Abstract
Hyperhomocysteinemia (HHcy) leads to several clinical manifestations including hepatic fibrosis. Excess deposition of extracellular matrix (ECM) components including collagen is the eponymous lesion of liver fibrosis. In this study, we demonstrated that elevated concentration of Hcy induced the expression of collagen type I in cultured human liver cells as well as in liver tissue of HHcy mice. Meanwhile, Hcy inhibited the expression of histone methyltransferase G9a. Mechanistically, silencing endogenous G9a by siRNA enhanced the promoter activity of COL1A1 in LO2 cells. Conversely, overexpressing G9a inhibited the promoter activity of COL1A1. CHIP assay demonstrated that G9a binds to the neuron-restrictive silencer element (NRSE) on the promoter of COL1A1. Hcy treatment decreased the binding of G9a on NRSE, which in turn decreased the level of H3K9me2 on the promoter of COL1A1, led to upregulation of COL1A1. Taken together, these results provide a novel mechanism on explaining how HHcy promotes ECM production.
Introduction
Homocysteine (Hcy) is a sulfur containing amino acid that is formed as a primary intermediate in the methionine cycle [1]. Methionine from dietary sources is converted to S-adenosyl methionine (SAM) by the enzyme methionine adenosyltransferase (MAT). SAM donates its methyl group to methyl acceptors including phospholipids, DNA, RNA, and protein, and then converted to S-adenosyl homocysteine (SAH), which is then hydrolyzed to Hcy and adenosine in hepatic tissue. Hcy can either be remethylated to methionine or converted to cystathionine by cystathionine- β-synthase (CBS) [2]. Elevated plasma Hcy concentrations, a condition known as hyperhomocysteinemia (HHcy) [3], is the consequence of enzymatic deficiencies and/or nutritional defects that interfere with the proper metabolism of methionine and/or Hcy [4]. The most common genetic cause associated with severe HHcy is homozygous CBS deficiency which is characterized by very high levels of plasma total Hcy (tHcy>200μM). The clinical manifestations of HHcy are very diverse, including mental retardation, cardiovascular problems, skeletal abnormalities, and hepatic compromise, with fatty accumulation and cirrhosis.
Hepatic fibrosis is a wound-healing response characterized by accumulation of extracellular matrix (ECM). In normal liver, ECM is a highly dynamic substratum with a precisely regulated balance between synthesis and degradation. During chronic liver injury, hepatic fibrosis develops as ECM production exceeds ECM degradation. Moreover, the composition of matrix is changed from collagens IV and VI to collagens I, III and fibronectin. There has been tremendous progress in revealing the regulatory mechanisms that control ECM-related gene expression during fibrosis, and research has focused primarily on transcriptional control pathways. TGF-β1 has been regarded as the most important growth factor implicated in collagen synthesis in hepatic fibrosis. TGF-β1 signals via its cognate receptors to Smad proteins to enhance collagen I gene transcription [5,6]. In addition, AP-2, NF-1 and c/EBP has been shown to regulate collagen gene expression [7].
Elevated Hcy levels have long been known to be linked to liver disease [8]. Fatty liver is a common finding in nutritionally induced HHcy due to methionine overload or folate deficiency. Patients with CBS deficiency also have hepatic steatosis which is accompanied by perisinusoidal or central venous fibrosis and fibrosis of hepatic arterioles [9]. It has been shown that CBS-deficient mice develop inflammation, fibrosis, and hepatic steatosis [10,11]. The upregulation of collagen I has been detected in the liver of CBS deficient mice and HHcy rats induced by chronic Hcy administration [12]. However, the molecular mechanism responsible for HHcy-induced collagen expression is unclear. Increased Hcy elevates the level of SAH which inhibits transferring methyl group from SAM to acceptors, leads to hypomethylation [13]. It has been proposed that Hcy-induced DNA hypomethylation is a biochemical mechanism by which Hcy induces vascular injury [14,15]. However, whether epigenetic modifications such as DNA methylation and histone modifications are involved in the pathogenesis of Hcy-induced hepatic fibrosis is largely unknown.
Neuron-Restrictive Silencing Factor/Repressor Element Silencing Transcription Factor (NRSF or REST) is a transcriptional regulator that regulates a network of genes by binding a 21–23 bp neuron-restrictive silencing element (NRSE) [16], which in turn recruits a silencing complex of chromatin remodeling proteins including SIN3A, histone deacetylases 1 and 2 [17], CoREST [18], methyl-CpG-binding protein 2, and histone methyltransferase G9a [19,20]. NRSF was originally perceived as a silencer of neuron-specific genes [17]. Later studies revealed that there are over 800 NRSE-containing genes in human genome, underscoring the importance of NRSF in controlling large programs of transcriptional regulation [19].
In the present study, we found a NRSE in the promoter of both human COL1A1 and mouse Col1α1, and G9a binds to the NRSE. Upon Hcy stimulation, the expression of G9a is decreased, results in reduced binding of G9a to the NRSE, which in turn decreased the level of H3K9me2 on the promoter of human COL1A1 and mouse Col1α1, led to upregulation of collagen I both in vitro and in vivo. This finding provides a novel epigenetic mechanism on explaining how elevated Hcy promotes liver fibrosis.
Results
Hcy induces collagen I expression in cultured normal human liver cells
Previous study has shown that HHcy causes liver fibrosis [10,21]. We thus examined whether Hcy affects the production of Col I, a major component of ECM. To this end, normal human liver cells (LO2) were incubated with Hcy (100μM) for indicated time period and then cell lysates were harvested for Western blotting. Fig 1A and 1B showed that the protein level of Col I was significantly increased in a time-dependent manner. We further treated LO2 cells with incremental concentrations of Hcy from 10 to 200μM for 48 h. As shown in Fig 1C and 1D, the level of Col I increased progressively with increased concentrations of Hcy. At 200μM of Hcy, the protein level of Col I was ~15 fold higher than that of untreated cells.
Fig 1. Hcy induces Col I expression in LO2 cells.
LO2 cells were incubated with 100μM of Hcy for indicated time period, or indicated concentration of Hcy for 48 h. The protein level of Col I was determined by Western blotting (A-D). β-actin was used to verify equivalent loading. Graphic representation of relative protein level of Col I normalized to β-actin (B, D). The mRNA level of COL1A1 gene was determined by real time PCR (E-F). Data are mean±SD of three independent experiments. *p<0.05 versus control cells.
