Abstract
Hydroxyapatite (HA) is the primary structural component of the skeleton and dentition. Under biological conditions, HA does not occur spontaneously and therefore must be actively synthesized by mineralizing cells such as osteoblasts. The mechanism(s) by which HA is actively synthesized by cells and deposited to create a mineralized matrix are not fully understood and the consequences of mineralization on cell function are even less well understood. HA can be chemically synthesized (HAp) and is therefore currently being investigated as a promising therapeutic biomaterial for use as a functional scaffold and implant coating for skeletal repair and dental applications. Here we investigated the biological effects of nano-HAp (10×100 nm) on the lineage commitment and differentiation of bone forming osteoblasts. Exposure of early stage differentiating osteoblasts resulted in dramatic and sustained changes in gene expression, both increased and decreased, whereas later stage osteoblasts were much less responsive. Analysis of the promoter region one of the most responsive genes, alkaline phosphatase, identified the stimulation of DNA methylation following cell exposure to nano-HAp. Collectively, the results reveal the novel epigenetic regulation of cell function by nano-HAp which has significant implication on lineage determination as well as identifying a novel potential therapeutic use of nanomaterials.
Keywords: hydroxyapatite, nanoparticle, gene expression, alkaline phosphatase, osteoblast, osteogenesis
1. Introduction
Mineral hydroxyapatite (HA) is naturally synthesized and makes up 70% of the skeleton and 90% of tooth enamel [1]. HA consists mainly of calcium and phosphate Ca5(PO4)3(OH) (or more often referred to as the two unit crystal form: Ca10(PO4)6(OH)2) at a ratio of 1.67 and is crystalline in form. Calcium (Ca) and phosphorus (P) represent two of the most common elements in nature as well as in the mammals [1]. Accordingly, these two ions have a particularly important relationship in biology and can exist in multiple forms (ratios) such as mono, di, and tri-calcium phosphate (ratios from 0.5 to 1.5) [2]. Recent studies have suggested that various forms of nano-Calcium-Phosphate (CaP) synthesized by the body are associated with functional mineralization of bone and pathological calcification of vascular smooth muscle [3–7]. Nano-sized hydroxyapatite (nano-HAp) is generated endogenously by osteoblasts in the form of matrix vesicles as the initiator of bone formation in the skeleton, [8] as well as in pathological calcification of cartilage and vasculature [9–12] and can be deposited in soft tissues in the form of dystrophic and metastatic calcifications, [13, 14] although the origins are not completely understood. Surprisingly, the effects of hydroxyapatite on cell function remain relatively unknown.
Synthesis of HA during functional mineralization is a controlled process by which differentiating osteoblasts concentrate calcium and phosphate to form HA which is then deposited on a collagen matrix. The concentration and deposition of HA is thought to occur through the secretion of matrix vesicles by osteoblasts. Matrix vesicles are lipid membrane vesicles of approximately 50 – 200 nm which contain HA and initiate matrix mineralization [8, 15, 16]. Osteoblasts derive from the mesenchyme lineage and start as multipotent bone marrow stromal cells and through a reasonably well defined differentiation process become mineralizing osteoblasts and ultimately become embedded in bone as mature osteocytes [17]. The osteoblast differentiation process can generally be divided into three distinct stages that are defined by 1) proliferation, 2) matrix maturation, and 3) mineralization [18, 19]. A number of marker genes/proteins are expressed at high levels for discrete periods of time during differentiation including, among others; alkaline phosphatase (ALP), bone sialoprotein (BSP), osteopontin (OPN), and osteocalcin (OSC) [20–22]. Early in the differentiation process (days 1 – 4), osteoblasts become confluent, exit the cell cycle, and respond to ascorbic acid with increased ALP RNA expression and enzymatic activity and deposition of a collagen matrix [18]. ALP is a membrane-bound ectoenzyme situated so that the catalytic subunit is extracellular. A number of functions have been proposed for the enzyme including cleavage of the mineralization inhibitor inorganic pyrophosphate (PPi) and generation of high local levels of inorganic phosphate [23]. ALP is critical for bone formation and mutations in or deletion of the gene (Alpl) results in bone defects in humans and mice [23–25]. An identified, but poorly understood aspect of lineage determination and progression is DNA methylation. A number of genes important for skeletal regulation, including ALP, have been suggested as targets of methylation [26–29] although the mechanisms and physiological relevance remain to be fully elucidated [30].
The use of nanotechnology is particularly well suited for use as biomaterials because control of structural properties and synthesis methods allow for manipulation of strength, shape, size, and surface for increased bioactivity and therapeutic benefit. Recently, HA is being investigated for use in various nanomaterials for its potential biocompatibility as well as osteoinductive properties [31–33]. Synthetic hydroxyapatite (HAp) is essentially identical to naturally occurring HA and has therefore become a commonly investigated biomaterial for use in skeletal, orthopedic, and dental repair, in part, by enhancing cell attachment, proliferation, and mineral formation [34–37]. Nano-HAp is now being investigated as a biomaterial to be used; directly as a therapeutic, a major component of bone graft substitutes, and an implant coating to improve biocompatibility and skeletal repair (reviewed in [36, 38–40]). However, in most cases these studies have focused on surface coatings and to date, the physicochemical properties of naturally occurring or engineered nano-HAp are not completely understood and the potential effects on genome regulation and cell function are only beginning to be explored.
To better understand the role of HAp in regulating the osteoblast lineage three different cell models were used; undifferentiated: bone marrow stromal cells (BMSCs) [41–43], pre-osteoblasts (MC3T3-E1) [20, 44], and terminally differentiated osteoblasts (osteocytes: MLO-Y4) [45]. Treatment of either bone marrow stromal cells or pre-osteoblasts with increasing concentrations of nano-HAp strongly altered expression of a number of genes including increased OPN, similar to our previous results with phosphate (Pi) alone [46]. Unexpectedly, nano-HAp stimulated a strong dose dependent suppression of the ALP, BSP and OSC RNA levels, which was different than Pi alone was identified. Significantly, the suppression was sustained for weeks even in the absence of nano-HAp suggesting a potentially permanent alteration in genomic output and implicative of epigenetic regulation. Quantification of methylation of a promoter region of ALP (gene: Alpl) in BMSCs defined a significant increase in response to nano-HAp treatment. Collectively, the results identify for the first time that nano-HAp is a potent regulator of the osteoblast lineage through changes in gene expression and identify methylation as a novel regulatory mechanism.
2. Materials and methods
2.1 Synthesis of hydroxyapatite nanomaterials
Pure phase of hydroxyapatite (HAp: Ca10(PO4)6(OH)2) was synthesized using a sonochemistry based precipitation method. 500 ml of 1 M calcium hydroxide (Ca(OH)2 at 99.0%, High Purity Chemical, Japan) aqueous solution was initially stirred vigorously using an overhead mechanical stirrer for 1 hour at room temperature. Then, 500 ml of a 0.6 M phosphoric acid aqueous solution (H3PO4, 85.0%, Junsei Chemical Co., Ltd., Japan) was slowly added drop wise into the cation solution with speed of 12.5 ml/min using a digital burette (876 Dosimat plus, Metrohm, Switzerland). During the whole reaction, sonication was given for complete mixing between Ca and P to prevent formation of nonstoichiometric HAp. The precipitates were aged for 24 hours for complete reaction. The HAp slurries were filter-pressed to collect precipitates and lyophilized to get non-agglomerated HAp nanoparticles (nano-HAp). To confirm that the stoichiometric HAp was synthesized, pH levels of the HAp slurry at the final stage of reaction and dried nano-HAP mixed in distilled water were measured by pH meter (Lab 860, Schott, Germany). The molar ratio between Ca and P in the synthesized nano-HAp was confirmed by inductively coupled plasma atomic emission spectrometer (Optima 4300 DV, PerkinElmer Inc., USA). In addition, homogeneous phase of nano-HAp was confirmed by X-ray Diffraction (M18XHFSRA, MAC Science Co., Japan) with monochromatic Cu Kα radiation (λ = 1.5405 Å). The morphology and the size of HAp nanoparticles were observed by high resolution transmission electron microscope (Tecnai F20, FEI, USA). Surface charge of HAp in distilled water was characterized by zeta potential (Otsuka Model ELS-Z, Otsuka Electronics Co., Ltd., Japan). Characteristics of vibration between metal and oxide were confirmed by Fourier transform infrared spectrometer (Nicolet 6700; Thermo Scientific, Waltham, MA) with attenuated total reflectance mode.
2.2 Synthesis of silica nanoparticles
Silica nanoparticles were synthesized by Stöber method [41, 47]. Tetraethyl orthosilicate (2.8 ml) (Sigma-Aldrich, St. Louis, MO) was added in 115 ml of ethanol anhydrous (Sigma-Aldrich), and 8 ml of 14% ammonia, diluted from a 28% concentrated stock (Sigma-Aldrich), was added with stirring for 16 hours. The silica nanoparticles were centrifuged, washed 4 times, and the nanoparticles were dispersed in sterilized water for cell culturing. The size and morphology was observed by transmission electron microscope, TEM (H-7600, Hitachi, Tokyo, Japan).
2.3 Calcium Oxide particles
Calcium Oxide particles were purchased from Nanoscale Materials Inc. (Manhattan, KS) as a dry powder and were resuspended in deionized water at a concentration of 10 mg/ml with vortexing and sonication.
2.4 Cell culture
Murine bone marrow stromal cells (BMSCs) were obtained from red marrow by centrifugation of the femur as described in [42] and cultured for up to 20 passages. BMSCs and pre-osteoblast MC3T3-E1 cells [20, 44] were cultured in α-Modified Eagle’s Medium (α-MEM; Thermo Scientific) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA) [43, 48]. Murine osteocyte, MLO-Y4 cells were kindly provided by Lynda F. Bonewald (University of Missouri–Kansas City, MO) and described in [45, 49] and cultured in α-MEM supplemented with 5% FBS and 5% calf serum (Gibco, Life technologies, Grand Island, NY) on collagen coated plates [45]. All growth medium were supplemented with 1% L-glutamine (Invitrogen, Life technologies), and 1% of penicillin/streptomycin (Thermo Scientific). All cell lines were cultured at 37° C in 5% CO2. Experimental medium consisted of growth medium with the addition of 50 µg/ml L-ascorbic acid (AA) (Sigma-Aldrich) as indicated with medium change twice weekly as described previously [21]. For in situ alkaline phosphatase staining, cell layers were overlaid with nitro blue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolyl-phosphate (BCIP) (Promega, Madison, WI) as described previously [20].
