Abstract
To identify polymorphisms associated with variability of iron overload severity in HFE-associated hemochromatosis, we performed exome sequencing of DNA from 35 male HFE C282Y homozygotes with either markedly increased iron stores (n=22; cases) or with normal or mildly increased iron stores (n=13; controls). The 35 participants, residents of the U.S., Canada, and Australia, reported no or light alcohol consumption. Sequencing data included 82,068 single nucleotide variants, and 10,337 genes were tested for a difference between cases and controls. A variant in the GNPAT gene showed the most significant association with severe iron overload (p = 3×10−6, p=0.033 by the likelihood ratio test after correction for multiple comparisons). Sixteen of 22 participants with severe iron overload had GNPAT polymorphism p.D519G (rs11558492) (15 heterozygotes, one homozygote). No control participant had this polymorphism. To examine functional consequences of GNPAT deficiency, we performed siRNA-based knockdown of GNPAT in the human liver-derived cell line HepG2/C3A. This knockdown resulted in a >17-fold decrease in expression of the mRNA encoding the iron regulatory hormone hepcidin. Conclusion: GNPAT p.D519G is associated with a high-iron phenotype in HFE C282Y homozygotes and may participate in hepcidin regulation.
Keywords: hemochromatosis, hepatic iron concentration, HepG2/C3A, iron overload, hepcidin
HFE-associated hemochromatosis (HH) is an autosomal recessive disorder in which excess iron reaches toxic levels by middle life in some patients.1 Organ damage occurs via oxidative tissue injury that may cause hepatic cirrhosis, arthropathy, hypogonadism, diabetes mellitus, and cardiomyopathy.1–4 These complications can be prevented by early diagnosis and phlebotomy therapy.5, 6 Approximately 90% of persons with HH are homozygous for a polymorphism in HFE (rs1800562), which causes substitution of tyrosine for cysteine in the HFE protein at position 282 (Cys282Tyr; p.C282Y).7 C282Y homozygotes with HH have excessive iron absorption due to inappropriately low levels of the iron regulatory hormone hepcidin.8, 9
Approximately five persons per 1,000 of Northern European descent are C282Y homozygotes.10–14 This includes approximately one million persons in the U.S.10 A minority of C282Y homozygotes accumulate enough iron to cause organ damage.15 Thus, it has been proposed that C282Y homozygosity is necessary but not sufficient to cause severe iron overload and that other genetic or environmental factors modify the phenotype.16 Mutations in other genes of iron metabolism have been associated with variations in iron stores among C282Y homozygotes and normal individuals.17–25 Such mutations demonstrate that modifier genes account for differences in severity of iron overload in some kinships but do not explain the variability in iron overload observed across most unrelated C282Y homozygotes.
We hypothesized that variants of genes other than HFE and those previously associated with hemochromatosis and iron overload phenotypes are involved in the regulation of iron metabolism and modulate expression of iron overload in HFE C282Y homozygotes. To examine this hypothesis, we studied HFE C282Y homozygotes at the extremes of phenotypic expression based on serum ferritin concentration, liver iron concentration, and the amount of iron removed by therapeutic phlebotomy to achieve iron depletion.
Materials and Methods
Study population
Consortium study sites identified HFE C282Y homozygotes in clinical practice settings or by population screening through the Hemochromatosis and Iron Overload Screening (HEIRS) Study and a study of the Prevalence of Iron Overload and Frequency of the Hemochromatosis Gene conducted at the Department of Veterans Affairs Long Beach Healthcare System. Approval for human studies was obtained from: University of California, Irvine; University of Western Ontario; QIMR Berghofer Medical Research Institute; Rochester General Health System; the Cancer Council Victoria; and Department of Veterans Affairs Long Beach Healthcare System. Written informed consent was obtained from all participants.
Clinical and laboratory data collection
Information on demographics, iron supplements, alcohol consumption, clinical observations, and laboratory biochemical tests was obtained from medical records of participants identified through clinical practice and from the NIH BioLincc biorepository for HEIRS Study participants.
Selection of participants with extreme iron phenotypes
Inclusion criteria were: 1) HFE C282Y homozygosity, 2) participant was unrelated to other subjects, 3) participant did not withdraw consent, 4) participant did not refuse blood storage, and 5) male sex. We excluded women to eliminate any effect of female sex on limiting iron accumulation. Criteria for selection of HFE C282Y homozygotes with increased iron stores included serum ferritin >1000 μg/L at diagnosis and either (a) hepatic iron concentration >236 μmol/g dry weight (reference range 0–36 μmol/g) or (b) mobilized body iron >10 g by quantitative phlebotomy.26 Criteria for HFE C282Y homozygotes with normal or mildly elevated iron stores included (a) serum ferritin <300 μg/L or either (a) age ≥40 y with ≤2.5 g iron removed by phlebotomy to achieve serum ferritin <50 μg/L, or (b) age ≥50 y with ≤3.0 g iron removed by phlebotomy. Alcohol consumption was characterized as “former drinker”, “never” or “light drinker” (less than 20 g alcohol per day), “moderate drinker” (20–60 g alcohol per day), and “heavy drinker” (>60 g alcohol per day).27 Participants who reported current or past moderate or heavy alcohol consumption were excluded to prevent confounding.
Exome sequencing
Quality control of sample DNA
Initial quality control performed on all samples included sample quantification using PicoGreen (Molecular Probes, Inc., Eugene, OR) and sex determination using a TaqMan assay (Applied Biosystems, Carlsbad, CA).28 All samples were genotyped using a high-frequency cosmopolitan 96-plex genotyping assay to ensure integrity of tracking throughout the sample preparation and sequencing pipeline.
Exome capture and sequencing
Approximately 1 μg of genomic DNA was used for a series of shotgun library construction steps, including acoustic fragmentation (Covaris, Inc., Woburn, MA), end-polishing and A-tailing, ligation of sequencing adaptors, and polymerase chain reaction (PCR) amplification. Sample shotgun libraries were captured for exome enrichment using the Roche/NimbleGen (Madison, WI) SeqCap EZ Cap v2 (~44 Mb) according to the manufacturer’s instructions. Enriched libraries were sequenced on an Illumina HiSeq 2000 (San Diego, CA) using paired-end 50bp runs.
Sequence Kernel Association Test
After quality control filtering, the dataset included 82,068 single nucleotide variants and 1,403 insertions/deletions (indels). Differences in the distribution of variants were tested between the case and control groups for each gene separately using the small sample adjusted sequence kernel association test (aSKAT-O) that includes rare and common variants but downweights the contribution of common variants to the test statistic.29 Only non-synonymous variants were included in the by-gene tests, and genes for which less than five individuals had polymorphisms were excluded because statistical power can be near zero for genes with such little variation in the sample. Principal components (PCs) were constructed from the exome variants to adjust for possible confounding by ancestry and to confirm no ancestral outliers. PCs 1 and 2 were included in the SKAT model. The likelihood ratio test (LRT) was applied for the most significant gene from the SKAT analysis.30 This test has more statistical power than SKAT but it is applied only to genes with low p-values detected by SKAT, as it requires Monte Carlo sampling with two million samples per gene to determine an accurate p-value with imbalanced sample sizes. Specifically, we used the parametric bootstrap to determine the null distribution of the LRT to obtain an accurate p-value and then adjusted for testing of multiple genes via the Bonferroni method. The positive False Discovery Rate (FDR) was estimated using the “qvalue” package in R (http://www.r-project.org).