Hcy could regulate Col I production at mRNA or post-translation level. To determine this, we examined the transcript of human COL1A1 in LO2 cells after Hcy treatment by real time PCR. As shown in Fig 1E and 1F, the mRNA level of COL1A1 was gradually increased in a time- and dose-dependent manner after Hcy treatment, indicating that Hcy upregulates COL1A1 gene transcription in cultured human liver cells.
Hcy induces Col1α1 expression in vivo
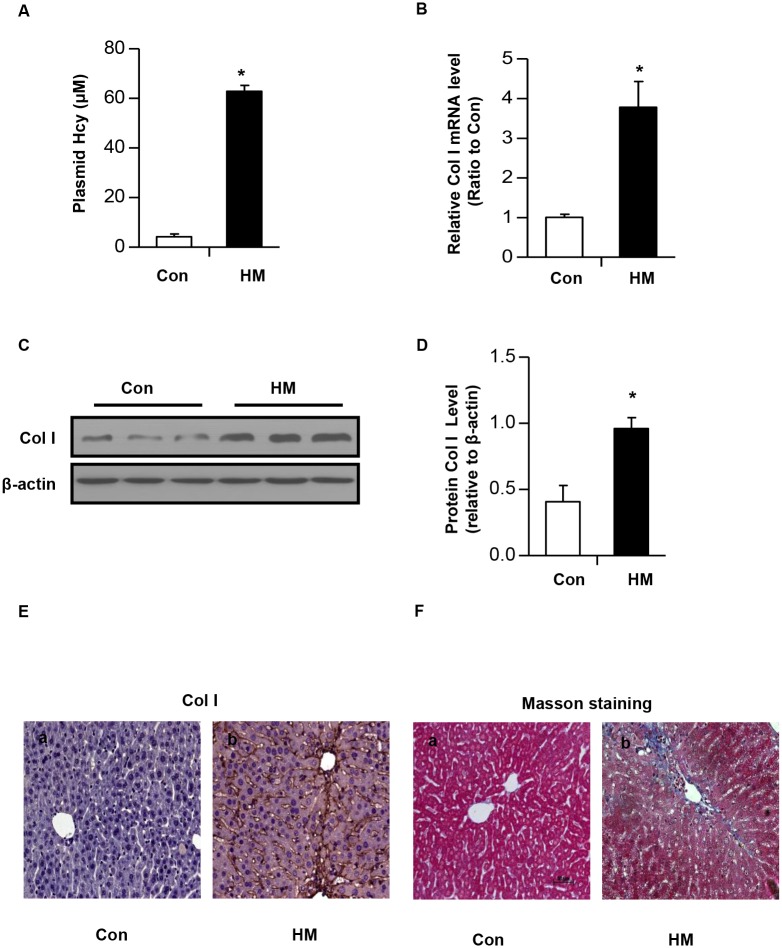
To further confirm the effect of Hcy on Col I production in vivo, we generated HHcy mice by feeding male C57BL/6 mice with a high methionine diet (HM) containing sufficient basal levels of B vitamins [22,23] for two weeks. The plasma level of Hcy was increased to 67μM±8.7μM, whereas the plasma level of Hcy is 5.5μM±0.7μM in control mice fed with regular rodent chow diet (Fig 2A). Liver tissues of HHcy mice and control mice were collected. Real time PCR showed an upregulation of Col1α1 gene (Fig 2B). Consistently, Western blotting and immunohistochemistry staining demonstrated that the protein level of Col I were significantly increased in the liver of HHcy mice (Fig 2C–2E). To confirm that HHcy causes hepatic fibrosis, Masson’s trichrome staining was performed. As shown in Fig 2F, compared with that of control mice, a dramatic increase in ECM accumulation was observed in the liver tissue of HHcy mice, which is consistent with previous studies [10].
Fig 2. Increased Col I expression in liver of HHcy mice.
(A) C57BL/6 mice were fed with regular rodent chow or a high methionine diet (HM) for two weeks, and then liver tissues were collected. (A) The plasma concentration of Hcy in mice. (B) The mRNA level of Col1α1 gene was determined by real time PCR. (C) The protein level of Col I was determined by western blotting. β-actin was used to verify equivalent loading. (D) Graphic representation of relative protein level of Col I normalized to β-actin. Data are expressed as mean±SD, n = 6. *p<0.05 versus mice fed with regular rodent chow. (E) Immunohistochemical staining for Col I in the liver of HHcy mice. (F) Representative micrographs show that HHcy caused hepatic fibrosis. Liver sections from either control mice or HHcy mice were subjected to Masson’s trichrome staining. Scale bar, 50μm.
G9a mediates Hcy-induced COL1A1 gene expression
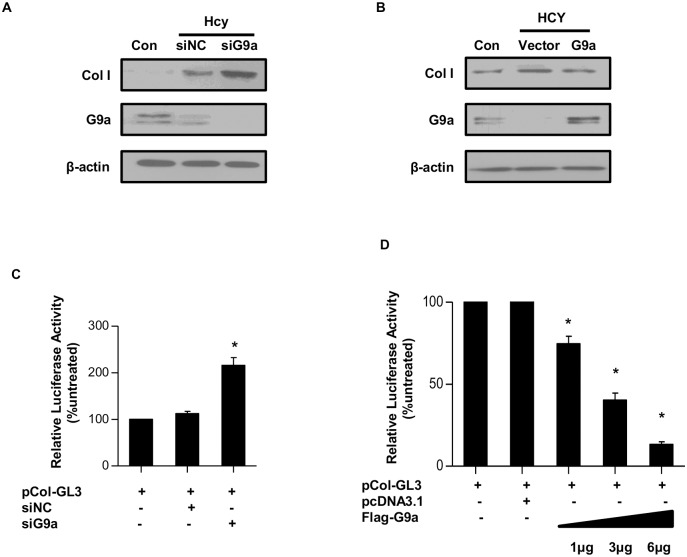
Since we have demonstrated that Hcy modulates COL1A1 gene expression, we determined to further explore the underlying mechanism. We assessed the promoter regions of both human COL1A1 and mouse Col1α1 gene using the online prediction service of NCBI and found a neuron restrictive silencing element (NRSE), which has been shown to recruit G9a to silence gene expression [19,20]. To evaluate the role of G9a in Hcy-induced COL1A1 gene expression, endogenous G9a was knocked down by siRNA (siG9a) in LO2 cells. LO2 cells were transfected with siG9a or control siRNA (siNC) and then treated with Hcy (100μM) for 48 h. Western blotting revealed that silencing endongenous G9a significantly increased the protein level of Col I when compared with control siRNA trasnfected cells (Fig 3A).
