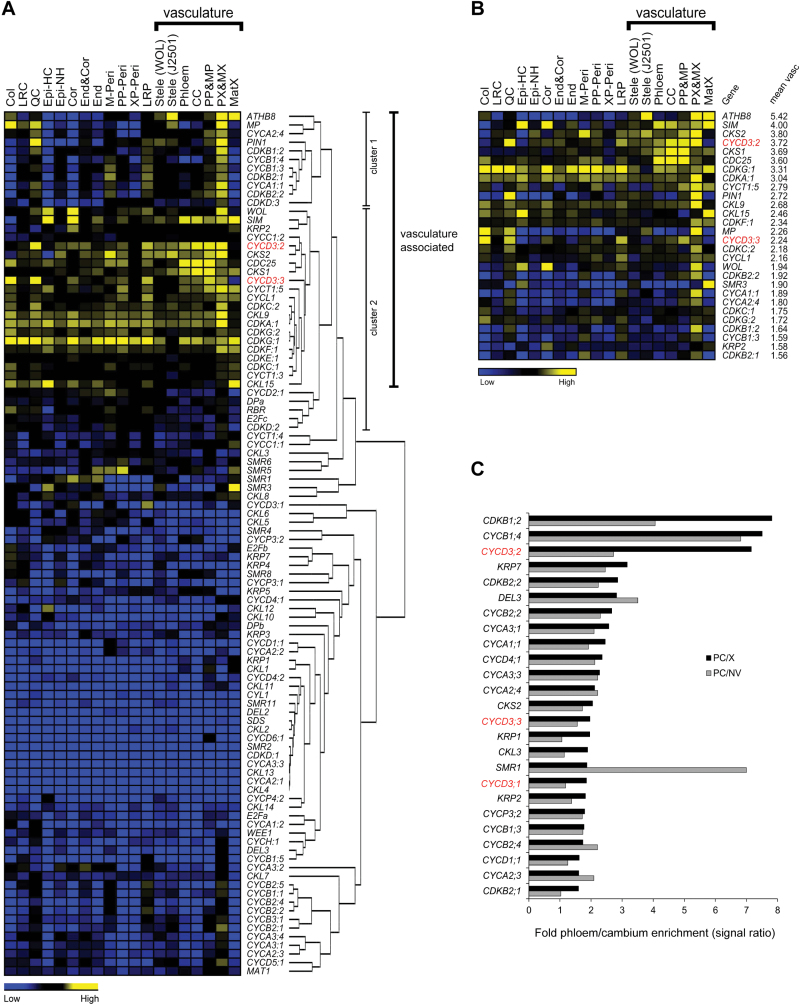
To explore the wider role of the cell cycle in vascular development and to identify candidate cell-cycle genes that promote cambial cell proliferation, the transcriptional regulation of all known core cell-cycle genes was examined using data from high resolution Arabidopsis cell- and tissue-specific microarray profiles derived from the root tip (Brady et al., 2007) and mature hypocotyl (Zhao et al., 2005). The combination of both datasets essentially provides broad coverage of cambium-associated expression during both primary and secondary vascular development. In Arabidopsis, 110 core cell-cycle genes have been recognized (Vandepoele et al., 2002; Menges et al., 2005; Van Leene et al., 2010) and, of these, 96 are represented by ATH1 array probes. Their transcription profiles weree first examined across 19 different cell and tissue types of the root tip (Brady et al., 2007), and included data on four classical markers of cambial activity (MP/ARF5, WOL, ATHB8, and PIN1; Hardtke and Berleth, 1998; Mähönen et al., 2000; Scarpella et al., 2004, 2006) in order to correlate cell-cycle gene transcriptional activity with vascular stem cell division. This showed different core cell-cycle regulators to have distinct expression patterns in the root, with the majority of genes showing low overall expression variation across all cell and tissue types and thus low cell- and tissue-specificity (see Supplementary Fig. S2 at JXB online). As expected, the four cambium markers showed enrichment in cells and tissues derived from the vasculature confirming that the root expression map (Brady et al., 2007) can serve as a reliable approximation of a vascular tissue-specific profile.
Cluster analysis of core cell-cycle and cambium marker gene expression profiles showed that genes exhibiting a relatively constant, low expression level across the root tip generally cluster together (comprising the major proportion of genes) and revealed several separate clusters comprising co-regulated genes sharing vascular tissue-associated activity (Fig. 1A). Outside the main cluster of genes with constant low expression, two clusters of regulated genes can be broadly defined; the first cluster contains genes clearly enriched in the vasculature, quiescent centre (QC), and lateral root primordia (LRP), and including ATHB8, MP, and PIN1, and a second cluster containing genes generally enriched in the vasculature but showing more variable expression peaks in other cell layers and including WOL (Fig. 1A). The second cluster also contained a small subset of genes which exhibited a more constant high level of expression across all cell layers, exemplified by CDKA;1 and CDKG;1 (Fig. 1A). Having a broadly shared pattern of specificity for the vasculature, cluster 1 and cluster 2 were designated, with the exception of CYCD2;1, DPa, RBR, E2Fc, and CDKD;2, as ‘vasculature-associated’ comprising 28 cell-cycle genes and four cambium marker genes. All 96 core cell-cycle genes and four cambium marker genes were ranked according to their average expression levels across all seven cell and tissue types representing the vasculature (Fig. 1B). The most highly vasculature-enriched genes included all four cambium markers, accompanied by representative gene members from almost all the main core cell cycle gene families (Vandepoele et al., 2002; Menges et al., 2005; Van Leene et al., 2010). Importantly, the vast majority of genes identified as ‘vasculature-associated’ (Fig. 1A), also had high overall expression in the vasculature (Fig. 1B), suggesting cell-cycle genes in this cluster have distinct roles in primary vascular development.
Fig. 1.
Expression patterns and clustering analysis of core cell-cycle genes in specific cell and tissue types of the root and mature hypocotyl. (A) Heat map representation of hierarchical clustering of 96 core cell-cycle genes and four cambium marker genes based on expression profile data across 19 different cell and tissue types of the root tip (Brady et al., 2007). The solid bar indicates the seven cell and tissue types comprising the vasculature. High and low levels of expression are indicated by yellow and blue, respectively. The expression levels shown are relative to zero based on global normalization method (Brady et al., 2007). Two distinct clusters of co-regulated genes showing broadly ‘vasculature-associated’ expression are indicated (cluster 1 and cluster 2). Cell and tissue types from left to right are columella (Col), lateral root cap (LRC), quiescent centre (QC), epidermis hair cell (Epi-HC), epidermis non-hair cell (Epi-NH), cortex (Cor), endodermis and cortex (End&Cor), endodermis (End), mature pericycle (M-Peri), phloem pole pericycle (PP-Peri), xylem pole pericycle (XP-Peri), lateral root primordia initials (LRP), stele using WOL marker [Stele (WOL)], stele using J2501 marker [Stele (J2501)], phloem and companion cell (Phloem), companion cell (CC), protophloem and metaphloem (PP&MP), protoxylem and metaxylem (PX&MX), mature xylem (MatX). (B) Ranking of 96 core cell-cycle genes and four cambium marker genes according to their average expression level across the seven cell and tissue types comprising the vasculature (indicated by the solid bar), based on expression profile data of the root tip (Brady et al., 2007). Only the top 28 highest vasculature-expressed genes are shown and are displayed with their corresponding expression patterns across the root. Abbreviations used are as shown in (A). (C) Ranking of core cell-cycle genes according to their degree of phloem/cambium (PC) enrichment calculated as fold-differences between mean array signal intensities in PC versus xylem (X) tissue (PC/X), and in PC versus non-vascular (NV) tissues (PC/NV), using microarray data from the mature hypocotyl (Zhao et al., 2005). Only the top 25 highest PC-enriched genes are shown.