Functional studies of GNPAT
GNPAT knockdown
A human liver-derived cell line, HepG2/C3A (CRL-10741, American Type Culture Collection, Manassas, VA), was used for functional studies. Cells were grown in MEM + Glutamax supplemented with 10% fetal calf serum (Life Technologies, Mulgrave, VIC, Australia). Control non-specific siRNA (si.NS) and siRNA specific for GNPAT (si.GNPAT; 10 pmol, GenePharma, Shanghai, China) were transfected into 1×105 HepG2/C3A cells in triplicate with Lipofectamine RNAimax (Life Technolgies) according to the manufacturer’s recommendations for reverse transfection at approximately 30% cell confluency. After 72 hours, RNA was isolated using Trizol (Life Technologies) and reverse transcribed with SuperScript III (Life Technologies). Quantitative PCR (qPCR) was performed on a Viia7 (Life Technologies) using SYBR green master mix (Roche, Castle Hill, NSW, Australia) for GNPAT, HAMP, ID1, and SMAD7, and expression levels were compared to the geometric means of reference genes ACTB and HPRT1 using 2−ΔCT. Primer pairs (Integrated DNA Technologies, Coralville, IA) used for quantitative PCR and GNPAT siRNA oligonucleotide sequences are shown in Table 1. For BMP6 treatment, cells were serum deprived for 6 hours in Opti-mem medium (Life Technologies), before the addition of recombinant human BMP6 (R&D Systems, Minneapolis, MN) (10 ng/mL; 4 hours) followed by harvesting.
Table 1.
Primer pairs used for quantitative PCR and GNPAT siRNA oligo sequences used for knockdowns.
| Gene | Sequence |
|---|---|
|
| |
| ACTB-forward | CAGGCACCAGGGCGTG |
| ACTB-reverse | GCCCACATAGGAATCCTTCTGA |
| HPRT1- forward | GAAAGGGTGTTTATTCCTCAT |
| HPRT1-reverse | CCCATCTCCTTCATCACAT |
| GNPAT- forward | ACATTT TTGTGCGCCCATCC |
| GNPAT-reverse | TCACGTAG AAGCGA AAG CA |
| HAMP- forward | CCACAACAGACGGGACAAC |
| HAMP-reverse | AAAATGCAGATGGGGAAGTG |
| ID1- forward | TGG AGC TGA ACT CGG AAT CCG |
| ID1-reverse | GAC ACA AGA TGC GAT CGT CCG |
| SMAD7- forward | TCACCTTAGCCGACTCTGCG |
| SMAD7-reverse | GTTTCAGCGGAGGAAGGCAC |
|
| |
| siRNA | Oligonucleotide sequence |
|
| |
| GNPAT- forward | GGGCUGUAUUCUCUGAAUATT |
| GNPAT-reverse | UAUUCAGAGAAUACAGCCCTT |
Western blotting
SMAD 1/5/8 phosphorylation was assessed as described previously,31 using anti-phospho-SMAD1/5/8 at 1:1000 (Cell Signaling Technology, Danvers, MA #9511S), and anti-actin at 1:35,000 (Sigma-Aldrich, #A2066).
Statistical analysis
Differences between groups for qPCR of si.NS vs si.GNPAT, proliferation and viability assays were assessed by independent sample t-tests. Differences between qPCR and Western quantification of si.NS vs si.GNPAT with or without BMP6 treatment were assessed using two-way analysis of variance (ANOVA) and post-hoc analyses were performed using the Bonferroni multiple comparisons procedure with multiplicity-adjusted P values. Fisher’s exact test was used to compare the allele frequencies of men with high- and low-iron phenotypes to that of European Americans. A comparison-wise significance level of 0.025 was used to maintain an overall significance level of 0.05.
Results
Clinical and laboratory data
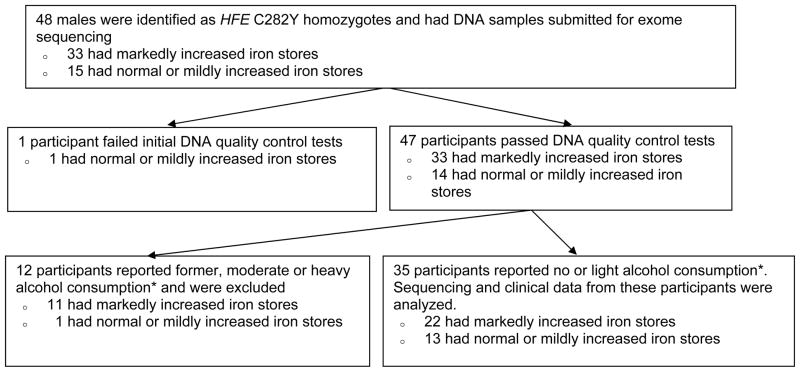
We identified participants with high-iron phenotypes (high expressers) and low-iron phenotypes (low expressers) through a unique international consortium of collaborating centers. Most high expressers and some low expressers were treated at associated clinics, whereas other low expressers were identified through population screening.10 The study schema is displayed in Figure 1. There were 48 males who met the basic inclusion criteria and had DNA submitted for exome sequencing. Of these, 47 met quality control tests. At diagnosis, 12 participants reported former alcohol consumption or current moderate or heavy alcohol consumption and were excluded. Of the remaining 35 participants who reported no or light alcohol consumption, 22 were C282Y male homozygotes with markedly increased iron stores (high expressers) and 13 were C282Y male homozygotes who had either normal or very mildly increased iron stores (low expressers). Based on iron parameters from C282Y homozygotes identified by population-based screening,10, 32 we estimate that, among light alcohol users or non-users, the present high expressers were among the highest 1% in terms of both serum ferritin and amount of iron removed to achieve iron depletion. The low expressers were among the lowest 4%.
Figure 1.
Study Schema
*Alcohol consumption was characterized as “former drinker”, “never” or “light drinker” (less than 20 g alcohol per day), “moderate drinker” (20–60 g alcohol per day), and “heavy drinker” (>60 g alcohol per day).