Fig 3. Hcy-induced Col I expression is mediated by G9a.
(A) LO2 cells were transfected with scramble siRNA (siNC) or siRNA targeting G9a (siG9a) and then treated with 100μM of Hcy for 48 h. Whole cell lysates were harvested, the protein levels of G9a and Col I were determined by Western blotting. β-actin was used to verify equivalent loading. (B) LO2 cells were transfected with Flag-tagged G9a or empty vector, and then treated with 100μM of Hcy for 48 h. The protein level of G9a and Col I was determined by Western blotting. β-actin was used to verify equivalent loading. (C) LO2 cells were transfected with either siNC or siG9a together with pCol I-luc plasmid and pRL. Relative luciferase activity was presented. *p <0.05 versus siNC transfected cells. (D) LO2 cells were transfected with empty vector or indicated amounts of Flag-tagged G9a together with pCol I-luc plasmid and pRL. Relative luciferase activity was presented. *p <0.05 versus empty vector transfected cells. Data are mean±SD of three independent experiments.
Alternatively, Flag-tagged G9a or empty vector was transfected into LO2 cells followed by Hcy treatment for 48 h. As shown in Fig 3B, Hcy significantly increased the protein level of Col I in empty vector transfected cells. Whereas, the protein level of Col I was significantly lower in G9a overexpressed cells than that of control cells (Fig 3B).
To examine whether G9a directly affect the promoter activity of COLA1 gene, we synthesized a reporter plasmid bearing a 1-kb fragment of the 5’-flanking region of human COL1A1 gene (pCol-GL3) and co-transfected it with siRNA targeting G9a (siG9a) or scramble siRNA (NC-siRNA). Compared with NC-siRNA, siG9a significantly enhanced the luciferase activity by 40% (Fig 3C). Next, pCol-GL3 was co-transfected with increasing amounts of Flag-tagged G9a into LO2 cells. As shown in Fig 3D, the promoter activity was gradually decreased as the amount of G9a plasmid was increased. A significant decrease was detected when 1μg of Flag-tagged G9a was transfected. These results support a role of G9a in modulating COL1A1 gene expression.
Hcy reduces the binding of G9a to the promoter of COL1A1
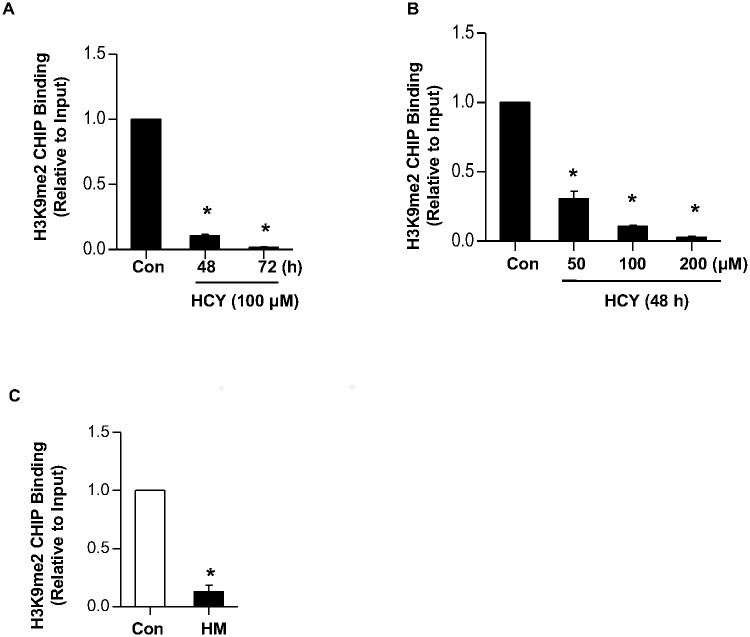
Since G9a overexpression inhibits the activity of COL1A1 promoter and there is a NRSE within the promoter region of COL1A1, we hypothesized that Hcy might reduce the binding of G9a to the NRSE on the promoter of COL1A1. To evaluate this hypothesis, chromatin immunoprecipitation (ChIP) assay was performed to determine the chromatin occupancy of G9a in the promoter of COL1A1 using anti-G9a antibody in LO2 cells and mice. ChIP-enriched DNA samples were analyzed by quantitative PCR (qPCR) using primers spanning human NRSE DNA sequences (Fig 4A). To evaluate the effect of Hcy on the binding activity of G9a, LO2 cells were treated with 100μM of Hcy for indicated time period. ChIP assays revealed that binding of G9a to the promoter of COL1A1 was decreased in a time-dependent manner (Fig 4B). Furthermore, we treated LO2 cells with indicated amount of Hcy for 48 h. Fig 4C showed that the binding of G9a to the promoter of COL1A1 was gradually decreased in a dose-dependent manner. Similarly, the binding of G9a to the promoter of Col1α1 was also significantly decreased in the liver of HHcy mice (Fig 4D). Collectively, these data indicate that a decrease of G9a binding to the promoter of COL1A1 might play a role in Hcy-induced COL1A1 expression.
Fig 4. Hcy decreased the binding of G9a to the COL1A1 promoter.
(A) Schematic representation of the promoters of COL1A1 and Col1α1 gene. NRSE region and primers used for ChIP assay were marked. (B-C) LO2 cells were treated with 100μM of Hcy for indicated time period (B), or indicated concentration of Hcy for 48 h (C), and then harvested for ChIP assay by using anti-G9a antibody. The changes in G9a on the COL1A1 promoter were examined by q-ChIP PCR. Data are means±SD of three independent experiments. *p<0.05 versus untreated cells. (D) Liver tissues of wild type mice and HHcy mice were collected for ChIP assay by using anti-G9a antibody. The changes in G9a on the Col1α1 promoter were examined by q-ChIP PCR. Data are expressed as mean±SD, n = 6. *p<0.05 versus mice fed with regular rodent chow.
Hcy reduces the level of H3K9me2 on the promoter of COL1A1
Since G9a is the primary enzyme for dimethylation of histone H3 lysine 9 (H3K9me2), we next examined whether histone H3 lysine methylation levels were also altered by Hcy. ChIP assay using H3K9me2-specific antibodies revealed that Hcy decreased the level of H3K9me2 on COL1A1 promoter in a time- and dose-dependent manner (Fig 5A and 5B). Similarly, the level of H3K9me2 on Col1α1 promoter was also dramatically decreased in the liver of HHcy mice (Fig 5C). These results suggest that upregulation of Col I might be caused, at least partially, by a loss of repressive epigenetic histone modification on its promoter.