The expression patterns of core cell-cycle genes were examined next using data from transcriptional profiling of the mature hypocotyl (Zhao et al., 2005). Calculations of fold-differences between mean array signal intensities in phloem/cambium (PC) versus xylem (X) tissue (PC/X), and in PC versus non-vascular (NV) tissues (PC/NV) (Zhao et al., 2005), revealed a group of 25 cell-cycle genes showing high specificity for phloem/cambium tissue suggesting distinct roles in secondary vascular development (Fig. 1C). A significant overlap was noted between the set of 25 PC-specific cell-cycle genes in the mature hypocotyl (Fig. 1C), and the set of 28 cell-cycle genes identified as ‘vasculature-associated’ in the root tip (Fig. 1A). Indeed, 11 cell-cycle genes were shared between both sets. This suggests that a common set of core cell-cycle genes are involved in both primary and secondary vascular development.
As we were primarily interested in identifying core cell-cycle genes that are involved in promoting cambial cell divisions, the root and hypocotyl microarray profiles (Fig. 1) were scrutinized for positive cell-cycle regulators having a clear vascular tissue-associated profile. Close inspection of the data shows that the CYCD3 class of D-type cyclins appear prominently among vasculature-enriched genes expressed during both primary and secondary vascular development (Fig. 1A–C; see Supplementary Fig. S2 at JXB online). The CYCD3 subgroup has already been shown to be an important positive cell-cycle regulator of cell division in developing leaves and embryos (Dewitte et al., 2007; Collins et al., 2012). Consistent with a proposed role for CYCD3 in vascular development, a partial expression analysis of CYCD3;1 using in situ hybridization revealed activity in the procambium of developing leaves and inflorescence stems (Dewitte et al., 2003). Strikingly, it was found that the CYCD3 genes also show peaks of activity in the QC and LRP, a pattern shared with known cambium regulators such as PIN1 and MP (Fig. 1A; Hardtke and Berleth, 1998; Scarpella et al., 2004, 2006), providing further confirmation of a close association between CYCD3 expression and vascular stem cell-associated activity. In light of these results, the decision was taken to focus our molecular investigations to elucidate the role of CYCD3 in vascular development.
CYCD3 genes are expressed in the cambium throughout development
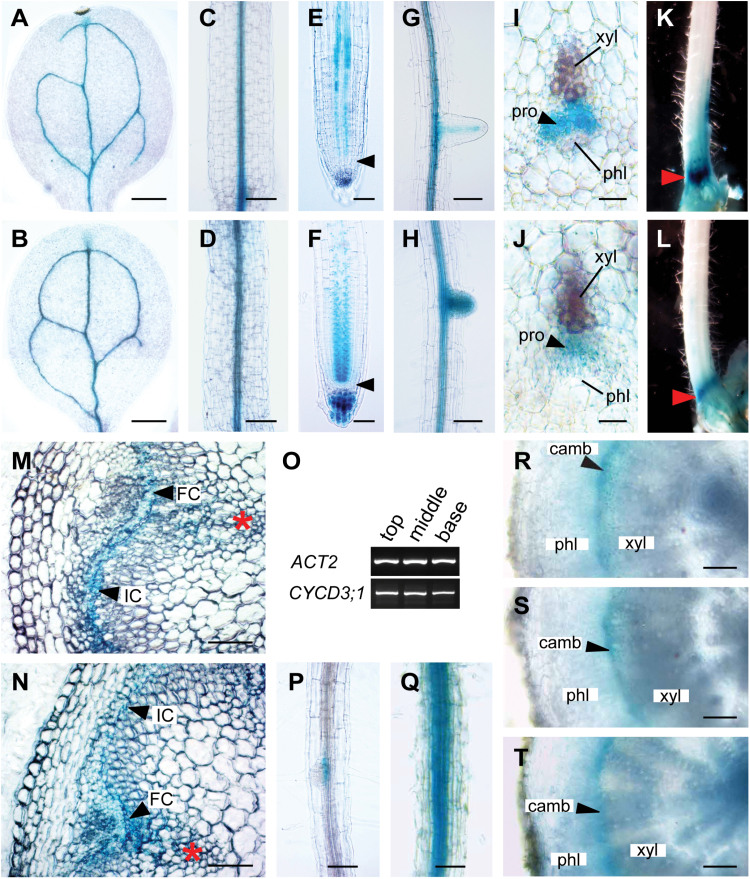
To investigate a functional role for CYCD3 in vascular development, an attempt was made to determine more precisely the in planta expression of CYCD3 genes using available GUS reporter transgenic plants. CYCD3;1 has already been shown to be expressed in the procambium of developing leaves and inflorescence stems (Dewitte et al., 2003). A preliminary expression analysis showed CYCD3;2 and CYCD3;3 to be expressed in seedling vascular tissue (Collins, 2008). However, this study did not achieve the spatial resolution required to determine whether these genes are specifically active in vascular stem cells, as found for CYCD3;1 (Dewitte et al., 2003). Therefore, we focused on determining the detailed tissue- and cell-specific expression of CYCD3;2 and CYCD3;3 during vascular tissue formation at different plant developmental stages using the GUS reporter lines pCYCD3;2:GUS and pCYCD3;3:GUS, respectively (Dewitte et al., 2007). Both genes shared a remarkable degree of overlap in their expression domains with activity closely associated with vascular tissue initiation from seedling to adult plant stages (Fig. 2). In 7-d-old seedlings, pCYCD3;2:GUS and pCYCD3;3:GUS showed strong activity in developing vasculature of cotyledons, leaves, hypocotyls, and roots (Fig. 2A–H). In roots, pCYCD3;2:GUS and pCYCD3;3:GUS expression marked the two procambial cell files, beginning first in the root apical meristem (RAM) with persistent activity in the elongation and maturation zones (Fig. 2E, F), and throughout the mature root zones and in emerging lateral roots (Fig. 2G, H). Examination of transverse sections from 5mm below the shoot apex of adult stems, corresponding to the zone where vascular development is initiated, showed pCYCD3;2:GUS and pCYCD3;3:GUS expression specifically in mitotically active procambial cells (Fig. 2I, J). In intact whole 30-cm-long stems, an accumulation of pCYCD3;2:GUS and pCYCD3;3:GUS staining was observed in a narrow region at the stem base immediately above the rosette leaves (Fig. 2K, L), corresponding to the zone of maximum interfascicular cambium (IC) initiation and secondary growth identified previously (Sehr et al., 2010). Transverse sections across this zone in both reporter lines confirmed GUS activity specifically in the fascicular cambium (FC) and IC (Fig. 2M, N).
Fig. 2.
