Abstract
Depression often accompanies the peri-menopausal transition and it often precedes overt symptomology in common neurodegenerative diseases (NDD; Alzheimer's, Parkinson's, Huntington, ALS). Serotonin dysfunction is frequently found in the different etiologies of depression. We have shown that ovariectomized (Ovx) monkeys treated with estradiol (E) for 28 days supplement with placebo or progesterone (P) on days 14-28 had reduced DNA fragmentation in serotonin neurons of the dorsal raphe nucleus; and long-term Ovx monkeys had fewer serotonin neurons than intact controls. We questioned the effect of E alone or E+P on gene expression related to DNA repair, protein folding (chaperones), the ubiquitin proteosome, axon transport, and NDD specific genes in serotonin neurons. Ovx macaques were treated with placebo, E or E+P (n=3/group) for 1 month. Serotonin neurons were laser captured and subjected to microarray analysis and qRT-PCR. Increases were confirmed with qRT-PCR in 5 genes that code for proteins involved in repair of strand breaks and nucleotide excision. NBN1, PCNA, GADD45A, RAD23A and GTF2H5 significantly increased with E or E+P treatment (all ANOVA p< 0.01). Chaperone genes HSP70, HSP60 and HSP27 significantly increased with E or E+P treatment (all ANOVA p<0.05). HSP90 showed a similar trend. Ubiquinase coding genes UBEA5, UBE2D3 and UBE3A (Parkin) increased with E or E+P (all ANOVA p<0.003). Transport related genes coding kinesin, dynein, and dynactin increased with E or E+P (all ANOVA p<0.03). SCNA (α synuclein) and ADAM10 (α secretase) increased (both ANOVA p<0.02), but PSEN1 (presenilin1) decreased (ANOVA p<0.02) with treatment. APP decreased 10-fold with E or E+P administration. Newman-Keuls posthoc comparisons indicated variation in the response to E alone versus E+P across the different genes. In summary, E or E+P increased gene expression for DNA repair mechanisms in serotonin neurons, thereby rendering them less vulnerable to stress-induced DNA fragmentation. In addition, E or E+P regulated 4 genes encoding proteins that are often misfolded or malfunctioning in neuronal populations subserving overt NDD symptomology. The expression and regulation of these genes in serotonergic neurons invites speculation that they may mediate an underlying disease process in NDDs, which in turn may be ameliorated or delayed with timely hormone therapy in women.
Keywords: DNA repair, chaperones, ubiquinases, transport, neurodegeneration, serotonin, estrogen, progesterone, dorsal raphe, macaques
Introduction
Depression, anxiety and cognitive loss accompany the onset of menopause in a subpopulation of women. We showed that ovarian steroids increase serotonin neural function in macaques at multiple levels including gene and protein expression (1), which correlate with changes in anxiety-like behaviors (2), that in turn model aspects of mental health in women after menopause (3-6). While these acute changes in serotonin neural function are very important, we recently found that the absence of ovarian steroids led to serotonin neuron degeneration and death in the absence of any overt trauma to the brain. This was preceded by marked progressive DNA fragmentation as detected with a TUNEL assay (7). In addition, ovariectomy of young female macaques in a semi-free ranging troop resulted in significantly fewer serotonin neurons 3 years later as detected with in situ hybridization for expression of Fev, the master gene that determines serotonin phenotype (2). These studies were consistent with our earlier observations that estradiol (E) with or without progesterone (P) supplementation suppressed gene expression in the caspase-independent apoptotic pathway, particularly expression of Apoptosis Inducing Factor (AIF) in serotonin neurons (8). Subsequently, we showed that E or E+P decreased AIF protein, AIF phosphorylation and AIF translocation to the nucleus (9). Upon apoptotic stimuli, AIF is released from the mitochondria and translocates to the nucleus where it triggers chromatin condensation and large DNA fragmentation, as observed in ovariectomized (Ovx) macaques (7). AIF nuclear translocation is thought to be a commitment point to neuronal cell death (10, 11). However, we observed recovery in terms of decreased TUNEL staining after one month of E+P treatment (7).
Altogether, these studies and reports in the literature, indicated that ovarian steroids may increase gene expression for DNA repair proteins and that the lack of E or E+P could lead to serotonin neuronal degeneration or apoptosis. Because depression often accompanies NDDs, which may be delayed with hormone therapy, we also reasoned that ovarian steroids might affect the expression of normal genes that have been implicated in neurodegenerative diseases. A number of NDDs involve translation of normal genes whose proteins subserve important cellular functions, but the proteins are mis-processed, mis-folded or subverted from normal function in various ways (12). NDDs may also be caused by genetic mutations that lead to similar problems or loss of functions (13, 14). Therefore, it was of additional interest to examine ovarian hormone effects on the expression of a subset of genes in serotonin neurons thought to be involved in NDDs in other systems.
Alterations in neuronal function can undergo recovery, but neuronal cell death is permanent in areas lacking stem cell replacement. Therefore, subsequent therapy intended to ameliorate serotonin deficits would not be effective if the neuronal targets were gone. This could apply to antidepressant treatments in NDD, as well as hormone replacement therapy in postmenopausal women.
Unfortunately, hormone therapy has been under assault for treatment of menopausal women since release of the negative results of the Women's Health Initiative, which administered non-human conjugated equine estrogens and synthetic androgenic medroxyprogesterone acetate to women approximately 10 years after onset of menopause (15). Estrogenic compounds can be used in women after complete hysterectomy but estrogenic plus progestogenic compounds should be used in women with a reproductive tract. Our model involves ovariectomy of adult rhesus macaques for a relatively short time, i.e. 5-12 months, followed by subcutaneous delivery of bioidentical estradiol (E) or estradiol supplemented with progesterone (days 14-28; E+P) for one month.
In this study we examined gene expression related to DNA repair and neurodegenerative diseases in laser captured serotonin neurons in the Ovx model administered clinically appropriate doses of E or E+P. Pivotal gene changes predicted by a microarray were examined by qRT-PCR.
Materials and Methods
This experiment was approved by the IACUC of the Oregon National Primate Research Center and conducted in accordance with the 2011 Eight Edition of the National Institute of Health Guide for the Care and Use of Laboratory Animals.
Animals and treatments
Nine adult female rhesus monkeys (Macaca mulatta) were oophorectomized (Ovx) by the surgical personnel of ONPRC ~8 months before assignment to this project according to accepted veterinary surgical protocol. All animals were born in China, were aged between 7-14 years by dental exam, weighed between 5 and 8 kg, and were in good health.
Animals were either treated with placebo (Ovx-control group; n=3), or treated with estradiol (E) for 28 days (E group; n=3), or treated with E for 28 days and then supplemented with progesterone (P) for the final 14 of the 28 days (E+P group; n=3). The placebo treatment of the spay-control monkeys consisted of implantation with empty Silastic capsules (s.c.). The E-treated monkeys were implanted with two 4.5-cm E-filled Silastic capsules (i.d. 0.132 in.; o.d. 0.183 in.; Dow Corning, Midland, MI). The capsule was filled with crystalline estradiol (1,3,5(10)-estratrien-3,17-b-diol; Steraloids, Wilton, NH). The E+P- treated group received E-filled capsules, and 14 days later, received one 6-cm capsule filled with crystalline progesterone (4-pregnen-3,20 dione; Steraloids). All capsules were placed in the periscapular area under ketamine anesthesia (ketamine HCl, 10mg/kg, s.c.; Fort Dodge Laboratories, Fort Dodge, IA).
The monkeys were euthanized at the end of the treatment periods according to procedures recommended by the 2013 Edition of the American Veterinary Medical Association Guidelines for the Euthanasia of Animals. Each animal was sedated with ketamine, administered pentobarbital (30 mg/kg, i.v.), and exsanguinated by severance of the descending aorta.
Steroid Hormone Assays
Assays for E and P were performed utilizing a Roche Diagnostics 2010 Elecsys assay instrument. Prior to these analyses, measurements of E and P on this platform were compared to traditional RIA's as previously reported (16). The E+P treatment regimen has been shown to cause differentiation of the uterine endometrium in a manner similar to a normal 28-day menstrual cycle (17).
Tissue preparation, Raphe Block and Laser Capture Dissection (n=9; 3 animals/treatment group)
The left ventricle of the heart was cannulated and the head of each animal was perfused with 3 liters of 1X cold RNA-later buffer (Ambion Inc., Austin, TX) plus 20% The brain was removed from the cranium, dissected into blocks and frozen at –80°C.
For laser capture, the pontine midbrain block was placed in a cryostat (Microm HM500OM) and brought to –20°C. Thin sections (7 μm) through the dorsal raphe nucleus were thaw mounted onto plain glass slides and frozen at –80°C. The next morning, the sections were processed in a rapid, RNAse free immunohistochemical assay for tryptophan hydroxylase (TPH). The sections were immersed in cold acetone for 1 minute, cold ethanol for 30 seconds, cold PBS for 3 minutes, and then covered with normal rabbit serum (NRS, 1/500) containing 1% RNAse inhibitor for 10 minutes. The NRS was removed and the sections were covered with sheep anti-TPH (1/300; Chemicon, Temecula, CA) containing 1% RNAse inhibitor for 30 minutes, then immersion washed in PBS for 3 minutes, and covered with biotinylated rabbit anti-sheep serum (1/120; Vector Laboratories, Burlingame, CA) containing 1% RNAse inhibitor for 20 minutes. The sections were then immersed in PBS for 3 minutes, covered with Vector ABC reagent for 30 minutes, immersed in cold 0.2 M Tris (pH 8.2), immersed in cold diaminobenzidine (DAB) containing H2O2 (30% solution diluted 1/5000) and dehydrated in 100% ethanol for 2 minutes. Finally, the slides were immersed in xylene for 2 minutes and then dried under vacuum for 1 hr prior to laser capture. Serotonin neurons appeared darkly stained and were captured with an Arcturus Laser Dissection Microscope (PixCell I). After capture to the microcap film (Capsure macro-211), the films were removed from the caps and immersed in lysis buffer. Up to 10 films with 1000-3000 captures/neurons each were collected into one microtube containing 200 μl of lysis buffer. Approximately 150,000 laser pulses were executed on each animal for an individual preparation (12-15 microtubes were pooled per animal). Hence, a preparation of captured cells was made for each animal and all animals were processed separately on the microarray and in the qRT-PCR assays. The variance around the group means represents animal-to-animal variation. Individuals interested in obtaining other areas of the brain for study are encouraged to contact the authors.
RNA extraction from laser captured neurons
Each animal produced multiple tubes with 10-12 films in each tube. The tubes containing the lysis buffer and films were vortexed to dislodge the captured material from the films. Each tube was adjusted to 350 μl of lysis buffer and then 350 μl of 70% ethanol was added and mixed well. The samples were extracted with the RNAeasy microRNA kit from Qiagen according to the directions. The final eluates of an individual animal were pooled and evaporated in a Speedvac, yielding one preparation per animal. The RNA was suspended in 12 μl of TE (0.01MTris+0.005M EDTA). The quantity of RNA in the resuspended sample was determined with the Ribogreen Quantitation Kit (Molecular Probes, Eugene, OR) or with a Nanodrop Spectrophotometer (ND 1000 V3.3, Wilmington, DE). The integrity of the RNA was examined with the Agilent Bioanalyzer using the pico-chip according to the directions of the manufacturer. This laser capture preparation has been used for examination of several pathways (18-21) and it was previously shown to be enriched approximately 7-fold for serotonin neuron-related mRNA (18).
The individual animal preparations were split. An aliquot was used for hybridization to the microarray and another aliquot was set aside for qRT-PCR. Two animals per group were used for hybridization and 3 animals per group were used to confirm gene changes with qRT-PCR.
Affymetrix hybridization
To screen a large number of genes the Rhesus Affymetrix GeneChip was utilized. Labeled target cRNA was prepared from the laser captured serotonin neurons of 6 animals (n=2 animals/treatment group). The cRNA from the laser capture pool of each individual animal was hybridized to Rhesus Affymetrix GeneChip arrays. The Affymetrix GeneChip® Rhesus Macaque Genome Array interrogates over 47,000 M. mulatta transcripts (small sequences from 3’ region of genes; 1-6 interrogated per gene). The array contains 52,024 rhesus probe sets and was designed using public data sources including data from the University of Nebraska (R. Norgren), the Baylor School of Medicine's rhesus macaque whole-genome shotgun assembly (October 1, 2004), and GenBank® STSs, ESTs, and mRNAs up to March 30, 2005. Additionally, probe sets were designed to interrogate rhesus transcripts orthologous to the 3’ end of human transcripts (GeneChip® Human Genome U133 Plus 2.0 Array and RefSeq sequences up to March 2005), sixteen viral sequences from human and other primate species, three different rhesus r(ribosomal) RNAs, and thirteen rhesus mitochondrial genes along with control and reporter sequences. Microarray assays were performed in the Affymetrix Microarray Core of the OHSU Gene Microarray Shared Resource.
