Abstract
Although PTP4A3 has been shown to be a very important factor in promoting cancer progression, the role of its close family member PTP4A2 is still largely unknown. Recent reports have shown contradicting results on the role of PTP4A2 in breast cancer progression. Considering this, we aimed to investigate the prognostic value of PTP4A2 in five independent breast cancer data sets (minimum 198 patients per cohort, totaling 1,124 patients) in the Gene Expression Omnibus Database. We found that high expression of PTP4A2 was a favorable prognostic marker in all five independent breast cancer data sets, as well as in the combined cohort, with a hazard ratio of 0.68 (95% confidence interval =0.56–0.83; P<0.001). Low PTP4A2 expression was associated with estrogen receptor-negative tumors and tumors with higher histological grading; furthermore, low expression was inversely correlated with the expression of genes involved in proliferation, including MKI67 and the MCM gene family encoding the minichromosome maintenance proteins. These findings suggest that PTP4A2 may play a role in breast cancer progression by dysregulating cell proliferation. PTP4A2 expression was positively correlated with ESR1, the gene encoding estrogen receptor-alpha, and inversely correlated with EGFR expression, suggesting that PTP4A2 may be involved in these two important oncogenic pathways. Together, our results suggest that expression of PTP4A2 is a favorable prognostic marker in breast cancer.
Keywords: breast cancer, PTP4A2, survival, prognostic marker, overexpression, minichromosome maintenance proteins
Introduction
The protein tyrosine phosphatase (PTP) family consists of PRL-1, PRL-2, and PRL-3, which are encoded by the PTP4A1, PTP4A2, and PTP4A3 genes, respectively. Zeng et al1 were the first to identify PTP4A2 and PTP4A3, owing to their homology to PTP4A1; subsequently, the same group2 showed that PTP4A1 and PTP4A3 could both promote cell migration, invasion, and metastasis. Recently, the molecular mechanisms for PTP4A3-mediated cancer progression have been studied and revealed,3 with roles in the promotion of epithelial–mesenchymal transition,4 angiogenesis,5 cell cycle regulation,6 and autophagy7 having been discovered. Although overexpression of PTP4A3 has been consistently shown to promote cancer progression in multiple types of cancer, there are very few reports on the role of PTP4A2 in cancer progression.8
Overexpression of PTP4A2 was shown to promote breast tumor formation in a mouse model.9 However, two studies10,11 have shown that PTP4A2 was not differentially expressed between normal, benign, and cancerous tissues of the breast, while two other recent publications12,13 demonstrated that increased expression of PTP4A2 was correlated with a favorable overall and disease-free survival. These contradictory findings on the role of PTP4A2 in breast cancer progression led us to investigate the prognostic significance of PTP4A2 gene expression in five publicly available breast cancer data sets, each comprising at least 198 patients. We have assessed the association between PTP4A2 expression and proliferation markers including MKI67 and the MCM gene family encoding the minichromosome maintenance proteins, which were previously shown to confer prognostic significance in breast cancer patients.14
Materials and methods
Extraction of clinical and microarray gene expression data from breast cancer patient data sets
Five breast cancer patient data sets, GSE2034,15 GSE3494,16 GSE7390,17 GSE11121,18 and GSE1227619 were identified in the Gene Expression Omnibus (GEO) database using the following inclusion criteria: data sets compiled using the HG-U133 microarray platform, comprising >180 patients, and for whom relapse or survival data were available. Microarray gene expression data were retrieved from the data matrices deposited in the GEO database by the original authors. R scripting was used to extract the expression values from probe sets of genes of interest, and the clinical data from the data matrices was downloaded from GEO as previously described.20
Correlations of gene expression levels and clinical data
All statistical analyses were performed using SPSS19.0. The associations between expression level of PTP4A2 and clinical characteristics of the tumor, including estrogen receptor (ER) status and histological grade, and that between expression level of PTP4A2 and expression of MCM2-7 were tested by analysis of variance. The correlations between expression levels of PTP4A2 and MKI67, PTP4A2 and ESR1 (the gene encoding ER-alpha), and PTP4A2 and EGFR (gene encoding epidermal growth factor receptor) were tested by the Spearman’s rank test. Expression levels of PTP4A2 were divided into high and low levels using lower quartile expression level as the cutoff point for survival analysis. For Kaplan–Meier survival analysis, results were compared by the Wilcoxon–Gehan test.
Identification of genes coexpressed with PTP4A2
Patients were stratified into two groups based on their expression levels of PTP4A2. The gene expression patterns of patients in the PTP4A2-low subgroup and those in the PTP4A2-high subgroup were compared. Probe sets that were differentially expressed between these two subgroups were identified by the two-sample Welch’s t-test. This test was used to avoid the type I error that can arise due to unequal variances of the values of probe sets between subgroups. Briefly, a Welch’s t-test was applied to each probe set corresponding to a certain gene in the data matrix using our own Java application MyStats. P-values and the differential expression in fold changes for all the probe sets were generated as tab-delimited worksheets of Excel for further analysis. The genes were prioritized by ascending P-values.
