Abstract
RNAs adopt diverse folded structures that are essential for function and thus play critical roles in cellular biology. A striking example of this is the ribosome, a complex, three-dimensionally folded macromolecular machine that orchestrates protein synthesis. Advances in RNA biochemistry, structural and molecular biology, and bioinformatics have revealed other non-coding RNAs whose functions are dictated by their structure. It is not surprising that aberrantly folded RNA structures contribute to disease. In this review, we provide a brief introduction into RNA structural biology and then describe how RNA structures function in cells and cause or contribute to neurological disease. Finally, we highlight successful applications of rational design principles to provide chemical probes and lead compounds targeting structured RNAs. Based on several examples of well-characterized RNA-driven neurological disorders, we demonstrate how designed small molecules can facilitate study of RNA dysfunction, elucidating previously unknown roles for RNA in disease, and provide lead therapeutics.
Introduction
The genomic revolution of 1990s and 2000s brought about the discovery of a wide variety of non-coding (nc)RNAs (Dunham et al., 2012), leading to increased attention on understanding their physiological functions. As with other biomolecules, the function of RNA is closely linked to its three-dimensional structure, hence the rising interest in RNA structural biology (Figure 1). Advances in the field culminated with remarkable results such as the structures of bacterial (Ban et al., 2000; Schluenzen et al., 2000; Schuwirth et al., 2005; Wimberly et al., 2000; Yusupova et al., 2001) and eukaryotic (Ben-Shem et al., 2011) ribosomes.
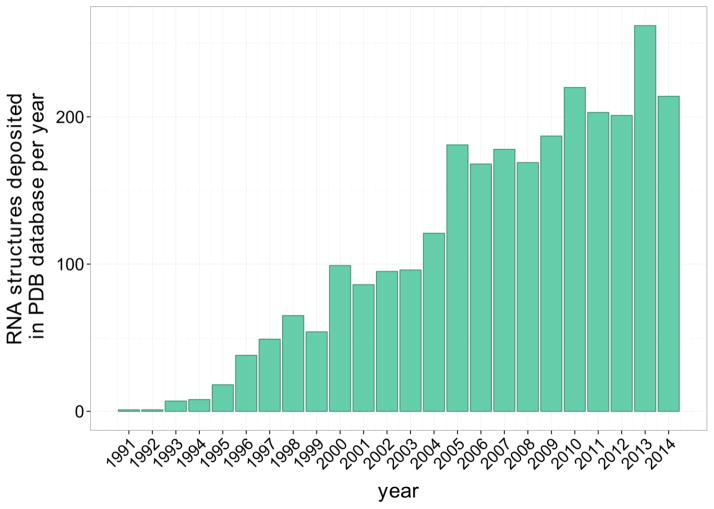
Figure 1.
Number of high-resolution structures containing RNA molecules deposited in PDB database per year since 1991. Source: PDB database http://www.rcsb.org (Berman et al., 2000).
As opposed to DNA’s double-stranded helix, RNA is most often single stranded and thus folds onto itself to minimize its free energy. RNA forms fully paired regions and non-canonically paired regions such as hairpins, internal loops, bulges, multibranch loops, and pseudoknots that dictate higher order folding patterns (i.e., tertiary structure, Figure 2). Both secondary and tertiary structural motifs serve as important recognition elements guiding RNA-RNA and RNA-protein interactions. The importance of proper RNA folding for executing its function was recognized early on from studies of rRNA and tRNA, which have highly conserved structural organizations (Korostelev et al., 2006). These structure-function relationships are not restricted to non-coding RNAs. The majority of eukaryotic protein-encoding RNAs (messenger RNA, mRNA) undergo splicing from pre-mRNA prior to translation (Li et al., 2007; Wang et al., 2008a), and secondary structural elements in proximity of intron-exon junctions determine the exact location of intron excision (Black and Grabowski, 2003; Buratti and Baralle, 2004; Buratti et al., 2004; De Conti et al., 2013; Hiller et al., 2007; Shepard and Hertel, 2008; Warf and Berglund, 2010). Moreover, the choice of splicing sites based on exonic and intronic splicing enhancers and silencers often depends on structural context (Hiller et al., 2007).
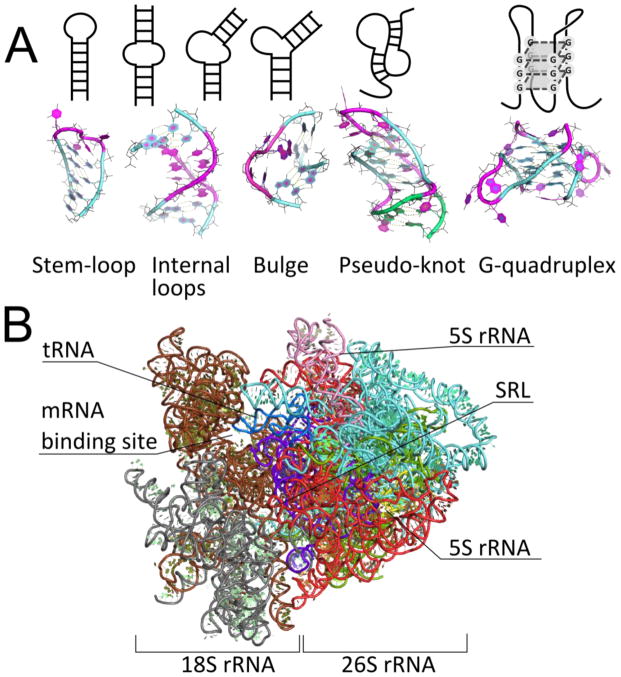
Figure 2.
Secondary structural elements found in RNA and the 3D structure of the ribosome. A) Schematics of secondary structural elements in RNA and representative three-dimensional structures from PDB. Nucleobases involved in Watson-Crick base pairing or G-quadruplexes are colored aquamarine and green; other bases are colored magenta. PDB IDs for 3D structures: stem-loop – 4TV0 (Zhang et al., 2014), internal loop – 2L8F (Lerman et al., 2011), bulge – 1AJL (Luebke et al., 1997), pseudo-knot – 2A43 (Pallan et al., 2005), G-quadruplex – 2KBP (Martadinata and Phan, 2009). B) Model of eukaryotic translation initiation complex. Proteins and mRNA have been removed for clarity. Individual RNA chains are represented in different colors (Voigts-Hoffmann et al., 2012). Downloaded and adapted from https://www.mol.biol.ethz.ch/groups/ban_group/Initiation. Structures are rendered with PyMOL (Schrödinger LLC, 2010).
An important feature of RNA secondary structures is their dynamic nature such that multiple structures with similar free energies can be adopted, allowing for conformational switching. Classic examples of large scale structural rearrangements include the ribosome’s structural changes that occur during translation (Korostelev et al., 2006) and the shape-shifting functions of riboswitches (Tucker and Breaker, 2005). Conformational changes upon interaction with other biomolecules are essential for other functional RNAs as well (Baumstark et al., 1997; Bugaut et al., 2012; Schultes and Bartel, 2000).
While proper folding of RNA is crucial to its normal function, it is natural that misfolding leads to dysregulation of cellular processes. In general, this pathology can arise either from loss-of-function or from gain-of-function (Mirkin, 2007; La Spada and Taylor, 2010). The former class of disease mechanisms usually denotes a mutation in sites that are crucial for proper folding and recognition of RNA by regulatory proteins. An altered folding equilibrium leads to dysregulation of cellular processes. An example are mutations in microtubule-associated protein tau (MAPT aka tau) pre-mRNA that destabilize the hairpin structure at the exon 10 – intron 10 junction (Figure 7), which alters its interaction with U1 snRNP and causes deregulation of alternative pre-mRNA splicing (Clark et al., 1998; Dumanchin et al., 1998; Hutton et al., 1998; Jiang et al., 2000; Spillantini et al., 1998a; Varani et al., 1999).
Figure 7.
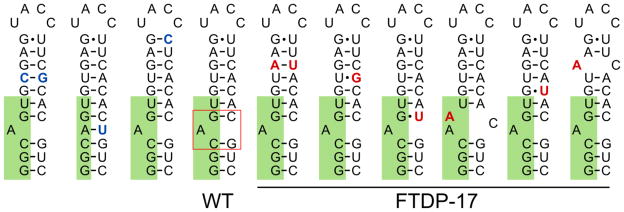
Secondary structure of the hairpin that regulates the alternative splicing of exon 10 in microtubule-associated protein tau (MAPT) pre-mRNA. Hairpin mutants are arranged in order of decreasing thermodynamic stability (left to right), according to published data (Donahue et al., 2006; Varani et al., 1999). Mutations increasing the number of intramolecular hydrogen bonds are highlighted in blue; mutations decreasing the number of Watson-Crick base pairs are highlighted in red. Exon nucleotides are highlighted in green. The bulge targeted by small molecules is boxed in red in the wild type (WT) hairpin (see main text for details). Mutations, which deregulate normal splicing of MAPT leading to frontotemporal dementia and Parkinsonism associated with chromosome 17 (FTDP-17), are underlined.
The gain-of-function class of disease mechanisms is triggered by emergence of aberrantly folded RNA structural motifs in locations where they are not normally present (Mirkin, 2007; Reddy and Housman, 1997; La Spada and Taylor, 2010). The most common cause of such pathologies is genetic mutations that lead to inclusions of pathogenic RNA fragments into gene transcripts, such as those observed in nucleotide repeat expansion disorders (La Spada et al., 1991; Verkerk et al., 1991). Depending on the location of a repeat expansion, there are a variety of downstream pathological mechanisms including misregulation of the RNA’s splicing in which the mutation is found (cis-regulation), production of non-functional or toxic proteins (Faustino and Cooper, 2003; Feng and Xie, 2013; Licatalosi and Darnell, 2006), and sequestration of essential proteins that corrupts their normal functions, which is typically processing of cellular RNAs (trans-regulation) (Krzyzosiak et al., 2012; Ranum and Cooper, 2006; Todd and Paulson, 2010).
Here, we provide a brief introduction into RNA structural biology, followed by a description of various RNA structures that perform biological functions and/or trigger pathological cascades in neurological diseases. Using several examples, we will exemplify how cellular and animal models of diseases help to understand pathogenic mechanisms. Finally, we will discuss the development and use of chemical tools (small molecules and oligonucleotides) that normalize deregulated RNA function, paving the road for potential RNA-targeting therapeutic interventions in neurological diseases in a precise and selective manner.
Brief overview of RNA structure
In contrast to DNA, RNA adopts a variety of secondary and tertiary structures. In base-paired regions, RNA adopts an A-type helical conformation, which is characterized by less compact folding than B-form DNA (11 base pairs per helical turn vs. 10.5 bp, respectively), a deeper major groove, and a shallower but wider minor groove. RNA’s 2′ hydroxyl group dictates a different sugar pucker, hydration state, and thermodynamic stability than DNA (Fohrer et al., 2006; Gyi et al., 1998). Hydrogen bonding between nucleobases plays a crucial role in the formation of RNA secondary and tertiary structures (SantaLucia et al., 1992; Turner et al., 1987); only 60–70% of bases in structured RNA form classic Watson-Crick contacts. Non-canonical Hoogsteen (Hoogsteen, 1963) and wobble pairs (Crick, 1966; Varani and McClain, 2000) are common in RNA and contribute to the diversity of folding and function (Schroeder et al., 2004).
RNA folding is hierarchical in that primary sequence defines secondary structure elements through nearest neighbor nucleotide effects (Brion and Westhof, 1997; Mathews et al., 1999, 2004). The most common secondary structures formed by RNA strands are base-paired regions, stem-loops (hairpins), internal loops, bulges (Hermann and Patel, 2000; Mathews et al., 1999, 2004), pseudoknots, kink-turns (Klein et al., 2001), complex multibranch loops, and G-quadruplexes (Gellert et al., 1962; Kim et al., 1991; Sundquist and Heaphy, 1993) (Figure 2). These secondary structural motifs in turn can stabilize each other through folding into more complex 3D patterns (Brion and Westhof, 1997). It is noteworthy that both secondary and tertiary structural elements can be dynamic and can interconvert depending on the presence of proteins, electrolytes, and small molecules (Schroeder et al., 2004), altering RNA function (Baumstark et al., 1997; Bugaut et al., 2012; Schultes and Bartel, 2000; Serganov et al., 2004).
The critical role of proper RNA folding for cellular functioning is best exemplified by the central place of RNA in controlling protein synthesis – from gene expression to splicing and translation of mRNA. Ribosomes and spliceosomes are comprised of highly structured RNA modules in complex with proteins (Figure 2B). At the same time, pre-mRNAs and often mRNAs themselves contain motifs that control efficiency of translation and splicing. For example, G-quadruplex structures in 5′ untranslated regions (UTRs) regulate translation of mRNA in context dependent manner: either by inhibition or by upregulation of cap-independent translation (Bugaut and Balasubramanian, 2012).
RNA’s function is often dependent on its interactions with other biomolecules and/or small molecules; those intermolecular interactions are dependent upon acquisition of the proper fold. These types of interactions, however, are far less studied than secondary structure. For RNA–RNA interactions, the most studied examples include “kissing” loop–loop and loop–stem contacts that were found to regulate viral (Chang and Tinoco, 1994, 1997; Nicholson and White, 2014), prokaryotic (Brunel et al., 2002; Marino et al., 1995), and eukaryotic RNAs (Sudarsan et al., 2003; Wachter, 2010). Stabilization of a kink-turn by long-distance contacts was found to be essential for the folding and cellular function of prokaryotic RNA (Lilley, 2012; Schroeder et al., 2011), particularly for ribosomal RNA (McPhee et al., 2014). Proteins can recognize their cognate RNA-binding partners structure-specifically, sequence-specifically, or via multiple modes of interaction (Serganov and Patel, 2008). The binding of proteins to RNA hairpins most often occurs with the apical loop rather than the helical stem region and resembles sequence-specific recognition of unstructured ssRNA. Therefore formation of such loops is a recurrent pattern of RNA folding that regulates interaction with regulatory proteins. The presence of similar structural motifs in two or more RNA molecules promiscuous protein binding, which can serve, for example, as a feedback loop for translational regulation by controlling ribosome loading (Serganov and Patel, 2008). Depending on context, the binding of RNA structural motifs to other biomolecules or structural domains can stabilize or destabilize its tertiary structure. Modulation of structural stability by small molecules underlies the biological function of folded RNA motifs, such as riboswitches (Thore et al., 2006; Zhang et al., 2010) and generally represents a potential strategy for targeting structured RNAs therapeutically, see e.g. (Childs et al., 2002; Meisner et al., 2004).
As of 2015, sequence complementarity is the most common way of targeting RNA. Mechanistically, an oligonucleotide hybridizes with a target strand (forms antiparallel base-paired duplex), thereby affecting its natural folding and interaction with cognate partners or recruiting intracellular machinery that cleaves the RNA (Bennett and Swayze, 2010). Such interaction, however, depends on the thermodynamic and kinetic barrier of unfolding the native conformation of both the target RNA and the antisense oligonucleotide (Freier et al., 1986; Mir and Southern, 1999; Walton et al., 1999), which can be prohibitively high (Li et al., 2008). Although there are RNA-binding proteins that facilitate RNA unfolding, the antisense-based strategy is still mostly applicable to non-structured or weakly structured RNAs. Hence, small molecules, which bind to folded RNA structures, e.g. loop regions, represent a complementary means for controlling RNA function to antisense oligonucleotides as small molecules are more apt to target folded RNAs.
