Abstract
Sex-biased genes are considered to account for most of phenotypic differences between males and females. In order to explore the sex-biased gene expression in crab, we performed the whole-body transcriptome analysis in male and female juveniles of the Chinese mitten crab Eriocheir sinensis using next-generation sequencing technology. Of the 23,349 annotated unigenes, 148 were identified as sex-related genes. A total of 29 candidate genes involved in primary sex determination pathways were detected, indicating the sex determination cascade of the mitten crab might be more complex than previously supposed. Differential expression analysis showed 448 differentially expressed genes (DEGs) between the two transcriptomes. Most of DEGs were involved in processes such as metabolism and immunity, and not associated with obvious sexual function. The pathway predominantly enriched for DEGs were related to lysosome, which might reflect the differences in metabolism between males and females. Of the immune DGEs, 18 up-regulated genes in females were humoral immune factors, and eight up-regulated genes in males were pattern recognition receptors, suggesting sex differences of immune defense might exist in the mitten crab. In addition, two reproduction-related genes, vitellogenin and insulin-like androgenic gland factor, were identified to express in both sexes but with significantly higher level in males. Our research provides the first whole-body RNA sequencing of sex-specific transcriptomes for juvenile E. sinensis and will facilitate further studies on molecular mechanisms of crab sexual dimorphism.
Introduction
Sexual dimorphism, which differentiates males and females in morphological, physiological and behavioral characteristics, is a common phenomenon in the animal kingdom. Based on the nearly identical genomes, the phenotypic differences between the sexes are thought to largely result from sex differences in gene expression [1,2]. So far, sex-biased gene expression has been well explored in mammals, birds, fish and insects, particularly in classic model organisms [3–6], however, little attention has been received to such studies in crustaceans.
A number of crustacean species exhibit significant sex differences in their biology and economic value [7]. For example, male crabs usually show higher growth rate and larger size than females when they reach maturation, while female crabs are of higher economic values than males [8,9]. Among the cultured crabs, the Chinese mitten crab Eriocheir sinensis (Henri Milne Edwards, 1854) is the most prevalent and commercially important species in China [10]. E. sinensis is a catadromous crustacean that spends most of its life in freshwater and migrates seawards to breed [11]. Due to the complex life cycle, mitten crabs might have unique regulatory mechanisms involved in crustacean reproduction or sexual development [12,13]. E. sinensis can serve as a model species of crustaceans to study sexual dimorphism and discover sex-biased gene expression.
Next-generation high-throughput sequencing technologies, such as Illumina/Solexa platform and Ion Proton system, have been used to generate large amounts of transcript sequences and gene expression data for non-model species without sequenced genomes [14,15]. Most studies of sex-biased transcriptome have focused on adult gonads in crustacean, and some sex-related genes have been identified, for example, Tra-2 [16] in the giant tiger shrimp Penaeus monodon, Dmrt, Fem-1 and vasa in the green mud crab Scylla paramamosain [17], and Dmc1 and DEAD box family genes in E. sinensis [12,18,19]. Currently, several studies have reported significant sex-biased gene expression in juvenile stages of fish and insects [20–22]. However, the sex-biased genes by comparing expression before reproductive maturity in crustaceans remain largely unexplored.
Here, we analyzed the whole-body transcriptomes of juvenile E. sinensis using Ion Proton sequencing technology. This study was designed to enrich the genetic resources for E. sinensis, to identify new candidate genes involved in sex determination and differentiation, and to detail the different expression patterns between the sexes. Furthermore, differentially expressed genes (DEGs) involved in metabolism, immunity and reproduction were determined and analyzed. Our results are important resources for future research on molecular mechanisms underlying the sexual dimorphism in E. sinensis and other crustaceans.
Materials and Methods
Ethics Statement
The Chinese mitten crabs were captured from Liao River in Panjin, China and no specific permission was required for the sampling area and species because of scientific research purpose. The sampling location is not privately owned or protected. No endangered or protected species were involved in the field sampling. The experiments were performed in accordance with the guidelines on the care and use of animals for scientific purpose set by the Institutional Animal Care and Use Committee (IACUC) of the Chinese Academy of Sciences (No. 2011-2). This study was specifically approved by the Committee on the Ethics of Animal Experiments of the Institute of Oceanology at the Chinese Academy of Sciences. All efforts were made to minimize the suffering of the animals.
Sample preparation and RNA isolation
Healthy mitten crabs were obtained from a local crab farm in Panjin, China. Five males (1.6 ± 0.4 g) and females (1.9 ± 0.2 g) at the third juvenile instar were pooled, as this stage is the earlier stage to distinguish their sex from morphology [23]. Total RNA was extract from the whole bodies using Trizol reagent (Invitrogen, USA) according to the manufacturer’s protocol. The quality and integrality were determined by Agilent2100 (Agilent, USA). The mRNA was purified from total RNA using Dynabeads mRNA Direct Micro kit (Ambion, USA). The final concentration was determined using a NanoDrop2000 spectrophotometer (Thermo Scientific, USA).
cDNA library construction and sequencing
Two libraries for male and female juveniles of E. sinensis were constructed in this study. For library preparation, equal amounts of purified mRNA samples from two biological replicates were pooled together for the cDNA synthesis. The cDNA libraries were prepared using an Ion Total RNA-seq kit v2 (Life Technologies, USA), with 100 ng fragmented mRNA. Adapter ligation, size selection, nick repair and amplification (12 cycles) were performed as described in the Ion Proton protocol associated with the kit. The resulting cDNA libraries were purified by AMPure beads (Beckman Coulter, USA), and their concentrations and sizes were determined by Agilent2100 (Agilent, USA). Emulsion PCR and enrichment steps were performed using an Ion PI Template OT2 200 kit v3 (Life Technologies) according to the manufacturer’s instructions. PI Chips were loaded according to the spinning protocol and sequencing was performed on Ion Proton Sequencer using the Proton 200 sequencing kit (Life Technologies). Base calls were collected with Torrent Suite using v4.0.2 software.
Sequence assembly
The raw data for each pool of samples were separately trimmed to remove adaptors and low quality regions (< Q20). Reads with a length of less than 50 bp were also discarded. The remaining reads without ambiguous bases (N) were de novo assembled in a unique file by Trinity (http://trinityrnaseq.sourceforge.net/) with k-mer length of 25 referring to the strategy of Grabherr et al. [24]. Trinity combining three independent software modules: Inchworm, Chrysalis and Butterfly, applied sequentially to process large volumes of RNA-seq reads into contigs, de Bruijn graphs and full-length transcripts.
Gene annotation
Gene functional annotation was performed by sequence comparison with public databases. All assembled transcripts were searched against the NCBI non-redundant (nr) protein database (http://www.ncbi.nlm.nih.gov/) using BlastX algorithm with an E-value cutoff of 1E-05. The unigenes were obtained after clustering the top hit results. Gene Ontology (GO) annotations were determined using Blast2GO to obtain a functional classification of the unigenes [25]. EggNOG (evolutionary genealogy of genes: Non-supervised Orthologous Groups) was performed to predict and classify potential functions of the unigenes based on known orthologous gene products [26]. EC (Enzyme Commission) terms and biochemical pathway information were generated by Kyoto Encyclopedia of Genes and Genomes (KEGG) database [27]. In addition, the unigenes associated with sex determination and differentiation were identified manually according to the annotation in consulting published literature and public datasets.
Identification of SNPs and SSRs
Single nucleotide polymorphisms (SNPs) were identified using SAMtools mpileup and VarScan (version 2.3.3) [28,29]. VarScan was run with the following parameters: “mpileup—p-value 0.01—min-avg-qual 20—min-reads2 2—min-var-freq 0.2-variants”. From these results, a set of high confidence SNPs (with coverage of 8 or more reads) were identified. All unigenes were searched for the presence of SSRs using MISA (http://pgrc.ipk-gatersleben.de/misa/) [30] with the following minimum length criteria (unit size/minimum repeat time): 2/6, 3/5, 4/5, 5/5 and 6/5. Compound microsatellites were defined as repeats interrupted by a non-repetitive sequence of a maximum of 100 nucleotides.
Differential expression analysis
The reads for a specific transcript were counted by mapping reads to assembled unigene sequences. The unigene expression was calculated using the reads per kb per million reads (RPKM) method [31]. Differentially expressed genes (DEGs) were identified by the DESeq program [32]. The fold change values > 2 and false discovery rates (FDR) adjusted significance values < 0.05 (-log10 (0.05) = 1.3) were considered as the threshold to judge the significance of unigene expression.
GO, eggNOG, KEGG Orthology (KO) and KEGG pathway enrichment analyses were used to categorize DEGs and evaluate DEGs in the potential biological pathways. Processes, functions or components in the GO and KEGG pathway enrichment analyses with p-values less than 0.05 (-log10 (0.05) = 1.3) were considered to be significantly different in the DEGs. Based on public databases and the published literatures, the crucial DEGs related to metabolism, immunity and reproduction were manually checked.