Data Analysis with GeneSifter Software
The microarray data was used as a screen for genes that had substantial literature support for roles in DNA repair, neurodegeneration and disease. The MAS5.0 data was processed with Affymetrix GCOS interface software and compressed into CHP files and uploaded to GeneSifter (VisX Labs, Seattle, WA). The software calculated the mean signal intensity and the standard error of the mean for each treatment group. It is common for a single gene to be represented by multiple probe sets that target different areas of the 3’ end of the gene. Probe sets that were undetectable in all treatment groups were eliminated. The remaining probe sets were further filtered and those probes sets exhibiting a 2-fold or greater change between the treatment groups were examined with Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis and Gene Navigation or Gene Function analysis. The use of a 2-fold filter was chosen based upon a review of fold change and statistical stringency (22).
Criteria for Validation
We chose to examine a technically reasonable number of genes in triplicate on a 384 Taqman qRT-PCR card that would be representative of complex functions. For example, the FANCF gene (Fanconi Anemia, complementation group F) plays a role in DNA repair and it is mutated in an autosomal recessive gene disorder of humans. It would be useless to examine its regulation in the monkey brain. Genes that had wide spread, peripheral, abundant or universal roles (eg., pertained to histones, chromatid adhesion, cell cycle, kinase activity, ATP activity, EST or open reading frame sequences) were not considered further.
The 28 specific genes chosen for verification with qRT-PCR were determined with different subroutines in GeneSifter and knowledge of the literature. We had evidence, as outlined in the Introduction, that DNA repair was likely targeted. Hence, we queried the database of probe sets showing 2-fold changes using the ‘Gene Function’ subroutine and the category called ‘DNA repair’. Of these, we chose to further examine 12 genes related to DNA repair enzymes that were identified by probes sets exhibiting robust signal intensity and pronounced changes with treatment.
Neurodegeneration studies show ‘common disease mechanisms’, ie., protein folding (chaperones) ubiquitin proteasome, and axon transport. Regarding chaperones, we focused on heat shock proteins due to our interest in steroid receptor activation where they play prominent roles. Therefore, we used the ‘Gene Navigation’ subroutine and searched the 2-fold database for ‘heat shock.’ For the heat shock genes, we chose to further verify individuals in the A, B, C and D families that exhibited robust signal intensities and marked changes with treatment.
To identify ubiquinases, we used the ‘Gene Navigation’ subroutine and searched the 2-fold database for ‘UBE.’ The ubiquinase pathway has been divided into activating (E1), conjugating (E2) and ligase (E3) proteins. For verification we chose an individual gene from each category that demonstrated robust signal intensity and a patent change with treatment.
To identify genes coding for axonal transport we used the ‘Gene Function’ subroutine and examined the 2-fold database for probe sets in the ‘axon cargo transport’ category. We also examined the ‘cytoskeletal-dependent intracellular transport’ category as well as individual genes noted to play prominent roles in the literature. We chose 4 genes that code for motor proteins and that were not overly abundant plus the gene that codes for a cytoskeletal protein comprising neurofibulary tangles.
Finally, we were interested in genes related to neurodegenerative diseases in humans. KEGG analysis revealed the altered probe sets related to Alzheimer's and Parkinson's. Based upon the literature, we chose several prominent genes that code for dysfunctional or toxic proteins in these 2 diseases.
Taqman qRT-PCR array
A custom Taqman qRT-PCR array was designed (Invitrogen Life Technologies, ABI, Foster City, CA) containing the pivotal genes related to DNA repair and NDDs or underlying pathologies determined as described above. This platform utilizes microfluidic distribution of samples into wells containing custom primers, which eliminated pipetting and sample-to-sample variation. The primers utilize a 5’ fluorescent reporter, FAM (Fluorescein amidite; Molecular Probes, Eugene, OR) and a 3’ quencher, TAMRA (tetramethylrhodamine), which improves sensitivity. The cards also contain the passive reference dye, ROX, which enables normalization. The genes examined were:
DNA repair- related - NSB1 (nibrin), NTHL1 (endonuclease), LIG4 (ligase), PCNA (proliferating nuclear antigen), POLG (polymerase), SHFM1 (homologous recombination), GADD45A (DNA damage inducible), RAD23A (DNA damage recognition), APEX1 (endonuclease), GTF2H5 (gene transcription factor), PARP2 (transferase), XRCC1 (single strand break repair), SHRPH (histone linker);
chaperones – heat shock protein (HSP)90, HSP70, HSP60, HSP27;
ubiquinases - UBEA5, UBE2D3, UBE3A (Parkin);
transport motors - KIF5B (kinesin), DNCLC1 (dynein), DCTN4 (dynactin), MAPT (tau); and
disease specific genes - SCNA (α synuclein), ADAM10 (α secretase), APP (amyloid beta precursor protein ) and PSEN1 (presenilin1).
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), GUSB and PIAA were measured for reference. GAPDH was used for normalization of the data. TPH2, which is known to increase with E or E+P treatment of Ovx macaques, was included on the Taqman array card as an additional regulatory control. Each sample was assayed in triplicate on the Taqman card.
Reverse transcription of laser captured samples followed by Taqman qPCR
Reverse transcription and complementary DNA (cDNA) synthesis was performed using Oligo-dT 15 and Random hexamer primers (Invitrogen Life Technologies, Carlsbad, CA) and Superscript III reverse transcriptase (200 U/μg of RNA, Invitrogen Life Technologies) at 42°C for 1 hr. cDNA was treated with RNAse H to disintegrate dsDNA. Total RNA (1.4 to 1.8 μg) from each laser capture pool and from a standard pool of rhesus tissues was transcribed and stored as cDNA at a concentration of 250 ng/μl. Then, 4 concentrations of the standard pool and the samples (7.5 ng) were loaded onto the custom Taqman cards for qPCR in 100 μl of reaction mix. There was a log linear increase in fluorescence detected as the concentration of amplified double-stranded product cDNA increased during the reaction. The fluorescence was detected as cycle threshold (Ct) with an ABI 7900 thermal cycler (Applied Biosystems Inc.) during 40 cycles. The slope of the standard curve was used to calculate the relative picograms of each transcript in the RNA extracted from the laser-captured pools. Then, the ratio of each transcript to GAPDH was calculated for each sample.
Primer selection
For our genes of interest, the ABI Rhesus Monkey primer inventory was examined. For the genes not present in the library, ABI designed and constructed new primers, which were then added to the Rhesus Monkey inventory. The exact primer sequences are proprietary, but the gene names, symbol, AB assay ID, context sequence, and NCBI gene reference information are shown in Table 1. The microarray signal intensity of a representative probe set is also shown.
Table 1.
Gene names, symbol, AB assay ID, context sequence, and NCBI gene reference information of genes included on the Taqman qRT-PCR card. The microarray signal intensity of a representative probe set is also shown.
| Gene Symbol | Gene Name | Function of encoded protein | Ref Seq | ABI assay ID | Array Signal | ||
|---|---|---|---|---|---|---|---|
| DNA Repair | Ovx | E | EP | ||||
| Increased | |||||||
| 1. NBN [NBS1 ] | nibrin | part of double strand break repair complex | XM_001085033.1 | RhO1039845 | 21 | 340 | 369 |
| 2. NTHL1* | Nth endonuclease Ill-like 1 | DNA N-glycosylase of the endonuclease III family that has apurinic/ apyrimidinic lyase activity | XM_001082772.2 | Rh00959765-m1 | 33 | 220 | 79 |
| 3. LIG4* | ligase IV, DNA, ATP-dependent | DNA ligase essential for DNA double-strand break (DSB) repair through nonhomologous end joining | XM_001084107 | Rh04269856 | 55 | 188 | 225 |
| 4. GTF2H5 | General transcription factor IIH, polypeptide5 | catalytic subunit of transcription/repair factor TFIIH, which functions in gene transcription and DNA repair | NM_001190969.1 | Rh02792678_m1 | 562 | 1240 | 1009 |
| 5. POLG2* | polymerase DNA directed γ2 | mitochondrial DNA polymerase; performs base excision repair (BER) | XM_001109823.2 | Rh00945165_m1 | 57 | 164 | 269 |
| 6. PCNA | proliferating cell nuclear antigen | BER gap filling | XM_001115746.2 | Rh02806147_m1 | 185 | 631 | 449 |
| 7. DSS1 (SHFMl)* | Split hand/foot malformation | homologus recombination double strand break | XM_001091523.2 | Rh02860294_m1 | 658 | 2137 | 1421 |
| 8. RAD23A | RAD23 homolog A | Involved in nucleotide excision repair (NER) | XM_001110103.2 | Rh00908422_m1 | 337 | 701 | 319 |
| 9. APEX1 (REFl)* | apurinic/apyrimidinic endonuclease 1 | multifunctional DNA repair enzyme | XM_001090240.2 | Rh02793202_m1 | 588 | 1356 | 928 |
| 10. PARP2* | poly(ADP-ribosyl)transferase-like 2 | capable of catalyzing a poly(ADP-ribosyl)ation reaction | XM_001088314.2 | Rh02857777_m1 | 13 | 172 | 39 |
| 11. XRCCl* | X-ray repair complementing | efficient repair of DNA single-strand breaks formed by exposure to Ionizing radiation and alkylating agents | XM_001100256.2 | Rh02860275_m1 | 79 | 305 | 115 |
| 12. SHPRH* | SNF2 histone linker | contains motifs characteristics of several DNA repair proteins, transcription factors, and helicases | XM_001086512.2 | Rh02797744_m1 | 188 | 568 | 425 |
| Decreased | |||||||
| 13. GADD45 | growth arrest-DNA-damage-inducible γ | Increases with treatment of DNA-damaging agents; activates p38/JNK pathway | XM_001095413.2 | RhOI 071483_m1 | 179 | 116 | 87 |
| Chaperones | |||||||
| 14. HSPCB (HSP90) | heat shock protein 90kD | molecular chaperone with key roles In signal transduction, protein folding, protein degradation | NM_001195524.1 | Rh02790147_m1 | 890 | 9712 | 2563 |
| 15. HSPA8 (HSP70) | heat shock 70kD protein 8 (?constitutive) | binds to nascent polypeptides to facilitate correct folding; ATPase in the disassembly of clathrin-coated | XM_002799828.1 | Rh04269846_g1 | 238 | 2390 | 425 |
| 16. HSPD1 (HSP60) | heat shock 60kD protein 1 | (chaperonin)essentlal for the folding and assembly of newly imported proteins in the mitochondria | NM_002156.4 | Hs01036753_g1 | 629 | 2657 | 1392 |
| 17. HSPB1 (HSP27) | heat shock 27kD protein 1 | stress resistance and actin organization;stress induced translocation from cytoplasm to nucleus | XM_001109274.2 | Rh02980144_s1 | 823 | 2680 | 1136 |
| Ubiquinases | |||||||
| 18. UBE1 [UBA5 ] | Ubiquitin-like modifier activating enzyme 1 | catalyzes the first step in ubiquitin conjugation to mark cellular proteins for degradation | NM_024818.3 | Hs01566989_m1 | 73 | 148 | 211 |
| 19. UBE2D3 | Ublquitin-conjugating enzyme E2D 3 | conjugating enzyme: functions in the ubiquitination of the tumor-suppressor protein p53 | NM_181892.2 | Hs01518517_m1 | 1089 | 4720 | 3063 |
| 20 UBE3A | ubiquitin protein ligase E3A | part of the ubiquitin protein degradation system; maternally expressed In brain | XM_002804686.1 | Rh00963674_m1 | 77 | 311 | 391 |
| Transport | |||||||
| 21. KIF5B | kinesin family member 5B | forward transport; axonal function | XM_002805607.1 | RhO1037194_m1 | 999 | 7049 | 4581 |
| 22. DCTN4 | dynactin 4 (p62) | retrograde microtubule-mediated trafficking | XM_001109230.2 | RhOI092757 | 296 | 1003 | 579 |
| 23. DNCLC1 | Dynein, light chain, LC8-type 1 | major molecular motor in retrograde axonal transport | NM_001037495.1 | Hs00867659_g1 | 4431 | 10043 | 7009 |
| 24. Tau (MAPT) | microtubule-associated protein tau | binds to microtubules and regulate the stability of these cytoskeletal structures | XM_001115803.2 | Rh04269822_m1 | 690 | 1906 | 854 |
| Disease | |||||||
| 25. APP | amyloid beta (A4) precursor protein | fragments promote transcriptional activation, or basis of amyloid plaques | XM_002803170.1 | Rh01552279_m1 | 15596 | 4570 | 8352 |
| 26. ADAM 10 | ADAM metallopeptidase domain 10 | alpha secretase cleavage of APP precludes amyloid beta formation | XM_001097016.2 | Rh01109565_m1 | 508 | 1338 | 911 |
| 27. PSEN1 | presenile 1 | fragments form gamma-secretase transmembrane protease that produces toxic a-beta 40-42 | XM_001088635.2 | Rh02826228_m1 | 431 | 1437 | 632 |
| 28. SNCA | alpha-synudein | Integrate presynaptic signaling and membrane trafficking; defects implicated in Parkinson's, Alzheimer's | XM_001095402.2 | Rhl103386_m1 | 528 | 2220 | 920 |
Statistical analysis
GeneSifter obtained the average signal intensity of the 2 animals in each group. The average signal intensities above threshold on the microarrays were further filtered for showing a 2-fold or greater change with E or E+P treatment compared to the Ovx-control group. Thus, included probe sets were detectable, and either the E or E+P group was different from the Ovx group by a factor of 2 or more. A pattern analysis was conducted on the filtered probe sets to determine the total number of probe sets that were increased or decreased by E alone, E and E+P, or by E+P alone. In addition, probe sets exhibiting 2-fold or greater changes were subjected to ‘Gene Navigation’ or ‘Gene Function’ subroutine searches. Representative genes were further examined with qRT-PCR. The average relative expression of the 3 groups (n=3 animals/group) from the qRT-PCR assays were compared with ANOVA followed by Newman Keul's posthoc pairwise comparison. Finally, the expression of each gene was correlated to all other genes. Variance between animals is not unusual for this type of preparation, and more animals would reduce the chance of making a type 2 error. Therefore, negative results need further confirmation. Comparisons were considered significantly different when there was greater than a 95% chance that the groups were different (p<0.05). The genes that exhibited a significant difference with ANOVA were further subjected to a multiple comparison False Discovery Rate procedure. GeneSifter was used for probe set filters, pattern analysis and searches; Prism 5.0 from Graph Pad (San Diego, CA) was used for statistical tests; Excel was used to obtain the correlation matrix and the Benjamini-Hochberg/Yekutieli test for False Discovery Rate was applied using MATLAB, Mathworks Central (http://www.mathworks.com/matlabcentral/fileexchange/27418-benjamini---hochberg-yekutieli-procedure-for-controlling-false-discovery-rate).
