Abstract
BRG1, the central ATPase of the human SWI/SNF complex, is critical for biological functions, including nuclear receptor (NR)-regulated transcription. Analysis of BRG1 mutants demonstrated that functional motifs outside the ATPase domain are important for transcriptional activity. In the course of experiments examining protein interactions mediated through these domains, Ku70 (XRCC6) was found to associate with a BRG1 fragment encompassing the conserved helicase-SANT-associated (HSA) and BRK domains of BRG1. Subsequent transcriptional activation assays and chromatin immunoprecipitation studies showed that Ku70/86 and components of the topoisomerase IIβ (TOP2β)/poly(ADP ribose) polymerase 1 (PARP1) complex are necessary for NR-mediated SWI/SNF-dependent transcriptional activation from endogenous promoters. In addition to establishing Ku-BRG1 binding and TOP2β/PARP1 recruitment by nuclear receptor transactivation, we demonstrate that the transient appearance of glucocorticoid receptor (GR)/BRG1-dependent, TOP2β-mediated double-strand DNA breaks is required for efficient GR-stimulated transcription. Taken together, these results suggest that a direct interaction between Ku70/86 and BRG1 brings together SWI/SNF remodeling capabilities and TOP2β activity to enhance the transcriptional response to hormone stimulation.
INTRODUCTION
The winding of DNA around histones creates nucleosomes that compact DNA into a dense chromatin structure that limits accessibility and represses transcription. This chromatin organization provides an additional level of control during certain nuclear processes, such as transcription, replication, recombination, and DNA damage repair. Proteins that alter DNA-histone contacts in chromatin by covalently modifying histones or by mechanically separating them from DNA have the power to activate or repress gene expression (1–5). Eukaryotes have evolved several such proteins and general biological processes dedicated to modulating chromatin architecture.
Two major classes of chromosome-modifying enzymatic activities alter nucleosomal structure; one class relies on covalent modification of histones, while the other uses the energy derived from ATP hydrolysis to alter the arrangement and stability of nucleosomes (6–9). While histone modifications do not substantially alter the nucleosome core particle, they can affect higher-order chromatin structures and gene expression (10–12).
ATP-dependent chromatin-remodeling complexes use energy derived from the hydrolysis of ATP to break histone-DNA contacts and reposition nucleosomes in a noncovalent manner (13). Generally, ATP-dependent chromatin remodelers are grouped into five major families, i.e., SWI/SNF, ISWI, Mi-2/NuRD, INO80, and SWR1, each being derived from the SNF2 helicase superfamily, which has a common structural core consisting of two RecA helicase-like domains that bind and hydrolyze ATP (1, 14–16).
The SWI/SNF family is highly divergent and can be present in multiple forms. Mammalian SWI/SNF exists as a large multiprotein complex that can have one of two possible central ATPase subunits, BRG1 (Brahma-related gene 1) or BRM (Brahma), that associate with several BAF (BRG1/BRM-associated factor) proteins to form a highly regulated multifunctional complex involved in numerous nuclear processes, including transcriptional regulation as well as DNA replication, repair, and recombination (17–19). However, most purified complexes contain core subunits BAF170, BAF155, and BAF47/INI1, as well as accessory subunits BAF250a/b or BAF180 and BAF200, BAF60a/b/c, BAF57, BAF53, and/or actin (18–20). Interestingly, SWI/SNF from purified mouse embryonic stem cells does not contain BAF170 and also does not contain BRM (21).
Over time, the number of BAF proteins identified has grown along with the number of distinct BAF complexes (17, 22). The differential configuration of SWI/SNF complexes suggests that the exchange of core and accessory subunits may help to refine the function of the central ATPase (BRG1 or BRM) so that it can regulate a variety of promoters with precise action and play a key regulatory role in numerous biological processes, including cell cycle progression, cell differentiation, immune response, and nuclear receptor (NR)-mediated signaling (21, 23–25).