CYCD3 genes are specifically expressed in the procambium and cambium during vascular development. (A–H) Images of 7-d-old seedlings showing pCYCD3;2:GUS (A, C, E, G) and pCYCD3;3:GUS (B, D, F, H) activity in the vasculature of the cotyledon (A, B), hypocotyl (C, D), root tip (E, F), and mature root zone and lateral root (G, H). In (E) and (F), the root apical meristem is indicated by arrowheads. (I, J) Transverse sections at 5mm below the shoot apex of 30-cm-long inflorescences showing pCYCD3;2:GUS (I) and pCYCD3;3:GUS (J) activity restricted to the procambium. (K, L) pCYCD3;2:GUS (K) and pCYCD3;3:GUS (L) staining accumulates at the base of 30-cm-long inflorescences (indicated by arrowheads) corresponding to the zone of maximum IC initiation and secondary growth. (M, N) Transverse sections at the base of 30-cm-long inflorescences immediately above the rosette reveal pCYCD3;2:GUS (M) and pCYCD3;3:GUS (N) activity restricted to fascicular cambium (FC) and interfascicular cambium (IC) (indicated by arrowheads). Asterisks indicate primary vascular bundles. (O) RT-PCR analysis reveal CYCD3;1 transcripts at the top, middle, and base of 30-cm-long inflorescences (ACT2 was used as endogenous control). (P, Q) pCYCD3;1:GUS expression was detected in root vascular tissue in 7-d-old seedlings (P), and in mature roots in 30-cm-tall plants (Q). (R-T) Transverse sections at the hypocotyl base from 30-cm-tall plants reveal pCYCD3;1:GUS (R), pCYCD3;2:GUS (S), and pCYCD3;3:GUS (T) expression restricted to the cambium. camb, cambium; FC, fascicular cambium; IC, interfascicular cambium; phl, phloem; pro, procambium; xyl, xylem. Scale bars=250 μm (A, B), 100 μm (C, D, G, H, M, N, P, Q, R, S, T), 50 μm (E, F), 25 μm (I, J).
Examination of the stem base from a GUS reporter line representing the CYCD3;1 gene, pCYCD3;1:GUS (Riou-Khamlichi et al., 1999), failed to show any staining (data not shown). However, consistent with previous expression studies of CYCD3;1 in the stem (Dewitte et al., 2003), CYCD3;1 transcripts were detected at the base (and top and middle) of mature stems using RT-PCR (Fig. 2O), suggesting that the pCYCD3;1:GUS line may not entirely recapitulate the true expression pattern for CYCD3;1 in the stem. Nevertheless, pCYCD3;1:GUS activity was observed in the root vasculature, which became progressively stronger as the roots reached mature stages, coinciding with the onset of root secondary growth (Fig. 2P, Q). Transverse sections of mature hypocotyls from 30-cm-tall plants revealed pCYCD3;1:GUS, pCYCD3;2:GUS, and pCYCD3;3:GUS expression specifically in the mitotically active cells of the vascular cambium, coinciding with the major phase of secondary growth in the hypocotyl (Fig. 2R–T). The expression patterns determined for all three CYCD3s using GUS reporters were in strong agreement with the results obtained from microarray profiling of the root tip and mature hypocotyl (Fig. 1). Together, the overlapping spatiotemporal patterns of CYCD3 transcriptional regulation observed confirm a close association of CYCD3 expression with active procambial/cambial cell proliferation throughout development, consistent with a distinct, subgroup-specific role for CYCD3 in vascular tissue formation.
Loss of CYCD3 causes reduced secondary growth and organ size
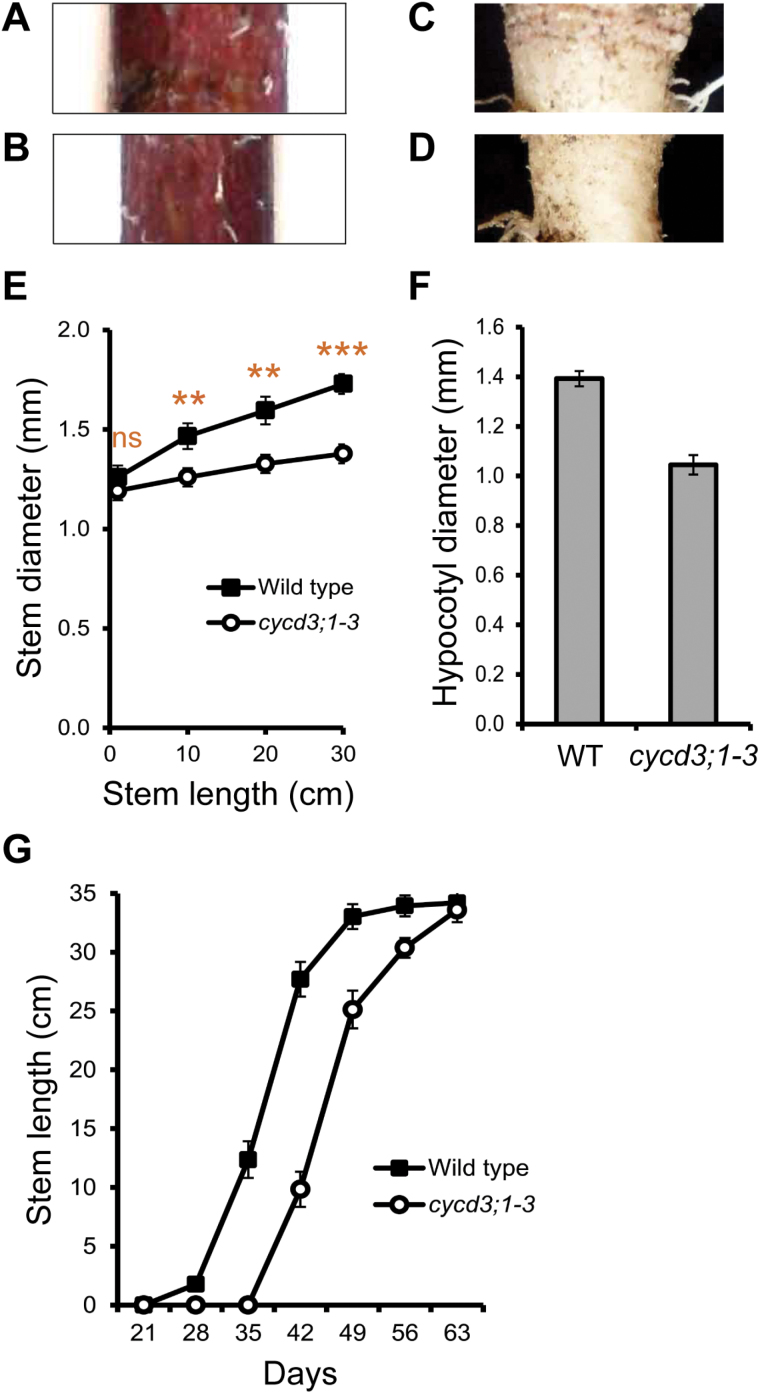
The highly overlapping patterns of expression observed for the three CYCD3 genes during vascular development suggested that the CYCD3-directed CYCD/RBR/E2F pathway (Dewitte et al., 2003; 2007) might play a specific role in regulating cambial cell proliferation. Therefore, the effect of reduced expression of CYCD3 on secondary growth was examined using a triple homozygous cycd3;1 cycd3;2 cycd3;3 (cycd3;1–3) loss-of-function mutant (Dewitte et al., 2007). Clear developmental defects were seen in cycd3;1–3 plants. At the base of adult stems and mature hypocotyls from 30-cm-tall plants, corresponding to major zones of prominent secondary growth, cycd3;1–3 was thinner compared with their wild-type equivalents (Fig. 3A–D). Radial growth at the stem base was monitored by measuring the diameter at different stages of stem elongation in wild-type and mutant plants. The cycd3;1–3 mutant showed a substantially reduced radial growth rate with an approximately 20% reduction (t test P <0.001) in thickening growth compared with the wild type at the stage where wild-type inflorescences had reached 30cm in length (Fig. 3E). At the base of mature hypocotyls, mutants showed an approximately 25% reduction (t test P <0.001) in thickening growth (Fig. 3F). The reduced stem and hypocotyl phenotypes in cycd3;1–3 closely resemble those reported in plants with mutations in known cambium regulators such as WOX4 and PXY/TDR (Fisher and Turner, 2007; Hirakawa et al., 2008, 2010; Ji et al., 2010), and can be considered diagnostic of plants with reduced cambium activity.
