Abstract
Superoxide dismutase 1 (SOD1) is the ubiquitously expressed and predominant dismutase in the cytoplasm. While transcriptional regulation of the SOD1 gene has been well characterized, post-transcriptional regulation of the gene remains largely unknown in eukaryotes. In this study, a full-length 3’UTR of the SOD1 transcript was cloned and characterized for its ability to regulate SOD1 gene expression in human cancer cells. Inclusion of the SOD1 3’UTR in the pGL3 reporter construct dramatically enhanced the reporter activity by 10–220-fold in various cell lines. RT-PCR analysis, however, indicated that the reporter gene mRNA levels were only modestly altered by the SOD1 3’UTR, suggesting that SOD1 3’UTR enhances the reporter gene activity not simply by stabilizing the mRNA but primarily through promoting translation of the protein. Bioinformatics analysis showed multiple stem and loop structures of the SOD1 3’UTR and alterations of this secondary structure led to remarkably reduced reporter gene activity. Importantly, introducing the SOD1 3’UTR to cancer cells attenuated endogenous SOD1 expression in a concentration-dependent manner, indicating the involvement of RNA trans-acting factors in this process. Using siRNA and RNA immunoprecipitation techniques, we identified AUF-1, a RNA binding protein, as a positive regulator of SOD1 expression through its 3’UTR. Consequently, AUF-1 was found to regulate redox balance in our cell model systems. Furthermore, in human ovarian, esophageal, and pancreatic cancer tissues, the expression of SOD1 was significantly correlated with that of AUF-1, further supporting the importance of AUF-1 in regulating SOD1 gene expression.
Keywords: SOD1, 3’UTR, cancer, AU-rich element, AUF-1
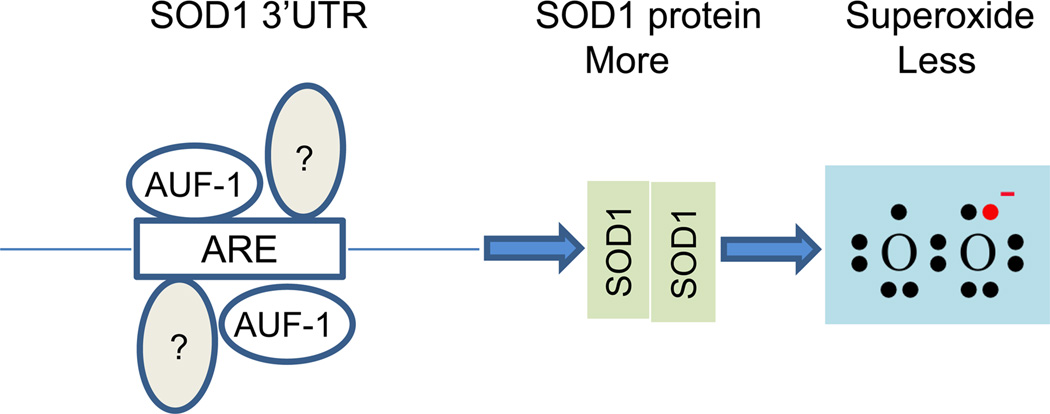
Graphical Abstract
The SOD1 3’UTR facilitates SOD1 protein production resulting in reduction of superoxide levels in human cancer cells.
Introduction
Superoxide dismutases (SODs) are enzymes that catalyze the degradation of superoxide into oxygen and hydrogen peroxide. The generated hydrogen peroxide is subsequently converted to water and oxygen by glutathione peroxidase (GPx) and/or catalase to maintain the redox homeostasis in a given cell system (1–3). Among the three SOD enzymes, SOD1 (also known as Cu/Zn superoxide dismutase) contributes to the majority of cellular SOD activity (4). Hence, SOD1 expression levels are important for the elimination of cellular superoxides. The human SOD1 gene is located on chromosome 21q22.11 and it produces a 16-kDa protein that is often homodimerized, and widely distributed in the cytosol, nucleus, peroxisome, and intermembrane space of the mitochondria (5,6). Mutations of the SOD1 gene have been linked to human diseases, such as familial amyotrophic lateral sclerosis (7,8). SOD1−/− mice are found to have a high rate of DNA mutations that occur at an early age and have an elevated susceptibility to oxidative stress and liver tumors (9,10). On the other hand, over-expression of SOD1 in human pancreatic (11), lung (12), and chemo-resistant breast cancer cells (13) has been observed although the mechanism behind remains unclear. What is known is that over-expression of SOD1 renders tumor cells more resistance to oxidative stress and chemotherapy (14) and the experimental evidence accumulated thus far supports the conclusion that SOD1 is a molecular target for cancer therapy (12,15).
How a gene is delicately regulated to produce the precise amount of protein to meet biological demand is a fundamental question in biology. In addition to transcriptional regulation, posttranscriptional regulation of gene expression fundamentally and rapidly modifies the gene expression process (16,17). In this context, the 3' untranslated region (3'UTR) of an mRNA is recognized to be heavily involved in mediating gene expression. The 3'UTR of a mRNA, which starts with the nucleotide immediately following the stop codon of the coding region (17,18), interacts with microRNAs (miRNAs) and RNA binding proteins through defined RNA elements to regulate mRNA expression or protein translation, thus altering gene expression levels (17,19–21).
Over the years transcriptional regulation of the SOD1 gene has been well characterized in different model systems (22,23). However, whether or not and how the SOD1 3'UTR contributes to expressional control of the SOD1 gene in human cancer cells is largely unknown. In the present study, we evaluated the role of SOD1 3’UTR in maintaining SOD1 expression level in human cancer cells. We found that the SOD1 3’UTR dramatically enhances SOD1 expression, with a magnitude that, to our knowledge, has not been previously described for any 3’UTR-mediated gene expression. Furthermore, we identified AUF-1, an established RNA binding protein (24), as a positive posttranscriptional regulator of SOD1 expression, providing a potential molecular mechanism for SOD1 over-expression in human cancer cells.
Materials and methods
Cell culture
The human pancreatic cancer cell line PANC1 and human esophageal cancer cell line TE-1 were maintained in DMEM supplemented with 10% FBS and antibiotics (100 units/ml penicillin G Sodium Salt and 100 units/ml streptomycin sulfate; Gibco, Grand Island, NY). The human hepatocellular carcinoma cell line HepG2 and ovarian cancer cell line A2780 were maintained in RPMI-1640 supplemented with 10% FBS and antibiotics. The cells were grown in a 37°C incubator with 5% CO2.
Reverse transcriptase PCR (RT-PCR) analysis of mRNA
Total RNA from cells was extracted with Trizol (Invitrogen, Carlsbad, CA) and reversely transcribed to cDNA using an oligo (dT)12 primer and Superscript II (Invitrogen). SYBR green dye (Takara Bio Inc., Shiga, Japan) was used for amplification of cDNA. mRNA levels of luciferase and the internal standard glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were measured by RT-PCR (Applied Biosystems, Foster City, CA). The following primers specific for luciferase and GAPDH were used: GAPDH: (forward) 5’-GAAGGTGAAGGTCGGAGTC-3’ and (reverse) 5’-GAAGATGGTGATGGGATTTC-3’; luciferase: (forward) 5’-GAGGTTCCATCTGCCAGGTA-3’ and (reverse) 5’-CACACAGTTCGCCTCTTTGA-3’. For real-time PCR analysis of luciferase and SOD1 mRNA levels, the synthesized cDNA was diluted in 2× iTaq Universal SYBR Green Supermix and combined with 4uM of each forward and reverse primer. Specific primers used for real-time PCR are as follows: SOD1, forward 5’-ACTGGTGGTCCATGAAAAAGC-3’ and reverse 5’-AACGACTTCCAGCGTTTCCT-3’; pGL3-firefly luciferase, forward 5’-CCGAGGGGGATGATAAACCG-3’ and reverse 5’-TCCCGGTATCCAGATCCACA-3’. PCR was run on the Bio-Rad CFX 96 Real-Time PCR instrument under the following conditions: 95°C for 15 seconds and 60°C for 1 min, 40 cycles. The absolute quantification method was used to quantify mRNA expression levels as described (http://www.gene-quantification.com/strategy.html). In brief, a standard curve of SOD1 expression was constructed using serially diluted samples of cDNA synthesized from total RNA that was extracted from an untreated sample. Using linear regression, the cycle threshold (Ct) value from the sample cDNAs were plotted onto the standard curve to obtain the quantity of amplified cDNA (equivalent to ng of cDNA used). The data are presented as the expression levels in the treated samples relative to the control samples.
Construction of reporter plasmids
To explore the influences of 3’UTR on the expression of SOD1, a fragment of the full-length SOD1 3’UTR was amplified by PCR using the cDNA made from A2780 cells. The amplified fragment was inserted into the pGL3-Promoter vector (Promega, Madison, WI) at the Xba I site in both sense and antisense directions. The pGL3-SOD1-Sense vector was used as a template for the cloning of serially deleted SOD1 3’UTR reporter vectors with primers described in Supplementary Table 1. These recombination plasmids were confirmed by DNA sequencing.
Cell transfection and luciferase assays
Chemically synthesized miRNA mimics and inhibitors were obtained from Ambion (Life Technologies, Carlsbad, CA). siRNA control and siRNA targeting HuR and AUF-1 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). AUF-1 cDNA was obtained from OriGene Technologies Inc. (Rockville, MD). Cells were transfected by Lipofectamine 2000 (Invitrogen) with 1 µg of each constructed vector or siRNA. In each transfection, 50 ng of pRL-TK (Promega, Madison, WI) was used to correct transfection efficiency. For co-transfection, vector was co-transfected with siRNA or plasmids. Luciferase activity was measured with the Dual-Luciferase Reporter Assay System (Promega, Madison, WI). Luciferase activities were evaluated as the ratio of Firefly luciferase to Renilla luciferase activities.