2.5 Cell viability
Cell viability was measured for all cell types using an XTT (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxylmethoxyphenyl)-2-(4-sulfonyl)-2H-tetrazolium) assay according to the manufacturer’s protocol (Promega). Cells were plated at ~ 5 × 103 cells per well (~ 50 – 60% confluent) in 96 well plates and nanoparticles were added after 24 hours as indicated. After 3 days the cells were analyzed using XTT (20 µl) assay reagent on a spectrophotometer (SpectraMAX250, Molecular Devices, CA, USA). The concentration of nanoparticles tested was 0, 10, 50, and 100 µg/ml, and the Pi (in the form of Na2HPO4, pH 6.8) was added at 0, 1, 4, and 8 mM (final concentration of 1, 2, 5, and 9 mM respectively as the medium contains 1 mM Pi).
2.6 RNA isolation, cDNA synthesis, and qRT-PCR
RNA was extracted using TRIzol reagent following the manufacturer’s protocol (Invitrogen). The RNA concentration was quantified by spectrophotometer (Nanodrop, Thermo Scientific) and complementary DNA (cDNA) was synthesized using QuantiTech Reverse Transcription kit (Qiagen, Valencia CA). qRT-PCR was performed using VeriQuest SYBR Green qPCR master mix (Affymetrix, Santa Clara CA) on a StepOnePlus thermocycler (Applied Biosystems, Life technologies). Primers were designed using qPrimerDepot software (http://mouseprimerdepot.nci.nih.gov/) and synthesized by Integrated DNA Technologies, Inc. (Coralville, IA) with sequences as follows; 18S (Rn18s) (control) (NR_003278): F-5’-TTGACGGAAGGGCACCACCAG-3’, R-5’-GCACCACCACCCACGGAATCG-3’. ALP (NM_007431): F-5’-ACAGACCCTCCCCACGAGT-3’, R-5’-TGTACCCTGAGATTCGTCCC-3’. BSP (NM_008318): F-5’-CGGCCACGCTACTTTCTTTA-3’, R-5’-CCTCTTCGGAACTATCGCAG-3’. OPN (NM_009263): F-5’-ATTTGCTTTTGCCTGTTTGG-3’, R-5’-TGGCTATAGGATCTGGGTGC-3’. OSC (NM_007541): F-5’-AAGCAGGAGGGCAATAAGGT-3’, R-5’-CAAGCAGGGTTAAGCTCACA-3’. Fold change was calculated using the 2-ΔΔCT method [50] and 18S values varied by less than 1 Ct (cycle threshold) across samples for individual experiments.
2.7 Genomic DNA isolation and methylation assay by MSREs
For the methylation study genomic DNA was extracted using DNeasy Blood & Tissue Kit (Qiagen). The genomic DNA concentration and quality were measured by spectrophotometer (Nanodrop, Thermo Scientific). The OneStep qMethyl™ Kit (Zymo Research Corporation, Irvine, CA) is used for the detection of region-specific DNA methylation via the selective amplification of a methylated region of DNA. This is accomplished by splitting any DNA to be tested into two parts: a "Test Reaction" and a "Reference Reaction". DNA in the Test Reaction is digested with Methylation Sensitive Restriction Enzymes (MSREs) while DNA in the Reference Reaction is not. The DNA from both samples is then amplified using real-time PCR in the presence of SYTO®9 fluorescent dye and quantitated [51]. The assay for the murine ALPL promoter was designed to the plus strand of the Alpl sequence in the UCSC browser, Assembly Dec.2011 (GRCm38/mm10). Murine Alkaline Phosphatase (Alpl) Promoter Region chr4:137795867-137796483-+strand resulting in a 253 bp amplicon with 9 cut sites. Forward Primer ATCATCCTGCCAGGCCACAAGAGAT (Sense). Reverse Primer CCAGAGTACGCTCCCGCCACTG (AntiSense) (Zymo).
3. Results
3.1 Synthesis and characterization of nano-HAp, SiO2, and CaO
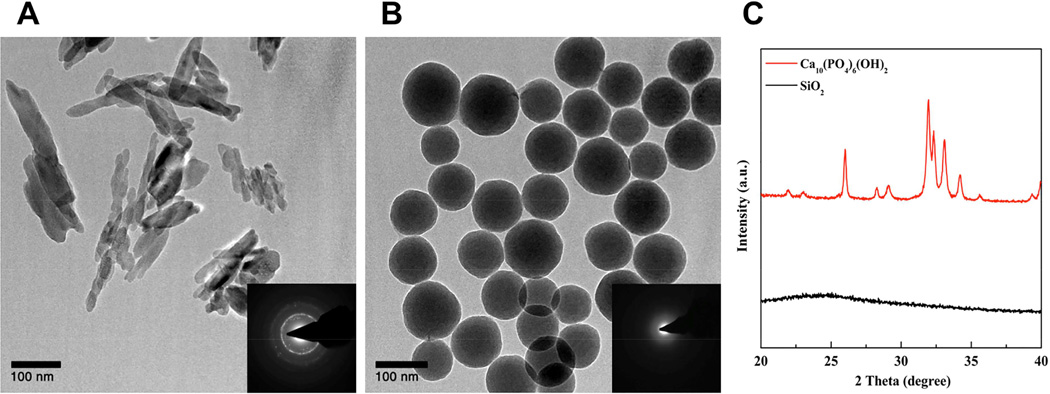
Nano-HAp was synthesized by a sonochemistry-based precipitation method. Calcium hydroxide and phosphoric acid were used as Ca and Pi source, respectively. After 24 hours of mixing, HAp was obtained by filter press and lyophilization. The nano-HAp was characterized by powder X-ray diffractometry (XRD), and its morphology, size, and surface charge were characterized by TEM and zeta potential. Nano-HAp particles were identified as nano-sized and rod-like shaped (Fig. 1) with crystallinity demonstrated by electron diffraction (Fig. 1-insets). TEM identified the size as approximately 100 nm in length and 10 nm in width (Fig. 1A) with a surface charge of −1.83 ± 0.52 mV in water. Powder XRD revealed that the synthesized material is hydroxyapatite, matching the JCPDS card (84–1998) (Fig. 1C). To determine the biological significance of the size of nano-HAp we chose 100 nm silica nanoparticles; a biomaterial with similar size but different composition. The silica nanoparticles were synthesized by the Stöber method as previously reported [52]. Their size was determined to be 113 ± 19 nm by zeta sizer (Supplementary S-Table 1), and surface charge was −41.7 ± 2.6 mV in water (Supplementary S-Table 1). Silica nanoparticles are amorphous when synthesized by the Stöber method and therefore X-ray diffraction did not produce dot patterns and powder XRD did not identify peaks, as expected. In addition, FT-IR analysis (Supplementary Fig. S1) revealed the characteristic peaks by P-O vibrations (1020 cm−1 and 960 cm−1) from HAp and by Si-O vibrations (1050 cm−1 for Si-O-Si, 945 cm−1 for Si-OH, and 795 cm−1 for Si-O) from silica nanoparticles.
Figure 1. Characterization of materials.
(A) Transmission Electron Microscopy (TEM) images of synthesized nano-HAp and (B) silica nanoparticles and (C) powder X-ray diffractograms. The inset of TEM images is electron diffraction patterns by TEM
3.2 Nano-HAp is not cytotoxic in osteoblast lineage cells
We first evaluated the general cytotoxicity of our new material. Nano-HAp was added in increasing concentrations for three days to murine cells representing three stages of osteoblast lineage commitment; BMSCs (bone marrow stromal cells - non-committed), MC3T3-E1 (pre-osteoblasts), and MLO-Y4 (osteocytes) and cell viability measured by XTT assay. Results identified cell viability of greater than 80% in all the tested cell types up to the maximum tested concentration of 100 µg/ml (Supplementary Fig. S2A). We also tested our reference materials (SiO2, CaO, and inorganic phosphate (Pi); in the form of Na2HPO4 (pH 6.8)) in BMSCs for 3 days. Silica is a well-known biocompatible material and we have previously shown 50 nm silica nanoparticles to stimulate osteoblast differentiation [48]. Here we used 100 nm silica nanoparticles to better approximate the size of our nano-HAp. CaO is an analog material with crystallinity to determine whether these biological effects are due to composition and or structure. Pi is a main component of nano-HAp and a critical ion for energy metabolism and bone tissues. Pi is necessary for mineralization as part of the osteoblastic differentiation process and has been extensively studied for its ability to stimulate changes in gene expression in mineralizing cells [18, 21]. Based on our theoretical calculation only 0.15 µmol of Pi and 0.25 µmol of Ca is necessary for the synthesis of 25 µg of nano-HAp, however, a 4 mM concentration was used to maximize the Pi effect, based on previous studies [42] and in addition to the 1 mM in the cell culture medium. No significant decrease in cell viability was identified (Supplementary Fig. S2B) and Pi, as expected, demonstrated a dose dependent increase in viability reflective of an increase in proliferation [22].
