Abstract
The development and regeneration of skeletal muscles require the fusion of mononulceated muscle cells to form multinucleated, contractile muscle fibers. Studies using a simple genetic model, Drosophila melanogaster, have discovered many evolutionarily conserved fusion-promoting factors in vivo. Recent work in zebrafish and mouse also identified several vertebrate-specific factors required for myoblast fusion. Here, we integrate progress in multiple in vivo systems and highlight conceptual advance in understanding how muscle cell membranes are brought together for fusion. We focus on the molecular machinery at the fusogenic synapse and present a three-step model to describe the molecular and cellular events leading to fusion pore formation.
Introduction
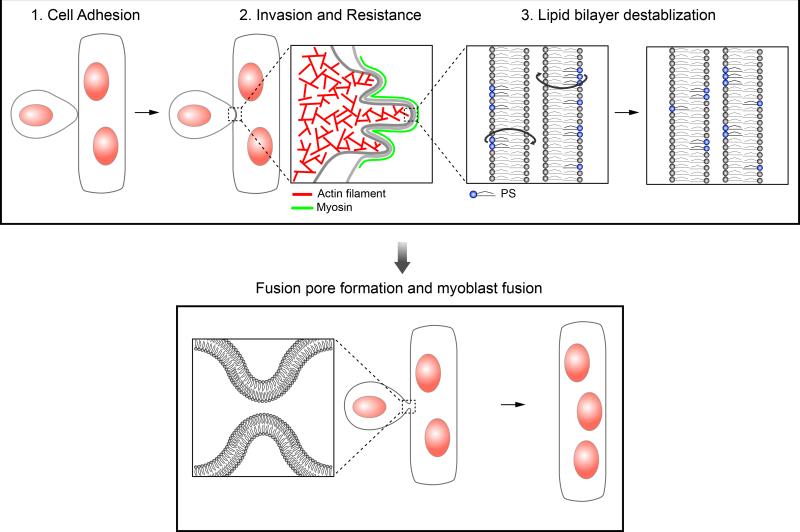
Skeletal muscle is a unique tissue composed of bundles of multinucleated muscle fibers. Each myofiber is the product of fusion of hundreds or thousands of mononucleated muscle cells known as myoblasts. Myoblast fusion is critical not only for skeletal muscle development during embryogenesis, but also for satellite cell-mediated muscle regeneration in adults [1,2]. For myoblast fusion to occur, two fusion partners must recognize each other, adhere their plasma membranes, open up fusion pores to allow cytoplasmic material exchange and, ultimately, merge into one cell. As with any membrane fusion event, the rate-limiting step for successful myoblast fusion is bringing two cell membranes into close proximity to facilitate fusion pore formation. Recent studies in multiple model organisms, including Drosophila, zebrafish and mouse, have uncovered many molecular components required for myoblast fusion in vivo [3]. Mechanistic studies of these components suggest that muscle cells take at least three consecutive steps toward fusion pore formation (Figure 1) – first, muscle cell adhesion mediated by cell adhesion molecules (CAMs); second, closer cell membrane apposition mediated by a pair of pushing and resisting forces from the two fusion partners; and third, destabilization of the lipid bilayers, which makes them prone to fusion. Here, we review the in vivo evidence from Drosophila, zebrafish, and mouse that supports the three-step model of myoblast fusion. Insights from myoblast fusion are likely to apply to other cell-cell fusion events, such as fusion between macrophages, osteoclasts, as well as sperm and egg.
Figure 1. The three-step model describing the cellular events leading to fusion pore formation in myoblast fusion.
Step 1 – The initial cell adhesion between a founder cell and an FCM is mediated by Ig domain-containing CAMs. Step 2 – Following cell adhesion, the FCM generates actin-propelled membrane protrusions to invade the apposing founder cell, the latter of which mounts a myosin II-mediated mechanosensory response to increase the cortical tension and therefore cortical resistance to the FCM invasion. Step 3 – once the two plasma membranes are brought into close proximity by the invasive and resistance forces, the lipid bilayers are destabilized, leading to the formation of a fusion pore. A hypothetical scenario depicted here is that PS could be flipped from the inner to the outer leaflet, resulting in a more disorganized outer leaflet prone to fusion.
The first step toward myoblast fusion – recognition and adhesion between muscle cells
Drosophila
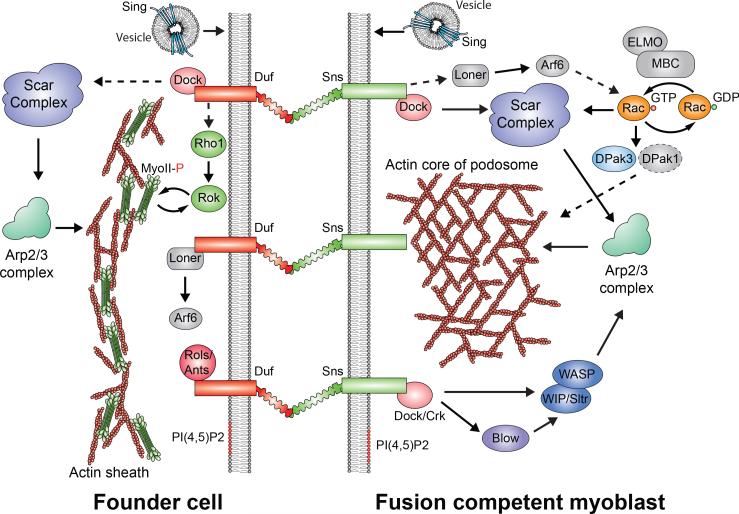
In Drosophila embryos, myoblast fusion occurs between two types of muscle cells, muscle founder cell and fusion competent myoblasts (FCMs), the fates of which are specified by the action of transcription factors [4,5]. Muscle founder cells act as “seeds” that attract the FCMs and ultimately determine the position, orientation, size, epidermal attachment and nerve innervation pattern of the future multinucleated muscle fibers. Recognition and adhesion between founder cells and FCMs are mediated by immunoglobulin (Ig) domain-containing CAMs (type I transmembrane protein) (Figure 2). In founder cells, two paralogs - Dumbfounded (Duf), also known as Kin-of-IrreC (Kirre), and Roughest (Rst), also known as IrreC - have redundant functions in myoblast fusion [6,7]. In FCMs, Sticks and stones (Sns) acts as the major CAM, whose function is partially compensated by its paralog Hibris (Hbs) [8-11]. Similar to muscle development in embryos, multinucleated Drosophila adult muscles are seeded by Duf-Rst-expressing founder cells, which attract the surrounding Sns-Hbs-expressing FCMs to fuse during the pupal stage [12,13].