The SWI/SNF chromatin-remodeling complex regulates nuclear receptor-stimulated transcription, and the BRG1 ATPase is recruited to hormone-responsive promoters upon stimulation with ligand (9, 26–28). The remodeling complex associates with various type I nuclear receptors, including glucocorticoid receptor (GR), estrogen receptor (ER), progesterone receptor (PR), and androgen receptor (AR), through critical interactions with BAF250a, BAF60a/c, and BAF57 (26, 29–36). Multiple interactions are involved in recruiting SWI/SNF to target promoters, and these interactions may be mediated and stabilized through direct and/or indirect associations involving one or more BAF subunits (18, 28).
The SWI/SNF complex has been shown to be integral to GR-mediated transcriptional regulation through detailed studies using the hormone-responsive mouse mammary tumor virus (MMTV) promoter (9, 27, 28, 37). This model promoter can be stably integrated into mammalian cells in order to study ligand-dependent nuclear receptor transcriptional activation. Such studies have demonstrated that with the localization of hormone-bound GR to the promoter, the SWI/SNF remodeling complex drives transcription by disrupting DNA-histone contacts and creating a loosened chromatin structure that facilitates the binding of transcription factors and the preinitiation machinery (38). This GR-dependent remodeling by the human SWI/SNF-like BAF complexes plays a key role in development, cell proliferation, and inflammation, and mutations or drugs that disrupt this process are associated with health problems, including cancer and heart disease (1, 18, 28).
Studies using the BRG1/BRM-null SW-13 cell line have established the requirement for BRG1 in hormone-dependent GR signaling from the chromosomally integrated MMTV promoter and from endogenous hormone-responsive promoters (38). Notably, studies using BRG1 mutants and chimeric proteins have shown that GR-induced transcriptional activation of target genes requires not only the BRG1 ATPase domain but also the BRG1 N terminus (39). Deletion or substitution of the BRG1 N terminus leads to a marked decrease in GR-mediated/BRG1-dependent transcriptional activation, with more targeted BRG1 mutants revealing the key N-terminal residues to be in the helicase-SANT-associated (HSA) domain region, which sits ∼300 residues to the N-terminal end of the BRG1 ATPase domain.
Deletion of the BRG1 HSA domain also results in the loss of BAF250a from the SWI/SNF chromatin-remodeling complex, resulting in an attenuated GR transactivation response (40). The HSA domain of BRG1 has also been shown to be important for the association between BRG1 and actin-related proteins such as BAF53 (28). Interestingly, HSA motifs have been found in numerous chromatin-remodeling proteins, including RSC, yeast and human SWI/SNF, SWR1, and INO80, where they have been shown to act as a general binding module for additional nuclear proteins that may help remodeling activity (41). Together these studies reveal the importance of the HSA domain as a critical interaction module, and determining which proteins interact through this motif may be an important step in elucidating the mechanism of SWI/SNF activity in nuclear processes such as transcriptional regulation (28, 40).
Numerous binding studies have been performed to identify proteins that associate with BRG1, and these studies have identified nuclear receptors, tumor suppressors, and transcriptional coactivators and corepressors (18). In this study, we performed a series of yeast two-hybrid (Y2H) screens of various human cDNA libraries using baits spanning the N terminus of BRG1, including the HSA and BRK regions, to identify novel protein interactions involving these domains that may play important roles in SWI/SNF function. These screens identified several proteins known to associate with the BRG1 N-terminal region that are also involved in processes such as DNA replication and damage repair, transcriptional regulation, cell cycle progression, apoptosis, and the structural maintenance of chromatin. One notable interaction our library screen identified was the association of a Ku70 (XRCC6/G22P1) fragment with the N-terminal HSA domain of BRG1.
Ku70 functions as a single-stranded DNA-dependent, ATP-dependent helicase and plays a key role in multiple nuclear processes such as DNA repair, chromosome maintenance, transcriptional regulation, and V(D)J recombination (42–44). The mechanism underlying the regulation of these diverse functions of Ku is still unclear. The Ku dimer consists of two tightly associated homologous subunits, Ku70 and Ku86, and was originally identified as an autoantigen recognized in the sera of patients with autoimmune disease (45). In eukaryotes, the Ku heterodimer contributes to genomic integrity through its ability to bind DNA double-strand breaks and facilitate repair by nonhomologous end joining (NHEJ). In mammalian cells, the Ku heterodimer recruits the catalytic subunit of DNA-dependent protein kinase (DNAPK), which is dependent on its association with the Ku70/86 heterodimer bound to DNA for its protein kinase activity (46).