Various methods have been developed to deduce RNA structure as understanding folding is foundational information that is used to generate structure-function hypotheses (Figure 4). Much of the activity in this area at present is deducing secondary structure, or a map of paired and unpaired regions. For example, much of the secondary structure of the ribosome was deduced by using phylogenetic comparison and the conservation of rRNA secondary structure (Figure 4A). Importantly, these studies have been used to assign the kingdoms of life and to elucidate new ones (Gutell et al., 1985; Noller and Woese, 1981). When the crystal structures of the ribosome became available, it was found that these phylogenetic secondary structures were highly accurate (Ban et al., 2000). When it is not possible to complete phylogenetic comparison because of a limited number of available RNA sequences, secondary structure prediction via free energy minimization is often used (Hofacker, 2003; Mathews et al., 1999, 2010; Zuker, 2003). The calculations provide the predicted lowest free energy structure and a series of suboptimal structures (Figure 4B). Alternatively, decomposition of RNA sequence into basic elements and reconstruction of the 3D folding based on known patterns for these elements proved to be helpful in spotting non-canonical base-pairs interactions (Parisien and Major, 2008). These approaches can be accurate for smaller RNAs, but accuracy diminishes as the size of an RNA increases. For larger RNAs, secondary structures can be deduced by using a combination of prediction and experimental constraints generated by structure probing (Mathews et al., 1999, 2010). RNA structure has been probed both in vitro and in cellulo with dimethyl sulfate (DMS) (Lempereur et al., 1985; Mathews et al., 2004; Tijerina et al., 2007; Wells et al., 2000), selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) (Kwok et al., 2013; Merino et al., 2005) (Figure 4C), and in a variety of nuclease protection assays (Ehresmann et al., 1987). This synergistic approach has been quite accurate to decode RNA structures, such as rRNA, viral RNAs, and others (Ding et al., 2014; Mathews et al., 2004; Merino et al., 2005; Weeks, 2010; Wilkinson et al., 2006). Likewise, methods have been developed to combine free energy minimization and sequence alignment (Bernhart et al., 2008; Mathews and Turner, 2002). Although secondary structure can provide important frameworks to develop hypotheses about the role of RNA structure, information about tertiary structure would also be helpful. Predictive approaches, in addition to the well-established experimental approaches such as X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy, are being developed to quickly and accurately decipher three-dimensional structure. It is likely that a combination of prediction and experiment, such as has been shown for secondary structure prediction, will allow for accurate models of RNA three dimensional structure to be generated quickly (Sripakdeevong et al., 2014).
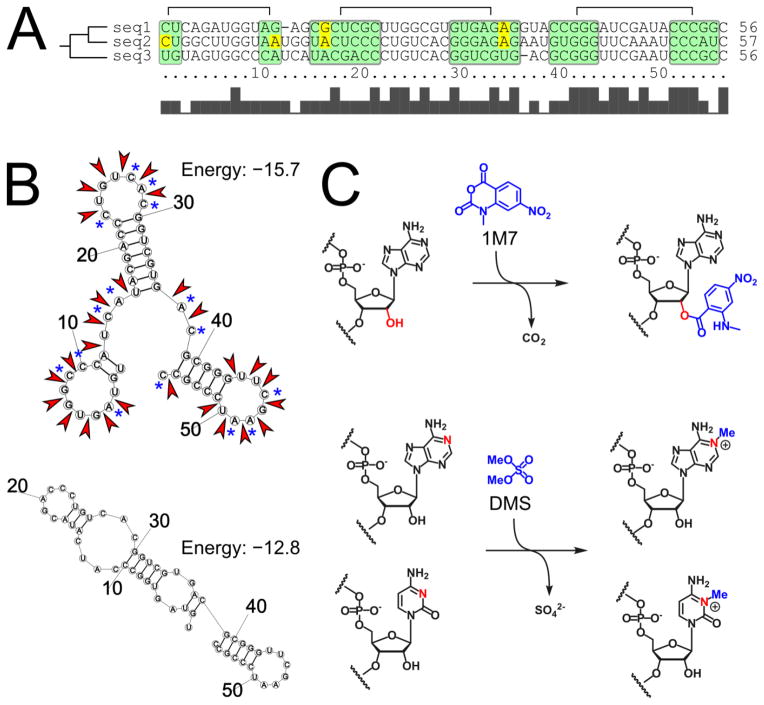
Figure 4.
RNA structure prediction and chemical mapping. A) Phylogenetic analysis of three sequences highlighting conserved base paired regions (green) and mismatches (yellow). B) Two alternative structures predicted for sequence 3 by RNAstructure (Reuter and Mathews, 2010). Red arrows indicate locations of reactive hydroxyl groups identified by 2′-hydroxyl acylation and primer extension (SHAPE). Blue asterisks indicate A and C residues susceptible for methylation by dimethyl sulfate (DMS). C) Chemical reactions underlying SHAPE and DMS mapping. 1M7 is 1-methyl-7-nitroisatoic anhydride.
The roles of structured RNAs in cellular biology
A great variety of functional, non-coding RNAs regulate neuronal development and physiological function (Qureshi and Mehler, 2012). Little is currently known, however, about the role of tertiary folding in most of these RNAs. Large non-coding RNAs (lncRNAs) are attracting attention due to the regulatory role they play in gene expression (Bond et al., 2009; Faghihi et al., 2008; Khalil et al., 2009; Ng et al., 2013; Tsai et al., 2010; Wahlestedt, 2013). Multiple enhancer RNA structures facilitate recruitment of CREB binding protein (CBP) to cognate genes and thereby regulate activity-dependent gene transcription in neurons, which underlies neuronal development and synaptic plasticity (Kim et al., 2010). Most of what is known about lncRNAs’ mechanism of action is antisense base-pairing with target genes, formation of DNA-RNA triplexes (Geisler and Coller, 2013), and allosteric recruitment of gene-repressing peptides to their target genes (Wang et al., 2008b) (Figure 5A).
Figure 5.
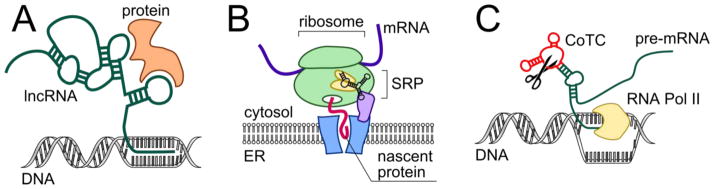
Representative functions of structured non-coding RNAs in cells. A) Schematic of a triplex formed between DNA and lncRNA that allosterically recruits gene-regulating proteins. B) Simplified representation of SRP RNA’s role in targeting membrane proteins to the endoplasmic reticulum (ER) for their proper folding. C) Schematic of co-transcriptional cleavage element regulating transcription termination of the β-globin gene. CoTC, co-transcriptional cleavage element; ER, endoplasmic reticulum; RNA Pol II, RNA polymerase II; SRP, signal recognition particle
It is estimated that >90% of human pre-mRNA is alternatively spliced, and this process is highly tissue-specific (Wang et al., 2008a). Many splicing regulators (proteins and ribonuclear proteins (RNPs)) apparently control their own syntheses (Li et al., 2007), providing important feedback loops. Some examples of splicing regulation by RNA-binding proteins and secondary structural elements are provided below. For more rigorous review of splicing regulation by pre-mRNA secondary structure, please see (De Conti et al., 2013; Warf and Berglund, 2010).
The neuro-oncological ventral antigen (NOVA) family of proteins regulates alternative splicing in neurons (Ule et al., 2006). Particularly, they control inhibitory neurotransmission in synapses by processing pre-mRNAs of glycine receptor subunit α2 (GlyRα2), GABAA receptor subunit γ2 (GABAARγ2), gephyrin, and Jnk2, amongst at least 50 other neuronal genes (Figure 3C) (Musunuru and Darnell, 2004; Ule et al., 2006). The sequence specificity of the RNA-binding K-homology (KH) domains of NOVA-1 protein was established by various methods such as systematic evolution of ligands by exponential enrichment (SELEX) (Musunuru and Darnell, 2004), UV cross-linking and immunoprecipitation (CLIP) (Ule et al., 2003), and individual-nucleotide resolution (i)CLIP (Sugimoto et al., 2012). NOVA-1 binds multiple repeats of UCAU intronic sequence in the GlyRα2 and GABAARγ2 pre-mRNAs. The binding of NOVA-1 KH-1–3 domains to RNA was studied extensively (Teplova et al., 2011) (Figure 3B), suggesting that multivalent binding of RNA-targeting motifs reorganizes the architecture of the RNA’s fold (Nicastro et al., 2015). Blockade of KH domains by antibodies disrupts NOVA–RNA interactions and causes neurodegeneration in patients with paraneoplastic opsoclonus-myoclonus-ataxia (POMA) (Luque et al., 1991).
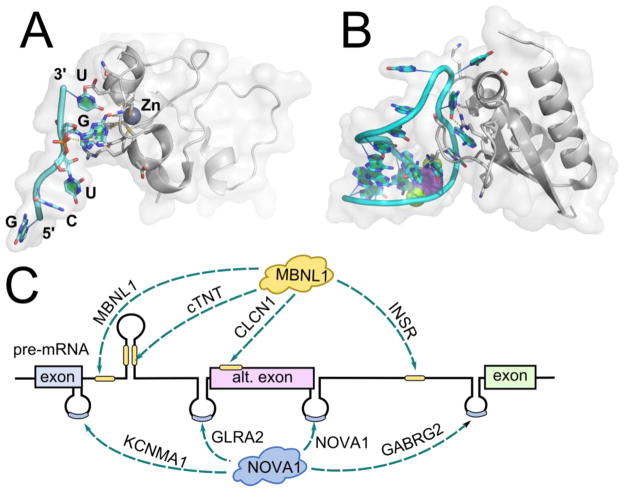
Figure 3.
Structure of splicing-regulating proteins MBNL1 and NOVA in complex with RNA recognition elements and location of splice sites in corresponding mRNAs. A) Fragment of MBNL1 complexed with a canonical recognition sequence of RNA, GCUGU, PDB ID 3D2S (Teplova and Patel, 2008). B) Fragment of NOVA1 complexed with a stem-loop of a cognate RNA, PDB ID 2ANN (Teplova et al., 2011). C) Locations of RNA recognition sites for MBNL1 and NOVA1, relative to alternatively spliced exons. Representative mRNA substrates are listed next to arrows denoting the RNA recognition element location (Konieczny et al., 2014; Ule et al., 2006). cTNT, cardiac troponin T, CLCN1, muscle-specific chloride ion channel 1; GABRG2, γ-aminobutyric acid (GABA) receptor γ2; GLRA2, glycine receptor α2; INSR, insulin receptor; KCNMA1, potassium large conductance calcium-activated channel, subfamily M, α1; MBNL1, muscleblind-like protein 1; NOVA1, neuro-oncological ventral antigen 1
Proteins from muscleblind-like family (MBNL), regulators of alternative pre-mRNA splicing, bind RNA with four zinc finger (ZnF) domains (Konieczny et al., 2014), determining which RNAs are MBNL substrates. MBNL1, the most studied member of the family, regulates alternative splicing of muscle-specific chloride ion channel (CLCN1) (Mankodi et al., 2002), insulin receptor (INSR) (Savkur et al., 2001), bridging integrator 1 (BIN1) (Fugier et al., 2011), cardiac troponin T (cTNT) (Philips et al., 1998), and several hundred other splicing events (Wang et al., 2012) (Figure 3C). By binding to a hairpin formed by canonical YGCY recognition sequence in cTNT pre-mRNA, MBNL1 triggers unwinding of the structure and subsequent splicing (Konieczny et al., 2014) (Figure 3A). Transcriptome-wide analysis revealed additional extranuclear roles of MBNL proteins such as targeting mRNAs to the rough endoplasmic reticulum and cellular membrane and regulation of local translation (Wang et al., 2012).
RNA structure can also regulate translation by controlling ribosomal loading. Over-expression of ribosomal proteins can cause binding to mRNA outside the context of a ribosome, thereby preventing translation. One example is L1 protein, the largest protein in the large ribosome subunit, which binds to its own mRNA (Tishchenko et al., 2008) by recognizing a domain that resembles rRNA fold (Nevskaya et al., 2005).
As mentioned above, a defined RNA structure also functions in the cellular delivery of proteins. Delivery of the translation complex to endoplasmic reticulum membrane is critical for maturation and targeting of secretory and membrane-bound proteins. This process is regulated by the signal recognition particle (SRP), a GTP-dependent ribonucleoprotein complex (Doudna and Batey, 2004). A U-turn forming Alu domain of SRP RNA plays the central role in assembly of the complex and binding to ribosome (Halic et al., 2004) and to SRP receptor in the endoplasmic membrane (Ataide et al., 2011) (Figure 5B). Mechanistic aspects of SRP function and the role of SRP RNA have been reviewed recently (Akopian et al., 2013).
Secondary structure elements in introns of mRNAs and in 5′ and 3′ UTRs have been identified as important markers for cellular splicing machinery (Buratti and Baralle, 2004), mRNA trafficking (e.g., axonal and dendritic transport (Gomes et al., 2014; Jung et al., 2012)), and translation initiation (Hughes, 2006). Stem-loops that contain canonical nucleotide sequences in their apical loop are markers for NOVA-1 recruiting. Notably, relative positioning of these structures determines NOVA-1-dependent skipping or inclusion of a particular exon (Figure 3) (Ule et al., 2006), similarly to MBNL-regulated mRNAs (Wang et al., 2012).
A stem-loop (with the canonical recognition pattern) in the 5′ UTR of ferritin-H and ferritin-L mRNA regulates expression of iron storage proteins (Thomson et al., 1999). The apical loop of the hairpin serves as a sensor for intracellular level of iron, hence the motif is called iron-responsive element (IRE). Interestingly, similar motifs were found in other mRNAs (Piccinelli and Samuelsson, 2007), including amyloid precursor protein (APP) (Rogers et al., 2002, 2011) and α-synuclein (Friedlich et al., 2007; Olivares et al., 2009). α-Synuclein is the prime toxic protein in Parkinson’s disease (PD) and other α-synucleinopathies (Lee and Trojanowski, 2006; Singleton et al., 2003). It forms fibrils that propagate across neurons in the brain and accumulate in Lewy bodies and Lewy neurites (Spillantini et al., 1997, 1998b). The expression level of α-synuclein is an important determinant in the rate of its fibrillization and neurotoxicity (El-Agnaf et al., 2006). Thus, down-regulating its expression is expected to be beneficial. The translation of some α-synuclein isoforms is regulated by a hairpin structure that is similar to the IRE in the 5′ UTR and was found to be iron-dependent (Febbraro et al., 2012).
Other common functional elements of structured mRNAs include three-way multibranch loops (junctions) (de la Peña et al., 2009), kink-turns (McPhee et al., 2014), hairpins, and G-quadruplexes (Bugaut and Balasubramanian, 2012). G-quadruplexes are often found in non-coding regions of mRNAs and provide structural domains recognized by functional proteins, e.g., fragile X mental retardation proteins (FMRP) (Melko and Bardoni, 2010). The structure selectivity of FMRP is different from other RNA-binding proteins containing KH domains (Siomi et al., 1993), as the RGG box binds G-quadruplex-containing mRNAs (including FMR1 mRNA which encodes FMRP) (Darnell et al., 2001). At the same time, another domain of FMRP binds between two ribosomal subunits and directly competes with eukaryotic translation elongation factors 1 alpha and 2 (eEF1A and eEF2) and aminoacyl-tRNA (Chen et al., 2014). Thereby, FMRP temporarily suspends the translation of the target mRNA and transports it from the nucleus to synapses. Phosphorylation of FMRP, due to external stimuli, decreases affinity of FMRP to the ribosome and de-represses transcription of its target mRNAs. Taken together, FMRP regulates activity-dependent local mRNA translation and influences cognitive processes at the cellular level (Antar et al., 2006).
Additional example of RNA-binding proteins essential for RNA transport is Staufen family (St Johnston et al., 1991). They regulate delivery of mRNA and thereby play central role in cellular differentiation and dendritic transport (Tang et al., 2001). Staufen proteins bind and stabilize base-paired sites in RNA secondary structures, which are abundant in 3′ UTR of mRNAs, (Sugimoto et al., 2015). Structural basis of double stranded (ds)RNA stem recognition by Staufen and other RNA-binding proteins has been recently reviewed (Gleghorn and Maquat, 2014).