Results
The association between PTP4A2 expression level and survival in breast cancer patients
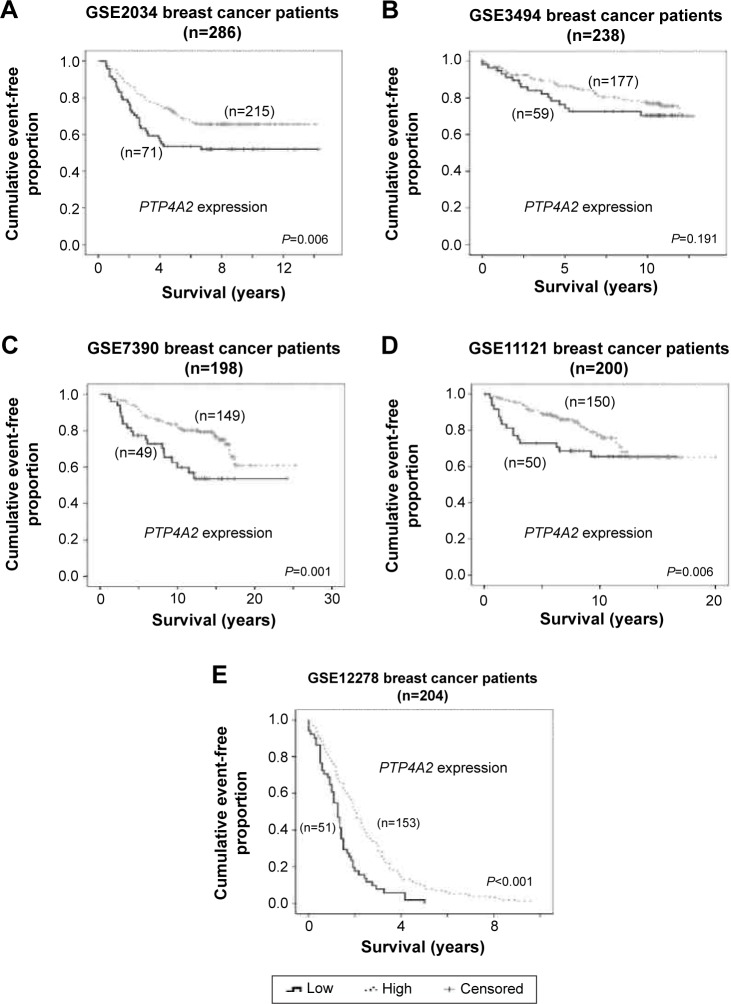
As shown in Figure 1, a low-level expression of PTP4A2 was consistently (and in four of the five cohorts, significantly) associated with a shorter survival time in the breast cancer data sets tested. In cohort GSE2034, patients whose tumors expressed PTP4A2 at a high level had a mean relapse-free survival time of 10.2 years, while those whose tumors expressed PTP4A2 at a low level had a mean relapse-free survival time of 8.4 years (Wilcoxon–Gehan test, P=0.006; Figure 1A). In cohort GSE3494, patients whose tumors expressed PTP4A2 at a high level had a mean disease-specific survival time of 10.7 years, while those whose tumors expressed PTP4A2 at a low level had a mean disease-specific survival time of 9.5 years (P=0.191; Figure 1B). In cohort GSE7390, patients whose tumors expressed PTP4A2 at a high level had a mean survival time of 19.7 years, while those whose tumors expressed PTP4A2 at a low level had a mean survival time of 15.7 years (P=0.001; Figure 1C). In cohort GSE11121, patients whose tumors expressed PTP4A2 at a high level had a mean distant metastasis-free survival time of 15.7 years, while those whose tumors expressed PTP4A2 at a low level had a mean distant metastasis-free survival time of 11.9 years (P=0.006; Figure 1D). In cohort GSE12276, patients whose tumors expressed PTP4A2 at a high level had a mean survival time of 2.4 years, while those whose tumors expressed PTP4A2 at a low level had a mean survival time of 1.4 years (P<0.001; Figure 1E). Our results suggest that a low-level expression of PTP4A2 is associated with a shorter survival time in breast cancer patients.
Figure 1.
The association between PTP4A2 expression and patient survival.
Notes: Kaplan–Meier curves for PTP4A2 expression in (A) GSE2034, (B) GSE3494, (C) GSE7390, (D) GSE11121, and (E) GSE12276 breast cancer data sets.
Abbreviation: PTP4A2, protein tyrosine phosphatase 4A2.
