Abstract
Numerous animals have invaded subterranean caverns and evolved remarkably similar features. These features include loss of vision and pigmentation, and gains in non-visual sensation. This broad convergence echoes smaller-scale convergence, in which members of the same species repeatedly evolve the same cave-associated phenotypes. The blind Mexican tetra of the Sierra de El Abra region of northeastern Mexico has a complex origin, having recurrently colonized subterranean environments through numerous invasions of surface-dwelling fish. These colonizations likely occurred ~1–5 MYa. Despite evidence of historical and contemporary gene flow between cave and surface forms, the cave-associated phenotype appears to remain quite stable in nature. This model system has provided insight to the mechanisms of phenotypic regression, the genetic basis for constructive trait evolution, and the origin of behavioral novelties. Here, we document the rise of this model system from its discovery by a Mexican surveyor in 1936, to a powerful system for cave biology and contemporary genetic research. The recently sequenced genome provides exciting opportunities for future research, and will help resolve several long-standing biological problems.
Keywords: Regressive phenotypic evolution, troglomorphy, cave biology
Introduction
Blind Mexican cavefish are remarkably well adapted to life underground. For unknown reasons, numerous ancestral surface-dwelling stocks invaded a geographically delimited karst region in northeastern Mexico numerous times over the past several million years (Ornelas-García et al., 2008; Bradic et al., 2012). Population genetic studies revealed that different epigean (surface-dwelling) stocks likely colonized each of three discrete areas: the older El Abra region, and the younger Guatemala and Micos regions (Bradic et al., 2012; Bradic et al., 2013; Coghill et al., 2014). Irrespective of geographic and phylogenetic distance, independent cavefish populations seem to move in the same phenotypic direction towards eye and pigmentation loss (Jeffery, 2006; 2009) and extra-visual sensory expansion (Bensouilah & Denizot, 1991; Yoshizawa et al., 2013). Several decades ago, breeding experiments revealed that cave and surface forms were capable of producing viable hybrid offspring (Şadoğlu, 1957a; Şadoğlu, 1957b), demonstrating the utility of these animals for laboratory investigations. This early discovery led to classical genetic studies revealing the participation of both Mendelian (albinism and brown; Şadoğlu & McKee, 1969) and complex cave-associated phenotypes (Wilkens, 1988) evolving in cave lineages. Complementation studies showed that cave populations from the same geographic region (e.g., the El Abra caves) evolved similar regressive phenotypes (such as eye loss) through different genetic loci (Wilkens, 1971; Borowsky, 2008). Recently, quantitative trait locus (QTL) analyses further clarified the architecture of simple (Protas et al., 2006; Gross et al., 2009) and complex traits (Borowsky & Wilkens, 2002; Kowalko et al., 2013a; Kowalko et al., 2013b; O'Quin et al., 2013) participating in cave evolution. Most recently, transcriptome profiling has revealed that diverse patterns of gene expression underlie many cave-associated phenotypes (Gross et al., 2013; Hinaux et al., 2013).
Several natural systems have been implemented as models for understanding the genetic basis for evolutionary change. Astyanax cavefish, however, are unique in several respects. First, unlike many cave-dwelling animals, related surface-dwelling morphs survive in the rivers and streams surrounding the cave network. Extant surface- and cave-dwelling forms evolved from common eyed, surface-fish like ancestors that invaded this region over a million years ago. The presence of both morphotypes has enabled a powerful comparative approach which has led to a number of molecular (Avise and Selander, 1972), genetic (Behrens et al., 1998), behavioral (Duboué et al., 2011), developmental (Alunni et al., 2007) and physiological (Breder & Rasquin, 1947) studies. The dark, nutrient-poor environment of the cave has led to extreme phenotypic changes, many of which are mediated by fixed genetic differences between cave and surface forms (Borowsky & Wilkens, 2002). Second, unlike many natural systems, cavefish likely diverged from their ancestral forms over the course several millions (rather than thousands) of years (Ornelas-García et al., 2008; Bradic et al., 2012; Coghill et al., 2014). This period of separation, however, depends on which cave populations are being studied (Peters et al., 1975; Dowling et al., 2002). This is because multiple surface-dwelling populations colonized multiple cave environments over the past several millions of years. This complicated demographic history therefore enables us to determine if the same trait evolves through the same genetic pathways in “older” versus “younger” cave populations (Strecker et al., 2012; Kowalko et al., 2013a). Finally, both cave and surface forms are highly tractable laboratory systems (Hinaux et al., 2011), enabling high-resolution studies at multiple levels of analysis.
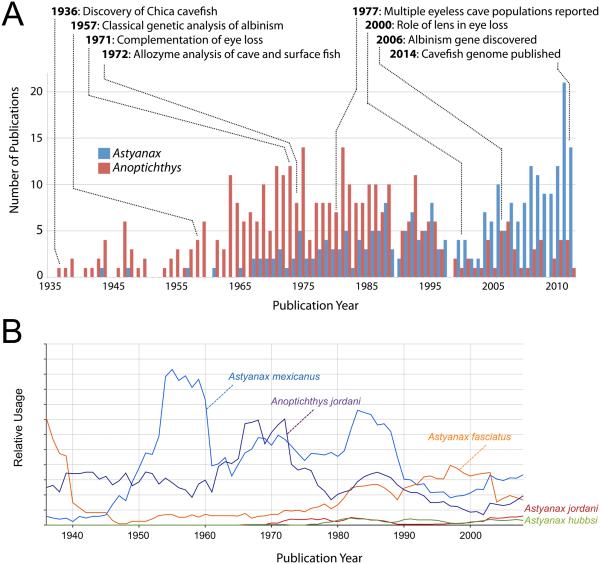
This intriguing evolutionary history mirrors an equally fascinating history as an experimental model system. Here, we chronicle use of the cavefish system from studies of taxonomic status and genus designation, to cutting-edge experimental observations and manipulation. A comprehensive survey of the scientific literature since their discovery demonstrates a clear growth of interest in this animal system – from humble origins in the commercial pet trade to a powerful genomic model for understanding evolutionary and phenotypic change (Fig.1A). The newly available physical genome opens further opportunities to provide unprecedented clarity to long-standing problems in the field of cave biology and beyond.
Figure 1.
Cave animals as models for biological research
Cave-dwelling organisms have long held the fascination of scientists and the general public. This is due to the dramatic and seemingly maladaptive traits (lack of eye and pigmentation) demonstrated by these intriguing creatures (Protas et al., 2011). Extreme characteristics of cave animals have been appreciated for over a century (Eigenmann, 1909), and Charles Darwin famously commented on the “problem” of trait loss for his theory of natural selection, attributing phenotypic regression in cave organisms to “disuse” rather than through obvious selective forces (Darwin, 1859). Three principal theories seek to explain loss of phenotypes in cave animals: a) the accumulation of neutral mutations affecting traits that lose relevance in the darkness of a cave (Wilkens, 1988); b) negative selection driven by energy conservation in nutrient-poor subterranean environments (Protas et al., 2007); or, c) indirect selection through antagonistic pleiotropic interactions between constructive traits and expendable (regressive) traits evolving in the cave (Wright, 1964). Although much progress has been made in last several decades through diverse experimental approaches, a consensus view as to how and why traits regress in cave animals has not been reached (Gross, 2012). Indeed, a combination of different mechanisms may ultimately participate together in the evolution of regressive phenotypes.