Figure 2. Signaling pathways leading to asymmetric F-actin and myosin accumulation in Drosophila myoblast fusion.
Trans-interaction between cell type-specific CAMs, such as Duf (founder cell; with 5 Ig domains) and Sns (FCM; with 8 Ig domains), triggers distinct signaling events in founder cells and FCMs. Briefly, in the founder cell, activation of Scar results in the formation of a thin sheath of F-actin at the fusogenic synapse. In addition, the Rho1-Rok-MyoII pathway amplifies the mechanosensitive accumulation of MyoII at the fusogenic synapse. In the FCM, activation of both Scar and WASP leads to the formation of an F-actin-enriched focus, which is part of an invasive podosome-like structure. For the functions of other components shown here, see the text.
Ectopic expression of either Duf or Rst in embryonic epithelial cells can redirect FCMs to these ectopic locations, demonstrating the ability of these Ig domain-containing CAMs to attract FCMs [6,7]. However, these CAMs are not sufficient to induce fusion, since overexpressing these proteins in heterologous cells causes cell adhesion but not cell fusion [14]. Consistent with this observation, recent X-ray crystallographic studies of the C. elegans homologs of Duf and Sns, SYG-1 and SYG-2, have shown that their trans-interaction is mediated by the most N-terminal Ig domains, and that their ectodomains form an L-shaped rigid structure propping the two apposing membranes ~45 nm apart [15], a distance too large for cell membrane fusion.
Vertebrates
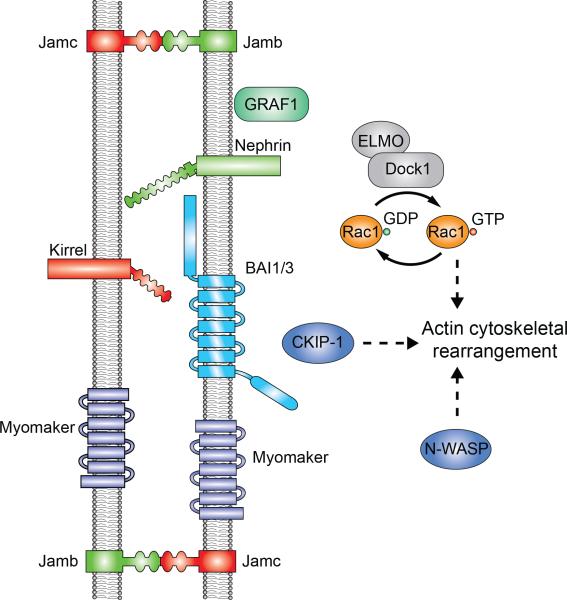
Although it remains unclear whether two types of muscle cells are involved in myoblast fusion in vertebrates, studies in zebrafish have implicated a function of a Duf homolog, Kirrel (also known as Kirrel3l), in the fusion of muscle cells to form the multinucleated fast twitch muscle fibers [16] (Figure 3). Kirrel is expressed in the fast muscle myotome, and morpholino treatment of kirrel resulted in mononucleated muscle cells. Notably, these mononucleated cells do not seem to properly attach to the myoseptal boundary as do multinucleated myofibers in wild-type embryos [16], suggesting that Kirrel may have an additional function in muscle cell attachment. Besides Kirrel, the potential function of the Sns homolog, Nephrin, in zebrafish muscle development has been explored [17]. However, the knockdown level of nephrin by morpholino was less than 50%, preventing a proper assessment of its loss-of-function phenotype [17]. On the other hand, cultured primary myoblasts from nephrin knockout mice have a decreased ability to fuse, suggesting that Nephrin may be involved in mouse myoblast fusion [17].
Figure 3. Signaling components required for vertebrate myoblast fusion in vivo.
Due to the lack of molecular and cellular markers for the fusogenic synapse in vertebrates, the localization and potential sidedness of these proteins are hypothetical.
In addition to Kirrel and Nephrin, a pair of vertebrate-specific Ig domain-containing transmembrane proteins, Jamb and Jamc (also known as Jam2a and Jam3b), have been shown to play a role in zebrafish myoblast fusion (Figure 3). Jamb and Jamc are highly expressed in the developing zebrafish myotome, and myoblast fusion fails to occur in mutant embryos lacking either Jamb or Jamc, as evidenced by the presence of mononucleated fast muscle fibers [18]. Transplantation experiments between jamb and jamc mutants demonstrate that trans-heterophilic interaction between the two CAMs is essential for myoblast fusion [18]. Similar to Duf and Sns, Jamb and Jamc induce strong cell adhesion but not cell-cell fusion when expressed in heterologous cells (Chen lab, unpublished). It remains to be determined whether Jamb and Jamc are localized and function at sites of fusion, how Jamb and Jamc relate to Kirrel, and what signaling components function downstream of Jamb and Jamc to regulate myoblast fusion.
Besides these Ig domain-containing CAMs, a cell-matrix adhesion molecule, beta1 integrin, is required for myoblast fusion in mouse embryos [19]. However, its localization relative to the fusion sites and its precise function during the fusion process remain unknown.
The second step toward myoblast fusion – enhancing cell membrane proximity
It is apparent that cell adhesion molecules are not sufficient to bring the membranes of fusing partners into close enough proximity required for fusion. Recent studies have revealed that the two fusion partners rearrange their actin cytoskeleton and the actomyosin network to achieve greater membrane proximity.