The association between the expression of PTP4A2 and ER status
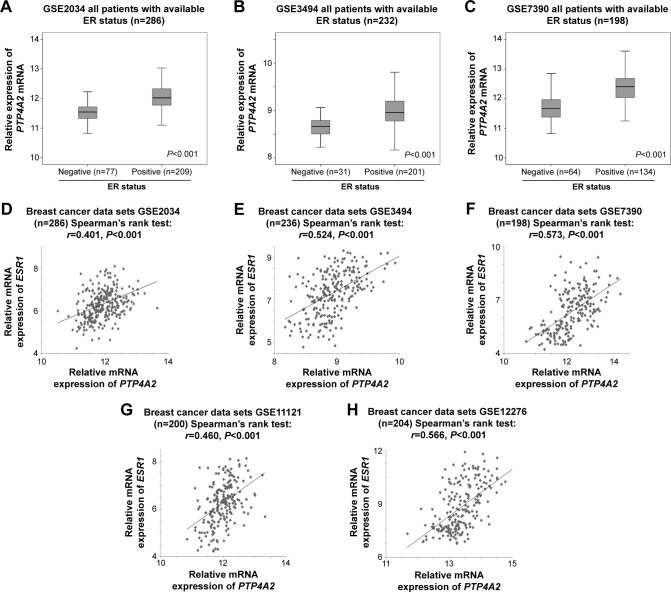
ER is a key biomarker in breast cancer, its status being associated with the efficacy of hormonal therapy. We analyzed all three cohorts for which patient ER status was known to investigate whether PTP4A2 expression level varied between patients with ER-positive and those with ER-negative disease. In cohort GSE2034, the 77 ER-negative tumors had significantly lower level of expression of PTP4A2 compared to the 209 ER-positive tumors (P<0.001; Figure 2A). In cohort GSE3494, the 31 ER-negative tumors had significantly lower level of expression of PTP4A2 compared to the 201 ER-positive tumors (P<0.001; Figure 2B). In cohort GSE7390, the 64 ER-negative tumors had significantly lower level of expression of PTP4A2 compared to the 134 ER-positive tumors (P<0.001; Figure 2C). Because ER-negative breast tumors are more aggressive and have a poorer prognosis,21 and considering that we have observed high PTP4A2 to be a favorable prognostic marker, it is unsurprising that low PTP4A2 correlates with ER-negative status.
Figure 2.
The association between PTP4A2 expression and the ER status and expression.
Notes: Box plots of PTP4A2 expression in tumors with different ER statuses in (A) GSE2034, (B) GSE3494, and (C) GSE7390 breast cancer data sets. Scatter plots for PTP4A2 expression against ESR1 expression in (A) GSE2034, (B) GSE3494, (C) GSE7390, (D) GSE2034, (E) GSE3494, (F) GSE7390, (G) GSE11121, and (H) GSE12276 breast cancer data sets.
Abbreviations: ER, estrogen receptor; ESR1, the gene encoding estrogen receptor-alpha; PTP4A2, protein tyrosine phosphatase 4A2.
Interestingly, ESR1 was identified as one of the genes being coexpressed with PTP4A2 in all the five breast cancer data sets. The expression levels of PTP4A2 and ESR1 were significantly correlated in GSE2034 (r=0.401, P<0.001; Figure 2D), GSE3494 (r=0.524, P<0.001; Figure 2E), GSE7390 (r=0.573, P<0.001; Figure 2F), GSE11121 (r=0.460, P<0.001; Figure 2G), and GSE12276 (r=0.566, P<0.001; Figure 2H) breast cancer data sets. Because ER-negative tumors had a lower expression of PTP4A2, while ESR1 and PTP4A2 expression was positively significantly correlated, our results suggest that ESR1 and PTP4A2 may be regulated in series or parallel in the same pathway.
The association between PTP4A2 expression level and histological grade
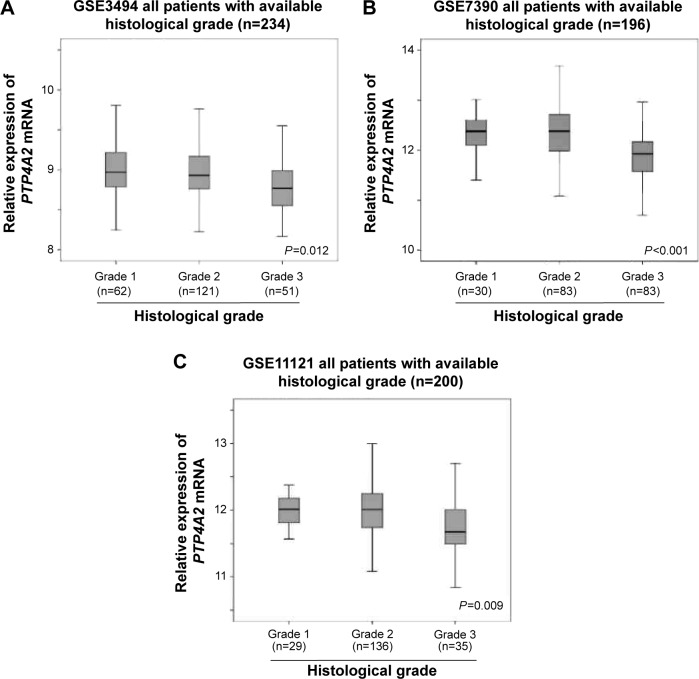
Histological grade is another important prognostic factor in breast cancer.22 Here, we also investigated the association between the expression level of PTP4A2 and the histological grading of the tumors in the three cohorts for which tumor grade had been documented. As shown in Figure 3, PTP4A2 expression was significantly lower in grade 3 tumors compared to those tumors with lower grading in GSE3494 (P=0.012; Figure 3A), GSE7390 (P<0.001; Figure 3B), and GSE11121 (P=0.009; Figure 3C), all the three breast cancer data sets with histological grading available in GEO database. The results suggest that PTP4A2 expression is downregulated in breast tumor with increasing histological grading.