3.3 Nano-HAp strongly influences gene expression in bone marrow stromal cells
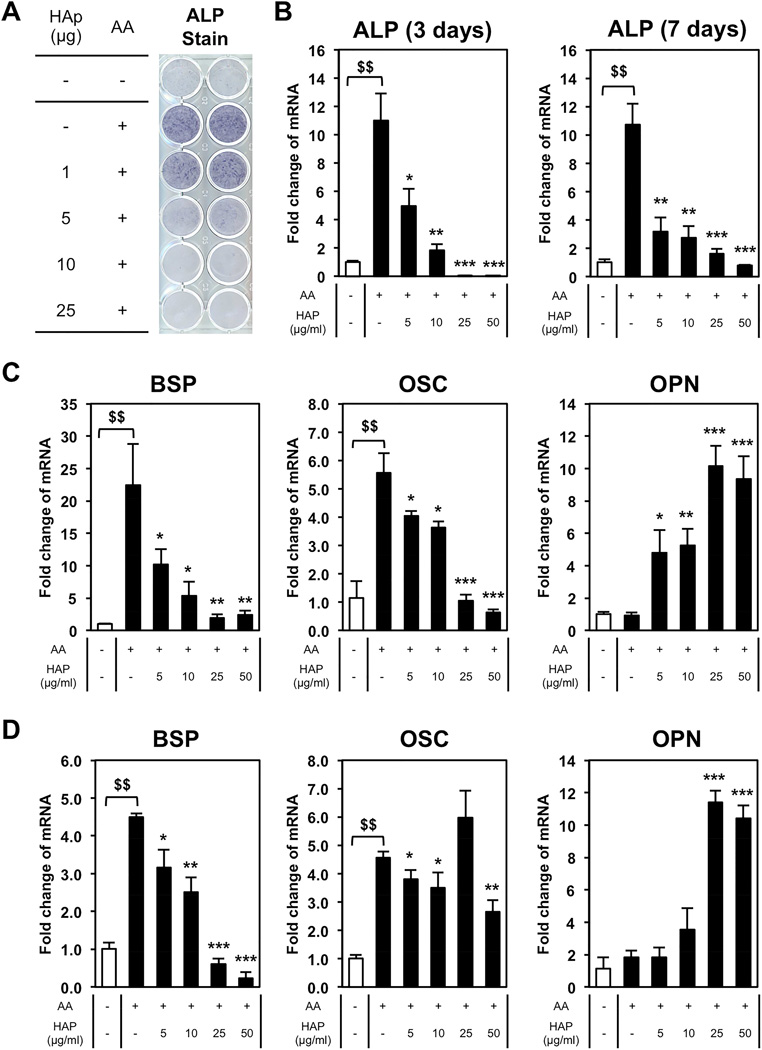
To determine the effect of Nano-HAp on osteoblast differentiation BMSCs were cultured in medium containing ascorbic acid (AA), required for differentiation and collagen matrix formation [22] and treated with increasing concentrations of nano-HAp. An increase in alkaline phosphatase (ALP) is one of the earliest markers of osteoblast differentiation and therefore cells were stained in situ for ALP enzyme activity after 7 days of treatment. The addition of AA resulted in the expected increase in ALP however, surprisingly, nano-HAp dose dependently decreased ALP activity (Fig. 2A). To determine if the effect was primarily on the protein or on gene expression, RNA was harvested from cells cultured in parallel and results revealed nano-HAp dose dependently decreased ALP RNA levels after 3 and 7 days of treatment (Fig. 2B). To investigate whether this effect is specific to ALP or represents a more general regulation of osteoblast genes we measured expression of other osteoblast markers; BSP, OSC, and OPN. Treatment with increasing concentrations of nano-HAp also resulted in a dose-dependent decrease in BSP and OSC expression, while OPN was dose-dependently increased at both time points (Fig. 2C, D).
Figure 2. Dose-dependency of hydroxyapatite in bone marrow stromal cells for 3 and 7 days.
BMSCs were cultured with or without 50 µg/ml of L-ascorbic acid (AA) and treated with nano-HAp (µg/ml) as indicated and incubated for 3 or 7 days. (A) Cells were stained for alkaline phosphatase (ALP) after 7 days. (B) Parallel cultures were harvested at 3 and 7 days and RNA levels measured by qRT-PCR. (C) The 3 day RNA samples described in (B) were analyzed by qRT-PCR for additional osteoblast marker genes. (D) The 7 day RNA samples described in (B) were analyzed by qRT-PCR for additional osteoblast marker genes. All results were normalized to 18S and are expressed as fold change from control +/− SD of 3 replicates. $$p<0.005 compared to untreated control; *p<0.05, **p<0.005, and ***p<0.0005 compared to the AA treated (Student’s t-test).
3.4 Nano-HAp induced changes in gene expression are different from SiO2, CaO, or phosphate
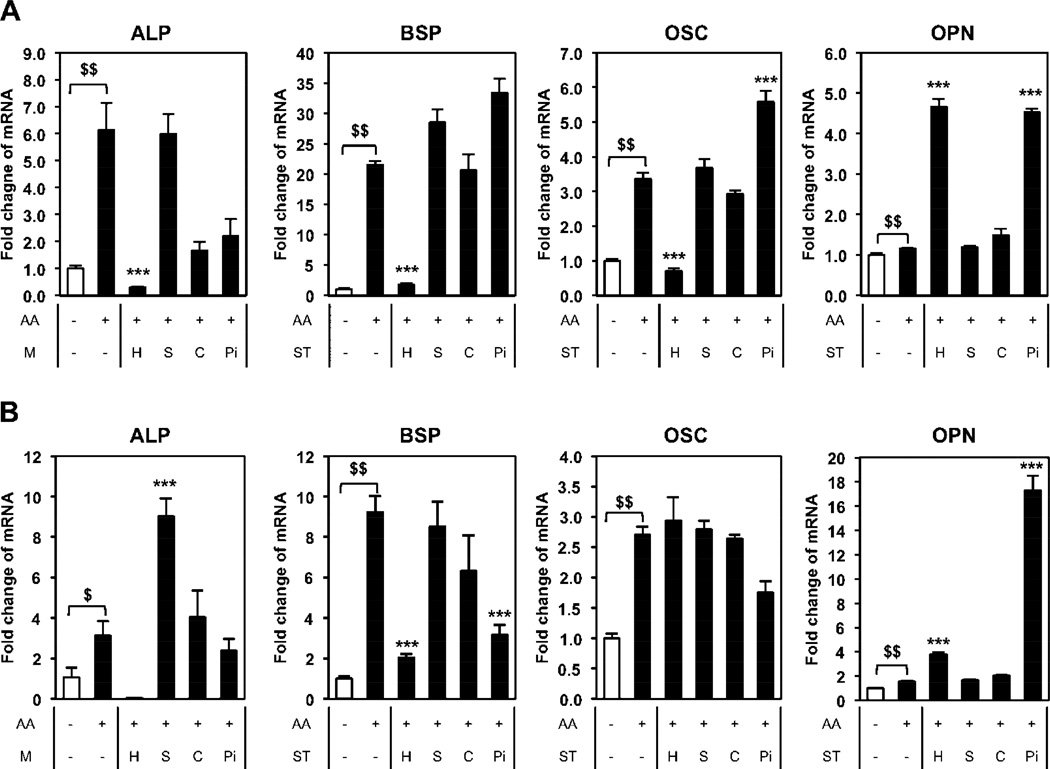
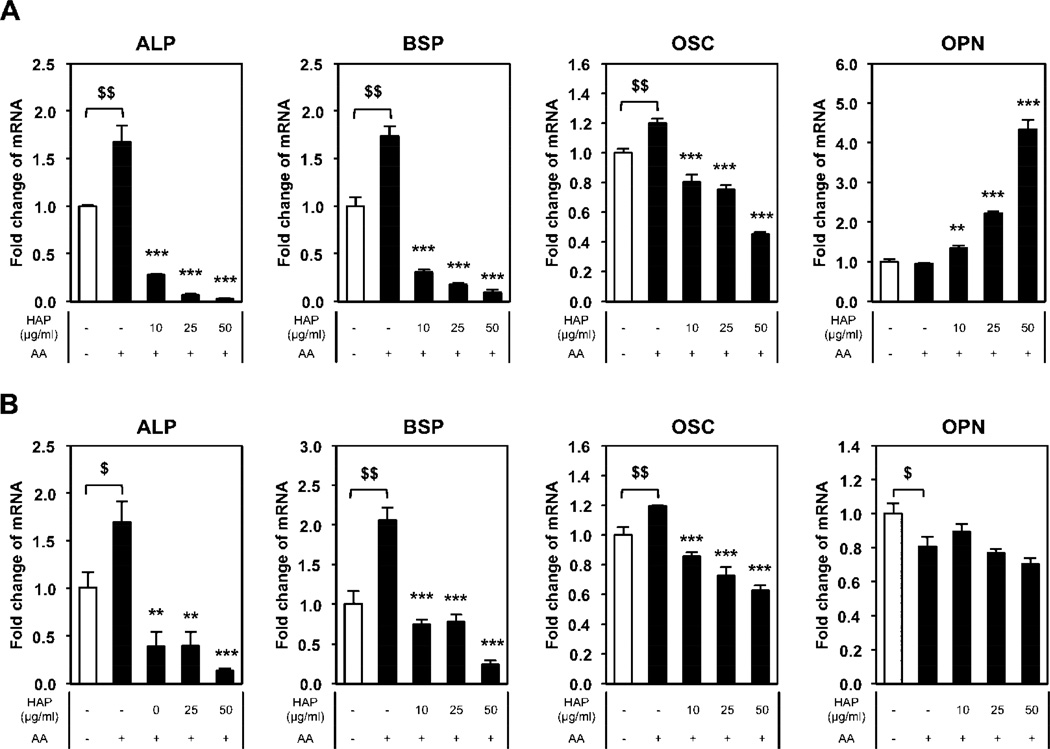
To begin to understand the physicochemical properties of nano-HAp required for the dramatic regulation of osteoblast gene expression we utilized nanoparticles of similar size (100 nm silica nanoparticles) and materials with slightly altered composition, calcium oxide (CaO) and phosphate (Pi) as described above. BMSCs were treated with the different materials for three (Fig. 3A) or seven (Fig. 3B) days and gene expression analyzed. Treatment with nano-HAp down regulated ALP, BSP, and OSC, whereas the SiO2 nanoparticles did not decrease any of the genes analyzed, and in fact significantly increased ALP at day 7. Although CaO and Pi decreased ALP RNA somewhat at day 3 there was little effect by day 7. Similar results were obtained with BSP and OSC with nano-HAp being the only material to produce a consistent decrease in the three genes over the two time points. OPN has been previously identified to respond to high Pi [46] and interestingly both nano-HAp and Pi produced a stimulation of OPN although Pi was much more potent at day 7 than day 3 (Fig. 3A, B). Collectively, the results identify a unique cellular response to nano-HAp and demonstrate that osteoblast lineage cells have the capacity to recognize and respond to different compositions of calcium and phosphorus.