FCM: Generating an F-actin-enriched invasive podosome-like structure
Drosophila
Genetic studies in Drosophila led to the identification of many actin cytoskeletal regulators in myoblast fusion. Interestingly, most of these regulators affect actin polymerization mediated by the Arp2/3 complex, a seven-subunit protein complex that nucleates branched actin filaments [20-22] (Figure 2). These include WASP and Scar, two actin nucleation-promoting factors (NPFs) of the Arp2/3 complex [20,21,23–25], the WASP-interacting protein, WIP/Solitary (Sltr) [20,24], and Kette/Nap1, a subunit of the pentameric Scar complex [26]. Upstream of the Scar complex are the small GTPase Rac [27,28], an activator of Scar, and Rac’s bipartite guanine nucleotide exchange factor (GEF) Mbc-Elmo [29–31]. The localization of Rac is controlled by the small GTPase Arf6 and its GEF Loner [32]. The Scar and WASP-WIP complexes are linked to the CAMs by SH2–SH3 domain-containing adaptor proteins, Crk and Dock [24,33,34], thus responding to the fusion signal in a temporally and spatially controlled manner. Strikingly, some of these molecular components function in a cell type-specific manner. For example, WASP-WIP and Mbc are only required in FCMs, whereas the Scar complex functions in both founder cells and FCMs [35,36].
Since genetic studies clearly demonstrated a role for Arp2/3-mediated actin polymerization in myoblast fusion, major efforts have been directed to understanding the precise function of actin cytoskeletal rearrangement in the fusion process. Immunohistochemical studies have revealed a dense, roundish F-actin-enriched focus at the site of fusion [21,24,37], and live imaging studies have demonstrated that each fusion event is mediated by an F-actin focus, a transient structure with an average life span of ~ 11 min [21,35]. Initially, it was suggested that the F-actin focus was symmetrically partitioned into the adherent founder cell and FCM at the site of fusion [21]. However, subsequent genetic, cell biological and electron microscopy studies unambiguously localized the actin focus in the FCM, thus revealing the striking asymmetry of F-actin accumulation at the site of fusion [35] (Figure 1). Similar F-actin asymmetry is observed in cultured Drosophila myoblasts [36] and in pupal muscle cells undergoing fusion [38]. Such cellular asymmetry results from the asymmetric requirement of the Arp2/3 NPFs. While Scar and WASP function in the FCM to generate the dense F-actin focus, Scar is required in the founder cell to generate a thin sheath of actin at the site of fusion [35]. Strikingly, the roundish actin focus observed using confocal microscopy causes an inward curvature on the founder cell membrane, and is actually composed of a cluster of invasive finger-like protrusions as revealed by electron microscopy [35]. When the confocal section is perpendicular to the axis of the invasive protrusions, the actin focus appears to be encircled by the cell adhesion molecules, Duf and Sns [35,37]. Such a bipartite configuration with an actin core encircled by adhesion molecules resembles that of a podosome, a dynamic cell–matrix adhesion structure mostly studied in cultured cells. Thus, this actin-enriched structure at the tip of the FCM is named a podosome-like structure (PLS), and the interface between two fusion partners, which comprises trans-interacting cell adhesion molecules and an invasive PLS from the FCM, is called a fusogenic synapse [35,39].
Genetic analyses have shown that proper PLS invasion is essential for fusion pore formation. In different fusion mutants where PLS invasion is compromised, fusion pores fail to form [33,35,40]. It has been proposed that PLS invasion increases the surface contact areas of the apposing fusion partners and pushes the two membranes into closer proximity to facilitate cell membranefusion [35, 39]. In support of this, actin-propelled invasive membrane protrusions have been shown to promote fusogenic protein engagement in a reconstituted cell fusion culture system [14]. Several proteins have been shown to regulate PLS invasion through distinct mechanisms. First, in wip/sltr mutant embryos, actin-enriched fingers form but fail to protrude into founder cells, suggesting that the WASP–WIP complex is required for PLS invasion [35]. Second, mutations in the PH domain containing FCM-specific protein Blown fuse (Blow) lead to the formation of shorter and smaller PLS protrusions [33]. Blow regulates the stability of the WASP–WIP complex by competing with WASP for WIP binding (Figure 2). The dissociation of the WASP–WIP complex, in turn, increases the dynamics of actin polymerization within the PLS to promote fusion [33]. Thus, it is the dynamics, rather than the amount, of actin polymerization that drives fusion pore formation. Third, mutations in the Drosophila group I p21-activated kinases (DPaks) also compromise PLS invasion [40]. DPak1 and DPak3 are redundantly required in FCMs for myoblast fusion with DPak3 playing a major role (Figure 2). The DPaks function downstream of the small GTPase Rac, and likely regulate the crosslinking and/or bundling of branched actin filaments within the PLS, although the substrate(s) of the DPaks in myoblast fusion remain unknown.
An additional way to regulate actin polymerization at the fusogenic synapse is by increasing the concentration of phospholipids. Phosphatidylinositol-(4,5)-bisphosphate, PI(4,5)P2, is enriched at both sides of the fusogenic synapse [41] (Figure 2). It has been reported that overexpression of the PH domain of PLCγ, which binds PI(4,5)P2, or a kinase-dead form of PI(4)-5-kinase in Drosophila embryos inhibited myoblast fusion, presumably by blocking the interaction between PI(4,5)P2 and its endogenous downstream targets or sequestering the wild-type PI(4)-5-kinase. In these abnormal embryos, actin regulators, such as Rac, WASP and Scar, show altered localization and the F-actin foci size is reduced, suggesting that PI(4,5)P2 promotes actin polymerization by recruiting actin regulators to the fusion sites [41]. However, it is unclear whether PI(4,5)P2 affects PLS invasion and loss-of-function studies of the PI(4)-5-kinases are required to validate the endogenous function of PI(4,5)P2 in myoblast fusion.