Figure 3.
The association between PTP4A2 expression and histological grading.
Notes: Box plots of PTP4A2 expression in tumors with different histological grades in (A) GSE3494, (B) GSE7390, and (C) GSE11121 breast cancer data sets.
Abbreviation: PTP4A2, protein tyrosine phosphatase 4A2.
The association between PTP4A2 expression and the expression of proliferation-related genes
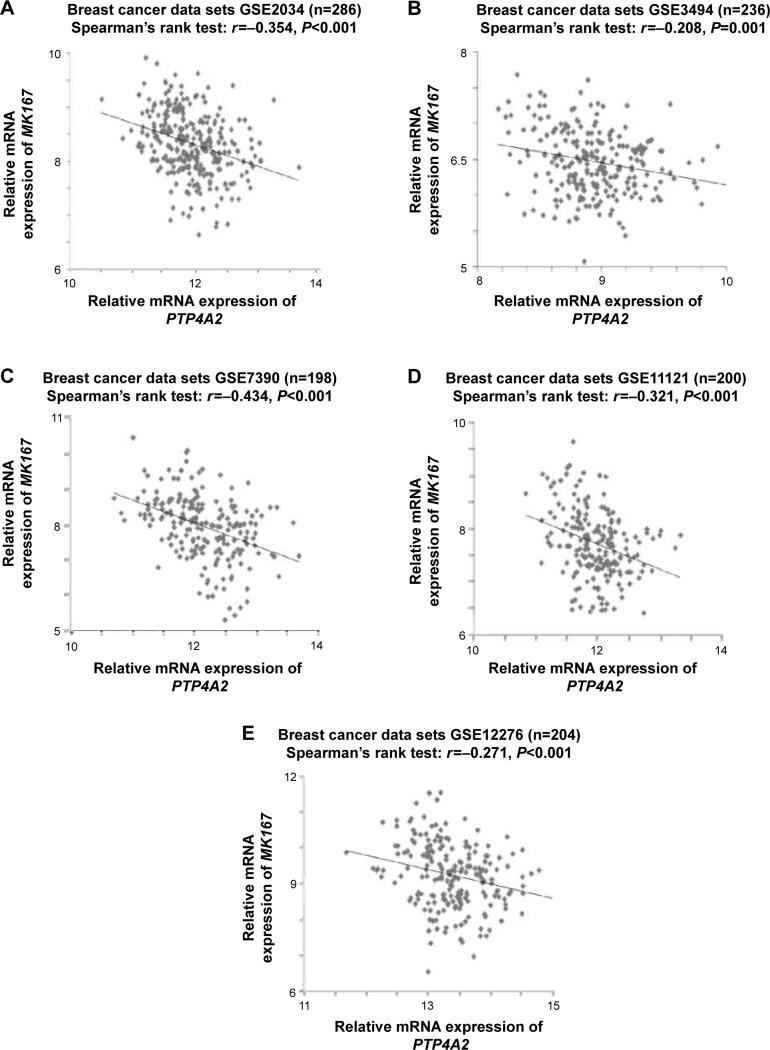
Next, to further elucidate the role of PTP4A2 in breast cancer progression, we investigated whether PTP4A2 expression is correlated with the expression of proliferation-related genes. It has been previously shown that MKI67 is an important prognostic and predictive biomarker in breast cancer.23 In the present study, we investigated the correlation between the expression levels of PTP4A2 and MKI67. As shown in Figure 4, the expression level of PTP4A2 was inversely correlated with the expression level of MKI67 in breast cancer specimens in GSE2034 (r=−0.354, P<0.001; Figure 4A), GSE3494 (r=−0.208, P=0.001; Figure 4B), GSE7390 (r=−0.434, P<0.001; Figure 4C), GSE11121 (r=−0.321, P<0.001; Figure 4D), and GSE12276 (r=−0.271, P<0.001; Figure 4E) breast cancer data sets.
Figure 4.
The correlations between PTP4A2 expression and MKI67 expression.
Notes: Scatter plots for PTP4A2 expression against MKI67 expression in (A) GSE2034, (B) GSE3494, (C) GSE7390, (D) GSE11121, and (E) GSE12276 breast cancer data sets.
Abbreviations: MKI67, marker of proliferation Ki-67; PTP4A2, protein tyrosine phosphatase 4A2.