Exome sequencing and statistical analysis
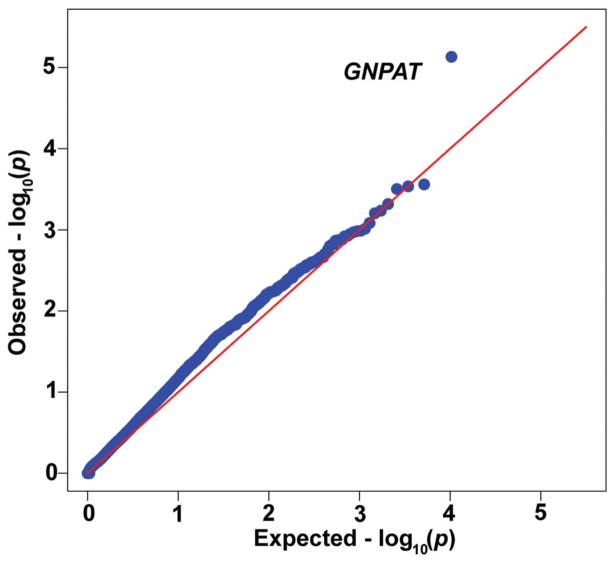
We tested 10,337 genes to determine if there were differences between iron phenotype expression groups. The most significant gene identified was GNPAT (also known as DHAPAT or DAPAT), encoding the peroxisomal enzyme glyceronephosphate O-acyltransferase (SKAT test p = 7.4×10−6; likelihood ratio test with bootstrap p = 3×10−6, p=0.033 corrected for multiple comparisons; estimated false discovery rate = 0.04).33 The quantile-quantile (Q-Q) plot (Figure 2) compares the observed distribution of −(log10p-values) from 10,337 genes to the expected uniform distribution if there were no variants modifying severity of iron phenotype.
Figure 2. Results of sequence kernel association test.
Quantile/quantile plot for SKAT p-values from the by-gene comparison of 13 low-iron expressers vs 22 high-iron expressers (all male C282Y homozygotes).
In this cohort, inspection of the two variants included in the test of GNPAT showed that 16 of 22 high-iron expressers carried GNPAT p.D519G (1556A>G; exon 11; chromosome 1q42; rs11558492). Fifteen were GNPAT p.D519G heterozygotes and the other was a homozygote. The GNPAT p.D519G homozygote presented at age 26 years with severe iron overload but no cirrhosis. One GNPAT p.D519G heterozygote presented at age 36 years with severe iron overload and cirrhosis. None of the low expressers had p.D519G. When SKAT was applied to each variant individually, GNPAT p.D519G was the most significantly different variant between groups with a nominal p-value similar to that of the by-gene analysis (p = 7.8×10−6), indicating that the by-gene signal is driven by this single variant. The geographic distribution of study sites contributing HFE C282Y homozygotes is displayed in Table 2. GNPAT p.D519G was present in 50%–85% of C282Y homozygotes with markedly increased iron stores submitted from four clinics in three countries and from HEIRS Study sites. C282Y homozygous low expressers, none of whom had p.D519G, were also identified at all study sites and in population screening studies.
Table 2.
HFE C282Y homozygotes
| Markedly Increased Iron Stores (N=22) | Normal or Mildly Increased Iron Stores (N=13) | |||
|---|---|---|---|---|
|
| ||||
| Study Site | Number of samples (%) | Number of samples with GNPAT p.A519G polymorphism (%) | Number of samples (%) | Number of samples with GNPAT p.A519G polymorphism (%) |
| Southern Iron Disorders Center, Birmingham, Alabama, USA | 7 (31.8%) | 6/7 (85.7%) | 1 (7.7%) | 0/1 (0%) |
| Royal Brisbane & Women’s Hospital, Brisbane, Australia | 5 (22.7%) | 3/5 (60.0%) | 3 (23.1%) | 0/3 (0%) |
| London Health Sciences Centre, London, Ontario, Canada | 4 (18.2%) | 3/4 (75.0%) | 1 (7.7%) | 0/1 (0%) |
| Rochester General Hospital, Rochester, New York, USA | 4 (18.2%) | 3/4 (75.0%) | 2 (15.4%) | 0/2 (0%) |
| Population screening, USA and Canada | 2 (9.1%)* | 1/2 (50.0%) | 6 (46.2%)† | 0/6 (0%) |
| TOTAL | 22 | 16 | 13 | 0 |
Two deidentified samples, collected from participants in the Hemochromatosis and Iron Overload Screening (HEIRS) Study, were obtained from the NHLBI Biologic Specimen and Data Repository Information Coordinating Center.
Four deidentified samples, collected from participants in the Hemochromatosis and Iron Overload Screening (HEIRS) Study, were obtained from the NHLBI Biologic Specimen and Data Repository Information Coordinating Center. Two samples were obtained from participants in a study of the Prevalence of Iron Overload and Frequency of the Hemochromatosis Gene conducted at the Department of Veterans Affairs Long Beach Health Care System.
Examination of fibrosis or cirrhosis as potential confounding variables
Because fibrosis/cirrhosis is expected to be uncommon among the low expressers and common among high expressers, we considered whether the GNPAT results might be driven by a causal association with fibrosis/cirrhosis. To assess this possibility, we tested for association between the GNPAT p.D519G variant and the presence of fibrosis/cirrhosis. Because only one of 13 low expressers had liver biopsy information, the association test was performed among high expressers only. Seventeen of 22 high expressers had liver biopsies; 6 of these 17 had fibrosis, without cirrhosis (35%), and 6 of 17 (35%) had cirrhosis (Table 3). The GNPAT p.D519G variant was observed in 3 of 6 with fibrosis (50%), 5 of 6 with cirrhosis (83%), and 4 of 5 without fibrosis/cirrhosis (80%) (p=0.53). Thus, the frequency of the variant is nearly equal in those with and without cirrhosis or fibrosis. We also observed no significant differences in frequency of the variant among high expressers with or without hepatomegaly or other clinical manifestations (Table 3).
Table 3.
Association between clinical characteristics and presence of GNPAT p.D519G in 22 HFE C282Y homozygotes with markedly increased iron stores
| Markedly Increased Iron Stores (N=22)
|
||||
|---|---|---|---|---|
| Characteristic | N (%) | Number with GNPAT p.D519G polymorphism (%) | Fisher’s Exact Test P-value | |
| Clinical Assessment | ||||
| Self-report of arthritis | Present | 13/22 (59.1%) | 9/13 (69.2%) | 1.00 |
| Absent | 9/22 (40.9%) | 7/9 (77.8%) | ||
|
| ||||
| Self-report of diabetes | Present | 3/22 (13.7%) | 1/3 (33.3%) | 0.17 |
| Absent | 19/22 (86.4%) | 15/19 (79.0%) | ||
|
| ||||
| Chronic fatigue/weakness | Present | 9/22 (40.9%) | 6/9 (66.7%) | 1.00 |
| Absent | 12/22 (54.6%) | 9/12 (75.0%) | ||
|
| ||||
| Physical Exam | ||||
| Hepatomegaly | Present | 9/22 (40.9%) | 8/9 (88.9%) | 0.33 |
| Absent | 13/22 (59.1%) | 8/13 (61.5%) | ||
|
| ||||
| Swollen or tender metacarpophalangeal joints | Present | 13/22 (59.1%) | 10/13 (76.9%) | 0.66 |
| Absent | 9/22 (40.9%) | 6/9 (66.7%) | ||
|
| ||||
| Related Conditions | ||||
| Fibrosis/Cirrhosis*† | Fibrosis | 6/17 (35.3%) | 3/6 (50.0%) | 0.53 |
| Cirrhosis | 6/17 (35.3%) | 5/6 (83.3%) | ||
| Absent | 5/17 (29.4%) | 4/5 (80.0%) | ||
Data from five patients were excluded because no liver biopsy had been performed.