Through the results presented here, we show Ku is recruited to BRG1 and is required for nuclear receptor (NR)-dependent SWI/SNF transcriptional activation of stably integrated MMTV promoters and other endogenous ligand-responsive promoters. Further investigation of the role of Ku in BRG1-regulated, NR-mediated transcription suggests that the enzymatic activity of TOP2β may also be involved. Using small interfering RNA (siRNA) protein knockdown and chromatin immunoprecipitation (ChIP) analyses, we have observed that components of the TOP2β/poly(ADP ribose) polymerase 1 (PARP1) complex, including TOP2β, DNAPK, Ku70/86, and PARP1, are recruited in a ligand-dependent manner to NR-responsive promoters and are involved in ligand-stimulated BRG1-mediated transcriptional activation. We also show that TOP2β-regulated double-strand DNA (dsDNA) breaks occur within chromatin-assembled MMTV upon addition of hormone and are necessary for regulated transcriptional activation. These data reveal the importance of the HSA domain of BRG1, via its interactions with Ku70, as a critical interactive surface that bridges key enzymatic activities involved in NR-mediated transcriptional initiation, enhancing our understanding of the underlying mechanisms.
MATERIALS AND METHODS
Y2H screen.
Yeast two-hybrid (Y2H) screening was carried out at Myriad Genetics Inc. (Salt Lake, UT) as described previously (47). In summary, BRG1-specific baits, corresponding to amino acids (aa) 1 to 250, 201 to 460, 329 to 507, 425 to 660, and 1053 to 1350, were cloned into the DNA-binding domain plasmids (pGBT.superB). Each bait plasmid was introduced into Myriad's ProNet yeast strain PNY200. The bait yeasts mated with ProNet MATa yeast cells containing three independent cDNA prey libraries prepared from human brain, spleen, or breast cancer/prostate cancer cell lines, fused to the C-terminal region of the Gal4 activation domain of the activation domain plasmid, pGAD.PN2. Each library was generated from random-primed, directionally cloned cDNA and represents over 5 million independent clones (average fragment size of 600 to 900 bp). Yeast transformants positive for prey-bait interaction were selected on SD−Ade/−His/−Leu/−Trp plates, and plasmid DNA isolated from positive clones was sequenced.
Cell culture.
SW-13 human adrenal carcinoma cells were maintained in normal growth medium containing Dulbecco's modified Eagle high-glucose medium (DMEM-H21) supplemented with 10% fetal bovine serum, 100 units/ml penicillin-streptomycin, and 2 mM l-glutamine. Parental MCF-7 and U2OS cells were maintained in accordance with ATCC specifications.
Transient transfection, drug treatment, and luciferase assay.
Transfections were carried out in antibiotic-free DMEM-H21 using Lipofectamine 2000 according to the manufacturer's instructions using the indicated expression plasmids: pSG5, pBJ5, and pcDNA3.1 (empty vectors) or pSG5.rGR, pBJ5/hBRG1a, pcDNA3.1/BRG1-3×Flag, pcDNA3.1D/Ku70-V5, pcDNA3.1D/BRG1-V5, pcDNA3.1D/BRG1.ΔHSA-V5, pcDNA3.1D/BRG1.ΔBRK-V5, and pcDNA3.1D/BRG1.Δ(475-656)-V5. Eighteen hours posttransfection, cells were treated with 100 nM dexamethasone (Dex) or an equal volume of vehicle (ethanol [EtOH]) for the indicated times in normal growth medium. For inhibition of TOP2, cells were treated with vehicle (dimethyl sulfoxide [DMSO]) or various concentrations of merbarone (Mer) (5-N-phenylcarboxamido-2-thiobarbituric acid) for 5 h prior to treatment with dexamethasone or vehicle for 16 h in normal growth medium. Following treatment, cells were assayed for luciferase activity as previously described (38). Relative light units (RLU) values were normalized to the total protein measured.