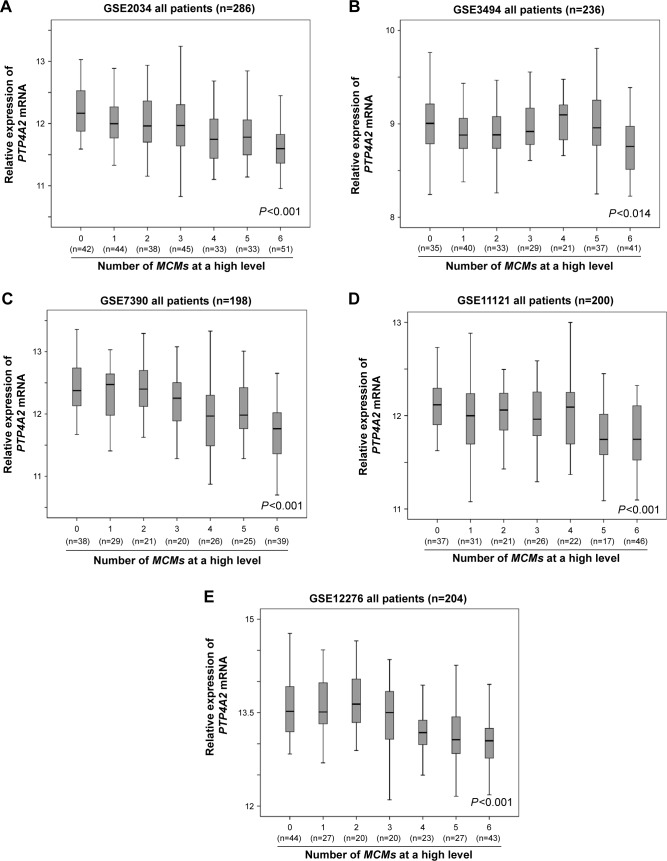
The MCM proteins play an important role in the regulation of cell proliferation and the initiation of DNA synthesis,24 and we have previously shown that members of the MCM gene family, when considered together, were a robust indicator of poor prognosis in breast cancer.14 In the present study, we investigated the correlation between the expression level of PTP4A2 and MCM gene family overexpression in the five breast cancer cohorts. As shown in Figure 5, the expression level of PTP4A2 decreased as the number of MCM genes expressed at a high level increased in cohorts GSE2034 (P<0.001; Figure 5A), GSE3494 (P=0.014; Figure 5B), GSE7390 (P<0.001; Figure 5C), GSE11121 (P<0.001; Figure 5D), and GSE12276 (P<0.001; Figure 5E).
Figure 5.
The association between PTP4A2 expression and the number of overexpressed MCM genes.
Notes: Box plots of PTP4A2 expression in tumors with increasing number of overexpressed MCM genes in (A) GSE2034, (B) GSE3494, (C) GSE7390, (D) GSE11121, and (E) GSE12276.
Abbreviations: MCM, minichromosome maintenance; PTP4A2, protein tyrosine phosphatase 4A2.
The prognostic significance of PTP4A2 in patients with different ER statuses or histological grading
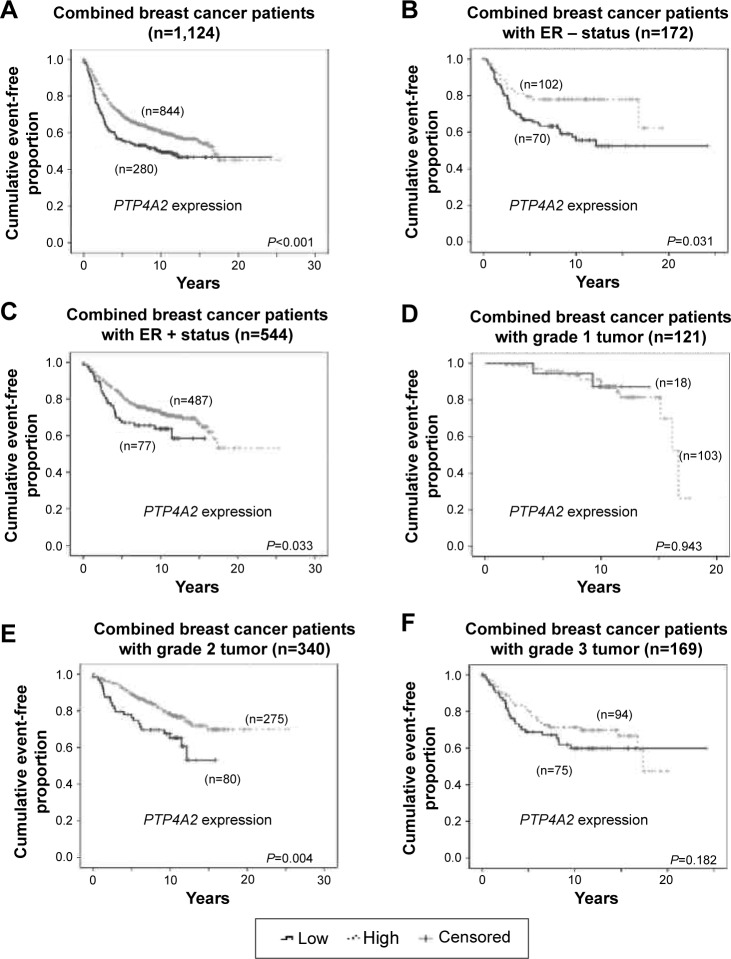
Because PTP4A2 expression was shown to be associated with survival and ER status, we further investigated whether the prognostic value of PTP4A2 is dependent on the ER status of the tumors. The five data sets were combined to increase the sample size and the statistical power to detect significant differences. In the combined data set, patients whose tumors expressed PTP4A2 at a low level had a mean survival time of 12.9 years, which was significantly shorter than those patients whose tumors expressed PTP4A2 at a high level, whose mean event-free survival time was 15.1 years (P<0.001; Figure 6A).