When data from biopsies showing fibrosis only (n=6) and those with cirrhosis (n=6) were combined, eight of the 12 (66.7%) had the GNPAT P.D519G polymorphism and there was still no significant association between presence of fibrosis and the GNPAT variant (Fisher’s Exact Test p-value = 1.00).
Allele frequencies of GNPAT p.D519G
We compared the allele frequencies of GNPAT p.D519G in the 22 men with high-iron phenotypes and the 13 men with normal or mildly increased iron phenotypes with that of 4300 European Americans in the NHLBI Exome Sequencing Project Exome Variant Server.34 The allele frequency in men with high-iron phenotypes was greater than that of European Americans (38.6% vs. 20.6%, respectively; Fisher’s exact test, nominal p = 0.0076). The allele frequency in men with normal or mildly increased iron phenotypes was significantly lower (0% vs. 20.6%, Fisher’s exact test, nominal p = 0.0054).
To determine whether other known mutations influence iron phenotypes, the exome data were used to screen for mutations in HAMP, HJV, TFR2, FPN1, and TMPRSS6. One high expresser was heterozygous for HJV p.G320V. He was among the six high expressers who did not have GNPAT p.D519G. No other known or probable mutations35–39 causing differences in expression between groups were found. The observed allele frequency of p.G320V in our cohort was 1 of 22 high expressers compared to 4 of 4300 European Americans (Fisher’s exact test, p = 0.025 with an odds ratio of 50.6). Because p.G320V possibly influences phenotype, we removed the data for this subject and performed an additional by-gene test, resulting in an increase in significance for GNPAT (p=1×10−6 by the LRT, p=0.0103 after adjustment for multiple testing).
We searched the Genome Variant Server (http://gvs.gs.washington.edu/GVS138) for variants in high linkage disequilibrium (LD) with GNPAT p.D519G that could potentially be causative variants that were tagged by GNPAT p.D519G. There are no non-synonymous coding variants within 250Kb of this site with r2 > 0.03. Our by-variant analysis of exome variants would have identified any coding variant in tight LD with GNPAT p.D519G that is statistically associated with the variant, but none were identified. Only four non-coding variants within 250Kb of GNPAT p.D519G had r2 > 0.30, and none were in GNPAT or its introns.
GNPAT p.D519G results in an aspartic acid (acidic) to glycine (nonpolar) substitution at position 519. Standard functional predictions conflict: SIFT prediction from ANNOVAR suggests that the change is deleterious, while Polyphen2 suggests that the change is benign. A more comprehensive functional prediction tool, Combined Annotation Dependent Depletion (CADD),40 predicts this variant to be among the 10% most deleterious substitutions in the human genome with a CADD score of 11.8.
Functional studies of GNPAT
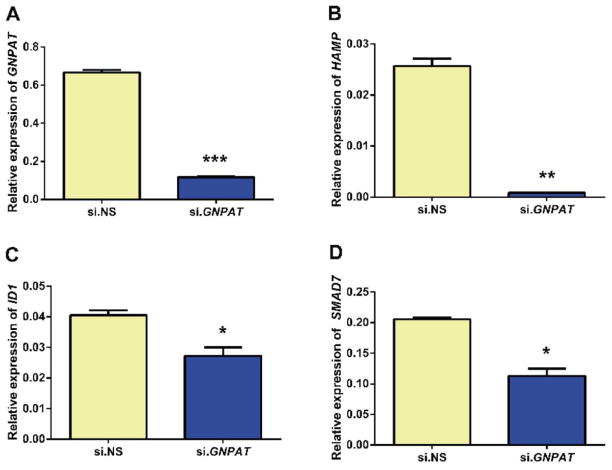
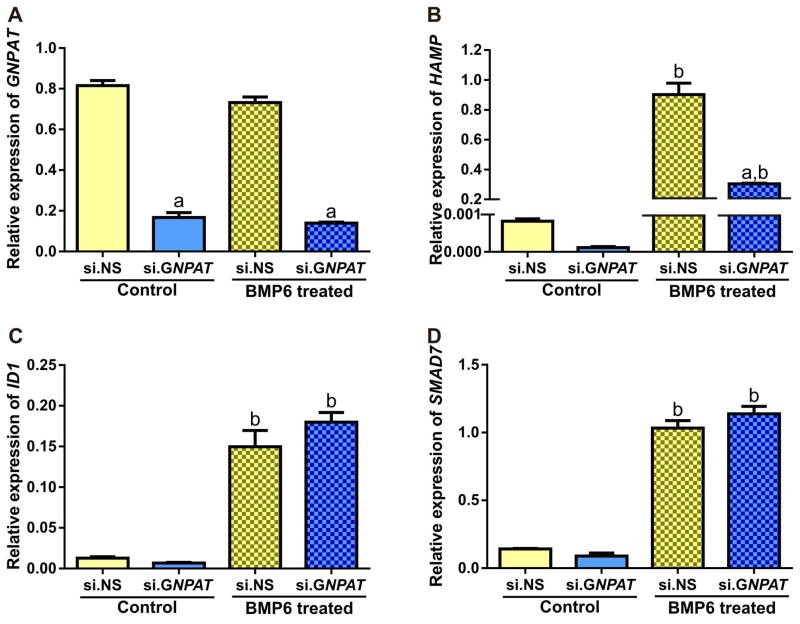
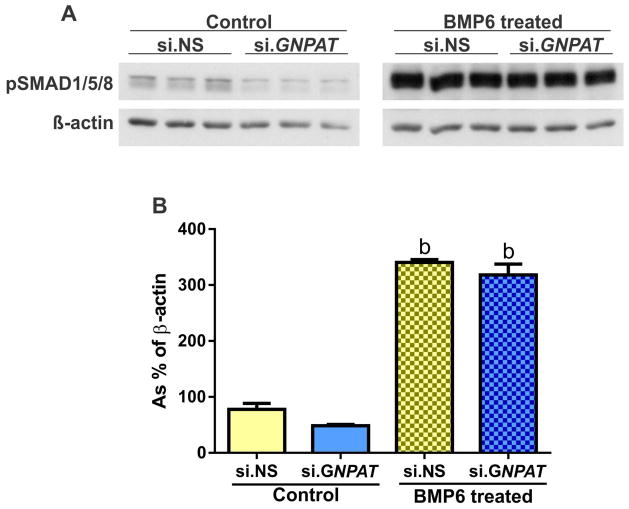
GNPAT was efficiently knocked down by its siRNA (~85% knockdown compared to control siRNA) (Figure 3A). This knockdown resulted in a >17-fold decrease in HAMP mRNA expression (Figure 3B). Expression of two genes co-ordinately regulated with HAMP, ID1 (inhibitor of DNA-binding protein 1) and SMAD7 (SMAD family member 7),41 was also decreased (Figure 3C and 3D), although the magnitude of the decrease was less. Cell proliferation and viability were similar in cells treated with si.GNPAT and those treated with control si.NS. Bone morphogenetic protein 6 (BMP6) is a major regulator of hepcidin.42 HepG2/C3A cells transfected with si.NS or si.GNPAT were serum starved, treated with or without BMP6, and analyzed for mRNA expression of HAMP, ID1 and SMAD7. We observed no effect of BMP6 treatment on GNPAT levels (Figure 4A). Treatment with BMP6 significantly induced expression of HAMP, ID1 and SMAD7, but only the induction HAMP was blunted by GNPAT knockdown (Figure 4B, 4C, 4D). To determine whether GNPAT exerted its effects on hepcidin through the BMP-SMAD pathway, we examined phospho-SMAD1/5/8 (pSMAD1/5/8) levels in cells treated with si.NS or si.GNPAT, with or without BMP6 (Figure 5). Knockdown of GNPAT under basal conditions (without BMP6) resulted in decreased pSMAD1/5/8, whereas GNPAT knockdown had no effect on pSMAD1/5/8 levels when cells were treated with BMP6.