Conversion of adenosine to inosine (i.e., RNA editing) is an important process in regulating splicing patterns of membrane receptors and ion channels, which has been proposed to be targeted therapeutically (Gott and Emeson, 2000; Morabito and Emeson, 2009). In general, it is the tertiary fold of the mRNA that determines the editing pattern (Bhalla et al., 2004; Ensterö et al., 2009; Rieder et al., 2013; Tian et al., 2011). Alternative editing of the serotonin receptor HTR2C pre-mRNA plays a major role in Prader-Willi syndrome pathology, a genetic disease associated with expression of serotonin receptor isoform with reduced constitutive activity and decreased efficiency of coupling to G-protein (Morabito et al., 2010).
Pathogenic RNA structures in neurological diseases: How can RNA structure cause disease?
Many of the functional secondary structural elements described above can spontaneously emerge as pathogenic agents as a result of point mutations, sequence deletions, and expansions. These newly formed, mutated RNA motifs interfere with normal interactions and initiate pathologic processes in cells. Examples of such gain-of-function include sequestration of RNA-binding proteins, activation of cryptic splicing sites, dysregulation of site-specific RNA editing by adenosine deaminase acting on RNA (ADAR), and formation of pseudo internal ribosome entry sites (IRES) and subsequent cap-independent translation (Mirkin, 2007), i.e., repeat associated non-ATG (RAN) translation (Zu et al., 2011). Interestingly, broad scale proteomics and transcriptome studies have shown that many peptides are produced without canonical AUG start codons (Lee et al., 2012; Slavoff et al., 2013; Stern-Ginossar et al., 2012). Thus, the biological implications of understanding RAN translation are not restricted to pathologic RNA structures.
Several causes of pathogenic RNA folding are known. In fact, most often it is the development of pathology that reveals the functional role of particular RNA structural motif. Single nucleotide polymorphisms (SNP) are a common cause of abnormal RNA folding. Mutations of a single nucleobase can change conformational stability of a secondary structure element, potentially disrupting a delicate equilibrium of RNA–protein interaction networks and causing downstream pathology. One of the most studied examples is MAPT (tau) mRNA, vide infra. For more detailed discussion on the role of mutated pre-mRNA secondary structures in splicing deregulation see (Warf and Berglund, 2010).
Retrotransposon insertions into intronic regions of pre-mRNAs contribute to protein isoform diversity by activation of cryptic splicing sites. This mechanism plays important roles in evolution, brain development, and cellular differentiation but often also contributes to genetic diseases (Baillie et al., 2011; Deininger and Batzer, 1999). For example, the structured retrotransposon Alu element of 7SL RNA plays a crucial role in the assembly of SRPs (Figure 5B). The Alu motif is the second most abundant retrotransposon in the human genome; its more than one million copies comprise roughly 11% of the entire genome (Lander et al., 2001). Insertions of Alu elements into intronic regions can activate cryptic exons, which leads to formation of unnatural protein isoforms (Pagani and Baralle, 2004; Vervoort et al., 1998). Abnormal recombination driven by Alu elements can also result in deletion of large gene fragments (Iida et al., 2012; Nakayama et al., 2010), and co-migration of other pathologic RNA fragments causes propagation of associated pathology (Clark et al., 2004; Kurosaki et al., 2009, 2012).
In the case of microsatellite repeat expansion disorders, particular DNA oligonucleotide fragments (repeated sequences) fold into stable hairpins, thereby causing strand ‘slipping’ during replication, repair, and recombination. This causes formation and elongation of such repetitive fragments (Gacy et al., 1995; López Castel et al., 2010). Transcription yields the corresponding single-stranded RNA that contains the expanded repeated sequence, which is aberrantly folded due to the presence of additional secondary structural elements (Figure 6A, B). Once stably incorporated into DNA sequence, expanded repeats are even more likely to fold into abnormal structures, which leads to gradual augmentation of pathology with age and in subsequent generations, a phenomenon known as repeat instability (Kovtun and McMurray, 2008; Liu et al., 2010; López Castel et al., 2010).
Figure 6.
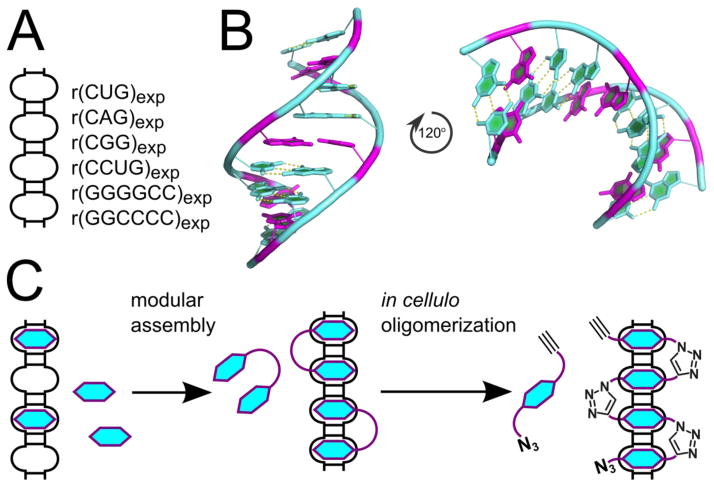
Targeting expanded repeating RNAs that cause or contribute to microsatellite disorders. A) Two-dimensional representation of a hairpin with multiple internal loops formed by expanded microsatellite repeats with examples of repetitive nucleotide sequences. B) Three-dimensional structure of r(CGG)3 fragment, PDB ID 3SJ2 (Kumar et al., 2011b). Coloring scheme is the same as for Figure 2. C) Targeting repetitive structural elements with small molecules, modularly assembled multivalent binders with increased affinity and selectivity, and in cellulo oligomerization as an ultimate strategy for precise targeting of expanded RNA repeats.
Microsatellite Repeat Expansion Disorders
As described above, RNA-binding proteins have sequence and structural binding preferences (Serganov and Patel, 2008). Some expanded repeats mimic these recognition elements and sequester proteins or RNA-processing cellular machinery away from their normal RNA targets (Echeverria and Cooper, 2012). Most often, GC-rich oligonucleotide sequences form expanded repeats: r(CAG)exp (where “exp” denotes an expanded repeating sequence) (Mangiarini et al., 1996), r(CUG)exp (Brook et al., 1992), r(CCUG)exp (Liquori et al., 2001), r(CGG)exp (Verkerk et al., 1991), and r(G4C2)exp (DeJesus-Hernandez et al., 2011) (Figure 6A). Downstream pathogenic processes in microsatellite repeat expansion disorders include deregulation of alternative pre-mRNA splicing (Philips et al., 1998; Ranum and Cooper, 2006), formation of insoluble nucleoprotein inclusions (foci) (Taneja et al., 1995), and RAN translation (Zu et al., 2011) into toxic peptides (reviewed in Krzyzosiak et al., 2012; Shin et al., 2009; Todd and Paulson, 2010). If a repeat expansion is located in a coding region, toxic proteins with a polypeptide chain encoded by the corresponding triplet codon are produced (La Spada and Taylor, 2010).
Two secondary structure elements are formed by expanded repeats: hairpins containing periodically repeating internal loops (Figure 6B) and G-quadruplexes. The latter is formed only by extremely G-rich sequences, e.g. r(G4C2)exp (Reddy et al., 2013). Some non-natural trinucleotide repeats (r(AGG) and r(UGG)) also fold into G-quadruplexes when expanded sufficiently (Sobczak et al., 2010). The central role of aberrant RNA folding in its gain-of-function has been extensively reviewed (Mirkin, 2007; Pearson, 2011; La Spada and Taylor, 2010). It is worth keeping in mind, however, that it is nearly impossible to delineate confounding role of sheer size of expanded repeats (and associated increase in stoichiometry of protein binding) from the role of their folding in RNA pathology.
r(CUG)exp and r(CCUG)exp: Myotonic dystrophy types 1 and 2
Formation of hairpins by r(CUG)exp and r(CCUG)exp, with multiple 1×1 and 2×2 internal loops in the stem, leads to the development of myotonic dystrophy (DM) types 1 and 2, respectively. DM1 is an autosomal dominant disease that is the most common form of adult muscular dystrophy with an incidence of 1:8000 (Brook et al., 1992). DM2 is also autosomal dominant but with less severe symptoms than DM1 (Liquori et al., 2001). While r(CUG)exp (DM1) resides in the 3′ UTR of dystrophia myotonica protein kinase (DMPK) mRNA, r(CCUG)exp (DM2) is located in the first intron of CCHC-type zinc finger nucleic acid binding protein (CNBP aka ZNF9) mRNA. There is overwhelming experimental evidence for protein sequestration as the major source of pathology in both diseases (Fardaei et al., 2002; Kanadia et al., 2006; Lu et al., 1999; Miller et al., 2000). MBNL proteins are sequestered in nuclear foci via dynamic interaction with r(CUG)exp or r(CCUG)exp (Fardaei et al., 2002; Mankodi et al., 2001). In DM1, sequestration leads to decreased nucleocytoplasmic transport and downregulation of DMPK. More importantly, protein sequestration deregulates alternative splicing of MBNL1- and MBNL2-dependent mRNAs (Charizanis et al., 2012; Ho et al., 2004; Philips et al., 1998). Inclusion of pathologic repeats in the native DMPK locus or a heterologous gene in transgenic mice is sufficient for development of disease phenotype, underscoring the central role of expanded repeat structure in DM pathology (Mankodi et al., 2000; Monckton et al., 1997). In addition to DM1, r(CUG)exp repeats are also characteristic of spinocerebellar ataxia type 8 (SCA8) (Daughters et al., 2009), and Huntington’s disease-like HDL2 (Rudnicki et al., 2007), where they also sequester MBNL proteins in nuclear foci and deregulate MBNL-dependent pre-mRNA splicing.
A great deal of effort has been expended to determine how MBNL proteins recognize r(CUG)exp and r(CCUG)exp, hence various studies into their secondary structures. Formation of r(CUG)exp and r(CCUG)exp hairpins has been confirmed in vitro (Gacy et al., 1995), and the stability of the RNA generally increases with repeat length (Napierała and Krzyzosiak, 1997; Tian et al., 2000). Importantly, these studies indicate that the hairpin stem contains non-canonically paired loops. A number of structures of r(CUG) repeat-containing constructs have been deposited in the Protein Data Bank (PDB) (Berman et al., 2000), which confirms formation of an imperfectly paired stem. The structures resemble an A-form helix but with deeper and narrower major grooves, reminiscent of A-form DNA. Further, some structures indicate loop dynamics (Coonrod et al., 2012; DeLorimier et al., 2014; Kiliszek et al., 2009; Kumar et al., 2011a; Mooers et al., 2005; Tamjar et al., 2012). The X-ray structure of r(CCUG)3 was recently published, providing insight into the structures of 5′UC3′/3′CU5′ present in the hairpin stem (Childs-Disney et al., 2014). In contrast to A-form RNA, the helical axis of r(CCUG)3 was bent by 18.5° at the central internal loop. Major and minor groove widening and narrowing, respectively, was observed in the internal loops. Molecular dynamics simulations suggested that 5′CU/3′UC internal loops exist in a dynamic equilibrium between two conformations (Childs-Disney et al., 2014).
A complex of tetrameric MBNL1 bound to a model RNA recognition site has been characterized by X-ray crystallography (Teplova and Patel, 2008) (Figure 3A); a complex of two MBNL1 zinc fingers complexed with two single stranded RNAs has also been refined from X-ray crystallographic data (Teplova and Patel, 2008). The specificity of MBNL1 binding to imperfect (with multiple U×U or CU×UC internal loops) rather than to fully base-paired stem-loop was demonstrated experimentally (Kino et al., 2004; Warf and Berglund, 2007). Taken together, these structural studies suggest that once bound to the DM1 and DM2 hairpins, MBNL1 unwinds their helical structure, which facilitates multivalent binding (Fu et al., 2012; Konieczny et al., 2014).
In addition to splicing deregulation, r(CUG)exp also recruits ribosomes and initiates RAN translation (Zu et al., 2011). This phenomenon was initially characterized in cells and tissue derived from SCA8 and DM1 patients. It was found that repeat length is a critical determinant of enabling RAN translation (Zu et al., 2011). These studies suggested that hairpins formed by r(CUG)exp serve as internal ribosome entry sites (IRES) and CUG binding protein (CUGBP1) acts as IRES translation-associated factor. After initial discovery of the stable hairpin structure formed by r(CUG)exp, experiments on other expanded trinucleotide repeats showed that the hairpins are formed by the vast majority (Sobczak et al., 2003), although not all trinucleotide sequences (Sobczak et al., 2010).
r(CGG)exp in FXS, FRAXE, & FXTAS
Expanded r(CGG) repeats form stable hairpins in the 5′ UTR of fragile X mental retardation mRNA (FMR1), which encodes fragile X mental retardation protein (FMRP), a key regulator of protein expression and trafficking in central nervous system. The repeats are major molecular pathogens in fragile X-associated tremor ataxia syndrome (FXTAS) (Hagerman et al., 2001; Jacquemont et al., 2003), fragile X syndrome (FXS) (Pieretti et al., 1991; Verkerk et al., 1991), and fragile XE syndrome (FRAXE) (Gecz et al., 1996; Knight et al., 1993). In FXTAS, a pre-mutation allele of r(CGG)exp (55–200 repeats (Jacquemont et al., 2003)) initiates RAN translation, leading to the premature start of FMR1 translation and production of non-functional FMRP with an N-terminal polyglycine chain (Todd et al., 2013). The repeats also bind and sequester various RNA-binding proteins in nuclear foci: heterogeneous ribonucleoprotein particle (hnRNP), MBNL1 (Iwahashi et al., 2006), DiGeorge syndrome critical region gene 8 protein (DGCR8) (Sellier et al., 2013), and Src-associated in mitosis, 68 kDa protein (Sam68) (Greco et al., 2006; Sellier et al., 2010) causing deregulation of alternative pre-mRNA splicing and microRNA processing (Arocena et al., 2005). Interestingly, stabilizing interruptions within the premutation allele of r(CGG)exp, such as AGG inserts, lead to branching of the hairpin structure. It was suggested that this branching precludes toxicity (Napierała et al., 2005).
FXS is caused by r(CGG) expansions of lengths >200, or the full mutation allele (Pieretti et al., 1991; Verkerk et al., 1991). Although it was known that the expansion leads to silencing of the FMR1 gene and hence loss of FMRP, the exact mechanism of silencing was only recently elucidated. Specifically, an RNA-DNA hybrid forms between r(CGG)exp and the FMR1 gene, silencing transcription via induction of chromatin remodeling (Colak et al., 2014). Stabilization of the r(CGG)exp hairpin by a small molecule prevents RNA unfolding and subsequent binding to FMR1 promoter region thus inhibiting gene silencing, vide infra.
Other hairpin-forming repeats include r(CAG)exp, which is most often located in coding regions and produce toxic polyglutamine-containing proteins as observed in Huntington’s disease (HD) (MacDonald et al., 1993). It was also suggested, however, that r(CAG)exp contributes to HD toxicity directly. Like r(CUG)exp, r(CAG)exp sequesters MBNL1 and binds with similar affinity (Mykowska et al., 2011). In addition, r(CAG)exp appears to sequester nucleolin to deregulate nuclear transport, recruit Dicer, and initiate RAN translation (Nalavade et al., 2013). In HD, the splicing factor SRSF6 binds to r(CAG)exp, which correlates with the observed deregulation of HTT (huntingtin) splicing in which exon 1 is aberrantly included. Exon 1 is followed by a premature stop codon, hence producing truncated huntingtin (protein) which is the major component of nuclear inclusions in HD (Sathasivam et al., 2013). The N-terminally truncated huntingtin has been shown to be highly pathogenic in a mouse model (Mangiarini et al., 1996). Together these findings suggest that aberrant splicing might be the major pathogenic cause of HD.