The DEG encoding insulin-like androgenic gland factor (IAG) was selected for further sequence and phylogenetic analysis. Multiple amino acid sequence alignment was performed using the Clustal X with the default settings [33]. Neighbor-joining tree with bootstrap values were constructed for phylogenetic analysis using MEGA 4.0 [34]. All the reference sequences for phylogenetic analysis were derived from GenBank.
Quantitative Real-time PCR (qRT-PCR) validation
To validate RNA-seq data and expression profiles obtained from DESeq analysis, mitten crabs at the same developmental stage, the independent samples of RNA-seq, were used for real-time PCR analyses. Three biological replicates were prepared for male and female juveniles. Approximately 2.5 μg of total RNA of each sample obtained as previously described were treated with DNase I (Ambion, USA) at 37°C for 1 h, reverse-transcribed using M-MLV reverse transcriptase (Promega, China) and amplified by qRT-PCR.
The SYBR Green I RT-PCR assay was carried out in an ABI PRISM 7300 Sequence Detection System (Applied Biosystems, USA). The gene-specific primers designed for the 12 genes are listed in S1 Table. These 12 genes all exhibited large significant differences in expression between male and female libraries. The β-actin from E. sinensis was chosen as reference gene for internal standardization. The PCR was carried out in a total volume of 10 μl, containing 5 μl of 2×SYBR Premix Ex Taq (TaKaRa, China), 0.2 μl 50×ROX Reference Dye, 2 μl of the diluted cDNA mix, 0.2 μl of each primer (10 μM), and 2.4 μl of sterile distilled H2O. The PCR program was 95°C for 5 min, followed by 40 cycles of 95°C for 5 s and 60°C for 31 s. Each sample was run in triplicate along with the internal control gene. To confirm that only one PCR product was amplified and detected, dissociation curve analysis of amplification products was performed at the end of each PCR reaction. After the PCR program, data were analyzed with ABI7300 SDS software (Applied Biosystems). Fold change for the gene expression relative to controls was determined by the 2-ΔΔCt method [35]. All data were given in terms of relative mRNA expression as mean ±S.E. The results were subjected to one-way analysis of variance (one way ANOVA) using SPSS 13.0, and the P values less than 0.05 and 0.01 were considered statistically significant.
Results
Transcriptome sequencing and assembly
Two cDNA libraries were generated with pooled mRNAs from the whole bodies of female and male juveniles of E. sinensis. After quality trimming and the removal of adapters, sequencing runs performed on Ion Proton platform produced a total of 42,979,050 reads for the female and 47,560,370 reads for the male. All data were deposited in NCBI Short Read Archive database with accession numbers SRX554564 and SR554562. Clustering and assembly of these reads resulted in 282,954 contigs with an average length of 274 bp. Further assembly analysis showed all contigs contributed to 151,128 transcripts with an average length of 614 bp (S2 Table).
Functional annotation
BlastX searches of all assembled transcripts against the NCBI nr database revealed 23,349 unigenes with significant matches to existing protein sequences above the preset cutoff value. The average length of these annotated unigenes was 988 bp with N50 length of 1,375 bp (S2 Table).
Based on GO analysis, 17,388 unigenes (74.5%) were assigned to one or more GO term. Finally, 77,697 GO assignments were obtained and fall into the three major functional categories (S1 Fig). Thereinto, 38,578 unigens (49.7%) were involved in biological process, 26,583 (34.2%) were cellular components and 12,518 (16.1%) have molecular functions. Moreover, 0.4% (323 unigenes) were assigned to reproduction of biological process.
To classify orthologous gene products, 22,101 unigenes were assigned to 25 function categories (S2 Fig). Of these, the cluster of ‘function unknown’ (4,825, 18.3%) represented the largest group, followed by ‘signal transduction mechanisms’ (3,757, 14.3%), ‘general function prediction only’ (3194, 12.1%) and ‘transcription’ (2,112, 8.0%). KEGG analysis revealed that 9,605 unigenes were assigned to six biochemical pathways, including metabolism, genetic information processing, environmental information processing, cellular processes, organismal systems and human diseases. A total of 3,743 unigenes had significant matches in the KEGG database with corresponding EC numbers (S2 Table).
SNP and SSR discovery
Using SAMtools/VarScan software, 48,753 SNPs and 15,271 indels were identified from E. sinensis unigenes. The overall frequency of all types of SNPs, including indels, was one per 360 bp (Fig 1A). Transition occurred 2.7 times more frequently than transversion. A/G was the most abundant transition (28.3%), and A/T was the most abundant transversion (5.3%). Indels were less frequent than transitions, with a frequency of one per 1,511 bp and a total proportion of around 23.9%. Of the identified SNPs and indels, 55,230 were found in both female and male E. sinensis unigenes. 3,411 SNPs and indels were found exclusively in female unigenes, while 5,383 were just present in male unigenes (Fig 1B).
Fig 1. Summary statistics of single nucleotide polymorphisms (SNPs) including indels from female and male Eriocheir sinensis unigenes.
(A) Classification of SNPs. Distribution (%) of each SNP and indel type. (B) Venn diagram of common and unique SNPs and indels of female and male unigenes.
In addition, using MISA, 3,928 perfect and 262 compound SSRs were detected from E. sinensis unigenes (S3 Table). Trinucleotide repeats were the most frequent type, counting a total number of 2,286 (54.6%) with AGG/CCT as a major motif accounted for 31.4% of all trinucleotide repeats. Dinucleotides repeats were the second most frequent type accounting for 37.3% of all SSRs with the abundance of AC/GT motif. And the other two distinguished types, compound and tetranucleotide repeats, accounted for less than 10% of all SSRs.
Genes related to sex determination and differentiation
Of the total unigenes, 148 were identified as sex-related genes based on sequence annotation and the published literature (S4 Table). These genes could be categorized into 36 groups and putatively involved in sex determination and differentiation, oogenesis, spermatogenesis and gonad development. Among these unigenes, 40 were classified as testis or sperm-specific genes, and 24 were ovary or oocyte-specific genes. Eleven unigenes annotated as SOX (SRY related HMG box) gene family were found, including SOX2, SOX5, SOX8, SOX14, SOXB1, SRY, SRY-box containing protein 8 and B2b2.
A total of 29 genes with similarity to those involved in primary sex determination pathway were identified (Table 1). Eight genes, including two fruitless (Fru), two transformer-2 (Tra-2) and four feminization-1 (Fem-1) genes, were associated with the sex determination in Drosophila melanogaster and Caenorhabditis elegans. Fourteen genes involved in mammalian female sexual development were detected: five genes belonging to the WNT signaling pathway (RSPO-1, WNT4 and β-catenin), a transcription factor (FOXL2), three estradiol receptors (ER), and five activin-binding proteins (FST). Seven genes involved in mammalian male sexual development were examined: a doublesex and mab-3 related transcription factor 2 (Dmrt2), three prostaglandin D synthases (PGDS), and three genes belonging SOX transcription factors (SRY and SOX8).
Table 1. Primary sex determination pathway related genes detected in Eriocheir sinensis transcriptomes.