A number of useful cave animal systems have emerged over the past century, including invertebrate systems such as Gammarus minus (Jones & Culver, 1989; Culver et al., 1995) and Asellus aquaticus (Kosswig & Kosswig, 1940; Protas et al., 2011). Gammarus minus is a freshwater, cave-adapted amphipod found in the eastern United States. Classical studies of multiple cave-adapted forms of Gammarus provided insight into natural selection and adaptation in cave-dwelling animals (Culver et al., 1995). More recently, this system has been adapted for genetic analyses. For instance, a recent report indicated no loss of functional constraint in opsin gene paralogs in cave-dwelling forms, but rather convergently reduced levels of gene expression. This loss of expression, without loss of gene function, may suggest previously pleiotropic roles for opsins (Carlini et al., 2013). Asellus aquaticus, a cave-adapted crustacean found in Slovenia, has emerged in recent years as a powerful invertebrate genetic model system. Using this system, Protas et al. (2011) demonstrated that, although significant morphological changes can be accomplished through one or a few genes, eye size variation within cave animals is likely mediated by multiple mechanisms even within the same population.
We propose that cave-dwelling organisms should be prioritized for further research. In addition to the question of how regressive changes occur, cave animals can also inform the genetic basis for constructive morphologies (Yoshizawa et al., 2010; Kowalko et al., 2013a; Yoshizawa et al., 2013). Cave-dwelling invertebrates evolve enhanced non-visual sensory systems such as longer antennae and limbs (Fong, 1989). Similarly, cave vertebrates evolve non-visual sensory expansions to their gustatory (Bensouilah & Denizot, 1991) and lateral line systems (Montgomery et al., 2001). Integrative studies of the genetic and developmental bases for regressive and constructive changes have the potential to inform how seemingly distinct phenotypes may be linked through shared genetic pathways. Further, closely related cave-dwelling animals frequently occupy the same geographic regions. Although similar phenotypes evolve across a localized landscape of cave-dwellers (Ornelas-García et al., 2008), they appear to do so through different genetic mechanisms (Borowsky, 2008; Gross et al., 2009). Thus, cave animals have potential to determine if certain genes or genetic pathways are favored in the evolution of particular traits evolving under similar cave conditions.
Cave systems can also be powerful models for human phenotypes of clinical relevance. For instance, the precise genetic basis for many visual system disorders in humans remains unknown (O'Quin et al., 2013). Evaluating analogous phenotypes in cave animals provides an opportunity for gene discovery in the context of a natural system subjected to intense environmental pressures (Albertson et al., 2009). Moreover, the tractability of this system permits unique functional genetic analyses wherein a derived (cave-associated) genetic variant can be directly tested in a surrogate “ancestral” (surface fish) organism (Elipot et al., 2014). Although many cave model systems have emerged over the years, one cave system is now poised to provide dramatic insights to many of these questions: the Astyanax cavefish of northeastern Mexico.
The rise of Astyanax cavefish in the scientific literature: 1936 - 2014
The first Astyanax cavefish were discovered at the Chica cave locality in the southern karst region of the Sierra de El Abra in 1936 (Hubbs & Innes, 1936). These fish were discovered and collected by Salvador Coronado, a Mexican surveyor. Coronado collected the specimens for a fish hobbyist, C. Basil Jordan, of the Texas Aquaria Fish Company in Dallas, Texas (Mitchell et al., 1977). This freshwater fish was assigned to the new genus Anoptichthys (lat. “bony fish with no eyes”) by renowned University of Michigan ichthyologist, Dr. Carl Hubbs. About a decade after the discovery of Chica cavefish, two subsequent expeditions to the El Abra region identified cavefish from the Sabinos and Pachón cave localities (Alvarez, 1946; Alvarez, 1947). Early studies sought to determine if the three distinct cave populations were related, originating from a single colonization of the subterranean environment by surface dwelling forms (reviewed in Mitchell et al., 1977). Based in part on morphological evidence (dermal bone fragmentation affecting the facial skeleton) the first three cave populations were each assigned as a distinct species of Anoptichthys (Alvarez, 1946; Alvarez, 1947).
The emergent field of Mexican cavefish biology experienced a conceptual shift in nomenclature in the 1950s when it was discovered that eyed surface-dwelling fish, common in the rivers and streams surrounding the limestone cave complex, could hybridize with cave-dwelling forms. Hybrid offspring from these crosses were viable, and could be interbred to produce experimental pedigrees (Şadoğlu, 1957a; Şadoğlu, 1957b). The ability to produce these hybrid pedigrees launched two key directions in Mexican cavefish research. First, the use of “Anoptichthys” to describe these cavefish gradually transitioned to “Astyanax” as cave and surface dwelling forms became regarded as sub-morphotypes of a single species (Schemmel, 1967). This shift was likely based on the biological species concept, as well as shared molecular similarity of cave and surface morphs informed by allozyme studies (Avise & Selander, 1972). However, not all Astyanax researchers agree on a singular taxonomic nomenclature. For example, the names Astyanax mexicanus (Protas et al., 2007), Astyanax fasciatus (Van Trump & McHenry, 2013), Astyanax hubbsi (Ornelas-García et al., 2008), and Astyanax jordani (Boudriot & Reutter, 2001) have all appeared in the recent literature referring to cave-adapted forms (Fig.1B).
In a humorous exchange, Avise (2001) recounted an interaction with Carl Hubbs, who coined the term “Anoptichthys” in 1936. Avise shared his assertion with Hubbs that cavefish and surface forms of Astyanax should be regarded as members of the same species owing to their molecular similarity (Avise & Selander, 1971). Hubbs replied that anyone who thinks blind Mexican cavefish and surface-dwelling Astyanax are members of the same genus, let alone the same species, “must be as blind as the fish themselves” (Avise, 2001; p.51). Nonetheless, the ability to hybridize different cave populations with surface-dwelling populations (as well as with other geographically-distinct cave populations) launched Astyanax as a classical genetic model system (Şadoğlu & McKee, 1969).
From the 1940s through the 1970s, 27 additional cave populations were discovered and named (Wilkens & Burns, 1972; Mitchell et al., 1977), including an additional Astyanax cavefish population in the southern Mexican state of Guerrero in 2001 (Espinasa et al., 2001). These population discoveries began to illustrate the common convergence on cave-associated characters in this system, as well as its utility for understanding the convergent evolution of extreme phenotypes.
Astyanax as a powerful model system for understanding mechanisms of biological diversity
Following the first formal scientific description of Mexican cavefish in 1936, subsequent reports from this decade focused on taxonomic assignment and descriptions of the natural history of these remarkable organisms (Innes, 1937; Breder, 1942). The following decade saw the first uses for cavefish as an experimental system. For instance, in addition to descriptive studies, the first histological studies of the degenerating eye in cave forms were performed (Gresser & Breder, 1940). Additionally, several behavioral phenotypes were first examined, including studies characterizing differential schooling and shoaling behaviors (Breder, 1943a; Morrow, 1948), alongside increased aggression studies (Breder, 1943b) in eyeless cave forms.