Western blot
Treated cells in a 6-well-culture cluster were washed twice with ice old PBS and directly lysed in 200 µl of 2×SDS cell lysis buffer in each well. The lysates were boiled, centrifuged at 10,000 rpm, and the supernatant was loaded onto a 12% SDS-PAGE gel. The samples were electrophoresed for 4 h and then transferred to a transfer membrane in Bio-Rad blot apparatus. The membranes were blotted with antibodies for SOD1, HuR, (Santa Cruz Biotechnology, Santa Cruz, CA) or AUF-1 (Abcam, Cambridge, UK)) followed with a secondary antibody linked to horseradish peroxidase. GAPDH was detected and served as the loading control as we recently described (25). The GAPDH antibody was from ProMab Biotechnologies, Inc. (Albany, CA).
Co-immunoprecipitation
Co-immunoprecipitation using AUF-1 and GAPDH antibody was performed as described previously (25). In short, PANC1 cells were washed with PBS and harvested by adding 150 µl of IP buffer (10 mM Tris-HCl, pH 7.4, 50 mM NaCl, 0.5 mM EDTA, 1 mM PMSF, and 1% Triton X-100). After sonicating the cells for 2 min on ice, insoluble material was removed by centrifugation. Supernatants were collected and pre-cleared by agarose-coupled protein A. The agarose beads were removed by centrifugation. AUF1 antibody and GAPDH antibody were added to the supernatants (1:100 ratio), and the reaction was incubated at 4°C overnight with gentle rotation. 50 µl of agarose-coupled protein A was added to capture the antibody-protein complexes by rotating for 2 h at 4°C. The supernatants were then removed by centrifuging at 2,000 × g for 5 min. Total RNA was isolated from the pellets using the Trizol reagent. Reverse transcription-PCR was performed as described earlier. The following primers specific for SOD1 3’UTR was used: (forward) 5’-ACATTCCCTTGGATGTAG-3’ and (reverse) 5’-TGACAAGTTTAATACC-3’.
Reactive oxygen species (ROS) measurement
The levels of ROS were determined using the ROS-sensitive dye 2’, 7’-dichlorofluorescein diacetate (DCF-DA), which was converted by ROS into the highly fluorescent 2’, 7’-dichlorofluorescein (DCF). Cells were washed with PBS buffer (pH 7.4) and incubated with DCF-DA (10 µM) for 30 min. The level of DCF fluorescence, which reflects the concentration of ROS, was measured by fluorescent microscopy. To induce ROS, cells were irradiated with 5 Gy X-ray irradiation using a 6-MV X-ray linear accelerator (Clinac 2100EX; Varian Medical Systems, Inc., CA) at a dose rate of 2 Gy/min; a 1.5-cm bolus was used as a compensator. After radiation, the medium was immediately replaced with fresh medium.
Superoxide anion measurement
PANC1 and A2780 cells were sham irradiated or exposed to 5 Gy irradiation. Cellular superoxide anions and SOD enzymatic activity were measured using the Superoxide Anion Detection Kit (Millipore, Billerica, MA). For superoxide anion assay, the cells (5.0 × 105) were suspended in 100 µl of the assay medium and combined with luminol (100 µM), enhancer (125 µM), and phorbol-12-myristate-13-acetate (PMA, 100 ng/ml). After a 30 min incubation at room temperature the light emission was recorded using a luminometer (Promega, Madison, WI). For SOD activity assay, the cell lysate was prepared using the lysate buffer from Promega (Madison, WI). In a polystyrene round-bottom tube, 100 µl of xanthine oxidase-luminol solution and 100 µl of the 50 µM xanthine solution (containing 25 µM xanthine, 0.1 mU/µl xanthine oxidase, and 100 µM luminal) were mixed. The cell lysate (5µg in 10 µl) that is suspected to contain SOD or 1 µl (25 units) of the provided SOD were added to the tube. The light emission was recorded at 30 seconds intervals for up to 2 min using a luminometer. The data were expressed as the levels relative to the light intensities detected in control groups.
Tissue samples
Ninety-three pancreatic cancer tissues were obtained from Shanghai Biochip Co. Ltd (Shanghai, China). All samples were reviewed histologically by hematoxylin and eosin staining and constructed into a tissue microarray. Twenty ovarian cancer tissues were obtained from surgical resections of patients between 2011–2012 at Changzhou Tumor Hospital Affiliated to Soochow University (Changzhou, China). Twenty normal esophageal tissues and 120 esophageal cancer tissues were obtained from surgical resections of patients between 2010–2012 at the Changzhou Tumor Hospital and Gastrointestinal Center, Jiangyin People’s Hospital (Jiangyin, China), as previously reported (26,27). Tissues were obtained before chemotherapy and radiation therapy. All patients gave signed, informed consent for their tissues to be used for scientific research. Ethical approval for the study was obtained from the Changzhou Tumor Hospital and Jiangyin People’s Hospital. All diagnoses were based on pathological and/or cytological evidence.
Immunohistochemistry (IHC)
IHC was performed using antibodies against human SOD1 and AUF-1. IHC staining and scoring were performed as reported previously (26,27): negative (−), < 10% of the whole tissue mass stained; weakly positive (+ 1), 10–25% of the whole tissue mass stained; moderately positive (+ 2), 25–75% of the whole tissue mass stained; and strongly positive (+ 3), > 75% of the tissue showed positive.
Statistical analyses
Data were expressed as the mean ± SEM from at least three independent experiments unless otherwise indicated. Statistical differences between the test and control values were analyzed by applying the Student’s t-test and Kruskal-Wallis one-way analysis. Correlation analysis between the IHC staining scores of SOD1 and AUF-1 was performed using Spearman's rho test. Statistical analysis was performed using SPSS software (Release 17.0, SPSS Inc.). Data were considered significant and indicated by “*” if the two-sided P < 0.05, “**” if P < 0.01. The log 2 median-centered SOD1 and AUF1 (HNRNPD) microarray expression levels were extracted from the Oncomine, v 4.4.4.4 (www.oncomine.org) database from the Collisson study (28) which consists of 20 pancreatic adenocarcinoma samples, and the Adib study (29) which consists of 16 ovarian serous adenocarcinoma samples.
Results
SOD1 3’UTR maintains high expression of the SOD1 gene in human cancer cells
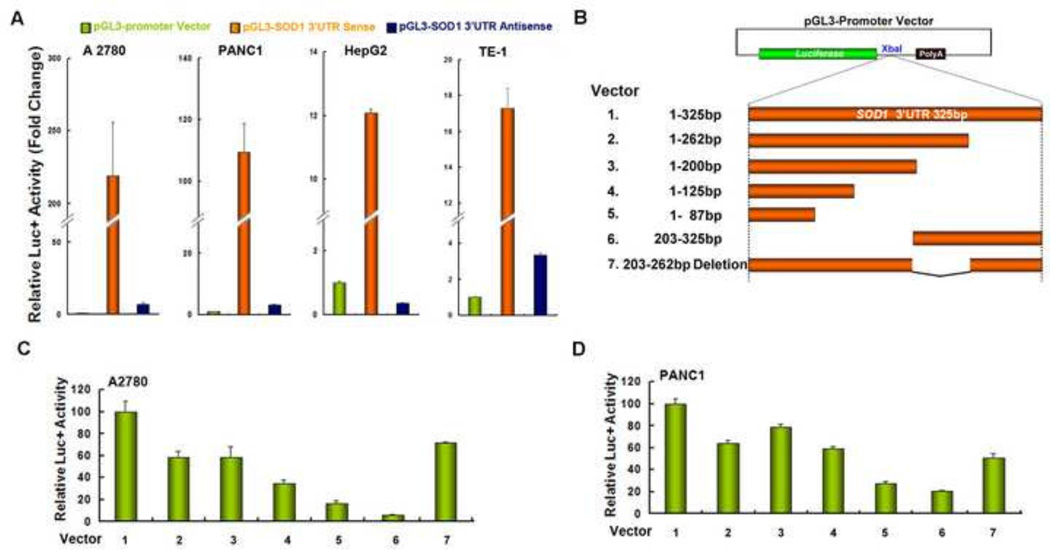
The nucleotide sequence of SOD1 3’UTR is rather conserved among different species (supplementary Figure 1). We cloned a full length SOD1 3’UTR from human A2780 cells and found that there is no mutation or deletion in this 326 base fragment. To investigate whether SOD1 3’UTR influences SOD1 expression, several human cell lines were transfected with the pGL3-Promoter vector, pGL3-promoter-SOD1 3’UTR sense or pGL3-promoter-SOD1 3’UTR antisense vector, respectively. Tranfection of the SOD1 3’UTR sense vector dramatically enhanced the reporter gene activity by 109-, 219-, 12- and 17-fold in PANC1, A2780, HepG2 and TE-1 cells, respectively (Figure 1A). In contrast, transfection of the SOD1 3’UTR antisense vector exhibited similar luciferase activity with that of the empty vector in all cell lines investigated (Figure 1A). These results suggested that SOD1 3’UTR maintains high expression of the SOD1 gene in human cancer cells. We next sought to determine which segment of the SOD1 3’UTR is critical for its function. Six reporter plasmids containing different fragments of the SOD1 3’UTR were constructed (Figure 1B). A2780 and PANC1 cells were transfected with the indicated constructs and assayed for luciferase activity 48 hours after transfection. As shown in Figure 1C and D, the truncated deletion of SOD1 3’UTR gradually decreased luciferase activity and deletion of any part of SOD1 3’UTR dramatically affects the reporter activity with the 87–325 fragment seemed to be most significant
Figure 1. Inclusion of the SOD1 3’UTR enhances luciferase reporter gene activity.
(A) Cells were transfected with pGL3-Promoter Vector, pGL3-promoter-SOD1 3’UTR sense or pGL3-promoter-SOD1 3’UTR antisense vector. 48 h after transfection, relative luciferase activity was analyzed. (B) Schematic representations of the different constructs used for luciferase assay. The firefly luciferase reporter gene (green box), the different segments of SOD1 3’UTR sequences (orange box) and the PolyA region (black box) are shown. Various regions of the SOD1 3’UTR were inserted downstream of the luciferase gene at the XbaI site. (C) A2780 and (D) PANC1 cells were transfected with each construct and assayed for luciferase activity after 48 hours. Data represent mean ± SEM of three independent experiments.