NU7026 was dissolved in DMSO as a 1 mM stock and stored at −20°C. SW-13/MMTV cells expressing GR and BRG1 were treated with 25 μM NU7026 or vehicle (DMSO) for 24 h prior to addition of hormone (48). After exposure, cells were treated with Dex or vehicle (EtOH) for 6 h, followed by total RNA extraction and cDNA synthesis. An equal amount of cDNA was used as the template for real-time PCR analysis using the indicated primers.
Immunoblot analysis.
Cells from subconfluent cultures were washed and scraped into phosphate-buffered saline (PBS) and pelleted by centrifugation. Whole-cell lysates were prepared and protein concentrations determined by Bradford assay as described previously (40). Samples were subjected to SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membranes, and blocked in Tris-buffered saline–Tween 20 (TTBS) containing either 5% nonfat dry milk or 5% normal donkey serum. Western blot analysis was performed as described previously using the indicated antibodies at the recommended concentrations (antibody information is available upon request) (40). To remove bound antibody, membranes were incubated in Restore Western blot stripping buffer (Pierce) according to the manufacturer's protocol.
IP assay.
SW-13, HeLa, or U2OS cells were transfected with BRG1, BRG1 mutant (ΔHSA, ΔBRK, or Δ475-656), or control expression plasmids. AT 24 h posttransfection, cells were harvested and lysed in immunoprecipitation (IP) buffer (50 mM HEPES [pH 7.0], 250 mM NaCl, 0.1% Nonidet P-40, 1 mM EDTA, 1 mM dithiothreitol [DTT]) containing protease inhibitor cocktail and benzonase nuclease (as indicated), followed by preclearance with Dynabead protein A/G magnetic beads (Invitrogen). Equal volumes of cleared lysate were immunoprecipitated overnight at 4°C with 5 μg of the indicated antibodies. Immunocomplexes were collected using protein G magnetic beads, washed extensively, and eluted in SDS loading buffer containing 2-mercaptoethanol. Coimmunoprecipitated proteins were identified by immunoblot analysis as described above. Antibody information is available upon request.
RNA isolation and RT-qPCR.
Total RNA was isolated from transfected SW-13 or parental MCF7 cells. At 24 h posttransfection, cells were scraped or treated with Dex, 17β-estradiol (E2), or vehicle (EtOH) for 4 h, and total RNA was extracted using TRIzol Plus (Invitrogen). DNase I-treated total RNA was reverse transcribed into single-stranded cDNA using oligo(dT) or random hexamers and SuperScript II (Invitrogen). An equal amount of cDNA from each experimental condition was amplified by real-time PCR using the Stratagene Mx3000P and Brilliant SYRB green quantitative PCR (qPCR) master mix with indicated gene-specific primer sets. For reverse transcription (RT)-qPCR, the average cycle threshold (CT) values were calculated and normalized to the CT values obtained from glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-specific primers. Optimized conditions for PCR amplification and ChIP and RT-PCR primer sequences are available on request.
siRNA transfection.
Small interfering RNA (siRNA) duplexes targeting the mRNA coding regions of Ku70, Ku86, DNAPK, PARP1, and TOP2β were designed using Web-based software and synthesized by Dharmacon. siRNAs targeting BAF155 and a nontargeting control (NTC) from Dharmacon were used as a positive control of SWI/SNF transcriptional activation knockdown or as a negative control of transcriptional activation knockdown, respectively. Sequence information regarding the chemically synthesized siRNA duplexes used in this study is available on request.
SW-13/MMTV or MCF7 parental cells were seeded into 6-well plates containing normal growth medium (minus antibiotics) at a density of 0.3 × 106 cells/well. Cells were cotransfected with GR and BRG1 expression plasmids (for SW-13/MMTV cells) or with 50 μM duplex siRNA using Lipofectamine 2000 in Opti-MEM (Invitrogen). At 48 h posttransfection, cells were treated with Dex (100 nM), 17β-estradiol (E2), or vehicle (EtOH) for 16 h in fresh normal growth medium. The extent of protein knockdown and expression of transfected plasmids were determined by immunoblotting at 48 h posttransfection. The effect of protein knockdown on transcriptional activation was evaluated by luciferase assay for SW-13/MMTV or by reverse transcription-real-time PCR to evaluate the transcription status from endogenous promoters using gene-specific primers.