Figure 6.
The association between PTP4A2 expression and survival in patients stratified on clinicopathological parameters.
Notes: Kaplan–Meier curves for PTP4A2 expression in the combined breast cancer data set for (A) the entire combined cohort, (B) patients with ER-negative tumors, (C) patients with ER-positive tumors, (D) patients with grade 1 tumors, (E) patients with grade 2 tumors, and (F) patients with grade 3 tumors.
Abbreviations: ER, estrogen receptor; PTP4A2, protein tyrosine phosphatase 4A2.
The prognostic significance of PTP4A2 was further analyzed in the 716 (out of 1,124) patients for whom the ER status was known. In the 172 patients with ER-negative tumors, those patients whose tumors expressed PTP4A2 at a low level had a significantly shorter event-free survival time than those whose tumors expressed PTP4A2 at a high level (P=0.031; Figure 6B). Similarly, in the 544 patients with ER-positive status, those patients whose tumors expressed PTP4A2 at a low level had a significantly shorter event-free survival time compared to those whose tumors expressed PTP4A2 at a high level (P=0.033; Figure 6C).
The prognostic significance of PTP4A2 was also analyzed in the 630 patients for whom tumor histological grade was known. In the 121 and 169 patients with grade 1 and 3 tumors, respectively, the event-free survival time was not significantly different between patients whose tumors expressed PTP4A2 at a high level and those whose tumors expressed PTP4A2 at a low level (Figure 6D and F). In the 340 patients with grade 2 tumors, those patients whose tumors expressed PTP4A2 at a low level had a significantly shorter event-free survival time compared to those whose tumors expressed PTP4A2 at a high level (P=0.004; Figure 6E).
The correlation between PTP4A2 and EGFR expression
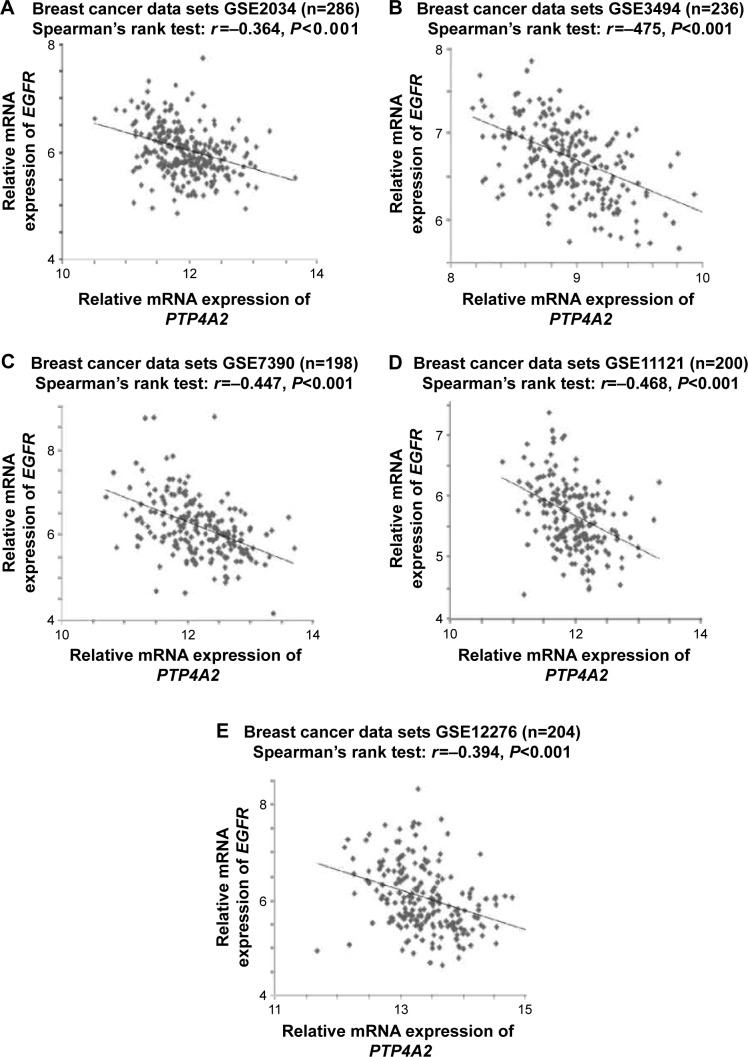
Considering that EGFR is known to regulate PTP4A3,25 we aimed to determine whether PTP4A2 and EGFR are coexpressed in breast cancer. As shown in Figure 7, the expression level of PTP4A2 was inversely correlated with the expression level of EGFR in all five breast cancer data sets analyzed, including GSE2034 (r=−0.364, P<0.001; Figure 7A), GSE3494 (r=−0.475, P<0.001; Figure 7B), GSE7390 (r=−0.447, P<0.001; Figure 7C), GSE11121 (r=−0.468, P<0.001; Figure 7D), and GSE12276 (r=−0.394, P<0.001; Figure 7E).