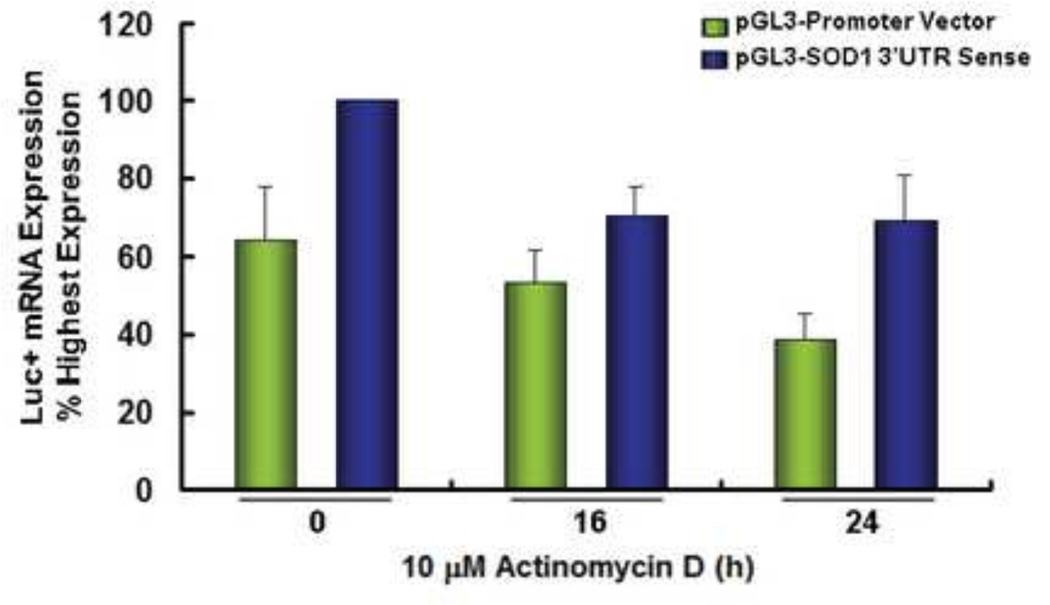
To understand whether the enhanced reporter gene activity is due to increased mRNA expression or protein translation, total RNA was extracted from A2780 cells 24 hours after transfection. Relative firefly luciferase mRNA levels were measured by real-time-RT-PCR after treatment with actinomycin D for 16–24 hours. As shown in Figure 2, inclusion of the SOD1 3’UTR modestly increased luciferase mRNA levels in both control and actinomycin D-treated cells, suggesting that the high luciferase activity is only partially attributed to the increased mRNA expression. A similar pattern of luciferase mRNA expression upon actinomycin D treatment was previously observed in HEK293 cells (30). Given the magnitude of the enhancement of reporter gene activity by SOD1 3’UTR, these results suggest that SOD1 3’UTR promotes translation of the SOD1 protein in our model system.
Figure 2. Inclusion of the SOD1 3’UTR modestly enhances luciferase mRNA expression.
A2780 Cells were transfected with pGL3-Promoter Vector or the pGL3-promoter-SOD1 3’UTR sense vector. Twenty-four hours after transfection, the cells were treated with actinomycin D (10 µM) for 16–24 h. RNA was extracted and relative firefly luciferase mRNA levels were analyzed by real-time RT-PCR. The luciferase mRNA level in pGL3-promoter-SOD1 3’UTR sense transfected cells (the highest level) was equivalent to that amplified from 1.8 ng cDNA. Data are representative of two individual experiments conducted in triplicate (mean ± SEM).
The integrity of SOD1 3’UTR is critical for its function
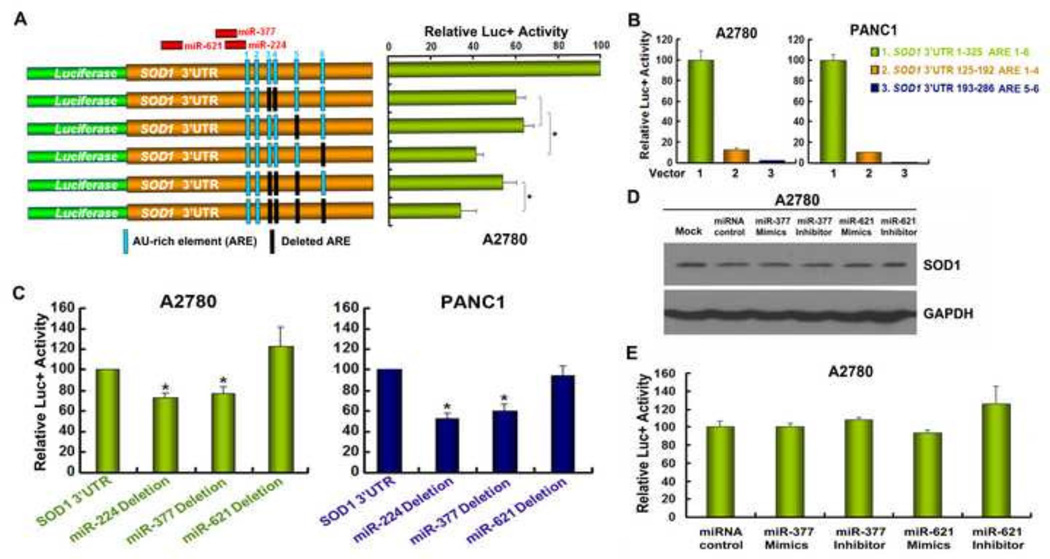
The interplay between cis-elements and trans-acting factors or miRNAs is critical for the function of 3’UTRs. The RNA motifs, such as AU-rich element (ARE), GC-rich and GU-rich elements, are known to play important roles for mRNA expression (18–21). There are six AREs in the SOD1 3’UTR predicted by a previous study (31). To test whether these AREs sustain the expression of SOD1, we constructed a series of vectors with deletion of four AREs individually or in combination (Figure 3A). A2780 cells were transfected with each construct and assayed for luciferase activity. We found that deletion of any of the four AREs remarkably reduces luciferase activity with the ARE 6 being the most important (Figure 3A), indicating that SOD1 expression is significantly sustained by the AREs in its 3’UTR. However, when the reporter vectors, containing only two or four of the putative AREs, were transfected to A2780 or PANC1 cells, luciferase activity was almost undetectable (Figure 3B), further suggesting that the importance of the AREs is related to their contribution to the integrity of the 3’UTR, which is essential for sustaining SOD1 expression.
Figure 3. The integrity of the SOD1 3’UTR sequence is critical for maintaining SOD1 expression.
(A) A diagram indicates the relative positions of the 6 AREs (column) and miRNA binding sites. Various reporter constructs with deletion of the AREs (black column) or miRNA binding sites in the SOD1 3’UTR were constructed. (B) A2780 and PANC1 cells were transfected with the indicated constructs and assayed for luciferase activity after 48 h. (C) A2780 and PANC1 cells were transfected with SOD1 3’UTR reporter constructs with miR-224, miR-377 or miR-621 binding sites deleted. Luciferase activity was determined 48 h after transfection. (D) A2780 cells were transfected with miR-377 or miR-621 mimics/inhibitors and SOD1 expression was analyzed by Western blot 48 h post transfection. (E) pGL3-SOD1 3’UTR sense vector was co-transfected with miR-377 or miR-621 mimics/inhibitor, respectively and assayed for relative luciferase activities 48 h post tranfection. Data represent mean ± SEM of three independent experiments, * P < 0.05, compared with pGL3-SOD1 3’UTR sense vector-transfected cells.
Bioinformatics tools (TargetScan and PicTar) consistently predicted that miR-224, miR-377 and miR-621 are possible regulators of SOD1 3’UTR (Figure 3A). A2780 and PANC1 cells were transfected with the wild-type SOD1 3’UTR reporter constructs or the constructs with deletion of the miRNA binding site for miR-224, miR-377 or miR-621. Interestingly, deletion of the binding site for miR-224 and miR-377, but not miR-621 in SOD1 3’UTR, significantly down-regulated luciferase reporter activity (Figure 3C), which was not in accordance with the general belief that miRNAs negatively regulate gene expression (17). In addition, application of miR-377 or miR-621 mimics/inhibitors did not affect endogenous SOD1 expression or the SOD1 3’UTR-driven reporter gene activity (Figure 3D and 3E), indicating that the SOD1 3’UTR’s function in sustaining SOD1 expression is less likely attributed to these miRNA species. Instead, the deletion of miR-224 and miR-377 binding sites may alter the integrity of the SOD1 3’UTR, given their close proximity to the ARE1, resulting in down-regulation of SOD1 3’UTR-driven reporter gene activity.