Results
General Expression Changes in Laser Captured Serotonin Neurons
The entire data set of the 6 Affymetrix microarray chips has been submitted to the Gene Expression Omnibus public database (www.ncbi.nlm.nih.gov/geo/info/linking.html) and assigned the GEO accession number GSE16169. After exclusion of probe sets that were below threshold or undetectable based upon internal controls on the chip (n= 32,761), the remaining probe sets (19,263) were filtered for 2-fold or greater differences between the groups leaving 10,493 probe sets for categorization. Of these probe sets, GeneSifter finds probe sets that are increased or decreased, which together are termed ‘altered’. Further analysis could be obtained by name (Gene Navigation subroutine) or function (Gene Function subroutine). KEGG analysis revealed the number of probe sets on the array and the number of those that were regulated in various pathways. The microarray contained 504 probe sets related to DNA repair of which 124 were detectable and showed a 2-fold or greater alteration (up or down) with treatment. KEGG analysis of ubiquitin mediated proteolysis found 67 of 137 probe sets altered 2-fold or more; and KEGG analysis of ALZ, Parkinson's, Huntington's and ALS-related genes indicated 310 of 555 probe sets were altered 2-fold or more by treatment, but there is redundancy between the diseases, meaning that this number of altered probe sets is over-estimated. Additional information from KEGG analysis indicated that the Z score for probe sets related to Huntington's was 5.81, Alzheimer's was 5.85, Parkinson's was 6.55, the proteasome was 5.36, the ribosome was 6.06, and cytoskeletal proteins was 2.47, indicating that changes in gene expression related to these pathways was significantly higher than expected in serotonin neurons from hormone-treated individuals.
Specific searches were performed on the 2-fold probe set database with different subroutines as described in the Methods. There was significant redundancy in the probe sets. The ‘DNA Repair ’ category contained 124 results that included 85 up-regulated and 30 down-regulated probe sets (supplemental material, S1). The ‘heat shock’ search produced 36 results that included 32 up-regulated and 4 down-regulated probe sets (supplemental material, S2). The ‘UBE’ search produced 49 results that included 39 up-regulated and 10 down-regulated probe sets. The ‘axon transport category’ produced 16 results that included 13 up-regulated and 3 down-regulated probe sets. The ‘cytoskeletal-dependent intracellular transport’ category showed 30 altered probes sets including 26 up-regulated and 4 down-regulated (supplemental material, S3). Based upon the literature, we searched individually for genes that code for proteins governing forward and reverse transport directions (merged with transport category genes in supplemental table, S4). Tubulin was excluded from further investigation due to abundance. E or E+P increased the expression of a majority of the genes in our searches. KEGG analysis indicated that 97 probe sets related to Alzeheimer's and 80 probe sets related to Parkinson's were altered (supplemental material, S5).
Within the different functional categories, we chose to verify genes with substantial support in the literature. At the same time, the number of genes verified needed to be reasonable for experimental execution. Table 1 shows the average signal intensity of the duplicate animals/microarray chips for the probe sets of genes that were further examined with qRT-PCR. This study reports 19 of the genes that were examined, plus our internal control, TPH2. Genes not shown exhibited maximum cycle times (40) in all groups and were considered undetectable in the 50μl sample. These genes included LIG4, NTHL1, PARP2, POLG2, SHPRH, XXRC1 and APEX. It was not clear why APEX was undetectable with qRT-PCR since the signal intensity was moderate on the array. Also not shown was SHFM1, which was detectable in the Ovx placebo group and undetectable in the E and E+P treated groups, and did not agree with the microarray signal intensities. However, SHFM1 was included in the correlation matrix below.
In addition, pattern analysis was performed and the results are shown in Table 2. There were 5060 probe sets that increased >2-fold by E alone of which 161 were significantly different (ANOVA, unequal variances, p<0.05). There were 6946 probe sets that increased >2 fold in both E and E+P treated groups of which 421 were significantly different. There were 1106 probe sets increased >2-fold by E+P treatment and not by E alone of which 18 were significantly different. There were 3011 probe sets that were decreased less than half by E alone, of which 263 were significantly different. There were 2807 probe sets that were decreased by half or more in both E and E+P treated groups, of which 297 were statistically different. Finally, there were 2417 probe sets that were decreased by half or more in the E+P treated group and not by E alone, of which 27 were statistically different.
Table 2.
Patterns of overall gene expression in enriched preparations of macaque serotonin neurons. Probe sets that were detectable according to Affymetrix internal controls were examined for different patterns of expression. The patterns of expression are listed at the top of the table. The number of probe sets that exhibited each pattern of expression and the number of probe sets that were different by ANOVA corrected for unequal variance at p<0.5 are shown for each pattern.
| Patterns | Ov=1; E>2; EP=1 | Ov=1; E>2; EP>2 | Ov=1; E=1: EP>2 | OV=1; E<0.5; EP=1 | Ov=1; E<0.5; EP<0.5 | OV=1; E=1; EP<0.5 |
|---|---|---|---|---|---|---|
| Probe sets | 5030 | 6943 | 1103 | 3011 | 2807 | 2471 |
| Probe sets, p<0.05 | 161 | 421 | 18 | 263 | 297 | 27 |
Validation of Expression Changes in Laser Captured Serotonin Neurons
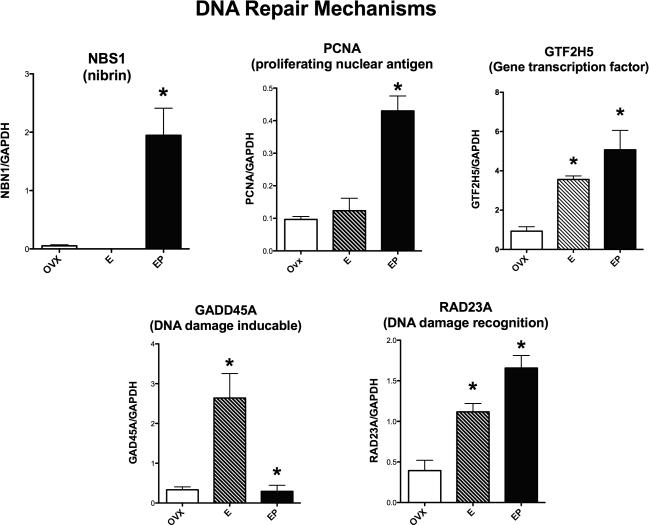
Figure 1 illustrates the relative expression of 5 genes that code for DNA repair. NBS1 (nibrin) was significantly different between the treatment groups (ANOVA, p = 0.0034). E+P treatment (EP) significantly increased NBS1 expression (Newman-Keuls’, p < 0.05) relative to the Ovx-placebo and E-treated groups (Newman-Keuls’, p < 0.05). PCNA (proliferating nuclear antigen) expression was significantly different between the groups (ANOVA, p = 0.0009). E+P treatment significantly increased PCNA expression relative to the Ovx-placebo and E-treated groups (Newman-Keuls’, p < 0.05). GTF2H5 (gene transcription factor 2H5) expression was significantly different between the groups (ANOVA, p = 0.0031). E and E+P treatment significantly increased GTF2H5 expression relative to the Ovx-placebo treated group (Newman-Keuls’, p < 0.05). The E and E+P treated groups were not different from each other. GADD45A (DNA damage inducible) expression was significantly different between the groups (ANOVA, p = 0.0062). E significantly increased GADD45A compared to the Ovx-placebo group (Newman-Keuls’, p < 0.05), but there was no difference between the Ovx-placebo group and the E+P-treated group, indicating that P suppressed the effect of E. RAD23A (DNA damage recognition) expression was significantly different between the groups (ANOVA p=0.0014). There was a significant increase in RAD23A with E and E+P treatment compared to the Ovx-placebo treated group (Newman-Keuls’, p < 0.05). In addition, E+P-treatment significantly increased RAD23A expression relative to E-treatment (Newman-Keuls’, p < 0.05).
Figure 1.
Histograms illustrating the expression of 5 genes that code for proteins that are involved in DNA repair. NBS1 (nibrin) and PCNA (proliferating nuclear antigen) were significantly increased with E+P treatment (EP on graph). GTF2H5 (gene transcription factor) and RAD23A (DNA damage recognition) were significantly increased by E or E+P treatment. GADD45A (DNA damage inducible) was significantly increased by E, but addition of P blocked the effect of E.
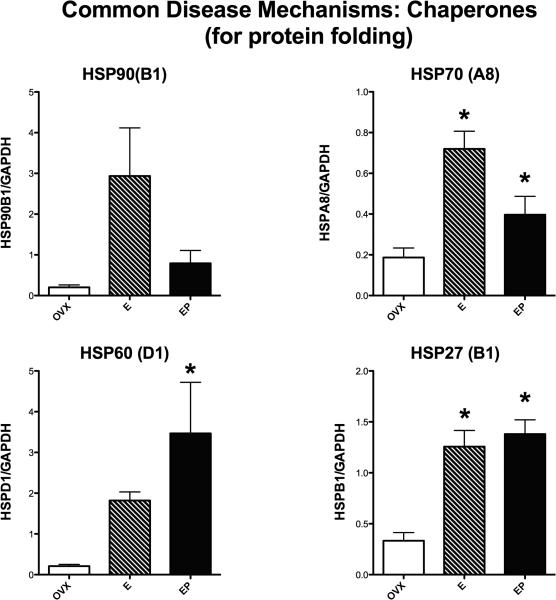
Figure 2 illustrates the relative expression of 4 genes that code for chaperone proteins, which play important roles in protein folding. HSP90(B1) expression was not different between the groups (ANOVA, p = 0.074), although there was a trend toward increased expression with E treatment relative to the Ovx-placebo group. Due to the small n, the increase may be real due to a type 2 statistical error. HSP70(A8) expression was significantly different between the groups (ANOVA, p = 0.0078). E treatment significantly increased HSP70(A8) expression relative to the Ovx-placebo treated group (Newman-Keuls’, p < 0.05). The E+P treated group was different from the E-treated group and not different from the Ovx-placebo group, suggesting that supplemental P treatment suppressed the E-induced increase in HSP70(A8). HSP60(D1) expression was significantly different between the groups (ANOVA, p = 0.054). E+P treatment significantly increased HSP60(D1) expression relative to the Ovx-placebo treated group (Newman-Keuls’, p < 0.05). However, the E+P group was not different from the E group, suggesting that with more animals, both E and E+P treatments would exhibit elevated HSP60(D1). HSP27(B1) expression was significantly different between the groups (ANOVA, p = 0.004). E and E+P treatment significantly increased HSP27(B1) expression relative to the Ovx-placebo treated group (Newman-Keuls’, p < 0.05).