| Gene | Unigene | Isoform name | Gene annotation [Matched species] | E-value |
|---|---|---|---|---|
| Genes involved in Drosophila melanogaster sex determination pathway | ||||
| Fru | comp31866_c0_seq1 | EsFru | fruitless [Chorthippus biguttulus] | 1.95863E-10 |
| comp21702_c0_seq1 | EsFru-like | sex determination protein fruitless-like [Ceratitis capitata] | 3.21181E-23 | |
| Tra-2 | comp74937_c1_seq33 | EsTra-2 | transformer-2 protein [Penaeus monodon] | 1.6998E-46 |
| comp74937_c1_seq2 | EsTra-2v | variant transformer-2 protein [Penaeus monodon] | 2.18253E-45 | |
| Genes involved in Caenorhabditis elegans sex determination pathway | ||||
| Tra-2 | comp74937_c1_seq33 | EsTra-2 | transformer-2 protein [Penaeus monodon] | 1.6998E-46 |
| comp74937_c1_seq2 | EsTra-2v | variant transformer-2 protein [Penaeus monodon] | 2.18253E-45 | |
| Fem-1 | comp73673_c0_seq4 | EsFem-1 | fem-1-like protein [Daphnia pulex] | 4.22441E-59 |
| comp62108_c0_seq1 | EsFem-1B | fem-1 homolog B-like protein [Locusta migratoria manilensis] | 0 | |
| comp73673_c0_seq1 | EsFem-1C | fem-1-like protein C [Crassostrea gigas] | 1.1536E-175 | |
| comp69377_c0_seq1 | EsFem-1hC | fem-1 homolog C-like protein [Locusta migratoria manilensis] | 7.84219E-12 | |
| Genes involved in mammalian sex determination and sexual differentiation pathway | ||||
| Ovarian development | ||||
| β-catenin | comp76922_c0_seq8 | beta-catenin [Parhyale hawaiensis] | 4.7035E-113 | |
| ER | comp66193_c2_seq1 | EsER1 | estradiol receptor-like protein 1 [Portunus trituberculatus] | 7.15127E-48 |
| comp55365_c1_seq1 | EsER2 | estradiol receptor-like protein 2 [Portunus trituberculatus] | 3.25746E-17 | |
| comp68632_c0_seq1 | EsER3 | estradiol receptor-like protein 3 [Portunus trituberculatus] | 0 | |
| FOXL2 | comp56545_c0_seq1 | forkhead box protein L2 [Astyanax mexicanus] | 3.7583E-26 | |
| FST | comp48310_c0_seq1 | EsFST | Follistatin [Acromyrmex echinatior] | 1.31862E-34 |
| comp43834_c0_seq1 | EsFSTp | Follistatin precursor [Pediculus humanus corporis] | 3.11653E-08 | |
| comp89797_c0_seq1 | EsFST5 | follistatin-like 5 [Scylla paramamosain] | 8.94175E-30 | |
| comp86047_c0_seq1 | EsFST5-like | follistatin-related protein 5-like [Nasonia vitripennis] | 1.52671E-56 | |
| comp35850_c0_seq1 | EsFST5-like2 | follistatin-related protein 5-like [Acyrthosiphon pisum] | 7.3293E-62 | |
| RSPO-1 | comp92691_c0_seq1 | EsRSPO-1 | R-spondin 1 [Glandirana rugosa] | 5.20609E-06 |
| comp26865_c0_seq1 | EsRSPO-1p | R-spondin-1 precursor [Oryzias latipes] | 9.0604E-25 | |
| WNT4 | comp30980_c0_seq1 | EsWNT4 | protein Wnt-4-like [Bos taurus] | 1.53802E-56 |
| comp9493_c0_seq1 | EsWNT4s | putative secreted signaling factor WNT4 [Daphnia pulex] | 5.87015E-09 | |
| Testis development | ||||
| Dmrt2 | comp18195_c0_seq1 | doublesex and mab-3 related transcription factor 2 [Xenopus laevis] | 2.46563E-10 | |
| PGDS | comp58387_c0_seq1 | EsPGDS | prostaglandin D synthase [Eriocheir sinensis] | 1.8541E-125 |
| comp53351_c0_seq1 | EsGPGDS | glutathione-dependent prostaglandin D synthase [Penaeus monodon] | 1.33867E-56 | |
| comp73984_c0_seq5 | EsHPGDS | hematopoietic prostaglandin D synthase [Penaeus monodon] | 2.16344E-48 | |
| SRY | comp34840_c0_seq1 | EsSRY | Sex-determining region Y protein [Pediculus humanus corporis] | 3.57723E-46 |
| SOX8 | comp21432_c0_seq1 | EsSRY8 | SRY-box containing gene 8 [Rattus norvegicus] | 6.87426E-11 |
| comp71497_c0_seq3 | EsSOX8 | Transcription factor SOX-8 [Camponotus floridanus] | 3.46753E-28 | |
The primary sex determination pathway related genes were expressed in both female and male E. sinensis transcriptomes (S3 Fig). EsPGDS was the most highly expressed transcript (RPKM value > 800), followed by EsER3 (RPKM value > 50) and EsHPGDS (RPKM value > 40). EsDmrt2, EsPGDS and EsSRY presented higher expression pattern in males (fold change values > 1.5), while EsFST5, EsRSPO-1p and EsFST5p showed preferential expression in females (fold change values > 1.5). The other 23 genes were similar expressed in females and males.
From the 148 unigenes annotated for sex-related genes, 18 SNPs were identified in female individuals and 23 in male individuals (S5 Table). Most unigenes had only one SNP, except for vitelline membrane outer layer protein I, SOX14, oocyte zinc finger protein, male reproductive-related microfibril-associated protein spermatogenesis-associated protein 5 and 20. In addition, five compound and 18 perfect SSRs were detected from the sex-related unigenes (S6 Table). The size of SSRs ranged from 12 to 242, with trinucleotide repeats as the most frequent type. Three sex-related genes, including SRY-box containing gene 8, SRY interacting protein 1 and zinc finger protein 76, were found to contain both SNPs and SSRs.
Differential expression between the sexes
The DESeq method identified 448 differentially expressed unigenes between the two transcriptomes, including 188 up-regulated in females and 260 up-regulated in males (S7 Table). The distribution of the significant changes detected was illustrated in a volcano plot, where the statistical significance of each unigene was plotted against fold change (S4 Fig). Sequences with the highest average differences between the sexes also had the smallest false discovery rate (FDR) values.
All of the DEGs were performed on GO function and pathway enrichment analysis. Based on GO analysis, a total of 340 DEGs with GO terms were categorized into three major functional groups (Fig 2). Within the biological processes, the highest percentage of DEGs involved in GO terms was ‘transport’ (10.7%), followed by ‘carbohydrate metabolic process’ (8.2%), ‘biosynthetic process’ (6.8%), ‘oxidoreductase activity’ (6.8%), ‘protein maturation’ (6.6%) and ‘carbohydrate metabolic process’ (6.5%) (Fig 2A). Of the cellular component categories, the majority of GO terms were related to ‘cell’ (23.1%), ‘intracellular’ (22.1%) and ‘cytoplasm’ (15.4%) (Fig 2B). As for molecular functions, the three major GO terms were ‘ion binding’ (64.6%), ‘RNA binding’ (8.9%) and ‘enzyme binding’ (6.8%) (Fig 2B). After the overall comparison was completed, the top ten significantly changed categories were obtained, including ‘hydrolase activity, acting on glycosyl bonds’, ‘carbohydrate metabolic process’, ‘extracellular region’, ‘oxidoreductase activity’, ‘protein maturation’, ‘transport’, ‘lyase activity’, ‘lipid metabolic process’, ‘immune system process’ and ‘ion binding’ (p < 0.05, Fig 2C).
Fig 2. GO distributions of differentially expressed genes (DEGs) from female and male Eriocheir sinensis transcriptomes.
(A) Different functional distribution of the DEGs involved with biological processes. (B) Different functional distribution of the DEGs involved with cellular components and molecular functions. (C) Differentially expressed functional processes. The horizontal line indicates the significance threshold (p < 0.05).
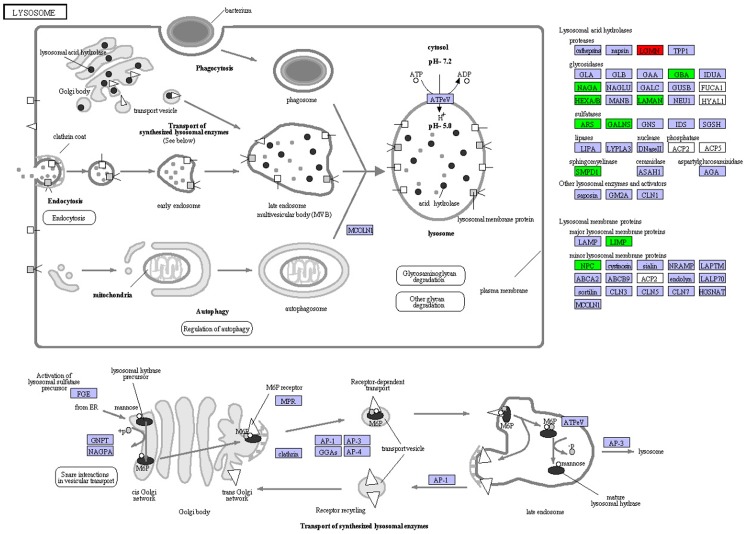
KEGG analysis revealed that 139 DEGs with KO terms were involved in 29 predicated biological pathways (S5 Fig). The most abundant pathways included ‘carbohydrate metabolism’ (36 DGEs), ‘glycan biosynthesis and metabolism’ (24 DGEs), ‘transport and catabolism’ (24 DGEs) and ‘lipid metabolism’ (16 DGEs). The significantly distinct categories were ‘carbohydrate metabolism’, ‘glycan biosynthesis and metabolism’, ‘metabolism of cofactors and vitamins’, ‘transport and catabolism’, ‘digestive system, ‘lipid metabolism’, ‘xenobiotics biodegradation and metabolism’ and ‘amino acid metabolism’ (p < 0.05, S5 Fig). Except for metabolic pathway, the pathway predominantly enriched for DEGs were related to lysosome (ko04142) of ‘transport and catabolism’ (Fig 3). There were 19 DEGs with ten KO terms in lysosomal pathway, including 18 up-regulated in males and one up-regulated in females. In addition, one important pathway identified as enriched for the up-regulated genes in males were associated with sphingolipid metabolism (ko00600) of ‘lipid metabolism’ (S6 Fig).
Fig 3. Expression pattern of genes involved in lysosomal pathway.