Vertebrates
Many of the actin polymerization regulators identified in Drosophila have been shown to play a conserved role in vertebrate myoblast fusion in vivo (Figure 3). These include the Mbc homolog DOCK1 and DOCK5 in zebrafish and mouse [42,43], the small GTPase Rac1 in zebrafish and mouse [42,44], and N-WASP in mouse [45], suggesting that vertebrate myoblast fusion may involve similar actin cytoskeletal rearrangements as in Drosophila, although the subcellular localization and the potential sidedness of these proteins have not been investigated. In addition, Baas et al. have identified a role for Casein kinase 2 interacting protein-1 (CKIP-1), a phospholipids binding protein, in fast muscle development in zebrafish embryos [46]. Morpholino knockdown of ckip-1 impairs fast muscle myoblast fusion and elongation without affecting muscle differentiation. Ckip-1 binds to actin capping protein and ArpC1A subunit of the Arp2/3 complex, which, in turn, regulates actin cytoskeletal dynamics. It remains unclear whether Ckip-1 regulates actin polymerization at the fusion sites and how it may coordinate the dynamics of phospholipids and the actin cytoskeleton in the fusion process.
Founder cell: increasing cortical tension via Myosin IImediated mechanosensory response
Studies of actin polymerization in FCMs have firmly established the concept of an “asymmetric fusogenic synapse” in myoblast fusion. However, it was not clear how founder cells respond to the PLS invasion from FCMs. Kim et al. have shown that the founder cell mounts a Myosin II (MyoII)-mediated mechanosensory response to its invasive fusion partner in Drosophila embryos and a reconstituted cell-fusion culture system [47] (Figure 1 and 2). MyoII acts as a mechanosensor, which directs its force-induced recruitment to the fusogenic synapse, and the mechanosensory response of MyoII is amplified by Duf-initiated chemical signaling through Rho1 and Rok. The accumulated MyoII, in turn, increases cortical tension and promotes fusion pore formation [47]. Thus, the protrusive and resisting forces from the FCM and the apposing founder cell, respectively, put the fusogenic synapse under high mechanical tension, which helps to overcome energy barriers for membrane apposition and drives cell membrane fusion. Interestingly, another myosin molecule, myosin 18, also accumulates at the fusogenic synapse in the founder cell [48], raising the possibility that the two myosins may function together to modulate cortical tension in founder cells. Future studies are required to investigate the potential functions of Rho, Rok and MyoII in vertebrate myoblast fusion in vivo.
The third step toward myoblast fusion – destabilizing the lipid bilayer
Once the two muscle cell membranes are brought into close proximity by the interplay between the protrusive and resisting forces from the two fusion partners, the lipid bilayer needs to be destabilized to facilitate fusion pore formation. Studies in Drosophila have yet to reveal any molecular components that directly facilitate this step of myoblast fusion. However, studies in cultured mouse C2C12 myoblasts in vitro suggested that transient exposure of phosphatidylserine (PS) on the cell surface may play a role in myoblast fusion [49,50]. Consistent with this hypothesis, Hochreiter-Hufford et al. identified a function for Brain-specific angiogenesis inhibitor 1 (BAI1), a cell surface receptor for PS, in myoblast fusion in vivo [51] (Figure 3). The BAI1 knockout mice exhibit a reduction in their ability to repair muscle without displaying major defects in muscle development. BAI1 is a member of the adhesion G-protein-coupled receptor family that recognizes PS exposure on apoptotic cells and initiates signaling cascade through the ELMO/Dock180/Rac1 pathway [52]. Recent studies of another BAI family member, BAI3, revealed a role for this protein in myoblast fusion during skeletal muscle development in chick embryos [53]. Similar to BAI1, BAI3 promotes myoblast fusion through its binding to ELMO. The functional requirement of BAI family members in skeletal muscle development and regeneration, therefore, indirectly supports a role for PS exposure in myoblast fusion in vivo. Given that myoblast fusion is a tightly regulated process, PS exposure is likely one of the many changes within the lipid bilayer leading to fusion pore formation. Future investigations are required to identify additional mechanisms underlying lipid destabilization and fusion pore formation.
Other cellular processes during myoblast fusion
Exocytosis
Electron microscopy (EM) and genetic analyses in Drosophila have indicated a role of exocytosis in myoblast fusion. EM analyses of Drosophila embryos revealed vesicles with electron-dense rims at muscle cell contact sites [20,22,24-26,54-56]. These vesicles appear to bud from the Golgi and traffic on microtubules [24]. Although similar vesicles have not been observed in the vicinity of actin-propelled membrane protrusions at the asymmetric fusogenic synapse, they may be involved in earlier exocytic events that transport cell adhesion molecules and/or fusogenic proteins to the plasma membrane. The molecular composition of these vesicles requires future investigation. Another piece of evidence to support the involvement of vesicle trafficking in myoblast fusion is the finding that a founder cell-specific adaptor protein, Antisocial/Rolling pebbles (Ants/Rols) [55,57,58], functions to replenish the cell adhesion molecule Duf by translocating Duf in exocytic vesicles [59]. Moreover, genetics studies have implicated Singles bar (Sing), a MARVEL domain-containing four-pass transmembrane protein, in myoblast fusion [56] (Figure 2). In sing mutant embryos, myoblast fusion is severely blocked, whereas muscle cell differentiation, migration and attachment appear normal. Since MARVEL domains are often found in lipid-associating proteins that are part of transport vesicles, Sing may be involved in vesicle trafficking during myoblast fusion. However, the precise localization and function of Sing in the fusion process remains to be explored.
Endocytosis
A role for endocytosis has been recently shown in myoblast fusion. Shin et al. depleted dynamin, a large GTPase responsible for endocytosis, in primary myoblasts isolated from dynamin 1 and 2 double conditional knockout mice and observed a significant decrease in myotube formation [60]. This result is consistent with those of RNAi knockdown of dynamin in cultured C2C12 cells and primary myoblasts [61]. Blocking endocytosis by other means also decreased C2C12 myoblast fusion [60]. Despite these findings, the specific function of endocytosis in myoblast fusion in vivo is yet to be established.