Figure 2.
Histograms illustrating the expression of 4 genes that code for chaperone proteins that are involved in protein folding. All of the examined chaperone genes showed an increase in expression with E or E+P treatment (E, EP on graph) to varying degrees of significance. HSP70 and HSP27 exhibited significant increases in expression with E and EP treatments. HSP60 was significantly increased by EP treatment and showed a strong trend toward an increase with E only treatment. HSP90 exhibited a trend toward increased expression with E or E+P treatment.
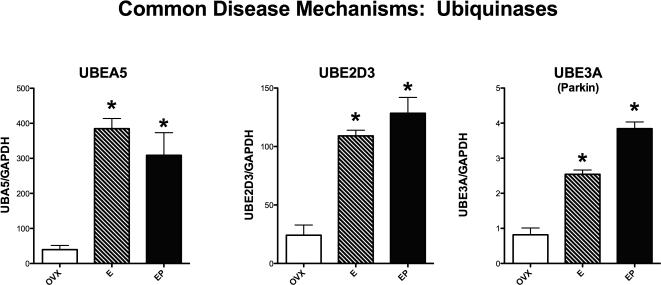
Figure 3 illustrates the effect of E or E+P administration on 3 ubiquinases (one each: activating, conjugating, and ligating), which are involved in protein transport to the lysosome for degradation. There was a significant difference between the groups in the expression of UBEA5, UBE2D3 and UBE3A, (ANOVA p= 0.0026, 0.0006 and 0.0001, respectively). E- and E+P-treatment significantly increased the expression of each ubiquinase compared to the Ovx-placebo group (Newman-Keuls’, p < 0.05).
Figure 3.
Histograms illustrating the expression of 3 genes that code for ubiquinases, which are proteins that designate and traffic other proteins for degradation. There was a significant increase in the expression of all 3 ubiquinase genes with E or E+P treatment (E, EP on graph).
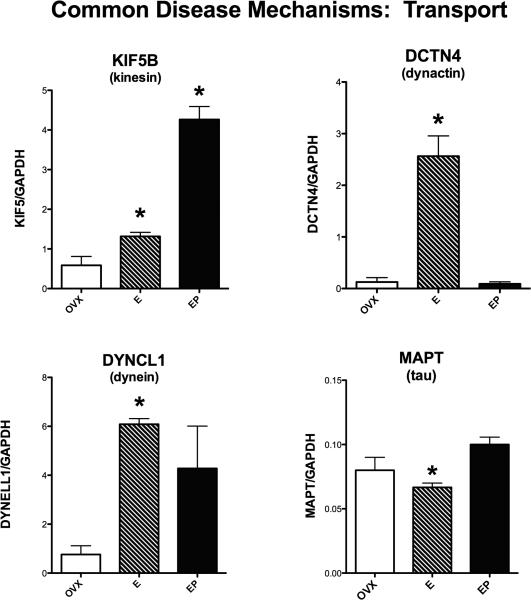
Figure 4 illustrates the effect of E or E+P on the expression of 3 cytoskeletal motor proteins involved in transport of material to or from the synapse, and the tau protein. There was a significant difference across the groups in expression of KIF5B (kinesin), DCTN4 (dynactin) and DYNCL1 (dynein) (ANOVA p=0.0001, 0.0004 and 0.027, respectively). There was a significant posthoc increase in kinesin with E and E+P treatment; a significant increase in dynactin with E only treatment, and a significant increase in dynein with E-only treatment (Newman-Keuls’, p < 0.05). MAPT (tau) was significantly different by ANOVA (p=0.039) with E+P greater than E (Newman-Keuls’ p<0.05). MAPT did not agree with the signal intensity reported on the microarray.
Figure 4.
Histograms illustrating the expression of 3 genes that code for transport motor proteins and MAPT that codes for a cytoskeletal protein present in neurofibrillary tangles. E and E+P (E, EP on graph) significantly increased expression of KIF5B (kinesin); and E alone significantly increased DCTN4 (dynactin) and DYNCL1 (dynein). E significantly suppressed MAPT compared to E+P.
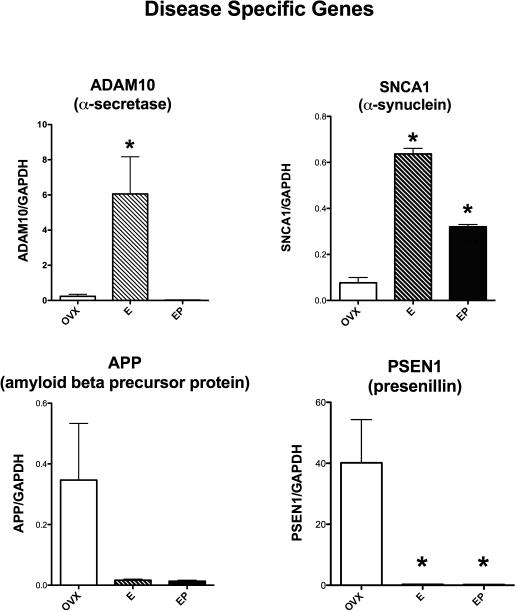
Figure 5 illustrates the effects of E or E+P administration on 4 genes related to specific neurodegenerative diseases. There was a significant difference between the groups in ADAM10 that codes for alpha-secretase (ANOVA p= 0.0213). E caused a significant posthoc increase in ADAM10 expression (Newman-Keuls’, p < 0.05). SNCA1 encodes α-synuclein. There was a significant difference between the groups in SNCA1 expression (ANOVA p=0.0001). E- and E+P-treatment significantly increased α -synuclein expression relative to placebo treatment, but E+P-treatment significantly reduced α -synuclein compared to E only treatment (Newman-Keuls’, p < 0.05). APP, amyloid beta (A4) precursor protein, decreased 10-fold with treatment from a ratio of 0.34±0.18 to 0.016±0.003 and 0.013±0.003 in Ovx, E and E+P treated groups, respectively. The variance in the Ovx group prevented statistically significant difference between the groups, but the difference is likely physiologically significant and the analysis would benefit from more animals. PSEN1 (presenillin) expression was significantly different between the groups (ANOVA p=0.0208). E and E+P-treatment significantly decreased PSEN1 expression relative to placebo treatment (Newman-Keuls’, p < 0.05).
Figure 5.
Histograms illustrating the expression of 4 genes that code for proteins known to be aberrant in different NDDs. ADAM10 (α-secretase) and SNCA1 (α -synuclein) were significantly increased by E or E+P treatments (E, EP on graph). PSEN1 (presenillin) was significantly decreased by E and E+P. APP (amyloid beta precursor protein) was not statistically different between the groups, but it exhibited a downward trend with E or E+P in a fashion similar to PSEN1.
Correlation Matrix
The expression of each gene across treatment groups was correlated with all of the other genes and the r2 values are shown in Table 3. A gene correlated with its’ self yields a perfect correlation of r2 equal to 1.00. Of note, the DNA repair gene, RAD23, exhibited nearly perfect correlation with 2 heat shock proteins and 2 ubiquinases. HSP90 showed high correlation with HSP70, UBEA5, dynactin (DCTN4) and dynein (DNCLC1), as well as α-secretase (ADAM10) and synuclein (SNCA). As expected, presenillin (PSEN1) and amyloid precursor protein (APP) showed moderate to strong inverse correlations with most of the other genes. SHFM1 (split hand and foot mutation1) showed a strong positive correlation with PSEN1 and APP.
Table 3.
Correlation matrix of the genes that were detectable in the qRT-PCR assay.
| NBN (NBSl) | GTF2H5 | PCNA | SHFM1 | RAD23A | GADD45 | HSP90 | HSP70 | HSP60 | HSP27 | UBA5 | UBE2DB | UBEBA | KIF5B | DCTN4 | DNCLC1 | MAPT | APP | ADAM10 | PSEN1 | SNCA | TPH2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NBN [NBS1] | 1.0000 | |||||||||||||||||||||
| GTF2H5 | 0.7626 | 1.0000 | ||||||||||||||||||||
| PCNA | 0.9954 | 0.8211 | 1.0000 | |||||||||||||||||||
| SHFM1 | −0.6758 | −0.9922 | −0.7433 | 1.0000 | ||||||||||||||||||
| RAD23A | 0.8072 | 0.9974 | 0.8601 | −0.9806 | 1.0000 | |||||||||||||||||
| GADD45 | −0.5333 | 0.1404 | −0.4498 | −0.2631 | 0.0688 | 1.0000 | ||||||||||||||||
| HSPCB (H5P90) | −0.3345 | 0.3544 | −0.2427 | −0.4685 | 0.2862 | 0.9756 | 1.0000 | |||||||||||||||
| H5PA8 (HSP70) | −0.1456 | 0.5289 | −0.0501 | −0.6308 | 0.4665 | 0.9145 | 0.9810 | 1.0000 | ||||||||||||||
| HSPD1 (HSP60) | 0.8571 | 0.9869 | 0.9026 | −0.9589 | 0.9959 | −0.0214 | 0.1986 | 0.3848 | 1.0000 | |||||||||||||
| HSPB1 (H5P27) | 0.5706 | 0.9664 | 0.6467 | −0.9909 | 0.9453 | 0.3903 | 0.5830 | 0.7294 | 0.9121 | 1.0000 | ||||||||||||
| UBE1 [UBA5] | 0.2847 | 0.8372 | 0.3752 | −0.8990 | 0.7956 | 0.6591 | 0.8082 | 0.9070 | 0.7378 | 0.9497 | 1.0000 | |||||||||||
| UBE2D3 | 0.6245 | 0.9814 | 0.6964 | −0.9977 | 0.9651 | 0.3276 | 0.5271 | 0.6818 | 0.9376 | 0.9977 | 0.9265 | 1.0000 | ||||||||||
| UBE3A | 0.8087 | 0.9972 | 0.8614 | −0.9801 | 1.0000 | 0.0662 | 0.2837 | 0.4642 | 0.9962 | 0.9445 | 0.7941 | 0.9644 | 1.0000 | |||||||||
| KIF5B | 0.9774 | 0.8820 | 0.9932 | −0.8162 | 0.9137 | −0.3427 | −0.1280 | 0.0666 | 0.9466 | 0.7312 | 0.4807 | 0.7753 | 0.9147 | 1.0000 | ||||||||
| DCTN4 | −0.5317 | 0.1423 | −0.4481 | −0.2649 | 0.0707 | 1.0000 | 0.9760 | 0.9153 | −0.0195 | 0.3921 | 0.6605 | 0.3295 | 0.0681 | −0.3408 | 1.0000 | |||||||
| DNCLC1 | 0.1588 | 0.7597 | 0.2527 | −0.8350 | 0.7109 | 0.7505 | 0.8773 | 0.9537 | 0.6446 | 0.9014 | 0.9917 | 0.8703 | 0.7091 | 0.3637 | 0.7518 | 1.0000 | ||||||
| MAPT | 0.9270 | 0.4643 | 0.8868 | −0.3500 | 0.5269 | −0.8117 | −0.6636 | −0.5061 | 0.6013 | 0.2209 | −0.0957 | 0.2859 | 0.5290 | 0.8269 | −0.8106 | −0.2232 | 1.0000 | |||||
| APP | −0.4867 | −0.9362 | −0.5682 | 0.9728 | −0.9085 | −0.4794 | −0.6604 | −0.7934 | −0.8671 | −0.9951 | −0.9760 | −0.9862 | −0.9074 | −0.6602 | −0.4811 | −0.9398 | −0.1234 | 1.0000 | ||||
| ADAM10 | −0.5470 | 0.1243 | −0.4643 | −0.2473 | 0.0525 | 0.9999 | 0.9719 | 0.9078 | −0.0377 | 0.3753 | 0.6467 | 0.3122 | 0.0500 | −0.3579 | 0.9998 | 0.7396 | −08211 | −04651 | 1.0000 | |||
| P5EN1 | −0.4801 | −0.9335 | −0.5620 | 0.9710 | −0.9053 | −0.4860 | −0.6660 | −0.7980 | −0.8633 | −0.9943 | −0.9776 | −0.9850 | −0.9042 | −0.6545 | −0.4877 | −0.9423 | −0.1160 | 1.0000 | −0.4717 | 1.0000 | ||
| SNCA | −0.0994 | 0.5678 | −0.0036 | −0.6663 | 0.5071 | 0.8947 | 0.9710 | 0.9989 | 0.4273 | 0.7604 | 0.9256 | 0.7151 | 0.5049 | 0.1130 | 0.8956 | 0.9666 | −04654 | −0.8209 | 0.8873 | −0.8252 | 1.0000 | |
| TPH2 | 0.2128 | 0.7943 | 0.3055 | −0.8640 | 0.7485 | 0.7130 | 0.8496 | 0.9357 | 0.6857 | 0.9238 | 0.9972 | 0.8961 | 0.7468 | 0.4144 | 0.7144 | 0.9985 | −0.1692 | −0.9572 | 0.7015 | −0.9593 | 0.9511 | 1.0000 |
Further comparisons
Of the 29 genes examined with qRT-PCR (including TPH2), 3 did not match the results of the microarray: GADD45, SHFM1, and MAPT which equals ~10%.