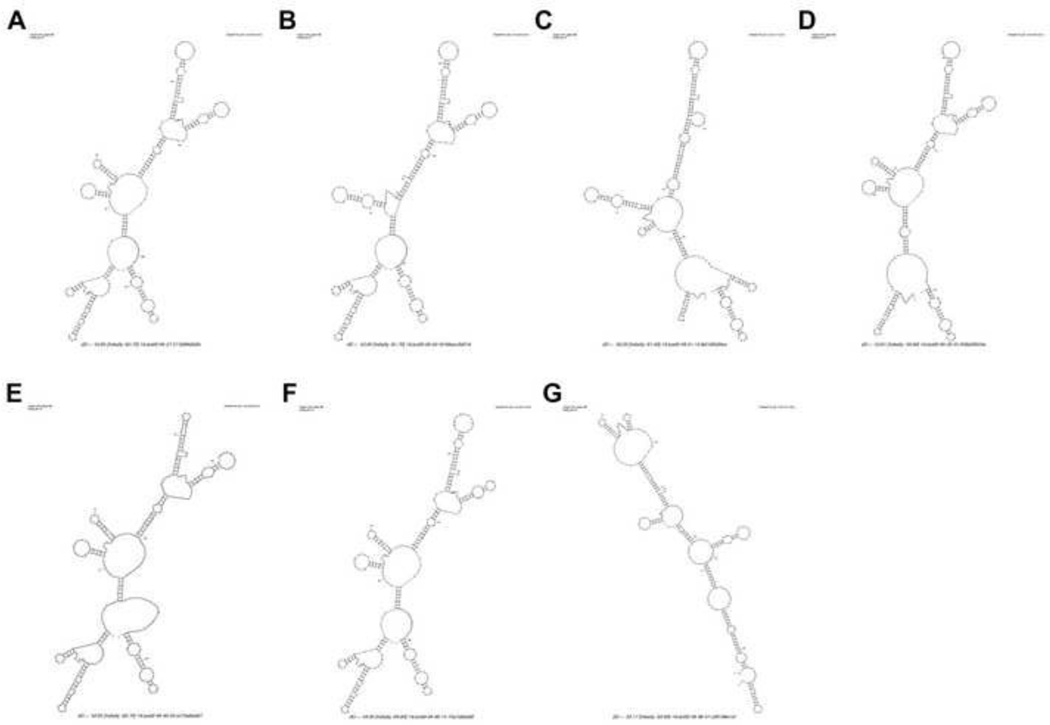
We then utilized RNAfold (32), a RNA secondary structure analysis tool, to predict the secondary structures of SOD1 3’UTR. As shown in Figure 4A, the predicted SOD1 3’UTR secondary structure contains multiple stem and loop structures. Deletion of the miR-224 and miR-377 but not miR-621 remarkably changed its original structure (Figure 4B–4D). The same is true when ARE6 was deleted from the 3’UTR (Figure 4G), whereas deletion of ARE 3/4, and 5 did not obviously alter the secondary structure (Figure 4E and 4F).
Figure 4. The predicted secondary structures of the SOD1 3’UTR.
The predicted secondary structures of (A) the full-length SOD1 3’UTR, (B) the SOD1 3’UTR with the miR-224 binding site deleted, (C) the SOD1 3’UTR with the miR-377 binding site deleted, (D) the SOD1 3’UTR with the miR-621 binding site deleted, (E) the SOD1 3’UTR with the ARE 3 and 4 deleted, (F) the SOD1 3’UTR with the ARE 5 deleted and (G) the SOD1 3’UTR with the ARE 6 deleted. The structures were predicted using the RNAFold computer algorithm (32).
Forced expression of SOD1 3’UTR reduces endogenous SOD1 expression
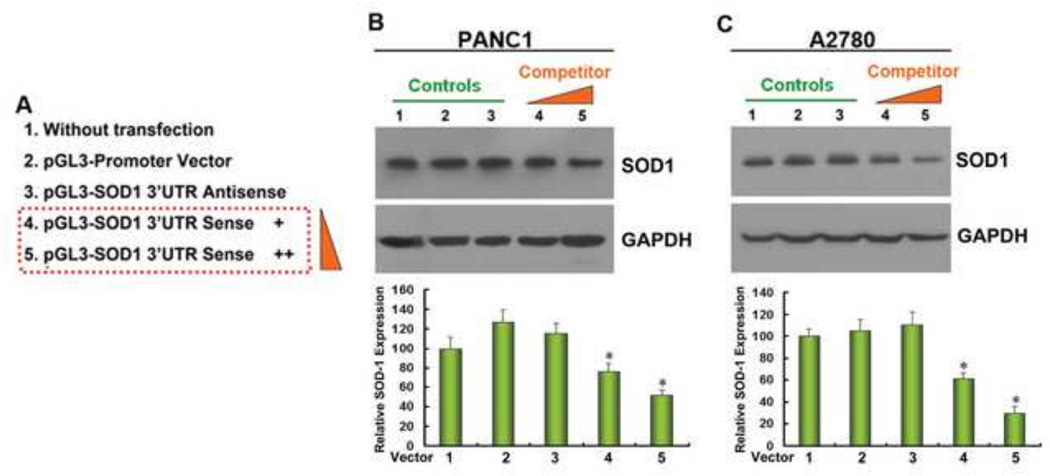
To understand whether trans-acting factors might interact with SOD1 3’UTR to sustain SOD1 gene expression in cancer cells, we first transfected the SOD1 3’UTR reporter constructs into A2780 and PANC1 cells and analyzed endogenous SOD1 protein expression by Western blot (Figure 5). We found that transfection of the pGL3 promoter vector or the vector with SOD1 3’UTR antisense did not affect endogenous SOD1 expression. However, transfection of SOD1 3’UTR sense vector, which shares identical sequence with endogenous SOD1 3’UTR, decreased endogenous SOD1 expression in a concentration-dependent manner, suggesting that the exogenous introduced SOD1 3’UTR competes with endogenous SOD1 3’UTR for the trans-acting factors that are required for maintaining SOD1 gene expression (Figure 5B and 5C).
Figure 5. Introducing exogenous SOD1 3’UTR attenuates endogenous SOD1 expression.
pGL3-SOD1 3’UTR sense vector was transfected in different amounts to determine its influence on endogenous SOD1 expression (A). 48 h after transfection, endogenous SOD1 protein was detected by Western blot in (B) PANC1 and (C) A2780 cells. Data represent mean ± SEM of three independent experiments. The relative SOD1 expression was quantified by densitometry. * P < 0.05, compared with untransfected control cells.
AUF-1 interacts with SOD1 3’UTR to regulate SOD1 expression
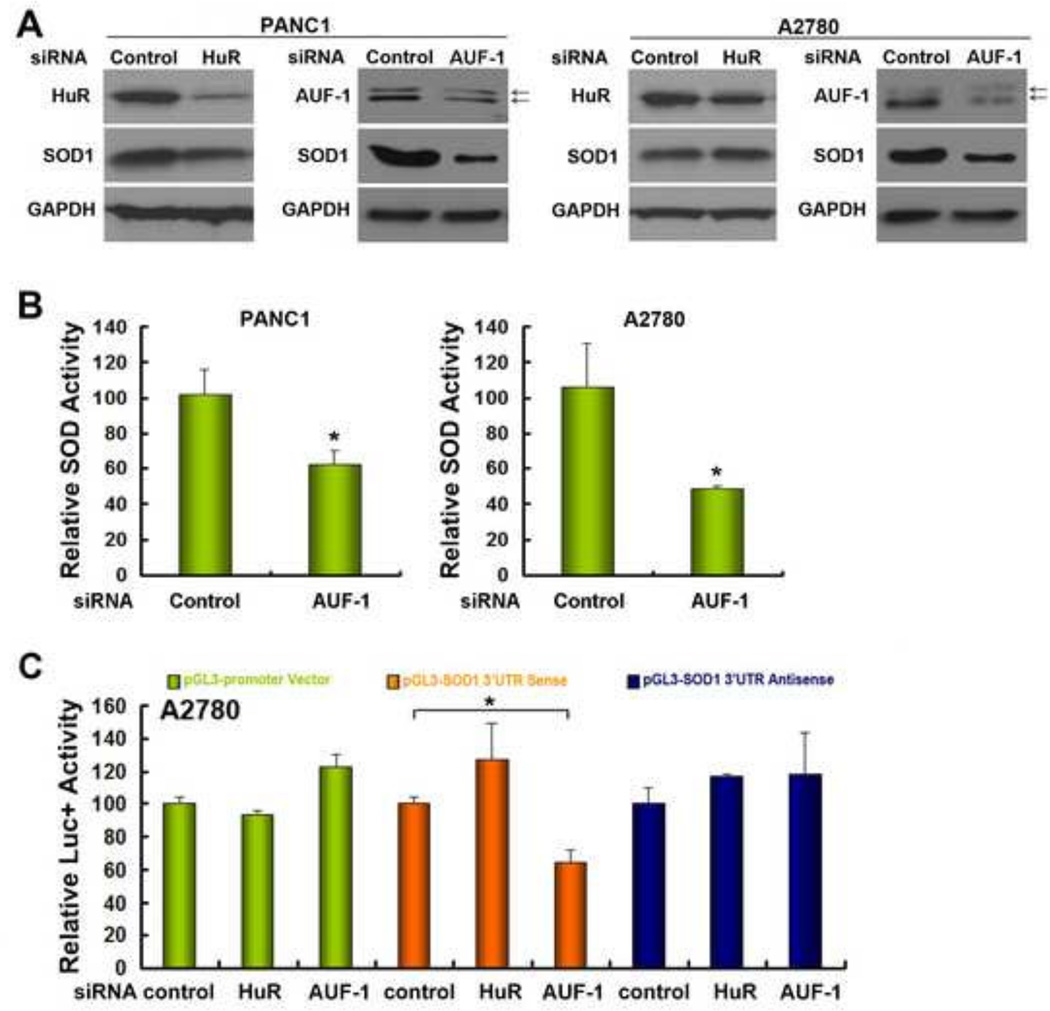
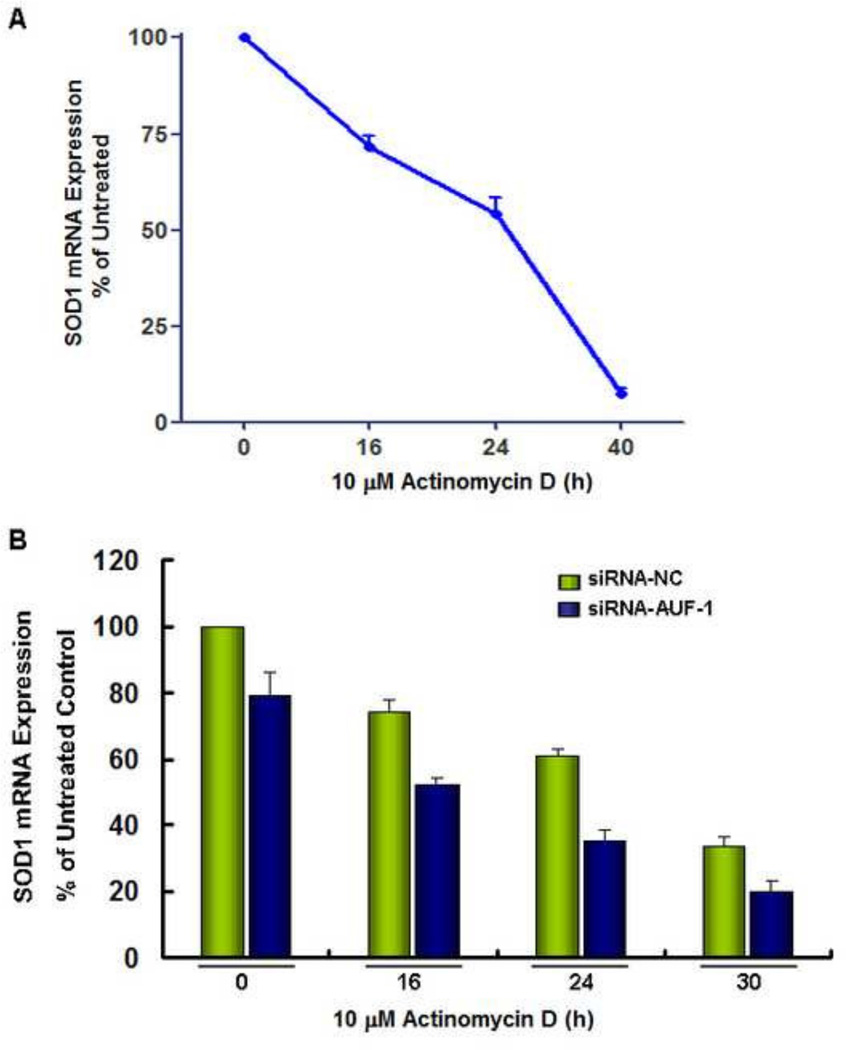
The above results suggest that the interaction of the SOD1 3’UTR with the trans-acting RNA binding factors is required to maintain SOD1 gene expression. HuR and AUF-1 are two ubiquitously expressed and well-established RNA binding proteins that are widely involved in regulating mammalian mRNA expression (24,33–35). To test whether these mRNA binding proteins are involved in mediating SOD1 expression, A2780 and PANC1 cells were transfected with scramble siRNA control or siRNA targeting HuR or AUF-1. The knockdown was confirmed by Western blot analysis (Figure 6A). We found that compared to that of HuR, knockdown of AUF-1 significantly reduces SOD1 protein expression levels (Figure 6A), which was accompanied by a corresponding decrease in SOD enzymatic activity (Figure 6B). Co-transfection of the SOD1 3’UTR reporter constructs with the AUF-1 siRNA significantly down-regulated the SOD1 3’UTR-driven reporter gene activity. In contrast, the reporter gene activity in cells transfected with control vector or the SOD1 3’UTR antisense vector was unaffected by AUF-1 knockdown, indicating that SOD1 3’UTR contains specific motifs for AUF-1 to interact with (Figure 6C). Furthermore, treatment of A2780 cells with actinomycin D indicated that knockdown of AUF-1 modestly lows down SOD1 mRNA expression (Figure 7); these results are consistent with those in Figure 2, suggesting that AUF-1 promotes SOD1 protein translation through its 3’UTR that leads to dramatic enhancement of the protein expression. The role of AUF-1 in regulating protein translation has been previously described (36,37).