Membrane curvature modulation
Given the presence of finger-like protrusions at the fusogenic synapse, it is conceivable that curvature-binding proteins may be required to stabilize these protrusions and promote myoblast fusion. Although a function of such proteins has not been revealed in Drosophila, a Rho- GTPase activating protein (GAP), GRAF1, which contains a lipid-binding BAR domain, a PH domain, a Rho-GAP domain and an SH3 domain, has been shown to play a role in mouse myoblast fusion in vivo [62] (Figure 3). GRAF1-depleted mice exhibit a significant and persistent reduction in cross-sectional area of post-natal muscles, indicative of a myoblast fusion defect. In contrast, overexpression of GRAF1 in cultured myoblasts induced robust fusion, which is dependent on both GAP and BAR domains. Whether the BAR domain in GRAF1 affects plasma membrane curvature in myoblast fusion is currently unknown.
Additional membrane-associated factor
Recent loss-of-function studies in mouse have identified an essential function for Myomaker (Tmem8c), a multi-pass transmembrane protein, in myoblast fusion [63] (Figure 3). Myomaker is exclusively expressed in the developing skeletal muscle. Embryonic skeletal muscle tissue in myomaker knockout mice contains only mononucleated myoblasts that are properly differentiated, demonstrating an indispensible function for Myomaker in myoblast fusion in vivo. Although Myomaker is undetectable in adult skeletal muscle, it is transiently induced to express in satellite cells after muscle injury [64]. Conditional knockout of Myomaker in satellite cells results in a complete block of muscle regeneration [64]. Thus, Myomaker is essential for both skeletal muscle development and regeneration. In addition, the Myomaker homolog in zebrafish has also been implicated in myoblast fusion, revealing a functional conservation of this protein in vertebrates [65]. Interestingly, although Myomaker is not sufficient to induce fusion in fibroblasts by itself, overexpressing Myomaker in fibroblasts makes these fibroblasts prone to fusion with myoblasts [63]. Future investigations are required to pinpoint Myomaker’s precise function in myoblast fusion.
Concluding remarks
The past decade has witnessed unprecedented progress in our understanding of myoblast fusion, owing to the application of multifaceted experimental approaches and studies in multiple genetically amenable model systems. The discovery of the asymmetric fusogenic synapse has overturned the conventional view that myoblast fusion is a symmetrical process with equal contributions from both fusion partners. A biophysical framework has emerged in that protrusive and resisting forces from the two fusion partners put the fusogenic synapse under high mechanical tension to drive cell membrane fusion. Many outstanding questions concerning the mechanisms of myoblast fusion still remain, For example, how exactly are the lipid bilayers destabilized at the fusogenic synapse? How are proteins and lipids organized into specific microdomains on the plasma membrane along the invasive protrusions? What is the identity of the elusive myoblast fusogen? Answers to these questions and many others posed throughout this review will continue to bring excitement to this research field in the next decade.
Acknowledgements
We thank members of the Chen lab for discussions and Khurts Shilagardi for comments on the manuscript. We apologize to colleagues whose original work is not cited because of space limits. Supported by NIH/NIAMS R01AR053173, NIH/NIGMS R01GM098816, the Muscular Dystrophy Association and a National Established Investigator Award from the American Heart Association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
*of special interest, **of outstanding interest
- 1.Buckingham M. Myogenic progenitor cells and skeletal myogenesis in vertebrates. Curr Opin Genet Dev. 2006;16:525–532. doi: 10.1016/j.gde.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 2.Wagers AJ, Conboy IM. Cellular and molecular signatures of muscle regeneration: current concepts and controversies in adult myogenesis. Cell. 2005;122:659–667. doi: 10.1016/j.cell.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 3*.Abmayr SM, Pavlath GK. Myoblast fusion: lessons from flies and mice. Development. 2012;139:641–656. doi: 10.1242/dev.068353. [A comprehensive review of components of myoblast fusion uncovered using Drosophila genetics, mouse genetics, and cultured mouse myoblasts.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen EH, Olson EN. Towards a molecular pathway for myoblast fusion in Drosophila. Trends Cell Biol. 2004;14:452–460. doi: 10.1016/j.tcb.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 5.Tixier V, Bataille L, Jagla K. Diversification of muscle types: recent insights from Drosophila. Exp Cell Res. 2010;316:3019–3027. doi: 10.1016/j.yexcr.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 6.Ruiz-Gomez M, Coutts N, Price A, Taylor MV, Bate M. Drosophila dumbfounded: a myoblast attractant essential for fusion. Cell. 2000;102:189–198. doi: 10.1016/s0092-8674(00)00024-6. [DOI] [PubMed] [Google Scholar]
- 7.Strunkelnberg M, Bonengel B, Moda LM, Hertenstein A, de Couet HG, Ramos RG, Fischbach KF. rst and its paralogue kirre act redundantly during embryonic muscle development in Drosophila. Development. 2001;128:4229–39. doi: 10.1242/dev.128.21.4229. [DOI] [PubMed] [Google Scholar]
- 8.Bour BA, Chakravarti M, West JM, Abmayr SM. Drosophila SNS, a member of the immunoglobulin superfamily that is essential for myoblast fusion. Genes Dev. 2000;14:1498–1511. [PMC free article] [PubMed] [Google Scholar]