Fig. 3.
Loss of CYCD3 causes retarded secondary growth and reduced organ size. (A, B) Representative examples of side views of the base of whole 30-cm-long inflorescences immediately above the rosette in wild-type (A) and cycd3;1–3 (B) plants showing a thin-stemmed phenotype in cycd3;1–3. (C, D) Representative examples of side views of the base of whole mature hypocotyls from 30-cm-tall wild-type (C) and cycd3;1–3 (D) plants showing a thin hypocotyl phenotype in cycd3;1–3. (E) Quantitative analysis of stem diameter at the base of 1, 10, 20, and 30-cm-long inflorescences of wild-type and cycd3;1–3 plants showing the reduced growth in cycd3;1–3. Error bars represent standard error. Significance levels are shown as the difference between wild type and cycd3;1–3 with P <0.01 and P <0.001 (n=10) indicated by double and triple asterisks, respectively (not statistically significant is indicated by ns). (F) Measurement of hypocotyl diameter in 30-cm-tall wild-type and cycd3;1–3 plants showing the reduced growth in cycd3;1–3. Error bars represent standard error. Student’s t test showed that the difference between wild type and cycd3;1–3 was extremely statistically significant (P <0.001; n=10). (G) Quantitative analysis of stem length in wild-type and cycd3;1–3 plants at 21, 28, 35, 42, 49, 56, and 63 d (days relative to date of sowing) showing equivalent rates of stem elongation in wild type and cycd3;1–3. Student’s t test showed that wild type and cycd3;1–3 were not significantly different in stem length at 63 d (n=10). Error bars represent standard error.
Measurements of apical growth of the stem showed that, despite the emergence of the inflorescence being delayed in cycd3;1–3 (see Supplementary Fig. S3 at JXB online), as previously reported (Dewitte et al., 2007), the rate of stem elongation was comparable between wild type and mutant, with cycd3;1–3 stems reaching the length of their wild-type equivalents toward the end of the growth period (Fig. 3G). These results show clearly that radial growth was more compromised than apical growth, suggesting that a decrease in the activity of the cambium was the major cause of the reduced thickening growth in the stem. Interestingly, delayed emergence and other perturbations in apical growth of the stem are characteristic features of wox4 and pxy mutants (Fisher and Turner, 2007; Ji et al., 2010). In summary, our data indicate an effect of loss of CYCD3 expression on cambium activity, leading to retarded development in the stem and hypocotyl, and reduced organ size.
CYCD3 promotes cambial cell proliferation in the inflorescence stem
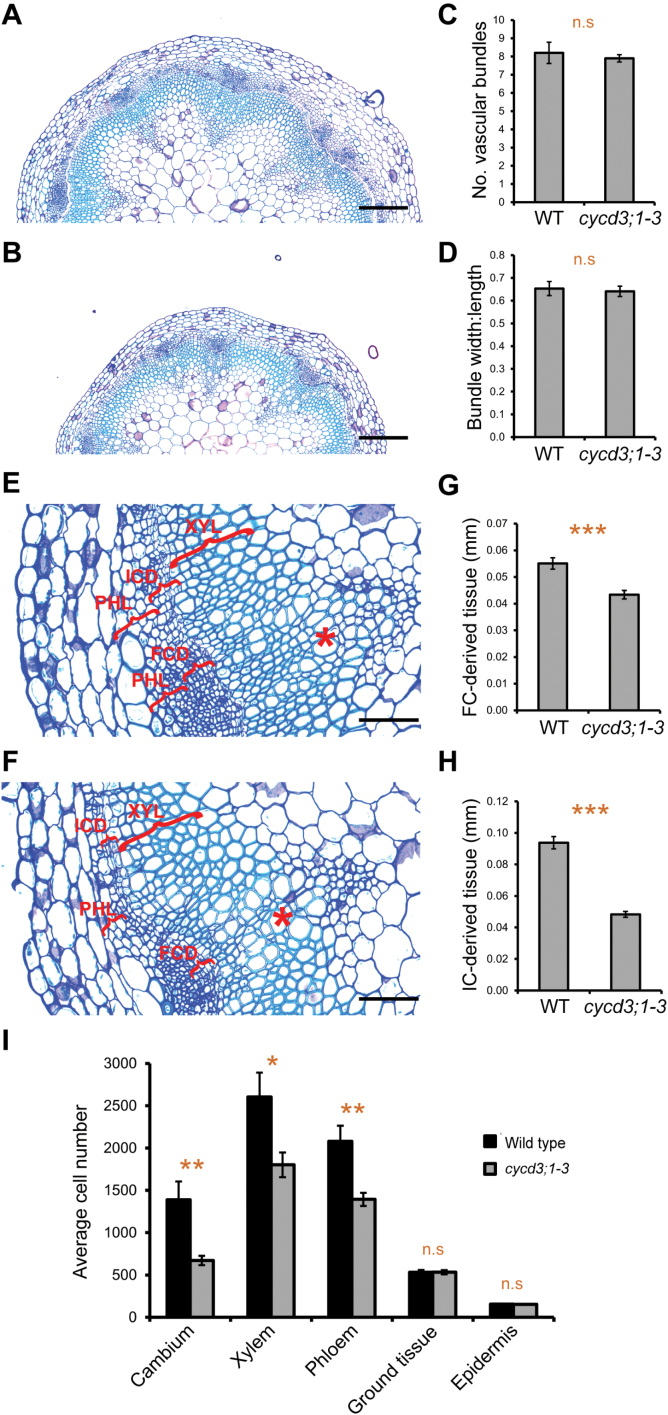
To determine more precisely the effect of reduced CYCD3 activity on cambium-related cell division and differentiation processes, the vascular anatomy of the stem base of 30-cm-tall cycd3;1–3 plants was analysed. At the base of adult stems, the vascular tissue is composed of radially oriented, closed cylinders of cambium (FC and IC) with an inner xylem and outer phloem which are, in turn, enclosed by a ground tissue (cortex and endodermis) and epidermis layers (Sehr et al., 2010; Sanchez et al., 2012). Different tissue layers are easily distinguished by the characteristic shapes and spatial division planes of the cells comprising each tissue. In terms of overall tissue patterning, the cycd3;1–3 mutant was indistinguishable from the wild type with a normal arrangement of vascular and non-vascular tissue (Fig. 4A, B), and there was no significant difference in the average number of vascular bundles between wild-type and mutant stems (Fig. 4C). Furthermore, calculations of the width:length (tangential:radial) ratio of vascular bundles, a measure of vascular organization (Fisher and Turner, 2007), showed no significant difference between the wild type and cycd3;1–3 (Fig. 4D). These results suggest that tissue patterning is not greatly affected upon loss of cycd3;1–3.
Fig. 4.