Figure 3. Comparison of nano-HAp (H) with silica nanoparticles (S), calcium oxide (C), and phosphate (Pi).
BMSCs were cultured with or without 50µg/ml of L-ascorbic acid (AA) as indicated and treated with 25 µg/ml of nano-HAp (H), silica nanoparticles (S), and calcium oxide (C) or 4 mM Pi (Pi) for (A) 3 days or (B) 7 days. The results were normalized to 18S and are expressed as fold change from control +/− SD of 3 replicates. $p<0.05 and $$p<0.005 compared to untreated control and ***p<0.0005 compared to the AA treated (Student’s t-test). M: Material.
3.5 Nano-HAp has less influence on more differentiated osteoblasts
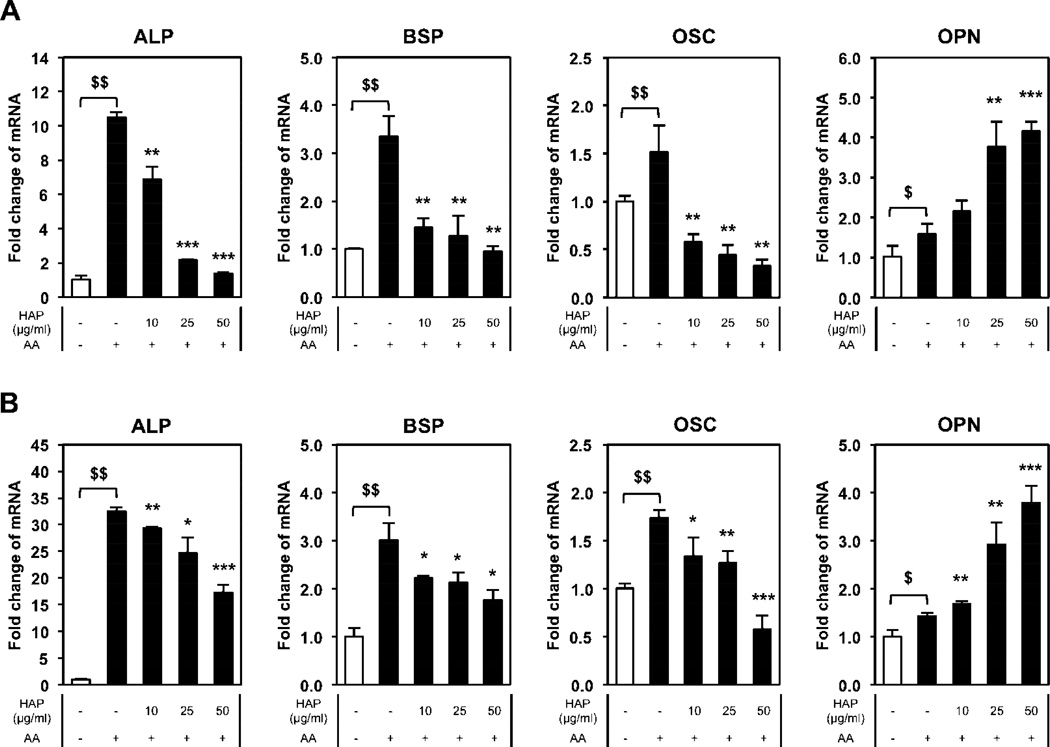
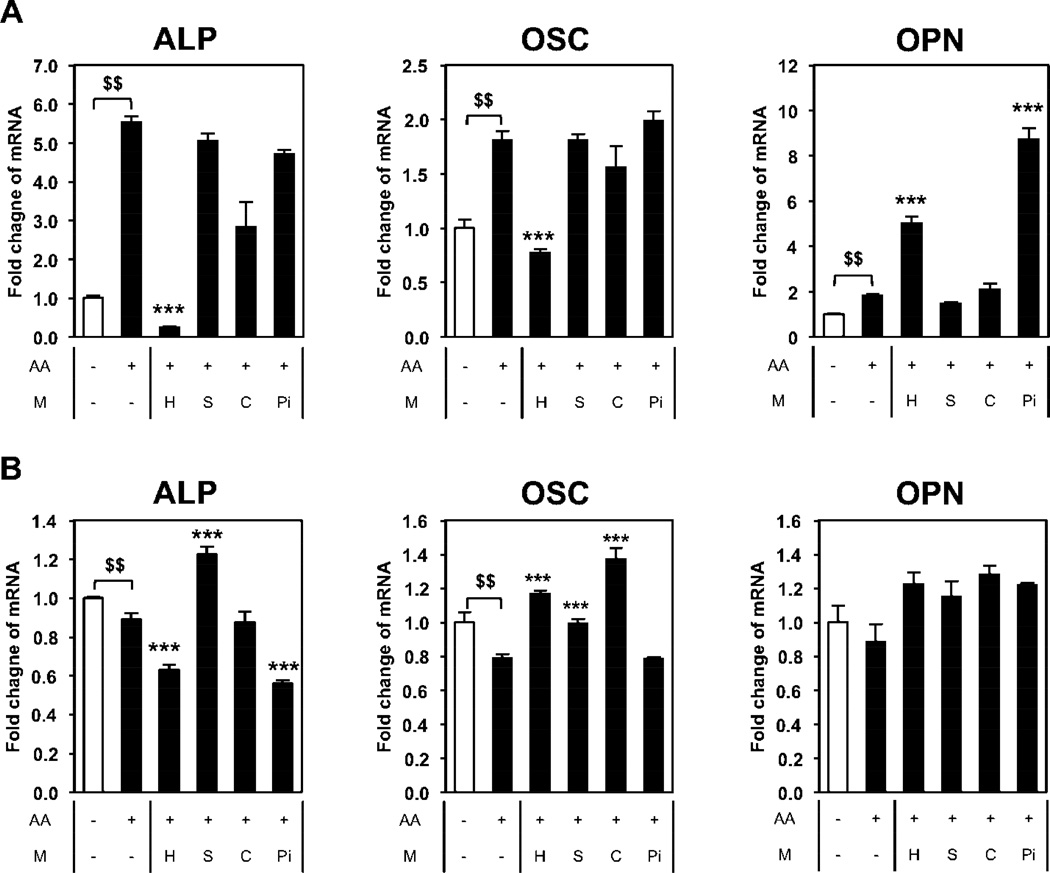
ALP is an important gene/protein in the process of osteoblast differentiation and is expressed relatively early in the process, starting before the cells mineralize and declining during the mineralization phase [20, 21]. To determine if nano-HAp has a similar or differential effect on cells depending on the state of differentiation, we differentiated BMSCs to osteoblasts for 4 or 21 days prior to treatment with nano-HAp for three days. In early stage osteoblasts (7 days) nano-HAp was effective at suppressing ALP gene expression however after 21 days of differentiation (mature osteoblasts) nano-HAp was much less effective at regulating ALP gene expression (Fig. 4A, B). To confirm that the effect of nano-HAp is different at different stages of osteoblast differentiation we utilized MC3T3-E1 cells which is a committed pre-osteoblast cell line and MLO-Y4 cells which represent terminally differentiated osteoblasts (osteocytes). Cells were treated with the different materials and RNA harvested after 3 days. MC3T3-E1 cells responded to the different materials similarly to BMSCs (Fig. 5A) as BMSCs although to somewhat lesser degree (Fig. 3A). However, the most differentiated cells, MLO-Y4, demonstrated a substantially blunted response to nano-HAp with ALP only partially responding and with no effect on OSC and OPN expression (Fig. 5B). The results suggest that nano-HAp produces less of an influence on gene expression as cells progress along the osteoblast lineage.
Figure 4. Nano-HAp has less influence on more differentiated osteoblast-like cells.
BMSCs were differentiated for total 7 days (A) and 24 days (B) with 50 µg/ml of L-ascorbic acid (AA). Cells were treated with nano-HAp (µg/ml) for last 3 days before harvesting RNA. $p<0.05 and $$p<0.005 compared to untreated control; *p<0.05, **p<0.005, and ***p<0.0005 compared to the AA treated (Student’s t-test).
Figure 5.
Cell specificity of nano-HAp. (A) MC3T3-E1 (pre-osteoblast) or (B) MLO-Y4 (osteocyte) were cultured with or without 50 µg/ml of L-ascorbic acid (AA) and treated with 25 µg/ml of nano-HAp (H), silica nanoparticles (S), and calcium oxide (C) or 4 mM Pi (Pi) for 3 days. The results were normalized to 18S and are expressed as fold change from control +/− SD of 3 replicates. $$p<0.005 compared to untreated control and ***p<0.0005 compared to the AA treated (Student’s t-test). M: Material.
3.6 A short exposure to nano-HAp results in long term suppression of ALP expression
The substantially blunted response of more differentiated cells to nano-HAp suggested the possibility that the mechanism(s) by which nano-HAp alters gene expression was no longer functional and therefore “turned off” as the cells differentiate. To investigate the temporal lineage dependent response to nano-HAp BMSCs were exposed to nano-HAp for 3 days “pulsed” followed by replacing the treated cells with non-nano-HAp containing medium and incubating for an additional 4 or 21 days (7 or 24 days total), with medium changes every 2 to 3 days. At 7 days the nano-HAp treatment resulted in dose-dependent decreases in osteoblastic marker genes; ALP, BSP, and OSC were down regulated, while OPN was up regulated (Fig. 6A). At the 21 day time point the cells demonstrated a similar suppression while the upregulated gene OPN was not significantly different from control (Fig. 6B). Taken together, the results reveal that even a short exposure to nano-HAp in the early stages of differentiation is sufficient to generate long-term changes in gene expression, suggestive of epigenetic regulation.