Figure 6. AUF-1 is involved in regulating SOD1 expression via the 3’UTR.
(A) Cells were transfected with the indicated siRNAs. 48 h after transfection, protein expression was detected by Western Blot in A2780 and PANC1 cells. (B) Relative SOD enzymatic activity after transfection of siRNA control or siRNA targeting AUF-1 in PANC1 and A2780 cells. (C) Luciferase reporters were co-transfected with scramble siRNA (control) or the siRNA targeting HuR or AUF-1 in A2780 cells. Luciferase activity was determined 48 h after transfection. Data represent mean ± SEM of three independent experiments, * P < 0.05, compared with scramble siRNA co-transfected cells.
Figure 7. AUF-1 regulates SOD1 mRNA expression in A2780 cells.
(A). Cells were treated with actinomycin D (10 µM) for 16–40 h. Total RNA was isolated and SOD1 mRNA levels were determined by real-time RT-PCR analysis. The SOD1 mRNA level in untreated control cells was equivalent to that amplified from 2.3 ng cDNA. (B). Cells with AUF-1 siRNA knockdown were treated with actinomycin D (10 µM) for 16–30 h. Total RNA was isolated and SOD1 mRNA levels were determined by real-time RT-PCR analysis. The SOD1 mRNA level in untreated control cells was equivalent to that amplified from 2.8 ng cDNA. Data (mean ± SEM) are representative of two individual experiments conducted in triplicate.
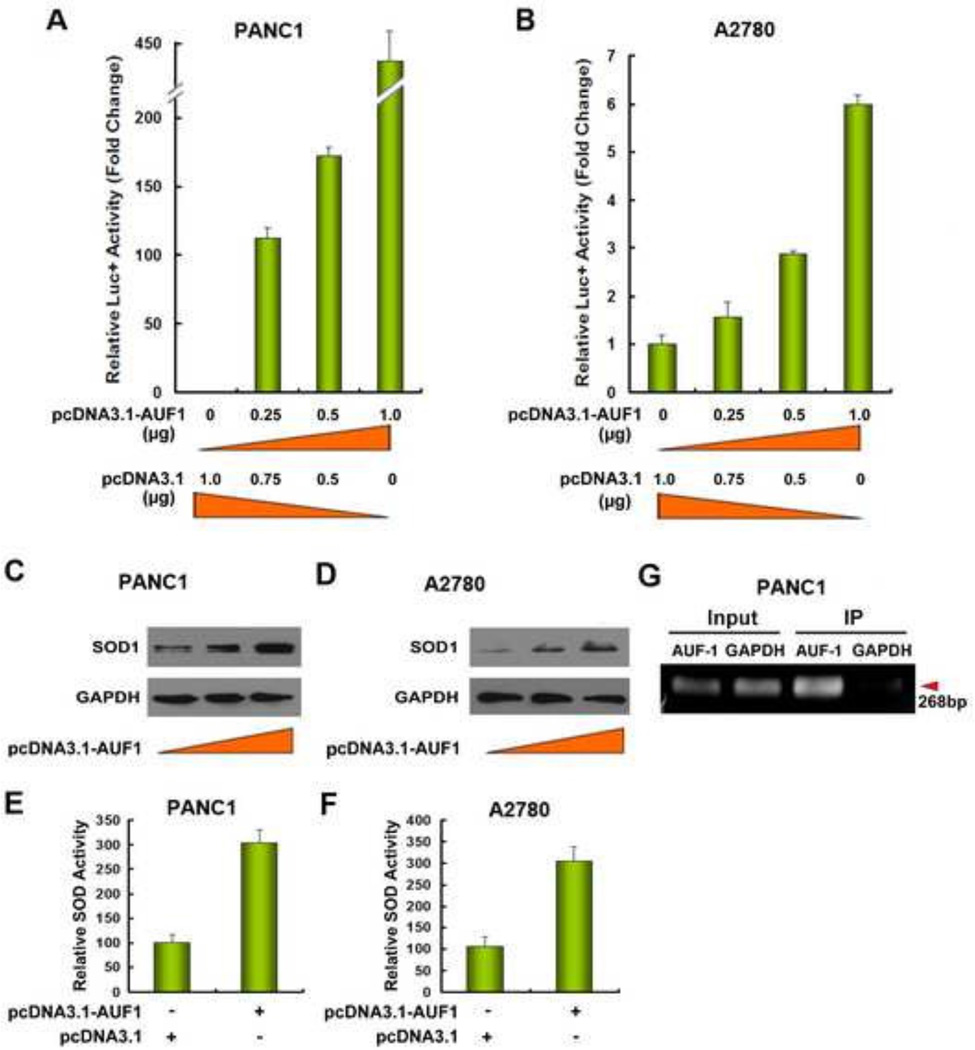
To further confirm the regulatory role of AUF-1 in SOD1 3’UTR-mediated gene expression, the pGL3-SOD1 3’UTR sense vector was co-transfected with different amount of pcDNA3.1-AUF1 vector in PANC1 and A2780 cells. As shown in Figure 8A and 8B, forced expression of AUF-1 increases the SOD1 3’UTR-driven reporter gene activity in a concentration-dependent manner in both cell lines, further indicating that AUF-1 is a positive regulator of SOD1 3’UTR-mediated gene expression. Western blot and SOD enzymatic analysis confirmed that forced expression of AUF-1 increased SOD1 protein expression (Figure 8C–8D) and enzyme activity in these cell lines (Figure 8E–8F).
Figure 8. AUF-1 overexpression increases SOD1 expression.
pGL3-SOD1-3’UTR vector was co-transfected with the indicated amount of AUF-1 expression vector (pcDNA3.1-AUF1) and/or control vector (pcDNA3.1) into (A) PANC1 and (B) A2780 cells. Luciferase activity was determined 24 h after transfection. Western blot was performed after transfection of differing amounts of AUF-1 expression vector (pcDNA3.1-AUF1) into (C) PANC1 and (D) A2780 cells. Relative SOD enzymatic activity after transfection of control vector or AUF-1 expression vector (pcDNA3.1-AUF1) into (E) PANC1 and (F) A2780 cells. Cell lysate from PANC1 cells were immunoprecipitated (IP) with the anti-AUF1 antibody or anti-GAPDH antibody, and RNA was isolated from the precipitants, reverse-transcribed, and PCR-amplified using primers covering SOD1 3’UTR mRNA (G).
We then immunoprecipitated AUF-1 using cell lysates prepared from PANC1 cells and analyzed SOD1 3’UTR expression in the precipitates. In contrast to GAPDH-precipitated sample, the presence of SOD1 3’UTR was only detected by RT-PCR in AUF-1-precipitated sample (Figure 8G), indicating a direct interaction of AUF-1 with SOD1 3’UTR in these cells.
