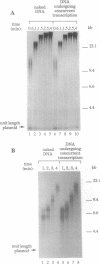
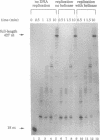
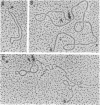
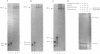Abstract
We have demonstrated elsewhere that a precisely placed, stalled Escherichia coli RNA polymerase ternary transcription complex (polymerase-RNA-DNA) stays on the DNA template after passage of a DNA replication fork. Moreover, the bypassed complex remains competent to resume elongation of its bound RNA chain. But the simplicity of our experimental system left several important questions unresolved: in particular, might the observation be relevant only to the particular ternary complex that we studied, and can the finding be generalized to a transcribing instead of a stalled RNA polymerase? To address these issues, we have created three additional ternary transcription complexes and examined their fates after passage of a replication fork. In addition, we have examined the fate of moving RNA polymerase molecules during DNA replication. The results suggest that our previous finding applies to all transcription intermediates of the E. coli RNA polymerase.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberts B. M., Frey L. T4 bacteriophage gene 32: a structural protein in the replication and recombination of DNA. Nature. 1970 Sep 26;227(5265):1313–1318. doi: 10.1038/2271313a0. [DOI] [PubMed] [Google Scholar]
- Arndt K. M., Chamberlin M. J. RNA chain elongation by Escherichia coli RNA polymerase. Factors affecting the stability of elongating ternary complexes. J Mol Biol. 1990 May 5;213(1):79–108. doi: 10.1016/S0022-2836(05)80123-8. [DOI] [PubMed] [Google Scholar]
- Barry J., Alberts B. In vitro complementation as an assay for new proteins required for bacteriophage T4 DNA replication: purification of the complex specified by T4 genes 44 and 62. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2717–2721. doi: 10.1073/pnas.69.9.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedinger P., Hochstrasser M., Jongeneel C. V., Alberts B. M. Properties of the T4 bacteriophage DNA replication apparatus: the T4 dda DNA helicase is required to pass a bound RNA polymerase molecule. Cell. 1983 Aug;34(1):115–123. doi: 10.1016/0092-8674(83)90141-1. [DOI] [PubMed] [Google Scholar]
- Brewer B. J. When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell. 1988 Jun 3;53(5):679–686. doi: 10.1016/0092-8674(88)90086-4. [DOI] [PubMed] [Google Scholar]
- Cha T. A., Alberts B. M. Studies of the DNA helicase-RNA primase unit from bacteriophage T4. A trinucleotide sequence on the DNA template starts RNA primer synthesis. J Biol Chem. 1986 May 25;261(15):7001–7010. [PubMed] [Google Scholar]
- Darst S. A., Edwards A. M., Kubalek E. W., Kornberg R. D. Three-dimensional structure of yeast RNA polymerase II at 16 A resolution. Cell. 1991 Jul 12;66(1):121–128. doi: 10.1016/0092-8674(91)90144-n. [DOI] [PubMed] [Google Scholar]
- Darst S. A., Kubalek E. W., Kornberg R. D. Three-dimensional structure of Escherichia coli RNA polymerase holoenzyme determined by electron crystallography. Nature. 1989 Aug 31;340(6236):730–732. doi: 10.1038/340730a0. [DOI] [PubMed] [Google Scholar]
- Dubochet J., Ducommun M., Zollinger M., Kellenberger E. A new preparation method for dark-field electron microscopy of biomacromolecules. J Ultrastruct Res. 1971 Apr;35(1):147–167. doi: 10.1016/s0022-5320(71)80148-x. [DOI] [PubMed] [Google Scholar]
- French S. Consequences of replication fork movement through transcription units in vivo. Science. 1992 Nov 20;258(5086):1362–1365. doi: 10.1126/science.1455232. [DOI] [PubMed] [Google Scholar]
- Helmstetter C. E. DNA synthesis during the division cycle of rapidly growing Escherichia coli B/r. J Mol Biol. 1968 Feb 14;31(3):507–518. doi: 10.1016/0022-2836(68)90424-5. [DOI] [PubMed] [Google Scholar]
- Kohlstaedt L. A., Wang J., Friedman J. M., Rice P. A., Steitz T. A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992 Jun 26;256(5065):1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- Krummel B., Chamberlin M. J. Structural analysis of ternary complexes of Escherichia coli RNA polymerase. Deoxyribonuclease I footprinting of defined complexes. J Mol Biol. 1992 May 20;225(2):239–250. doi: 10.1016/0022-2836(92)90918-a. [DOI] [PubMed] [Google Scholar]
- Krummel B., Chamberlin M. J. Structural analysis of ternary complexes of Escherichia coli RNA polymerase. Individual complexes halted along different transcription units have distinct and unexpected biochemical properties. J Mol Biol. 1992 May 20;225(2):221–237. doi: 10.1016/0022-2836(92)90917-9. [DOI] [PubMed] [Google Scholar]
- Liu B., Wong M. L., Tinker R. L., Geiduschek E. P., Alberts B. M. The DNA replication fork can pass RNA polymerase without displacing the nascent transcript. Nature. 1993 Nov 4;366(6450):33–39. doi: 10.1038/366033a0. [DOI] [PubMed] [Google Scholar]
- Meyer T. F., Geider K., Kurz C., Schaller H. Cleavage site of bacteriophage fd gene II-protein in the origin of viral strand replication. Nature. 1979 Mar 22;278(5702):365–367. doi: 10.1038/278365a0. [DOI] [PubMed] [Google Scholar]
- Morris C. F., Hama-Inaba H., Mace D., Sinha N. K., Alberts B. Purification of the gene 43, 44, 45, and 62 proteins of the bacteriophage T4 DNA replication apparatus. J Biol Chem. 1979 Jul 25;254(14):6787–6796. [PubMed] [Google Scholar]
- Morris C. F., Moran L. A., Alberts B. M. Purification of gene 41 protein of bacteriophage T4. J Biol Chem. 1979 Jul 25;254(14):6797–6802. [PubMed] [Google Scholar]
- Nossal N. G. RNA priming of DNA replication by bacteriophage T4 proteins. J Biol Chem. 1980 Mar 10;255(5):2176–2182. [PubMed] [Google Scholar]
- Shermoen A. W., O'Farrell P. H. Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell. 1991 Oct 18;67(2):303–310. doi: 10.1016/0092-8674(91)90182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surratt C. K., Milan S. C., Chamberlin M. J. Spontaneous cleavage of RNA in ternary complexes of Escherichia coli RNA polymerase and its significance for the mechanism of transcription. Proc Natl Acad Sci U S A. 1991 Sep 15;88(18):7983–7987. doi: 10.1073/pnas.88.18.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOSHIKAWA H., O'SULLIVAN A., SUEOKA N. SEQUENTIAL REPLICATION OF THE BACILLUS SUBTILIS CHROMOSOME. 3. REGULATION OF INITIATION. Proc Natl Acad Sci U S A. 1964 Oct;52:973–980. doi: 10.1073/pnas.52.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]