Figure 3. Representative graphs of siRNA mediated knockdown of GNPAT in HepG2/C3A cells transfected with either 10 pmol non-specific siRNA (si.NS) or 10 pmol GNPAT-specifc siRNA (si.GNPAT) for 72 hr.
Experiments were performed four times. Relative expressions of (A) GNPAT, (B) HAMP, (C) ID1 and (D) SMAD7 mRNA were analyzed by using qPCR and normalized to the geometric mean of ACTB and HPRT1. Error bars represent the standard error of the mean (SEM). P-values for fold change were calculated using Student’s two-tailed t-test; * indicates a p-value <0.05, ** p <0.01 and *** p < 0.001.
Figure 4. Representative graphs of siRNA mediated knockdown of GNPAT in HepG2/C3A either treated with or without BMP6.
HepG2/C3A cells were treated with control (si.NS) or GNPAT-specific siRNA (si.GNPAT). Cells were serum starved for 6 hours and then treated with vehicle or BMP6 (10ng/mL for four hours). Experiments were performed four times. Relative expressions of (A) GNPAT, (B) HAMP, (C) ID1 and (D) SMAD7 mRNA were analyzed using qPCR and normalized to the geometric means of ACTB and HPRT1. Error bars represent the standard error of the mean (SEM). P-values for fold change were calculated using two-way ANOVA with a Bonferroni multiple comparison test post-hoc with multiplicity adjusted P-values; a = p<0.0001 comparing si.GNPAT to si.NS within treatment group, b = p<0.001 comparing control to BMP6 treated within si.RNA group.
Figure 5. Western blot of phosphoSMAD1/5/8 and associated quantification following siRNA mediated knockdown of GNPAT in HepG2/C3A either treated with or without BMP6.
Experiments were performed twice. HepG2/C3A cells were treated with control (si.NS) or GNPAT-specific siRNA (si.GNPAT). Cells were serum starved for 6 hours and then treated with vehicle or BMP6 (10ng/mL for four hours). (A) Representative Western Blot of phosphoSMAD1/5/8. (B) Quantification of phosphoSMAD1/5/8 relative to the reference gene β-actin. Error bars represent the standard error of the mean (SEM). P-values for fold change were calculated using two-way ANOVA with a Bonferroni multiple comparison test post-hoc with multiplicity adjusted P-values; b = p<0.01 comparing control to BMP6 treated within si.RNA group.
Discussion
In light of the marked differences in clinical penetrance among HFE C282Y homozygotes, the existence of a common genetic variant that interacts with the C282Y mutation or C282Y protein to produce more severe penetrance is plausible. In the present study, GNPAT p.D519G was found in 16 of 22 HFE C282Y homozygotes with high-iron phenotypes who did not consume large quantities of ethanol and in none of the C282Y homozygotes with low-iron phenotypes. Most of the men with high-iron phenotypes were GNPAT p.D519G heterozygotes, although one was a GNPAT p.D519G homozygote. The allele frequency of GNPAT p.D519G in men with high-iron phenotypes was significantly greater than in 4300 European Americans, and the allele frequency in men with normal or mildly increased iron phenotypes was significantly lower than that of European Americans. This is consistent with the postulate that GNPAT p.D519G is associated with high-iron phenotypes in C282Y homozygotes. It is also possible that a haplotype tightly linked with GNPAT is involved, but we did not identify any potentially deleterious mutations in LD with GNPAT p.D519G. Among the HFE C282Y homozygotes with high-iron phenotypes who had undergone liver biopsy, there was no statistically significant association between fibrosis/cirrhosis and the presence of GNPAT p.D519G.
The increased intestinal iron absorption in hemochromatosis is mediated at the level of mucosal transfer of iron into the circulation.43 This is attributable to increased ferroportin concentration on the basolateral membrane of duodenal enterocytes, resulting from a deficiency of hepcidin, which normally inhibits iron absorption by binding to ferroportin and causing its internalization and degradation.8 Hepcidin is central to the regulation of iron homeostasis, and decreased hepcidin levels are associated with most inherited iron overload disorders.8 In HH, upregulation of the hepcidin gene (HAMP) does not occur despite significant hepatic iron loading.9 Our observations suggest that loss of GNPAT expression or single nucleotide variants which affect GNPAT expression/activity affect HAMP expression and that GNPAT is a potential genetic modifier of hemochromatosis iron phenotypes. Although hepcidin is downregulated in HFE C282Y homozygotes, hepcidin expression is lower in C282Y homozygotes who also have HAMP, HJV, or TFR2 mutations,44 implying that further downregulation is possible. Thus, GNPAT variants such as GNPAT p.D519G identified in the current study could cause further downregulation of hepcidin.
Rare deleterious GNPAT point mutations or deletions in either homozygous or compound heterozygous configuration (but not GNPAT p.D519G) result in rhizomelic chondrodysplasia punctata (RCDP) type 2, (MIM#222765). Severe anomalies present at birth in RCDP type 2 include skeletal dysplasia, facial deformities, cataracts, and severe mental retardation. Iron overload has not been reported to be part of this group of rare disorders. The combined incidence of RCDP types 1–3 is <1:100,000. Extensive in vitro studies based on two siblings with RCDP type 2 revealed that GNPAT is required for ether-lipid and plasmalogen synthesis and that GNPAT deficiency impairs cholesterol distribution and formation of caveolae and clathrin-coated pits, resulting in reduced transferrin receptor recycling.45 Other GNPAT mutations/deletions have been found in persons with peroxisomal disease, a class of disorders in which increased hepatic iron is observed.46
GNPAT knockdown did not have any clear influence on the BMP-SMAD pathway. The BMP-SMAD pathway appears to be operating normally in this situation, because treatment with exogenous BMP6 increases phosphorylation of SMAD1/5/8 and ID1 and SMAD7 mRNA expression. GNPAT silencing had no effect on these components of the pathway. HAMP mRNA expression was significantly lower in GNPAT silenced cells treated with BMP6, suggesting that GNPAT-mediated effects on HAMP may be independent of the BMP-SMAD pathway. HAMP is regulated by many factors, such as hypoxia, erythroid factors and inflammation that act through signaling pathways independent of BMP-SMAD.47–50 It is possible that GNPAT knockdown modifies components of any or a combination of these pathways, with consequent downstream effects on hepcidin and ultimately iron homeostasis, such as the regulation of intestinal iron absorption.