ChIP and reChIP assays.
Transfected SW-13/MMTV or parental MCF7 cells were treated with 100 nM Dex, 17β-estradiol (E2), or vehicle (EtOH) for 1 h. After treatment, cells were cross-linked with 1% formaldehyde at 37°C for 10 min, washed twice with PBS, sedimented, and homogenized in SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris [pH 8.1], proteases inhibitors) on ice for 15 min. Cellular lysates were sonicated to obtain sheared DNA fragments ranging from ∼200 to 500 bp. After centrifugation, the supernatant was diluted in chromatin immunoprecipitation (ChIP) dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.1], 167 mM NaCl, protease inhibitors) and precleared with Dynabead protein G magnetic beads (Invitrogen) at 4°C for 30 min. Cleared lysates were incubated using the indicated antibodies overnight at 4°C. Immunocomplexes were collected with protein G beads, washed extensively with various ChIP buffers, reverse cross-linked overnight at 65°C, and digested with proteinase K (10 mg/ml) for 2 h at 45°C. Immunopurified DNA fragments were eluted from beads by incubation in elution buffer (1% SDS, 0.1 M NaHCO3) at room temperature for 30 min and purified using PCR purification columns (Qiagen).
The reChIP assays were performed following the protocol described previously (49). Specifically, nuclei from transfected SW-13/MMTV cells were isolated after formaldehyde cross-linking, and the complexes were eluted with 50 μl of 10 mm dithiothreitol at 37°C for 30 min. The eluted immunocomplexes were diluted 20 times with reChIP buffer (1% Triton X-100, 2 mm EDTA, 150 mm NaCl, 20 mm Tris-HCl [pH 8.0]), and the ChIP procedure described above was repeated again. DNA fragments were detected by real-time PCR using the indicated promoter-specific primers. Average cycle threshold (CT) amplification values were calculated and presented as percentage of sample input. Primer sequences are available on request.
DNA break/reiterative primer extension.
Detection of transient DNA strand breaks was performed using a modified reiterative primer extension technique. SW-13/MMTV cells were transfected with GR and pBJ5 (empty vector) or BRG1 expression plasmids. At 24 h posttransfection, cells were treated with Dex (100 nM) for the indicated times or with Mer (200 μM) for 5 h prior to treatment with Dex for 15 min. Nuclei were immediately isolated in buffer containing Streck cell fixative and lysed in proteinase K buffer, and genomic DNA was purified by phenol-chloroform extraction and sodium acetate precipitation as previously described (50). Equal amounts of genomic DNA were digested to completion using restriction endonuclease AvaII or HaeIII to detect transient breaks within the lower and upper DNA strands, respectively. Twenty-five micrograms of purified DNA was used as the template for reiterative primer extension with 32P-labeled oligonucleotides specific for the nucleosome B region of MMTV. Extension products were analyzed by using linear Taq polymerase amplification with 32P-labeled oligonucleotides (lower strand, 5′-TCGTCACTTATCCTTCACTTTCCAGA-3′; upper strand, 5′-CAGTGGCTGGACTAATAGAAC-3′) specific for the nucleosome B region of MMTV. Purified products were analyzed on polyacrylamide denaturing gels. Detection and quantification of strand-specific DNA breaks were performed using ImageQuant analysis software.
Restriction enzyme hypersensitivity assay.
SW-13/MMTV cells were transiently transfected with expression plasmids for GR and BRG1. Cells were pretreated with 100 μM merbarone, followed by treatment with Dex (100 nM) or vehicle for the times indicated. Nuclei were isolated and subject to digestion with SstI (in vivo) and HaeIII (in vitro) as previously described (37). Equal amounts of purified digestion products were analyzed by linear Taq polymerase amplification with a 32P-labeled oligonucleotide specific for MMTV (38). Purified amplification products were resolved on polyacrylamide denaturing gels.
RESULTS
Ku protein interacts with the chromatin-remodeling factor BRG1.