Fig 5. Hcy decreased the level of H3K9me2 on the COL1A1 promoter.
(A) (A-B) LO2 cells were treated with 100μM of Hcy for indicated time period (A), or indicated concentration of Hcy for 48 h (B), and then harvested for ChIP assay by using anti-H3K9me2 antibody. The changes of H3K9me2 on the COL1A1 promoter were examined by q-ChIP PCR. Data are means±SD of three independent experiments. *p<0.05 versus untreated cells. (C) Liver tissues of wild type mice and HHcy mice were collected for ChIP assay by using anti-H3K9me2 antibody. The changes in H3K9me2 on the Col1α1 promoter were examined by q-ChIP PCR. Data are expressed as mean±SD, n = 6. *p<0.05 versus mice fed with regular rodent chow.
Hcy downregulates G9a expression both in vitro and in vivo
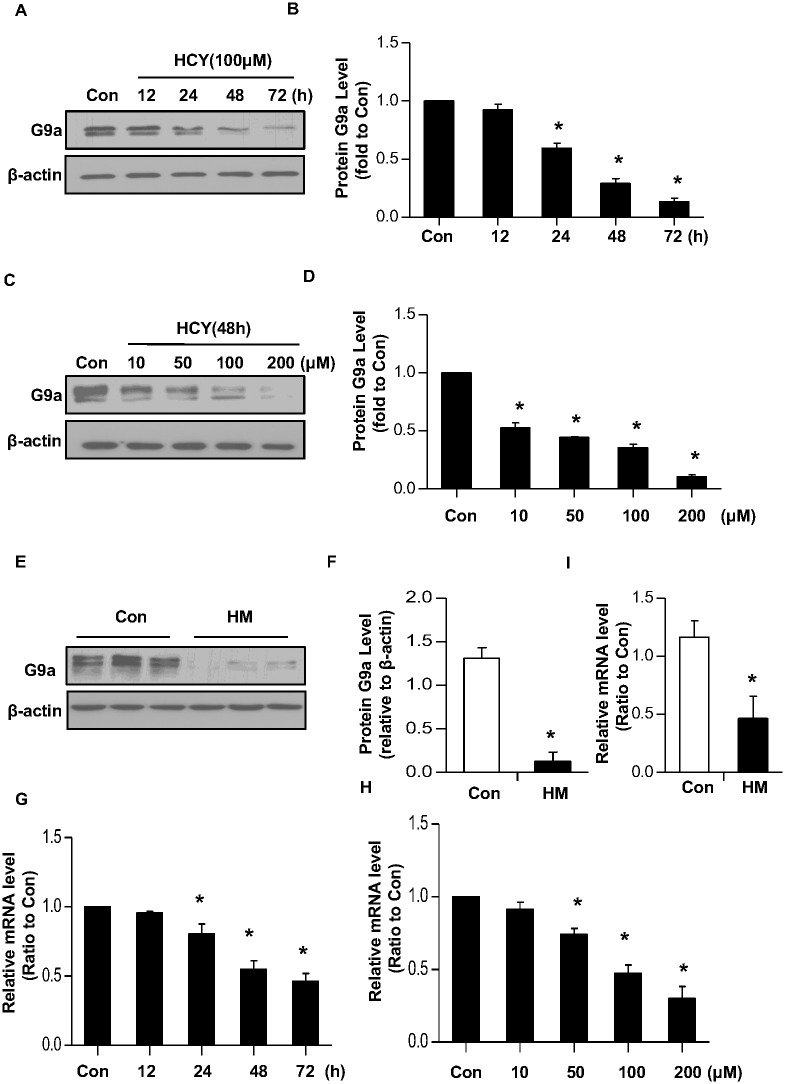
The decreased binding of G9a to the promoter of both human COL1A1 and mice Col1α1 might be caused by a reduced protein level of G9a. To test this possibility, LO2 cells were treated with various concentrations of Hcy for indicated time period. Western blotting revealed that Hcy gradually decreased the protein level of G9a in a time- and dose-manner (Fig 6A–6D). Furthermore, the protein level of G9a in liver tissue of HHcy was also significantly lower than that of control mice (Fig 6E and 6F).
Fig 6. Homocysteine decreases G9a expression both in vitro and in vivo.
LO2 cells were incubated with 100μM of Hcy for indicated time period (A-B), or indicated concentration of Hcy for 48 h (C-D). The protein level of G9a was determined by Western blotting. β-actin was used to verify equivalent loading. Graphic representation of relative protein level of G9a normalized to β-actin (B, D). Data are mean±SD of three independent experiments. *p<0.05 versus control cells. (E) Liver tissues of HHcy mice were collected from HHcy mice or control mice fed with regular rodent chow. The protein level of G9a in liver was determined by Western blotting. β-actin was used to verify equivalent loading. (F) Graphic representation of relative protein level of G9a normalized to β-actin. (G-H) LO2 cells were incubated with 100μM of Hcy for indicated time period (G), or indicated concentration of Hcy for 48 h (H). The mRNA level of G9a was determined by real time PCR. (I) Liver tissues were collected from HHcy mice or control mice fed with regular rodent chow. The mRNA level of G9a in the liver was determined by real time PCR. Data are expressed as mean±SD, n = 6. *p<0.05 versus control mice fed with regular rodent chow.
To examine whether Hcy inhibits G9a expression at the transcription level, LO2 cells were treated with various concentrations of Hcy with indicated time period and then mRNA was extracted for real time PCR. As shown in Fig 6G and 6H, the mRNA level of G9a was gradually decreased in a time- and dose-dependent manner after Hcy treatment. Moreover, the mRNA level of G9a in the liver tissue of HHcy mice was also significantly lower than that of control mice (Fig 6I), indicating that Hcy suppresses the transcription of G9a gene.