The pathway is based on a KEGG pathway analysis. The up-regulated genes in male and female are labeled by green and red, respectively, and the purple color represents genes with no expression differences between female and male transcriptomes of Eriocheir sinensis.
Candidate DEGs involved in metabolism, immunity and reproduction
By sequence annotation and functional classification, we identified 61 differentially expressed unigenes related to metabolism, immunity and reproduction. The metabolism-related DGEs could be categorized into four classes, including five female-biased genes in amino acid metabolism, ten male-biased genes in lipid metabolism, six male-biased genes in glycan metabolism, and three male-biased genes in cytochrome P450 (CYP450) superfamily (Table 2). A total of 30 immunity-related DGEs were found, including 18 up-regulated humoral immune factors in females, eight up-regulated pattern recognition receptors (PRRs) (CLEC and LGBP) and four other up-regulated immune factors in males (Table 3). The female-biased immune DGEs were largely involved in antimicrobial peptide synthesis (Lyz, Crustin and Carcinin), prophenoloxidase (proPO) system (SP, PPAF and Kazal) and antioxidant system (SOD and Trx). Of the reproduction-related DGEs, two genes involved in ovarian development showed female-biased expression, while five genes, including two Vg homologs, VMO1, IAG and NPC2, showed male-biased expression (Table 4).
Table 2. List of differentially expressed genes related to metabolism in female and male Eriocheir sinensis transcriptomes.
| Gene | Unigene | Gene annotation [Matched species] | Fold change (Female: Male) |
|---|---|---|---|
| Up-regulated genes in females | |||
| Amino acid metabolism-related genes | |||
| AspRS | comp59739_c0_seq1 | aspartate-tRNA ligase, mitochondrial-like [Takifugu rubripes] | 3.17 |
| Ark | comp72982_c2_seq1 | arginine kinase [Eriocheir sinensis] | 2.32 |
| Arg1 | comp74968_c0_seq2 | arginase-1 isoform 2 [Trichechus manatus latirostris] | 2.08 |
| NIT2 | comp56883_c0_seq1 | Nit protein 2-like [Saccoglossus kowalevskii] | 7.86 |
| comp48414_c0_seq2 | Omega-amidase NIT2-B [Crassostrea gigas] | 6.85 | |
| Up-regulated genes in males | |||
| Lipid metabolism-related genes | |||
| ACAC | comp77110_c0_seq6 | Acetyl-CoA carboxylase [Harpegnathos saltator] | 0.47 |
| ASAH2 | comp73699_c0_seq4 | ceramidase [Danaus plexippus] | 0.42 |
| ARSA | comp74420_c0_seq3 | arylsulfatase A-like [Strongylocentrotus purpuratus] | 0.29 |
| FASN | comp75785_c0_seq2 | fatty acid synthase [Tribolium castaneum] | 0.43 |
| comp76291_c0_seq1 | fatty acid synthase-like isoform 1 [Nasonia vitripennis] | 0.46 | |
| GBA | comp74162_c1_seq1 | glucosylceramidase-like [Ciona intestinalis] | 0.46 |
| comp63076_c0_seq1 | glucosylceramidase-like [Monodelphis domestica] | 0.47 | |
| Se-Gpx | comp66076_c0_seq2 | selenium-dependent glutathione peroxidase [Procambarus clarkii] | 0.21 |
| SMPD1 | comp72185_c1_seq1 | acid sphingomyelinase 1 [Glossina morsitans morsitans] | 0.36 |
| PLRP2 | comp75117_c0_seq1 | pancreatic lipase-related protein 2-like [Acyrthosiphon pisum] | 0.34 |
| Glycan metabolism-related genes | |||
| A4GALT | comp9581_c0_seq1 | lactosylceramide 4-alpha-galactosyltransferase [Rattus norvegicus] | 0.33 |
| GALNS | comp76195_c0_seq5 | n-acetylgalactosamine-6-sulfatase-like [Monodelphis domestica] | 0.40 |
| comp76195_c0_seq6 | galactosamine (N-acetyl)-6-sulfate sulfatase [Xenopus (Silurana) tropicalis] | 0.49 | |
| DHDH | comp63990_c0_seq2 | Trans-1,2-dihydrobenzene-1,2-diol dehydrogenase [Acromyrmex echinatior] | 0.44 |
| HEXA_B | comp75397_c0_seq1 | beta-hexosaminidase subunit alpha-like [Ornithorhynchus anatinus] | 0.43 |
| LAMAN | comp60626_c0_seq1 | lysosomal alpha-mannosidase precursor [Danio rerio] | 0.40 |
| Cytochrome P450 superfamily | |||
| CYP3A4 | comp59688_c0_seq1 | cytochrome P450, family 3, subfamily A, polypeptide 4 [Callorhinchus milii] | 0.08 |
| CYP9Z7 | comp75041_c0_seq1 | cytochrome P450 9Z7 [Tribolium castaneum] | 0.16 |
| CYP2B19 | comp69705_c0_seq2 | cytochrome P450 2B19-like [Ciona intestinalis] | 0.48 |
Table 3. List of differentially expressed genes related to immunity in female and male Eriocheir sinensis transcriptomes.
| Gene | Unigene | Gene annotation [Matched species] | Fold change (Female: Male) |
|---|---|---|---|
| Up-regulated genes in females | |||
| Lyz | comp67931_c0_seq2 | lysozyme [Scylla paramamosain] | 7.21 |
| A2ML2 | comp75726_c0_seq17 | alpha2 macroglobulin isoform 2 [Fenneropenaeus chinensis] | 2.15 |
| Carcinin | comp61957_c0_seq1 | carcinin-like protein [Carcinus maenas] | 2.72 |
| Crustin | comp54631_c0_seq1 | crustin [Scylla tranquebarica] | 3.94 |
| comp66850_c0_seq1 | crustin 2 [Portunus trituberculatus] | 2.06 | |
| CPAMD8 | comp74090_c0_seq4 | C3 and PZP-like alpha-2-macroglobulin domain-containing protein 8-like [Metaseiulus occidentalis] | 2.07 |
| HPT factor 9 | comp58745_c0_seq1 | HPT factor 9 [Pacifastacus leniusculus] | 2.25 |
| Kazal | comp67267_c0_seq3 | Kazal-type protease inhibitor [Eriocheir sinensis] | 2.25 |
| PPAF | comp72920_c0_seq1 | prophenoloxidase activating factor serine proteinase [Scylla serrata] | 7.33 |
| comp57767_c0_seq1 | phenoloxidase activating factor [Portunus trituberculatus] | 2.32 | |
| SP | comp67291_c1_seq3 | clip domain serine proteinase 3 [Portunus trituberculatus] | 2.63 |
| comp71922_c0_seq2 | clip domain serine proteinase [Portunus trituberculatus] | 2.30 | |
| comp63300_c1_seq1 | clip domain serine protease [Eriocheir sinensis] | 2.11 | |
| comp67168_c0_seq1 | serine proteinase [Portunus trituberculatus] | 2.06 | |
| comp67235_c0_seq1 | trypsin-like serine protease [Eriocheir sinensis] | 2.16 | |
| SOD | comp66967_c0_seq1 | superoxidase dismutase [Eisenia fetida] | 2.28 |
| PDGF/VEGF-related factor 1 | comp75589_c0_seq4 | PDGF/VEGF-related factor 1 [Eriocheir sinensis] | 2.26 |
| Trx1 | comp61426_c0_seq2 | Trx1 [Eriocheir sinensis] | 2.08 |
| Up-regulated genes in males | |||
| CLEC | comp62386_c0_seq1 | C-type lectin [Eriocheir sinensis] | 0.29 |
| comp74386_c0_seq1 | C-type lectin [Eriocheir sinensis] | 0.37 | |
| comp69162_c2_seq4 | C-type lectin 1 [Marsupenaeus japonicus] | 0.34 | |
| comp18326_c0_seq1 | C-type lectin-1 [Litopenaeus vannamei] | 0.45 | |
| comp22144_c0_seq1 | C-type lectin-2 [Litopenaeus vannamei] | 0.37 | |
| CLR | comp45203_c0_seq1 | C-type lectin receptor protein [Eriocheir sinensis] | 0.46 |
| HC6 | comp67750_c0_seq2 | hemocyanin subunit 6 [Eriocheir sinensis] | 0.50 |
| LGBP | comp56987_c0_seq1 | lipopolysaccharide and beta-1,3-glucan binding protein [Procambarus clarkii] | 0.19 |
| comp72288_c0_seq1 | lipopolysaccharide and beta-1,3-glucan binding protein [Eriocheir sinensis] | 0.29 | |
| MIF | comp54328_c1_seq1 | macrophage migration inhibitory factor [Anisakis simplex] | 0.40 |
| Peritrophin | comp54270_c0_seq1 | peritrophin [Macrobrachium nipponense] | 0.39 |
| PLGRKT | comp59036_c0_seq2 | plasminogen receptor (KT) [Dasypus novemcinctus] | 0.40 |
Table 4. List of differentially expressed genes related to reproduction in female and male Eriocheir sinensis transcriptomes.
