Abstract
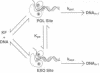
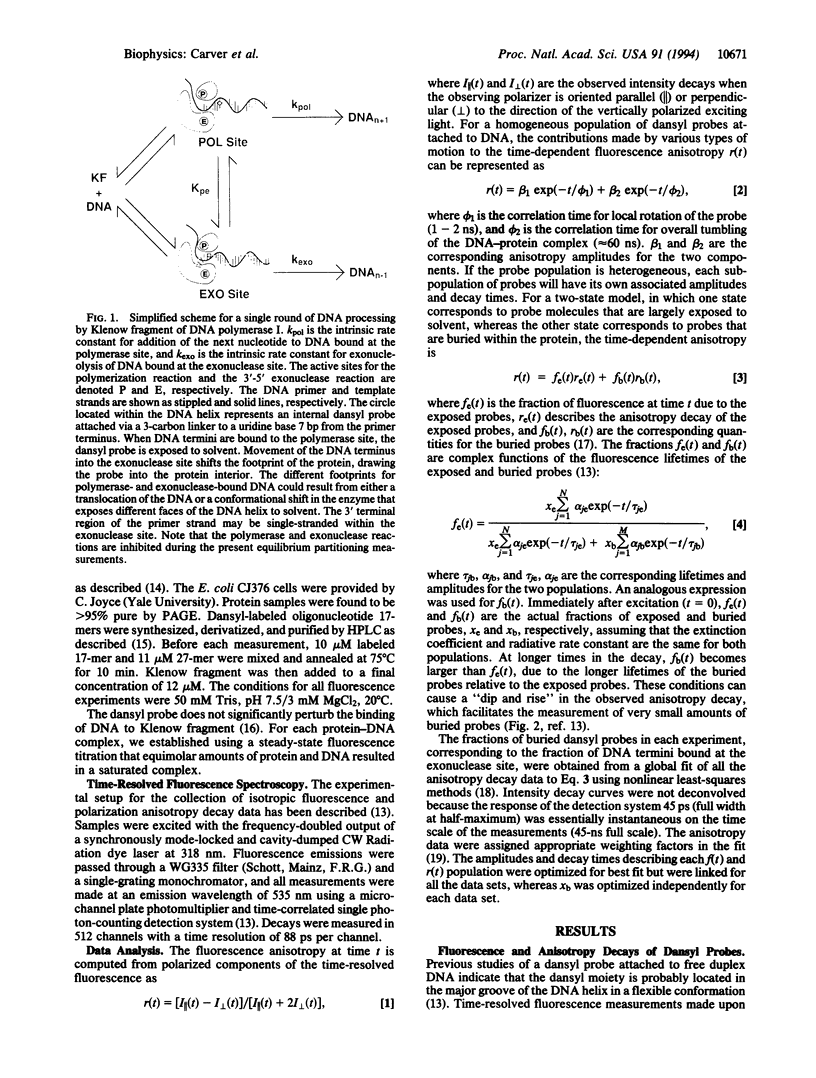
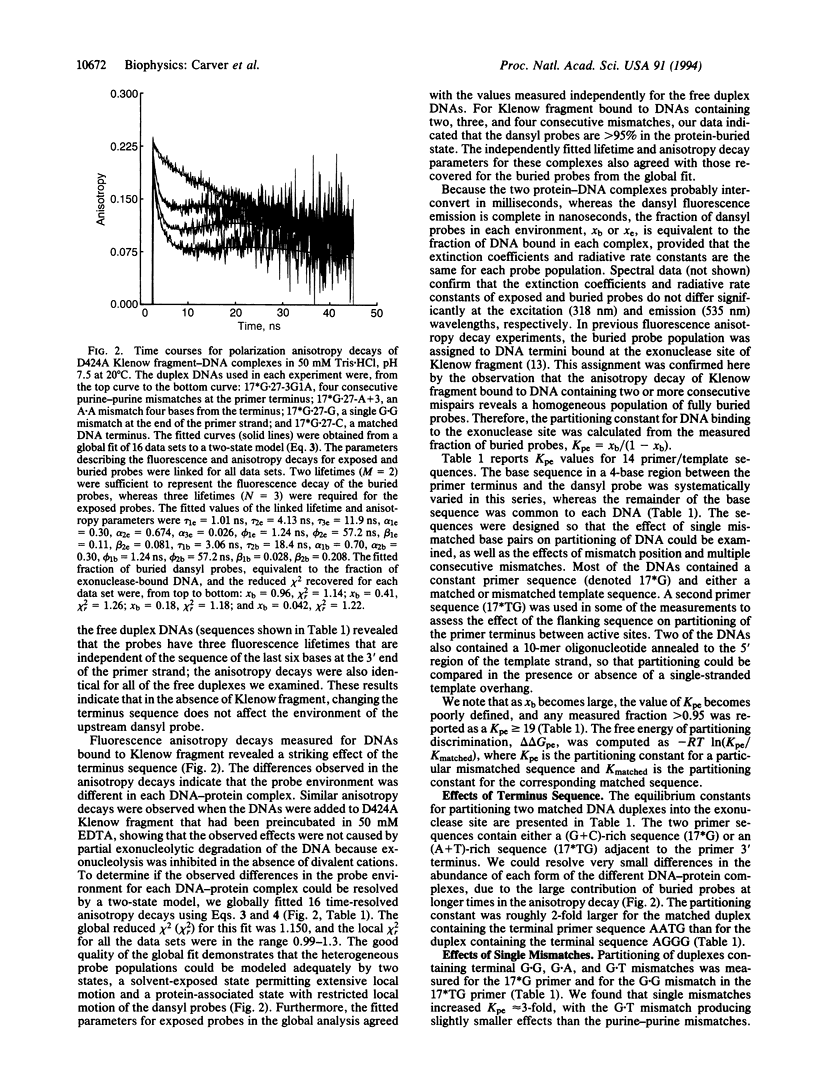
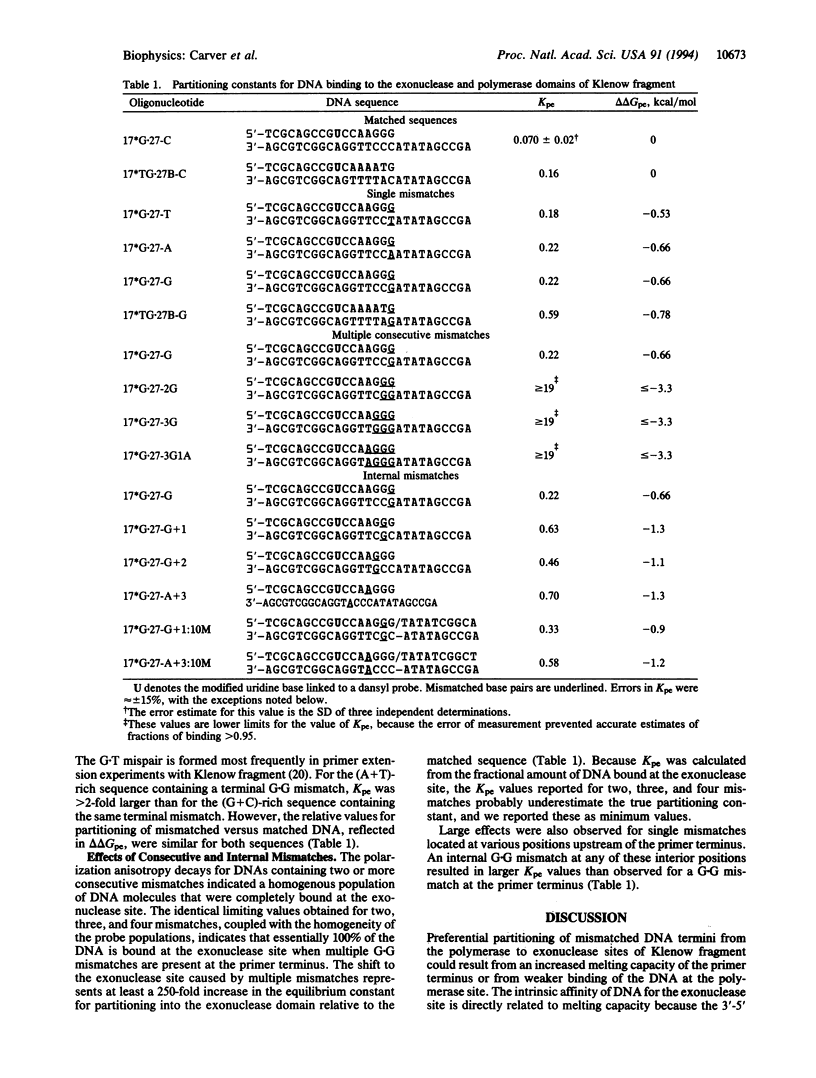
Fluorescence depolarization decays were measured for 5-dimethylaminonaphthalene-1-sulfonyl (dansyl) probes attached internally to 17-mer.27-mer oligonucleotides bound to Klenow fragment of DNA polymerase I. The time-resolved motions of the dansyl probes were sensitive indicators of DNA-protein contacts, showing that the protein binds to DNA with two footprints, corresponding to primer termini at either the polymerase or 3'-5' exonuclease sites. We examined complexes of Klenow fragment with DNAs containing various base mismatches. Single mismatches at the primer terminus caused a 3- to 4-fold increase in the equilibrium partitioning of DNA into the exonuclease site; the largest effects were observed for purine-purine mismatches. Two or more consecutive G.G mismatches caused the DNA to bind exclusively at the exonuclease site, with a partitioning constant at least 250-fold greater than that of the corresponding matched DNA sequence. Internal single mismatches produced larger effects than the same mismatch at the primer terminus, with a delta delta G relative to the matched sequence of -1.1 to -1.3 kcal/mol for mismatches located 2, 3, or 4 bases from the primer terminus. Although part of the observed effects may be attributed to the increased melting capacity of the DNA, it appears that the polymerase site also promotes movement of DNA into the exonuclease site by rejecting aberrant primer termini. These effects suggest that the polymerase and exonuclease sites act together to recognize specific errors that distort the primer terminus, such as frameshifts, in addition to proofreading misincorporated bases.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen D. J., Benkovic S. J. Resonance energy transfer measurements between substrate binding sites within the large (Klenow) fragment of Escherichia coli DNA polymerase I. Biochemistry. 1989 Dec 12;28(25):9586–9593. doi: 10.1021/bi00451a006. [DOI] [PubMed] [Google Scholar]