- 9.Dworak HA, Charles MA, Pellerano LB, Sink H. Characterization of Drosophila hibris, a gene related to human nephrin. Development. 2001;128:4265–4276. doi: 10.1242/dev.128.21.4265. [DOI] [PubMed] [Google Scholar]
- 10.Artero RD, Castanon I, Baylies MK. The immunoglobulin-like protein Hibris functions as a dose-dependent regulator of myoblast fusion and is differentially controlled by Ras and Notch signaling. Development. 2001;128:4251–64. doi: 10.1242/dev.128.21.4251. [DOI] [PubMed] [Google Scholar]
- 11.Shelton C, Kocherlakota KS, Zhuang S, Abmayr SM. The immunoglobulin superfamily member Hbs functions redundantly with Sns in interactions between founder and fusion-competent myoblasts. Development. 2009;136:1159–1168. doi: 10.1242/dev.026302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dutta D, Anant S, Ruiz-Gomez M, Bate M, VijayRaghavan K. Founder myoblasts and fibre number during adult myogenesis in Drosophila. Development. 2004;131:3761–3772. doi: 10.1242/dev.01249. [DOI] [PubMed] [Google Scholar]
- 13.Gildor B, Schejter ED, Shilo BZ. Bidirectional Notch activation represses fusion competence in swarming adult Drosophila myoblasts. Development. 2012;139:4040–4050. doi: 10.1242/dev.077495. [DOI] [PubMed] [Google Scholar]
- 14**.Shilagardi K, Li S, Luo F, Marikar F, Duan R, Jin P, Kim JH, Murnen K, Chen EH. Actin-propelled invasive membrane protrusions promote fusogenic protein engagement during cell-cell fusion. Science. 2013;340:359–363. doi: 10.1126/science.1234781. [The first reconstitution of high efficiency cell-cell fusion using a non-fusing cell line, revealing that actin-propelled membrane protrusions are used as a general mechanism in cell-cell fusion.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ozkan E, Chia PH, Wang RR, Goriatcheva N, Borek D, Otwinowski Z, Walz T, Shen K, Garcia KC. Extracellular architecture of the SYG-1/SYG-2 adhesion complex instructs synaptogenesis. Cell. 2014;156:482–494. doi: 10.1016/j.cell.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srinivas BP, Woo J, Leong WY, Roy S. A conserved molecular pathway mediates myoblast fusion in insects and vertebrates. Nat Genet. 2007;39:781–786. doi: 10.1038/ng2055. [DOI] [PubMed] [Google Scholar]
- 17.Sohn RL, Huang P, Kawahara G, Mitchell M, Guyon J, Kalluri R, Kunkel LM, Gussoni E. A role for nephrin, a renal protein, in vertebrate skeletal muscle cell fusion. Proc Natl Acad Sci U S A. 2009;106:9274–9279. doi: 10.1073/pnas.0904398106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Powell GT, Wright GJ. Jamb and jamc are essential for vertebrate myocyte fusion. PLoS Biol. 2011;9:e1001216. doi: 10.1371/journal.pbio.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwander M, Leu M, Stumm M, Dorchies OM, Ruegg UT, Schittny J, Muller U. Beta1 integrins regulate myoblast fusion and sarcomere assembly. Dev Cell. 2003;4:673–685. doi: 10.1016/s1534-5807(03)00118-7. [DOI] [PubMed] [Google Scholar]
- 20.Massarwa R, Carmon S, Shilo BZ, Schejter ED. WIP/WASp-based actin-polymerization machinery is essential for myoblast fusion in Drosophila. Dev Cell. 2007;12:557–569. doi: 10.1016/j.devcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 21.Richardson BE, Beckett K, Nowak SJ, Baylies MK. SCAR/WAVE and Arp2/3 are crucial for cytoskeletal remodeling at the site of myoblast fusion. Development. 2007;134:4357–4367. doi: 10.1242/dev.010678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berger S, Schafer G, Kesper DA, Holz A, Eriksson T, Palmer RH, Beck L, Klambt C, Renkawitz-Pohl R, Onel SF. WASP and SCAR have distinct roles in activating the Arp2/3 complex during myoblast fusion. J Cell Sci. 2008;121:1303–1313. doi: 10.1242/jcs.022269. [DOI] [PubMed] [Google Scholar]
- 23.Schafer G, Weber S, Holz A, Bogdan S, Schumacher S, Muller A, Renkawitz-Pohl R, Onel SF. The Wiskott-Aldrich syndrome protein (WASP) is essential for myoblast fusion in Drosophila. Dev Biol. 2007;304:664–674. doi: 10.1016/j.ydbio.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 24.Kim S, Shilagardi K, Zhang S, Hong SN, Sens KL, Bo J, Gonzalez GA, Chen EH. A critical function for the actin cytoskeleton in targeted exocytosis of prefusion vesicles during myoblast fusion. Dev Cell. 2007;12:571–586. doi: 10.1016/j.devcel.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 25.Gildor B, Massarwa R, Shilo BZ, Schejter ED. The SCAR and WASp nucleation-promoting factors act sequentially to mediate Drosophila myoblast fusion. EMBO Rep. 2009 doi: 10.1038/embor.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schroter RH, Lier S, Holz A, Bogdan S, Klambt C, Beck L, Renkawitz-Pohl R. kette and blown fuse interact genetically during the second fusion step of myogenesis in Drosophila. Development. 2004;131:4501–4509. doi: 10.1242/dev.01309. [DOI] [PubMed] [Google Scholar]
- 27.Luo L, Liao YJ, Jan LY, Jan YN. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- 28.Hakeda-Suzuki S, Ng J, Tzu J, Dietzl G, Sun Y, Harms M, Nardine T, Luo L, Dickson BJ. Rac function and regulation during Drosophila development. Nature. 2002;416:438–442. doi: 10.1038/416438a. [DOI] [PubMed] [Google Scholar]
- 29.Rushton E, Drysdale R, Abmayr SM, Michelson AM, Bate M. Mutations in a novel gene, myoblast city, provide evidence in support of the founder cell hypothesis for Drosophila muscle development. Development. 1995;121:1979–1988. doi: 10.1242/dev.121.7.1979. [DOI] [PubMed] [Google Scholar]