Figure 6. Nano-HAp produces sustained changes in gene expression.
BMSCs were cultured with or without 50 µg/ml of L-ascorbic acid (AA) as indicated and treated with nano-HAp (µg/ml) for 3 days at the start of culturing followed by removal of nano-HAp containing medium and continued culturing in non-nano-HAp containing medium for an additional (A) 4 days (7 days total) or (B) 21 days (24 days total) and harvested for RNA analysis. The results were normalized to 18S and are expressed as fold change from control +/− SD of 3 replicates. $p<0.05 and $$p<0.005 compared to untreated control; *p<0.05, **p<0.005, and ***p<0.0005 compared to the AA treated (Student’s t-test).
3.7 Nano-HAp stimulates ALP (Alpl) DNA methylation
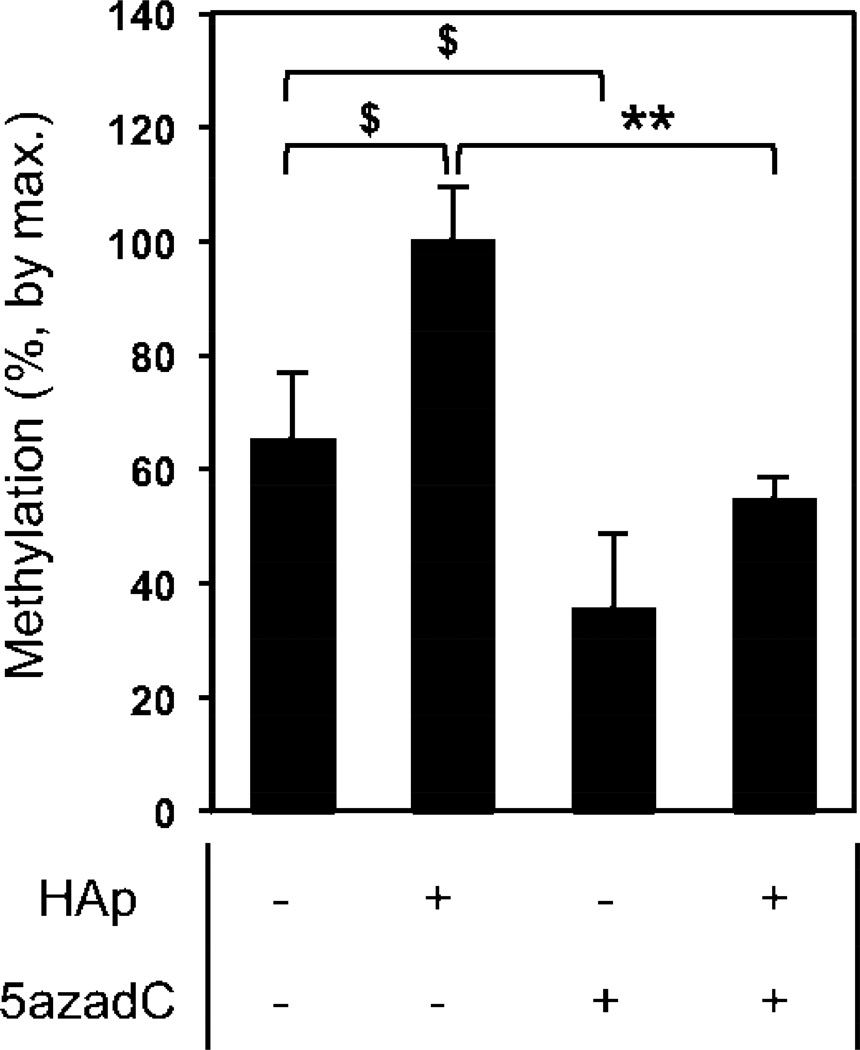
Epigenetic regulation can involve a number of different events such as histone acetylation and changes in chromatin remodeling, however one of the most common causes of long-term changes is cytosine methylation of DNA [53]. This occurs at CpG dinucleotide “islands” by the transfer of a methyl group from S-adenosyl-L-methionine (SAM) to the fifth carbon position of cytosine by a methyltransferase (DNA (Cytosine-5-)-Methyltransferase – (DNMT)) often in the promoter and first exon region of the gene. The sustained suppression of ALP in BMSCs after treatment with nano-HAp suggested the possibility the response was due to increased DNA methylation. We first performed an in silico analysis of the ALP gene (Alpl) and identified a significant CpG island predicted in the 5 prime untranslated region (promoter) and into the first exon. This is in agreement with recent studies identifying regulation of Alpl in the same general region [27–29]. To determine if the methylation of this CpG island was altered by cell stimulation by nano-HAp an assay consisting of Methylation Specific Restriction Enzyme digestion in combination with quantitative RT-PCR was used. The results revealed that exposure of BMSCs to nano-HAp significantly increased ALP promoter methylation, relative to AA alone, in this region by approximately 35% (Fig. 7). The DNMT inhibitor 5-aza-2-dexoycytidine was used as a control [54, 55] and as expected decreased promoter methylation (Fig. 7). The results identify a significant increase in ALP promoter methylation in response to treatment with nano-HAp.
Figure 7. Nano-HAp alters DNA methylation of the alkaline phosphatase gene (ALPL).
Methylation Sensitive Restriction Enzymes in combination with real-time PCR was used to calculate the percent methylation of a 253 base pair amplicon of the ALP promoter region. BMSCs were treated with 50 µg/ml of L-ascorbic acid (AA) and nano-HAp (25 µg/ml) as indicated for 4 days. The DNMT inhibitor, 5-aza-2-deoxycytidine (5 µM) was used as a positive control. The results are expressed as percent change from nano-HAp treated +/− SD of 3 replicates. Data are representative of three independent experiments. $p<0.05 and **p<0.005 comparisons as indicated (Student’s t-test).
4. Discussion
BMSCs are capable of differentiating along the osteoblastic lineage and in culture, the addition of L-ascorbic acid (AA) results in the formation of a collagen matrix, increase ALP activity, and with the addition of β-glycerophosphate (βGP) provides a source of Pi for formation of HA and mineralization of the matrix [20]. Although the molecular and cellular events required for formation of a mineralized matrix are generally understood [43], the effects of matrix mineralization on osteoblasts are surprisingly poorly understood. In this study we created a novel model to study the potential effects of matrix mineralization on the osteoblast lineage. To eliminate the endogenous formation of mineral, βGP was not added to experiments and therefore, the addition of AA stimulated differentiation and the formation of a collagen matrix without mineral formation. To specifically study the role of HA we synthesized nano-HAp which could then be exogenously added to the cultures at specific times and concentrations.
Nano-HAp can be generally synthesized by pyrolysis and precipitation. The latter is often used in laboratory preparations because of ease of use and precise control of size, shape, and phase. The filter press and lyophilization were used for the purification of nano-HAp. The surface charge was close to neutral and therefore the particles, although stable in water did not exhibit long-term mono-dispersion. This also resulted in the size detected by the zeta sizer as being much larger than by TEM. To completely disperse nano-HAp in water prior to use in cell culture the solution was sonicated and vortexed. To improve stability in solution, a surface modification like PEGylation could enhance stability in water and increase circulation time for study in vivo [56]. Other surface modifications such as a thin silica shell [57] or polymer grafting [58] could also be used [59]. However, it is possible that these surface modifications could alter Ca/P ratio [59] or result in the cell recognizing the particles as silica or polymer resulting in a different biological response. These potential outcomes will have to be determined empirically and could vary by cell type.
HAp is one of a number of basic Calcium-Phosphate forms (Ca/P), which also include; octacalcium phosphate (OCP, Ca/P=1.33), tricalcium phosphate (TCP, Ca/P=1.50), and HAp (Ca/P=1.67). Each phase has a unique Ca/P ratio and shows different properties; TCP can be dissolved/degraded whereas HAP is very stable in neutral and basic conditions [60, 61] such as cell culture medium which is generally pH 7.4. Since the Ca/P ratio by the precipitation method mainly varies with pH condition and homogeneity, the pH was monitored in real-time and adjusted to create the desired phase of Ca/P. Sonication was used to maintain calcium hydroxide and phosphoric acid in solution and assist the generation of the seed and subsequent growth ending in creating nano-size, monodispersed hydroxyapatites. The nano-HAp described in this study is the same phase as the mineral generated during the osteoblastic differentiation (reviewed in [62]).
Previous studies have mainly focused on hydroxyapatite as an osteo-promoting surface coating however few studies have investigated the effects of free hydroxyapatite which can be generated by differentiating osteoblasts and possibly internalized. Using our cell models in combination with exogenously synthesized nano-HAp, we identify herein that nano-HAp has the capacity to produce sustained changes in gene expression. In Figure 2, the expression levels of the osteoblast differentiation marker genes ALP, BSP and OSC were strongly and dose-dependently decreased in response to exposure to nano-HAp. As demonstrated in Figure 6, exposure of cells to a “pulse” of nano-HAp for only three days in the early stages of differentiation resulted in ALP suppression even after 21 days in the absence of nano-HAp. ALP and BSP are first activated at an early stage of differentiation coordinated with cell cycle exit and corresponding with formation of collagen matrices and therefore the results suggest a negative feedback loop by which early genes are “turned off” by progression of differentiation to mineralization. Interestingly, not all genes were down regulated. Expression of OPN, a gene associated with the later stages of differentiation and mineralization was strongly stimulated at short time points. Further, OPN was not regulated in the long-term experiment suggesting that nano-HAP can generate both temporary and permanent changes in gene expression and identifies the possibility at least two distinct mechanisms of action. Additionally, this series of experiments revealed that cells have the capacity to rapidly detect the existence of nano-HAp in the extracellular environment and modulate genes expression accordingly.