Loss of CYCD3 leads to reduced cambial activity and vascular cell number in the inflorescence stem. (A, B) Histological representations at the base of 30-cm-long inflorescences immediately above the rosette showing tissue organization in wild-type (A) and cycd3;1–3 (B) plants. (C) Measurement of vascular bundle number at the base of wild-type and cycd3;1–3 stems. Error bars represent standard error. Student’s t test showed that wild type and cycd3;1–3 were not significantly different (n=5). n.s, not statistically significant. (D) Measurement of the width:length (tangential:radial) ratio of vascular bundles at the base of wild-type and cycd3;1–3 stems. Error bars represent standard error. Student’s t test showed that wild type and cycd3;1–3 were not significantly different (n=15). n.s, not statistically significant. (E, F) Higher magnification histological representations of vascular tissue at the base of wild-type (E) and cycd3;1–3 (F) stems. The lateral extension of the FC-derived (FCD) and IC-derived (ICD) tissue, and of the phloem (PHL) and xylem (XYL) tissues are indicated by red brackets. Asterisks indicate primary vascular bundles. (G, H) Measurement of lateral extension of the FCD (G) and ICD (H) tissues at the base of wild-type and cycd3;1–3 stems. Error bars represent standard error. Student’s t tests showed that the differences between wild type and cycd3;1–3 in both the FCD (G) and ICD (H) tissues were extremely statistically significant (P <0.001; n=15) indicated by triple asterisks. (I) Quantification of average cell number of different cell types in tissue sections at the base of wild-type and cycd3;1–3 stems. Error bars represent standard error. Significance levels are shown as the difference between wild type and cycd3;1–3 with P <0.05 and P <0.01 indicated by single and double asterisks, respectively (n=15). n.s, not statistically significant. Scale bars=200 μm (A, B), 100 μm (E, F).
The FC and its immediate population of daughter cells have been defined as FC-derived (FCD) tissue, whereas the IC and its immediate population of daughter cells were defined as IC-derived (ICD) tissue and both can be used as a measure of cambium activity (Sehr et al., 2010). The effect of loss of CYCD3 function on cambium activity in mature cycd3;1–3 stems was analysed. Microscopic analysis of FCD and ICD in wild-type stems identified the small, uniform, flattened, and undifferentiated cells of the FC and IC organized into radial cell files (resulting from continuous periclinal cell divisions), surrounded by more rounded and less uniformly shaped, differentiating xylem, and phloem and associated parenchyma cells (not organized into radial cell files) (Fig. 4E). Observations in the cycd3;1–3 mutant showed that the equivalent cell types were present and were phenotypically indistinguishable from the wild type (Fig. 4E, F), indicating that CYCD3 is not essential for the establishment and identity of the cambium. By contrast, quantification of the lateral extension of the cambium-derived tissue in the stem base showed cycd3;1–3 to have an approximately 21% reduction (t test P <0.001) in FCD, and an approximately 48% reduction (t test P <0.001) in ICD, compared with the wild type (Fig. 4G, H), indicating that loss of CYCD3 causes a substantial decrease in the activity of the cambium.
The reduced cambium activity observed in cycd3;1–3 suggested that both cambial cell number and the number of daughter cells in the xylem and phloem lineages could be affected. Quantification of cell numbers in the cambium, xylem, and phloem of cycd3;1–3 and wild-type stems (see Supplementary Fig. S4 at JXB online; Fig. 4E, F) showed that the number of cells comprising the cambium in cycd3;1–3 was reduced by approximately 52% (t test P <0.01) compared with the wild type, with the mutant also showing respective reductions in xylem and phloem cell number of approximately 31% (t test P <0.05) and 33% (t test P <0.01) (Fig. 4I), confirming that loss of CYCD3 causes a marked decrease in cambium activity, limiting cell number in the xylem and phloem cell lineages. Cell numbers were also quantified in the surrounding ground tissue (cortex plus endodermis) and epidermis (see Supplementary Fig. S4 at JXB online) tissue layers which are established during primary growth from the apical meristem. The ground tissue and epidermis layers were easily identified in cross-sections by their characteristic large cells, with a single cell layer of endodermis enclosed by a 4–5 cell thick cortex, in turn, enclosed by a single cell-layered epidermis (see Supplementary Fig. S4 at JXB online). No significant difference was found in the number of cells comprising both the ground tissue and epidermis between the wild type and cycd3;1–3 (Fig. 4I), supporting the proposition that decreased cambium activity, as opposed to a defective apical meristem, was the major cause of the reduced vascular cell number and overall radial organ size in cycd3;1–3 stems.
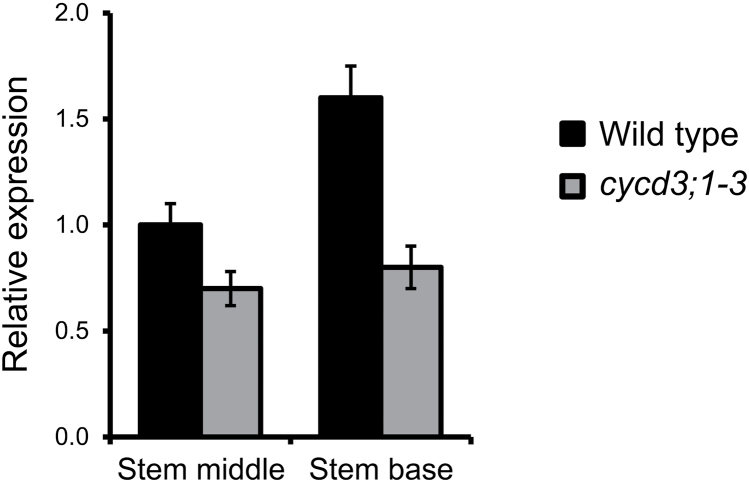
Importantly, the vascular cell populations which exhibited the phenotypic effects upon loss of CYCD3 overlapped the domains in which CYCD3 genes were shown to be expressed (Fig. 2). To assess the status of mitotic cell cycle activity in developing vascular tissue in cycd3;1–3 stems, reverse transcription quantitative PCR (RT-qPCR) was performed to monitor the expression of the mitotic B-type cyclin gene CYCB1;1, a commonly-used marker of mitotic cell cycle activity during plant development (Coloń-Carmona et al., 1999). Consistent with reduced cambial cell division in cycd3;1–3 stems, mutants showed reduced CYCB1;1 expression compared with the wild type in the middle and base of stems, an effect that was more pronounced in the stem base (Fig. 5), confirming that reduced cambial cell number in cycd3;1–3 stems is associated with reduced mitotic cell cycle activity. Taken together, our results demonstrate that CYCD3 plays a significant positive role in the regulation of cambium activity in the stem and that its loss leads to retarded secondary growth and reduced organ size.
Fig. 5.
Expression analysis of the CYCB1;1 mitotic marker gene in the inflorescence stem. Reverse transcription quantitative PCR was used to assay CYCB1;1 transcript abundance in the middle and base of 30-cm-long stems of wild-type and cycd3;1–3 plants using ACTIN2 as the endogenous reference gene. Relative transcript abundance is scaled to expression in the middle stem of the wild type. Error bars represent standard error.