- Allen D. J., Darke P. L., Benkovic S. J. Fluorescent oligonucleotides and deoxynucleotide triphosphates: preparation and their interaction with the large (Klenow) fragment of Escherichia coli DNA polymerase I. Biochemistry. 1989 May 30;28(11):4601–4607. doi: 10.1021/bi00437a014. [DOI] [PubMed] [Google Scholar]
- Bebenek K., Joyce C. M., Fitzgerald M. P., Kunkel T. A. The fidelity of DNA synthesis catalyzed by derivatives of Escherichia coli DNA polymerase I. J Biol Chem. 1990 Aug 15;265(23):13878–13887. [PubMed] [Google Scholar]
- Beese L. S., Derbyshire V., Steitz T. A. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science. 1993 Apr 16;260(5106):352–355. doi: 10.1126/science.8469987. [DOI] [PubMed] [Google Scholar]
- Brenowitz S., Kwack S., Goodman M. F., O'Donnell M., Echols H. Specificity and enzymatic mechanism of the editing exonuclease of Escherichia coli DNA polymerase III. J Biol Chem. 1991 Apr 25;266(12):7888–7892. [PubMed] [Google Scholar]
- Brutlag D., Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. 36. A proofreading function for the 3' leads to 5' exonuclease activity in deoxyribonucleic acid polymerases. J Biol Chem. 1972 Jan 10;247(1):241–248. [PubMed] [Google Scholar]
- Carroll S. S., Cowart M., Benkovic S. J. A mutant of DNA polymerase I (Klenow fragment) with reduced fidelity. Biochemistry. 1991 Jan 22;30(3):804–813. doi: 10.1021/bi00217a034. [DOI] [PubMed] [Google Scholar]
- Cowart M., Gibson K. J., Allen D. J., Benkovic S. J. DNA substrate structural requirements for the exonuclease and polymerase activities of procaryotic and phage DNA polymerases. Biochemistry. 1989 Mar 7;28(5):1975–1983. doi: 10.1021/bi00431a004. [DOI] [PubMed] [Google Scholar]