The N-terminal region of BRG1 contains several domains of conserved sequence homology whose functions and biological relevance remain unclear, including a QLQ region, an HSA domain, and a BRK domain (18). Recent studies indicate that these domains are important for interactions between BRG1 and other subunits of the SWI/SNF chromatin remodeling complex as well as proteins involved in transcriptional regulation (40). To identify possible interactions involving the N-terminal region, a series of yeast two-hybrid (Y2H) screens was conducted using three independent cDNA libraries prepared from various human tissues or oncogenic cell lines and baits spanning the N-terminal region of BRG1. Of the 103 open reading frames identified as BRG1 binding partners, several have been previously described. These include BRD7 (51), CBX5 (52), SMARCC1 and SMARCC2 (53), SSXT (54), TAL1 (55, 56), and TMF1 (57). Our Y2H screen also revealed 13 novel interactions which require the BRG1 N-terminal region (Table 1). These novel BRG1-associating proteins have diverse functions and participate in various nuclear processes, including nucleosome assembly, chromatin maintenance, transcriptional regulation, and DNA repair.
TABLE 1.
BRG1-interacting protein as identified in yeast two-hybrid screena
| No. | Bait |
Prey |
||
|---|---|---|---|---|
| Name | Amino acid coordinates | Name | Amino acid coordinates | |
| 1* | BRG1 | 329–507 | BAG1 | 111–281 |
| 2 | BRG1 | 1–250 | BRD7 | 158–652 |
| 3* | BRG1 | 201–507 | CBX1 | 104–186 |
| 4 | BRG1 | 201–460 | CBX3 | 42–182 |
| 5 | BRG1 | 205–507 | CBX5 | 8–192 |
| 6* | BRG1 | 1375–1648 | CHD5 | 16–399 |
| 7* | BRG1 | 329–507 | DDX3X | 87–285 |
| 8* | BRG1 | 425–660 | XRCC6 (Ku70) | 415–555 |
| 9* | BRG1 | 205–450 | ING5 | 1–241 |
| 10* | BRG1 | 329–507 | IPO7 | 595–772 |
| 11* | BRG1 | 329–507 | IPO5 | 324–812 |
| 12* | BRG1 | 600–800 | NAP1L1 | 2–358 |
| 13 | BRG1 | 205–507 | SMARCC1 | 536–693 |
| 14 | BRG1 | 201–507 | SMARCC2 | 215–915 |
| 15* | BRG1 | 329–507 | SPDEF | 103–326 |
| 16* | BRG1 | 1–250 | SSXT | 8–392 |
| 17 | BRG1 | 1053–1350 | TAL1 | 2–193 |
| 18* | BRG1 | 329–507 | THOC2 | 1134–1380 |
| 19 | BRG1 | 201–460 | TMF1 | 879–1094 |
| 20* | BRG1 | 425–660 | TPR | 339–571 |
| 21* | BRG1 | 205–507 | TRIM11 | 6–279 |
The identity of prey clones was derived from a screen using BRG1-specific baits. BRG1-specific baits were cloned into a DNA-binding domain vector (pGBT.superB) and transformed into PNY200 yeast. Bait yeasts were mated with MATa yeast containing cDNA prey libraries prepared from human brain, spleen, or breast cancer/prostate cancer. Bold, interaction surface encompassing the BRG1-HSA domain; *, novel interaction.