Figure 7.
The correlations between PTP4A2 expression and EGFR expression.
Notes: Scatter plots for PTP4A2 expression against EGFR expression in (A) GSE2034, (B) GSE3494, (C) GSE7390, (D) GSE11121, and (E) GSE12276 breast cancer data sets.
Abbreviations: EGFR, epidermal growth factor receptor; PTP4A2, protein tyrosine phosphatase 4A2.
Discussion
In the present study, we have demonstrated that PTP4A2 expression is a favorable prognostic marker for breast cancer patients. A low-level expression of PTP4A2 was associated with ER-negative tumor status, while PTP4A2 expression level was positively correlated with ESR1 expression level, suggesting that PTP4A2 may be regulated by the ER pathway. However, the prognostic significance of PTP4A2 was independent of tumor ER status. In addition, we found that PTP4A2 expression level was significantly lower in breast cancers with high histological grade, and that PTP4A2 expression was only significantly predictive of survival in patients with grade 2 tumors. On the other hand, PTP4A2 expression was inversely correlated with that of proliferation-associated genes, including MKI67 and MCM2–7. Finally, we have putatively identified EGFR as a gene coexpressed with PTP4A2 in all five breast cancer data sets, suggesting that PTP4A2 may be involved in the EGFR pathway.
The prognostic significance of PTP4A2 is controversial because some reports suggest that PTP4A2 promotes breast cancer progression,9 while others suggest that PTP4A2 plays an opposite role suppressing breast cancer progression.12,13 Considering these contradicting reports, we tested the prognostic significance of PTP4A2 in five large breast cancer data sets available in the GEO database. The results are highly consistent between these five breast cancer data sets, showing that a low-level expression of PTP4A2 mRNA was associated with a shorter event-free survival time, therefore suggesting that PTP4A2 expression is a favorable prognostic marker.
The regulation of PTP4A2 during breast cancer progression is largely unknown. In the present study, we have revealed two pathways that could potentially regulate PTP4A2. The first one is the ER pathway, wherein PTP4A2 expression was significantly higher in ER-positive tumors than in ER-negative tumors, and its expression was positively correlated with that of ER-alpha gene (ESR1). On the other hand, PTP4A2 expression was inversely correlated with EGFR. Our results warrant further analysis to investigate in vitro whether PTP4A2 is downstream of these two important pathways.
The observation of an association between increased expression of PTP4A2 and decreased expression of proliferation genes provides further insight into the possible mechanism for the role of PTP4A2 during cancer progression; alternatively, it is possible that PTP4A2 is a driver that inhibits proliferation of breast cancer cells, while it is also possible that downregulation of PTP4A2 is only a by-product of the dysregulated proliferation in breast cancer. Nonetheless, this result was consistent with the observation that PTP4A2 expression is a favorable prognostic marker in breast cancer.
One limitation of our study is that, as protein expression data are not available in the tested cohorts, our study is limited to the analysis of mRNA levels of PTP4A2; investigation on PRL-2 protein expression in a breast cancer cohort is required to validate our results and confirm the prognostic significance of PTP4A2.
In conclusion, the prognostic significance of PTP4A2 has been demonstrated with a high consistency. Because of the conflicting data presented previously, our study has provided convincing information that PTP4A2 expression is a favorable prognostic marker. Nevertheless, further investigations on how PTP4A2 is regulated and the molecular mechanisms driven by PTP4A2 in breast cancer progression are required to fully elucidate the role of PTP4A2 in breast cancer progression.