GNPAT p.D519G is common among people of European descent (allele frequency 20.6%).34 Our data indicate that co-inheritance of GNPAT p.D519G in some men with C282Y homozygosity is associated with high-iron phenotypes. The participants in this study represented a wide geographic distribution. GNPAT p.D519G was present in high expressers at study sites in Australia, Canada, and the United States, and 50–85% of the high expressers from the clinical practice sites and screening studies in these areas were heterozygous or homozygous for the GNPAT variant. It is important to confirm the association of GNPAT p.D519G with high-iron phenotypes in male HFE C282Y homozygotes from other geographic areas and in female C282Y homozygotes. Based on the effect of GNPAT knockdown on hepcidin expression in a cell system, it is plausible that GNPAT p.D519G has sufficient functional effect, perhaps on membrane receptor localization or recycling, to alter the hemochromatosis phenotype by indirectly stimulating iron absorption. GNPAT p.D519G has been reported by Thai and colleagues,45 who found that introduction of this mutation into control DHAPAT (GNPAT) cDNA resulted in a 70% reduction of enzyme activity.
A limitation of the current study is a lack of available serum samples collected at study entry that would be suitable for measurement of hepcidin levels. Thus, we could not study steady-state, pre-treatment hepcidin levels in the present cohort.
Statistical power to discover variants of interest in cohorts of 13 and 22 subjects per group is relatively low. A post-hoc power calculation showed that the power to find GNPAT p.D519G is about 42% at a Bonferroni-corrected p-value of 0.05, assuming the distributions are as observed.
Although this is not a prospective study, our results demonstrate that GNPAT p.D519G is associated with greater iron overload and may be relevant to severity and prognosis. Identifying GNPAT p.D519G in young C282Y homozygotes could predict the potential severity of iron overload later in life and inform clinical recommendations regarding initiation of phlebotomy therapy and long-term follow-up of iron stores and related manifestations. More work will be needed to confirm a prognostic role for GNPAT p.D519G by testing specificity and sensitivity in a larger sample including subjects with intermediate iron phenotype, a large proportion of C282Y homozygotes. It is possible that GNPAT variants influence other iron-related conditions including iron deficiency, anemia of chronic disease, and secondary iron overload, although this is unproven. Six of the 22 C282Y homozygotes with a high-iron phenotype that we studied did not have GNPAT p.D519G, consistent with the possibility that other modifier alleles exist. The present evidence for GNPAT p.D519G as a significant phenotype modifier of HFE C282Y homozygosity indicates that common modifier alleles could be a major source of variable phenotype penetrance.
Supplementary Material
Acknowledgments
We sincerely thank the patients identified through clinical practice sites who agreed to contribute samples and data to iron-related investigations. We thank the HEIRS Study participants who volunteered for this study and all HEIRS Study investigators.10 This manuscript was prepared using HEIRS Research Materials obtained from the NHLBI Biologic Specimen and Data Repository Information Coordinating Center and does not necessarily reflect the opinions or views of the HEIRS Study investigators or the NHLBI. We thank Jeannette Dixon for assistance with identification of samples and data, and Daniel Wallace and Lesa Ostini for technical assistance. We thank Wen-Pin Chen of the Biostatistical Shared Resource, Chao Family Comprehensive Cancer Center, University of California, Irvine, for assistance with statistical analyses and graphics. Our work was supported in part by grant 1R24DK093433-01 from the National Institute of Diabetes and Digestive and Kidney Diseases, grant P30 CA-62203 from the National Cancer Institute, and funds from the Department of Veterans Affairs. This work was also supported in part by a Project Grant (APP1031325) from the National Health and Medical Research Council (NHMRC) of Australia to V.N.S.; V.N.S., G.A.R and G.J.A are supported by Senior Research Fellowships from the NHMRC of Australia.
Abbreviations
- FDR
false discovery rate
- GWAS
genome wide association studies
- HH
HFE-associated hemochromatosis
- LRT
likelihood ratio test
- LD
linkage disequilibrium
- SKAT
sequence kernel association test
- aSKAT-O
small sample adjusted sequence kernel association test
- SEM
standard error of the mean
Footnotes
Authorship
Contribution: C.E.M, M.J.E., V.N.S, P.D.P., J.C.B., P.C.A., L.W.P., L.C.G., K.J.A., D.A.N., G.A.R., G.J.A., and G.D.M. designed the research. P.D.P, P.C.A, J.C.B., L.W.P, V.N.S. and G.D.M. contributed samples and data. D.A.N. supervised exome sequencing. M.J.E. and T.L. analyzed data. J.B.G. and C.J.M. performed functional studies. C.E.M, M.J.E., V.N.S, P.D.P., J.C.B., P.C.A, J.B.G., C.J.M., L.C.G, K.J.A., G.A.R., and G.D.M. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Contributor Information
Christine E. McLaren, Email: cmclaren@uci.edu.
Mary J. Emond, Email: emond@u.washington.edu.
V. Nathan Subramaniam, Email: nathan.subramaniam@qimrberghofer.edu.au.
Pradyumna D. Phatak, Email: Prad.Phatak@rochesterregional.org.
James C. Barton, Email: ironmd@isp.com.
Paul C. Adams, Email: padams@uwo.ca.
Justin B. Goh, Email: justin.goh@qimrberghofer.edu.au.
Cameron J. McDonald, Email: Cameron.McDonald@qimrberghofer.edu.au.
Lawrie W. Powell, Email: l.powell@uq.edu.au.
Lyle C. Gurrin, Email: lgurrin@unimelb.edu.au.
Katrina J. Allen, Email: Katie.Allen@rch.org.au.
Deborah A. Nickerson, Email: debnick@u.washington.edu.
Tin Louie, Email: tinlouie@u.washington.edu.
Grant A. Ramm, Email: grant.ramm@qimrberghofer.edu.au.
Gregory J. Anderson, Email: greg.anderson@qimrberghofer.edu.au.
Gordon D. McLaren, Email: gordon.mclaren@va.gov.