RNA hairpins and G-quadruplexes in ALS/FTD
G-rich strands of RNA and DNA can form G-quadruplex structures (Sundquist and Klug, 1989). The pathogenic expanded hexanucleotide r(G4C2)exp in C9ORF72, associated with amyotrophic lateral sclerosis and frontotemporal dementia (or c9ALS/FTD) (Akimoto et al., 2014; DeJesus-Hernandez et al., 2011), folds into two structures that are in equilibrium: a hairpin displaying periodically repeating G-rich internal loops and a G-quadruplex (Haeusler et al., 2014; Reddy et al., 2013; Su et al., 2014). r(G4C2)exp is the most frequent cause of familial amyotrophic ALS/FTD (Akimoto et al., 2014; DeJesus-Hernandez et al., 2011). Akin to other repeat expansions, r(G4C2)exp forms ribonuclear foci that sequester RNA-binding proteins, deregulates gene expression (Donnelly et al., 2013; Haeusler et al., 2014; Xu et al., 2013), and initiates RAN-translation (Ash et al., 2013; Donnelly et al., 2013). Formation of RNA-DNA hybrids (R-loops) underlies dysregulation of gene expression by r(G4C2)exp via different mechanisms (Haeusler et al., 2014; Wang et al., 2015). The RAN translation products of C9ORF72 are dipeptide repeat (DPR) proteins: poly(GlyPro), poly(GlyArg), poly(GlyAla), poly(ProAla), and poly(ProArg). Among them arginine-rich products appear to be the most toxic (Mizielinska et al., 2014). DPR proteins penetrate cellular membranes, accumulate in nucleoli, and impair pre-mRNA splicing and biogenesis of rRNA (Kwon et al., 2014; Mizielinska et al., 2014; Xu et al., 2013). Replacement of r(G4C2)exp by synonymous sequences encoding poly(GlyArg) and poly(ProArg) proteins causes neurodegeneration in a Drosophila model of ALS/FTD, which suggests a prevalent role for RAN translation in r(G4C2)exp-mediated pathology (Mizielinska et al., 2014).
Interestingly, C9ORF72 is bidirectionally transcribed; the antisense strand contains r(G2C4)exp which also forms nuclear foci and toxic RAN peptides (Gendron et al., 2013; Zu et al., 2013). Moreover, recent data suggest its importance in c9ALS/FTD pathogenesis, which should be taken into account in therapeutic treatment strategies (Lagier-Tourenne et al., 2013). Such considerations of both sense and antisense strands to toxicity is not unique to c9ALS/FTD as shown by the Ranum laboratory (Daughters et al., 2009).
Tauopathies: Tau mutations affect stability of RNA structure at exon-intron junctions
The stability of other RNA hairpins, not associated with repeat expansions, regulates normal alternative pre-mRNA splicing. For example, a stable stem-loop hairpin structure between exon 10 and intron 10 of MAPT dictates exon 10 inclusion or exclusion (Hutton et al., 1998; Jiang et al., 2000) (Figure 7). Mutations that destabilize the hairpin lead to more frequent inclusion of the exon and upregulation of a longer isoform of tau protein that contains four microtubule-binding domains (Liu and Gong, 2008). Overproduction of this mutant tau protein leads to frontotemporal dementia and Parkinsonism associated with chromosome 17 (FTDP-17) (Goedert et al., 1998). In contrast, stabilization of the hairpin leads to exon skipping (Donahue et al., 2006). NMR structural data confirmed that mutation of +3 G residue to A severely affects the conformation of the stem-loop (Varani et al., 1999), which regulates access of U1 snRNP to the splice site and exon inclusion (Jiang et al., 2000).
Leveraging RNA Structure to Design Chemical Probes of Function
The most challenging aspect of targeting RNA structural motifs with chemical probes is achieving the required level of specificity in the presence of high concentrations of bystander RNAs. rRNA and tRNA comprise ~80% and ~15% of total cellular RNA, respectively; the level of an individual mRNA is ≪1% (Johnson et al., 1975, 1977). Thus, from an abundance standpoint, a given structured RNA motif is most likely to be found in rRNA or tRNA. A modular approach in which multiple motifs are targeted simultaneously is one manner to overcome this problem (Figure 6C). That is, selectivity is much improved with a multivalent compound because, although there may be a large number of single targetable RNA motifs in the transcriptome, there are far fewer RNAs that have two targetable sites separated by a specific distance (Kumar et al., 2011a).
Pathogenic RNAs are traditionally targeted with antisense oligonucleotide probes. RNAs that are highly structured, however, are difficult to target with antisense oligonucleotides as the target RNA’s structure must first be unfolded. In contrast, small molecules are more apt to bind structured RNAs by binding to discrete motifs. Development of RNA-targeting small molecules has long been hampered by absence of rational principles of ligand design. Structure-based approaches have been complicated by RNA’s high flexibility and low-barrier for dynamic rearrangement of secondary structure elements. High throughput screening libraries are optimized for protein targets, leading to low hit rates in RNA-targeting screening campaigns. Despite these challenges, significant advances have been made in the development of small molecules that target RNA (Disney, 2013; Disney et al., 2014; Gallego and Varani, 2001; Guan and Disney, 2012; Shortridge and Varani, 2015; Thomas and Hergenrother, 2008; Velagapudi et al., 2014). Below, we summarize the present state of the development of chemical tools to study structured RNAs and targeting it therapeutically.
Identifying and designing chemical probes
Several key advances have pushed the RNA-targeting field forward including those in RNA structural biology, structure-based approaches including modeling of dynamic ensembles, and identification of RNA-binding modules (Batey et al., 2004; Childs-Disney et al., 2014; Davidson et al., 2009; Disney, 2013; Disney et al., 2014; Gallego and Varani, 2001; Jahromi et al., 2013a; Lee et al., 2010; Montange and Batey, 2006; Ofori et al., 2012; Palde et al., 2010; Parkesh et al., 2011; Shortridge and Varani, 2015; Stelzer et al., 2011; Trausch et al., 2011; Yildirim et al., 2013). High-resolution structures of ribosomes and other RNA-protein complexes combined with modeling of RNA dynamics have enabled structure-based approaches to develop new antibiotics and antivirals. Such studies have also enabled a fragment-based approach to drug design (Garavís et al., 2014). We recently reported a computational approach to design small molecules that bind RNA named Inforna (Velagapudi et al., 2014). Inforna uses a database of privileged RNA motif-small molecule interactions derived from a small molecule library-vs.-RNA motif library screen (2-Dimensional Combinatorial Screening; 2DCS (Childs-Disney et al., 2007; Disney et al., 2008)). This approach opens a new opportunity to overcome the selectivity problem of RNA-targeting small molecule ligands. While the number of structural building blocks is limited for RNA and targetable motifs can be present in many cellular RNAs, the probability of co-localization of two structural motifs in close proximity is much lower (see discussion in Kumar et al., 2011a). We demonstrated that a modular assembly approach using multivalent compounds that bind two motifs in close proximity increases selectively and potency (Lee et al., 2009a; Pushechnikov et al., 2009; Tran et al., 2014). Modular assembly is particularly attractive for targeting expanded repeats because of their own modular organization; by combining several identical RNA-targeting fragments in one molecular entity, a significant gain in affinity and selectivity can be achieved (Childs-Disney et al., 2012a) (Figure 6C). Unfortunately, as the valency of modularly assembled compounds increases, so does molecular weight, which hampers cellular permeability. To solve this problem, we demonstrated that a pathogenic RNA could template the assembly of RNA-targeting oligomers inside cells, using a disease-causing RNA as a catalysts for inhibitor synthesis at the required site of action (Rzuczek et al., 2014) (Figure 6C).
Another challenge in designing chemical probes for RNA is validation of a target in vivo. Techniques developed for resolving RNA-protein interactions (CLIP, iCLIP, etc.) are not directly applicable to small molecules. To solve this problem, we developed a technique named Chemical Cross-Linking and Isolation by Pull-down (Chem-CLIP) (Guan and Disney, 2013). It is based on selective covalent modification of a target RNA by attaching a reactive module and biotin to an RNA-binding scaffold. Reacted RNAs are subsequently captured with streptavidin-coated beads and analyzed by qRT-PCR. Extension of this technique termed ChemCLIP-Map allows one to locate the binding site of the small molecule (Yang et al., 2015). After reaction in cells, total RNA is treated with an antisense oligonucleotide and RNase H. Cleaved RNA fragments that reacted with the small molecule are then isolated with streptavidin beads, thereby establishing the region of the mRNA to which small molecule binds (Yang et al., 2015).
Targeting the RNAs that cause the myotonic dystrophies
Several approaches have been used to improve downstream defects caused by r(CUG)exp (DM1). To inhibit inclusion of exon 7a in CLCN1 mRNA, a 25-mer morpholino antisense oligonucleotides (ASO) targeting CLCN1 3′ and 5′ splice sites were administered to transgenic mice expressing r(CUG)250 in the 3′ UTR of human skeletal actin gene (HSALR mice (Mankodi et al., 2002)). The oligonucleotides improved splicing of CLCN1 and eliminated associated channelopathy (Wheeler et al., 2007). Similar effects together with correction of other DM1-associated pathologies were achieved with 25-mer morpholino ASO targeting r(CUG)exp in the same mouse model of DM1 (Wheeler et al., 2009, 2012). The ASO corrected splicing of MBNL1-dependent mRNAs but also downregulated the Taxilin beta (Txlnb) gene, which contains r(CUG)9. Finally, phase I and II clinical trials of a gapmer targeting mutant DMPK mRNA has been recently initiated (Isis Pharmaceuticals, 2014). A number of small molecule probes have also been developed for targeting r(CUG)exp that displace MBNL1 and improve downstream defects (Arambula et al., 2009; Childs-Disney et al., 2012a, 2012b, 2013; Hoskins et al., 2014; Jahromi et al., 2013a, 2013b; Parkesh et al., 2012). These compounds were either identified from screening, designed from the structure of r(CUG) repeats, or designed from privileged RNA motif-small molecule interactions including modularly assembled compounds thereof.
Several examples of small molecules that target r(CCUG)exp and improve DM2-associated defects have been reported (Childs-Disney et al., 2014; Lee et al., 2009b; Nguyen et al., 2014; Rzuczek et al., 2014). Our group reported that the aminoglycoside kanamycin A selectively binds 2×2 5′CU3′/3′UC5′ internal loops. From the X-ray structure and molecular dynamics simulations we identified that dynamic equilibrium between two conformations facilitates recognition of the small molecule. Once bound, a kanamycin derivative stabilizes one conformational state of the loop, thereby stabilizing the whole structure (Childs-Disney et al., 2014). When assembled into dimers and higher order oligomers, kanamycin exhibited high affinity and selectivity for r(CCUG) repeats (Lee et al., 2009a) and improved DM2-associated defects in a cellular model (Childs-Disney et al., 2014). During the course of our in cellulo studies, the structure of r(CCUG) repeats was refined and the binding of a dimeric kanamycin ligand modeled (Childs-Disney et al., 2014). This enabled the design of a kanamycin derivative that oligomerized in cellulo upon binding to r(CCUG)exp (Rzuczek et al., 2014). The derivative contained both azide and alkyne groups that were precisely positioned within the aminoglycoside as determined by modeling studies. Upon binding adjacent 5′CU3′/3′UC5′ in r(CCUG)exp, the alkyne group of one kanamycin is brought into close proximity to the azide group of an adjacent kanamycin. The otherwise unreactive groups react to form a stable triazole via a Huisgen dipolar cycloaddition reaction, a variant of click chemistry (Kolb et al., 2001). The in cellulo assembled oligomers exhibited potency (nM) far greater than pre-assembled oligomers and assembly only occurs in DM2-affected cells, not in healthy cells (Rzuczek et al., 2014). These studies also advance a new strategy in drug design, where a drug is synthesized on-site by using disease affected cell as a reactor and a disease causing biomolecule as a drug synthesis catalyst (Figure 6C).
Targeting r(CGG)exp, a pathogenic agent in FXTAS and FXS
Following the same rational approach to the design of ligands targeting r(CUG)exp and r(CCUG)exp, our group has developed selective compounds that target r(CGG)exp in both FXTAS and FXS (Colak et al., 2014; Disney et al., 2012; Tran et al., 2014). The small molecule 1a was identified to bind 5′CGG3′/3′GGC5′ using chemical similarity searching of a known RNA binder (Disney et al., 2012). A high-throughput FRET-based screen was used to identify compounds that disrupt the binding of r(CGG) repeats to an RNA-binding protein (DGCR8Δ) (Disney et al., 2012). Small molecule 1a inhibits formation of nuclear foci and improves FXTAS-associated alternative pre-mRNA splicing defects (Disney et al., 2012). Importantly, 1a was essential in elucidating the mechanism of FMRP silencing in FXS (Colak et al., 2014). The molecule stabilizes r(CGG)exp and prevents its unfolding – a crucial step in formation of the RNA-DNA hybrid that leads to inhibition of gene expression (Colak et al., 2014).
Modular assembly of a benzimidazole derivative that binds 5′CGG3′/3′GGC5′ also improves FXTAS-associated defects (Tran et al., 2014). Notably, binding of the compound to r(CGG)exp in cellulo does not affect translation of a downstream open reading frame (ORF). This is significant as the FMR1 ORF encodes FMRP, the loss of which causes FXS. An antisense oligonucleotide also improves FXTAS-associated defects but inhibits translation of the downstream ORF (Tran et al., 2014). The analogous molecule that binds r(CGG)exp irreversibly, via the installation of a nucleic acid-reactive group, inhibits RAN translation and normalizes alternative pre-mRNA splicing patterns (Yang et al., 2015). In contrast to antisense oligonucleotide targeting r(CGG)exp, the molecule did not affect loading of ribosomes onto mRNA or inhibit translation of the downstream ORF. By using a reactive compound, the binding sites for the small molecules were mapped to r(CGG)exp in cells using Chem-CLIP-Map (Yang et al., 2015).
Small molecule probes of r(G4C2)exp in c9ALS/FTD
Interestingly, the structures of r(CGG)exp and r(G4C2)exp have a shared motif – 1×1 nucleotide GG internal loops. We therefore completed a chemical similarity search of 1a to construct a library of small molecules that might bind r(G4C2)exp and alleviate ALS-associated symptoms. We identified three small molecules that bind r(G4C2)exp in cellulo (via Chem-CLIP), two of which inhibit r(G4C2)exp foci formation and RAN translation in c9ALS cellular model, including patient-derived iNeurons (Su et al., 2014). One was further tested in ALS patient-derived cells (iNeurons) and shown to be bioactive (Su et al., 2014). In another study, a porphyrin-derived G-quadruplex binder, TMPyP4, was shown in vitro to disrupt G-quadruplexes formed by r(G4C2)exp and inhibit its interaction with RNA-binding proteins in vitro (Zamiri et al., 2014).
Targeting MAPT (tau) mutations
As aforementioned, MAPT mutations can cause destabilization of RNA secondary structure, leading to deregulation of alternative splicing via altered interaction with U1 snRNP (Jiang et al., 2000). Stabilization of the RNA’s structure restores normal splicing patterns (Donahue et al., 2006) (Figure 7). A high throughput, FRET-based screen identified that the intercalator mitoxantrone (MTX, LDN-13978) stabilizes the mutated MAPT hairpin (Donahue et al., 2007). Importantly, mere intercalating ability is not sufficient for stabilization (Donahue et al., 2007). A structural study of MTX complexed with the MAPT hairpin by NMR spectroscopy revealed that MTX binds the bulge region of the stem-loop (Zheng et al., 2009). A number of attempts were made to optimize MTX by means of classic medicinal chemistry (Yang et al., 2009; Zheng et al., 2009) or conjugating it to aminoglycosides (Artigas and Marchán, 2015; Artigas et al., 2015). In addition, alternative chemotypes were actively sought via dynamic combinatorial chemistry (Lõpez-Senín et al., 2011; Ofori et al., 2012), and ‘Janus’-type compounds were designed to recognize the GU wobble base pair created by the mutations (Artigas and Marchán, 2013).