Acknowledgments
This study was supported by the University of Macau Start-Up Research Grant (SRG2014-00006-FHS), the Multi-Year Research Grant (MYRG2015-00065-FHS), the National Science Foundation for Young Scientists of China (grant #31301172), the Natural Science Foundation of Fujian Province (grant #2014J01122), and the National Undergraduate Training Programs for Innovation and Entrepreneurship at Fujian Normal University (grant #201410394019).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Zeng Q, Hong W, Tan YH. Mouse PRL-2 and PRL-3, two potentially prenylated protein tyrosine phosphatases homologous to PRL-1. Biochem Biophys Res Commun. 1998;244(2):421–427. doi: 10.1006/bbrc.1998.8291. [DOI] [PubMed] [Google Scholar]
- 2.Zeng Q, Dong JM, Guo K, et al. PRL-3 and PRL-1 promote cell migration, invasion, and metastasis. Cancer Res. 2003;63(11):2716–2722. [PubMed] [Google Scholar]
- 3.Al-Aidaroos AQ, Zeng Q. PRL-3 phosphatase and cancer metastasis. J Cell Biochem. 2010;111(5):1087–1098. doi: 10.1002/jcb.22913. [DOI] [PubMed] [Google Scholar]
- 4.Wang H, Quah SY, Dong JM, Manser E, Tang JP, Zeng Q. PRL-3 down-regulates PTEN expression and signals through PI3K to promote epithelial-mesenchymal transition. Cancer Res. 2007;67(7):2922–2926. doi: 10.1158/0008-5472.CAN-06-3598. [DOI] [PubMed] [Google Scholar]
- 5.Guo K, Li J, Wang H, et al. PRL-3 initiates tumor angiogenesis by recruiting endothelial cells in vitro and in vivo. Cancer Res. 2006;66(19):9625–9635. doi: 10.1158/0008-5472.CAN-06-0726. [DOI] [PubMed] [Google Scholar]
- 6.Basak S, Jacobs SB, Krieg AJ, et al. The metastasis-associated gene Prl-3 is a p53 target involved in cell-cycle regulation. Mol Cell. 2008;30(3):303–314. doi: 10.1016/j.molcel.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang YH, Al-Aidaroos AQ, Yuen HF, et al. A role of autophagy in PTP4A3-driven cancer progression. Autophagy. 2014;10(10):1787–1800. doi: 10.4161/auto.29989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stephens BJ, Han H, Gokhale V, Von Hoff DD. PRL phosphatases as potential molecular targets in cancer. Mol Cancer Ther. 2005;4(11):1653–1661. doi: 10.1158/1535-7163.MCT-05-0248. [DOI] [PubMed] [Google Scholar]
- 9.Hardy S, Wong NN, Muller WJ, Park M, Tremblay ML. Overexpression of the protein tyrosine phosphatase PRL-2 correlates with breast tumor formation and progression. Cancer Res. 2010;70(21):8959–8967. doi: 10.1158/0008-5472.CAN-10-2041. [DOI] [PubMed] [Google Scholar]
- 10.Dumaual CM, Sandusky GE, Soo HW, Werner SR, Crowell PL, Randall SK. Tissue-specific alterations of PRL-1 and PRL-2 expression in cancer. Am J Transl Res. 2012;4(1):83–101. [PMC free article] [PubMed] [Google Scholar]
- 11.Radke I, Gotte M, Kersting C, Mattsson B, Kiesel L, Wulfing P. Expression and prognostic impact of the protein tyrosine phosphatases PRL-1, PRL-2, and PRL-3 in breast cancer. Br J Cancer. 2006;95(3):347–354. doi: 10.1038/sj.bjc.6603261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andres SA, Wittliff JL, Cheng A. Protein tyrosine phosphatase 4A2 expression predicts overall and disease-free survival of human breast cancer and is associated with estrogen and progestin receptor status. Horm Cancer. 2013;4(4):208–221. doi: 10.1007/s12672-013-0141-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andres SA, Brock GN, Wittliff JL. Interrogating differences in expression of targeted gene sets to predict breast cancer outcome. BMC Cancer. 2013;13:326. doi: 10.1186/1471-2407-13-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwok HF, Zhang SD, McCrudden CM, et al. Prognostic significance of minichromosome maintenance proteins in breast cancer. Am J Cancer Res. 2015;5(1):52–71. [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 16.Miller LD, Smeds J, George J, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A. 2005;102(38):13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desmedt C, Piette F, Loi S, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13(11):3207–3214. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt M, Böhm D, von Törne C, et al. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008;68(13):5405–5413. doi: 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- 19.Bos PD, Zhang XH, Nadal C, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459(7249):1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuen HF, Gunasekharan VK, Chan KK, et al. RanGTPase: a candidate for Myc-mediated cancer progression. J Natl Cancer Inst. 2013;105(7):475–488. doi: 10.1093/jnci/djt028. [DOI] [PubMed] [Google Scholar]
- 21.Barcellos-Hoff MH. Does microenvironment contribute to the etiology of estrogen receptor-negative breast cancer? Clin Cancer Res. 2013;19(3):541–548. doi: 10.1158/1078-0432.CCR-12-2241. [DOI] [PubMed] [Google Scholar]
- 22.Rakha EA, Reis-Filho JS, Baehner F, et al. Breast cancer prognostic classification in the molecular era: the role of histological grade. Breast Cancer Res. 2010;12(4):207. doi: 10.1186/bcr2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yerushalmi R, Woods R, Ravdin PM, Hayes MM, Gelmon KA. Ki67 in breast cancer: prognostic and predictive potential. Lancet Oncol. 2010;11(2):174–183. doi: 10.1016/S1470-2045(09)70262-1. [DOI] [PubMed] [Google Scholar]
- 24.Ryu S, Driever W. Minichromosome maintenance proteins as markers for proliferation zones during embryogenesis. Cell Cycle. 2006;5(11):1140–1142. doi: 10.4161/cc.5.11.2779. [DOI] [PubMed] [Google Scholar]
- 25.Al-Aidaroos AQ, Yuen HF, Guo K, et al. Metastasis-associated PRL-3 induces EGFR activation and addiction in cancer cells. J Clin Invest. 2013;123(8):3459–3471. doi: 10.1172/JCI66824. [DOI] [PMC free article] [PubMed] [Google Scholar]