CYCD3 restrains cell expansion and differentiation of cambial cells into xylem cells
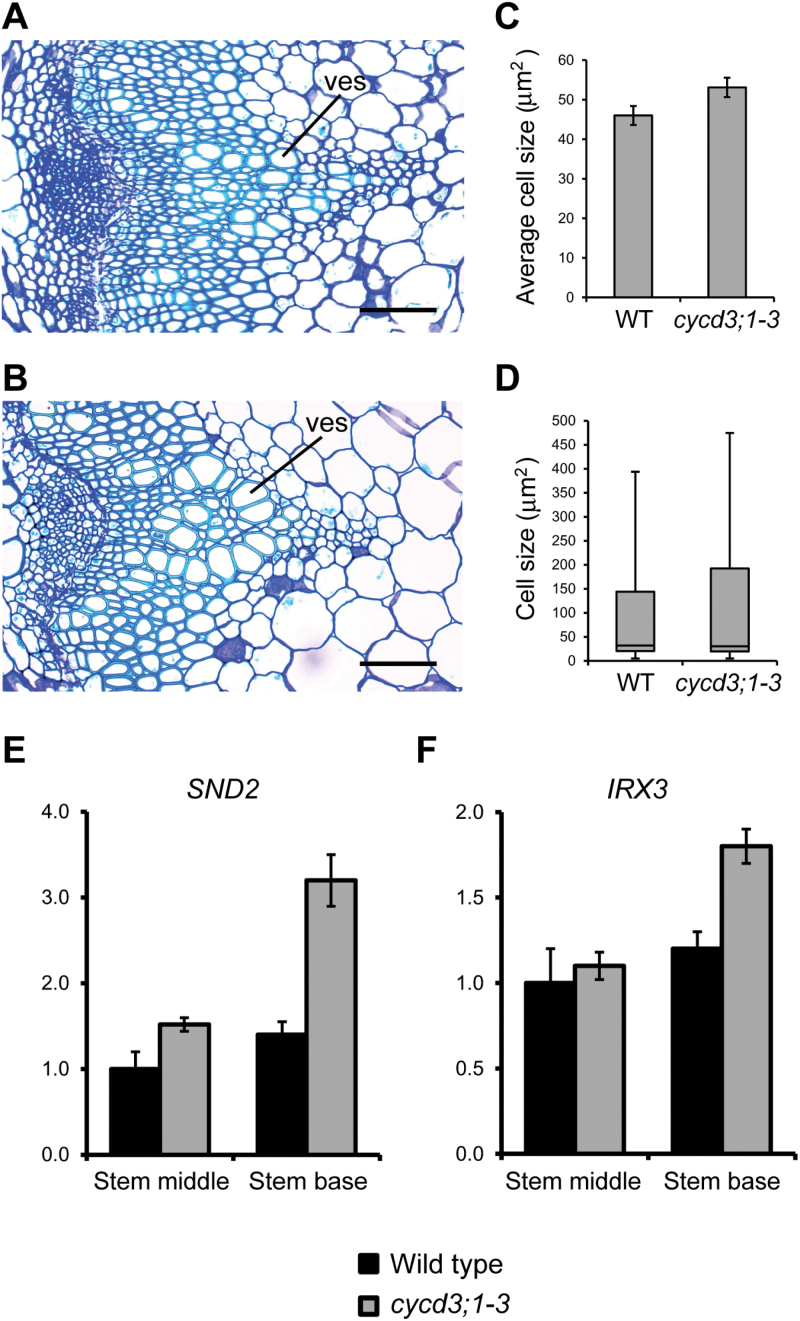
An additional phenotype that was noted at the stem base of cycd3;1–3 mutants was the presence of enlarged xylem cells, particularly those of vessels (Fig. 6A, B). Arabidopsis CYCD3 genes have been shown previously to mediate the balance between cell division and cell expansion during leaf development by regulating the duration of the ‘mitotic window’ and the onset of cell expansion and associated endoreduplication events (Dewitte et al., 2007). To determine the overall effects of loss of CYCD3 function on cell expansion in the xylem cell lineage, xylem cell sizes in the stem base of 30-cm-tall cycd3;1–3 and wild-type plants was quantified. Overall, it was found that average xylem cell size in cycd3;1–3 was increased by approximately 13% (t test P <0.05) compared with the wild type (Fig. 6C). Consistent with an overall shift to increased xylem cell size, the mutant showed a greater range of cell sizes with a significant number of cells reaching sizes not seen in the wild type (Fig. 6D). These results indicate that loss of CYCD3 causes a significant enhancement in cell growth in developing xylem cells. The apparent enlargement of xylem cells observed in cycd3;1–3 stems suggested that the cellular transition from division to differentiation pathways in the xylem could be affected. To evaluate further the effects of loss of CYCD3 on the progression of vascular differentiation in stems, RT-qPCR was used to determine the expression of two well-characterized markers of xylem cell differentiation, SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN2 (SND2) and IRREGULAR XYLEM3 (IRX3) which, respectively, encode a transcription factor and a cellulose synthase gene that are tightly associated with xylem secondary cell wall biosynthesis (Brown et al., 2005; Zhong et al., 2008). Consistent with an overall shift toward differentiation in the developing xylem of cycd3;1–3 stems, mutants showed an up-regulation of SND2 and IRX3 expression compared with the wild type of approximately 2.3-fold (130%) and 1.5-fold (50%), respectively, an effect that was more pronounced in the stem base (Fig. 6E, F).
Fig. 6.
Loss of CYCD3 leads to increased xylem cell size and differentiation in the inflorescence stem. (A, B) High magnification histological representations at the base of 30-cm-long inflorescences showing xylem cells in primary and secondary vascular tissue in wild-type (A) and cycd3;1–3 (B) plants. (C) Measurement of xylem cell size at the base of 30-cm-tall wild-type and cycd3;1–3 plants showing the increased average cell size in cycd3;1–3. Error bars represent standard error. Student’s t test showed that the difference between the wild type and cycd3;1–3 was statistically significant (P <0.05; n >2000). (D) Box plot representation of xylem cell size at the base of 30-cm-tall plants showing the distribution of xylem cell sizes in wild-type and cycd3;1–3 stems. Displayed are the maximum and minimum values, median, and 30th and 95th percentiles. Error bars represent standard error. (E, F) Expression analysis of the xylem cell differentiation marker genes SND2 (E) and IRX3 (F) in the middle and base of 30-cm-long stems of wild-type and cycd3;1–3 plants by RT-qPCR. Transcript abundance was normalized using ACTIN2 as the endogenous reference gene. Relative transcript abundance is scaled to expression in the middle stem of the wild type. Error bars represent standard error. ves, xylem vessels. Scale bars=100 μm (A, B).
To exclude the possibility that the increased IRX3 and SND2 expression observed in the mutant was the result of an increase in the proportion of xylem cells (relative to the total of all other cell types) in the stem, the proportional contribution of xylem cells was calculated using data derived from stem cross-sections presented in Fig. 4I. The wild type had an average xylem cell number of 2605 and an average total number of all cell types of 6757.2, whereas the mutant had respective numbers of 1801.6 and 4553.2. Calculation of the proportion of xylem cells in stems showed both lines to be almost identical, with the wild type having 39% xylem (2605/6757.2) and the mutant having 40% xylem (1801.6/4553.2), ruling out an increase in the proportion of xylem cells as the cause of increased SND2 and IRX3 expression.
Importantly, the CYCD3 genes were shown to be specifically expressed in the cambium during stem vascular development and absent from older xylem cells (Fig. 2M, N), suggesting that a key point of regulation of cell expansion/differentiation processes by CYCD3 may occur in xylem cell precursors and their immediate progeny cells. Together, our data suggest that CYCD3 plays an important additional role in stem vascular development, in restraining cell expansion and differentiation in developing xylem cells.