A Benjamimi-Hochberg/Yekutieli test for multiple comparisons was conducted on the qRT-PCR data using MATLAB. Twenty-one p-values were entered (20 graphed genes and TPH). The False Discovery Rate Procedure indicated that the critical p was equal to 0.0239 using a false discovery rate of 0.05. This substantiated the majority of our individual results. However, 4 of our significant individual results had p values that ranged from 0.028 to 0.05. These genes included HSPD1, UBEA5, MAPT, DYNCL1.
TPH2 Internal control and GAPHD
TPH2 was determined in the laser-captured samples (n=3/treatment) on the same Taqman card and at the same time. The TPH2/GAPDH ratio significantly increased in the E and E+P-treated groups, respectively, over the placebo-control group (ANOVA p=0.0001; Newman-Keul's P<0.05) indicating that serotonin neurons responded to the treatments. In turn, the increases in TPH2 support the validity of the regulation of other genes. GAPDH was not significantly altered by treatment.
Microarray Discoveries
The entire probe sets from the different searches that showed 2-fold or greater regulation by E or E+P are shown in the Supplemental Material. Supplementary information is available at the Molecular Psychiatry website.
Steroid Hormone Verification
The concentration of E and P in a serum sample obtained from each animal at necropsy was obtained to verify the efficacy of the Silastic implants. The concentration of E in the serum of the E and E+P-treated animals was 136.5±15 pg/ml and the concentration of P in the serum of the E+P-treated animals was 8.17±1.15 ng/ml. The concentration of E is similar to that observed in the mid-follicular or mid-luteal phase and the concentration of P is similar to that observed in the mid-luteal phase (23). The concentrations of E and P in the serum of the placebo-control animals were 11.0 ± 0.1 pg/ml and 0.16±0.13 ng/ml, respectively (significantly different from treated animals by ANOVA, p=0.01).
Discussion
Quality of life surveys have found that depression frequently precedes overt symptomology in neurodegenerative diseases (NDD) regardless of type (24, 25). Depression may inaugurate Alzheimer's or appear in later phases (26), and depression has been observed in the premotor phase in patients affected by Parkinson's disease (27, 28). Dysfunction in the serotonin system often underlies depression and different evidential approaches indicate that the serotonin system is affected in Parkinson's (29). Indeed, serotonergic antidepressants are the first-line treatment for depression in AD (30). However, SSRI administration in Parkinson's may worsen motor symptoms (27); and recent clinical trial data and meta-analysis found inconclusive or no benefit of SSRI's in Alzheimer's patients (30, 31). A postmortem study of patients with Alzheimer's and Parkinson's found fewer serotonin neurons in the raphe nuclei (32, 33) as well as neurofibrillary tangles and loss of large neurons (34, 35). It has also been proposed that serotonin neurodegeneration plays a role in depression not related to NDDs (36-38).
Reports of beneficial effects of estradiol (E) on Parkinson's and Alzheimer's diseases have appeared (39-41) with supporting information from animal models (42-44). The relative risk of developing Alzheimer's diminished by about one third in postmenopausal women who had taken E as compared with age-matched women who had not (45). However, there appears to be an important window of opportunity for the beneficial effects of estrogen therapy (41, 46).
Neurodegenerative disease is usually thought of in the context of severe deficits in motor or cognitive function whereas depression and mood disorders have traditionally been viewed as neurochemical deficits. However, it has recently been suggested that even the psychopathologies may involve functional degeneration of critical central neural systems and many are thought to have a serotonergic etiology (47, 48). There is evidence that depression is accompanied by marked changes in the number or size of neurons and glia in discrete brain regions (49). In tree shrews, stress increased the incidence of apoptosis in the temporal cortex. Moreover, antidepressant treatment reduced apoptotic neurons to control unstressed levels (50). The midbrain has not received the attention of the forebrain areas, yet it is the location of the serotonin cell bodies, and these cells control many forebrain functions. Thus, any loss or degeneration of serotonin neurons could have profound ramifications. We have accumulated a body of evidence that the steroid hormones, E and P, play a role in serotonin neuron resilience (7, 18, 51). This study shows that E or E+P increases gene expression related to DNA repair, protein folding, protein degradation and axonal transport. In addition, E or E+P decreased expression of genes that code for potentially toxic proteins and increased expression of α-secretase, which prevents production of toxic Aβ fragments.
Of note, the results of the FDR procedure suggested that the significant difference detected between the treatment groups of 4 genes discussed below may be false positives. On the other hand, the small n raises the risk that rejecting these genes as ‘not different’ or not regulated by E or E+P could be a type 2 statistical error. We are often faced with this issue in primate research and in the past we have found that with more animals, differences become significant (see (16) versus (52)). Therefore, they are included in the discussion. Furthermore, accepting 4 out of 19 genes as false positives does not change the overall message.
DNA repair minimally requires lesion recognition, single strand excision, lesion removal, gap-filling synthesis, and finally ligation. Repair is divided into BER, or single base excision repair, or NER with excision of 28 nucleotides around the lesion in one strand and gap repair using the other strand as the template (53). In addition, DNA repair can be divided into subpathways: global genome repair (GGR), transcription-coupled repair (TCR), and transcription domain-associated repair (DAR) (54). Neurons generally do not replicate their genomic DNA, and can therefore dispense with the task of removing DNA damage from the non-essential bulk of their genome, as long as they are able to maintain the integrity of the genes that must be expressed, or TCR (55, 56).
This study of DNA repair-related gene expression showed a marked effect of ovarian steroids on the expression of 5 relevant genes. NBS1, codes for nibrin, which is involved in DNA double-strand break repair and DNA damage-induced checkpoint activation. Nibrin is a member of the MRE11/RAD50 double-strand break repair complex and the observed increased expression with E+P would support DNA repair (57). RAD23A codes a protein involved in DNA damage recognition in nucleotide excision repair (NER) (53). The significant increase in RAD23A with E and E+P indicates improved NER would follow. PCNA codes for proliferating cell nuclear antigen, which contributes to leading strand synthesis during DNA replication and is a cofactor of DNA polymerase delta (58). The increase in PCNA with E+P suggests that more DNA repair is underway. PCNA-positive cells also increased in embryonic chicken hypothalamic following E treatment (59). GTF2H5 codes for a subunit of transcription/repair factor TFIIH, which functions in gene transcription and DNA repair. This protein stimulates ATPase activity to trigger DNA opening during DNA repair (60). The increase in GTF2H5 expression with E and E+P could increase DNA opening for repair. All of these actions would be beneficial to neurons subjected to stresses that lead to DNA damage, such as pro-inflammatory cytokines or environmental toxins. Although GADD45 (growth arrest and DNA damage 45) showed a decrease with E and E+P on the microarray, it was increased by E treatment with qRT-PCR. GADD45 transcript levels are increased following treatment with DNA-damaging agents or cellular stress, and in neurons, they can play a protective role after injury (61). In general, GADDs are regulators of cell cycle, senescence, survival, and apoptosis. GADD45 proteins also promote DNA methylation, an epigenetic function (62). Nonetheless, the disagreement between the microarray and qRT-PCR has been rare in our experience.
Chaperones are pivotal for proper protein folding, without which multiple cellular functions go awry. Of significance to Parkinson's disease, chaperones are capable of preventing α-synuclein misfolding, oligomerization, and aggregate formation that leads to Lewy Bodies (63-65). HSP27 particularly modulates intermediate filament organization under conditions of physiological stress and neurodegenerative disease (66). We found E or E+P increased gene expression for HSP70, HSP60 and HSP27 and with more power, E or E+P would have stimulated HSP90 as well. E increased HSP90 and HSP70 in rat hypothalamus, which is consistent with our observations (67). Several of the HSP family members also play important roles in steroid receptor activation, needed for mediating the actions of E and P (68).
The modification of proteins with ubiquitin is an important cellular mechanism for targeting abnormal or short-lived proteins for degradation (69-71). Ubiquitination involves at least three classes of enzymes: ubiquitin-activating enzymes, or E1s, ubiquitin-conjugating enzymes, or E2s, and ubiquitin-protein ligases, or E3s (72). UBEA5 encodes a member of the E1-type ubiquitin-activating enzyme family. UBE2D5 encodes a member of the E2 ubiquitinconjugating enzyme family and functions in the ubiquitination of the tumor-suppressor protein p53, which is induced by an E3 ubiquitin-protein ligase such as UBE3A. UBE3A is an E3 ubiquitin-protein ligase, the third part of the ubiquitin protein degradation system. It functions similarly to Parkin, which is inactivated in Parkinson's (73). E, with or without supplemental P, significantly increased gene expression for these 3 ubiquinases. In fact, supplemental P significantly increased gene expression for UBE3A over induction by E alone, further supporting a neuroprotective effect of P as well as E.
Axonal transport defects also play a role in neurodegeneration (74-76). We recently discovered that axonal transport of serotonin is severely compromised by aromatase inhibition, indicating that it was supported by E (77). E administration to ovariectomized monkeys also markedly elevated gene transcription underlying RhoGTPases and actin remodeling in serotonin neurons (19). These observations prompted examination of the expression of genes related to axonal transport. Herein we show that KIF5B, which codes for kinesin (anterograde); DYNCL1, which codes for dynein (retrograde) and DCTN4, which codes for dynactin (dynein adaptor), were all increased by E ± P. These 3 proteins are critical motors for the transport of vesicles through axons. An important caveat is that serotonin is not packaged in vesicles until prior to release; and it is thought to diffuse through the cytoplasm. However, the movement of serotonin over long distances is not compatible with diffusion. Classical fast axonal transport involves moving cargo vesicles along actin fibers, whereas slow axonal transport moves hundreds of soluble or cytoplasmic proteins. The slow transport may involve highly kinetic association/disassociation of soluble factors with a mobile carrier (78). However, studies have been limited to soluble proteins and not biogenic amines. Nonetheless, VMAT, the vescicular monoamine transporter, is moved by fast axonal transport in vesicles and serotonin might associate with this cargo.
Depression can occur prior to, or coincident with, symptomology and diagnosis in NDDs (24, 79). Moreover, inflammation, mitochondrial dysfunction and neurodegeneration of serotonin neurons has been linked to depression (36, 37). Based upon the KEGG analysis and Z-scores related to NDDs in laser–captured serotonin neurons, there was reason to further validate the effect of E and P on several genes directly related to NDD, even though the serotonin system has not been proven to be a first order target in most cases. In addition, the major motor neurodegenerative diseases have not been observed in nonhuman primates nor do rhesus monkeys have plaques or tangles associated with ALZ. Therefore, the actions of E or E+P are assumed to be executed on normally expressed genes in macaques without NDD.