The 1950s witnessed the first formal classical genetic studies in which surface and cave forms were hybridized to produce experimental F2 pedigrees (Şadoğlu, 1956; Şadoğlu, 1957b). The first evaluated phenotype was albinism, which demonstrated a nearly perfect (1:3) Mendelian ratio (Şadoğlu, 1957a). This segregation pattern was later shown to occur roughly equally in both males and females (i.e., not sex-linked), for both albinism and another reduced pigmentation phenotype called brown (melanin reduction; Şadoğlu & McKee, 1969). The 1960s was marked by expanded research directions focusing on endocrine differences between cave and surface forms (Mattheij, 1968), as well as studies characterizing enhanced chemoreceptive and lateral line sensitivity (Humbach, 1960) in cave forms.
During the 1970s, several authors returned to the question of which evolutionary mechanism(s) explain the loss of phenotypic characters in cavefish (Wilkens, 1971; Peters & Peters, 1973). In addition, attention turned to the central nervous system (Voneida & Sligar, 1976), and early husbandry and breeding studies (Şadoğlu, 1979), which set the stage for future studies utilizing this model as a system for understanding evolution and development. The 1980s represented a period marked by growth in the number of investigations using Astyanax cavefish. Schemmel (1980) performed one of the first classical genetic analyses that advanced from a morphological trait to a behavior (feeding) in cavefish (Schemmel, 1980). Additional discoveries during this decade included the discovery of increased lipid storage (Rose & Mitchell, 1982), decreased oxygen consumption (Hüppop, 1986), putative alterations to the circadian clock (Erckens & Martin, 1982a; Erckens & Martin, 1982b), and acutely sensitive olfactory systems (Quinn, 1980) in cave-dwelling forms. Several studies advocated the importance of neutral mutation as a principal mechanism driving regressive phenotypic loss in cavefish (Wilkens, 1988). This set the stage for ongoing debates seeking to identify the mechanism(s) for trait loss as occurring through neutral forces, negative selection, indirect selection (pleiotropy) or a combination thereof (reviewed in Gross, 2012).
During the 1990s, a number of integrative studies focused on the intersection of behavior (Langecker et al., 1993), constructive phenotypic evolution (e.g., neuromast and taste bud structure; Bensouilah & Denizot, 1991) and cave adaptation (Langecker et al., 1993). Reports also began to focus on the mechanistic basis for increased sensitivity to the environment (Langecker et al., 1993). This decade also witnessed a dramatic growth in the application of molecular and genetic tools for studying cave evolution. These included genetic sequence analyses of opsins (Yokoyama & Yokoyama, 1990a; Yokoyama & Yokoyama, 1990b; Langecker et al., 1993), crystallins (Langecker et al., 1995; Behrens et al., 1998), and a key regulator of eye development, pax6 (Behrens et al., 1997; Jeffery & Martasian, 1998).
Further genetic studies continued into the 2000s, including analysis of Prox1. This gene is a critical regulator of lens development, but is also expressed strongly in taste buds and neuromasts – two key constructive tissue types that are greatly expanded in cavefish (Jeffery et al., 2000). Genetic information was also used to clarify the relationships and origins of cave-dwelling forms (Espinasa & Borowsky, 2001; Strecker et al., 2003). Several population genetic studies were performed during this decade, using both mitochondrial (Dowling et al., 2002; Strecker et al., 2004) and nuclear markers (Ornelas-García et al., 2008), to understand the highly complex evolutionary origin of these remarkable creatures.
A landmark developmental study in 2000 demonstrated the inductive role of the lens in generating the adult structural eye in cave/surface heterospecific grafting experiments (Yamamoto & Jeffery, 2000). In 2006, another important study by Protas et al. discovered that the gene Oca2 governs albinism in multiple independent cave populations. This study represented the first identification of a causative gene underlying a cave-associated QTL (Protas et al., 2006). This decade also marked a significant expansion in the study of Astyanax cavefish, with several labs around the world adopting this model system for studies of evolution (Jeffery, 2001; Wilkens & Strecker, 2003), development (Yamamoto et al., 2003; Menuet et al., 2007), genetics (Strickler et al., 2002; Hooven et al., 2004) and behavior (Yoshizawa & Jeffery, 2008; Sharma et al., 2009). Additionally, the first formal international Astyanax community meeting was held in Ciudad Valles, Mexico in the spring of 2009.
The field of Astyanax biology continues to grow. Over the past four years, a developmental staging table has been produced (Hinaux et al., 2011), two transcriptomic studies have been published (Hinaux et al., 2013; Gross et al., 2013), several QTL analyses of complex behavioral phenotypes have been performed (Yoshizawa et al., 2010; Kowalko et al., 2013a; O'Quin et al., 2013; Yoshizawa et al., 2013), and a first-draft physical genome has become available (McGaugh et al., 2014). In addition to new insights on classic problems (e.g., the evolutionary and developmental mechanism of vision loss; Ma et al., 2014), recent studies have forged new ground. For instance, Astyanax cavefish have provided a powerful natural example of the potential role for heat shock protein chaperones as a buffer for phenotypic variation in the natural world (Rohner et al., 2013). Further, this model system has provided new insights to the evolution of sleep (Duboué et al., 2011; Duboué et al., 2012), the potential selective value of albinism (Bilandžija et al., 2013), and the evolutionary underpinnings of aggression in cavefish (Elipot et al., 2013). Many advancements have been made since the discovery of Astyanax cavefish (Wiley & Mitchell, 1971), and many additional discoveries are waiting to be unveiled in light of these powerful new genomic resources.
Future directions for Astyanax biology
The expansion of research directions, in concert with the availability of a physical genome, suggests that we are poised for many great discoveries in the field of Astyanax biology. For instance, genomic tools provide a powerful template upon which genome scans for selection (Fumagalli et al., 2011) and genome-wide association studies (GWAS; Timmann et al., 2012) can now be implemented as tools for understanding the evolution of these unique organisms. QTL studies enabled us to explore the genetic architecture for trait evolution (Gross et al., 2014), however this approach is limited by natural recombination occurring within an experimental pedigree. GWAS will enable a more flexible approach (Clarke et al., 2013), with increased precision (Button et al., 2013), for detecting genetic associations. GWAS can also be expanded to multiple different cave populations to examine the genetic basis for traits evolving in geographically distinct populations (Dowling et al., 2002; Gross & Wilkens, 2013).
Contemporary genome editing tools, such as Tol2- and I-SceI-mediated transgenesis (Elipot et al., 2014) and TALENs (Ma et al., 2015), will accelerate functional validation of promising candidate genes. These tools will allow us to determine if certain genetic lesions recapitulate cave-associated phenotypes in surface-dwelling fish. Additionally, transcriptomic profiling will provide high-resolution information on changes in gene expression occurring as consequences of subterranean colonization. For example, are the same genes (as with albinism; Protas et al., 2006; Gross & Wilkens, 2013) targeted for expression level changes in distantly related cave populations evolving the same trait? Alternatively, are different genes (as is implied by complementation crosses for vision; Wilkens, 1971; Borowsky, 2008) recruited to mediate the evolution of cave-associated phenotypes? In addition to these discovery-based questions, a number of additional long-standing questions now have hope for increased clarity.
Have karyotypic changes accompanied recurrent adaptation to the cave environment?