Discussion
In contrast to our relatively good understanding of the regulation of primary meristems, remarkably little is known concerning the molecular mechanisms controlling cell division in the cambial meristem and its integration with vascular differentiation. In this work, the wider role of the cell cycle in vascular development was investigated and the Arabidopsis CYCD3 subgroup of cell-cycle genes were identified as being key regulators of cambial cell proliferation and secondary growth. The CYCD3 genes were shown previously to control the balance between cell division and cell expansion during leaf and petal development (Dewitte et al., 2007), and CYCD3 is also required for normal cell division and developmental rate during embryogenesis (Collins et al., 2012). In a mutant lacking all three CYCD3 genes (cycd3;1–3), it was found that radial organ size was severely affected with mutant stems and hypocotyls showing a marked reduction in diameter linked to reduced cell division in the cambium, leading to decreased xylem and phloem cell numbers. Conversely, it was found that the reduced xylem cell number was accompanied by an increase in xylem cell size and differentiation. Interestingly, perturbation of CYCD3 levels in transgenic plants through the constitutive over-expression of CYCD3;1 was shown to retard secondary xylem formation resulting from an inhibition of cell differentiation associated with reduced radial growth (Dewitte et al., 2003). This apparent reciprocal phenotype to the CYCD3 triple loss-of-function mutant phenotype analysed in this study, which exhibits enhanced xylem cell differentiation, indicates that correct levels of CYCD3 activity is required for normal xylem development and radial organ growth. Importantly, all three CYCD3 genes showed activity in the procambium and cambium, linking CYCD3 expression to the stem cell populations that drive both primary and secondary vascular development. Our data show that developmental stage- and cell-type-specific regulation of CYCD3 is required to promote mitotic cell-cycle activity in the cambium and that its down-regulation is required for the progression of cell expansion and differentiation of vascular stem cells into xylem cells, providing the first conclusive evidence of a direct link between the cell cycle and vascular development in controlling plant organ growth.
The vascular development phenotypes observed in cycd3;1–3 closely resemble those reported in plants where the expression of known cambium regulators were manipulated, including wox4 and pxy/tdr (Fisher and Turner, 2007; Hirakawa et al., 2008, 2010; Ji et al., 2010; Suer et al., 2011), hca2 (Guo et al., 2009), rul1 (Agusti et al., 2011), or wox14 (Etchells et al., 2013). In these mutants, insufficient cambial cell divisions contributed to the inhibited thickening growth in different organs. Significantly, no effects on the orientation of cell division or overall vascular organization were observed in the cycd3;1–3 mutant, indicating a distinct role for CYCD3 in processes that specifically control the rate of cambial cell division. In this respect, the cycd3;1–3 mutant phenotype most closely resembles those seen in mutants of WOX4 and WOX14, which have already been shown to regulate stem cell proliferation independently of processes that control vascular organization (Etchells et al., 2013).
A number of developmental, hormonal, and environmental signals are known to regulate procambium/cambium activity (Miyashima et al., 2013), which presumably must ultimately impinge on cell-cycle regulation. The phytohormones auxin and cytokinin play particularly important roles during both primary and secondary growth by controlling cambial cell proliferation (Little et al., 2002; Scarpella et al., 2006; Matsumoto-Kitano et al., 2008). Several lines of experimental evidence support a role for CYCD3 in the integration of multiple input signals to control procambium/cambium activity. CYCD3;1 responds positively to auxin and cytokinin in line with its proposed role as a regulator of the G1/S phase transition in response to external signals (Riou-Khamlichi et al., 1999; Oakenfull et al., 2002; Dewitte et al., 2007). Consistent with a role downstream of hormone signalling, the expression patterns revealed for CYCD3s in the procambium and cambium of the stem (Fig. 2) overlap domains of activity of the DR5 reporter, a commonly used marker of localized auxin accumulation (Ibañes et al., 2009). Recent work has shown CYCD3;3 as a direct target of DNA-binding with one finger 3.4/OBF-binding protein 1 (DOF3.4/OBP1), a member of the DOF group of plant-specific transcription factors (DOF TFs) proposed to have central roles in vascular development (Skirycz et al., 2008; Le Hir and Bellini, 2013). DOF3.4/OBP1 was found directly to bind and regulate the CYCD3;3 promoter in vitro and, like all three CYCD3s, was shown to be expressed in the procambium of the stem (Skirycz et al., 2008). Another DOF TF, DOF5.6/HCA2, was shown to promote cell proliferation in the interfascicular cambium (Guo et al., 2009). Intriguingly, a number of DOF TFs including DOF3.4/OBP1 are known to be regulated by auxin (Kang and Singh, 2000). Thus, CYCD3 could provide a link between hormone signalling and cell-cycle activation in cambial cells through distinct DOF TFs and CYCD3 genes.
The additional phenotype observed in the cycd3;1–3 mutant of increased xylem cell expansion was associated with the up-regulation of xylem secondary cell wall biosynthetic genes, suggesting that CYCD3 is required to restrain cell expansion/differentiation in the xylem cell lineage. As expression of CYCD3 was observed specifically in the cambium during vascular development (Fig. 2), the most likely scenario is that CYCD3 exerts control over cell expansion/differentiation in xylem cell precursors and their immediate progeny cells and that subsequent CYCD3 down-regulation in cells displaced from the cambium is required for continued cell expansion and differentiation in xylem cells. This is supported by observations of xylem development in plants over-expressing CYCD3;1, where elevated CYCD3;1 levels lead to inhibited differentiation and retarded xylem formation (Dewitte et al., 2003). The proposed role for CYCD3 in restraining cell expansion/differentiation of vascular cells appears to parallel its role in restraining cell expansion/differentiation during leaf and petal development (Dewitte et al., 2007). In leaves, CYCD3 acts to inhibit endocycles, and its loss results in unrestrained endocycles, associated with enhanced cell expansion and differentiation (Dewitte et al., 2007). It is notable that xylem cells in different plant species are known to have both endocycles and substantial cell growth, to enable them to perform their specialized functions (Martίnez-Pérez et al., 2001; Prieto et al., 2004). Further work will be required to elucidate the precise mode of action of CYCD3 in restraining cell expansion and differentiation in the xylem cell lineage, and whether CYCD3 interacts with the endocycle in this regulation, as it does in leaves (Dewitte et al., 2007).
It is concluded that CYCD3 is an important regulator of cambial cell division, determining the number of proliferating cells and their progeny in the xylem and phloem, and the size of xylem cells during vascular development. Tight control of the balance between cell division and cell differentiation/expansion mediated by CYCD3 is critical for proper vascular development and secondary growth, which ultimately determines radial organ size.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Confirmation of the genotype of the cycd3;1–3 line by RT-PCR.
Supplementary Fig. S2. Radial distribution of expression levels of 96 core cell-cycle genes and four cambium marker genes based on expression profile data across 19 different cell and tissue types of the root tip (Brady et al., 2007).
Supplementary Fig. S3. Illustration of delayed flowering in the cycd3;1–3 line compared with the wild type.
Supplementary Fig. S4. Example of cell counting using transverse sections at the base of 30-cm-long inflorescences immediately above the rosette.
Acknowledgements
The authors would like to thank Todd Bass of the Veterinary Diagnostic Laboratories (Colorado State University, USA) for excellent histological support, Jim Murray (Cardiff, UK) for kindly providing the cycd3;1–3, pCYCD3;1:GUS, pCYCD3;2:GUS, and pCYCD3;3:GUS lines, and Thomas Greb (Vienna, Austria) for helpful comments. This work was supported by funds from University of Greenwich (UK) RAE and Colorado State University (USA), Colorado Agricultural Experiment Station.