| Gene | Unigene | Gene annotation [Matched species] | Fold change (Female: Male) |
|---|---|---|---|
| Up-regulated genes in females | |||
| Vg receptor | comp76488_c0_seq9 | vitellogenin receptor [Macrobrachium rosenbergii] | 2.18 |
| Pxt | comp58063_c0_seq3 | chorion peroxidase-like [Acyrthosiphon pisum] | 2.16 |
| Up-regulated genes in males | |||
| Vg | comp10184_c0_seq1 | vitellogenin [Crassostrea gigas] | 0.19 |
| comp77225_c0_seq2 | vitellogenin [Charybdis feriata] | 0.47 | |
| VMO1 | comp67371_c0_seq1 | vitelline membrane outer layer protein 1 homolog isoform 1 [Oryzias latipes] | 0.38 |
| IAG | comp61312_c0_seq3 | insulin-like androgenic gland factor [Callinectes sapidus] | 0.25 |
| NPC2 | comp55602_c0_seq1 | epididymal secretory protein E1-like [Hydra magnipapillata] | 0.36 |
A partial sequence of 741 bp (comp61312_c0_seq3), without an ATG codon but with a TAA stop codon at position 526 bp, was identified to encode an IAG ortholog (EsIAG, S7 Fig). The deduced amino acid sequence was 132 aa in length and contained B chain and A chain, with three disulfide bridges (between CB9 and CA12, CB20 and CA25, CA11 and CA16, respectively). Multiple sequence alignment of EsIAG and six decapod IAGs showed the conserved cysteine residues shared by all sequences (S7A Fig). EsIAG displayed 36.3% amino acid identity with Callinectes sapidus IAG1, 32.5% with C. sapidus IAG2 and 33.1% with Scylla paramamosain IAG. The phylogenetic tree showed that the IAGs formed two major clades: one with three isopods and the other containing five subclades from the decapods (S7B Fig). The crab IAGs were separated in two gruops. EsIAG was clustered with IAGs from the Atlantic blue crab C. sapidus and the mud crab S. paramamosain to form one group. Another group included IAG from the blue swimmer crab Portunus pelagicus, which had a closer relationship with two IAGs from crayfish.
qRT-PCR validation of RNA-seq data
To verify the result of RNA-seq analysis, 12 DGEs based on RNA-seq were selected for qRT-PCR to further investigate the expression profiles. The expression patterns from qRT-PCR showed general agreement with those from the RNA-seq (Fig 4). Five genes including AspRS, NIT2, Lyz, Crustin and PPAF were up-regulated in females, while Vg, ARSA, Se-Gpx, CYP3A4, CLEC, LGBP and IAG were up-regulated in males. Among these genes, NIT2 showed the largest up-regulation in females and CYP3A4 manifested the largest up-regulation in males, which was consistent with RNA-seq results. The consistent expression between qRT-PCR and RNA-seq analyses confirmed the accuracy of Proton results.
Fig 4. qRT-PCR validation of selected differentially expressed genes identified by RNA-seq.
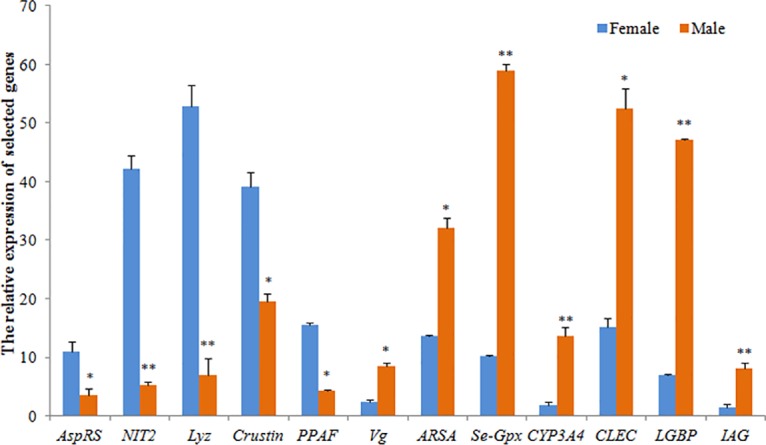
Vertical bars represent the mean ± S.E. (n = 3). Significant differences are indicated with an asterisk at P < 0.05, and two asterisks at P < 0.01.
Discussion
Whole-body reference transcriptome of male and female Eriocheir sinensis
Compared with the available data from database and the previously reported transcriptomes from male gonads, the present study, providing more than 10 Gb clean data, is the first whole-body RNA sequencing of sex-specific transcriptomes for juvenile Chinese mitten crab E. sinensis. This resource expands the limited amount of sex-related sequence data of E. sinensis, which will facilitate further molecular investigation on sexual dimorphism of crab.
Similar to recent studies in E. sinensis [36–38], only about 15% of all assembled transcripts are successfully matched in the nr database, probably due to the limited number of crustacean sequences in public database. However, this percentage is higher than that in gonad transcriptomes of S. paramamosain [17], supposedly because the whole bodies of juvenile are used and more extensive sequencing depth are applied in our transcriptomes. From our sequencing effort, a large number of SNPs and SSRs are detected for future genetics studies. This database, especially the sex-related markers, will play important roles in the exploration and utilization of sex-related genes, and may provide powerful tools for early gender identification and breeding in this commercially important species.
Candidate genes involved in sex determination and differentiation
Though several sex-related genes were obtained previously [39–41], little information about sex determination cascades is available in the mitten crab. GO analysis and orthology prediction supply gene function classification labels and an overall framework for the sex-specific transcriptomes.
Two Tra-2 and two Fru genes are identified in our transcriptome database. The Tra-2 and Fru genes, by promoting female sexual development or directing male sexual behavior, have been proved to participate in the Drosophila sex determination cascade [42,43]. Together with the previously cloned sex-lethal (Sxl) [41] and our identified double sex (dsx) [44], most of ortholog genes in ‘Sxl-Tra/Tra-2-Dsx/Fru’ pathway are detected in the mitten crab. As suggested in the penaeid shrimp [16,45], the present data provides a hint that the mitten crab might adopt a similar sex determination pathway with that in Drosophila. However, the absence of Tra and lack of a sex specificity of Sxl in the mitten crab, also reported in the silkworm Bombyx mori [46–48], imply that genetic regulation for sex determination potentially does not initiate from Sxl.
We discover four homologs of Fem-1 that have not been identified previously in E. sinensis. In C. elegans, Fem-1, encoding an ankyrin repeat protein Fem-1, is a component of the signal transduction pathway controlling sex determination [49]. In our high density linkage map of E. sinensis, another ankyrin repeat-containing gene is found to be located on the putative sex chromosome, suggesting its possible role in sex determination [44]. However, in the present study, the four homologs of Fem-1 are expressed similarly in both males and females. Whether Fem-1 participates in the process of sex differentiation in the crab has yet to be established.
Besides the above genes, many genes involved in mammalian sex determination cascade are reported in E. sinensis, further indicating the complex sex determination system of the mitten crab. In mammals, the presence of the male-determining SRY gene directs the undifferentiated gonad to develop into testes by promoting the expression of SOX9 [50]. Early ovarian development has long been considered to be a default pathway switched on passively by the absence of SRY gene. However, recent reports have revealed that FOXL2-leading pathway and RSPO-1-activating signaling pathway act independently and complementary to each other to promote ovarian development [51–53]. In invertebrates, orthologs of FOXL2 have been characterized, but without a good understanding of their role in reproduction [54–57]. Though the orthologs genes in ‘RSPO-1/WNT4/β-catenin’ signaling pathway are detected, further investigations are needed to determine whether this pathway exists in the mitten crab.
Patterns of gene expression between the sexes
Differential expression analysis reveals 188 and 260 significantly expressed unigenes in female and male transcriptomes, respectively. These results show a slight imbalance in favor of the male sequences. This tendency has also been reported in adult transcriptomes of Caligus rogercresseyi [58] and gonad transcriptomes of the green mud crab [17] and other species such as Acipenser fulvescens [59] and Haliotis rufescens [60]. Coinciding with that in male and female rainbow trout embryos [20], most DEGs in this study are not related to sexual function. Further GO and KEGG analyses reveal these DEGs are largely involved in biological processes, such as lipid metabolism, glycan metabolism, transport and catabolism, and immune system process. This suggests that there are inherent and broad differences in the transcriptomes of male and female mitten crab, and that these differences are present before sexual maturation.