- Derbyshire V., Grindley N. D., Joyce C. M. The 3'-5' exonuclease of DNA polymerase I of Escherichia coli: contribution of each amino acid at the active site to the reaction. EMBO J. 1991 Jan;10(1):17–24. doi: 10.1002/j.1460-2075.1991.tb07916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echols H., Goodman M. F. Fidelity mechanisms in DNA replication. Annu Rev Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- Fersht A. R., Knill-Jones J. W., Tsui W. C. Kinetic basis of spontaneous mutation. Misinsertion frequencies, proofreading specificities and cost of proofreading by DNA polymerases of Escherichia coli. J Mol Biol. 1982 Mar 25;156(1):37–51. doi: 10.1016/0022-2836(82)90457-0. [DOI] [PubMed] [Google Scholar]
- Freemont P. S., Friedman J. M., Beese L. S., Sanderson M. R., Steitz T. A. Cocrystal structure of an editing complex of Klenow fragment with DNA. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8924–8928. doi: 10.1073/pnas.85.23.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M. F., Creighton S., Bloom L. B., Petruska J. Biochemical basis of DNA replication fidelity. Crit Rev Biochem Mol Biol. 1993;28(2):83–126. doi: 10.3109/10409239309086792. [DOI] [PubMed] [Google Scholar]
- Guest C. R., Hochstrasser R. A., Dupuy C. G., Allen D. J., Benkovic S. J., Millar D. P. Interaction of DNA with the Klenow fragment of DNA polymerase I studied by time-resolved fluorescence spectroscopy. Biochemistry. 1991 Sep 10;30(36):8759–8770. doi: 10.1021/bi00100a007. [DOI] [PubMed] [Google Scholar]
- Joyce C. M., Sun X. C., Grindley N. D. Reactions at the polymerase active site that contribute to the fidelity of Escherichia coli DNA polymerase I (Klenow fragment). J Biol Chem. 1992 Dec 5;267(34):24485–24500. [PubMed] [Google Scholar]
- Kuchta R. D., Benkovic P., Benkovic S. J. Kinetic mechanism whereby DNA polymerase I (Klenow) replicates DNA with high fidelity. Biochemistry. 1988 Sep 6;27(18):6716–6725. doi: 10.1021/bi00418a012. [DOI] [PubMed] [Google Scholar]