APP codes for amyloid beta (A4) precursor protein, and it was decreased 10-fold by E or E+P. Ovariectomy increased APP in the hippocampus of rats, which is consistent with our data (80). Decreasing expression of this potentially toxic protein should be beneficial (81). ADAM10 encodes the alpha (α) secretase enzyme, which under normal conditions cleaves APP within the luminal/extracellular domain to yield soluble APP derivatives and membrane tethered a or bcarboxyl-terminal fragments that are not toxic. Its expression is upregulated during neuronal differentiation and after neural injury and the APP/α-secretase derivative peptides have proposed roles in cell signaling, long-term potentiation, and cell adhesion (82). The induction of normal ADAM10/alpha-secretase gene expression by E suggests another mechanism by which E is neuroprotective and agrees with ex vivo assessments in guinea pigs (43). In Ovx rats, administration of E prior to Aβ injection decreased cholinergic neuron loss and partly prevented fiber degeneration (83).
SCNA1 encodes for α -synuclein; although, the normal function of α-synuclein is not well understood. Studies suggest that it plays an important role in maintaining a supply of synaptic vesicles in presynaptic terminals (14). The increase in SCNA1 observed with E indicates a mechanism by which E could facilitate synaptic transmission. Mutations in SNCA1 are associated with early onset of Parkinson's disease and mis-folded α-synuclein is also a sizeable component of Lewy bodies (14).
The expression of tau (MAPT gene) was increased by E+P, but the difference was small and it surfaced as a false positive. Tau protein is a highly soluble microtubule-associated protein (MAP) that is active primarily in the distal portions of axons where it interacts with tubulin to stabilize microtubules and promote tubulin assembly into microtubules. Neither short- nor long-term E treatment affected Tau protein expression in the prefrontal cortex of rats (84). It is possible that tau is regulated at the protein level or differentially compartmentalized. However, other genes encoding important transport motor proteins, kinesin, dynactin and dynein, were clearly increased by E and/or E+P.
Notably, E or E+P severely suppressed PSEN1, which encodes presenillin, best known for its role in ALZ. Cleaved fragments of presenillin form the proteolytic subunit of gamma (γ)-secretase. This enzyme is responsible for a toxic cleavage of APP that yields amyloid- β40−42 or Aβ components of ALZ plaques (85). The dramatic inhibition of PSEN1 gene expression by E or E+P further suggests that repression of γ-secretase activity is important for neuronal viability. Zheng and colleagues demonstrated that E administration reduced Aβ levels in transgenic models of amyloidosis (86) and E treatment of neuronal cultures also decreased expression of PSEN1 (87). Thus several studies suggest that in the absence of E, an elevation in PSEN1 leads to the production and precipitation of Aβ in neurons that in turn, could contribute to dysfunction. Our data suggest that serotonin neurons are susceptible to this pathology.
It may be noted that supplemental P treatment caused different patterns of expression in different genes. In previous studies, as in this study, supplemental P may either (1) augment the effect of E, or (2) suppress the effect of E or (3) be neutral so that the effect of E+P is equal to the effect of E alone. We have observed all of these patterns in both in situ hybridization assays and in qRT-PCR assays. At this time, there does not seem to be an overarching explanation for the different interactions of P with E. We showed that serotonin neurons in female macaques contain ERβ and PR (88, 89). While these receptors bind to cognate response elements, they can also block other transcription factors (90) and compete for common co-regulators (91-94). We can only guess that PR has various mechanisms of action in a gene specific manner.
In a similar vein, we observed strong correlation between several genes from the same or different categories in this study. It is attractive to speculate that the steroid receptor mechanisms operate on the correlated genes in a like manner. It is also possible that the correlated genes are impacting one another. The DNA repair gene, RAD23A, exhibited nearly perfect correlation with HSPs 60 and 27, as well as ubiquinases UBE2D3 and UBE3A, which have conjugating and ligase activities, respectively. This is consistent with the need for ubiquitinproteosome activity for NER. RAD23A has a ubiquitin-binding domain for UBE2D3, which is required for optimal NER. In this case, the UBE2 domain of RAD23 is a conserved stabilization signal that allows RAD23 to interact with the proteasome without destruction (95, 96). HSP chaperones are not only important for protein folding, but also play critical roles in DNA repair (97-101). HSP90 showed high correlation with HSP70, UBEA5, DCTN4 (dynactin) and DNCLC1 (dynein), as well as ADAM10 (α-secretase) and SNCA (synuclein). This is consistent with HSP90 and HSP70 inhibition of aggregation and related functions (102), as well as signaling protein movement (103). PSEN1 (presenillin) and APP (amyloid precursor protein), were both decreased by treatments; they strongly correlated with each other, and each showed moderate to strong inverse correlations with most of the other genes. The coded protein of PSEN1 is a subunit of γ-secretase, which acts on APP to produce Aβ. The coordinated decrease in expression of both genes by ovarian steroids would be decidely neuroprotective.
The changes in gene expression reported in this study are part of a larger effect of E and E+P on gene expression in multiple pathways of serotonin neurons. We previously reported the effect of E and E+P on genes related to function and neurotransmission (1), apoptosis (18), spinogenesis (19), synaptogenesis (21) and glutamate neurotransmission (20). Many other pathways contain genes that were altered as well, including but not limited to those coding for adhesion, extracellular matrix, intracellular scaffolds, microtubule associated proteins, membrane proteins, growth factors, vesicles, receptors, channels, etc. Almost any pathway involved in the health, maintenance, or function of serotonin neurons likely has genes that are altered by ovarian hormones. E and/or E+P altered (up and down) by 2-fold or more ~10,000 probe sets. The number of probe sets that were altered by E or E+P may seem high, but in our first report of gene expression in laser captured neurons (18), we described a vast improvement in the detection of expression changes compared to a block of tissue. We speculated that even a small block of tissue would contain many other cell types that could obscure expression changes in serotonin neurons. In addition, this number of probe sets likely has some false positives, and many of the probe sets did not exhibit robust signal intensity. It is also important to remember that any one gene may have 1-6 probe sets on the array with an average of 2-3 probe sets/gene. If ~3000 genes were up or down regulated by either or both of the treatment groups, this does not approach the number of active genes in a cell at a given moment (104). This number may also be lower due to the noise in microarrays. In addition, ERα and ERβ are expressed by phenotypically diverse neurons throughout the brain (105). This means that E, acting via the transcription factor, ER, has an important role in the whole brain, and not just in serotonin neurons.
Nonetheless, this study provides further molecular support to the concept that the pathologies of NDDs may also manifest in serotonin neurons and lead to the onset of depression prior to, or coincident with, overt symptomology. The data also support the idea that serotonin neurodegeneration plays a role in depression, which has been previously proposed (36-38). However, the concept that ovarian hormone loss acts through gene expression and leads to serotonin neurodegeneration, which contributes to postmenopausal and age-associated depression, as well as depression associated with NDDs, provides crucial integration of a loose association of available information. It makes sense that administration of hormone therapy or antidepressants long after menopause or onset of NDD would be useless if the target neurons were gone.
The ability of ovarian hormones to prevent serotonin neurodegeneration in the Ovx monkey model encourages early hormonal intervention in perimenopausal women as proposed by Schmidt and Rubinow (106). Moreover, serotonergic antidepressants appear to have some neuroprotective/ neurotropic qualities as well (107), and limited data suggest that serotonergic antidepressants are more efficacious in women who have used hormone therapy (108).
In conclusion, we found that ovarian steroid hormones had a number of beneficial effects related to serotonin neuron viability. E or E+P increased gene expression associated with DNA repair, protein folding, protein degradation, axonal transport and the expression of 2 normal genes that become pathological in neurodegenerative disease. E or E+P also decreased expression of presenillin and APP, potentially pathological proteins of Alzheimer's disease. Together, the data indicate that hormone replacement therapy acts in numerous ways to promote serotonin neuronal viability and perhaps protect from NDDs. The genes examined also suggest a mechanism whereby NDD pathologies may manifest in serotonin neurons.
Supplementary Material
Acknowledgements
We are deeply grateful to the dedicated staff of the Division of Animal Resources including the staff of the Departments of Surgery and Pathology for their expertise and helpfulness in all aspects of monkey management. The staff of the OHSU Gene Microarray Shared Resource was essential for this study.
Supported by NIH grants: MH62677 to CLB, U54 contraceptive Center Grant HD 18185, P51 OD 011092 for the operation of ONPRC
Footnotes
Supplementary information is available at the Molecular Psychiatry website.
Literature Cited
- 1.Bethea CL, Lu NZ, Gundlah C, Streicher JM. Diverse actions of ovarian steroids in the serotonin neural system. Frontiers in Neuroendocrinology. 2002:2341–100. doi: 10.1006/frne.2001.0225. [DOI] [PubMed] [Google Scholar]
- 2.Coleman K, Robertson ND, Bethea CL. Long-term ovariectomy alters social and anxious behaviors in semi-free ranging Japanese macaques. Behavioral Brain Res. 2011;225:317–327. doi: 10.1016/j.bbr.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kugaya A, Epperson CN, Zoghbi S, van Dyck CH, Hou Y, Fujita M, Staley JK, Garg PK, Seibyl JP, Innis RB. Increase in prefrontal cortex serotonin 2A receptors following estrogen treatment in postmenopausal women. Am J Psychiatry. 2003;160(8):1522–4. doi: 10.1176/appi.ajp.160.8.1522. [DOI] [PubMed] [Google Scholar]
- 4.Soares CN, Almeida OP, Joffe H, Cohen LS. Efficacy of estradiol for the treatment of depressive disorders in perimenopausal women: a double-blind, randomized, placebo- controlled trial. Arch Gen Psychiatry. 2001;58(6):529–34. doi: 10.1001/archpsyc.58.6.529. [DOI] [PubMed] [Google Scholar]
- 5.Heikkinen J, Vaheri R, Timonen U. A 10-year follow-up of postmenopausal women on long-term continuous combined hormone replacement therapy: Update of safety and quality-of-life findings. J Br Menopause Soc. 2006;12(3):115–25. doi: 10.1258/136218006778234093. [DOI] [PubMed] [Google Scholar]
- 6.Steinberg EM, Rubinow DR, Bartko JJ, Fortinsky PM, Haq N, Thompson K, Schmidt PJ. A cross-sectional evaluation of perimenopausal depression. J Clin Psychiatry. 2008;69(6):973–80. doi: 10.4088/jcp.v69n0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lima FB, Bethea CL. Ovarian steroids decrease DNA fragmentation in the serotonin neurons of non-injured rhesus macaques. Mol Psychiatry. 2010;15:657–68. doi: 10.1038/mp.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bethea CL, Reddy AP. Effect of ovarian hormones on survival genes in laser captured serotonin neurons from macaques. J Neurochem. 2008;105(4):1129–43. doi: 10.1111/j.1471-4159.2008.05213.x. [DOI] [PubMed] [Google Scholar]