Numerous small- and large-scale chromosomal alterations have likely accompanied the transition from a lighted environment to the darkness of a cave. Astyanax cavefish have undergone a series of natural replicate “experiments” of cave colonization (Mitchell et al., 1977). We can now understand these processes, using the surrogate “ancestral” surface fish genome for comparison, to understand chromosomal dynamics at the population level. For example, do closely related cave populations demonstrate higher similarity to one another compared to distantly related populations? Are certain regions of the genome consistent “targets” of cave evolution? How flexible is the Astyanax karyotype? Do “labile” regions of the Astyanax karyotype co-localize with genomic regions identified from earlier linkage mapping studies? Prior studies have characterized extensive synteny between the Astyanax and Danio rerio genomes (Gross et al., 2008; Gross et al., 2013; O'Quin et al., 2013; Carlson et al., 2015). Future approaches aimed at linking genomes from multiple cave and surface genomes will clarify how the karyotype has evolved in distinct populations, and identify the consequences of these changes.
How have the same phenotypes convergently evolved in numerous distinct cave populations?
One of the most intriguing discoveries in Astyanax biology over the past several decades is the finding that geographically and phylogenetically distinct populations have evolved eye loss through different genes. In 1971, Wilkens discovered that fish bred from two different eyeless populations had offspring with a larger structural eye than either of the parental populations (Wilkens, 1971). This complementation study indicates that distinct genetic mechanisms mediate trait loss, even when two cave populations arose from the same ancestral epigean stock. At present, however, the identity of these genes remains unknown. Genomic analyses provide the opportunity to identify candidate sequence variants impacting eye loss within lineages (McGaugh et al., 2014). Subsequent comparisons between populations will identify eye loss loci shared between distantly related cavefish, in addition to a subset of loci that may also be shared.
Transcriptional profiling carried out in different cave populations will identify the genetic pathways impacted by the evolution of trait gains and losses. For instance, even if distinct eye loss genes are altered in independent cave populations, do these genes converge on the same downstream genetic networks? To answer this question, it will be crucial to determine if certain gene expression patterns are universally associated with cave adaptation – irrespective of the time since colonization. Or, alternatively are entirely different genes recruited recurrently in geographically distinct populations? Evaluating the mRNA architecture of whole embryos, tissues or cells (using a comprehensive genomic template) promises extraordinary insight to this intriguing and unanswered question.
These experimental approaches, evaluated in the context of advanced population genetic studies (Ornelas-García et al., 2008; Hausdorf et al., 2011; Bradic et al., 2012; Strecker et al., 2012; Bradic et al., 2013; Coghill et al., 2014), will help inform the long-standing question of what mechanism(s) mediate the extinction of phenotypes from certain lineages.
Are regressive phenotypes evolving neutrally, through selection, or pleiotropy?
The question of why traits are lost within lineages is long-standing (Eigenmann, 1909). Darwin commented on the problem of trait loss for the theory of natural selection – a phenomenon he illustrated using cave animals (Darwin, 1859; Ha & Nehm, 2014). Over 150 years later, we still do not have a comprehensive understanding of how and why trait loss occurs (Gross, 2012). Some evidence suggests that traits, such as vision, can be lost through selection for constructive phenotypes. For instance, Yamamoto et al. (2004) first reported a role for shh signaling in cavefish eye degeneration. Specifically, an expanded shh signaling domain was found to negatively impact expression of critical vision genes. It was later shown that this expanded shh signaling domain is also developmentally linked to increased numbers of taste buds, which is likely under selection (Yamamoto et al., 2009). Since vision is expendable in the darkness of the cave, these two phenotypes are united through pleiotropic consequences of shh expression, a form of indirect selection (Jeffery, 2010).
By performing genomic scans for selection in the genomic intervals surrounding shh and other candidate genes (McGaugh et al., 2014), we can determine whether recent selection within populations is the causative mechanism leading to convergent trait loss. By combining these genomic approaches with sophisticated methods of genome editing using cavefish embryos, we will better understand if and how alternative phenotypic effects arise from targeted genetic changes. Finally, evaluating phenotypic consequences of gene editing experiments in vivo will help ascertain if certain genes may offer a (cryptic) selective advantage in the complete darkness of a cave through recapitulation of cave-associated phenotypes in surface fish.
Conclusions
Since the discovery of depigmented and eyeless fish at the Chica cave in 1936 (Hubbs & Innes, 1936), Astyanax has been an intriguing and powerful system for contemporary biology. Our historical analysis of the literature indicates that with each technical improvement, Astyanax has been used to catalyze important discoveries for the fields of evolution, development, genetics and behavior. The ability to breed cave and surface fish to create viable offspring (Şadoğlu, 1957b) led to the use of this model system for classical (Şadoğlu, 1956), and later quantitative (Protas et al., 2008), genetic studies. Experimental embryology procedures, such as lentectomy (Yamamoto et al., 2003), led to the discovery of the lens as a key inductive tissue. Knockdown analyses revealed a role for pleiotropy (Yamamoto et al., 2009) in driving the development of both regressive and constructive features. The application of QTL analyses led to the identification of genes that govern simple traits (Gross et al., 2009), and revealed the complex genetic basis for a variety of multifactorial traits (Kowalko et al., 2013a; Kowalko et al., 2013b). Transcriptomic studies have identified the sequence and expression of unique genetic features for the cave and surface forms (Gross et al., 2013; Hinaux et al., 2013). This historical pattern suggests that we are once again at an exciting stage in Astyanax biology. With the newly available sequenced genome (McGaugh et al., 2014), Astyanax is poised to make even further contributions to our understanding of traits with human relevance (Gross et al., 2014), the genomic basis for cave adaptation (Bradic et al., 2013), and help inform our understanding of why certain lineages discard phenotypes over evolutionary time (Cronk, 2009). While much has been done to this point, much more is waiting to be discovered, and this powerful natural system is on the cusp of providing exciting discoveries for many years to come.
Methods
We quantified primary literature, focused on Astyanax cavefish, through individual-year searches from 1936 through 2014 using the search terms “Anoptichthys” and “Astyanax”. All literature searches were performed using Google Scholar (scholar.google.com) and titles were retained for each positive hit. Results were filtered, and irrelevant uses of each term were discarded. Search hits were screened to avoid redundant documentation of an earlier-published report. The results of this analysis are presented in Fig.1A.
Nomenclature usage in catalogued books was evaluated for several Astyanax and Anoptichthys species names using the Google NGram Viewer (books.google.com/ngrams). This open source search tool provides quantitative comparisons of term usage over delimited periods of time. We searched for species names associated with Astyanax cave and surface fish (e.g., mexicanus, jordani, hubbsi, fasciatus, and jordani) as well as various Anoptichthys species names (jordani, hubbsi, and antrobius). Although our search query ranged from 1936 through 2014, information through the Google NGram Viewer database is currently available only through the year 2008 (Fig.1B). All default settings were retained for this analysis (e.g., smoothing value = 3). Although several forms of Astyanax returned a positive hit, only one Anoptichthys species name (jordani) successfully returned a hit. The results of this analysis are presented in Fig.1B.