Lysosomes are membrane-enclosed organelles that contain an array of enzymes capable of breaking down all types of biological polymers-proteins, nucleic acids, carbohydrates and lipids [61]. Of DEGs involved in lysosomal pathway, one gene encoding protease legumain is up-regulated in females, while other genes encoding glycosidases, sulfatases, sphingomyelinase and membrane proteins are up-regulated in males. Together with the identified DEGs related to metabolism, this differential expression pattern might due to the higher level of amino acid metabolism in females as well as higher levels of glycan and lipid metabolisms in males. Three metabolism-related CYP450 genes, CYP2B19, CYP3A4 and CYP9Z7, were identified as male-biased DGEs. Enzymes in CYP2 and CYP3 families, especially CYP3A4, have important roles in steroid biosynthesis and metabolism in human [62]. The higher transcripts of CYP450 enzymes in males might indicate male mitten crabs require a large amount of steroid during early juvenile stages.
Sex differences in the immune defense, where females show greater immunity or resistance to infection, have been demonstrated for several arthropods [63–67]. In many cases, females have higher levels of hemocytes or PO activity than males in the absence of infection. Most of these studies have focused on insects such as butterflies, crickets, dragonflies and scorpionflies, with relatively few studies on crustaceans. Here, we first report some genes that potentially contribute to the sex differences in the immune system of the mitten crab.
Humoral immune responses that mainly occur in hemolymph include proPO system, clotting cascade and a wide array of antimicrobial peptides [68]. The identified female-biased humoral immune factors indicate females have greater lysozyme and PO activities in hemocytes, which is in agreement with those studies in insects [64–66]. PRRs, as a set of germline-encoded receptors, can interact with pathogen associated molecular pattern (PAMP) and activate innate immune response [69]. The higher transcripts of PRRs in males might suggest male mitten crabs could trigger quick and effective defense responses in the presence of pathogens infection. Sex differences in immunity appear to be related to differential reproductive strategies and the resulting resource trade-offs in life history. The identified immune DEGs provide a framework for future research to unravel the mechanism of six-biased immune regulation in crab.
Among the reproduction-related DGEs, two special genes vitellogenin (Vg) and insulin-like androgenic gland factor (IAG) were identified. Vg, usually considered as a female specific protein, could serve as energy resource for embryonic development [70]. Apart from its nutritional function, Vg has been shown to play important roles in innate immunity by acting as a multivalent pattern recognition receptor, a bactericidal molecule or an acute phase protein [71]. The Vg gene is normally silent in males, but can be activated by estrogen exposure [72,73]. Interestingly, we detected the expression of Vg in the normal physiological conditions of male E. sinensis, which is consistent with the finding in European honey bee Apis mellifera [74,75] but contrary to most studies in crustaceans [76–78]. This male-biased expression suggests that Vg might have functions in addition to its roles in oocyte maturation and energy supply for embryogenesis. IAG, a key regulator of male sex differentiation in crustaceans [79], is first discovered from E. sinensis transcriptomes. EsIAG shows remarkable structural similarity and sequence homology with other IAGs, suggesting that it is a member of the insulin/insulin-like growth factor family. By qRT-PCR validation, EsIAG is expressed in both sexes with a significantly higher level in males. It suggests that EsIAG might be not expressed exclusively in the male AG, which is also reported in S. paramamosain [79], C. sapidus [80] and Fenneropenaeus chinensis [81].
In conclusion, this is the first whole-body, sex-specific transcriptomes of juvenile E. sinensis using RNA-seq technology. More than 90 million clean reads were obtained, and some candidate genes in sex determination and differentiation were found. A large number of differentially expressed genes between the sexes were identified, and most of them had no obvious sexual function. Many potential SNPs and SSRs were detected that could be used for further gender identification and genetic breeding studies. This study will not only provide valuable genetic resources for the understanding of sexual dimorphism in E. sinensis, but also facilitate further investigations of functional genomics for this species and other closely related species.
Supporting Information
(TIF)
(TIF)
(TIF)
For each unigene, the ratio of expression levels (Female vs. Male) is plotted against the-log error rate. The horizontal line indicates the significance threshold (FDR adjusted < 0.05), and the vertical lines indicate the two fold change threshold. Non-differentially expressed genes are shown with orange dots, and DEGs are shown with blue dots.
(TIF)
The horizontal line indicates the significance threshold (P < 0.05).
(TIF)
The up-regulated in male are labeled by green, and the purple color represents genes with no expression differences between female and male Eriocheir sinensis transcriptomes.
(TIF)
B and A chains are marked in yellow and green boxes, respectively. The six conserved cysteine residues are highlighted with dark red background and the predicted disulfide bridges are drawn. The species and the GenBank accession numbers are as follow: Callinectes sapidus IAG1(AEI72263), C. sapidus IAG2 (AHM93481), Cherax destructor (ACD91988), Cherax quadricarinatus (ABH07705), Fenneropenaeus chinensis IAG1(AFU60548), F. chinensis IAG2 (AFU60549), Jasus edwardsii (AIM55892), Litopenaeus vannamei (AIR09497), Macrobrachium lar (BAJ78349), Macrobrachium nipponense IAG1 (AGB56976), Macrobrachium nipponense IAG2 (AHA33389), Macrobrachium rosenbergii (ACJ38227), Macrobrachium vollenhovenii (AHZ34725), Marsupenaeus japonicus (BAK20460), Palaemon pacificus (BAJ84109), Palaemon paucidens (BAJ84108), Penaeus monodon (ADA67878), Portunus pelagicus (ADK46885), Sagmariasus verreauxi (AHY99679) and Scylla paramamosain (AIF30295). Three isopods Armadillidium vulgare (BAA86893), Porcellio dilatatus (BAC57013), Porcellio scabar (BAC57012) served as outgroups.
(TIF)
(DOC)
(DOC)
(DOC)
(XLS)
(DOC)
(DOC)
(XLS)
Data Availability
All data were deposited into the Sequence Read Archive (SRA) database (http://www.ncbi.nlm.nih.gov/Traces/sra/) under accession numbers SRX554564 and SR554562 and all other relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from the Chinese National "863" Project (No. 2012AA10A409, http://www.most.gov.cn/eng/) and National Natural Science Foundation of China (41276165 and 41206147, http://www.nsfc.gov.cn/publish/portal1/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Connallon T, Knowles LL (2005) Intergenomic conflict revealed by patterns of sex-biased gene expression. Trends in Genetics 21: 495–499. [DOI] [PubMed] [Google Scholar]
- 2. Assis R, Zhou Q, Bachtrog D (2012) Sex-biased transcriptome evolution in Drosophila . Genome Biology and Evolution 4: 1189–1200. 10.1093/gbe/evs093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yang X, Schadt EE, Wang S, Wang H, Arnold AP, Ingram-Drake L, et al. (2006) Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Research 16: 995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Small CM, Carney GE, Mo Q, Vannucci M, Jones AG (2009) A microarray analysis of sex-and gonad-biased gene expression in the zebrafish: evidence for masculinization of the transcriptome. BMC Genomics 10: 579 10.1186/1471-2164-10-579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mank JE, Hultin-Rosenberg L, Webster MT, Ellegren H (2008) The unique genomic properties of sex-biased genes: insights from avian microarray data. BMC Genomics 9: 148 10.1186/1471-2164-9-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Parisi M, Nuttall R, Edwards P, Minor J, Naiman D, Lü J, et al. (2004) A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biology 5: R40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hartnoll RG (1982) Growth In: Bliss DE, editor. The Biology of Crustacea. New York: Academic Press; 111–197 p. [Google Scholar]
- 8. Lee HH, Hsu CC (2003) Population biology of the swimming crab Portunus sanguinolentus in the waters off Northern Taiwan. Journal of Crustacean Biology 23: 691–699. [Google Scholar]
- 9. Mokhtari M, Savari A, Rezai H, Kochanian P, Bitaab A (2008) Population ecology of fiddler crab, Uca lactea annulipes (Decapoda: Ocypodidae) in Sirik mangrove estuary, Iran. Estuarine, Coastal and Shelf Science 76: 273–281. [Google Scholar]