AUF-1 modulates the generation of ROS in cancer cells
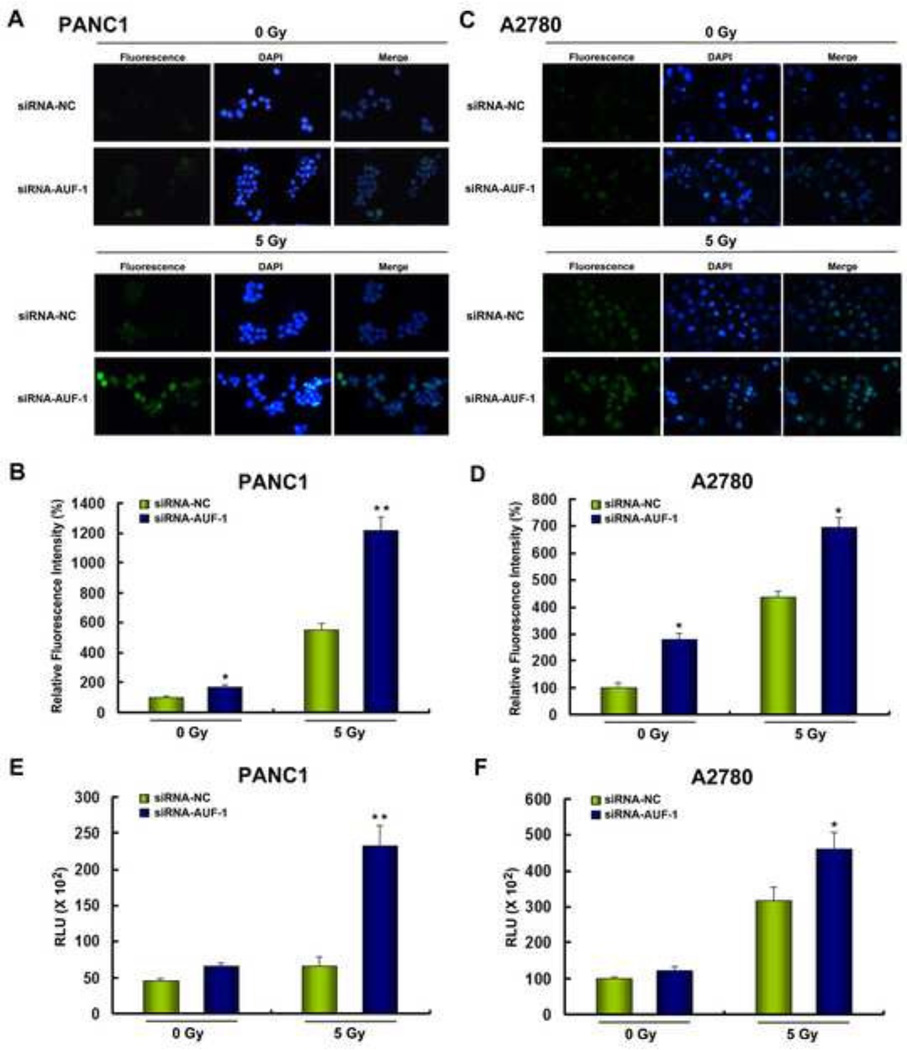
SOD1 is an established antioxidant enzyme that acts in concert with other antioxidant molecules to maintain redox homeostasis. To determine whether AUF-1, as a SOD1 expression regulator, might affect cellular ROS levels, PANC1 and A2780 cells were transfected with siRNA control (siRNA-NC) or siRNA targeting AUF-1 (siRNA-AUF-1). Two days after transfection, the ROS levels in the cells were analyzed using the DCF-DA probe. Transfection of siRNA-AUF-1 to PANC1 cells was found to promote basal ROS generation compared with that of siRNA-NC transfection (Figure 9A). Furthermore, transfection of siRNA-AUF-1 significantly enhanced radiation-induced ROS levels (Figure 9A). Figure 9B shows the quantification and statistical analysis of the relative ROS levels that were significantly increased after AUF-1 knockdown in basal as well as the irradiated state of PANC1 cells. Similar results were obtained in A2780 cells with or without irradiation (Figure 9C and 9D). Because SOD1 is a superoxide dismutase, the high ROS level after AUF-1 knockdown is most likely due to an accumulation of cellular superoxides. This assumption was confirmed by direct measurement of superoxide levels in both A2780 and PANC1 cells (Figure 9E–9F).
Figure 9. AUF-1 regulates ROS levels in PANC1 and A2780 cells.
(A) Determination of ROS levels of PANC1 cells transfected with scrambled siRNA (siRNA-NC) or siRNA-AUF1. To induce ROS, cells were exposed to 5 Gy X-ray irradiation. Six hours after irradiation, the ROS levels were determined using the ROS-sensitive dye 2’, 7’-dichlorofluorescein diacetate (DCF-DA). Fluorescent signals, reflecting the concentration of ROS, were analyzed by fluorescent microscopy. (B) Relative ROS level in the indicated group of cells as calculated by ImageJ image analysis software. (C) Determination of ROS levels of A2780 cells transfected with scrambled siRNA or siRNA-AUF1 with or without 5 Gy irradiation. (D) Relative ROS levels in the indicated group of cells as calculated by ImageJ image analysis software, *P < 0.01. Superoxide anion levels were determined in (E) PANC1 and (F) A2780 cells. Cells were transfected with siRNA-NC or siRNA-AUF-1 and treated with or without 5 Gy irradiation. RLU, relative light unit; *P < 0.05, **P < 0.01. Data (mean ± SEM) are representative of three experiments.
The expression of SOD1 correlates with that of AUF-1 in human tissues
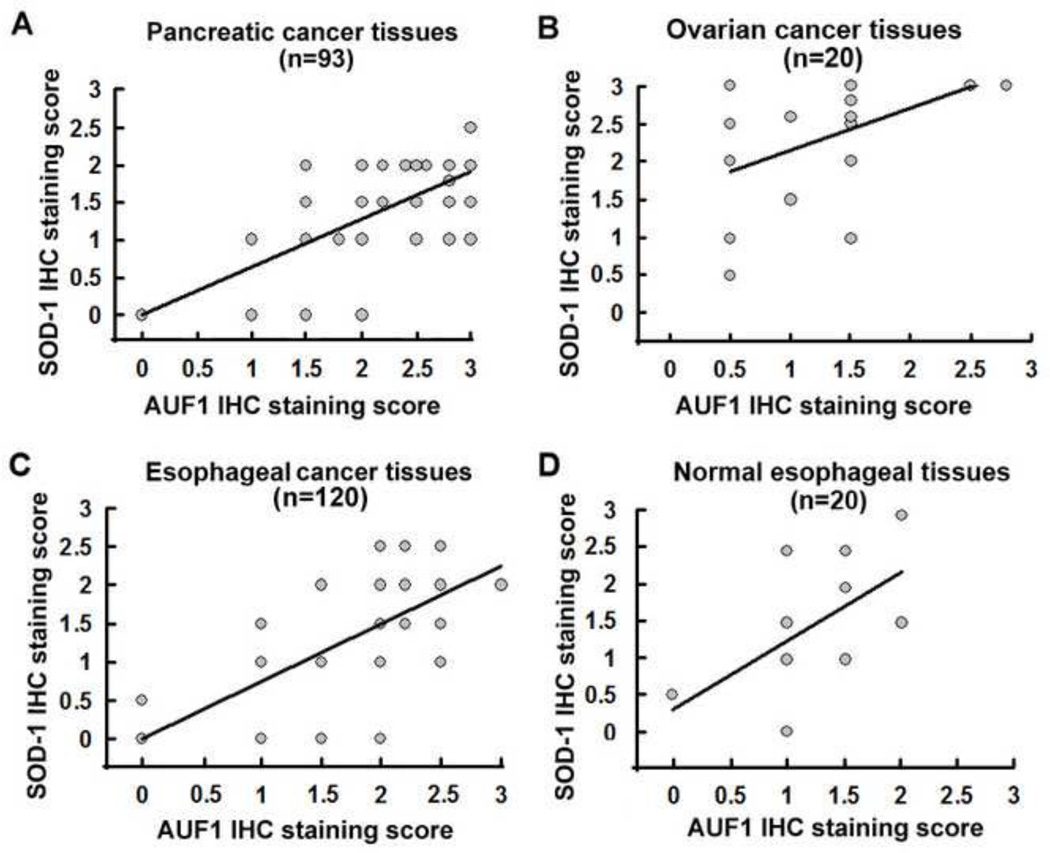
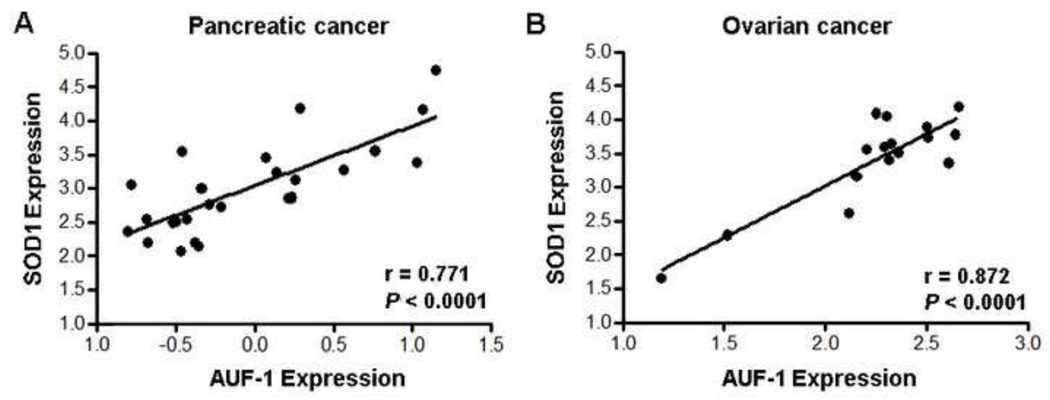
To understand whether SOD1 expression is associated with that of AUF-1, we analyzed expression of these genes in human cancer tissue samples (93 cases of pancreatic cancer tissues, 20 cases of ovarian cancer tissues, 120 cases of esophageal cancer tissues and 20 cases of normal esophageal tissues). Immunohistochemistry analysis showed that, in most cancer tissues examined, expression of SOD1 is significantly correlated to AUF-1 expression levels (rho = 0.558, P < 0.001 in 93 pancreatic cancer tissues, Figure 10A; rho = 0.169, P = 0.47 in 20 ovarian cancer tissues, Figure 10B; rho = 0.659, P < 0.001 in 120 esophageal cancer tissues, Figure 10C; rho = 0.565, P = 0.044 in 20 normal esophageal tissues, Figure 10D). These results strongly support the involvement of AUF-1 in positively regulating SOD1 gene expression in cancer tissues. The less significant association of the gene expression in ovarian cancer tissues may be due to the relatively small sample size. To further confirm the association of AUF-1 and SOD1 expression in cancer tissues, an Oncomine database search was performed. The search revealed a significant correlation of AUF-1 and SOD1 expression in both ovarian cancer and pancreatic cancer tissues (Figure 11). The survival curves in association with SOD1 and AUF-1 expression levels in pancreatic carcinoma were also constructed (Supplement Figure 3).