Acknowledgements
The authors wish to thank Drs. Kimberly Cooper and Michael Shapiro for the invitation to contribute to this special issue. We are also grateful to Brian Carlson, Amanda Powers, Bethany Stahl and the members of the international Astyanax community for helpful conversations that stimulated many of the topics discussed in this commentary. We thank two anonymous reviewers for indispensible comments on an earlier draft of this manuscript. The Gross lab gratefully acknowledges support from the National Institute for Dental and Craniofacial Research (NIH), Award number DE022403.
Grant Sponsor NIH/NIDCR, Grant Number DE022403
References
- Albertson RC, Cresko W, Detrich HW, III, Postlethwait JH. Evolutionary mutant models for human disease. Trends Genet. 2009;25:74–81. doi: 10.1016/j.tig.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alunni A, Menuet A, Candal E, Pénigault J, Jeffery WR, Rétaux S. Developmental mechanisms for retinal degeneration in the blind cavefish Astyanax mexicanus. J Comp Neurol. 2007;505:221–233. doi: 10.1002/cne.21488. [DOI] [PubMed] [Google Scholar]
- Alvarez J. Revisión del género Anoptichthys con descripción de una especie nueva (Pisces, Characidae) An Esc Nac Cien Biol México. 1946;4:263–282. [Google Scholar]
- Alvarez J. Descripción de Anoptichthys hubbsi caracínido ciego de La Cueva de Los Sabinos, S. L. P. Rev Soc Mexicana Hist Nat. 1947;8:215–219. [Google Scholar]
- Avise JC, Selander RK. Evolutionary genetics of cave-dwelling fishes of the genus Astyanax. Evolution. 1972;26:1–19. doi: 10.1111/j.1558-5646.1972.tb00170.x. [DOI] [PubMed] [Google Scholar]
- Avise JC. Captivating Life. Smithsonian Institute Press; Washington D.C.: 2001. p. 212. [Google Scholar]
- Behrens M, Langecker TG, Wilkens H, Schmale H. Comparative analysis of Pax-6 sequence and expression in the eye development of blind cave fish Astyanax fasciatus and its epigean conspecific. Mol Biol Evol. 1997;14:299–308. doi: 10.1093/oxfordjournals.molbev.a025765. [DOI] [PubMed] [Google Scholar]
- Behrens M, Wilkens H, Schmale H. Cloning of the αA-crystallin genes of a blind cave form and the epigean form of Astyanax fasciatus: A comparative analysis of structure, expression and evolutionary conservation. Gene. 1998;216:319–326. doi: 10.1016/s0378-1119(98)00346-1. [DOI] [PubMed] [Google Scholar]
- Bensouilah M, Denizot J-P. Taste buds and neuromasts of Astyanax jordani: Distribution and immunochemical demonstration of co-localized substance P and enkephalins. Eur J Neurosci. 1991;3:407–414. doi: 10.1111/j.1460-9568.1991.tb00828.x. [DOI] [PubMed] [Google Scholar]
- Bilandžija H, Ma L, Parkhurst A, Jeffery WR. A potential benefit of albinism in Astyanax cavefish: Downregulation of the oca2 gene increases tyrosine and catecholamine levels as an alternative to melanin synthesis. PLoS One. 2013;8:e80823. doi: 10.1371/journal.pone.0080823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowsky R. Restoring sight in blind cavefish. Curr Biol. 2008;18:R23–4. doi: 10.1016/j.cub.2007.11.023. [DOI] [PubMed] [Google Scholar]
- Borowsky R, Wilkens H. Mapping a cave fish genome: Polygenic systems and regressive evolution. J Hered. 2002;93:19–21. doi: 10.1093/jhered/93.1.19. [DOI] [PubMed] [Google Scholar]
- Boudriot F, Reutter K. Ultrastructure of the taste buds in the blind cave fish Astyanax jordani (“Anoptichthys”) and the sighted river fish Astyanax mexicanus (Teleostei, Characidae) J Comp Neurol. 2001;434:428–444. doi: 10.1002/cne.1185. [DOI] [PubMed] [Google Scholar]
- Bradic M, Beerli P, García-de León FJ, Esquivel-Bobadilla S, Borowsky RL. Gene flow and population structure in the Mexican blind cavefish complex (Astyanax mexicanus) BMC Evol Biol. 2012;12:9. doi: 10.1186/1471-2148-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradic M, Teotónio H, Borowsky RL. The population genomics of repeated evolution in the blind cavefish Astyanax mexicanus. Mol Biol Evol. 2013;30:2383–2400. doi: 10.1093/molbev/mst136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breder CM. Descriptive ecology of La Cueva Chica, with especial reference to the blind fish, Anoptichthys. Zoologica. 1942;27:7–15. [Google Scholar]
- Breder CM, Rasquin P. Comparative studies in the light sensitivity of blind characins from a series of Mexican caves. Bull Amer Mus Nat Hist. 1947;89:325–351. [Google Scholar]
- Breder CM. Apparent changes in phenotypic ratios of the characins at the type locality of Anoptichthys jordani Hubbs and Innes. Copeia. 1943a;1943:26–30. [Google Scholar]
- Breder CM. A note on erratic viciousness in Astyanax mexicanus (Phillipi) Copeia. 1943b;1943:82–84. [Google Scholar]
- Button KS, Ioannidis JP, Mokrysz C, Nosek BA, Flint J, et al. onfidence and precision increase with high statistical power. Nature Rev Neurosci. 2013;14:585. doi: 10.1038/nrn3475-c4. [DOI] [PubMed] [Google Scholar]
- Carlini DB, Satish S, Fong DW. Parallel reduction in expression, but no loss of functional constraint, in two opsin paralogs within cave populations of Gammarus minus (Crustacea: Amphipoda) BMC Evol Biol. 2013;13:89. doi: 10.1186/1471-2148-13-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson BM, Onusko SW, Gross JB. A high-density linkage map for Astyanax mexicanus using genotyping-by-sequencing technology. G3. 2015 doi: 10.1534/g3.114.015438. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke GM, Rivas MA, Morris AP. A flexible approach for the analysis of rare variants allowing for a mixture of effects on binary or quantitative traits. PLoS Genet. 2013;9:e1003694. doi: 10.1371/journal.pgen.1003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghill LM, Hulsey CD, Chaves-Campos J, García-de León FJ, Johnson SG. Next generation phylogeography of cave and surface Astyanax mexicanus. Mol Phylogenet Evol. 2014;79:368–374. doi: 10.1016/j.ympev.2014.06.029. [DOI] [PubMed] [Google Scholar]
- Cronk QCB. Evolution in reverse gear: The molecular basis of loss and reversal. Cold Spring Harb Symp Quant Biol. 2009;74:259–266. doi: 10.1101/sqb.2009.74.034. [DOI] [PubMed] [Google Scholar]
- Culver DC, Kane TC, Fong DW. Adaptation and natural selection in caves: The evolution of Gammarus minus. Harvard University Press; Cambridge, MA: 1995. p. 240. [Google Scholar]
- Darwin C. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. John Murray; London, UK: 1859. p. 502. [PMC free article] [PubMed] [Google Scholar]