References
- 1.Edwards CQ, Barton JC. Hemochromatosis. In: Greer JP, Arber DA, Glader B, List AF, Means RT, Paraskevas F, et al., editors. Wintrobe’s Clinical Hematology. 13. Philadelphia, PA: Lippincott Williams & Wilkins; 2013. pp. 663–381. [Google Scholar]
- 2.Tavill AS. Diagnosis and management of hemochromatosis. Hepatology. 2001;33(5):1321–8. doi: 10.1053/jhep.2001.24783. [DOI] [PubMed] [Google Scholar]
- 3.Adams PC, Barton JC. How I treat hemochromatosis. Blood. 2010;116(3):317–25. doi: 10.1182/blood-2010-01-261875. [DOI] [PubMed] [Google Scholar]
- 4.Bacon BR, Adams PC, Kowdley KV, Powell LW, Tavill AS. Diagnosis and management of hemochromatosis: 2011 practice guideline by the American Association for the Study of Liver Diseases. Hepatology. 2011;54(1):328–43. doi: 10.1002/hep.24330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niederau C, Fischer R, Sonnenberg A, Stremmel W, Trampisch HJ, Strohmeyer G. Survival and causes of death in cirrhotic and in noncirrhotic patients with primary hemochromatosis. N Engl J Med. 1985;313(20):1256–62. doi: 10.1056/NEJM198511143132004. [DOI] [PubMed] [Google Scholar]
- 6.Niederau C, Fischer R, Purschel A, Stremmel W, Haussinger D, Strohmeyer G. Long-term survival in patients with hereditary hemochromatosis. Gastroenterology. 1996;110(4):1107–19. doi: 10.1053/gast.1996.v110.pm8613000. [DOI] [PubMed] [Google Scholar]
- 7.Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, et al. A Novel MHC Class I-Like Gene Is Mutated in Patients with Hereditary Haemochromatosis. Nat Genet. 1996;13(4):399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 8.Ganz T. Systemic iron homeostasis. Physiol Rev. 2013;93(4):1721–41. doi: 10.1152/physrev.00008.2013. [DOI] [PubMed] [Google Scholar]
- 9.Bridle KR, Frazer DM, Wilkins SJ, Dixon JL, Purdie DM, Crawford DH, et al. Disrupted hepcidin regulation in HFE-associated haemochromatosis and the liver as a regulator of body iron homoeostasis. Lancet. 2003;361(9358):669–73. doi: 10.1016/S0140-6736(03)12602-5. [DOI] [PubMed] [Google Scholar]
- 10.Adams PC, Reboussin DM, Barton JC, McLaren CE, Eckfeldt JH, McLaren GD, et al. Hemochromatosis and iron-overload screening in a racially diverse population. N Engl J Med. 2005;352(17):1769–78. doi: 10.1056/NEJMoa041534. [DOI] [PubMed] [Google Scholar]
- 11.Adams PC, Gregor JC, Kertesz AE, Valberg LS. Screening blood donors for hereditary hemochromatosis: decision analysis model based on a 30-year database. Gastroenterology. 1995;109(1):177–88. doi: 10.1016/0016-5085(95)90283-x. [DOI] [PubMed] [Google Scholar]
- 12.McLaren CE, Gordeuk VR, Looker AC, Hasselblad V, Edwards CQ, Griffen LM, et al. Prevalence of heterozygotes for hemochromatosis in the white population of the United States. Blood. 1995;86(5):2021–7. [PubMed] [Google Scholar]
- 13.Witte DL, Crosby WH, Edwards CQ, Fairbanks VF, Mitros FA. Practice guideline development task force of the College of American Pathologists. Hereditary hemochromatosis. Clin Chim Acta. 1996;245(2):139–200. doi: 10.1016/0009-8981(95)06212-2. [DOI] [PubMed] [Google Scholar]
- 14.Pietrangelo A. Hereditary hemochromatosis--a new look at an old disease. N Engl J Med. 2004;350(23):2383–97. doi: 10.1056/NEJMra031573. [DOI] [PubMed] [Google Scholar]
- 15.Allen KJ, Gurrin LC, Constantine CC, Osborne NJ, Delatycki MB, Nicoll AJ, et al. Iron-overload-related disease in HFE hereditary hemochromatosis. N Engl J Med. 2008;358(3):221–30. doi: 10.1056/NEJMoa073286. [DOI] [PubMed] [Google Scholar]
- 16.Waalen J, Nordestgaard BG, Beutler E. The penetrance of hereditary hemochromatosis. Best Pract Res Clin Haematol. 2005;18(2):203–20. doi: 10.1016/j.beha.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 17.Le Gac G, Ferec C. The molecular genetics of haemochromatosis. Eur J Hum Genet. 2005;13(11):1172–85. doi: 10.1038/sj.ejhg.5201490. [DOI] [PubMed] [Google Scholar]
- 18.Pietrangelo A, Caleffi A, Henrion J, Ferrara F, Corradini E, Kulaksiz H, et al. Juvenile hemochromatosis associated with pathogenic mutations of adult hemochromatosis genes. Gastroenterology. 2005;128(2):470–9. doi: 10.1053/j.gastro.2004.11.057. [DOI] [PubMed] [Google Scholar]
- 19.Constantine CC, Anderson GJ, Vulpe CD, McLaren CE, Bahlo M, Yeap HL, et al. A novel association between a SNP in CYBRD1 and serum ferritin levels in a cohort study of HFE hereditary haemochromatosis. Br J Haematol. 2009;147(1):140–9. doi: 10.1111/j.1365-2141.2009.07843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barton JC, Lafreniere SA, Leiendecker-Foster C, Li H, Acton RT, Press RD, et al. HFE, SLC40A1, HAMP, HJV, TFR2, and FTL mutations detected by denaturing high-performance liquid chromatography after iron phenotyping and HFE C282Y and H63D genotyping in 785 HEIRS Study participants. Am J Hematol. 2009;84(11):710–4. doi: 10.1002/ajh.21524. [DOI] [PubMed] [Google Scholar]
- 21.Island ML, Jouanolle AM, Mosser A, Deugnier Y, David V, Brissot P, et al. A new mutation in the hepcidin promoter impairs its BMP response and contributes to a severe phenotype in HFE related hemochromatosis. Haematologica. 2009;94(5):720–4. doi: 10.3324/haematol.2008.001784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nai A, Pagani A, Silvestri L, Campostrini N, Corbella M, Girelli D, et al. TMPRSS6 rs855791 modulates hepcidin transcription in vitro and serum hepcidin levels in normal individuals. Blood. 2011;118(16):4459–62. doi: 10.1182/blood-2011-06-364034. [DOI] [PubMed] [Google Scholar]
- 23.Valenti L, Fracanzani AL, Rametta R, Fraquelli M, Soverini G, Pelusi S, et al. Effect of the A736V TMPRSS6 polymorphism on the penetrance and clinical expression of hereditary hemochromatosis. J Hepatol. 2012;57(6):1319–25. doi: 10.1016/j.jhep.2012.07.041. [DOI] [PubMed] [Google Scholar]
- 24.Pelucchi S, Mariani R, Calza S, Fracanzani AL, Modignani GL, Bertola F, et al. CYBRD1 as a modifier gene that modulates iron phenotype in HFE p.C282Y homozygous patients. Haematologica. 2012;97(12):1818–25. doi: 10.3324/haematol.2012.062661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Tayrac M, Roth MP, Jouanolle AM, Coppin H, le Gac G, Piperno A, et al. Genome-wide association study identifies TF as a significant modifier gene of iron metabolism in HFE hemochromatosis. J Hepatol. doi: 10.1016/j.jhep.2014.10.017. Prepublished on December 3, 2014, as doi: http://dx.doi.org/10.1016/j.jhep.2014.10.017. [DOI] [PubMed]