Figure 10. Correlation of SOD1 and AUF-1 expression in human tissue samples.
IHC staining of SOD1 protein showed a significant positive correlation with AUF-1 in (A) 93 pancreatic cancer tissues, (B) 20 ovarian cancer tissues, (C) 120 esophageal cancer tissues and (D) 20 normal esophageal tissues.
Figure 11. Correlation of AUF-1 and SOD1 expression in pancreatic and ovarian cancer tissues.
The log 2 median-centered expression levels of AUF1 were plotted against the expression levels of SOD1 from the Collisson pancreas study (28) (pancreatic carcinoma, left) and the Adib ovarian study (29) (ovarian serous adenocarcinoma, right) obtained from Oncomine and analyzed by Pearson correlation analysis (two-tailed) using GraphPad Prism version 4.00 for Windows (GraphPad Software, San Diego California USA).
Discussion
Eukaryotic gene expression is tightly coordinated through transcriptional and post-transcriptional regulation. The 3’UTRs of mRNAs are targeted by trans-acting factors or miRNAs to post-transcriptionally regulate gene expression. The versatile function of 3’UTRs is realized together with the complexity of the potential RNA-interacting factors (38). In the present study, we demonstrated that the SOD1 3’UTR significantly regulates SOD1 expression that has not been previously described, which reveals a novel cellular mechanism of SOD1 gene expression regulation in human cancer cells.
Our previous studies and reports from other groups, have demonstrated that inclusion of a 3’UTR may lead to unchanged or decreased reporter gene activity due to miRNA targeting of the 3’UTR resulting in degradation of the transcripts or suppression of protein translation (17,18,25,39). This is clearly not the case for SOD1 3’UTR as we observed that inclusion of the SOD1 3’UTR in a well-established luciferase reporter gene construct dramatically enhanced the reporter gene activity in several human cancer cell lines. To our knowledge, this magnitude of the up-regulation of reporter gene activity by a 3’UTR has never been previously reported. The increased luciferase activity is only partially attributed to an increase in mRNA expression as evidenced by the analysis of the reporter gene mRNA levels, suggesting that SOD1 protein translation is affected by SOD1 3’UTR. The relevance of the SOD1 3’UTR to endogenous SOD1 expression is confirmed by introducing the SOD1 3’UTR reporter constructs to cancer cells that leads to a down-regulation of endogenous SOD1 expression. This indicates that trans-acting factors or miRNAs in these model systems interact with SOD1 3’UTR thereby maintaining a high level expression of the gene. Therefore, when SOD1 3’UTR is exogenously introduced into the cells, it competes for these trans-acting factors or miRNAs, thereby leading to a decrease in endogenous SOD1 expression level.
Given the large number of miRNAs annotated in the human genome, 30%–80% of human genes are predicted to be influenced by miRNAs (40,41). However, a direct involvement of miRNA regulation in maintaining high expression of the SOD1 gene is less likely to occur in our cancer cell model systems due to the following considerations. First, miRNAs are best known to bind to complementary sequences of the 3‘UTRs of target mRNAs to cause degradation of the transcripts and/or repression of protein translation (17,18). A dramatic increase of the reporter gene activity by SOD1 3’UTR does not support a direct involvement of miRNAs in this event. Second, even though bioinformatics predicted the presence of miRNA binding sites for miR-621, miR-224 and miR-377, deletion of the miRNA binding site for miR-621 from the SOD1 3’UTR construct had no effect on the reporter gene activity, and deletion of miR-224 and miR-377 binding sites led to a reduction of the reporter gene activity, opposite to what was expected, which may be explained by the fact that these two adjacent miRNA binding sites are close to the ARE1 site that is important for the integrity of the SOD1 3’UTR. Third, direct application of miR-224, miR-377 or miR-621 mimics/inhibitors did not alter the SOD1 3’UTR-driven reporter gene activity, further suggesting that SOD1 3’UTR is not targeted by these miRNAs in our model system. Note that SOD1 was reported to be regulated by miR-377 in human and mouse neuronal cells (42). The discrepancy between our results and the previous report could be explained by the fact that miRNA regulation of gene expression is tightly controlled in a cell type-dependent manner (43).
A serial deletion and mutation analysis of the SOD1 3’UTR, along with bioinformatics analysis of its secondary structure indicate the critical role of the SOD1 3’UTR sequence integrity in regulating SOD1 gene expression. Using RNAfold, a web-based RNA secondary structure analytic tool, we predicted the potential impact of deletion or mutation of the SOD1 3’UTR sequence on the SOD1 3’UTR secondary structure and SOD1 3’UTR-driven reporter gene activity (Figure 4). This prediction was tested by experimentation. Most notably, deletion of the miR-224/377 binding sites from the SOD1 3’UTR reporter construct significantly altered its secondary structure and the reporter gene activity, whereas deletion of the miR-621 binding site did not dramatically disrupt the secondary structure or the reporter gene activity. Furthermore, single deletion of the ARE 6 remarkably altered the secondary structure (Figure 4G) and more significantly the reporter gene activity compared to the deletion of ARE 3/4 and ARE 5. Deletion of any large fragment from various parts of the SOD1 3’UTR dramatically disrupted the secondary structure (data not shown) and the reporter gene activity. The importance of the SOD1 3’UTR sequence integrity in regulating SOD1 gene expression was further confirmed by the observation that the AREs sequence alone cannot sustain the SOD1 3’UTR-driven reporter gene activity (Figure 2B). Thus, the SOD1 3’UTR sequence integrity is critical for its secondary structure and function.
An important observation of the present study is the physical and functional association of AUF-1, an established RNA binding protein, with SOD1 3’UTR. AUF-1, also called heterogeneous nuclear ribonucleoprotein D (hnRNPD), is ubiquitously expressed in eukaryotic cell and binds to AREs contained in the 3’UTR of many short-lived mRNAs. This binding has been shown in vitro to control the stability of ARE containing mRNAs, including proto-oncogenes, cytokines, or other signaling molecules (44). It is known that AUF-1 not only regulates mRNA stability (45) but also protein translation (36,37). In the present study, we found that whereas knockdown of HuR had no effect on SOD1 expression in cancer cells, knockdown of AUF-1 attenuated endogenous SOD1 expression and SOD1 3’UTR-driven reporter gene activity, indicating its involvement in regulating SOD1 gene expression via its 3’UTR. Indeed, AUF-1 directly binds to the SOD1 3’UTR as demonstrated by co-immunoprecipitation analysis (Figure 10G). AUF-1 clearly plays a role for SOD1 expression, as evidenced by attenuated SOD1 expression after AUF-1 siRNA knockdown and enhanced SOD1 expression after forced expression of AUF-1 in A2780 and PANC1 cells. Considering the magnitude of the reporter gene activity and the mRNA levels driven by the SOD1 3’UTR, our results suggest that AUF-1 binds to SOD1 3’UTR and primarily promote SOD1 protein translation, leading to an enhanced SOD1 protein expression, a concept consistent with previous observations (46). Consequently, AUF-1 was found to regulate ROS level in these cancer cell model systems. To our knowledge, this is the first observation that AUF-1 is involved in regulating the antioxidant system in eukaryotic cells. More interestingly, we found that expression of AUF-1 is highly associated with that of SOD1 in ovarian, pancreatic and esophageal tissues, and the association was further confirmed by an Oncomine database search. These observations, along with results from our mechanistic study, suggest that AUF-1 regulation of SOD1 expression is a novel cellular mechanism that may explain why SOD1 is overexpressed in human cancer cells (11–13). The correlation in expression of these two genes in normal esophageal tissues may indicate that AUF-1 regulation of SOD1 expression is not limited to cancer cells. Further characterization of AUF-1 interaction with the SOD1 3’UTR is warranted to better understanding its contribution to SOD1 gene expression in human cells.
Supplementary Material
Highlights.
The SOD1 3’UTR was characterized for its ability to regulate SOD1 gene expression
The SOD1 3’UTR maintains high expression of the SOD1 gene independent of miRNAs
AUF-1 binds to the SOD1 3’UTR to positively regulate SOD1 expression
AUF-1 regulates the redox balance in human cancer cells
The expression of SOD1 is correlated to that of AUF-1 in human cancer tissues
Acknowledgements
This work is supported in part by grants from the American Cancer Society (CNE-117557); the Susan G. Komen for the Cure Foundation (KG081083); the NIH OK-INBRE program (3P20RR016478-09S2); the Oklahoma Center for the Advancement of Science and Technology (HR14-147); the National Natural Science Foundation of China (81472920 and 81372433) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure
The authors have no conflict of interest to disclose.