- 10. Chen DW, Zhang M, Shrestha S (2007) Compositional characteristics and nutritional quality of Chinese mitten crab (Eriocheir sinensis). Food Chemistry 103: 1343–1349. [Google Scholar]
- 11. Veilleux É, De Lafontaine Y (2007) Biological synopsis of the Chinese mitten crab (Eriocheir sinensis). Canadian Manuscript Report of Fisheries and Aquatic Sciences 2812: vi + 45p. [Google Scholar]
- 12. He L, Wang Q, Jin X, Wang Y, Chen L, Liu L, et al. (2012) Transcriptome profiling of testis during sexual maturation stages in Eriocheir sinensis using Illumina sequencing. PloS one 7: e33735 10.1371/journal.pone.0033735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rudnick DA, Hieb K, Grimmer KF, Resh VH (2003) Patterns and processes of biological invasion: the Chinese mitten crab in San Francisco Bay. Basic and Applied Ecology 4: 249–262. [Google Scholar]
- 14. Metzker ML (2010) Sequencing technologies—the next generation. Nature Reviews Genetics 11: 31–46. 10.1038/nrg2626 [DOI] [PubMed] [Google Scholar]
- 15. Garber M, Grabherr MG, Guttman M, Trapnell C (2011) Computational methods for transcriptome annotation and quantification using RNA-seq. Nature Methods 8: 469–477. 10.1038/nmeth.1613 [DOI] [PubMed] [Google Scholar]
- 16. Leelatanawit R, Sittikankeaw K, Yocawibun P, Klinbunga S, Roytrakul S, Aoki T, et al. (2009) Identification, characterization and expression of sex-related genes in testes of the giant tiger shrimp Penaeus monodon . Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology 152: 66–76. [DOI] [PubMed] [Google Scholar]
- 17. Gao J, Wang X, Zou Z, Jia X, Wang Y, Zhang Z (2014) Transcriptome analysis of the differences in gene expression between testis and ovary in green mud crab (Scylla paramamosain). BMC Genomics 15: 585 10.1186/1471-2164-15-585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. He L, Jiang H, Cao D, Liu L, Hu S, Wang Q (2013) Comparative transcriptome analysis of the accessory sex gland and testis from the Chinese mitten crab (Eriocheir sinensis). PloS one 8: e53915 10.1371/journal.pone.0053915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang W, Wan H, Jiang H, Zhao Y, Zhang X, Hu S, et al. (2011) A transcriptome analysis of mitten crab testes (Eriocheir sinensis). Genetics and Molecular Biology 34: 136–141. 10.1590/S1415-47572010005000099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hale MC, Xu P, Scardina J, Wheeler PA, Thorgaard GH, Nichols KM (2011) Differential gene expression in male and female rainbow trout embryos prior to the onset of gross morphological differentiation of the gonads. BMC Genomics 12: 404 10.1186/1471-2164-12-404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhao M, Zha XF, Liu J, Zhang WJ, He NJ, Cheng DJ, et al. (2011) Global expression profile of silkworm genes from larval to pupal stages: Toward a comprehensive understanding of sexual differences. Insect Science 18: 607–618. [Google Scholar]
- 22. Perry JC, Harrison PW, Mank JE (2014) The ontogeny and evolution of sex-biased gene expression in Drosophila melanogaster . Molecular Biology and Evolution 31: 1206–1219. 10.1093/molbev/msu072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee T-H, Yamauchi M, Yamazaki F (1994) Sex differentiation in the crab Eriocheir japonicus (Decapoda, Grapsidae). Invertebrate Reproduction and Development 25: 123–137. [Google Scholar]
- 24. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology 29: 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- 26. Powell S, Szklarczyk D, Trachana K, Roth A, Kuhn M, Muller J, et al. (2012) eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Research 40: D284–D289. 10.1093/nar/gkr1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M (2004) The KEGG resource for deciphering the genome. Nucleic Acids Research 32: D277–D280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, et al. (2009) VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25: 2283–2285. 10.1093/bioinformatics/btp373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thiel T, Michalek W, Varshney R, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theoretical and Applied Genetics 106: 411–422. [DOI] [PubMed] [Google Scholar]
- 31. Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods 5: 621–628. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 32. Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biology 11: R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- 35. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 36. Hui M, Liu Y, Song C, Li Y, Shi G, Cui Z (2014) Transcriptome changes in Eriocheir sinensis megalopae after desalination provide insights into osmoregulation and stress adaption in larvae. PloS one 9: e114187 10.1371/journal.pone.0114187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Song C, Cui Z, Hui M, Liu Y, Li Y, Li X (2015) Comparative transcriptomic analysis provides insights into the molecular basis of brachyurization and adaptation to benthic lifestyle in Eriocheir sinensis . Gene 558: 88–98. 10.1016/j.gene.2014.12.048 [DOI] [PubMed] [Google Scholar]
- 38. Li Y, Hui M, Cui Z, Liu Y, Song C, Shi G (2015) Comparative transcriptomic analysis provides insights into the molecular basis of the metamorphosis and nutrition metabolism change from zoeae to megalopae in Eriocheir sinensis . Comparative Biochemistry and Physiology Part D: Genomics and Proteomics 13: 1–9. [DOI] [PubMed] [Google Scholar]
- 39. Zhang EF, Qiu GF (2010) A novel Dmrt gene is specifically expressed in the testis of Chinese mitten crab, Eriocheir sinensis . Development Genes and Evolution 220: 151–159. 10.1007/s00427-010-0336-2 [DOI] [PubMed] [Google Scholar]
- 40. Wang Q, Fang D-A, Sun J-L, Wang Y, Wang J, Liu L-H (2012) Characterization of the vasa gene in the Chinese mitten crab Eriocheir sinensis: A germ line molecular marker. Journal of Insect Physiology 58: 960–965. 10.1016/j.jinsphys.2012.04.012 [DOI] [PubMed] [Google Scholar]
- 41. Shen H, Hu Y, Zhou X (2014) Sex-lethal gene of the Chinese mitten crab Eriocheir sinensis: cDNA cloning, induction by eyestalk ablation, and expression of two splice variants in males and females. Development Genes and Evolution 224: 97–105. 10.1007/s00427-014-0467-y [DOI] [PubMed] [Google Scholar]
- 42. Inoue K, Hoshijima K, Higuchi I, Sakamoto H, Shimura Y (1992) Binding of the Drosophila transformer and transformer-2 proteins to the regulatory elements of doublesex primary transcript for sex-specific RNA processing. Proceedings of the National Academy of Sciences 89: 8092–8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ryner LC, Goodwin SF, Castrillon DH, Anand A, Villella A, Baker BS, et al. (1996) Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell 87: 1079–1089. [DOI] [PubMed] [Google Scholar]
- 44. Cui Z, Hui M, Liu Y, Song C, Li X, Li Y et al. (2015) High density linkage mapping aided by transcriptomics documents ZW sex determination system in the Chinese mitten crab Eriocheir sinensis . Heredity 10.1038/hdy.2015.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li S, Li F, Wen R, Xiang J (2012) Identification and characterization of the sex-determiner transformer-2 homologue in Chinese shrimp, Fenneropenaeus chinensis . Sexual Development 6: 267–278. 10.1159/000341377 [DOI] [PubMed] [Google Scholar]
- 46. Mita K, Kasahara M, Sasaki S, Nagayasu Y, Yamada T, Kanamori H, et al. (2004) The genome sequence of silkworm, Bombyx mori . DNA Research 11: 27–35. [DOI] [PubMed] [Google Scholar]
- 47. Niimi T, Sahara K, Oshima H, Yasukochi Y, Ikeo K, Traut W (2006) Molecular cloning and chromosomal localization of the Bombyx Sex-lethal gene. Genome 49: 263–268. [DOI] [PubMed] [Google Scholar]
- 48. Suzuki MT (2010) Sex determination: insights from the silkworm. Journal of Genetics 89: 357–363. [DOI] [PubMed] [Google Scholar]
- 49. Doniach T, Hodgkin J (1984) A sex-determining gene, fem-1, required for both male and hermaphrodite development in Caenorhabditis elegans . Developmental Biology 106: 223–235. [DOI] [PubMed] [Google Scholar]
- 50. Kashimada K, Koopman P (2010) Sry: the master switch in mammalian sex determination. Development 137: 3921–3930. 10.1242/dev.048983 [DOI] [PubMed] [Google Scholar]
- 51. Garcia-Ortiz JE, Pelosi E, Omari S, Nedorezov T, Piao Y, Karmazin J, et al. (2009) Foxl2 functions in sex determination and histogenesis throughout mouse ovary development. BMC Developmental Biology 9: 36 10.1186/1471-213X-9-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kocer A, Pinheiro I, Pannetier M, Renault L, Parma P, Radi O, et al. (2008) R-spondin1 and FOXL2 act into two distinct cellular types during goat ovarian differentiation. BMC Developmental Biology 8: 36 10.1186/1471-213X-8-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhou L, Charkraborty T, Yu X, Wu L, Liu G, Mohapatra S, et al. (2012) R-spondins are involved in the ovarian differentiation in a teleost, medaka (Oryzias latipes). BMC Developmental Biology 12: 36 10.1186/1471-213X-12-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Adell T, Müller WE (2004) Isolation and characterization of five Fox (Forkhead) genes from the sponge Suberites domuncula . Gene 334: 35–46. [DOI] [PubMed] [Google Scholar]