- 26.Powell LW, Dixon JL, Ramm GA, Purdie DM, Lincoln DJ, Anderson GJ, et al. Screening for hemochromatosis in asymptomatic subjects with or without a family history. Arch Intern Med. 2006;166(3):294–301. doi: 10.1001/archinte.166.3.294. [DOI] [PubMed] [Google Scholar]
- 27.Olynyk JK, Knuiman MW, Divitini ML, Bartholomew HC, Cullen DJ, Powell LW. Effects of HFE gene mutations and alcohol on iron status, liver biochemistry and morbidity. J Gastroenterol Hepatol. 2005;20(9):1435–41. doi: 10.1111/j.1440-1746.2005.03967.x. [DOI] [PubMed] [Google Scholar]
- 28.Yu L, Martinez FD, Klimecki WT. Automated high-throughput sex-typing assay. Biotechniques. 2004;37(4):662–4. doi: 10.2144/04374DD01. [DOI] [PubMed] [Google Scholar]
- 29.Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, et al. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet. 2012;91(2):224–37. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris AP, Zeggini E. An evaluation of statistical approaches to rare variant analysis in genetic association studies. Genet Epidemiol. 2010;34(2):188–93. doi: 10.1002/gepi.20450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDonald CJ, Wallace DF, Ostini L, Subramaniam VN. Parenteral vs. oral iron: influence on hepcidin signaling pathways through analysis of Hfe/Tfr2-null mice. Am J Physiol Gastrointest Liver Physiol. 2014;306(2):G132–9. doi: 10.1152/ajpgi.00256.2013. [DOI] [PubMed] [Google Scholar]
- 32.Gordeuk VR, Reboussin DM, McLaren CE, Barton JC, Acton RT, McLaren GD, et al. Serum ferritin concentrations and body iron stores in a multicenter, multiethnic primary-care population. Am J Hematol. 2008;83(8):618–26. doi: 10.1002/ajh.21179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Storey JD. A direct approach to false discovery rates. Journal of the Royal Statistical Society Series B-Statistical Methodology. 2002;64:479–98. [Google Scholar]
- 34.Exome Variant Server. NHLBI GO Exome Sequencing Project (ESP) Seattle WA: Jan, 2013. (URL: http://evs.gs.washington.edu/EVS/) [Google Scholar]
- 35.Mattman A, Huntsman D, Lockitch G, Langlois S, Buskard N, Ralston D, et al. Transferrin receptor 2 (TfR2) and HFE mutational analysis in non-C282Y iron overload: identification of a novel TfR2 mutation. Blood. 2002;100(3):1075–7. doi: 10.1182/blood-2002-01-0133. [DOI] [PubMed] [Google Scholar]
- 36.Biasiotto G, Belloli S, Ruggeri G, Zanella I, Gerardi G, Corrado M, et al. Identification of new mutations of the HFE, hepcidin, and transferrin receptor 2 genes by denaturing HPLC analysis of individuals with biochemical indications of iron overload. Clin Chem. 2003;49(12):1981–8. doi: 10.1373/clinchem.2003.023440. [DOI] [PubMed] [Google Scholar]
- 37.Beutler E, Van Geet C, te Loo DM, Gelbart T, Crain K, Truksa J, et al. Polymorphisms and mutations of human TMPRSS6 in iron deficiency anemia. Blood Cells Mol Dis. 2010;44(1):16–21. doi: 10.1016/j.bcmd.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sato T, Iyama S, Murase K, Kamihara Y, Ono K, Kikuchi S, et al. Novel missense mutation in the TMPRSS6 gene in a Japanese female with iron-refractory iron deficiency anemia. Int J Hematol. 2011;94(1):101–3. doi: 10.1007/s12185-011-0881-0. [DOI] [PubMed] [Google Scholar]
- 39.Del-Castillo-Rueda A, Moreno-Carralero MI, Cuadrado-Grande N, Alvarez-Sala-Walther LA, Enriquez-de-Salamanca R, Mendez M, et al. Mutations in the HFE, TFR2, and SLC40A1 genes in patients with hemochromatosis. Gene. 2012;508(1):15–20. doi: 10.1016/j.gene.2012.07.069. [DOI] [PubMed] [Google Scholar]
- 40.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–5. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kautz L, Meynard D, Monnier A, Darnaud V, Bouvet R, Wang RH, et al. Iron regulates phosphorylation of Smad1/5/8 and gene expression of Bmp6, Smad7, Id1, and Atoh8 in the mouse liver. Blood. 2008;112(4):1503–9. doi: 10.1182/blood-2008-03-143354. [DOI] [PubMed] [Google Scholar]
- 42.Meynard D, Kautz L, Darnaud V, Canonne-Hergaux F, Coppin H, Roth MP. Lack of the bone morphogenetic protein BMP6 induces massive iron overload. Nat Genet. 2009;41(4):478–81. doi: 10.1038/ng.320. [DOI] [PubMed] [Google Scholar]
- 43.McLaren GD, Nathanson MH, Jacobs A, Trevett D, Thomson W. Regulation of intestinal iron absorption and mucosal iron kinetics in hereditary hemochromatosis. J Lab Clin Med. 1991;117(5):390–401. [PubMed] [Google Scholar]
- 44.Roy CN. An update on iron homeostasis: make new friends, but keep the old. Am J Med Sci. 2013;346(5):413–9. doi: 10.1097/MAJ.0000000000000190. [DOI] [PubMed] [Google Scholar]
- 45.Thai TP, Rodemer C, Jauch A, Hunziker A, Moser A, Gorgas K, et al. Impaired membrane traffic in defective ether lipid biosynthesis. Hum Mol Genet. 2001;10(2):127–36. doi: 10.1093/hmg/10.2.127. [DOI] [PubMed] [Google Scholar]
- 46.Wanders RJ, Ferdinandusse S, Brites P, Kemp S. Peroxisomes, lipid metabolism and lipotoxicity. Biochim Biophys Acta. 2010;1801(3):272–80. doi: 10.1016/j.bbalip.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 47.Pietrangelo A, Dierssen U, Valli L, Garuti C, Rump A, Corradini E, et al. STAT3 is required for IL-6-gp130-dependent activation of hepcidin in vivo. Gastroenterology. 2007;132(1):294–300. doi: 10.1053/j.gastro.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 48.Sonnweber T, Nachbaur D, Schroll A, Nairz M, Seifert M, Demetz E, et al. Hypoxia induced downregulation of hepcidin is mediated by platelet derived growth factor BB. Gut. 2014;63(12):1951–9. doi: 10.1136/gutjnl-2013-305317. [DOI] [PubMed] [Google Scholar]
- 49.Subramaniam VN, Wallace DF. Linking hypoxia and iron homeostasis: a ‘plate’ full of factors. Gut. 2014;63(12):1840–2. doi: 10.1136/gutjnl-2014-306970. [DOI] [PubMed] [Google Scholar]
- 50.Kautz L, Jung G, Valore EV, Rivella S, Nemeth E, Ganz T. Identification of erythroferrone as an erythroid regulator of iron metabolism. Nat Genet. 2014;46(7):678–84. doi: 10.1038/ng.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.