- Dowling TE, Martasian DP, Jeffery WR. Evidence for multiple genetic forms with similar eyeless phenotypes in the blind cavefish, Astyanax mexicanus. Mol Biol Evol. 2002;19:446–455. doi: 10.1093/oxfordjournals.molbev.a004100. [DOI] [PubMed] [Google Scholar]
- Duboué ER, Borowsky RL, Keene AC. β-adrenergic signaling regulates evolutionarily derived sleep loss in the Mexican cavefish. Brain Behav Evol. 2012;80:233–243. doi: 10.1159/000341403. [DOI] [PubMed] [Google Scholar]
- Duboué ER, Keene AC, Borowsky RL. Evolutionary convergence on sleep loss in cavefish populations. Curr Biol. 2011;21:671–676. doi: 10.1016/j.cub.2011.03.020. [DOI] [PubMed] [Google Scholar]
- Eigenmann CH. Cave vertebrates of America: A study in degenerative evolution. The Carnegie Institution of Washington; Washington D.C.: 1909. p. 336. [Google Scholar]
- Elipot Y, Legendre L, Pére S, Sohm F, Rétaux S. Astyanax transgenesis and husbandry: How cavefish enters the laboratory. Zebrafish. 2014;11:291–299. doi: 10.1089/zeb.2014.1005. [DOI] [PubMed] [Google Scholar]
- Elipot Y, Hinaux H, Callebert J, Rétaux S. Evolutionary shift from fighting to foraging in blind cavefish through changes in the serotonin network. Curr Biol. 2013;23:1–10. doi: 10.1016/j.cub.2012.10.044. [DOI] [PubMed] [Google Scholar]
- Erckens W, Martin W. Exogenous and endogenous control of swimming activity in Astyanax mexicanus (Characidae, Pisces) by direct light response and by a circadian oscillator. I. Analyses of the time-control systems of an epigean river population. Z Naturforsch. 1982a;37:1253–1265. [Google Scholar]
- Erckens W, Martin W. Exogenous and endogenous control of swimming activity in Astyanax mexicanus (Characidae, Pisces) by direct light response and by a circadian oscillator. II. Features of time-controlled behavior of a cave population and their comparison to an epigean ancestral form. Z Naturforsch. 1982b;37:1266–1273. [Google Scholar]
- Espinasa L, Borowsky R. Origins and relationship of cave populations of the blind Mexican tetra, Astyanax fasciatus, in the Sierra de El Abra. Environ Biol Fishes. 2001;62:233–237. [Google Scholar]
- Espinasa L, Rivas-Manzano P, Pérez HE. A new blind cave fish population of genus Astyanax: Geography, morphology and behavior. Environ Biol Fishes. 2001;62:339–344. [Google Scholar]
- Fong DW. Morphological evolution of the amphipod Gammarus minus in caves: Quantitative genetic analysis. Am Midl Nat. 1989;121:361–378. [Google Scholar]
- Fumagalli M, Sironi M, Pozzoli U, Ferrer-Admetlla A, Pattini L, et al. Signatures of environmental genetic adaptation pinpoint pathogens as the main selective pressure through human evolution. PLoS Genet. 2011;7:e1002355. doi: 10.1371/journal.pgen.1002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresser EB, Breder CM. The histology of the eye of the cave characin, Anoptichthys. Zoologica. 1940;25:113–116. [Google Scholar]
- Gross JB. The complex origin of Astyanax cavefish. BMC Evol Biol. 2012;12:105. doi: 10.1186/1471-2148-12-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JB, Furterer A, Carlson BC, Stahl BA. An integrated transcriptome-wide analysis of cave and surface dwelling Astyanax mexicanus. PLoS One. 2013;8:e55659. doi: 10.1371/journal.pone.0055659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JB, Wilkens H. Albinism in phylogenetically and geographically distinct populations of Astyanax cavefish arises through the same loss-of-function Oca2 allele. Heredity. 2013;111:122–30. doi: 10.1038/hdy.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JB, Borowsky R, Tabin CJ. A novel role for Mc1r in the parallel evolution of depigmentation in independent populations of the cavefish Astyanax mexicanus. PLoS Genet. 2009;5:e1000326. doi: 10.1371/journal.pgen.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JB, Krutzler AJ, Carlson BM. Complex craniofacial changes in blind cave-dwelling fish are mediated by genetically symmetric and asymmetric loci. Genetics. 2014;196:1303–1319. doi: 10.1534/genetics.114.161661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JB, Protas M, Conrad M, Scheid PE, Vidal O, et al. Synteny and candidate gene prediction using an anchored linkage map of Astyanax mexicanus. Proc Natl Acad Sci U S A. 2008;105:20106–20111. doi: 10.1073/pnas.0806238105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Nehm RH. Darwin’s difficulties and students’ struggles with trait loss: Cognitive-historical parallelisms in evolutionary explanation. Sci Educ. 2014;23:1051–1074. [Google Scholar]
- Hausdorf B, Wilkens H, Strecker U. Population genetic patterns revealed by microsatellite data challenge the mitochondrial DNA based taxonomy of Astyanax in Mexico (Characidae, Teleostei) Mol Phylogenet Evol. 2011;60:89–97. doi: 10.1016/j.ympev.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Hinaux H, Pottin K, Chalhoub H, Pére S, Elipot Y, et al. A developmental staging table for Astyanax mexicanus surface fish and Pachón cavefish. Zebrafish. 2011;8:155–165. doi: 10.1089/zeb.2011.0713. [DOI] [PubMed] [Google Scholar]
- Hinaux H, Poulain J, Da Silva C, Noirot C, Jeffery WR, et al. De novo sequencing of Astyanax mexicanus surface fish and Pachón cavefish transcriptomes reveals enrichment of mutations in cavefish putative eye genes. PLoS One. 2013;8:e53553. doi: 10.1371/journal.pone.0053553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooven TA, Yamamoto Y, Jeffery WR. Blind cavefish and heat shock protein chaperones: A novel role for hsp90α in lens apoptosis. Int J Dev Biol. 2004;48:731–738. doi: 10.1387/ijdb.041874th. [DOI] [PubMed] [Google Scholar]
- Hubbs CL, Innes WT. The first known blind fish of the family Characidae: A new genus from Mexico. Occas Papers Mus Zool, Univ Michigan. 1936;342:1–7. [Google Scholar]
- Humbach I. Geruch und geschmack bei den augenlosen höhlenfischen Anoptichthys jordani, Hubbs und Innes und Anoptichthys hubbsi, Alvarez. Naturwiss. 1960;47:551–551. [Google Scholar]
- Hüppop K. Oxygen consumption of Astyanax fasciatus (Characidae, Pisces): A comparison of epigean and hypogean populations. Environ Biol Fishes. 1986;17:299–308. [Google Scholar]
- Innes WT. A cavern characin Anoptichthys jordani, Hubbs and Innes. Aquarium. 1937;5:200–202. [Google Scholar]
- Jeffery WR. Cavefish as a model system in evolutionary developmental biology. Dev Biol. 2001;231:1–12. doi: 10.1006/dbio.2000.0121. [DOI] [PubMed] [Google Scholar]
- Jeffery WR. Regressive evolution of pigmentation in the cavefish Astyanax. Isr J Ecol Evol. 2006;52:405–422. [Google Scholar]