- 30.Erickson MR, Galletta BJ, Abmayr SM. Drosophila myoblast city encodes a conserved protein that is essential for myoblast fusion, dorsal closure, and cytoskeletal organization. J Cell Biol. 1997;138:589–603. doi: 10.1083/jcb.138.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geisbrecht ER, Haralalka S, Swanson SK, Florens L, Washburn MP, Abmayr SM. Drosophila ELMO/CED-12 interacts with Myoblast city to direct myoblast fusion and ommatidial organization. Dev Biol. 2008;314:137–149. doi: 10.1016/j.ydbio.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen EH, Pryce BA, Tzeng JA, Gonzalez GA, Olson EN. Control of myoblast fusion by a guanine nucleotide exchange factor, loner, and its effector ARF6. Cell. 2003;114:751–762. doi: 10.1016/s0092-8674(03)00720-7. [DOI] [PubMed] [Google Scholar]
- 33*.Jin P, Duan R, Luo F, Zhang G, Hong SN, Chen EH. Competition between Blown Fuse and WASP for WIP Binding Regulates the Dynamics of WASP-Dependent Actin Polymerization In Vivo. Dev Cell. 2011;20:623–638. doi: 10.1016/j.devcel.2011.04.007. [First study to demonstrate that the actin dynamics but not the total amount of actin polymeirzation is critical for generating invasive protrusions to induce myoblast fusion.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaipa BR, Shao H, Schafer G, Trinkewitz T, Groth V, Liu J, Beck L, Bogdan S, Abmayr SM, Onel SF. Dock mediates Scar- and WASp-dependent actin polymerization through interaction with cell adhesion molecules in founder cells and fusion-competent myoblasts. J Cell Sci. 2013;126:360–372. doi: 10.1242/jcs.113860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35**.Sens KL, Zhang S, Jin P, Duan R, Zhang G, Luo F, Parachini L, Chen EH. An invasive podosome-like structure promotes fusion pore formation during myoblast fusion. J Cell Biol. 2010;191:1013–1027. doi: 10.1083/jcb.201006006. [First study to reveal invasive podosome-like structures in fusion competent myoblasts, demonstrating that myoblast fusion is an asymmetric process. Also demonstrated that podosome invasion promote fusion pore formation and clarified the morphology of fusion pores.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haralalka S, Shelton C, Cartwright HN, Katzfey E, Janzen E, Abmayr SM. Asymmetric Mbc, active Rac1 and F-actin foci in the fusion-competent myoblasts during myoblast fusion in Drosophila. Development. 2011;138:1551–1562. doi: 10.1242/dev.057653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kesper DA, Stute C, Buttgereit D, Kreiskother N, Vishnu S, Fischbach KF, Renkawitz-Pohl R. Myoblast fusion in Drosophila melanogaster is mediated through a fusion-restricted myogenic-adhesive structure (FuRMAS). Dev Dyn. 2007;236:404–415. doi: 10.1002/dvdy.21035. [DOI] [PubMed] [Google Scholar]
- 38.Mukherjee P, Gildor B, Shilo BZ, VijayRaghavan K, Schejter ED. The actin nucleator WASp is required for myoblast fusion during adult Drosophila myogenesis. Development. 2011;138:2347–2357. doi: 10.1242/dev.055012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39*.Chen EH. Invasive podosomes and myoblast fusion. Curr Top Membr. 2011;68:235–258. doi: 10.1016/B978-0-12-385891-7.00010-6. [A review focusing on the molecular components of the myoblast fusion machinery at the asymmetric fusogenic synapse.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40*.Duan R, Jin P, Luo F, Zhang G, Anderson N, Chen EH. Group I PAKs function downstream of Rac to promote podosome invasion during myoblast fusion in vivo. J Cell Biol. 2012;199:169–185. doi: 10.1083/jcb.201204065. [First study to suggest that the dense orgnization of the actin filaments within the PLS is critical for PLS invasion and fusion pore formation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41*.Bothe I, Deng S, Baylies M. PI(4,5)P2 regulates myoblast fusion through Arp2/3 regulator localization at the fusion site. Development. 2014;141:2289–2301. doi: 10.1242/dev.100743. [First study to show a potential function of PIP2 at the fusogenic synapse in vivo.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moore CA, Parkin CA, Bidet Y, Ingham PW. A role for the Myoblast city homologues Dock1 and Dock5 and the adaptor proteins Crk and Crk-like in zebrafish myoblast fusion. Development. 2007;134:3145–3153. doi: 10.1242/dev.001214. [DOI] [PubMed] [Google Scholar]
- 43.Laurin M, Fradet N, Blangy A, Hall A, Vuori K, Cote JF. The atypical Rac activator Dock180 (Dock1) regulates myoblast fusion in vivo. Proc Natl Acad Sci U S A. 2008;105:15446–15451. doi: 10.1073/pnas.0805546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vasyutina E, Martarelli B, Brakebusch C, Wende H, Birchmeier C. The small G-proteins Rac1 and Cdc42 are essential for myoblast fusion in the mouse. Proc Natl Acad Sci U S A. 2009;106:8935–8940. doi: 10.1073/pnas.0902501106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gruenbaum-Cohen Y, Harel I, Umansky KB, Tzahor E, Snapper SB, Shilo BZ, Schejter ED. The actin regulator N-WASp is required for muscle-cell fusion in mice. Proc Natl Acad Sci U S A. 2012;109:11211–11216. doi: 10.1073/pnas.1116065109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baas D, Caussanel-Boude S, Guiraud A, Calhabeu F, Delaune E, Pilot F, Chopin E, Machuca-Gayet I, Vernay A, Bertrand S, et al. CKIP-1 regulates mammalian and zebrafish myoblast fusion. J Cell Sci. 2012;125:3790–3800. doi: 10.1242/jcs.101048. [DOI] [PubMed] [Google Scholar]