- 9.Tokuyama Y, Reddy AP, Bethea CL. Neuroprotective actions of ovarian hormones without insult in the raphe region of rhesus macaques. Neuroscience. 2008;154(2):720–31. doi: 10.1016/j.neuroscience.2008.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miramar MD, Costantini P, Ravagnan L, Saraiva LM, Haouzi D, Brothers G, Penninger JM, Peleato ML, Kroemer G, Susin SA. NADH oxidase activity of mitochondrial apoptosis-inducing factor. J Biol Chem. 2001;276(19):16391–8. doi: 10.1074/jbc.M010498200. [DOI] [PubMed] [Google Scholar]
- 11.Krantic S, Mechawar N, Reix S, Quirion R. Apoptosis-inducing factor: a matter of neuron life and death. Prog Neurobiol. 2007;81(3):179–96. doi: 10.1016/j.pneurobio.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 12.Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416(6880):507–11. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 13.Mullan M, Crawford F. The molecular genetics of Alzheimer's disease. Mol Neurobiol. 1994;9(1-3):15–22. doi: 10.1007/BF02816100. [DOI] [PubMed] [Google Scholar]
- 14.Dawson TM, Dawson VL. Molecular pathways of neurodegeneration in Parkinson's disease. Science. 2003;302(5646):819–22. doi: 10.1126/science.1087753. [DOI] [PubMed] [Google Scholar]
- 15.Bethea CL. MPA: medroxy-progesterone acetate contributes to much poor advice for women. Endocrinology. 2011;152(2):343–5. doi: 10.1210/en.2010-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bethea CL, Pau FK, Fox S, Hess DL, Berga SL, Cameron JL. Sensitivity to stress- induced reproductive dysfunction linked to activity of the serotonin system. Fertil Steril. 2005;83(1):148–55. doi: 10.1016/j.fertnstert.2004.06.051. [DOI] [PubMed] [Google Scholar]
- 17.Brenner RM, Slayden OD. Cyclic changes in the primate oviduct and endometrium. In: Knobil E, Neill JD, editors. The Physiology of Reproduction. Second Edition. 2 ed. Raven Press, Ltd.; New York: 1994. pp. 541–69. [Google Scholar]
- 18.Bethea C, Reddy A. Effect of ovarian hormones on survival genes in laser captured serotonin neurons from macaques. J Neurochem. 2008:1051129–43. doi: 10.1111/j.1471-4159.2008.05213.x. [DOI] [PubMed] [Google Scholar]
- 19.Bethea CL, Reddy AP. Effect of ovarian hormones on genes promoting dendritic spines in laser-captured serotonin neurons from macaques. Mol Psychiatry. 2010;15(10):1034–44. doi: 10.1038/mp.2009.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bethea CL, Reddy AP. Ovarian steroids increase glutamatergic related gene expression in serotonin neurons of macaques. Mol Cell Neurosci. 2012;49(3):251–62. doi: 10.1016/j.mcn.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bethea CL, Reddy AP. Effect of ovarian steroids on gene expression related to synapse assembly in serotonin neurons of macaques. J Neurosci Res. 2012;90(7):1324–34. doi: 10.1002/jnr.23004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dalman MR, Deeter A, Nimishakavi G, Duan ZH. Fold change and p-value cutoffs significantly alter microarray interpretations. BMC bioinformatics. 2012;13(Suppl 2):S11. doi: 10.1186/1471-2105-13-S2-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hotchkiss J, Knobil K. The menstrual cycle and its neuroendocrine control. In: Knobil E, Neill JD, editors. The Physiology of Reproduction. 2 ed. Raven Press; New York: 1994. pp. 711–50. [Google Scholar]
- 24.Kessing LV. Depression and the risk for dementia. Curr Opin Psychiatry. 2012;25(6):457–61. doi: 10.1097/YCO.0b013e328356c368. [DOI] [PubMed] [Google Scholar]
- 25.Hidasi Z, Salacz P, Csibri E. [Depression in neuropsychiatric diseases]. Ideggyogyaszati szemle. 2012;65(1-2):6–15. [PubMed] [Google Scholar]
- 26.Gallarda T. [Alzheimer's disease and depression]. L'Encephale. 1999 25 Spec No 514-7.; discussion 8. [PubMed] [Google Scholar]
- 27.Vajda FJ, Solinas C. Current approaches to management of depression in Parkinson's Disease. J Clin Neurosci. 2005;12(7):739–43. doi: 10.1016/j.jocn.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Berg D. Biomarkers for the early detection of Parkinson's and Alzheimer's disease. Neuro-degenerative diseases. 2008;5(3-4):133–6. doi: 10.1159/000113682. [DOI] [PubMed] [Google Scholar]
- 29.Politis M, Loane C. Serotonergic dysfunction in Parkinson's disease and its relevance to disability. Scientific World J. 2011;11:1726–34. doi: 10.1100/2011/172893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banerjee S, Hellier J, Romeo R, Dewey M, Knapp M, Ballard C, Baldwin R, Bentham P, Fox C, Holmes C, Katona C, Lawton C, Lindesay J, Livingston G, McCrae N, Moniz-Cook E, Murray J, Nurock S, Orrell M, O'Brien J, Poppe M, Thomas A, Walwyn R, Wilson K, Burns A. Study of the use of antidepressants for depression in dementia: the HTA-SADD trial--a multicentre, randomised, double-blind, placebo-controlled trial of the clinical effectiveness and cost-effectiveness of sertraline and mirtazapine. Health Technol Assess. 2013;17(7):1–166. doi: 10.3310/hta17070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leong C. Antidepressants for depression in patients with dementia: a review of the literature. Consultant Pharmacist. 2014;29(4):254–63. doi: 10.4140/TCP.n.2014.254. [DOI] [PubMed] [Google Scholar]
- 32.Chen CP, Eastwood SL, Hope T, McDonald B, Francis PT, Esiri MM. Immunocytochemical study of the dorsal and median raphe nuclei in patients with Alzheimer's disease prospectively assessed for behavioural changes. Neuropathol Applied Neurobiol. 2000;26(4):347–55. doi: 10.1046/j.1365-2990.2000.00254.x. [DOI] [PubMed] [Google Scholar]
- 33.Politis M, Wu K, Loane C, Quinn NP, Brooks DJ, Oertel WH, Bjorklund A, Lindvall O, Piccini P. Serotonin neuron loss and nonmotor symptoms continue in Parkinson's patients treated with dopamine grafts. Sci Translational Med. 2012;4(128):128ra41. doi: 10.1126/scitranslmed.3003391. [DOI] [PubMed] [Google Scholar]
- 34.Yamamoto T, Hirano A. Nucleus raphe dorsalis in Alzheimer's disease: neurofibrillary tangles and loss of large neurons. Ann Neurol. 1985;17(6):573–7. doi: 10.1002/ana.410170608. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto T, Hirano A. Nucleus raphe dorsalis in parkinsonism-dementia complex of Guam. Acta neuropathologica. 1985;67(3-4):296–9. doi: 10.1007/BF00687815. [DOI] [PubMed] [Google Scholar]
- 36.Myint AM, Kim YK. Cytokine-serotonin interaction through IDO: a neurodegeneration hypothesis of depression. Med Hypotheses. 2003;61(5-6):519–25. doi: 10.1016/s0306-9877(03)00207-x. [DOI] [PubMed] [Google Scholar]
- 37.Gardner A, Boles RG. Beyond the serotonin hypothesis: mitochondria, inflammation and neurodegeneration in major depression and affective spectrum disorders. Prog Neuropsychopharmacol Biol Psychiatry. 2011;35(3):730–43. doi: 10.1016/j.pnpbp.2010.07.030. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez JJ, Noristani HN, Verkhratsky A. The serotonergic system in ageing and Alzheimer's disease. Prog Neurobiol. 2012;99(1):15–41. doi: 10.1016/j.pneurobio.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 39.Tsang KL, Ho SL, Lo SK. Estrogen improves motor disability in parkinsonian postmenopausal women with motor fluctuations. Neurology. 2000;54(12):2292–8. doi: 10.1212/wnl.54.12.2292. [DOI] [PubMed] [Google Scholar]
- 40.Henderson VW. Estrogen, cognition, and a woman's risk of Alzheimer's disease. Am J Med. 1997;103(3A):11S–8S. doi: 10.1016/s0002-9343(97)00261-1. [DOI] [PubMed] [Google Scholar]
- 41.Henderson VW. Estrogen-containing hormone therapy and Alzheimer's disease risk: understanding discrepant inferences from observational and experimental research. Neuroscience. 2006;138(3):1031–9. doi: 10.1016/j.neuroscience.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 42.Simpkins JW, Green PS, Gridley KE, Singh M, de Fiebre NC, Rajakumar G. Role of estrogen replacement therapy in memory enhancement and the prevention of neuronal loss associated with Alzheimer's disease. Am J Med. 1997;103(3A):19S–25S. doi: 10.1016/s0002-9343(97)00260-x. [DOI] [PubMed] [Google Scholar]
- 43.Petanceska SS, Nagy V, Frail D, Gandy S. Ovariectomy and 17beta-estradiol modulate the levels of Alzheimer's amyloid beta peptides in brain. Neurology. 2000;54(12):2212–7. doi: 10.1212/wnl.54.12.2212. [DOI] [PubMed] [Google Scholar]
- 44.Bourque M, Dluzen DE, Di Paolo T. Signaling pathways mediating the neuroprotective effects of sex steroids and SERMs in Parkinson's disease. Front Neuroendocrinol. 2012;33(2):169–78. doi: 10.1016/j.yfrne.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 45.Tang MX, Jacobs D, Stern Y, Marder K, Schofield P, Gurland B, Andrews H, Mayeux R. Effect of oestrogen during menopause on risk and age at onset of Alzheimer's disease. Lancet. 1996;348(9025):429–32. doi: 10.1016/S0140-6736(96)03356-9. [DOI] [PubMed] [Google Scholar]
- 46.Imtiaz B, Tuppurainen M, Tiihonen M, Kivipelto M, Soininen H, Hartikainen S, Tolppanen AM. Oophorectomy, Hysterectomy, and Risk of Alzheimer's Disease: A Nationwide Case-Control Study. J Alzh Disease. JAD. 2014 doi: 10.3233/JAD-140336. [DOI] [PubMed] [Google Scholar]
- 47.Tejani-Butt SM, Yang J, Pawlyk AC. Altered serotonin transporter sites in Alzheimer's disease raphe and hippocampus. NeuroReport. 1995;6:1207–10. doi: 10.1097/00001756-199505300-00033. [DOI] [PubMed] [Google Scholar]
- 48.Benninghoff J, Schmitt A, Mossner R, Kesch K-P. When cells become depressed: focus on neural stems cells in novel treatment strategies against depression. J Neural Transm. 2002;109:947–62. doi: 10.1007/s007020200078. [DOI] [PubMed] [Google Scholar]
- 49.Manji HK, Duman RS. Impairments of neuroplasticity and cellular resilience in severe mood disorders: implications for the development of novel therapeutics. Psychopharmacol Bull. 2001;35(2):5–49. [PubMed] [Google Scholar]
- 50.Alfonso J, Pollevick GD, Van Der Hart MG, Flugge G, Fuchs E, Frasch AC. Identification of genes regulated by chronic psychosocial stress and antidepressant treatment in the hippocampus. Eur J Neurosci. 2004;19(3):659–66. doi: 10.1111/j.1460-9568.2004.03178.x. [DOI] [PubMed] [Google Scholar]
- 51.Bethea CL, Smith AW, Centeno ML, Reddy AP. Long-term ovariectomy decreases serotonin neuron number and gene expression in free ranging macaques. Neuroscience. 2012;49:251–62. doi: 10.1016/j.neuroscience.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lima FB, Centeno ML, Costa ME, Reddy AP, Cameron JL, Bethea CL. Stress sensitive female macaques have decreased fifth Ewing variant (Fev) and serotonin-related gene expression that is not reversed by citalopram. Neuroscience. 2009;164(2):676–91. doi: 10.1016/j.neuroscience.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dantuma NP, Heinen C, Hoogstraten D. The ubiquitin receptor Rad23: at the crossroads of nucleotide excision repair and proteasomal degradation. DNA Repair (Amst) 2009;8(4):449–60. doi: 10.1016/j.dnarep.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 54.Nouspikel T. DNA repair in mammalian cells : Nucleotide excision repair: variations on versatility. Cell Mol Life Sci. 2009;66(6):994–1009. doi: 10.1007/s00018-009-8737-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nouspikel T, Hanawalt PC. Terminally differentiated human neurons repair transcribed genes but display attenuated global DNA repair and modulation of repair gene expression. Mol Cell Biol. 2000;20(5):1562–70. doi: 10.1128/mcb.20.5.1562-1570.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9(12):958–70. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 57.Carney JP. Chromosomal breakage syndromes. Curr Opin Immunol. 1999;11(4):443–7. doi: 10.1016/S0952-7915(99)80074-0. [DOI] [PubMed] [Google Scholar]
- 58.Zhu Q, Chang Y, Yang J, Wei Q. Post-translational modifications of proliferating cell nuclear antigen: A key signal integrator for DNA damage response (Review). Oncology letters. 2014;7(5):1363–9. doi: 10.3892/ol.2014.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cao A, Zhang C. Sex-specific effects of androgen and estrogen on proliferation of the embryonic chicken hypothalamic neurons. Endocrine. 2007;31(2):161–6. doi: 10.1007/s12020-007-0026-7. [DOI] [PubMed] [Google Scholar]