The cellular response to nano-HAp appears to be mainly the result of its intrinsic materials property. The longitudinal axis of our synthesized nano-HAp is around 100 nm and therefore we synthesized spherical silica nanoparticles at a similar 100 nm size with similar hydration volume. Nano-HAp produced a very distinct response relative to the 100 nm silica nanoparticles. Whereas nano-HAp suppressed ALP, BSP, and OSC at 3 and 7 days, the silica nanoparticles produced either no change or a stimulation of the three genes at days 3 and 7 suggesting a limited role of material size in the response. The stimulation of osteoblast gene expression by silica nanoparticles is in agreement with previous studies using 50 nm particles [41, 48, 63]. The composition and crystallinity effects were considered by using 4 µm CaO material and Pi. Previous studies have demonstrated the capability of Pi to alter osteoblast cell phenotype through specific changes in signal transduction pathways, cellular processes, and gene expression [20–22, 42, 46, 64, 65] and we were interested in determining if HAp acted in the same manner. Although less dramatic than the silica particle the sources of Ca and Pi produced overlapping but also distinct changes in gene expression relative to nano-HAp. All three sources suppressed ALP at day 3 however only Nano-HAp resulted in sustained suppression demonstrated at day 7. Whereas CaO and Pi had little effect on BSP or OSC at day 3, nano-HAp produced a profound suppression of these genes. OPN was equally stimulated by HAp and Pi at day 3, however at 7 days of Pi stimulation expression was almost 5 fold higher than HAp. The results revealed that Pi has overlapping but also distinct effects and suggests cells retain the ability to recognize different forms of Pi and CaP [66]. Further, the different responses between nano-HAp and CaO suggest crystallinity is not a primary factor in the response.
As demonstrated in the “pulse” experiments nano-HAp has the capacity to produce sustained changes in gene expression. One regulatory process by which external stimuli can alter gene expression is through epigenetic changes. Epigenetic regulatory mechanisms such as histone or transcription factor post-translational medication are generally considered temporary [67], if stimuli are removed gene expressions return to baseline. However, DNA methylation is considered permanent, capable of being passed to daughters cells during division [68]. Cytosine methylation of DNA[53] occurs at CpG dinucleotide “islands” by the transfer of a methyl group from S-adenosyl-L-methionine (SAM) to the fifth carbon position of cytosine by a methyltransferase (DNA (Cytosine-5-)-Methyltransferase – (DNMT)) often in the promoter region of the gene [69]. A number of genes important for skeletal regulation, including Alpl, have been suggested as targets of methylation [26–29], although the mechanisms and physiological relevance remain to be fully elucidated. Here we used the ALP promoter region to determine that nano-HAp stimulates DNA methylation. BSP was also tightly coordinated with ALP suggesting the possibility that nano-HAp generates a signal capable of coordinately stimulating a regulatory “program” to change cell behavior. The idea that nanomaterials, in general, might epigenetically alter cells has recently gained attention, although few clear examples have been demonstrated and little is known about the mechanism of action in those cases [30]. Taken together, results presented herein identify nano-HAp as capable of strongly regulating expression of a key early osteoblast lineage marker gene through epigenetic regulation making this one of the first studies to provide a clear and robust example that nanomaterials can stimulate epigenetic regulation of gene expression through methylation of DNA.
The dramatic effect of nano-HAP on gene expression appears to be dependent on the degree of osteoblast lineage differentiation. The most dramatic effect on gene expression was identified in the least differentiated BMSCs with an intermediate response in the partially committed MC3T3-E1 cells and only very modest changes were detected in the most differentiated MLO-Y4 cells. MLO-Y4 cells originate from the same osteoblastic lineage but are cells that have reached a terminally differentiated state and are embedded in the hydroxyapatite of bone. These cells therefore represent a stage of differentiation which has already been exposed to HAp. The results reveal a regulatory mechanism by which the generation of a mineralized matrix by differentiating osteoblasts is a signal to alter the cell to a terminally differentiated state. Terminal differentiation is an important step for most cells as this is considered to require both tissue specific gene expression as well as cell cycle exit [70]. The result is a cell that is committed to perform a highly specific function. The lack of proper terminal differentiation or a reversal is often associated with a cancer phenotype in which a cell reverts to cell division at a spatially and temporally inappropriate time. Further, OPN is a secreted protein known to bind calcium, phosphate and mineral [71] and the strong stimulation by HAp and its ions may represent a mechanism by which the cell attempts to modulate mineral growth and possibly protect the cell from the negative effects of high ion concentration [72, 73]. Our results provide a logical mechanism by which the generation of HA mineral is a key signal to turn off the early stages of osteoblast differentiation involved in matrix preparation and turn on genes involved in regulation of calcification creating a terminally differentiated cell not capable of reverting to an early phenotype.
5. Conclusions
Here we investigated the possibility that nano-hydroxyapatite can alter osteoblast behavior through specific molecular and cellular mechanisms similar to phosphate ions. To this end we synthesized 10×100 nm nano-HAp and investigated the effects on three different cells representing different stages of the osteoblast lineage to determine the cellular and molecular response. Unexpectedly, treatment of cells with nano-HAp resulted in dramatic changes in gene expression both up and down regulated. The results identify a specific gene expression regulatory program modulated by nano-HAp in differentiating osteoblasts involving promoter methylation to suppress pro-osteoblastic marker genes ALP and possibly BSP, and OSC. These results are relevant to osteoblast lineage determination, demonstrating that the generation of a mineralized matrix, a normal aspect of osteoblast differentiation, is a strong signal to permanently alter the cell phenotype through epigenetic regulation. Further, this study identifies a possible mechanism for manipulating cells for trans- or de-differentiation, the ability to convert a differentiated cell to behave as a different cell type or more stem cell like. Additionally, these results suggest that care should be taken in using HA based biomaterials that might produce nanoparticles from wear or degradation. Findings herein also have important implications for human health. HA and related CaP compositions are produced endogenously and therefore our results may have implications relative to the etiology of diseases of calcium phosphate homeostasis such as chronic kidney disease [74–76] and hydroxyapatite crystal deposition disease [77], in addition to functional mineralization associated with bone homeostasis [78]. Finally, the results have important implications for the field of nanotechnology by providing an increased understanding of important cellular and molecular regulatory functions that can be influenced by nanomaterials and define epigenetic regulation as a mechanism by which cell behavior can be modified. These studies raise the interesting potential that nanomaterials may be specifically engineered to manipulate the expression of genes involved in diverse diseases through epigenetic regulation.
Supplementary Material
Acknowledgements
This study was supported in part by grants to G.R Beck Jr. from Biomedical Laboratory Research & Development Service Award Number I01BX002363 from the VA Office of Research and Development and a grant from the NCI (CA136716). H. L. Jang and K. T. Nam appreciate the support International Research & Development Program of the National Research Foundation of Korea (NRF, No. 2013K1A3A1A32035536). The content is solely the responsibility of the authors and does not represent the official views of the Department of Veterans Affairs, National Institutes of Health, or the United States Government.
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.biomaterials
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
References