- 47**.Kim JH, Ren Y, Ng WP, Li S, Kee YS, Son S, Zhang S, Zhang G, Fletcher DA, Robinson DN, Chen EH. Mechanical tension drives cell membrane fusion. Dev Cell. 2015 doi: 10.1016/j.devcel.2015.01.005. online Feb. 12. [First study to demonstate the MyoII-mediated mechanosensory response in founder cells, which leads to increased cortical tension counteracting the PLS invasion. Established a biophysical framework for myoblast fusion.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bonn BR, Rudolf A, Hornbruch-Freitag C, Daum G, Kuckwa J, Kastl L, Buttgereit D, Renkawitz-Pohl R. Myosin heavy chain-like localizes at cell contact sites during Drosophila myoblast fusion and interacts in vitro with Rolling pebbles 7. Exp Cell Res. 2013;319:402–416. doi: 10.1016/j.yexcr.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 49.van den Eijnde SM, van den Hoff MJ, Reutelingsperger CP, van Heerde WL, Henfling ME, Vermeij-Keers C, Schutte B, Borgers M, Ramaekers FC. Transient expression of phosphatidylserine at cell-cell contact areas is required for myotube formation. J Cell Sci. 2001;114:3631–3642. doi: 10.1242/jcs.114.20.3631. [DOI] [PubMed] [Google Scholar]
- 50.Jeong J, Conboy IM. Phosphatidylserine directly and positively regulates fusion of myoblasts into myotubes. Biochem Biophys Res Commun. 2011;414:9–13. doi: 10.1016/j.bbrc.2011.08.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51*.Hochreiter-Hufford AE, Lee CS, Kinchen JM, Sokolowski JD, Arandjelovic S, Call JA, Klibanov AL, Yan Z, Mandell JW, Ravichandran KS. Phosphatidylserine receptor BAI1 and apoptotic cells as new promoters of myoblast fusion. Nature. 2013;497:263–267. doi: 10.1038/nature12135. [First study to demonstrate a function of a PS receptor in myoblast fusion during muscle regeneration.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park D, Tosello-Trampont AC, Elliott MR, Lu M, Haney LB, Ma Z, Klibanov AL, Mandell JW, Ravichandran KS. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature. 2007;450:430–434. doi: 10.1038/nature06329. [DOI] [PubMed] [Google Scholar]
- 53*.Hamoud N, Tran V, Croteau LP, Kania A, Cote JF. G-protein coupled receptor BAI3 promotes myoblast fusion in vertebrates. Proc Natl Acad Sci U S A. 2014;111:3745–3750. doi: 10.1073/pnas.1313886111. [First study to demonstrate a function of a PS receptor in myoblast fusion during muscle development in vivo.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doberstein SK, Fetter RD, Mehta AY, Goodman CS. Genetic analysis of myoblast fusion: blown fuse is required for progression beyond the prefusion complex. J Cell Biol. 1997;136:1249–1261. doi: 10.1083/jcb.136.6.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rau A, Buttgereit D, Holz A, Fetter R, Doberstein SK, Paululat A, Staudt N, Skeath J, Michelson AM, Renkawitz-Pohl R. rolling pebbles (rols) is required in Drosophila muscle precursors for recruitment of myoblasts for fusion. Development. 2001;128:5061–73. doi: 10.1242/dev.128.24.5061. [DOI] [PubMed] [Google Scholar]
- 56.Estrada B, Maeland AD, Gisselbrecht SS, Bloor JW, Brown NH, Michelson AM. The MARVEL domain protein, Singles Bar, is required for progression past the pre-fusion complex stage of myoblast fusion. Dev Biol. 2007;307:328–339. doi: 10.1016/j.ydbio.2007.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen EH, Olson EN. Antisocial, an intracellular adaptor protein, is required for myoblast fusion in Drosophila. Dev Cell. 2001;1:705–715. doi: 10.1016/s1534-5807(01)00084-3. [DOI] [PubMed] [Google Scholar]
- 58.Menon SD, Chia W. Drosophila rolling pebbles: a multidomain protein required for myoblast fusion that recruits D-Titin in response to the myoblast attractant Dumbfounded. Dev Cell. 2001;1:691–703. doi: 10.1016/s1534-5807(01)00075-2. [DOI] [PubMed] [Google Scholar]
- 59.Menon SD, Osman Z, Chenchill K, Chia W. A positive feedback loop between Dumbfounded and Rolling pebbles leads to myotube enlargement in Drosophila. J Cell Biol. 2005;169:909–920. doi: 10.1083/jcb.200501126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60*.Shin NY, Choi H, Neff L, Wu Y, Saito H, Ferguson SM, De Camilli P, Baron R. Dynamin and endocytosis are required for the fusion of osteoclasts and myoblasts. J Cell Biol. 2014;207:73–89. doi: 10.1083/jcb.201401137. [First study to suggest a potential function of endocytosis in myoblast fusion.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leikina E, Melikov K, Sanyal S, Verma SK, Eun B, Gebert C, Pfeifer K, Lizunov VA, Kozlov MM, Chernomordik LV. Extracellular annexins and dynamin are important for sequential steps in myoblast fusion. J Cell Biol. 2013;200:109–123. doi: 10.1083/jcb.201207012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lenhart KC, Becherer AL, Li J, Xiao X, McNally EM, Mack CP, Taylor JM. GRAF1 promotes ferlin-dependent myoblast fusion. Dev Biol. 2014;393:298–311. doi: 10.1016/j.ydbio.2014.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63**.Millay DP, O'Rourke JR, Sutherland LB, Bezprozvannaya S, Shelton JM, Bassel-Duby R, Olson EN. Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature. 2013;499:301–305. doi: 10.1038/nature12343. [Discovery of a skeletal muscle-specific transmembrane protein required for mouse myoblast fusion.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Millay DP, Sutherland LB, Bassel-Duby R, Olson EN. Myomaker is essential for muscle regeneration. Genes Dev. 2014;28:1641–1646. doi: 10.1101/gad.247205.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Landemaine A, Rescan PY, Gabillard JC. Myomaker mediates fusion of fast myocytes in zebrafish embryos. Biochem Biophys Res Commun. 2014;451:480–484. doi: 10.1016/j.bbrc.2014.07.093. [DOI] [PubMed] [Google Scholar]