Discussion
Gene regulation by extracellular stimuli involves not only transcription factors binding to their cognate DNA binding sites but also epigenetic changes in chromatin [24]. The transcriptional mechanisms on the overproduction of ECM during the process of organ fibrosis have been extensively studied. Collagen I is a downstream target gene of TGF-β/Smads pathway and its expression is induced by TGF-β in fibrotic organs [25]. Previous study showed that CBS-deficient mice (tHcy 205μM) develop liver fibrosis concomitant with an enhanced expression of Col I and TGF-β [8,9], suggesting Hcy-induced TGF-β expression is a mechanism for Col1α1 gene induction. In the present study, we provided evidence that the transcription of Col I gene is regulated by histone methyltransferase G9a. ChIP assay demonstrated that G9a binds to the promoter of human COL1A1 and mice Col1α1, catalyzes local H3K9me2 to maintain Col I expression at low level under normal condition. Whereas, Hcy downregulates G9a expression, which in turn decreased the level of H3K9me2 at the promoter of COL1A1, leading to COL1A1 gene induction. This finding suggests that changes of histone modifications contribute to Hcy-regulated gene expression. Previous study demonstrates that TGF-β increases the recruitment of SET7/9, a H3K4 mono-methyltransferase, to the promoter of Col1α1, increases H3K4me1 level at the promoter of Col1α1, results in Col1α1 gene induction in rat mesangial cells [26]. Moreover, histone deacetylase inhibitor trichostatin A abrogated the stimulatory effect of TGF-β on collagen I transcription in skin fibroblasts [27,28] and renal proximal tubular cells [29]. Since TGF-β1 is upregulated in the liver of HHcy mice, it is possible that TGF-β-induced histone modifications also contributes to HHcy-modulated ECM genes expression.
G9a, a key methyltransferase responsible for H3K9me2 at euchromatin and facultative heterochromatin, does not contain a DNA binding sequence [30]. Previous studies demonstrate that recruitment of G9a by NRSF is important in the repression of neuronal genes outside of the nervous system [31,32]. In the present study, we found, for the first time, that there is a NRSE in the promoter of COL1A1, and G9a is recruited to the COL1A1 promoter via the NRSE. This finding adds COL1A1 gene as a new member to the family of NRSE-containing genes, and supports the role of NRSF in controlling large programs of transcriptional regulation.
G9a is widely expressed in most tissues including fetal liver, bone marrow, peripheral blood leukocytes, thymus, lymph node, spleen and developing skeletal muscles [33]. Cancer transcriptome analysis revealed that G9a is overexpressed in many different types of tumors including hepatocellular, colon, prostate, lung and invasive transitional cell carcinomas and in B cell chronic lymphocytic leukemia, and responsible for various aspects of tumorigenesis, including cellular differentiation, proliferation and epithelial to mesenchymal transition [34]. Therefore, it is important to identify factors that could modulate the expression of G9a. Previous study showed that repeated administration of cocaine decreases G9a mRNA levels in nucleus accumbens, and results in increased gene expression [35,36]. In the present study, we identified Hcy as another factor which inhibits G9a gene expression. Previous study reported that Hcy accelerates protein degradation [37,38]. We treated LO2 cells with proteasome inhibitor MG132, and lysosome inhibitor chloroquine, but did not observed any obvious effect on Hcy-induced G9a suppression (data not shown), indicating that Hcy does not promote G9a degradation. We next measured mRNA level of G9a and found that Hcy decreased G9a mRNA in a time and dose-dependent manner, suggesting that Hcy modulates G9a expression at the transcription level.
The transcription regulation of G9a has been much less explored. Previous study showed that transcription factor C/EBPβ binds to the promoter of G9a, and C/EBPβ activates G9a expression during preadipocyte differentiation. Increased G9a expression results in enhanced H3K9 dimethylation at the C/EBPα promoter, which in turn represses C/EBPα expression [39]. Using the online prediction service of NCBI, we found 6 potential binding sites of C/EBPα in the proximal promoter region (from -1000bp to the transcription start site) of G9a gene. It is therefore possible that HHcy downregulates G9a expression through regulating the expression of C/EBPα and/or C/EBPβ. In supporting this hypothesis, previous studies have shown that both C/EBPα and C/EBPβ are expressed at high levels in the liver and play decisive roles in hepatocyte proliferation and differentiation [40]. Moreover, the cebpa promoter region contains CpG islands and cebpa gene expression is elevated in dnmt1-deficient embryos [41]. Since increased Hcy elevates the level of SAH which inhibits transferring methyl group to acceptors, Hcy might induce C/EBPα expression through inhibiting DNA hypomethylation at the promoter of C/EBPα, which in turn modulates G9a expression. Noteworthy, we found that Hcy treatment caused a more dramatic decrease in G9a protein than that of mRNA, suggesting that post-transcriptional mechanism is also involved in Hcy-modulated G9a expression.
Hcy-induced DNA hypomethylation has been proposed as a biochemical mechanism by which Hcy modulates gene expression. However, results have been inconsistent in many animal studies. Hcy increased cellular SAH concentration in endothelial cells, but not in vascular smooth muscle cells [42]. Moreover,in CBS null mice, higher SAH concentrations were detected in all tissues studied, but lower DNA methylation status was only detected in the liver [43]. These studies suggest other mechanisms might be implicated in Hcy-modulated gene expression. Our study showed that Hcy downregulated the expression of histone methyltransferase G9a, which in turn decreased the level of H3K9me2 at the promoter of COL1A1, suggesting the involvement of histone modifications in Hcy-modulated gene expression. In line with our finding, previous study demonstrated that Hcy reduced the binding of methyl CpG binding protein 2 (MeCP2) and increased the bindings of acetylated histone H3 and H4 at the cyclin A promoter [15]. Increasing evidence demonstrates that H3K9 hypermethylation serves as a docking site for the chromatin modifier protein heterochromatin protein 1(HP1), which in turn recruits DNMT1 and stimulates its activity leading to DNA hypermethylation in the surrounding area [44,45]. By using the online prediction service of NCBI, we found a couple of CpG islands in the promoters of both COL1A1 and Col1α1 gene. Therefore, Hcy-mediated G9a repression might lead to DNA hypomethylation to reinforce the epigenetic activation of gene transcription. Consistently, it has been shown that disrupting the function and expression of G9a by its inhibitor and siRNA resulted in a marked reduction in DNA methylation at the COX-2 promoter and led to the restoration of COX-2 expression [46,47]. Conversely, co-recruitment of G9a, DNMT1, and HP1 to the promoter of the survivin gene stimulates H3K9me2 and DNA hypermethylation [44,45].
Taken together, our study showed that Hcy downregulates G9a expression, which in turn decreases the binding of G9a and the level of H3K9me2 on the promoter collagen I gene, leads to its upregulation. Because G9a specifically catalyzes H3K9me2, it is conceivable that Hcy-mediated G9a repression causes a decrease in global euchromatic H3K9me2, which might alter the expression of numerous genes. Therefore, it will be of great interest to further explore the involvement of G9a-mediated histone modification in HHcy-induced multi-organs damage.