- 1.Palmer LC, Newcomb CJ, Kaltz SR, Spoerke ED, Stupp SI. Biomimetic Systems for Hydroxyapatite Mineralization Inspired by Bone and Enamel. Chem Rev. 2008;108:4754–4783. doi: 10.1021/cr8004422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bohner M, Tadier S, van Garderen N, de Gasparo A, Dobelin N, Baroud G. Synthesis of Spherical Calcium Phosphate Particles for Dental and Orthopedic Applications. Biomatter. 2013;3:e25103-1–e25103-15. doi: 10.4161/biom.25103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunter LW, Charlesworth JE, Yu S, Lieske JC, Miller VM. Calcifying Nanoparticles Promote Mineralization in Vascular Smooth Muscle Cells: Implications for Atherosclerosis. Int J Nanomedicine. 2014;9:2689–2698. doi: 10.2147/IJN.S63189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dai XY, Zhao MM, Cai Y, Guan QC, Zhao Y, Guan Y, Kong W, Zhu WG, Xu MJ, Wang X. Phosphate-Induced Autophagy Counteracts Vascular Calcification by Reducing Matrix Vesicle Release. Kidney Int. 2013;83:1042–1051. doi: 10.1038/ki.2012.482. [DOI] [PubMed] [Google Scholar]
- 5.Shanahan CM, Crouthamel MH, Kapustin A, Giachelli CM. Arterial Calcification in Chronic Kidney Disease: Key Roles for Calcium and Phosphate. Circ Res. 2011;109:697–711. doi: 10.1161/CIRCRESAHA.110.234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirsch T, Nah HD, Shapiro IM, Pacifici M. Regulated Production of Mineralization-Competent Matrix Vesicles in Hypertrophic Chondrocytes. J Cell Biol. 1997;137:1149–1160. doi: 10.1083/jcb.137.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belem LC, Zanetti G, Souza AS, Jr, Hochhegger B, Guimaraes MD, Nobre LF, Rodrigues RS, Marchiori E. Metastatic Pulmonary Calcification: State-of-the-Art Review Focused on Imaging Findings. Respir Med. 2014;108:668–676. doi: 10.1016/j.rmed.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 8.Wu LN, Genge BR, Dunkelberger DG, LeGeros RZ, Concannon B, Wuthier RE. Physicochemical Characterization of the Nucleational Core of Matrix Vesicles. J Biol Chem. 1997;272:4404–4411. doi: 10.1074/jbc.272.7.4404. [DOI] [PubMed] [Google Scholar]
- 9.Bertazzo S, Gentleman E, Cloyd KL, Chester AH, Yacoub MH, Stevens MM. Nano-Analytical Electron Microscopy Reveals Fundamental Insights into Human Cardiovascular Tissue Calcification. Nat Mater. 2013;12:576–583. doi: 10.1038/nmat3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reynolds JL, Joannides AJ, Skepper JN, McNair R, Schurgers LJ, Proudfoot D, Jahnen-Dechent W, Weissberg PL, Shanahan CM. Human Vascular Smooth Muscle Cells Undergo Vesicle-Mediated Calcification in Response to Changes in Extracellular Calcium and Phosphate Concentrations: A Potential Mechanism for Accelerated Vascular Calcification in Esrd. J Am Soc Nephrol. 2004;15:2857–2867. doi: 10.1097/01.ASN.0000141960.01035.28. [DOI] [PubMed] [Google Scholar]
- 11.Lee SY, Kim HY, Gu SW, Kim HJ, Yang DH. 25-Hydroxyvitamin D Levels and Vascular Calcification in Predialysis and Dialysis Patients with Chronic Kidney Disease. Kidney Blood Press Res. 2012;35:349–354. doi: 10.1159/000335952. [DOI] [PubMed] [Google Scholar]
- 12.Villa-Bellosta R, Millan A, Sorribas V. Role of Calcium-Phosphate Deposition in Vascular Smooth Muscle Cell Calcification. Am J Physiol Cell Physiol. 2011;300:C210–C220. doi: 10.1152/ajpcell.00229.2010. [DOI] [PubMed] [Google Scholar]
- 13.Dalinka MK, Melchior EL. Soft Tissue Calcifications in Systemic Disease. Bull N Y Acad Med. 1980;56:539–563. [PMC free article] [PubMed] [Google Scholar]
- 14.Tanabe H, Akashi T, Kawachi H, Andou N, Eishi Y, Takizawa T, Koike M, Ichinose S. Identification of Hydroxyapatite Deposits in the Smooth Muscle Cells and Ganglion Cells of Autopsied Small Intestines. J Med Dent Sci. 2004;51:129–138. [PubMed] [Google Scholar]
- 15.Anderson HC, Garimella R, Tague SE. The Role of Matrix Vesicles in Growth Plate Development and Biomineralization. Front Biosci. 2005;10:822–837. doi: 10.2741/1576. [DOI] [PubMed] [Google Scholar]
- 16.Xiao Z, Camalier CE, Nagashima K, Chan KC, Lucas DA, de la Cruz MJ, Gignac M, Lockett S, Issaq HJ, Veenstra TD, Conrads TP, Beck GR., Jr Analysis of the Extracellular Matrix Vesicle Proteome in Mineralizing Osteoblasts. J Cell Physiol. 2007;210:325–335. doi: 10.1002/jcp.20826. [DOI] [PubMed] [Google Scholar]
- 17.Bonewald LF. The Amazing Osteocyte. J Bone Miner Res. 2011;26:229–238. doi: 10.1002/jbmr.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beck GR., Jr Inorganic Phosphate as a Signaling Molecule in Osteoblast Differentiation. J Cell Biochem. 2003;90:234–243. doi: 10.1002/jcb.10622. [DOI] [PubMed] [Google Scholar]
- 19.Aubin JE. Regulation of Osteoblast Formation and Function. Rev Endocr Metab Disord. 2001;2:81–94. doi: 10.1023/a:1010011209064. [DOI] [PubMed] [Google Scholar]
- 20.Beck GR, Jr, Sullivan EC, Moran E, Zerler B. Relationship between Alkaline Phosphatase Levels, Osteopontin Expression, and Mineralization in Differentiating Mc3t3-E1 Osteoblasts. J Cell Biochem. 1998;68:269–280. doi: 10.1002/(sici)1097-4644(19980201)68:2<269::aid-jcb13>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 21.Beck GR, Jr, Moran E, Knecht N. Inorganic Phosphate Regulates Multiple Genes During Osteoblast Differentiation, Including Nrf2. Exp Cell Res. 2003;288:288–300. doi: 10.1016/s0014-4827(03)00213-1. [DOI] [PubMed] [Google Scholar]
- 22.Conrads KA, Yi M, Simpson KA, Lucas DA, Camalier CE, Yu LR, Veenstra TD, Stephens RM, Conrads TP, Beck GR., Jr A Combined Proteome and Microarray Investigation of Inorganic Phosphate-Induced Pre-Osteoblast Cells. Mol Cell Proteomics. 2005;4:1284–1296. doi: 10.1074/mcp.M500082-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Whyte MP. Hypophosphatasia and the Role of Alkaline Phosphatase in Skeletal Mineralization. Endocr Rev. 1994;15:439–461. doi: 10.1210/edrv-15-4-439. [DOI] [PubMed] [Google Scholar]
- 24.Rathburn JC. Hypophosphatasia, a New Developmental Anomaly. Am J Dis Child. 1948;75:822–831. doi: 10.1001/archpedi.1948.02030020840003. [DOI] [PubMed] [Google Scholar]
- 25.Fedde KN, Blair L, Silverstein J, Coburn SP, Ryan LM, Weinstein RS, Waymire K, Narisawa S, Millan JL, MacGregor GR, Whyte MP. Alkaline Phosphatase Knock-out Mice Recapitulate the Metabolic and Skeletal Defects of Infantile Hypophosphatasia. J Bone Miner Res. 1999;14:2015–2026. doi: 10.1359/jbmr.1999.14.12.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collas P. Epigenetic States in Stem Cells. Biochim Biophys Acta. 2009;1790:900–905. doi: 10.1016/j.bbagen.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 27.Delgado-Calle J, Sanudo C, Sanchez-Verde L, Garcia-Renedo RJ, Arozamena J, Riancho JA. Epigenetic Regulation of Alkaline Phosphatase in Human Cells of the Osteoblastic Lineage. Bone. 2011;49:830–838. doi: 10.1016/j.bone.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 28.Escalante-Alcalde D, Recillas-Targa F, Hernandez-Garcia D, Castro-Obregon S, Terao M, Garattini E, Covarrubias L. Retinoic Acid and Methylation Cis-Regulatory Elements Control the Mouse Tissue Non-Specific Alkaline Phosphatase Gene Expression. Mech Dev. 1996;57:21–32. doi: 10.1016/0925-4773(96)00524-2. [DOI] [PubMed] [Google Scholar]
- 29.Cho YD, Yoon WJ, Kim WJ, Woo KM, Baek JH, Lee G, Ku Y, van Wijnen AJ, Ryoo HM. Epigenetic Modifications and Canonical Wingless/Int-1 Class (Wnt) Signaling Enable Trans-Differentiation of Nonosteogenic Cells into Osteoblasts. J Biol Chem. 2014;289:20120–20128. doi: 10.1074/jbc.M114.558064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoccoro A, Karlsson HL, Coppede F, Migliore L. Epigenetic Effects of Nano-Sized Materials. Toxicology. 2013;313:3–14. doi: 10.1016/j.tox.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 31.Furuzono T, Yasuda S, Kimura T, Kyotani S, Tanaka J, Kishida A. Nano-Scaled Hydroxyapatite/Polymer Composite Iv. Fabrication and Cell Adhesion Properties of a Three-Dimensional Scaffold Made of Composite Material with a Silk Fibroin Substrate to Develop a Percutaneous Device. J Artif Organs. 2004;7:137–144. doi: 10.1007/s10047-004-0264-x. [DOI] [PubMed] [Google Scholar]
- 32.Ito Y, Tanaka N, Fujimoto Y, Yasunaga Y, Ishida O, Agung M, Ochi M. Bone Formation Using Novel Interconnected Porous Calcium Hydroxyapatite Ceramic Hybridized with Cultured Marrow Stromal Stem Cells Derived from Green Rat. J Biomed Mater Res A. 2004;69:454–461. doi: 10.1002/jbm.a.30014. [DOI] [PubMed] [Google Scholar]
- 33.Itoh S, Kikuchi M, Koyama Y, Takakuda K, Shinomiya K, Tanaka J. Development of a Hydroxyapatite/Collagen Nanocomposite as a Medical Device. Cell Transplant. 2004;13:451–461. doi: 10.3727/000000004783983774. [DOI] [PubMed] [Google Scholar]
- 34.Sun D, Chen Y, Tran RT, Xu S, Xie D, Jia C, Wang Y, Guo Y, Zhang Z, Guo J, Yang J, Jin D, Bai X. Citric Acid-Based Hydroxyapatite Composite Scaffolds Enhance Calvarial Regeneration. Sci Rep. 2014;4:6912. doi: 10.1038/srep06912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li L, Pan HH, Tao JH, Xu XR, Mao CY, Gu XH, Tang RK. Repair of Enamel by Using Hydroxyapatite Nanoparticles as the Building Blocks. J Mater Chem. 2008;18:4079–4084. [Google Scholar]
- 36.Zhou H, Lee J. Nanoscale Hydroxyapatite Particles for Bone Tissue Engineering. Acta Biomater. 2011;7:2769–2781. doi: 10.1016/j.actbio.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 37.Zhang JZ, Liu GY, Wu QO, Zuo JL, Qin YG, Wang JC. Novel Mesoporous Hydroxyapatite/Chitosan Composite for Bone Repair. Journal of Bionic Engineering. 2012;9:243–251. [Google Scholar]
- 38.Park JW, Suh JY, Chung HJ. Effects of Calcium Ion Incorporation on Osteoblast Gene Expression in Mc3t3-E1 Cells Cultured on Microstructured Titanium Surfaces. J Biomed Mater Res A. 2008;86:117–126. doi: 10.1002/jbm.a.31618. [DOI] [PubMed] [Google Scholar]