- Lai M. D., Beattie K. L. Influence of DNA sequence on the nature of mispairing during DNA synthesis. Biochemistry. 1988 Mar 8;27(5):1722–1728. doi: 10.1021/bi00405a051. [DOI] [PubMed] [Google Scholar]
- Ludescher R. D., Peting L., Hudson S., Hudson B. Time-resolved fluorescence anisotropy for systems with lifetime and dynamic heterogeneity. Biophys Chem. 1987 Oct;28(1):59–75. doi: 10.1016/0301-4622(87)80075-3. [DOI] [PubMed] [Google Scholar]
- Maldonado-Rodriguez R., Beattie K. L. In vitro mutagenesis in the lacI gene of Escherichia coli: fate of 3'-terminal mispairs versus internal base mispairs in a transfection assay. Mutat Res. 1991 Mar;247(1):5–18. doi: 10.1016/0027-5107(91)90028-m. [DOI] [PubMed] [Google Scholar]
- Ollis D. L., Brick P., Hamlin R., Xuong N. G., Steitz T. A. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. 1985 Feb 28-Mar 6Nature. 313(6005):762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- Petruska J., Goodman M. F., Boosalis M. S., Sowers L. C., Cheong C., Tinoco I., Jr Comparison between DNA melting thermodynamics and DNA polymerase fidelity. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6252–6256. doi: 10.1073/pnas.85.17.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatzky-Schwartz M., Hiller Y., Reich Z., Ghirlando R., Weinberger S., Minsky A. Attenuation of DNA-protein interactions associated with intrinsic, sequence-dependent DNA curvature. Biochemistry. 1992 Mar 3;31(8):2339–2346. doi: 10.1021/bi00123a019. [DOI] [PubMed] [Google Scholar]
- Weiss S. J., Fisher P. A. Interaction of Drosophila DNA polymerase alpha holoenzyme with synthetic template-primers containing mismatched primer bases or propanodeoxyguanosine adducts at various positions in template and primer regions. J Biol Chem. 1992 Sep 15;267(26):18520–18526. [PubMed] [Google Scholar]