Materials and Methods
Cell culture and treatment
Human hepatic cell lines (LO2) (American Type Culture Collection, Manassas, VA) were cultured in DMEM supplemented with 10% FBS (Life Technologies, Gaithersburg, MD) at 37°C in a 5% CO2 incubator. Fresh medium containing DL-Hcy was replaced every 24 h. DL-Hcy was purchased from Sigma-Aldrich (St Louis, MO).
Plasmid constructs
Human G9a were amplified and subcloned into pcDNA3.1 vector. To generate pCol-GL3 reporter plasmid, a fragment containing 1500bp upstream of the transcriptional start site of human COL1A1 promoter was amplified and subcloned into pGL3 vector (Promega, Madison, WI). Primers used were as follow: Forward 5’-CCCGGTACCAGAGAAATGAACAGGGCA-3’, Reverse 5’-CCCCTCGAGACTGGCCCGGGCCCCTTT-3’. All expression constructs were generated by standard PCR-based cloning strategies, and all expression constructs were verified by DNA sequencing.
RNA Interference
Oligonucleotide siRNA duplex was synthesized by Shanghai Gene Pharma (Shanghai, China). RNAi oligonucleotides were transfected into LO2 cells using the Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. The sequences of RNAi oligonucleotides were as follows: scramble siRNA: UUCUCCGAACGUGUCACGU; G9a siRNA: CCAUGCUGUCAACUACCAUGG
Real-time PCR
Total RNA was extracted from cells or tissues using Trizol reagent (Invitrogen,Carlsbad, CA). cDNA was synthesized from 2μg of RNA using the SuperScript kit (Invitrogen). Reaction was performed on a 7500 Sequence Detection System (Applied Biosystems). Primers used in this study were as follows:
human G9a Forward 5’-ACAGAGGAAGAGGTAGGCCC-3’, Reverse 5’-CCATGAACTCTCTCGGTGGC-3’;
human COL1A1: Forward 5’-GAGATGATGGGGAAGCTGGA-3’, Reverse 5’-GCACCATCATTTCCACGAGC-3’;
human GADPH Forward 5’-AGAAGGCTGGGGCTCATTTG-3’, Reverse 5’-AGGGGCCATCCACAGTCTTC-3’;
mouse G9a Forward 5’-TTCCTTGTCTCCCCTCCCAG-3’, Reverse 5’-CTATGAACTCTCTCGGCGGC-3’;
mouse Col1α1: Forward 5’-GAGAGGTGAACAAGGTCCCG-3’, Reverse 5’-AAACCTCTCTCGCCTCTTGC-3’;
mouse GADPH Forward 5’-GGTGAAGGTCGGTGTGAACG-3’, Reverse 5’-CTCGCTCCTGGAAGATGGTG-3’.
Western blotting
Cells and liver tissues were lysed with immunoprecipitation assay buffer (25mM Tris-HCl, pH 7.4, 150mM KCl, 5mM EDTA, 0.5% Na deoxycholate, 0.1% SDS, 1% NP-40). Lysates were subjected to Western blotting using method described previously [19]. The following primary antibodies were used in this study: anti-G9a (Abcam ab40542), anti-Col I (Abcam ab34710), anti-dimethyl-H3K9 (Abcam ab1220) and β-actin (Huatesheng Biotechnolgy, Fushun, China).
ChIP assay and q-ChIP PCR
Chromatin immunoprecipitation (ChIP) assay was performed as described previously [19]. Briefly, 2×107cells were fixed with 1% formaldehyde at 37°C for 10 min and were then lysed on ice for 15 min. These lysed extracts were subjected to shearing by sonication. After centrifugation at 14,000 rpm for 15 min, the soluble chromatin was subjected to immunoprecipitation with indicated antibodies, and then the complexes were drawn off with protein A-agarose beads and washed sequentially with low-salt, high-salt, LiCl, and Tris-EDTA buffers and were finally extracted with freshly prepared 1% SDS-0.1 M NaHCO3. Heating the samples at 65°C for 6 h, and then DNA was purified with a Qiagen DNA extraction kit. Primers used in this study were as follows: human COL1A1: Forward 5’-GCTGGGAAGGAGGGTCTCTA-3’, Reverse 5’-TGAGAGATGGAGTGGGGAGG-3’; mouse Col1α1 Forward 5’-CATGGCCAGGAGGACCTTTT-3’, Reverse 5’-TTGATGGAGAGCTGGGAGGA-3’.
Animals and Experimental Design
Wild-type C57BL/6 mice, obtained from and housed in Southern Medical University animal facility,at the age of 6–9 weeks, were fed with either standard rodent chow or HM diet containing 19.56g/kg (2%) methionine and sufficient basal levels of B vitamins [22]. After 2 weeks on the diet, mice were sacrificed. Blood was collected by cardiac puncture. Liver tissues were removed, flash-frozen, and stored at -80°C. All experiments were approved by the Southern Medical University Ethics Committee for Animal Experiments and strictly adhered to the guidelines for animal experiments of Southern Medical University. All surgery was performed under pentobarbital sodium, and all efforts were made to minimize suffering.
Immunohistochemistry
Liver was fixed overnight in 4%paraformaldehyde solution in phosphate-buffered saline. Sections (4 μm thickness) were deparaffinized with xylene, followed by rehydration in ethanol. Hydrogen peroxide (3%) was used to eliminate endogenous peroxidase. Sections were incubated overnight at 4°C with primary antibodies against Col I (Abcam 34710). After extensive washing in PBS buffer, sections were then incubated for 30 minutes with secondary antibodies (Dako, Carpinteria, CA). The immunostaining was examined by an Olympus BX51 microscope (Olympus, Tokyo, Japan). Positive stains were quantified using image analysis software (Image Pro-Plus, Media Cybernetics, Silver Spring, MD).
Masson Trichrome Staining
Selected liver sections were stained using the Masson Trichrome Stain Kit (Richard-Allan Scientific, Kalamazoo, MI) according to the manufacturer’s protocols.
Hcy measurement
Total plasma Hcy concentration was measured using Homocysteine-EIA Kit (Axis Shield, Scotland) according to the manufacturer’s instructions. Hcy measurement was calibrated to the NIST standard reference material SRM1955.