- 39.Noor Z. Nanohydroxyapatite Application to Osteoporosis Management. J Osteoporos. 2013;679025:1–6. doi: 10.1155/2013/679025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nandi SK, Roy S, Mukherjee P, Kundu B, De DK, Basu D. Orthopaedic Applications of Bone Graft & Graft Substitutes: A Review. Indian J Med Res. 2010;132:15–30. [PubMed] [Google Scholar]
- 41.Ha SW, Weitzmann MN, Beck GR., Jr Bioactive Silica Nanoparticles Promote Osteoblast Differentiation through Stimulation of Autophagy and Direct Association with Lc3 and P62. ACS Nano. 2014;8:5898–5910. doi: 10.1021/nn5009879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Camalier CE, Yi M, Yu LR, Hood BL, Conrads KA, Lee YJ, Lin Y, Garneys LM, Bouloux GF, Young MR, Veenstra TD, Stephens RM, Colburn NH, Conrads TP, Beck GR., Jr An Integrated Understanding of the Physiological Response to Elevated Extracellular Phosphate. J Cell Physiol. 2013;228:1536–1550. doi: 10.1002/jcp.24312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aubin JE, Triffitt JT. Mesenchymal Stem Cells and Osteoblast Differentiation. 2nd ed. San Diego: Academic press; 2002. [Google Scholar]
- 44.Sudo H, Kodama HA, Amagai Y, Yamamoto S, Kasai S. In Vitro Differentiation and Calcification in a New Clonal Osteogenic Cell Line Derived from Newborn Mouse Calvaria. J Cell Biol. 1983;96:191–198. doi: 10.1083/jcb.96.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bonewald LF. Establishment and Characterization of an Osteocyte-Like Cell Line, Mlo-Y4. J Bone Miner Metab. 1999;17:61–65. doi: 10.1007/s007740050066. [DOI] [PubMed] [Google Scholar]
- 46.Beck GR, Jr, Zerler B, Moran E. Phosphate Is a Specific Signal for Induction of Osteopontin Gene Expression. Proc Natl Acad Sci U S A. 2000;97:8352–8357. doi: 10.1073/pnas.140021997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stöber W, Fink A, Bohn E. Controlled Growth of Monodisperse Silica Spheres in the Micron Size Range. J Colloid Interface Sci. 1968:62–69. [Google Scholar]
- 48.Beck GR, Jr, Ha SW, Camalier CE, Yamaguchi M, Li Y, Lee JK, Weitzmann MN. Bioactive Silica-Based Nanoparticles Stimulate Bone-Forming Osteoblasts, Suppress Bone-Resorbing Osteoclasts, and Enhance Bone Mineral Density in Vivo. Nanomedicine. 2012;8:793–803. doi: 10.1016/j.nano.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosser J, Bonewald LF. Studying Osteocyte Function Using the Cell Lines Mlo-Y4 and Mlo-A5. Methods Mol Biol. 2012;816:67–81. doi: 10.1007/978-1-61779-415-5_6. [DOI] [PubMed] [Google Scholar]
- 50.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative Pcr and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 51.Hashimoto K, Kokubun S, Itoi E, Roach HI. Improved Quantification of DNA Methylation Using Methylation-Sensitive Restriction Enzymes and Real-Time Pcr. Epigenetics. 2007;2:86–91. doi: 10.4161/epi.2.2.4203. [DOI] [PubMed] [Google Scholar]
- 52.Ha SW, Camalier CE, Beck GR, Jr, Lee JK. New Method to Prepare Very Stable and Biocompatible Fluorescent Silica Nanoparticles. Chem Commun (Camb) 2009:2881–2883. doi: 10.1039/b902195g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miranda TB, Jones PA. DNA Methylation: The Nuts and Bolts of Repression. J Cell Physiol. 2007;213:384–390. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- 54.Delgado-Calle J, Arozamena J, Perez-Lopez J, Bolado-Carrancio A, Sanudo C, Agudo G, de la Vega R, Alonso MA, Rodriguez-Rey JC, Riancho JA. Role of Bmps in the Regulation of Sclerostin as Revealed by an Epigenetic Modifier of Human Bone Cells. Mol Cell Endocrinol. 2013;369:27–34. doi: 10.1016/j.mce.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 55.Vaes BL, Lute C, van der Woning SP, Piek E, Vermeer J, Blom HJ, Mathers JC, Muller M, de Groot LC, Steegenga WT. Inhibition of Methylation Decreases Osteoblast Differentiation Via a Non-DNA-Dependent Methylation Mechanism. Bone. 2010;46:514–523. doi: 10.1016/j.bone.2009.09.033. [DOI] [PubMed] [Google Scholar]
- 56.Jokerst JV, Lobovkina T, Zare RN, Gambhir SS. Nanoparticle Pegylation for Imaging and Therapy. Nanomedicine. 2011;6:715–728. doi: 10.2217/nnm.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Borum L, Wilson OC. Surface Modification of Hydroxyapatite. Part Ii. Silica. Biomaterials. 2003;24:3681–3688. doi: 10.1016/s0142-9612(03)00240-0. [DOI] [PubMed] [Google Scholar]
- 58.Choi HW, Lee HJ, Kim KJ, Kim HM, Lee SC. Surface Modification of Hydroxyapatite Nanocrystals by Grafting Polymers Containing Phosphonic Acid Groups. J Colloid Interface Sci. 2006;304:277–281. doi: 10.1016/j.jcis.2006.05.069. [DOI] [PubMed] [Google Scholar]
- 59.Tanaka H, Futaoka M, Hino R. Surface Modification of Calcium Hydroxyapatite with Pyrophosphoric Acid. J Colloid Interface Sci. 2004;269:358–363. doi: 10.1016/j.jcis.2003.07.039. [DOI] [PubMed] [Google Scholar]
- 60.Koerten HK, van der Meulen J. Degradation of Calcium Phosphate Ceramics. J Biomed Mater Res. 1999;44:78–86. doi: 10.1002/(sici)1097-4636(199901)44:1<78::aid-jbm9>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 61.Chow LC. Next Generation Calcium Phosphate-Based Biomaterials. Dent Mater J. 2009;28:1–10. doi: 10.4012/dmj.28.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Farbod K, Nejadnik MR, Jansen JA, Leeuwenburgh SC. Interactions between Inorganic and Organic Phases in Bone Tissue as a Source of Inspiration for Design of Novel Nanocomposites. Tissue Eng Part B Rev. 2014;20:173–188. doi: 10.1089/ten.TEB.2013.0221. [DOI] [PubMed] [Google Scholar]
- 63.Ha SW, Sikorski JA, Weitzmann MN, Beck GR., Jr Bio-Active Engineered 50nm Silica Nanoparticles with Bone Anabolic Activity: Therapeutic Index, Effective Concentration, and Cytotoxicity Profile in Vitro. Toxicol In Vitro. 2013;28:354–364. doi: 10.1016/j.tiv.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beck GR, Jr, Knecht N. Osteopontin Regulation by Inorganic Phosphate Is Erk1/2-, Protein Kinase C-, and Proteasome-Dependent. J Biol Chem. 2003;278:41921–41929. doi: 10.1074/jbc.M304470200. [DOI] [PubMed] [Google Scholar]
- 65.Camalier CE, Young MR, Bobe G, Perella CM, Colburn NH, Beck GR., Jr Elevated Phosphate Activates N-Ras and Promotes Cell Transformation and Skin Tumorigenesis. Cancer Prev Res (Phila) 2010;3:359–370. doi: 10.1158/1940-6207.CAPR-09-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pasch A, Farese S, Graber S, Wald J, Richtering W, Floege J, Jahnen-Dechent W. Nanoparticle-Based Test Measures Overall Propensity for Calcification in Serum. J Am Soc Nephrol. 2012;23:1744–1752. doi: 10.1681/ASN.2012030240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stein GS, Zaidi SK, Stein JL, Lian JB, van Wijnen AJ, Montecino M, Young DW, Javed A, Pratap J, Choi JY, Ali SA, Pande S, Hassan MQ. Transcription-Factor-Mediated Epigenetic Control of Cell Fate and Lineage Commitment. Biochem Cell Biol. 2009;87:1–6. doi: 10.1139/o08-094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cheng XD, Hashimoto H, Horton JR, Zhang X. Mechanisms of DNA Methylation, Methyl-Cpg Recognition, and Demethylation in Mammals. Handbook of Epigenetics: The New Molecular and Medical Genetics. 2011:9–24. [Google Scholar]
- 69.Jeltsch A, Jurkowska RZ. New Concepts in DNA Methylation. Trends Biochem Sci. 2014;39:310–318. doi: 10.1016/j.tibs.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 70.Buttitta LA, Edgar BA. Mechanisms Controlling Cell Cycle Exit Upon Terminal Differentiation. Curr Opin Cell Biol. 2007;19:697–704. doi: 10.1016/j.ceb.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hunter GK. Role of Osteopontin in Modulation of Hydroxyapatite Formation. Calcif Tissue Int. 2013;93:348–354. doi: 10.1007/s00223-013-9698-6. [DOI] [PubMed] [Google Scholar]
- 72.Steitz SA, Speer MY, McKee MD, Liaw L, Almeida M, Yang H, Giachelli CM. Osteopontin Inhibits Mineral Deposition and Promotes Regression of Ectopic Calcification. Am J Pathol. 2002;161:2035–2046. doi: 10.1016/S0002-9440(10)64482-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boskey AL, Maresca M, Ullrich W, Doty SB, Butler WT, Prince CW. Osteopontin-Hydroxyapatite Interactions in Vitro: Inhibition of Hydroxyapatite Formation and Growth in a Gelatin-Gel. Bone Miner. 1993;22:147–159. doi: 10.1016/s0169-6009(08)80225-5. [DOI] [PubMed] [Google Scholar]
- 74.Block GA, Hulbert-Shearon TE, Levin NW, Port FK. Association of Serum Phosphorus and Calcium X Phosphate Product with Mortality Risk in Chronic Hemodialysis Patients: A National Study. Am J Kidney Dis. 1998;31:607–617. doi: 10.1053/ajkd.1998.v31.pm9531176. [DOI] [PubMed] [Google Scholar]
- 75.Ketteler M, Schlieper G, Floege J. Calcification and Cardiovascular Health: New Insights into an Old Phenomenon. Hypertension. 2006;47:1027–1034. doi: 10.1161/01.HYP.0000219635.51844.da. [DOI] [PubMed] [Google Scholar]
- 76.Jono S, McKee MD, Murry CE, Shioi A, Nishizawa Y, Mori K, Morii H, Giachelli CM. Phosphate Regulation of Vascular Smooth Muscle Cell Calcification. Circ Res. 2000;87:E10–E17. doi: 10.1161/01.res.87.7.e10. [DOI] [PubMed] [Google Scholar]
- 77.Garcia GM, McCord GC, Kumar R. Hydroxyapatite Crystal Deposition Disease. Semin Musculoskelet Radiol. 2003;7:187–193. doi: 10.1055/s-2003-43229. [DOI] [PubMed] [Google Scholar]
- 78.Clarke B. Normal Bone Anatomy and Physiology. Clin J Am Soc Nephrol. 2008;3(Suppl 3):S131–S139. doi: 10.2215/CJN.04151206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.