- 55. Magie CR, Pang K, Martindale MQ (2005) Genomic inventory and expression of Sox and Fox genes in the cnidarian Nematostella vectensis . Development Genes and Evolution 215: 618–630. [DOI] [PubMed] [Google Scholar]
- 56. Shimeld SM, Boyle MJ, Brunet T, Luke GN, Seaver EC (2010) Clustered Fox genes in lophotrochozoans and the evolution of the bilaterian Fox gene cluster. Developmental Biology 340: 234–248. 10.1016/j.ydbio.2010.01.015 [DOI] [PubMed] [Google Scholar]
- 57. Teaniniuraitemoana V, Huvet A, Levy P, Klopp C, Lhuillier E, Gaertner-Mazouni N, et al. (2014) Gonad transcriptome analysis of pearl oyster Pinctada margaritifera: identification of potential sex differentiation and sex determining genes. BMC Genomics 15: 491 10.1186/1471-2164-15-491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Farlora R, Araya-Garay J, Gallardo-Escárate C (2014) Discovery of sex-related genes through high-throughput transcriptome sequencing from the salmon louse Caligus rogercresseyi . Marine Genomics 15: 85–93. 10.1016/j.margen.2014.02.005 [DOI] [PubMed] [Google Scholar]
- 59. Hale MC, Jackson JR, DeWoody JA (2010) Discovery and evaluation of candidate sex-determining genes and xenobiotics in the gonads of lake sturgeon (Acipenser fulvescens). Genetica 138: 745–756. 10.1007/s10709-010-9455-y [DOI] [PubMed] [Google Scholar]
- 60. Valenzuela‐Muñoz V, Bueno-Ibarra MA, Escárate CG (2014) Characterization of the transcriptomes of Haliotis rufescens reproductive tissues. Aquaculture Research 45: 1026–1040. [Google Scholar]
- 61. Cooper GM (2000) The Cell: A Molecular Approach 2nd edition Boston University. Sunderland (MA): Sinauer Associates. [Google Scholar]
- 62. El-Sankary W, Bombail V, Gibson GG, Plant N (2002) Glucocorticoid-mediated induction of CYP3A4 is decreased by disruption of a protein: DNA interaction distinct from the pregnane X receptor response element. Drug Metabolism and Disposition 30: 1029–1034. [DOI] [PubMed] [Google Scholar]
- 63. Radhika M, Nazar AA, Munuswamy N, Nellaiappan K (1998) Sex-linked differences in phenol oxidase in the fairy shrimp Streptocephalus dichotomus Baird and their possible role (Crustacea: Anostraca). Hydrobiologia 377: 161–164. [Google Scholar]
- 64. Kurtz J, Wiesner A, Götz P, Sauer KP (2000) Gender differences and individual variation in the immune system of the scorpionfly Panorpa vulgaris (Insecta: Mecoptera). Developmental and Comparative Immunology 24: 1–12. [DOI] [PubMed] [Google Scholar]
- 65. Kurtz J, Sauer KP (2001) Gender differences in phenoloxidase activity of Panorpa vulgaris hemocytes. Journal of Invertebrate Pathology 78: 53–55. [DOI] [PubMed] [Google Scholar]
- 66. Rolff J (2001) Effects of age and gender on immune function of dragonflies (Odonata, Lestidae) from a wild population. Canadian Journal of Zoology 79: 2176–2180. [Google Scholar]
- 67. Lindsey E, Altizer S (2009) Sex differences in immune defenses and response to parasitism in monarch butterflies. Evolutionary Ecology 23: 607–620. [Google Scholar]
- 68. Iwanaga S, Lee BL (2005) Recent advances in the innate immunity of invertebrate animals. BMB Reports 38: 128–150. [DOI] [PubMed] [Google Scholar]
- 69. Akira S, Uematsu S, Takeuchi O (2006) Pathogen recognition and innate immunity. Cell 124: 783–801. [DOI] [PubMed] [Google Scholar]
- 70. Raikhel A, Brown M, Belles X (2005) Hormonal control of reproductive processes. Comprehensive Molecular Insect Science 3: 433–491. [Google Scholar]
- 71. Zhang S, Wang S, Li H, Li L (2011) Vitellogenin, a multivalent sensor and an antimicrobial effector. The International Journal of Biochemistry & Cell Biology 43: 303–305. [DOI] [PubMed] [Google Scholar]
- 72. Flouriot G, Pakdel F, Ducouret B, Valotaire Y (1995) Influence of xenobiotics on rainbow trout liver estrogen receptor and vitellogenin gene expression. Journal of Molecular Endocrinology 15: 143–151. [DOI] [PubMed] [Google Scholar]
- 73. Palmer BD, Palmer SK (1995) Vitellogenin induction by xenobiotic estrogens in the red-eared turtle and African clawed frog. Environmental Health Perspectives 103: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Piulachs M, Guidugli K, Barchuk A, Cruz J, Simoes Z, Belles X (2003) The vitellogenin of the honey bee, Apis mellifera: structural analysis of the cDNA and expression studies. Insect Biochemistry and Molecular Biology 33: 459–465. [DOI] [PubMed] [Google Scholar]
- 75. Guidugli KR, Piulachs MD, Bellés X, Lourenço AP, Simões ZL (2005) Vitellogenin expression in queen ovaries and in larvae of both sexes of Apis mellifera. Archives of Insect Biochemistry and Physiology 59: 211–218. [DOI] [PubMed] [Google Scholar]
- 76. Tsutsui N, Kawazoe I, Ohira T, Jasmani S, Yang W-J, Wilder MN, et al. (2000) Molecular characterization of a cDNA encoding vitellogenin and its expression in the hepatopancreas and ovary during vitellogenesis in the kuruma prawn, Penaeus japonicus . Zoological Science 17: 651–660. 10.2108/zsj.15.651 [DOI] [PubMed] [Google Scholar]
- 77. Shechter A, Aflalo ED, Davis C, Sagi A (2005) Expression of the reproductive female-specific vitellogenin gene in endocrinologically induced male and intersex Cherax quadricarinatus crayfish. Biology of Reproduction 73: 72–79. [DOI] [PubMed] [Google Scholar]
- 78. Zmora N, Trant J, Chan S-M, Chung JS (2007) Vitellogenin and its messenger RNA during ovarian development in the female blue crab, Callinectes sapidus: gene expression, synthesis, transport, and cleavage. Biology of Reproduction 77: 138–146. [DOI] [PubMed] [Google Scholar]
- 79. Huang X, Ye H, Huang H, Yang Y, Gong J (2014) An insulin-like androgenic gland hormone gene in the mud crab, Scylla paramamosain, extensively expressed and involved in the processes of growth and female reproduction. General and Comparative Endocrinology 204: 229–238. 10.1016/j.ygcen.2014.06.002 [DOI] [PubMed] [Google Scholar]
- 80. Chung JS, Manor R, Sagi A (2011) Cloning of an insulin-like androgenic gland factor (IAG) from the blue crab, Callinectes sapidus: Implications for eyestalk regulation of IAG expression. General and Comparative Endocrinology 173: 4–10. 10.1016/j.ygcen.2011.04.017 [DOI] [PubMed] [Google Scholar]
- 81. Li S, Li F, Sun Z, Xiang J (2012) Two spliced variants of insulin-like androgenic gland hormone gene in the Chinese shrimp, Fenneropenaeus chinensis . General and Comparative Endocrinology 177: 246–255. 10.1016/j.ygcen.2012.04.010 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
For each unigene, the ratio of expression levels (Female vs. Male) is plotted against the-log error rate. The horizontal line indicates the significance threshold (FDR adjusted < 0.05), and the vertical lines indicate the two fold change threshold. Non-differentially expressed genes are shown with orange dots, and DEGs are shown with blue dots.
(TIF)
The horizontal line indicates the significance threshold (P < 0.05).
(TIF)
The up-regulated in male are labeled by green, and the purple color represents genes with no expression differences between female and male Eriocheir sinensis transcriptomes.
(TIF)
B and A chains are marked in yellow and green boxes, respectively. The six conserved cysteine residues are highlighted with dark red background and the predicted disulfide bridges are drawn. The species and the GenBank accession numbers are as follow: Callinectes sapidus IAG1(AEI72263), C. sapidus IAG2 (AHM93481), Cherax destructor (ACD91988), Cherax quadricarinatus (ABH07705), Fenneropenaeus chinensis IAG1(AFU60548), F. chinensis IAG2 (AFU60549), Jasus edwardsii (AIM55892), Litopenaeus vannamei (AIR09497), Macrobrachium lar (BAJ78349), Macrobrachium nipponense IAG1 (AGB56976), Macrobrachium nipponense IAG2 (AHA33389), Macrobrachium rosenbergii (ACJ38227), Macrobrachium vollenhovenii (AHZ34725), Marsupenaeus japonicus (BAK20460), Palaemon pacificus (BAJ84109), Palaemon paucidens (BAJ84108), Penaeus monodon (ADA67878), Portunus pelagicus (ADK46885), Sagmariasus verreauxi (AHY99679) and Scylla paramamosain (AIF30295). Three isopods Armadillidium vulgare (BAA86893), Porcellio dilatatus (BAC57013), Porcellio scabar (BAC57012) served as outgroups.
(TIF)
(DOC)
(DOC)
(DOC)
(XLS)
(DOC)
(DOC)
(XLS)
Data Availability Statement
All data were deposited into the Sequence Read Archive (SRA) database (http://www.ncbi.nlm.nih.gov/Traces/sra/) under accession numbers SRX554564 and SR554562 and all other relevant data are within the paper and its Supporting Information files.