Luciferase reporter assays
LO2 cells were transfected with pCol-GL3 reporter plasmid, pRL null together with siG9a or Flag-tagged G9a using Lipofectamine 2000 (Invitrogen). Cells were harvested 48 h after transfection and the luciferase activities were analyzed by the luciferase reporter assay system (Promega, Madison, WI) on a GloMax 96 MicroplateLuminometer (Promega). Renilla luciferase activity was normalized to firefly luciferase expression for each sample.
Statistical analyses
Data were expressed as means ± SD. Comparisons between two groups were conducted using the two-tailed t test. Differences among more than two groups were compared using one-way ANOVA. P<0.05 was considered statistically significant.
Data Availability
All relevant data are within the paper.
Funding Statement
National 973 Program (2012CB517700) JN. Nature and Science Foundation of China (81288001) JN. Natural Science Foundation of Guangdong province (S2013020012748) JN. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Finkelstein JD. Pathways and regulation of homocysteine metabolism in mammals. Semin Thromb Hemost. 2000; 26: 219–25 [DOI] [PubMed] [Google Scholar]
- 2. Kraus JP, Janosik M, Kozich V, Mandell R, Shih V, Sperandeo MP, et al. Cystathionine beta-synthase mutations in homocystinuria. Hum Mutat. 1999; 13: 362–75 [DOI] [PubMed] [Google Scholar]
- 3. Gellekink H, den Heijer M, Heil SG, Blom HJ. Genetic determinants of plasma total homocysteine. Semin Vasc Med. 2005; 5:98–109 [DOI] [PubMed] [Google Scholar]
- 4. Faraci FM. Hyperhomocysteinemia: a million ways to lose control. Arterioscler Thromb Vasc Biol. 2003; 23: 371–3 [DOI] [PubMed] [Google Scholar]
- 5. Yoshida K, Murata M, Yamaguchi T, Matsuzaki K. TGF-beta/Smad signaling during hepatic fibro-carcinogenesis (review). Int J Oncol. 2014; 45: 1363–71 10.3892/ijo.2014.2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Qu Y, Zong L, Xu M, Dong Y, Lu L. Effects of 18alpha-glycyrrhizin on TGF-beta1/Smad signaling pathway in rats with carbon tetrachloride-induced liver fibrosis. Int J Clin Exp Pathol. 2015; 8: 1292–301 [PMC free article] [PubMed] [Google Scholar]
- 7. Cho MK, Cho YH, Lee GH, Kim SG. Induction of cyclooxygenase-2 by bovine type I collagen in macrophages via C/EBP and CREB activation by multiple cell signaling pathways. Biochem Pharmacol. 2004; 67: 2239–50 [DOI] [PubMed] [Google Scholar]
- 8. Bosy-Westphal A, Petersen S, Hinrichsen H, Czech N, Müller MJ. Increased plasma homocysteine in liver cirrhosis. Hepatol Res. 2001; 20: 28–38 [DOI] [PubMed] [Google Scholar]
- 9. Robert K, Nehme J, Bourdon E, Pivert G, Friguet B, Delcayre C, et al. Cystathionine beta synthase deficiency promotes oxidative stress, fibrosis, and steatosis in mice liver. Gastroenterology. 2005; 128: 1405–15 [DOI] [PubMed] [Google Scholar]
- 10. Garcia-Tevijano ER, Berasain C, Rodriguez JA, Corrales FJ, Arias R, Martin-Duce A, et al. Hyperhomocysteinemia in liver cirrhosis: mechanisms and role in vascular and hepatic fibrosis. Hypertension. 2001; 38: 1217–21 [DOI] [PubMed] [Google Scholar]
- 11. Maclean KN, Sikora J, Kozich V, Jiang H, Greiner LS, Kraus E, et al. Cystathionine beta-synthase null homocystinuric mice fail to exhibit altered hemostasis or lowering of plasma homocysteine in response to betaine treatment. Mol Genet Metab. 2010; 101: 163–71 10.1016/j.ymgme.2010.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maclean KN, Sikora J, Kozich V, Jiang H, Greiner LS, Kraus E, et al. A novel transgenic mouse model of CBS-deficient homocystinuria does not incur hepatic steatosis or fibrosis and exhibits a hypercoagulative phenotype that is ameliorated by betaine treatment. Mol Genet Metab. 2010; 101: 153–62 10.1016/j.ymgme.2010.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sibani S, Melnyk S, Pogribny IP, Wang W, Hiou-Tim F, Deng L, et al. Studies of methionine cycle intermediates (SAM, SAH), DNA methylation and the impact of folate deficiency on tumor numbers in Min mice. Carcinogenesis. 2002; 23: 61–5 [DOI] [PubMed] [Google Scholar]
- 14. Wang H, Jiang X, Yang F, Chapman GB, Durante W, Sibinga NE, et al. Cyclin A transcriptional suppression is the major mechanism mediating homocysteine-induced endothelial cell growth inhibition. Blood. 2002; 99: 939–45 [PMC free article] [PubMed] [Google Scholar]
- 15. Jamaluddin MD, Chen I, Yang F, Jiang X, Jan M, Liu X, et al. Homocysteine inhibits endothelial cell growth via DNA hypomethylation of the cyclin A gene. Blood. 2007; 110: 3648–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995; 267: 1360–3 [DOI] [PubMed] [Google Scholar]
- 17. Zuccato C, Tartari M, Crotti A, Goffredo D, Valenza M, Conti L, et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet. 2003; 35: 76–83 [DOI] [PubMed] [Google Scholar]
- 18. Fahrner JA, Baylin SB. Heterochromatin: stable and unstable invasions at home and abroad. Genes Dev. 2003; 17: 1805–12 [DOI] [PubMed] [Google Scholar]
- 19. Roopra A, Qazi R, Schoenike B, Daley TJ, Morrison JF. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol Cell. 2004; 14: 727–38 [DOI] [PubMed] [Google Scholar]
- 20. Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005; 19: 815–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Roblin X, Pofelski J, Zarski JP. Steatosis, chronic hepatitis virus C infection and homocysteine. Gastroenterol Clin Biol. 2007; 31: 415–20 [DOI] [PubMed] [Google Scholar]
- 22. Waterland RA. Assessing the effects of high methionine intake on DNA methylation. J Nutr. 2006; 136: 1706S–1710S [DOI] [PubMed] [Google Scholar]
- 23. Tyagi N, Moshal KS, Sen U, Vacek TP, Kumar M, Hughes WJ, et al. H2S protects against methionine-induced oxidative stress in brain endothelial cells. Antioxid Redox Signal. 2009; 11: 25–33 10.1089/ars.2008.2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu S, Tao Y. Interplay between chromatin modifications and paused RNA polymerase II in dynamic transition between stalled and activated genes. Biol Rev Camb Philos Soc. 2013; 88: 40–8 10.1111/j.1469-185X.2012.00237.x [DOI] [PubMed] [Google Scholar]