References
- 1.Fridovich I. The biology of oxygen radicals. Science. 1978;201:875–880. doi: 10.1126/science.210504. [DOI] [PubMed] [Google Scholar]
- 2.Imlay JA, Fridovich I. Assay of metabolic superoxide production in Escherichia coli. J Biol Chem. 1991;266:6957–6965. [PubMed] [Google Scholar]
- 3.Fridovich I. Superoxide radical and superoxide dismutases. Annu Rev Biochem. 1995;64:97–112. doi: 10.1146/annurev.bi.64.070195.000525. [DOI] [PubMed] [Google Scholar]
- 4.Crapo JD, Oury T, Rabouille C, Slot JW, Chang LY. Copper,zinc superoxide dismutase is primarily a cytosolic protein in human cells. Proc Natl Acad Sci U S A. 1992;89:10405–10409. doi: 10.1073/pnas.89.21.10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liochev SI, Fridovich I. Mechanism of the peroxidase activity of Cu, Zn superoxide dismutase. Free Radic Biol Med. 2010;48:1565–1569. doi: 10.1016/j.freeradbiomed.2010.02.036. [DOI] [PubMed] [Google Scholar]
- 6.Halliwell B, Gutteridge JM. Role of free radicals and catalytic metal ions in human disease: an overview. Methods Enzymol. 1990;186:1–85. doi: 10.1016/0076-6879(90)86093-b. [DOI] [PubMed] [Google Scholar]
- 7.Zelko IN, Mariani TJ, Folz RJ. Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic Biol Med. 2002;33:337–349. doi: 10.1016/s0891-5849(02)00905-x. [DOI] [PubMed] [Google Scholar]
- 8.Gaudette M, Hirano M, Siddique T. Current status of SOD1 mutations in familial amyotrophic lateral sclerosis. Amyotrophic lateral sclerosis and other motor neuron disorders : official publication of the World Federation of Neurology, Research Group on Motor Neuron Diseases. 2000;1:83–89. doi: 10.1080/14660820050515377. [DOI] [PubMed] [Google Scholar]
- 9.Elchuri S, Oberley TD, Qi W, Eisenstein RS, Jackson Roberts L, Van Remmen H, Epstein CJ, Huang TT. CuZnSOD deficiency leads to persistent and widespread oxidative damage and hepatocarcinogenesis later in life. Oncogene. 2005;24:367–380. doi: 10.1038/sj.onc.1208207. [DOI] [PubMed] [Google Scholar]
- 10.Busuttil RA, Garcia AM, Cabrera C, Rodriguez A, Suh Y, Kim WH, Huang TT, Vijg J. Organ-specific increase in mutation accumulation and apoptosis rate in CuZn-superoxide dismutase-deficient mice. Cancer Res. 2005;65:11271–11275. doi: 10.1158/0008-5472.CAN-05-2980. [DOI] [PubMed] [Google Scholar]
- 11.Crnogorac-Jurcevic T, Efthimiou E, Nielsen T, Loader J, Terris B, Stamp G, Baron A, Scarpa A, Lemoine NR. Expression profiling of microdissected pancreatic adenocarcinomas. Oncogene. 2002;21:4587–4594. doi: 10.1038/sj.onc.1205570. [DOI] [PubMed] [Google Scholar]
- 12.Somwar R, Erdjument-Bromage H, Larsson E, Shum D, Lockwood WW, Yang G, Sander C, Ouerfelli O, Tempst PJ, Djaballah H, et al. Superoxide dismutase 1 (SOD1) is a target for a small molecule identified in a screen for inhibitors of the growth of lung adenocarcinoma cell lines. Proc Natl Acad Sci U S A. 2011;108:16375–16380. doi: 10.1073/pnas.1113554108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aird KM, Allensworth JL, Batinic-Haberle I, Lyerly HK, Dewhirst MW, Devi GR. ErbB1/2 tyrosine kinase inhibitor mediates oxidative stress-induced apoptosis in inflammatory breast cancer cells. Breast Cancer Res Treat. 2012;132:109–119. doi: 10.1007/s10549-011-1568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown DP, Chin-Sinex H, Nie B, Mendonca MS, Wang M. Targeting superoxide dismutase 1 to overcome cisplatin resistance in human ovarian cancer. Cancer Chemother Pharmacol. 2009;63:723–730. doi: 10.1007/s00280-008-0791-x. [DOI] [PubMed] [Google Scholar]
- 15.Huang P, Feng L, Oldham EA, Keating MJ, Plunkett W. Superoxide dismutase as a target for the selective killing of cancer cells. Nature. 2000;407:390–395. doi: 10.1038/35030140. [DOI] [PubMed] [Google Scholar]
- 16.Wu X, Brewer G. The regulation of mRNA stability in mammalian cells: 2.0. Gene. 2012;500:10–21. doi: 10.1016/j.gene.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuersten S, Goodwin EB. The power of the 3' UTR: translational control and development. Nat Rev Genet. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- 19.de Moor CH, Meijer H, Lissenden S. Mechanisms of translational control by the 3' UTR in development and differentiation. Seminars in cell & developmental biology. 2005;16:49–58. doi: 10.1016/j.semcdb.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 21.Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends in genetics : TIG. 2008;24:416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Yoo HY, Chang MS, Rho HM. Induction of the rat Cu/Zn superoxide dismutase gene through the peroxisome proliferator-responsive element by arachidonic acid. Gene. 1999;234:87–91. doi: 10.1016/s0378-1119(99)00176-6. [DOI] [PubMed] [Google Scholar]
- 23.Seo SJ, Kim HT, Cho G, Rho HM, Jung G. Sp1 and C/EBP-related factor regulate the transcription of human Cu/Zn SOD gene. Gene. 1996;178:177–185. doi: 10.1016/0378-1119(96)00383-6. [DOI] [PubMed] [Google Scholar]
- 24.Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. Embo J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, Yu H, Lou JR, Zheng J, Zhu H, Popescu NI, Lupu F, Lind SE, Ding WQ. MicroRNA-19 (miR-19) regulates tissue factor expression in breast cancer cells. J Biol Chem. 2011;286:1429–1435. doi: 10.1074/jbc.M110.146530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu P, Zhou J, Zhu H, Xie L, Wang F, Liu B, Shen W, Ye W, Xiang B, Zhu X, et al. VEGF-C promotes the development of esophageal cancer via regulating CNTN-1 expression. Cytokine. 2011;55:8–17. doi: 10.1016/j.cyto.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 27.Luo J, Zhou X, Ge X, Liu P, Cao J, Lu X, Ling Y, Zhang S. Upregulation of Ying Yang 1 (YY1) suppresses esophageal squamous cell carcinoma development through heme oxygenase-1. Cancer Sci. 2013 doi: 10.1111/cas.12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, Cooc J, Weinkle J, Kim GE, Jakkula L, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adib TR, Henderson S, Perrett C, Hewitt D, Bourmpoulia D, Ledermann J, Boshoff C. Predicting biomarkers for ovarian cancer using gene-expression microarrays. Br J Cancer. 2004;90:686–692. doi: 10.1038/sj.bjc.6601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Putnik M, Zhao C, Gustafsson JA, Dahlman-Wright K. Effects of two common polymorphisms in the 3' untranslated regions of estrogen receptor beta on mRNA stability and translatability. BMC genetics. 2009;10:55. doi: 10.1186/1471-2156-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milani P, Gagliardi S, Cova E, Cereda C. SOD1 Transcriptional and Posttranscriptional Regulation and Its Potential Implications in ALS. Neurology research international. 2011;2011:458427. doi: 10.1155/2011/458427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lorenz R, Bernhart SH, Honer Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. ViennaRNA Package 2.0. Algorithms for molecular biology : AMB. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trojanowicz B, Dralle H, Hoang-Vu C. AUF1 and HuR: possible implications of mRNA stability in thyroid function and disorders. Thyroid research. 2011;4(Suppl 1):S5. doi: 10.1186/1756-6614-4-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan YX, Chen H, Kilberg MS. Interaction of RNA-binding proteins HuR and AUF1 with the human ATF3 mRNA 3'-untranslated region regulates its amino acid limitation-induced stabilization. J Biol Chem. 2005;280:34609–34616. doi: 10.1074/jbc.M507802200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sureban SM, Murmu N, Rodriguez P, May R, Maheshwari R, Dieckgraefe BK, Houchen CW, Anant S. Functional antagonism between RNA binding proteins HuR and CUGBP2 determines the fate of COX-2 mRNA translation. Gastroenterology. 2007;132:1055–1065. doi: 10.1053/j.gastro.2006.12.031. [DOI] [PubMed] [Google Scholar]
- 36.Liao B, Hu Y, Brewer G. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat Struct Mol Biol. 2007;14:511–518. doi: 10.1038/nsmb1249. [DOI] [PubMed] [Google Scholar]
- 37.Laroia G, Cuesta R, Brewer G, Schneider RJ. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- 38.Matoulkova E, Michalova E, Vojtesek B, Hrstka R. The role of the 3' untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA biology. 2012;9:563–576. doi: 10.4161/rna.20231. [DOI] [PubMed] [Google Scholar]
- 39.Zhang S, Chen H, Zhao X, Cao J, Tong J, Lu J, Wu W, Shen H, Wei Q, Lu D. REV3L 3'UTR 460 T>C polymorphism in microRNA target sites contributes to lung cancer susceptibility. Oncogene. 2013;32:242–250. doi: 10.1038/onc.2012.32. [DOI] [PubMed] [Google Scholar]
- 40.Lu J, Clark AG. Impact of microRNA regulation on variation in human gene expression. Genome Res. 2012;22:1243–1254. doi: 10.1101/gr.132514.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang T, Kumar S, Shen Y, Lu J, Wu ML, Shi S, Li WH, Wu CI. Adverse interactions between micro-RNAs and target genes from different species. Proc Natl Acad Sci U S A. 2010;107:12935–12940. doi: 10.1073/pnas.1007591107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Q, Wang Y, Minto AW, Wang J, Shi Q, Li X, Quigg RJ. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. FASEB J. 2008;22:4126–4135. doi: 10.1096/fj.08-112326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci U S A. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gouble A, Grazide S, Meggetto F, Mercier P, Delsol G, Morello D. A new player in oncogenesis: AUF1/hnRNPD overexpression leads to tumorigenesis in transgenic mice. Cancer Res. 2002;62:1489–1495. [PubMed] [Google Scholar]
- 45.Sela-Brown A, Silver J, Brewer G, Naveh-Many T. Identification of AUF1 as a parathyroid hormone mRNA 3'-untranslated region-binding protein that determines parathyroid hormone mRNA stability. J Biol Chem. 2000;275:7424–7429. doi: 10.1074/jbc.275.10.7424. [DOI] [PubMed] [Google Scholar]
- 46.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.