References
- Agusti J, Lichtenberger R, Schwarz M, Nehlin L, Greb T. 2011. Characterization of transcriptome remodeling during cambium formation identifies MOL1 and RUL1 as opposing regulators of secondary growth. PLoS Genetics 7, e1001312. doi:10.1371/journal.pgen.1001312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. 2007. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806. [DOI] [PubMed] [Google Scholar]
- Brown DM, Zeef LAH, Ellis J, Goodacre R, Turner SR. 2005. Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. The Plant Cell 17, 2281–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C. 2008. CYCD gene function in Arabidopsis seed development . PhD thesis, University of Cambridge. [Google Scholar]
- Collins C, Dewitte W, Murray JAH. 2012. D-type cyclins control cell division and developmental rate during Arabidopsis seed development. Journal of Experimental Botany 63, 3571–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coloń-Carmona A, You R, Haimovitch-Gal T, Doerner P. 1999. Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. The Plant Journal 20, 503–508. [DOI] [PubMed] [Google Scholar]
- De Hoon MJL, Imoto S, Nolan J, Miyano S. 2004. Open source clustering software. Bioinformatics 20, 1453–1454. [DOI] [PubMed] [Google Scholar]
- Dewitte W, Riou-Khamlichi C, Scofield S, Healy JM, Jacqmard A, Kilby NJ, Murray JA. 2003. Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. The Plant Cell 15, 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W, Scofield S, Alcasabas AA, et al. 2007. Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proceedings of the National Academy of Sciences, USA 104, 14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. 1998. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences, USA 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchells JP, Provost CM, Mishra L, Turner SR. 2013. WOX4 and WOX14 act downstream of the PXY receptor kinase to regulate plant vascular proliferation independently of any role in vascular organisation. Development 140, 2224–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchells JP, Turner SR. 2010. The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development 137, 767–774. [DOI] [PubMed] [Google Scholar]
- Fisher K, Turner S. 2007. PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Current Biology 17, 1061–1066. [DOI] [PubMed] [Google Scholar]
- Gonzalez N, Vanhaeren H, Inzé D. 2012. Leaf size control: complex coordination of cell division and expansion. Trends in Plant Science 17, 332–340. [DOI] [PubMed] [Google Scholar]
- Guo Y, Qin G, Gu H, Qu L-J. 2009. Dof5.6/HCA2, a Dof transcription factor gene, regulates interfascicular cambium formation and vascular tissue development in Arabidopsis . The Plant Cell 21, 3518–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T. 1998. The Arabidopsis gene MONOPTERUS encodes a transcription factor mediating embryo axis formation and vascular development. The EMBO Journal 17, 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y, Kondo Y, Fukuda H. 2010. TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis . The Plant Cell 22, 2618–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y, Shinohara H, Kondo Y, Inoue A, Nakanomyo I, Ogawa M, Sawa S, Ohashi-Ito K, Matsubayashi Y, Fukuda H. 2008. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proceedings of the National Academy of Sciences, USA 105, 15208–15213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibañes M, Fábregas N, Chory J, Caño-Delgado AI. 2009. Brassinosteroid signalling and auxin transport are required to establish the periodic pattern of Arabidopsis shoot vascular bundles. Proceedings of the National Academy of Sciences, USA 106, 13630–13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J, Strable J, Shimizu R, Koenig D, Sinha N, Scanlon MJ. 2010. WOX4 promotes procambial development. Plant Physiology 152, 1346–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H-G, Singh KB. 2000. Characterization of salicylic acid-responsive, Arabidopsis Dof domain proteins: overexpression of OBP3 leads to growth defects. The Plant Journal 21, 329–339. [DOI] [PubMed] [Google Scholar]
- Le Hir R, Bellini C. 2013. The plant-specific Dof transcription factors family: new players involved in vascular system development and functioning in Arabidopsis . Frontiers in Plant Science 4, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little CHA, MacDonald JE, Olsson O. 2002. Involvement of indole-3-acetic acid in fascicular and interfascicular cambial growth and interfascicular extraxylary fiber differentiation in Arabidopsis thaliana inflorescence stems. International Journal of Plant Sciences 163, 519–529. [Google Scholar]
- Mähönen AP, Bonke M, Kauppinen L, Riikonen M, Benfey PN, Helariutta Y. 2000. A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes and Development 14, 2938–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto-Kitano M, Kusumoto T, Tarkowski P, Kinoshita-Tsujimura K, Václavíková K, Miyawaki K, Kakimoto T. 2008. Cytokinins are central regulators of cambial activity. Proceedings of the National Academy of Sciences, USA 105, 20027–20031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martίnez-Pérez E, Shaw P, Moore G. 2001. The Ph1 locus is needed to ensure specific somatic and meiotic centromere association. Nature 411, 204–207. [DOI] [PubMed] [Google Scholar]
- Menges M, De Jager SM, Gruissem W, Murray JAH. 2005. Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. The Plant Journal 41, 546–566. [DOI] [PubMed] [Google Scholar]
- Menges M, Pavesi G, Morandini P, Bögre L, Murray JAH. 2007. Genomic organization and evolutionary conservation of plant D-type cyclins. Plant Physiology 145, 1558–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashima S, Sebastian J, Lee JY, Helariutta Y. 2013. Stem cell function during plant vascular development. The EMBO Journal 32, 178–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakenfull EA, Riou-Khamlichi C, Murray JAH. 2002. Plant D-type cyclins and the control of G1 progression. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 357, 749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research 29, 2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto P, Santos AP, Moore G, Shaw P. 2004. Chromosomes associate premeiotically and in xylem vessel cells via their telomeres and centromeres in diploid rice (Oryza sativa). Chromosoma 112, 300–307. [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Huntley R, Jacqmard A, Murray JAH. 1999. Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283, 1541–1544. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. 2004. Java Treeview: extensible visualization of microarray data. Bioinformatics 20, 3246–3248. [DOI] [PubMed] [Google Scholar]
- Sanchez P, Nehlin L, Greb T. 2012. From thin to thick: major transitions during stem development. Trends in Plant Science 17, 113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella E, Francis P, Berleth T. 2004. Stage-specific markers define early steps of procambium development in Arabidopsis leaves and correlate termination of vein formation with mesophyll differentiation. Development 131, 3445–3455. [DOI] [PubMed] [Google Scholar]
- Scarpella E, Marcos D, Friml J, Berleth T. 2006. Control of leaf vascular patterning by polar auxin transport. Genes and Development 20, 1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz M, Smith R, Ellis B. 2013. Xylem tissue specification, patterning, and differentiation mechanisms. Journal of Experimental Botany 64, 11–31. [DOI] [PubMed] [Google Scholar]
- Sehr EM, Agusti J, Lehner R, Farmer EE, Schwarz M, Greb T. 2010. Analysis of secondary growth in the Arabidopsis shoot reveals a positive role of jasmonate signalling in cambium formation. The Plant Journal 63, 811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A, Radziejwoski A, Busch W, et al. 2008. The DOF transcription factor OBP1 is involved in cell cycle regulation in Arabidopsis thaliana . The Plant Journal 56, 779–792. [DOI] [PubMed] [Google Scholar]
- Suer S, Agusti J, Sanchez P, Schwarz M, Greb T. 2011. WOX4 imparts auxin responsiveness to cambium cells in Arabidopsis . The Plant Cell 23, 3247–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Raes J, De Veylder L, Rouzé P, Rombauts S, Inzé D. 2002. Genome-wide analysis of core cell-cycle genes in Arabidopsis . The Plant Cell 14, 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J, Hollunder J, Eeckhout D, et al. 2010. Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana . Molecular Systems Biology 6, 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Craig JC, Petzold HE, Dickerman AW, Beers EP. 2005. The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiology 138, 803–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, Zhou J, McCarthy RL, Ye Z-H. 2008. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis . The Plant Cell 20, 2763–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.