- 25. Liu Q, Zhang Y, Mao H, Chen W, Luo N, Zhou Q, et al. A crosstalk between the Smad and JNK signaling in the TGF-beta-induced epithelial-mesenchymal transition in rat peritoneal mesothelial cells. PLoS One. 2012; 7: e32009 10.1371/journal.pone.0032009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sun G, Reddy MA, Yuan H, Lanting L, Kato M, Natarajan R. Epigenetic histone methylation modulates fibrotic gene expression. J Am Soc Nephrol. 2010; 21: 2069–80 10.1681/ASN.2010060633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Glenisson W, Castronovo V, Waltregny D. Histone deacetylase 4 is required for TGFbeta1-induced myofibroblastic differentiation. Biochim Biophys Acta. 2007; 1773: 1572–82 [DOI] [PubMed] [Google Scholar]
- 28. Diao JS, Xia WS, Yi CG, Wang YM, Li B, Xia W, et al. Trichostatin A inhibits collagen synthesis and induces apoptosis in keloid fibroblasts. Arch Dermatol Res. 2011; 303: 573–80 10.1007/s00403-011-1140-1 [DOI] [PubMed] [Google Scholar]
- 29. Yoshikawa M, Hishikawa K, Marumo T, Fujita T. Inhibition of histone deacetylase activity suppresses epithelial-to-mesenchymal transition induced by TGF-beta1 in human renal epithelial cells. J Am Soc Nephrol. 2007; 18: 58–65 [DOI] [PubMed] [Google Scholar]
- 30. Chen X, Skutt-Kakaria K, Davison J, Ou YL, Choi E, Malik P, et al. G9a/GLP-dependent histone H3K9me2 patterning during human hematopoietic stem cell lineage commitment. Genes Dev. 2012; 26: 2499–511 10.1101/gad.200329.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bruce AW, Donaldson IJ, Wood IC, Yerbury SA, Sadowski MI, Chapman M, et al. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc Natl Acad Sci U S A. 2004; 101: 10458–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zuccato C, Belyaev N, Conforti P, Ooi L, Tartari M, Papadimou E, et al. Widespread disruption of repressor element-1 silencing transcription factor/neuron-restrictive silencer factor occupancy at its target genes in Huntington's disease. J Neurosci. 2007; 27: 6972–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Davison JM, Mellott TJ, Kovacheva VP, Blusztajn JK. Gestational choline supply regulates methylation of histone H3, expression of histone methyltransferases G9a (Kmt1c) and Suv39h1 (Kmt1a), and DNA methylation of their genes in rat fetal liver and brain. J Biol Chem. 2009; 284: 1982–9 10.1074/jbc.M807651200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dong C, Wu Y, Yao J, Wang Y, Yu Y, Rychahou PG, et al. G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J Clin Invest. 2012; 122: 1469–86 10.1172/JCI57349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maze I, Covington HR, Dietz DM, LaPlant Q, Renthal W, Russo SJ, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010; 327: 213–6 10.1126/science.1179438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Covington HR, Maze I, Sun H, Bomze HM, DeMaio KD, Wu EY, et al. A role for repressive histone methylation in cocaine-induced vulnerability to stress. Neuron. 2011; 71: 656–70 10.1016/j.neuron.2011.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jakubowski H, Zhang L, Bardeguez A, Aviv A. Homocysteine thiolactone and protein homocysteinylation in human endothelial cells: implications for atherosclerosis. Circ Res. 2000; 87: 45–51 [DOI] [PubMed] [Google Scholar]
- 38. Ji C, Kaplowitz N. ER stress: can the liver cope? J Hepatol. 2006; 45: 321–33 [DOI] [PubMed] [Google Scholar]
- 39. Li SF, Guo L, Qian SW, Liu Y, Zhang YY, Zhang ZC, et al. G9a is transactivated by C/EBPbeta to facilitate mitotic clonal expansion during 3T3-L1 preadipocyte differentiation. Am J Physiol Endocrinol Metab. 2013; 304: E990–8 10.1152/ajpendo.00608.2012 [DOI] [PubMed] [Google Scholar]
- 40. Michalopolous G. Terminating hepatocyte proliferation during liver regeneration: the roles of two members of the same family (CCAAT-enhancer-binding protein alpha and beta) with opposing actions. Hepatology. 2015; 61: 32–4 10.1002/hep.27329 [DOI] [PubMed] [Google Scholar]
- 41. Liu X, Jia X, Yuan H, Ma K, Chen Y, Jin Y, et al. DNA methyltransferase 1 functions through C/ebpa to maintain hematopoietic stem and progenitor cells in zebrafish. J Hematol Oncol. 2015; 8: 15 10.1186/s13045-015-0115-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Castro R, Rivera I, Struys EA, Jansen EE, Ravasco P, Camilo ME, et al. Increased homocysteine and S-adenosylhomocysteine concentrations and DNA hypomethylation in vascular disease. Clin Chem. 2003; 49: 1292–6 [DOI] [PubMed] [Google Scholar]
- 43. Heil SG, Riksen NP, Boers GH, Smulders Y, Blom HJ. DNA methylation status is not impaired in treated cystathionine beta-synthase (CBS) deficient patients. Mol Genet Metab. 2007; 91: 55–60 [DOI] [PubMed] [Google Scholar]
- 44. Eskeland R, Eberharter A, Imhof A. HP1 binding to chromatin methylated at H3K9 is enhanced by auxiliary factors. Mol Cell Biol. 2007; 27: 453–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ebert A, Lein S, Schotta G, Reuter G. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res. 2006; 14: 377–92 [DOI] [PubMed] [Google Scholar]
- 46. St-Germain ME, Gagnon V, Parent S, Asselin E. Regulation of COX-2 protein expression by Akt in endometrial cancer cells is mediated through NF-kappaB/IkappaB pathway. Mol Cancer. 2004; 3: 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Keum YS, Kim HG, Bode AM, Surh YJ, Dong Z. UVB-induced COX-2 expression requires histone H3 phosphorylation at Ser10 and Ser28. Oncogene. 2013; 32: 444–52 10.1038/onc.2012.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.