- 60.Giglia-Mari G, Coin F, Ranish JA, Hoogstraten D, Theil A, Wijgers N, Jaspers NG, Raams A, Argentini M, van der Spek PJ, Botta E, Stefanini M, Egly JM, Aebersold R, Hoeijmakers JH, Vermeulen W. A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat Genet. 2004;36(7):714–9. doi: 10.1038/ng1387. [DOI] [PubMed] [Google Scholar]
- 61.Sultan FA, Sweatt JD. The role of the Gadd45 family in the nervous system: a focus on neurodevelopment, neuronal injury, and cognitive neuroepigenetics. Adv Exp Med Biol. 2013:79381–119. doi: 10.1007/978-1-4614-8289-5_6. [DOI] [PubMed] [Google Scholar]
- 62.Schafer A. Gadd45 proteins: key players of repair-mediated DNA demethylation. Adv Exp Med Biol. 2013:79335–50. doi: 10.1007/978-1-4614-8289-5_3. [DOI] [PubMed] [Google Scholar]
- 63.Muller P, Ruckova E, Halada P, Coates PJ, Hrstka R, Lane DP, Vojtesek B. C-terminal phosphorylation of Hsp70 and Hsp90 regulates alternate binding to co-chaperones CHIP and HOP to determine cellular protein folding/degradation balances. Oncogene. 2013;32(25):3101–10. doi: 10.1038/onc.2012.314. [DOI] [PubMed] [Google Scholar]
- 64.Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nat Rev Mol Cell Biol. 2013;14(10):630–42. doi: 10.1038/nrm3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dimant H, Ebrahimi-Fakhari D, McLean PJ. Molecular chaperones and co-chaperones in Parkinson disease. Neuroscientist. 2012;18(6):589–601. doi: 10.1177/1073858412441372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Head MW, Goldman JE. Small heat shock proteins, the cytoskeleton, and inclusion body formation. Neuropathol Applied Neurobiol. 2000;26(4):304–12. doi: 10.1046/j.1365-2990.2000.00269.x. [DOI] [PubMed] [Google Scholar]
- 67.Olazabal UE, Pfaff DW, Mobbs CV. Sex differences in the regulation of heat shock protein 70 kDa and 90 kDa in the rat ventromedial hypothalamus by estrogen. Brain Res. 1992;596(1-2):311–4. doi: 10.1016/0006-8993(92)91563-t. [DOI] [PubMed] [Google Scholar]
- 68.Chambraud B, Berry M, Redeuilh G, Chambon P, Baulieu EE. Several regions of human estrogen receptor are involved in the formation of receptor-heat shock protein 90 complexes. J Biol Chem. 1990;265(33):20686–91. [PubMed] [Google Scholar]
- 69.Berke SJ, Paulson HL. Protein aggregation and the ubiquitin proteasome pathway: gaining the UPPer hand on neurodegeneration. Current Opinion Genetics Develop. 2003;13(3):253–61. doi: 10.1016/s0959-437x(03)00053-4. [DOI] [PubMed] [Google Scholar]
- 70.Ross CA, Pickart CM. The ubiquitin-proteasome pathway in Parkinson's disease and other neurodegenerative diseases. Trends Cell Biol. 2004;14(12):703–11. doi: 10.1016/j.tcb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 71.Hol EM, Fischer DF, Ovaa H, Scheper W. Ubiquitin proteasome system as a pharmacological target in neurodegeneration. Expert Rev Neurotherapeutics. 2006;6(9):1337–47. doi: 10.1586/14737175.6.9.1337. [DOI] [PubMed] [Google Scholar]
- 72.Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2(3):169–78. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 73.Walden H, Martinez-Torres RJ. Regulation of Parkin E3 ubiquitin ligase activity. Cell Mol Life Sci. 2012;69(18):3053–67. doi: 10.1007/s00018-012-0978-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chevalier-Larsen E, Holzbaur EL. Axonal transport and neurodegenerative disease. Biochim Biophys Acta. 2006;1762(11-12):1094–108. doi: 10.1016/j.bbadis.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 75.Duncan JE, Goldstein LS. The genetics of axonal transport and axonal transport disorders. PLoS genetics. 2006;2(9):e124. doi: 10.1371/journal.pgen.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eschbach J, Dupuis L. Cytoplasmic dynein in neurodegeneration. Pharmacol Ther. 2011;130(3):348–63. doi: 10.1016/j.pharmthera.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 77.Bethea CL, Coleman K, Phu K, Reddy AP, Phu A. Relationships between androgens, serotonin gene expression and innervation in male macaques. Neuroscience. 2014:274341–56. doi: 10.1016/j.neuroscience.2014.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Henry JP, Sagne C, Bedet C, Gasnier B. The vesicular monoamine transporter: from chromaffin granule to brain. Neurochem Int. 1998;32(3):227–46. doi: 10.1016/s0197-0186(97)00092-2. [DOI] [PubMed] [Google Scholar]
- 79.Tsuno N, Homma A. What is the association between depression and Alzheimer's disease? Expert Rev Neurotherapeutics. 2009;9(11):1667–76. doi: 10.1586/ern.09.106. [DOI] [PubMed] [Google Scholar]
- 80.Anukulthanakorn K, Malaivijitnond S, Kitahashi T, Jaroenporn S, Parhar I. Molecular events during the induction of neurodegeneration and memory loss in estrogen-deficient rats. Gen Comp Endocrinol. 2013:181316–23. doi: 10.1016/j.ygcen.2012.07.034. [DOI] [PubMed] [Google Scholar]
- 81.Gandy S, Petanceska S. Regulation of Alzheimer beta-amyloid precursor trafficking and metabolism. Biochim Biophys Acta. 2000;1502(1):44–52. doi: 10.1016/s0925-4439(00)00031-4. [DOI] [PubMed] [Google Scholar]
- 82.Zheng H, Koo EH. The amyloid precursor protein: beyond amyloid. Mol Neurodegen. Jul. 2006;3:1–5. doi: 10.1186/1750-1326-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Szego EM, Csorba A, Janaky T, Kekesi KA, Abraham IM, Morotz GM, Penke B, Palkovits M, Murvai U, Kellermayer MS, Kardos J, Juhasz GD. Effects of estrogen on beta- amyloid-induced cholinergic cell death in the nucleus basalis magnocellularis. Neuroendocrinology. 2011;93(2):90–105. doi: 10.1159/000321119. [DOI] [PubMed] [Google Scholar]
- 84.Camacho-Arroyo I, Gonzalez-Arenas A, Espinosa-Raya J, Pina-Medina AG, Picazo O. Short- and long-term treatment with estradiol or progesterone modifies the expression of GFAP, MAP2 and Tau in prefrontal cortex and hippocampus. Life Sci. 2011;89(3-4):123–8. doi: 10.1016/j.lfs.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 85.Acx H, Chavez-Gutierrez L, Serneels L, Lismont S, Benurwar M, Elad N, De Strooper B. Signature amyloid beta profiles are produced by different gamma-secretase complexes. J Biol Chem. 2014;289(7):4346–55. doi: 10.1074/jbc.M113.530907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zheng H, Xu H, Uljon SN, Gross R, Hardy K, Gaynor J, Lafrancois J, Simpkins J, Refolo LM, Petanceska S, Wang R, Duff K. Modulation of A(beta) peptides by estrogen in mouse models. J Neurochem. 2002;80(1):191–6. doi: 10.1046/j.0022-3042.2001.00690.x. [DOI] [PubMed] [Google Scholar]
- 87.Nord LC, Sundqvist J, Andersson E, Fried G. Analysis of oestrogen regulation of alpha-, beta- and gamma-secretase gene and protein expression in cultured human neuronal and glial cells. Neuro-degenerative diseases. 2010;7(6):349–64. doi: 10.1159/000282279. [DOI] [PubMed] [Google Scholar]
- 88.Bethea CL. Colocalization of progestin receptors with serotonin in raphe neurons of macaque. Neuroendocrinology. 1993:571–6. doi: 10.1159/000126334. [DOI] [PubMed] [Google Scholar]
- 89.Gundlah C, Lu NZ, Mirkes SJ, Bethea CL. Estrogen receptor beta (ERb) mRNA and protein in serotonin neurons of macaques. Mol Brain Research. 2001:9114–22. doi: 10.1016/s0169-328x(01)00108-5. [DOI] [PubMed] [Google Scholar]
- 90.Cerillo G, Rees A, Manchanda N, Reilly C, Brogan I, White A, Needham M. The oestrogen receptor regulates NFkappaB and AP-1 activity in a cell-specific manner. J Steroid Biochem Mol Biol. 1998;67:79–88. doi: 10.1016/s0960-0760(98)00078-8. [DOI] [PubMed] [Google Scholar]
- 91.Meyer ME, Gronemeyer H, Turcotte B, Bocquel MT, Tasset D, Chambon P. Steroid hormone receptors compete for factors that mediate their enhancer function. Cell. 1989;57(3):433–42. doi: 10.1016/0092-8674(89)90918-5. [DOI] [PubMed] [Google Scholar]
- 92.Torchia J, Rose DW, Inostroza J, Kamei Y, Westin S, Glass CK, Rosenfeld MG. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387(6634):677–84. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 93.Charlier TD, Ball GF, Balthazart J. Inhibition of steroid receptor coactivator-1 blocks estrogen and androgen action on male sex behavior and associated brain plasticity. J Neurosci. 2005;25(4):906–13. doi: 10.1523/JNEUROSCI.3533-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Johnson AB, O'Malley BW. Steroid receptor coactivators 1, 2, and 3: critical regulators of nuclear receptor activity and steroid receptor modulator (SRM)-based cancer therapy. Mol Cell Endocrinol. 2012;348(2):430–9. doi: 10.1016/j.mce.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Heessen S, Masucci MG, Dantuma NP. The UBA2 domain functions as an intrinsic stabilization signal that protects Rad23 from proteasomal degradation. Mol Cell. 2005;18(2):225–35. doi: 10.1016/j.molcel.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 96.Gillette TG, Yu S, Zhou Z, Waters R, Johnston SA, Reed SH. Distinct functions of the ubiquitin-proteasome pathway influence nucleotide excision repair. EMBO J. 2006;25(11):2529–38. doi: 10.1038/sj.emboj.7601120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Franklin TB, Krueger-Naug AM, Clarke DB, Arrigo AP, Currie RW. The role of heat shock proteins Hsp70 and Hsp27 in cellular protection of the central nervous system. Int J Hypertherm. 2005;21(5):379–92. doi: 10.1080/02656730500069955. [DOI] [PubMed] [Google Scholar]
- 98.Latchman DS. HSP27 and cell survival in neurones. Int J Hypertherm. 2005;21(5):393–402. doi: 10.1080/02656730400023664. [DOI] [PubMed] [Google Scholar]
- 99.Kotoglou P, Kalaitzakis A, Vezyraki P, Tzavaras T, Michalis LK, Dantzer F, Jung JU, Angelidis C. Hsp70 translocates to the nuclei and nucleoli, binds to XRCC1 and PARP-1, and protects HeLa cells from single-strand DNA breaks. Cell Stress Chaperones. 2009;14(4):391–406. doi: 10.1007/s12192-008-0093-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Niu P, Liu L, Gong Z, Tan H, Wang F, Yuan J, Feng Y, Wei Q, Tanguay RM, Wu T. Overexpressed heat shock protein 70 protects cells against DNA damage caused by ultraviolet C in a dose-dependent manner. Cell Stress Chaperones. 2006;11(2):162–9. doi: 10.1379/CSC-175R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Quanz M, Herbette A, Sayarath M, de Koning L, Dubois T, Sun JS, Dutreix M. Heat shock protein 90alpha (Hsp90alpha) is phosphorylated in response to DNA damage and accumulates in repair foci. J Biol Chem. 2012;287(12):8803–15. doi: 10.1074/jbc.M111.320887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Daturpalli S, Waudby CA, Meehan S, Jackson SE. Hsp90 inhibits alpha-synuclein aggregation by interacting with soluble oligomers. J Mol Biol. 2013;425(22):4614–28. doi: 10.1016/j.jmb.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 103.Pratt WB, Galigniana MD, Harrell JM, DeFranco DB. Role of hsp90 and the hsp90- binding immunophilins in signalling protein movement. Cell Signal. 2004;16(8):857–72. doi: 10.1016/j.cellsig.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 104.Ekstrand MI, Nectow AR, Knight ZA, Latcha KN, Pomeranz LE, Friedman JM. Molecular profiling of neurons based on connectivity. Cell. 2014;157(5):1230–42. doi: 10.1016/j.cell.2014.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shughrue PJ, Lane MV, Merchenthaler I. Comparative distribution of estrogen receptor-a and -b mRNA in the rat central nervous system. J Comp Neurol. 1997:388507–25. doi: 10.1002/(sici)1096-9861(19971201)388:4<507::aid-cne1>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 106.Schmidt PJ, Rubinow DR. Sex hormones and mood in the perimenopause. Ann N Y Acad Sci. 2009:117970–85. doi: 10.1111/j.1749-6632.2009.04982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Taler M, Miron O, Gil-Ad I, Weizman A. Neuroprotective and procognitive effects of sertraline: in vitro and in vivo studies. Neurosci Lett. 2013:55093–7. doi: 10.1016/j.neulet.2013.06.033. [DOI] [PubMed] [Google Scholar]
- 108.Schneider LS, Small GW, Hamilton SH, Bystritsky A, Nemeroff CB, Meyers BS. Estrogen replacement and response to fluoxetine in a multicenter geriatric depression trial. Fluoxetine Collaborative Study Group. Am J Geriatr Psychiatry. 1997:597–106. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.