- Jeffery WR. Pleiotropy and eye degeneration in cavefish. Heredity. 2010;105:495–496. doi: 10.1038/hdy.2010.7. [DOI] [PubMed] [Google Scholar]
- Jeffery WR, Strickler AG, Guiney S, Heyser DG, Tomarev SI. Prox 1 in eye degeneration and sensory organ compensation during development and evolution of the cavefish Astyanax. Dev Genes Evol. 2000;210:223–230. doi: 10.1007/s004270050308. [DOI] [PubMed] [Google Scholar]
- Jeffery WR. Regressive evolution in Astyanax cavefish. Annu Rev Genet. 2009;43:25–47. doi: 10.1146/annurev-genet-102108-134216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery WR, Martasian DP. Evolution of eye regression in the cavefish Astyanax: Apoptosis and the pax-6 gene. Am Zool. 1998;38:685–696. [Google Scholar]
- Jones R, Culver DC. Evidence for selection on sensory structures in a cave population of Gammarus minus (Amphipoda) Evolution. 1989;43:688–693. doi: 10.1111/j.1558-5646.1989.tb04267.x. [DOI] [PubMed] [Google Scholar]
- Kosswig C, Kosswig L. Die variabilität bei Asellus aquaticus unter besonderer berucksichtigung der variabilität in isolierten unter-und aberirdischen population. Rev Fac Sci (Istanbul) B. 1940;5:1–55. [Google Scholar]
- Kowalko JE, Rohner N, Linden TA, Rompani SB, Warren WC, et al. Convergence in feeding posture occurs through different genetic loci in independently evolved cave populations of Astyanax mexicanus. Proc Natl Acad Sci U S A. 2013a;110:16933–16938. doi: 10.1073/pnas.1317192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalko J, Rohner N, Rompani S, Peterson B, Linden T, et al. Loss of schooling behavior in cavefish through sight-dependent and sight-independent mechanisms. Curr Biol. 2013b;23:1874–1883. doi: 10.1016/j.cub.2013.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langecker TG, Schmale H, Wilkens H. Transcription of the opsin gene in degenerate eyes of cave-dwelling Astyanax fasciatus (Teleostei, Characidae) and of its conspecific epigean ancestor during early ontogeny. Cell Tissue Res. 1993;273:183–192. [Google Scholar]
- Langecker TG, Wilkens H, Schmale H. Developmental constraints in regressive evolution: Studies of the expression of the γs-crystallin gene in the developing lens of cave-dwelling Astyanax fasciatus (Cuvier, 1819) (Teleostei, Characidae) by in situ hybridization. J Zool Syst Evol Res. 1995;33:123–128. [Google Scholar]
- Ma L, Parkhurst A, Jeffery W. The role of a lens survival pathway including sox2 and αA-crystallin in the evolution of cavefish eye degeneration. EvoDevo. 2014;5:28. doi: 10.1186/2041-9139-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Jeffery WR, Essner JJ, Kowalko JE. Genome editing using TALENs in blind Mexican cavefish, Astyanax mexicanus. Under review. 2015 doi: 10.1371/journal.pone.0119370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattheij JAM. The cell types in the adenohypophysis of the blind Mexican cave fish, Anoptichthys jordani (Hubbs and Innes) Z Zellforsch. 1968;90:542–553. doi: 10.1007/BF00339502. [DOI] [PubMed] [Google Scholar]
- McGaugh SE, Gross JB, Aken B, Blin M, Borowsky R, et al. The cavefish genome reveals candidate genes for eye loss. Nat Comm. 2014;5:5307. doi: 10.1038/ncomms6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menuet A, Alunni A, Joly J, Jeffery WR, Rétaux S. Expanded expression of sonic hedgehog in Astyanax cavefish: Multiple consequences on forebrain development and evolution. Development. 2007;134:845–855. doi: 10.1242/dev.02780. [DOI] [PubMed] [Google Scholar]
- Mitchell RW, Russell WH, Elliott WR. Mexican eyeless characin fishes, genus Astyanax: Environment, distribution, and evolution. Vol. 89. Texas Tech Press; Lubbock, Texas: 1977. [Google Scholar]
- Montgomery JC, Coombs S, Baker CF. The mechanosensory lateral line system of the hypogean form of Astyanax fasciatus. Environ Biol Fishes. 2001;62:87–96. [Google Scholar]
- Morrow JE. Schooling behavior in fishes. Q Rev Biol. 1948;23:27–38. doi: 10.1086/396078. [DOI] [PubMed] [Google Scholar]
- O'Quin KE, Yoshizawa M, Doshi P, Jeffery WR. Quantitative genetic analysis of retinal degeneration in the blind cavefish Astyanax mexicanus. PloS One. 2013;8:e57281. doi: 10.1371/journal.pone.0057281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornelas-García CP, Domínguez-Domínguez O, Doadrio I. Evolutionary history of the fish genus Astyanax Baird & Girard (1854) (Actinopterygii, Characidae) in Mesoamerica reveals multiple morphological homoplasies. BMC Evol Biol. 2008;8:340. doi: 10.1186/1471-2148-8-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters N, Peters G. Genetic problems in the regressive evolution of cavernicolous fish. In: Schröder JH, editor. Genetics and Mutagenesis of Fish. Springer; Berlin: 1973. pp. 187–201. [Google Scholar]
- Peters VN, Scholl A, Wilkens H. Der Micos-Fisch, Höhlenfisch in statu nascendi oder Bastard? Ein Beitrag zur Evolution der Höhlentiere. J Zool Syst Evol Res. 1975;13:110–124. [Google Scholar]
- Protas M, Conrad M, Gross JB, Tabin C, Borowsky R. Regressive evolution in the Mexican cave tetra, Astyanax mexicanus. Curr Biol. 2007;17:452–454. doi: 10.1016/j.cub.2007.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protas ME, Trontelj P, Patel NH. Genetic basis of eye and pigment loss in the cave crustacean, Asellus aquaticus. Proc Natl Acad Sci U S A. 2011;108:5702–5707. doi: 10.1073/pnas.1013850108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protas ME, Hersey C, Kochanek D, Zhou Y, Wilkens H, et al. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat Genet. 2006;38:107–111. doi: 10.1038/ng1700. [DOI] [PubMed] [Google Scholar]
- Protas M, Tabansky I, Conrad M, Gross JB, Vidal O, et al. Multi-trait evolution in a cave fish, Astyanax mexicanus. Evol Dev. 2008;10:196–209. doi: 10.1111/j.1525-142X.2008.00227.x. [DOI] [PubMed] [Google Scholar]
- Quinn TP. Locomotor responses of juvenile blind cave fish, Astyanax jordani, to the odors of conspecifics. Behav Neural Biol. 1980;29:123–127. doi: 10.1016/s0163-1047(80)92568-6. [DOI] [PubMed] [Google Scholar]
- Rohner N, Jarosz DF, Kowalko JE, Yoshizawa M, Jeffery WR, et al. Cryptic variation in morphological evolution: HSP90 as a capacitor for loss of eyes in cavefish. Science. 2013;342:1372–1375. doi: 10.1126/science.1240276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose FL, Mitchell RW. Comparative lipid values of epigean and cave-adapted Astyanax. Southwest Nat. 1982;27:357–358. [Google Scholar]