By using Inforna, a compound was identified by our laboratory that binds to the A-bulge in the MAPT mutant RNA hairpin (Luo and Disney, 2014). The molecule binds the same A-bulge, which was targeted by MTX, and increases thermal stability of the mutant MAPT hairpin but not of the wild type (Figure 7). Mutation of the A-bulge to an AU pair ablates small molecule binding, thus indicating secondary structure-specific mode of action (Luo and Disney, 2014). The compound also affects exon 10 inclusion in cellular models of disease.
Targeting IRE-like hairpins in Alzheimer’s (AD) and Parkinson’s Diseases (PD)
The free concentration of intracellular iron has a profound effect on conformation of IREs in several mRNAs, including APP (Alzheimer’s) and α-synuclein (Parkinson’s) (Friedlich et al., 2007; Rogers et al., 2011). The IRE-like hairpins regulate the expression level of the corresponding proteins (Febbraro et al., 2012; Rogers et al., 2002). High-throughput screening yielded a number of active compounds that regulate iron-dependent APP (Bandyopadhyay et al., 2006) and α-synuclein (Rogers et al., 2011) expression levels. It is plausible that an iron-dependent RNA conformational switch may serve as a mechanistic link connecting iron levels and protein-induced pathologies (Fine et al., 2015). Targeting the IRE-like hairpin directly is thus an attractive alternative therapeutic option in AD and PD.
Summary & Conclusions
The expanding functions of ncRNAs open up new opportunities for drug discovery. Structural biology and biophysical studies provide rational design principles for RNA-targeting ligands. There is no doubt that the number of functionally active RNA structural motifs will grow due to development of bioinformatic tools, our expanding knowledge of RNA structural biology, and the sheer size and diversity of the human genome. At the same time, the complexity of newly discovered RNA regulatory networks poses a significant challenge on the validation of bioinformatic transcriptome-wide analyses, and hence on the design of selective chemical tools and therapeutics. Variable tissue-specific patterns of pre-mRNA splicing, emerging functional role of RNA localization, as well as complex tangles of feedback loops are just few examples of ‘known unknowns’ of RNA cellular biology. In addition, despite substantial progress in understanding of RNA biochemistry, biophysics and structure, the relevance of these discoveries for in vivo biology remains disputable and requires rigorous validation. Nevertheless, in our opinion, it is an unaffordable luxury to disregard RNA as a potential drug target. Various therapeutically relevant structured ncRNA targets were outlined in this review, and strategies for targeting them provide a broad range of opportunities in drug development for currently incurable diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akimoto C, Volk AE, van Blitterswijk MM, Van den Broeck M, Leblond CS, Lumbroso S, Camu W, Neitzel B, Onodera O, van Rheenen W, et al. A blinded international study on the reliability of genetic testing for GGGGCC-repeat expansions in C9orf72 reveals marked differences in results among 14 laboratories. J Med Genet. 2014;51:419–424. doi: 10.1136/jmedgenet-2014-102360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akopian D, Shen K, Zhang X, Shan S. Signal recognition particle: an essential protein-targeting machine. Annu Rev Biochem. 2013;82:693–721. doi: 10.1146/annurev-biochem-072711-164732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antar LN, Li C, Zhang H, Carroll RC, Bassell GJ. Local functions for FMRP in axon growth cone motility and activity-dependent regulation of filopodia and spine synapses. Mol Cell Neurosci. 2006;32:37–48. doi: 10.1016/j.mcn.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Arambula JF, Ramisetty SR, Baranger AM, Zimmerman SC. A simple ligand that selectively targets CUG trinucleotide repeats and inhibits MBNL protein binding. Proc Natl Acad Sci U S A. 2009;106:16068–16073. doi: 10.1073/pnas.0901824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arocena DG, Iwahashi CK, Won N, Beilina A, Ludwig AL, Tassone F, Schwartz PH, Hagerman PJ. Induction of inclusion formation and disruption of lamin A/C structure by premutation CGG-repeat RNA in human cultured neural cells. Hum Mol Genet. 2005;14:3661–3671. doi: 10.1093/hmg/ddi394. [DOI] [PubMed] [Google Scholar]
- Artigas G, Marchán V. Synthesis of Janus compounds for the recognition of G-U mismatched nucleobase pairs. J Org Chem. 2013;78:10666–10677. doi: 10.1021/jo401684j. [DOI] [PubMed] [Google Scholar]
- Artigas G, Marchán V. Synthesis and Tau RNA binding evaluation of ametantrone-containing ligands. J Org Chem. 2015;80:2155–2164. doi: 10.1021/jo502661j. [DOI] [PubMed] [Google Scholar]
- Artigas G, López-Senín P, González C, Escaja N, Marchán V. Ametantrone-based compounds as potential regulators of Tau pre-mRNA alternative splicing. Org Biomol Chem. 2015;13:452–464. doi: 10.1039/c4ob01925c. [DOI] [PubMed] [Google Scholar]
- Ash PEA, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, Rademakers R, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ataide SF, Schmitz N, Shen K, Ke A, Shan S, Doudna JA, Ban N. The crystal structure of the signal recognition particle in complex with its receptor. Science. 2011;331:881–886. doi: 10.1126/science.1196473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, Brennan PM, Rizzu P, Smith S, Fell M, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S, Ni J, Ruggiero A, Walshe K, Rogers MS, Chattopadhyay N, Glicksman MA, Rogers JT. A high-throughput drug screen targeted to the 5′ untranslated region of Alzheimer amyloid precursor protein mRNA. J Biomol Screen Off J Soc Biomol Screen. 2006;11:469–480. doi: 10.1177/1087057106287271. [DOI] [PubMed] [Google Scholar]
- Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- Baumstark T, Schröder ARW, Riesner D. Viroid processing: Switch from cleavage to ligation is driven by a change from a tetraloop to a loop E conformation. EMBO J. 1997;16:599–610. doi: 10.1093/emboj/16.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, de Loubresse N, Melnikov S, Jenner L, Yusupova GZ, Yusupov MM, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova GZ, et al. The Structure of the Eukaryotic Ribosome at 3.0 A Resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhart SH, Hofacker IL, Will S, Gruber AR, Stadler PF. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinformatics. 2008;9:474. doi: 10.1186/1471-2105-9-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla T, Rosenthal JJC, Holmgren M, Reenan RA. Control of human potassium channel inactivation by editing of a small mRNA hairpin. Nat Struct Mol Biol. 2004;11:950–956. doi: 10.1038/nsmb825. [DOI] [PubMed] [Google Scholar]
- Black DL, Grabowski PJ. Alternative pre-mRNA splicing and neuronal function. In: Jeanteur P, editor. Regulation of Alternative Splicing. Springer; Berlin Heidelberg: 2003. pp. 187–216. [DOI] [PubMed] [Google Scholar]
- Bond AM, Vangompel MJW, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, Kohtz JD. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009;12:1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brion P, Westhof E. Hierarchy and dynamics of RNA folding. Annu Rev Biophys Biomol Struct. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion JP, Hughes TA, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- Brunel C, Marquet R, Romby P, Ehresmann C. RNA loop-loop interactions as dynamic functional motifs. Biochimie. 2002;84:925–944. doi: 10.1016/s0300-9084(02)01401-3. [DOI] [PubMed] [Google Scholar]
- Bugaut A, Balasubramanian S. 5′-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012;40:4727–4741. doi: 10.1093/nar/gks068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugaut A, Murat P, Balasubramanian S. An RNA hairpin to G-quadruplex conformational transition. J Am Chem Soc. 2012;134:19953–19956. doi: 10.1021/ja308665g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol Cell Biol. 2004;24:10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Muro AF, Giombi M, Gherbassi D, Iaconcig A, Baralle FE. RNA folding affects the recruitment of SR proteins by mouse and human polypurinic enhancer elements in the fibronectin EDA exon. Mol Cell Biol. 2004;24:1387–1400. doi: 10.1128/MCB.24.3.1387-1400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KY, Tinoco I. Characterization of a “kissing” hairpin complex derived from the human immunodeficiency virus genome. Proc Natl Acad Sci U S A. 1994;91:8705–8709. doi: 10.1073/pnas.91.18.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KY, Tinoco I. The structure of an RNA “kissing” hairpin complex of the HIV TAR hairpin loop and its complement. J Mol Biol. 1997;269:52–66. doi: 10.1006/jmbi.1997.1021. [DOI] [PubMed] [Google Scholar]
- Charizanis K, Lee KY, Batra R, Goodwin M, Zhang C, Yuan Y, Shiue L, Cline M, Scotti MM, Xia G, et al. Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron. 2012;75:437–450. doi: 10.1016/j.neuron.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E, Sharma MR, Shi X, Agrawal RK, Joseph S. Fragile X mental retardation protein regulates translation by binding directly to the ribosome. Mol Cell. 2014;54:407–417. doi: 10.1016/j.molcel.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs JL, Disney MD, Turner DH. Oligonucleotide directed misfolding of RNA inhibits Candida albicans group I intron splicing. Proc Natl Acad Sci U S A. 2002;99:11091–11096. doi: 10.1073/pnas.172391199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs-Disney JL, Wu M, Pushechnikov A, Aminova O, Disney MD. A small molecule microarray platform to select RNA internal loop-ligand interactions. ACS Chem Biol. 2007;2:745–754. doi: 10.1021/cb700174r. [DOI] [PubMed] [Google Scholar]
- Childs-Disney JL, Parkesh R, Nakamori M, Thornton CA, Disney MD. Rational design of bioactive, modularly assembled aminoglycosides targeting the RNA that causes myotonic dystrophy type 1. ACS Chem Biol. 2012a;7:1984–1993. doi: 10.1021/cb3001606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs-Disney JL, Hoskins JW, Rzuczek SG, Thornton CA, Disney MD. Rationally designed small molecules targeting the RNA that causes myotonic dystrophy type 1 are potently bioactive. ACS Chem Biol. 2012b;7:856–862. doi: 10.1021/cb200408a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs-Disney JL, Stepniak-Konieczna E, Tran T, Yildirim I, Park H, Chen CZ, Hoskins JW, Southall N, Marugan JJ, Patnaik S, et al. Induction and reversal of myotonic dystrophy type 1 pre-mRNA splicing defects by small molecules. Nat Commun. 2013;4:2044. doi: 10.1038/ncomms3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs-Disney JL, Yildirim I, Park H, Lohman JR, Guan L, Tran T, Sarkar P, Schatz GC, Disney MD. Structure of the myotonic dystrophy type 2 RNA and designed small molecules that reduce toxicity. ACS Chem Biol. 2014;9:538–550. doi: 10.1021/cb4007387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark LN, Poorkaj P, Wszolek Z, Geschwind DH, Nasreddine ZS, Miller B, Li D, Payami H, Awert F, Markopoulou K, et al. Pathogenic implications of mutations in the tau gene in pallido-ponto-nigral degeneration and related neurodegenerative disorders linked to chromosome 17. Proc Natl Acad Sci U S A. 1998;95:13103–13107. doi: 10.1073/pnas.95.22.13103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RM, Dalgliesh GL, Endres D, Gomez M, Taylor J, Bidichandani SI. Expansion of GAA triplet repeats in the human genome: Unique origin of the FRDA mutation at the center of an Alu. Genomics. 2004;83:373–383. doi: 10.1016/j.ygeno.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Colak D, Zaninovic N, Cohen MS, Rosenwaks Z, Yang WY, Gerhardt J, Disney MD, Jaffrey SR. Promoter-bound trinucleotide repeat mRNA drives epigenetic silencing in fragile X syndrome. Science. 2014;343:1002–1005. doi: 10.1126/science.1245831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Conti L, Baralle M, Buratti E. Exon and intron definition in pre-mRNA splicing. Wiley Interdiscip Rev RNA. 2013;4:49–60. doi: 10.1002/wrna.1140. [DOI] [PubMed] [Google Scholar]
- Coonrod LA, Lohman JR, Berglund JA. Utilizing the GAAA tetraloop/receptor to facilitate crystal packing and determination of the structure of a CUG RNA helix. Biochemistry. 2012;51:8330–8337. doi: 10.1021/bi300829w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick FH. Codon anticodon pairing: the wobble hypothesis. J Mol Biol. 1966;19:548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Daughters RS, Tuttle DL, Gao W, Ikeda Y, Moseley ML, Ebner TJ, Swanson MS, Ranum LPW. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5:e1000600. doi: 10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson A, Leeper TC, Athanassiou Z, Patora-Komisarska K, Karn J, Robinson JA, Varani G. Simultaneous recognition of HIV-1 TAR RNA bulge and loop sequences by cyclic peptide mimics of Tat protein. Proc Natl Acad Sci U S A. 2009;106:11931–11936. doi: 10.1073/pnas.0900629106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA. Alu repeats and human disease. Mol Genet Metab. 1999;67:183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLorimier E, Coonrod LA, Copperman J, Taber A, Reister EE, Sharma K, Todd PK, Guenza MG, Berglund JA. Modifications to toxic CUG RNAs induce structural stability, rescue mis-splicing in a myotonic dystrophy cell model and reduce toxicity in a myotonic dystrophy zebrafish model. Nucleic Acids Res. 2014;42:12768–12778. doi: 10.1093/nar/gku941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- Disney MD. Rational design of chemical genetic probes of RNA function and lead therapeutics targeting repeating transcripts. Drug Discov Today. 2013;18:1228–1236. doi: 10.1016/j.drudis.2013.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disney MD, Labuda LP, Paul DJ, Poplawski SG, Pushechnikov A, Tran T, Velagapudi SP, Wu M, Childs-Disney JL. Two-dimensional combinatorial screening identifies specific aminoglycoside-RNA internal loop partners. J Am Chem Soc. 2008;130:11185–11194. doi: 10.1021/ja803234t. [DOI] [PubMed] [Google Scholar]
- Disney MD, Liu B, Yang WY, Sellier C, Tran T, Charlet-Berguerand N, Childs-Disney JL. A small molecule that targets r(CGG)(exp) and improves defects in fragile X-associated tremor ataxia syndrome. ACS Chem Biol. 2012;7:1711–1718. doi: 10.1021/cb300135h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disney MD, Yildirim I, Childs-Disney JL. Methods to enable the design of bioactive small molecules targeting RNA. Org Biomol Chem. 2014;12:1029–1039. doi: 10.1039/c3ob42023j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue CP, Muratore C, Wu JY, Kosik KS, Wolfe MS. Stabilization of the tau exon 10 stem loop alters pre-mRNA splicing. J Biol Chem. 2006;281:23302–23306. doi: 10.1074/jbc.C600143200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue CP, Ni J, Rozners E, Glicksman MA, Wolfe MS. Identification of tau stem loop RNA stabilizers. J Biomol Screen. 2007;12:789–799. doi: 10.1177/1087057107302676. [DOI] [PubMed] [Google Scholar]
- Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, Fines DM, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Batey RT. Structural insights into the signal recognition particle. Annu Rev Biochem. 2004;73:539–557. doi: 10.1146/annurev.biochem.73.011303.074048. [DOI] [PubMed] [Google Scholar]
- Dumanchin C, Camuzat A, Campion D, Verpillat P, Hannequin D, Dubois B, Saugier-Veber P, Martin C, Penet C, Charbonnier F, et al. Segregation of a missense mutation in the microtubule-associated protein tau gene with familial frontotemporal dementia and parkinsonism. Hum Mol Genet. 1998;7:1825–1829. doi: 10.1093/hmg/7.11.1825. [DOI] [PubMed] [Google Scholar]
- Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverria GV, Cooper TA. RNA-binding proteins in microsatellite expansion disorders: mediators of RNA toxicity. Brain Res. 2012;1462:100–111. doi: 10.1016/j.brainres.2012.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehresmann C, Baudin F, Mougel M, Romby P, Ebel JP, Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987;15:9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Agnaf OMA, Salem SA, Paleologou KE, Curran MD, Gibson MJ, Court JA, Schlossmacher MG, Allsop D. Detection of oligomeric forms of α-synuclein protein in human plasma as a potential biomarker for Parkinson’s disease. FASEB J. 2006;20:419–425. doi: 10.1096/fj.03-1449com. [DOI] [PubMed] [Google Scholar]
- Ensterö M, Daniel C, Wahlstedt H, Major F, Öhman M. Recognition and coupling of A-to-I edited sites are determined by the tertiary structure of the RNA. Nucleic Acids Res. 2009;37:6916–6926. doi: 10.1093/nar/gkp731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of β-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fardaei M, Rogers MT, Thorpe HM, Larkin K, Hamshere MG, Harper PS, Brook JD. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum Mol Genet. 2002;11:805–814. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- Febbraro F, Giorgi M, Caldarola S, Loreni F, Romero-Ramos M. α-Synuclein expression is modulated at the translational level by iron. Neuroreport. 2012;23:576–580. doi: 10.1097/WNR.0b013e328354a1f0. [DOI] [PubMed] [Google Scholar]
- Feng D, Xie J. Aberrant splicing in neurological diseases. Wiley Interdiscip Rev RNA. 2013;4:631–649. doi: 10.1002/wrna.1184. [DOI] [PubMed] [Google Scholar]
- Fine JM, Renner DB, Forsberg AC, Cameron RA, Galick BT, Le C, Conway PM, Stroebel BM, Frey WH, Hanson LR. Intranasal deferoxamine engages multiple pathways to decrease memory loss in the APP/PS1 model of amyloid accumulation. Neurosci Lett. 2015;584:362–367. doi: 10.1016/j.neulet.2014.11.013. [DOI] [PubMed] [Google Scholar]
- Fohrer J, Hennig M, Carlomagno T. Influence of the 2′-hydroxyl group conformation on the stability of A-form helices in RNA. J Mol Biol. 2006;356:280–287. doi: 10.1016/j.jmb.2005.11.043. [DOI] [PubMed] [Google Scholar]
- Freier SM, Kierzek R, Jaeger JA, Sugimoto N, Caruthers MH, Neilson T, Turner DH. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlich AL, Tanzi RE, Rogers JT. The 5′-untranslated region of Parkinson’s disease α-synuclein messengerRNA contains a predicted iron responsive element. Mol Psychiatry. 2007;12:222–223. doi: 10.1038/sj.mp.4001937. [DOI] [PubMed] [Google Scholar]
- Fu Y, Ramisetty SR, Hussain N, Baranger AM. MBNL1-RNA recognition: contributions of MBNL1 sequence and RNA conformation. Chembiochem. 2012;13:112–119. doi: 10.1002/cbic.201100487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugier C, Klein AF, Hammer C, Vassilopoulos S, Ivarsson Y, Toussaint A, Tosch V, Vignaud A, Ferry A, Messaddeq N, et al. Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat Med. 2011;17:720–725. doi: 10.1038/nm.2374. [DOI] [PubMed] [Google Scholar]
- Gacy AM, Goellner G, Juranić N, Macura S, McMurray CT. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell. 1995;81:533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- Gallego J, Varani G. Targeting RNA with small-molecule drugs: therapeutic promise and chemical challenges. Acc Chem Res. 2001;34:836–843. doi: 10.1021/ar000118k. [DOI] [PubMed] [Google Scholar]
- Garavís M, López-Méndez B, Somoza A, Oyarzabal J, Dalvit C, Villasante A, Campos-Olivas R, González C. Discovery of selective ligands for telomeric RNA G-quadruplexes (TERRA) through 19F-NMR based fragment screening. ACS Chem Biol. 2014;9:1559–1566. doi: 10.1021/cb500100z. [DOI] [PubMed] [Google Scholar]
- Gecz J, Gedeon AK, Sutherland GR, Mulley JC. Identification of the gene FMR2, associated with FRAXE mental retardation. Nat Genet. 1996;13:105–108. doi: 10.1038/ng0596-105. [DOI] [PubMed] [Google Scholar]
- Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc Natl Acad Sci U S A. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PEA, Caulfield T, Daughrity L, Dunmore JH, Castanedes-Casey M, Chew J, et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013;126:829–844. doi: 10.1007/s00401-013-1192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleghorn ML, Maquat LE. “Black sheep” that don’t leave the double-stranded RNA-binding domain fold. Trends Biochem Sci. 2014;39:328–340. doi: 10.1016/j.tibs.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M, Crowther RA, Spillantini MG. Tau mutations cause frontotemporal dementias. Neuron. 1998;21:955–958. doi: 10.1016/s0896-6273(00)80615-7. [DOI] [PubMed] [Google Scholar]
- Gomes C, Merianda TT, Lee SJ, Yoo S, Twiss JL. Molecular determinants of the axonal mRNA transcriptome. Dev Neurobiol. 2014;74:218–232. doi: 10.1002/dneu.22123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gott JM, Emeson RB. Functions and mechanisms of RNA editing. Annu Rev Genet. 2000;34:499–531. doi: 10.1146/annurev.genet.34.1.499. [DOI] [PubMed] [Google Scholar]
- Greco CM, Berman RF, Martin RM, Tassone F, Schwartz PH, Chang A, Trapp BD, Iwahashi CK, Brunberg J, Grigsby J, et al. Neuropathology of fragile X-associated tremor/ataxia syndrome (FXTAS) Brain. 2006;129:243–255. doi: 10.1093/brain/awh683. [DOI] [PubMed] [Google Scholar]
- Guan L, Disney MD. Recent advances in developing small molecules targeting RNA. ACS Chem Biol. 2012;7:73–86. doi: 10.1021/cb200447r. [DOI] [PubMed] [Google Scholar]
- Guan L, Disney MD. Covalent small-molecule-RNA complex formation enables cellular profiling of small-molecule-RNA interactions. Angew Chem Int Ed Engl. 2013;52:10010–10013. doi: 10.1002/anie.201301639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell RR, Weiser B, Woese CR, Noller HF. Comparative anatomy of 16-S-like ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- Gyi JI, Lane AN, Conn GL, Brown T. The orientation and dynamics of the C2′-OH and hydration of RNA and DNA·RNA hybrids. Nucleic Acids Res. 1998;26:3104–3110. doi: 10.1093/nar/26.13.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler AR, Donnelly CJ, Periz G, Simko EaJ, Shaw PG, Kim MS, Maragakis NJ, Troncoso JC, Pandey A, Sattler R, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman RJ, Leehey M, Heinrichs W, Tassone F, Wilson R, Hills J, Grigsby J, Gage B, Hagerman PJ. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology. 2001;57:127–130. doi: 10.1212/wnl.57.1.127. [DOI] [PubMed] [Google Scholar]
- Halic M, Becker T, Pool MR, Spahn CMT, Grassucci RA, Frank J, Beckmann R. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004;427:808–814. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- Hermann T, Patel DJ. RNA bulges as architectural and recognition motifs. Structure. 2000;8:47–54. doi: 10.1016/s0969-2126(00)00110-6. [DOI] [PubMed] [Google Scholar]
- Hiller M, Zhang Z, Backofen R, Stamm S. Pre-mRNA secondary structures influence exon recognition. PLoS Genet. 2007;3:2147–2155. doi: 10.1371/journal.pgen.0030204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TH, Charlet-Berguerand N, Poulos MG, Singh G, Swanson MS, Cooper TA. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogsteen K. The crystal and molecular structure of a hydrogen-bonded complex between 1-methylthymine and 9-methyladenine. Acta Crystallogr. 1963;16:907–916. [Google Scholar]
- Hoskins JW, Ofori LO, Chen CZ, Kumar A, Sobczak K, Nakamori M, Southall N, Patnaik S, Marugan JJ, Zheng W, et al. Lomofungin and dilomofungin: inhibitors of MBNL1-CUG RNA binding with distinct cellular effects. Nucleic Acids Res. 2014;42:6591–6602. doi: 10.1093/nar/gku275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TA. Regulation of gene expression by alternative untranslated regions. Trends Genet. 2006;22:119–122. doi: 10.1016/j.tig.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- Iida A, Hosono N, Sano M, Kamei T, Oshima S, Tokuda T, Nakajima M, Kubo M, Nakamura Y, Ikegawa S. Novel deletion mutations of OPTN in amyotrophic lateral sclerosis in Japanese. Neurobiol Aging. 2012;33:1843.e19–e1843.e24. doi: 10.1016/j.neurobiolaging.2011.12.037. [DOI] [PubMed] [Google Scholar]
- Isis Pharmaceuticals. ClinicalTrials.gov. Bethesda (MD): National Library of Medicine (USA); 2014. A Safety and Tolerability Study of Multiple Doses of ISIS-DMPKRx in Adults With Myotonic Dystrophy Type 1. [Internet] NLM identifier: NCT02312011. Available from: https://clinicaltrials.gov/ct2/show/NCT02312011. [Google Scholar]
- Iwahashi CK, Yasui DH, An HJ, Greco CM, Tassone F, Nannen K, Babineau B, Lebrilla CB, Hagerman RJ, Hagerman PJ. Protein composition of the intranuclear inclusions of FXTAS. Brain. 2006;129:256–271. doi: 10.1093/brain/awh650. [DOI] [PubMed] [Google Scholar]
- Jacquemont S, Hagerman RJ, Leehey M, Grigsby J, Zhang L, Brunberg JA, Greco C, Des Portes V, Jardini T, Levine R, et al. Fragile X premutation tremor/ataxia syndrome: molecular, clinical, and neuroimaging correlates. Am J Hum Genet. 2003;72:869–878. doi: 10.1086/374321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahromi AH, Fu Y, Miller KA, Nguyen L, Luu LM, Baranger AM, Zimmerman SC. Developing bivalent ligands to target CUG triplet repeats, the causative agent of myotonic dystrophy type 1. J Med Chem. 2013a;56:9471–9481. doi: 10.1021/jm400794z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahromi AH, Nguyen L, Fu Y, Miller KA, Baranger AM, Zimmerman SC. A novel CUG(exp)·MBNL1 inhibitor with therapeutic potential for myotonic dystrophy type 1. ACS Chem Biol. 2013b;8:1037–1043. doi: 10.1021/cb400046u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Cote J, Kwon JM, Goate AM, Wu JY. Aberrant splicing of tau pre-mRNA caused by intronic mutations associated with the inherited dementia frontotemporal dementia with parkinsonism linked to chromosome 17. Mol Cell Biol. 2000;20:4036–4048. doi: 10.1128/mcb.20.11.4036-4048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LF, Williams JG, Abelson HT, Green H, Penman S. Changes in RNA in relation to growth of the fibroblast. III Posttranscriptional regulation of mRNA formation in resting and growing cells. Cell. 1975;4:69–75. doi: 10.1016/0092-8674(75)90135-x. [DOI] [PubMed] [Google Scholar]
- Johnson LF, Abelson HT, Penman S, Green H. The relative amounts of the cytoplasmic RNA species in normal, transformed and senescent cultured cell lines. J Cell Physiol. 1977;90:465–470. doi: 10.1002/jcp.1040900310. [DOI] [PubMed] [Google Scholar]
- Jung H, Yoon BC, Holt CE. Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nat Rev Neurosci. 2012;13:514–514. doi: 10.1038/nrn3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanadia RN, Shin J, Yuan Y, Beattie SG, Wheeler TM, Thornton CA, Swanson MS. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc Natl Acad Sci U S A. 2006;103:11748–11753. doi: 10.1073/pnas.0604970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W. Structural insights into CUG repeats containing the “stretched U-U wobble”: implications for myotonic dystrophy. Nucleic Acids Res. 2009;37:4149–4156. doi: 10.1093/nar/gkp350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Cheong C, Moore PB. Tetramerization of an RNA oligonucleotide containing a GGGG sequence. Nature. 1991;351:331–332. doi: 10.1038/351331a0. [DOI] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino Y, Mori D, Oma Y, Takeshita Y, Sasagawa N, Ishiura S. Muscleblind protein, MBNL1/EXP, binds specifically to CHHG repeats. Hum Mol Genet. 2004;13:495–507. doi: 10.1093/hmg/ddh056. [DOI] [PubMed] [Google Scholar]
- Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SJ, Flannery aV, Hirst MC, Campbell L, Christodoulou Z, Phelps SR, Pointon J, Middleton-Price HR, Barnicoat A, Pembrey ME. Trinucleotide repeat amplification and hypermethylation of a CpG island in FRAXE mental retardation. Cell. 1993;74:127–134. doi: 10.1016/0092-8674(93)90300-f. [DOI] [PubMed] [Google Scholar]
- Kolb HC, Finn MG, Sharpless KB. Click chemistry: diverse chemical function from a few good reactions. Angew Chem Int Ed Engl. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Konieczny P, Stepniak-Konieczna E, Sobczak K. MBNL proteins and their target RNAs, interaction and splicing regulation. Nucleic Acids Res. 2014;42:10873–10887. doi: 10.1093/nar/gku767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- Kovtun IV, McMurray CT. Features of trinucleotide repeat instability in vivo. Cell Res. 2008;18:198–213. doi: 10.1038/cr.2008.5. [DOI] [PubMed] [Google Scholar]
- Krzyzosiak WJ, Sobczak K, Wojciechowska M, Fiszer A, Mykowska A, Kozlowski P. Triplet repeat RNA structure and its role as pathogenic agent and therapeutic target. Nucleic Acids Res. 2012;40:11–26. doi: 10.1093/nar/gkr729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Park H, Fang P, Parkesh R, Guo M, Nettles KW, Disney MD. Myotonic dystrophy type 1 RNA crystal structures reveal heterogeneous 1 × 1 nucleotide UU internal loop conformations. Biochemistry. 2011a;50:9928–9935. doi: 10.1021/bi2013068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Fang P, Park H, Guo M, Nettles KW, Disney MD. A crystal structure of a model of the repeating r(CGG) transcript found in fragile X syndrome. Chembiochem. 2011b;12:2140–2142. doi: 10.1002/cbic.201100337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T, Matsuura T, Ohno K, Ueda S. Alu-mediated acquisition of unstable ATTCT pentanucleotide repeats in the human ATXN10 gene. Mol Biol Evol. 2009;26:2573–2579. doi: 10.1093/molbev/msp172. [DOI] [PubMed] [Google Scholar]
- Kurosaki T, Ueda S, Ishida T, Abe K, Ohno K, Matsuura T. The unstable CCTG repeat responsible for myotonic dystrophy type 2 originates from an AluSx element insertion into an early primate genome. PLoS One. 2012;7:e38379. doi: 10.1371/journal.pone.0038379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok CK, Ding Y, Tang Y, Assmann SM, Bevilacqua PC. Determination of in vivo RNA structure in low-abundance transcripts. Nat Commun. 2013;4:2971. doi: 10.1038/ncomms3971. [DOI] [PubMed] [Google Scholar]
- Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T, Kim J, Yun J, Xie Y, McKnight SL. Poly-dipeptides encoded by the C9ORF72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014;345:1139–1145. doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Peña M, Dufour D, Gallego J. Three-way RNA junctions with remote tertiary contacts: a recurrent and highly versatile fold. RNA. 2009;15:1949–1964. doi: 10.1261/rna.1889509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Baughn M, Rigo F, Sun S, Liu P, Li HR, Jiang J, Watt AT, Chun S, Katz M, et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci U S A. 2013;110:E4530–E4539. doi: 10.1073/pnas.1318835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lee VMY, Trojanowski JQ. Mechanisms of Parkinson’s disease linked to pathological α-Synuclein: new targets for drug discovery. Neuron. 2006;52:33–38. doi: 10.1016/j.neuron.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Lee MK, Gal M, Frydman L, Varani G. Real-time multidimensional NMR follows RNA folding with second resolution. Proc Natl Acad Sci U S A. 2010;107:9192–9197. doi: 10.1073/pnas.1001195107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MM, Pushechnikov A, Disney MD. Rational and modular design of potent ligands targeting the RNA that causes myotonic dystrophy 2. ACS Chem Biol. 2009a;4:345–355. doi: 10.1021/cb900025w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MM, Childs-Disney JL, Pushechnikov A, French JM, Sobczak K, Thornton CA, Disney MD. Controlling the specificity of modularly assembled small molecules for RNA via ligand module spacing: targeting the RNAs that cause myotonic muscular dystrophy. J Am Chem Soc. 2009b;131:17464–17472. doi: 10.1021/ja906877y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Liu B, Lee S, Huang SX, Shen B, Qian SB. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci U S A. 2012;109:E2424–E2432. doi: 10.1073/pnas.1207846109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempereur L, Nicoloso M, Riehl N, Ehresmann C, Ehresmann B, Bachellerie JP. Conformation of yeast 18S rRNA. Direct chemical probing of the 5′ domain in ribosomal subunits and in deproteinized RNA by reverse transcriptase mapping of dimethyl sulfate-accessible sites. Nucleic Acids Res. 1985;13:8339–8357. doi: 10.1093/nar/13.23.8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerman YV, Kennedy SD, Shankar N, Parisien M, Major F, Turner DH. NMR structure of a 4 × 4 nucleotide RNA internal loop from an R2 retrotransposon: Identification of a three purine-purine sheared pair motif and comparison to MC-SYM predictions. RNA. 2011;17:1664–1677. doi: 10.1261/rna.2641911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PTX, Vieregg J, Tinoco I. How RNA unfolds and refolds. Annu Rev Biochem. 2008;77:77–100. doi: 10.1146/annurev.biochem.77.061206.174353. [DOI] [PubMed] [Google Scholar]
- Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nat Rev Neurosci. 2007;8:819–831. doi: 10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. Splicing regulation in neurologic disease. Neuron. 2006;52:93–101. doi: 10.1016/j.neuron.2006.09.017. [DOI] [PubMed] [Google Scholar]
- Lilley DMJ. The structure and folding of kink turns in RNA. Wiley Interdiscip Rev RNA. 2012;3:797–805. doi: 10.1002/wrna.1136. [DOI] [PubMed] [Google Scholar]
- Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, Day JW, Ranum LP. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- Liu F, Gong CX. Tau exon 10 alternative splicing and tauopathies. Mol Neurodegener. 2008;3:8. doi: 10.1186/1750-1326-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Chen X, Bissler JJ, Sinden RR, Leffak M. Replication-dependent instability at (CTG)·(CAG) repeat hairpins in human cells. Nat Chem Biol. 2010;6:652–659. doi: 10.1038/nchembio.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López Castel A, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat Rev Mol Cell Biol. 2010;11:165–170. doi: 10.1038/nrm2854. [DOI] [PubMed] [Google Scholar]
- Lõpez-Senín P, Gõmez-Pinto I, Grandas A, Marchán V. Identification of ligands for the Tau exon 10 splicing regulatory element RNA by using dynamic combinatorial chemistry. Chem - A Eur J. 2011;17:1946–1953. doi: 10.1002/chem.201002065. [DOI] [PubMed] [Google Scholar]
- Lu X, Timchenko NA, Timchenko LT. Cardiac elav-type RNA-binding protein (ETR-3) binds to RNA CUG repeats expanded in myotonic dystrophy. Hum Mol Genet. 1999;8:53–60. doi: 10.1093/hmg/8.1.53. [DOI] [PubMed] [Google Scholar]
- Luebke KJ, Landry SM, Tinoco I. Solution conformation of a five-nucleotide RNA bulge loop from a group I intron. Biochemistry. 1997;36:10246–10255. doi: 10.1021/bi9701540. [DOI] [PubMed] [Google Scholar]
- Luo Y, Disney MD. Bottom-up design of small molecules that stimulate Exon 10 skipping in mutant MAPT pre-mRNA. ChemBioChem. 2014;15:2041–2044. doi: 10.1002/cbic.201402069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque FA, Furneaux HM, Ferziger R, Rosenblum MK, Wray SH, Schold SC, Glantz MJ, Jaeckle KA, Biran H, Lesser M. Anti-Ri: an antibody associated with paraneoplastic opsoclonus and breast cancer. Ann Neurol. 1991;29:241–251. doi: 10.1002/ana.410290303. [DOI] [PubMed] [Google Scholar]
- MacDonald ME, Ambrose CM, Duyao MP, Myers RH, Lin C, Srinidhi L, Barnes G, Taylor SA, James M, Groot N, et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- Mankodi A, Logigian E, Callahan L, McClain C, White R, Henderson D, Krym M, Thornton CA. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- Mankodi A, Urbinati CR, Yuan QP, Moxley RT, Sansone V, Krym M, Henderson D, Schalling M, Swanson MS, Thornton CA. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum Mol Genet. 2001;10:2165–2170. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- Mankodi A, Takahashi MP, Jiang H, Beck CL, Bowers WJ, Moxley RT, Cannon SC, Thornton CA. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- Marino JP, Gregorian RS, Csankovszki G, Crothers DM. Bent helix formation between RNA hairpins with complementary loops. Science. 1995;268:1448–1454. doi: 10.1126/science.7539549. [DOI] [PubMed] [Google Scholar]
- Martadinata H, Phan AT. Structure of propeller-type parallel-stranded RNA G-quadruplexes, formed by human telomeric RNA sequences in K+ solution. J Am Chem Soc. 2009;131:2570–2579. doi: 10.1021/ja806592z. [DOI] [PubMed] [Google Scholar]
- Mathews DH, Turner DH. Dynalign: an algorithm for finding the secondary structure common to two RNA sequences. J Mol Biol. 2002;317:191–203. doi: 10.1006/jmbi.2001.5351. [DOI] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci U S A. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Moss WN, Turner DH. Folding and finding RNA secondary structure. Cold Spring Harb Perspect Biol. 2010;2:a003665. doi: 10.1101/cshperspect.a003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhee SA, Huang L, Lilley DMJ. A critical base pair in k-turns that confers folding characteristics and correlates with biological function. Nat Commun. 2014;5:5127. doi: 10.1038/ncomms6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner NC, Hackermüller J, Uhl V, Aszódi A, Jaritz M, Auer M. mRNA openers and closers: Modulating AU-rich element-controlled mRNA stability by a molecular switch in mRNA secondary structure. ChemBioChem. 2004;5:1432–1447. doi: 10.1002/cbic.200400219. [DOI] [PubMed] [Google Scholar]
- Melko M, Bardoni B. The role of G-quadruplex in RNA metabolism: Involvement of FMRP and FMR2P. Biochimie. 2010;92:919–926. doi: 10.1016/j.biochi.2010.05.018. [DOI] [PubMed] [Google Scholar]
- Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J Am Chem Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir KU, Southern EM. Determining the influence of structure on hybridization using oligonucleotide arrays. Nat Biotechnol. 1999;17:788–792. doi: 10.1038/11732. [DOI] [PubMed] [Google Scholar]
- Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- Mizielinska S, Grönke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IOC, Pietrzyk J, et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014;345:1131–1135. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monckton DG, Coolbaugh MI, Ashizawa KT, Siciliano MJ, Caskey CT. Hypermutable myotonic dystrophy CTG repeats in transgenic mice. Nat Genet. 1997;15:193–196. doi: 10.1038/ng0297-193. [DOI] [PubMed] [Google Scholar]
- Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- Mooers BHM, Logue JS, Berglund JA. The structural basis of myotonic dystrophy from the crystal structure of CUG repeats. Proc Natl Acad Sci U S A. 2005;102:16626–16631. doi: 10.1073/pnas.0505873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morabito MV, Emeson RB. RNA editing as a therapeutic target for CNS disorders. Neuropsychopharmacology. 2009;34:246–246. doi: 10.1038/npp.2008.159. [DOI] [PubMed] [Google Scholar]
- Morabito MV, Abbas AI, Hood JL, Kesterson RA, Jacobs MM, Kump DS, Hachey DL, Roth BL, Emeson RB. Mice with altered serotonin 2C receptor RNA editing display characteristics of Prader-Willi syndrome. Neurobiol Dis. 2010;39:169–180. doi: 10.1016/j.nbd.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musunuru K, Darnell RB. Determination and augmentation of RNA sequence specificity of the Nova K-homology domains. Nucleic Acids Res. 2004;32:4852–4861. doi: 10.1093/nar/gkh799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mykowska A, Sobczak K, Wojciechowska M, Kozlowski P, Krzyzosiak WJ. CAG repeats mimic CUG repeats in the misregulation of alternative splicing. Nucleic Acids Res. 2011;39:8938–8951. doi: 10.1093/nar/gkr608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T, Ogiwara I, Ito K, Kaneda M, Mazaki E, Osaka H, Ohtani H, Inoue Y, Fujiwara T, Uematsu M, et al. Deletions of SCN1A 5′ genomic region with promoter activity in dravet syndrome. Hum Mutat. 2010;31:820–829. doi: 10.1002/humu.21275. [DOI] [PubMed] [Google Scholar]
- Nalavade R, Griesche N, Ryan DP, Hildebrand S, Krauss S. Mechanisms of RNA-induced toxicity in CAG repeat disorders. Cell Death Dis. 2013;4:e752. doi: 10.1038/cddis.2013.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napierała M, Krzyzosiak WJ. CUG repeats present in myotonin kinase RNA form metastable “slippery” hairpins. J Biol Chem. 1997;272:31079–31085. doi: 10.1074/jbc.272.49.31079. [DOI] [PubMed] [Google Scholar]
- Napierała M, Michalowski D, de Mezer M, Krzyzosiak WJ. Facile FMR1 mRNA structure regulation by interruptions in CGG repeats. Nucleic Acids Res. 2005;33:451–463. doi: 10.1093/nar/gki186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevskaya N, Tishchenko S, Gabdoulkhakov A, Nikonova E, Nikonov O, Nikulin A, Platonova O, Garber M, Nikonov S, Piendl W. Ribosomal protein L1 recognizes the same specific structural motif in its target sites on the autoregulatory mRNA and 23S rRNA. Nucleic Acids Res. 2005;33:478–485. doi: 10.1093/nar/gki194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SY, Lin L, Soh BS, Stanton LW. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet. 2013;29:461–468. doi: 10.1016/j.tig.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Nguyen L, Lee J, Wong CH, Zimmerman SC. Small molecules that target the toxic RNA in myotonic dystrophy type 2. ChemMedChem. 2014;9:2455–2462. doi: 10.1002/cmdc.201402095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro G, Taylor IA, Ramos A. KH RNA interactions: back in the groove. Curr Opin Struct Biol. 2015;30:63–70. doi: 10.1016/j.sbi.2015.01.002. [DOI] [PubMed] [Google Scholar]
- Nicholson BL, White KA. Functional long-range RNA-RNA interactions in positive-strand RNA viruses. Nat Rev Microbiol. 2014;12:493–504. doi: 10.1038/nrmicro3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller H, Woese C. Secondary structure of 16S ribosomal RNA. Science. 1981;212:403–411. doi: 10.1126/science.6163215. [DOI] [PubMed] [Google Scholar]
- Ofori LO, Hoskins JW, Nakamori M, Thornton CA, Miller BL. From dynamic combinatorial “hit” to lead: in vitro and in vivo activity of compounds targeting the pathogenic RNAs that cause myotonic dystrophy. Nucleic Acids Res. 2012;40:6380–6390. doi: 10.1093/nar/gks298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares D, Huang X, Branden L, Greig NH, Rogers JT. Physiological and pathological role of alpha-synuclein in Parkinson’s disease through iron mediated oxidative stress; the role of a putative iron-responsive element. Int J Mol Sci. 2009;10:1226–1260. doi: 10.3390/ijms10031226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani F, Baralle FE. Genomic variants in exons and introns: identifying the splicing spoilers. Nat Rev Genet. 2004;5:389–396. doi: 10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- Palde PB, Ofori LO, Gareiss PC, Lerea J, Miller BL. Strategies for recognition of stem-loop RNA structures by synthetic ligands: Application to the HIV-1 frameshift stimulatory sequence. J Med Chem. 2010;53:6018–6027. doi: 10.1021/jm100231t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallan PS, Marshall WS, Harp J, Jewett FC, Wawrzak Z, Brown BA, Rich A, Egli M. Crystal structure of a luteoviral RNA pseudoknot and model for a minimal ribosomal frameshifting motif. Biochemistry. 2005;44:11315–11322. doi: 10.1021/bi051061i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisien M, Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- Parkesh R, Fountain M, Disney MD. NMR spectroscopy and molecular dynamics simulation of r(CCGCUGCGG)2 reveal a dynamic UU internal loop found in myotonic dystrophy type 1. Biochemistry. 2011;50:599–601. doi: 10.1021/bi101896j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkesh R, Childs-Disney JL, Nakamori M, Kumar A, Wang E, Wang T, Hoskins JW, Tran T, Housman DE, Thornton CA, et al. Design of a bioactive small molecule that targets the myotonic dystrophy type 1 RNA via an RNA motif-ligand database and chemical similarity searching. J Am Chem Soc. 2012;134:4731–4742. doi: 10.1021/ja210088v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CE. Repeat associated non-ATG translation initiation: one DNA, two transcripts, seven reading frames, potentially nine toxic entities! PLoS Genet. 2011;7:e1002018. doi: 10.1371/journal.pgen.1002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- Piccinelli P, Samuelsson T. Evolution of the iron-responsive element. RNA. 2007;13:952–966. doi: 10.1261/rna.464807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieretti M, Zhang FP, Fu YH, Warren ST, Oostra BA, Caskey CT, Nelson DL. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991;66:817–822. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- Pushechnikov A, Lee MM, Childs-Disney JL, Sobczak K, French JM, Thornton CA, Disney MD. Rational design of ligands targeting triplet repeating transcripts that cause RNA dominant disease: application to myotonic muscular dystrophy type 1 and spinocerebellar ataxia type 3. J Am Chem Soc. 2009;131:9767–9779. doi: 10.1021/ja9020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat Rev Neurosci. 2012;13:528–541. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranum LPW, Cooper TA. RNA-mediated neuromuscular disorders. Annu Rev Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- Reddy PS, Housman DE. The complex pathology of trinucleotide repeats. Curr Opin Cell Biol. 1997;9:364–372. doi: 10.1016/s0955-0674(97)80009-9. [DOI] [PubMed] [Google Scholar]
- Reddy K, Zamiri B, Stanley SYR, Macgregor RB, Pearson CE. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J Biol Chem. 2013;288:9860–9866. doi: 10.1074/jbc.C113.452532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder LE, Staber CJ, Hoopengardner B, Reenan RA. Tertiary structural elements determine the extent and specificity of messenger RNA editing. Nat Commun. 2013;4:2232. doi: 10.1038/ncomms3232. [DOI] [PubMed] [Google Scholar]
- Rogers JT, Randall JD, Cahill CM, Eder PS, Huang X, Gunshin H, Leiter L, McPhee J, Sarang SS, Utsuki T, et al. An iron-responsive element type II in the 5′-untranslated region of the Alzheimer’s amyloid precursor protein transcript. J Biol Chem. 2002;277:45518–45528. doi: 10.1074/jbc.M207435200. [DOI] [PubMed] [Google Scholar]
- Rogers JT, Mikkilineni S, Cantuti-Castelvetri I, Smith DH, Huang X, Bandyopadhyay S, Cahill CM, Maccecchini ML, Lahiri DK, Greig NH. The alpha-synuclein 5′ untranslated region targeted translation blockers: anti-alpha synuclein efficacy of cardiac glycosides and Posiphen. J Neural Transm. 2011;118:493–507. doi: 10.1007/s00702-010-0513-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicki DD, Holmes SE, Lin MW, Thornton CA, Ross CA, Margolis RL. Huntington’s disease-like 2 is associated with CUG repeat-containing RNA foci. Ann Neurol. 2007;61:272–282. doi: 10.1002/ana.21081. [DOI] [PubMed] [Google Scholar]
- Rzuczek SG, Park H, Disney MD. A toxic RNA catalyzes the in cellulo synthesis of its own inhibitor. Angew Chem Int Ed Engl. 2014;53:10956–10959. doi: 10.1002/anie.201406465. [DOI] [PubMed] [Google Scholar]
- SantaLucia J, Kierzek R, Turner DH. Context dependence of hydrogen bond free energy revealed by substitutions in an RNA hairpin. Science. 1992;256:217–219. doi: 10.1126/science.1373521. [DOI] [PubMed] [Google Scholar]
- Sathasivam K, Neueder A, Gipson TA, Landles C, Benjamin AC, Bondulich MK, Smith DL, Faull RLM, Roos RAC, Howland D, et al. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc Natl Acad Sci U S A. 2013;110:2366–2370. doi: 10.1073/pnas.1221891110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savkur RS, Philips AV, Cooper TA. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, et al. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Schrödinger LLC. The PyMOL Molecular Graphics System, Version 1.3r1 2010 [Google Scholar]