- Şadoğlu P. A preliminary report on the genetics of the Mexican cave characins. Copeia. 1956;1956:113–114. [Google Scholar]
- Şadoğlu P. A Mendelian gene for albinism in natural cave fish. Cell Mol Life Sci. 1957a;13:394–394. [Google Scholar]
- Şadoğlu P. Mendelian inheritance in the hybrids between the Mexican blind cave fishes and their overground ancestor. Verh Dtsch Zool Ges Graz. 1957b;1957:432–439. [Google Scholar]
- Şadoğlu P. A breeding method for blind Astyanax mexicanus based on annual spawning patterns. Copeia. 1979;1979:369–371. [Google Scholar]
- Şadoğlu P, McKee A. A second gene that affects eye and body color in Mexican blind cave fish. J Hered. 1969;60:10–14. doi: 10.1093/oxfordjournals.jhered.a107917. [DOI] [PubMed] [Google Scholar]
- Schemmel C. Comparative studies of the cutaneous sense organs in epigean and hypogean forms of Astyanax with regard to the evolution of cavernicoles. Z Morphol Tiere. 1967;61:255–316. [Google Scholar]
- Schemmel C. Studies on the genetics of feeding behaviour in the cave fish Astyanax mexicanus f. Anoptichthys. An example of apparent monofactorial inheritance by polygenes. Z Tierpsychol. 1980;53:9–22. doi: 10.1111/j.1439-0310.1980.tb00730.x. [DOI] [PubMed] [Google Scholar]
- Sharma S, Coombs S, Patton P, de Perera TB. The function of wall-following behaviors in the Mexican blind cavefish and a sighted relative, the Mexican tetra (Astyanax) J Comp Physiol A. 2009;195:225–240. doi: 10.1007/s00359-008-0400-9. [DOI] [PubMed] [Google Scholar]
- Strecker U, Bernatchez L, Wilkens H. Genetic divergence between cave and surface populations of Astyanax in Mexico (Characidae, Teleostei) Mol Ecol. 2003;12:699–710. doi: 10.1046/j.1365-294x.2003.01753.x. [DOI] [PubMed] [Google Scholar]
- Strecker U, Faúndez VH, Wilkens H. Phylogeography of surface and cave Astyanax (Teleostei) from Central and North America based on cytochrome b sequence data. Mol Phylogenet Evol. 2004;33:469–481. doi: 10.1016/j.ympev.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Strecker U, Hausdorf B, Wilkens H. Parallel speciation in Astyanax cave fish (Teleostei) in northern Mexico. Mol Phylogenet Evol. 2012;62:62–70. doi: 10.1016/j.ympev.2011.09.005. [DOI] [PubMed] [Google Scholar]
- Strickler AG, Famuditimi K, Jeffery WR. Retinal homeobox genes and the role of cell proliferation in cavefish eye degeneration. Int J Dev Biol. 2002;46:285–294. [PubMed] [Google Scholar]
- Timmann C, Thye T, Vens M, Evans J, May J, et al. Genome-wide association study indicates two novel resistance loci for severe malaria. Nature. 2012;489:443–446. doi: 10.1038/nature11334. [DOI] [PubMed] [Google Scholar]
- Van Trump WJ, McHenry MJ. The lateral line system is not necessary for rheotaxis in the Mexican blind cavefish (Astyanax fasciatus) Integr Comp Biol. 2013;53:799–809. doi: 10.1093/icb/ict064. [DOI] [PubMed] [Google Scholar]
- Voneida TJ, Sligar CM. A comparative neuroanatomic study of retinal projections in two fishes: Astyanax hubbsi (the blind cave fish), and Astyanax mexicanus. J Comp Neurol. 1976;165:89–105. doi: 10.1002/cne.901650108. [DOI] [PubMed] [Google Scholar]
- Wilkens H. Genetic interpretation of regressive evolutionary processes: Studies on hybrid eyes of two Astyanax populations (Characidae, Pisces) Evolution. 1971;25:530–544. doi: 10.1111/j.1558-5646.1971.tb01913.x. [DOI] [PubMed] [Google Scholar]
- Wilkens H. Evolution and genetics of epigean and cave Astyanax fasciatus (Characidae, Pisces): Support for the neutral mutation theory. In: Hecht MK, Wallace B, editors. Evolutionary Biology. Plenum Press; New York: 1988. pp. 271–367. [Google Scholar]
- Wilkens H, Burns RJ. A new Anoptichthys cave population. Ann Spéléol. 1972;27:263–270. [Google Scholar]
- Wilkens H, Strecker U. Convergent evolution of the cavefish Astyanax (Characidae, Teleostei): Genetic evidence from reduced eye-size and pigmentation. Biol J Linn Soc. 2003;80:545–554. [Google Scholar]
- Wiley S, Mitchell RW. A bibliography of the Mexican eyeless characin fishes of the genus Astyanax. In: Reddell JR, Mitchell RW, editors. Studies of the Cavernicole Fauna of Mexico, AMCS Bulletin 4. The Speleo Press; Austin: 1971. pp. 231–239. [Google Scholar]
- Wright S. Pleiotropy in the evolution of structural reduction and of dominance. Am Nat. 1964;98:65–70. [Google Scholar]
- Yamamoto Y, Espinasa L, Stock DW, Jeffery WR. Development and evolution of craniofacial patterning is mediated by eye-dependent and -independent processes in the cavefish Astyanax. Evol Dev. 2003;5:435–446. doi: 10.1046/j.1525-142x.2003.03050.x. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Stock DW, Jeffery WR. Hedgehog signalling controls eye degeneration in blind cavefish. Nature. 2004;431:844–847. doi: 10.1038/nature02864. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Byerly MS, Jackman WR, Jeffery WR. Pleiotropic functions of embryonic sonic hedgehog expression link jaw and taste bud amplification with eye loss during cavefish evolution. Dev Biol. 2009;330:200–211. doi: 10.1016/j.ydbio.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Jeffery WR. Central role for the lens in cave fish eye degeneration. Science. 2000;289:631–633. doi: 10.1126/science.289.5479.631. [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Yokoyama S. Isolation, DNA sequence and evolution of a color visual pigment gene of the blind cave fish Astyanax fasciatus. Vision Res. 1990a;30:807–816. doi: 10.1016/0042-6989(90)90049-q. [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Yokoyama S. Convergent evolution of the red- and green-like visual pigment genes in fish, Astyanax fasciatus, and human. Proc Natl Acad Sci U S A. 1990b;87:9315–9318. doi: 10.1073/pnas.87.23.9315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa M, Gorički S, Soares D, Jeffery WR. Evolution of a behavioral shift mediated by superficial neuromasts helps cavefish find food in darkness. Curr Biol. 2010;20:1631–1636. doi: 10.1016/j.cub.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa M, Jeffery WR, van Netten SM, McHenry MJ. The sensitivity of lateral line receptors and their role in the behavior of Mexican blind cavefish (Astyanax mexicanus) J Exp Biol. 2013;217:886–895. doi: 10.1242/jeb.094599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa M, Jeffery WR. Shadow response in the blind cavefish Astyanax reveals conservation of a functional pineal eye. J Exp Biol. 2008;211:292–299. doi: 10.1242/jeb.012864. [DOI] [PMC free article] [PubMed] [Google Scholar]