- Schroeder KT, Daldrop P, Lilley DMJ. RNA tertiary interactions in a riboswitch stabilize the structure of a kink turn. Structure. 2011;19:1233–1240. doi: 10.1016/j.str.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder R, Barta A, Semrad K. Strategies for RNA folding and assembly. Nat Rev Mol Cell Biol. 2004;5:908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- Schultes EA, Bartel DP. One sequence, two ribozymes: implications for the emergence of new ribozyme folds. Science. 2000;289:448–452. doi: 10.1126/science.289.5478.448. [DOI] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JHD. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Sellier C, Rau F, Liu Y, Tassone F, Hukema RK, Gattoni R, Schneider A, Richard S, Willemsen R, Elliott DJ, et al. Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J. 2010;29:1248–1261. doi: 10.1038/emboj.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellier C, Freyermuth F, Tabet R, Tran T, He F, Ruffenach F, Alunni V, Moine H, Thibault C, Page A, et al. Sequestration of DROSHA and DGCR8 by expanded CGG RNA Repeats Alters microRNA processing in fragile X-associated tremor/ataxia syndrome. Cell Rep. 2013;3:869–880. doi: 10.1016/j.celrep.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Patel DJ. Towards deciphering the principles underlying an mRNA recognition code. Curr Opin Struct Biol. 2008;18:120–129. doi: 10.1016/j.sbi.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, Hertel KJ. Conserved RNA secondary structures promote alternative splicing Conserved RNA secondary structures promote alternative splicing. Rna. 2008;14:1463–1469. doi: 10.1261/rna.1069408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Charizanis K, Swanson MS. Pathogenic RNAs in microsatellite expansion disease. Neurosci Lett. 2009;466:99–102. doi: 10.1016/j.neulet.2009.07.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortridge MD, Varani G. Structure based approaches for targeting non-coding RNAs with small molecules. Curr Opin Struct Biol. 2015;30:79–88. doi: 10.1016/j.sbi.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, et al. α-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- Siomi H, Siomi MC, Nussbaum RL, Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993;74:291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- Slavoff SA, Mitchell AJ, Schwaid AG, Cabili MN, Ma J, Levin JZ, Karger AD, Budnik BA, Rinn JL, Saghatelian A. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat Chem Biol. 2013;9:59–64. doi: 10.1038/nchembio.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczak K, de Mezer M, Michlewski G, Krol J, Krzyzosiak WJ. RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res. 2003;31:5469–5482. doi: 10.1093/nar/gkg766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczak K, Michlewski G, de Mezer M, Kierzek E, Krol J, Olejniczak M, Kierzek R, Krzyzosiak WJ. Structural diversity of triplet repeat RNAs. J Biol Chem. 2010;285:12755–12764. doi: 10.1074/jbc.M109.078790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Spada AR, Taylor JP. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat Rev Genet. 2010;11:247–258. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. α-Synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- Spillantini MG, Murrell JR, Goedert M, Farlow MR, Klug A, Ghetti B. Mutation in the tau gene in familial multiple system tauopathy with presenile dementia. Proc Natl Acad Sci U S A. 1998a;95:7737–7741. doi: 10.1073/pnas.95.13.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillantini MG, Crowther RA, Jakes R, Hasegawa M, Goedert M. α-Synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with Lewy bodies. Proc Natl Acad Sci U S A. 1998b;95:6469–6473. doi: 10.1073/pnas.95.11.6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripakdeevong P, Cevec M, Chang AT, Erat MC, Ziegeler M, Zhao Q, Fox GE, Gao X, Kennedy SD, Kierzek R, et al. Structure determination of noncanonical RNA motifs guided by 1H NMR chemical shifts. Nat Methods. 2014;11:413–416. doi: 10.1038/nmeth.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D, Beuchle D, Nüsslein-Volhard C. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 1991;66:51–63. doi: 10.1016/0092-8674(91)90138-o. [DOI] [PubMed] [Google Scholar]
- Stelzer AC, Frank AT, Kratz JD, Swanson MD, Gonzalez-Hernandez MJ, Lee J, Andricioaei I, Markovitz DM, Al-Hashimi HM. Discovery of selective bioactive small molecules by targeting an RNA dynamic ensemble. Nat Chem Biol. 2011;7:553–559. doi: 10.1038/nchembio.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N, Weisburd B, Michalski A, Le VTK, Hein MY, Huang SX, Ma M, Shen B, Qian SB, Hengel H, et al. Decoding human cytomegalovirus. Science. 2012;338:1088–1093. doi: 10.1126/science.1227919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Zhang Y, Gendron TF, Bauer PO, Chew J, Yang WY, Fostvedt E, Jansen-West K, Belzil VV, Desaro P, et al. Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron. 2014;83:1043–1050. doi: 10.1016/j.neuron.2014.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Barrick JE, Breaker RR. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 2003;9:644–647. doi: 10.1261/rna.5090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y, König J, Hussain S, Zupan BB, Curk TT, Frye M, Ule J, Konig J, Hussain S, Zupan BB, et al. Analysis of CLIP and iCLIP methods for nucleotide-resolution studies of protein-RNA interactions. Genome Biol. 2012;13:R67. doi: 10.1186/gb-2012-13-8-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y, Vigilante A, Darbo E, Zirra A, Militti C, D’Ambrogio A, Luscombe NM, Ule J. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature. 2015;519:491–494. doi: 10.1038/nature14280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundquist WI, Heaphy S. Evidence for interstrand quadruplex formation in the dimerization of human immunodeficiency virus 1 genomic RNA. Proc Natl Acad Sci U S A. 1993;90:3393–3397. doi: 10.1073/pnas.90.8.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundquist WI, Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- Tamjar J, Katorcha E, Popov A, Malinina L. Structural dynamics of double-helical RNAs composed of CUG/CUG- and CUG/CGG-repeats. J Biomol Struct Dyn. 2012;30:505–523. doi: 10.1080/07391102.2012.687517. [DOI] [PubMed] [Google Scholar]
- Taneja KL, McCurrach ME, Schalling M, Housman DE, Singer RH. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J Cell Biol. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang SJ, Meulemans D, Vazquez L, Colaco N, Schuman E. A role for a rat homolog of staufen in the transport of RNA to neuronal dendrites. Neuron. 2001;32:463–475. doi: 10.1016/s0896-6273(01)00493-7. [DOI] [PubMed] [Google Scholar]
- Teplova M, Patel DJ. Structural insights into RNA recognition by the alternative-splicing regulator muscleblind-like MBNL1. Nat Struct Mol Biol. 2008;15:1343–1351. doi: 10.1038/nsmb.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplova M, Malinina L, Darnell JC, Song J, Lu M, Abagyan R, Musunuru K, Teplov A, Burley SK, Darnell RB, et al. Protein-RNA and protein-protein recognition by dual KH1/2 domains of the neuronal splicing factor Nova-1. Structure. 2011;19:930–944. doi: 10.1016/j.str.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JR, Hergenrother PJ. Targeting RNA with small molecules. Chem Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- Thomson AM, Rogers JT, Leedman PJ. Iron-regulatory proteins, iron-responsive elements and ferritin mRNA translation. Int J Biochem Cell Biol. 1999;31:1139–1152. doi: 10.1016/s1357-2725(99)00080-1. [DOI] [PubMed] [Google Scholar]
- Thore S, Leibundgut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- Tian B, White RJ, Xia T, Welle S, Turner DH, Mathews MB, Thornton CA. Expanded CUG repeat RNAs form hairpins that activate the double-stranded RNA-dependent protein kinase PKR. RNA. 2000;6:79–87. doi: 10.1017/s1355838200991544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian N, Yang Y, Sachsenmaier N, Muggenhumer D, Bi J, Waldsich C, Jantsch MF, Jin Y. A structural determinant required for RNA editing. Nucleic Acids Res. 2011;39:5669–5681. doi: 10.1093/nar/gkr144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijerina P, Mohr S, Russell R. DMS footprinting of structured RNAs and RNA-protein complexes. Nat Protoc. 2007;2:2608–2623. doi: 10.1038/nprot.2007.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishchenko S, Kljashtorny V, Kostareva O, Nevskaya N, Nikulin A, Gulak P, Piendl W, Garber M, Nikonov S. Domain II of Thermus thermophilus ribosomal protein L1 hinders recognition of its mRNA. J Mol Biol. 2008;383:301–305. doi: 10.1016/j.jmb.2008.08.058. [DOI] [PubMed] [Google Scholar]
- Todd PK, Paulson HL. RNA-mediated neurodegeneration in repeat expansion disorders. Ann Neurol. 2010;67:291–300. doi: 10.1002/ana.21948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd PK, Oh S, Krans A, He F, Sellier C, Frazer M, Renoux AJ, Chen KC, Scaglione KM, Basrur V, et al. CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron. 2013;78:440–455. doi: 10.1016/j.neuron.2013.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran T, Childs-Disney JL, Liu B, Guan L, Rzuczek SG, Disney MD. Targeting the r(CGG) repeats that cause FXTAS with modularly assembled small molecules and oligonucleotides. ACS Chem Biol. 2014;9:904–912. doi: 10.1021/cb400875u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trausch JJ, Ceres P, Reyes FE, Batey RT. The structure of a tetrahydrofolate-sensing riboswitch reveals two ligand binding sites in a single aptamer. Structure. 2011;19:1413–1423. doi: 10.1016/j.str.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker BJ, Breaker RR. Riboswitches as versatile gene control elements. Curr Opin Struct Biol. 2005;15:342–348. doi: 10.1016/j.sbi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Turner DH, Sugimoto N, Kierzek R, Dreikert SD. Free energy increments for hydrogen bonds in nucleic acid base pairs. J Am Chem Soc. 1987;109:3783–3785. [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- Varani G, McClain WH. The G·U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000;1:18–23. doi: 10.1093/embo-reports/kvd001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani L, Hasegawa M, Spillantini MG, Smith MJ, Murrell JR, Ghetti B, Klug A, Goedert M, Varani G. Structure of tau exon 10 splicing regulatory element RNA and destabilization by mutations of frontotemporal dementia and parkinsonism linked to chromosome 17. Proc Natl Acad Sci U S A. 1999;96:8229–8234. doi: 10.1073/pnas.96.14.8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi SP, Gallo SM, Disney MD. Sequence-based design of bioactive small molecules that target precursor microRNAs. Nat Chem Biol. 2014;10:291–297. doi: 10.1038/nchembio.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerk AJMH, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DPA, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang F, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Vervoort R, Gitzelmann R, Lissens W, Liebaers I. A mutation (IVS8+0.6kbdelTC) creating a new donor splice site activates a cryptic exon in an Alu-element in intron 8 of the human β-glucuronidase gene. Hum Genet. 1998;103:686–693. doi: 10.1007/pl00008709. [DOI] [PubMed] [Google Scholar]
- Voigts-Hoffmann F, Klinge S, Ban N. Structural insights into eukaryotic ribosomes and the initiation of translation. Curr Opin Struct Biol. 2012;22:768–777. doi: 10.1016/j.sbi.2012.07.010. [DOI] [PubMed] [Google Scholar]
- Wachter A. Riboswitch-mediated control of gene expression in eukaryotes. RNA Biol. 2010;7:67–76. doi: 10.4161/rna.7.1.10489. [DOI] [PubMed] [Google Scholar]
- Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov. 2013;12:433–446. doi: 10.1038/nrd4018. [DOI] [PubMed] [Google Scholar]
- Walton SP, Stephanopoulos GN, Yarmush ML, Roth CM. Prediction of antisense oligonucleotide binding affinity to a structured RNA target. Biotechnol Bioeng. 1999;65:1–9. [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008a;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Cody NAL, Jog S, Biancolella M, Wang TT, Treacy DJ, Luo S, Schroth GP, Housman DE, Reddy S, et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Haeusler AR, Simko EA. Emerging role of RNA DNA hybrids in C9orf72-linked neurodegeneration. Cell Cycle. 2015;14:526–532. doi: 10.1080/15384101.2014.995490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008b;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warf MB, Berglund JA. MBNL binds similar RNA structures in the CUG repeats of myotonic dystrophy and its pre-mRNA substrate cardiac troponin T. RNA. 2007;13:2238–2251. doi: 10.1261/rna.610607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warf MB, Berglund JA. Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem Sci. 2010;35:169–178. doi: 10.1016/j.tibs.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks KM. Advances in RNA structure analysis by chemical probing. Curr Opin Struct Biol. 2010;20:295–304. doi: 10.1016/j.sbi.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells SE, Hughes JM, Igel AH, Ares M. Use of dimethyl sulfate to probe RNA structure in vivo. Methods Enzymol. 2000;318:479–493. doi: 10.1016/s0076-6879(00)18071-1. [DOI] [PubMed] [Google Scholar]
- Wheeler TM, Lueck JD, Swanson MS, Dirksen RT, Thornton CA. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J Clin Invest. 2007;117:3952–3957. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TM, Sobczak K, Lueck JD, Osborne RJ, Lin X, Dirksen RT, Thornton CA. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, Cheng SH, Wentworth BM, Bennett CF, Thornton CA. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- Xu Z, Poidevin M, Li X, Li Y, Shu L, Nelson DL, Li H, Hales CM, Gearing M, Wingo TS, et al. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc Natl Acad Sci U S A. 2013;110:7778–7783. doi: 10.1073/pnas.1219643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Peacey E, Dickson J, Donahue CP, Zheng S, Varani G, Wolfe MS. Mitoxantrone analogues as ligands for a stem-loop structure of tau Pre-mRNA. J Med Chem. 2009;52:6523–6526. doi: 10.1021/jm9013407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WY, Wilson HD, Velagapudi SP, Disney MD. Inhibition of non-ATG translational events in cells via covalent small molecules targeting RNA. J Am Chem Soc. 2015;137:5336–5345. doi: 10.1021/ja507448y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim I, Park H, Disney MD, Schatz GC. A dynamic structural model of expanded RNA CAG repeats: a refined X-ray structure and computational investigations using molecular dynamics and umbrella sampling simulations. J Am Chem Soc. 2013;135:3528–3538. doi: 10.1021/ja3108627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupova GZ, Yusupov MM, Cate JHD, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- Zamiri B, Reddy K, Macgregor RB, Pearson CE. TMPyP4 porphyrin distorts RNA G-quadruplex structures of the disease-associated r(GGGGCC)n repeat of the C9orf72 gene and blocks interaction of RNAbinding proteins. J Biol Chem. 2014;289:4653–4659. doi: 10.1074/jbc.C113.502336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Lau MW, Ferré-D’Amaré AR. Ribozymes and riboswitches: Modulation of RNA function by small molecules. Biochemistry. 2010;49:9123–9131. doi: 10.1021/bi1012645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Tan D, DeRose EF, Perera L, Dominski Z, Marzluff WF, Tong L, Hall TMT. Molecular mechanisms for the regulation of histone mRNA stem-loop-binding protein by phosphorylation. Proc Natl Acad Sci U S A. 2014;111:E2937–E2946. doi: 10.1073/pnas.1406381111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Chen Y, Donahue CP, Wolfe MS, Varani G. Structural basis for stabilization of the Tau pre-mRNA splicing regulatory element by novantrone (mitoxantrone) Chem Biol. 2009;16:557–566. doi: 10.1016/j.chembiol.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, Margolis J, Peterson M, Markowski TW, Ingram MAC, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Liu Y, Bañez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci U S A. 2013;110:E4968–E4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]