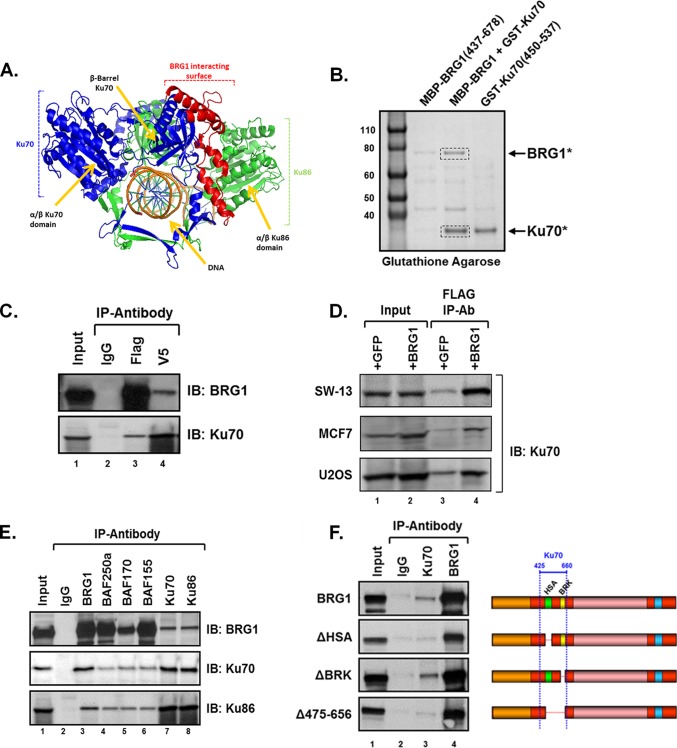
From this Y2H screen, we identified a region of the double-strand break repair protein Ku70 (XRCC6) as a binding partner for a BRG1 fragment encompassing two domains of conserved homology, HSA and BRK motifs, that lie adjacent to BRG1 in the ATPase domain. The segment of Ku70 involved in the BRG1 interaction includes the central DNA-binding beta-barrels, polypeptide rings, and C-terminal arm of the protein (58) (Fig. 1A). To validate the Y2H interaction results obtained for Ku70 with BRG1, we performed in vitro pulldowns with the domains identified in our screen. Following incubation, the Ku70-bound glutathione S-transferase (GST) beads were extensively washed, and the remaining proteins were eluted and subjected to SDS-PAGE analysis. We found that BRG1-MBP selectively binds GST-Ku70, with the identity of each protein verified by mass spectrometry analysis, validating the Y2H observation (Fig. 1B).
FIG 1.
Ku associates with the BRG1-SWI/SNF complex. Protein pulldown studies and immunoprecipitation assays were performed to validate the identified yeast two-hybrid interaction between Ku and BRG1. (A) Schematic representation of the Ku dimer with the putative BRG1 interaction surface indicated. Ku86 is shown in green and Ku70 displayed in blue and red. The amino acid residues 425 to 660 of Ku70, marked in red, were found to associate with the HSA-BRK region of BRG1. (B) A purified BRG1 fragment containing the HSA and BRK (aa 437 to 678) domains interacts with a purified Ku70 fragment (aa 450 to 537). Bacterially expressed MBP-BRG1 and GST-Ku proteins were used for glutathione-Sepharose pulldown analysis. Stained SDS-PAGE shows that the BRG1 protein associates with the Ku70 fragment. Mass spectrometry analysis confirms the identity of BRG1, Ku70, or nonspecific bands. (C) Coimmunoprecipitation assays were performed using whole-cell lysates from parental SW-13 cells, expressing BRG1-3×Flag and Ku70-V5 proteins, and antibodies specific to the indicated epitope tag. Immunopurified complexes were resolved by 7.5% SDS-PAGE and analyzed by Western blotting (IB) using anti-BRG1 or -Ku70 antibodies. (D) Ku70 associates with BRG1 in various cell types. Immunoprecipitation assays were performed using whole-cell lysates IPs were performed as described above. Immunopurified complexes were washed, resolved by SDS-PAGE, and analyzed by Western blotting using Ku70-specific antibody. (E) Complexes containing endogenous Ku and BRG1 were immunoprecipitated from transfected SW-13 cells expressing BRG1, using antibodies specific for SWI/SNF subunit BRG1, BAF250a, BAF170 or BAF155, as well as for Ku70 or Ku86. Immunopurified proteins were resolved by SDS-PAGE and immunoblotted using BRG1, Ku70, Ku86, BAF250a, BAF170, or BAF155 antibodies. (F) The HSA domain of BRG1 is required for Ku binding. Whole-cell lysates from SW-13 cells expressing BRG1 or mutant BRG1 proteins ΔHSA, or ΔBRK were immunoprecipitated, in the presence of benzonase nuclease, using antibodies specific for BRG1 or Ku70. Purified proteins were resolved by SDS-PAGE and analyzed by immunoblotting with anti-BRG1 antibody. For each immunoprecipitation, input represents 10% of the total lysate used and normal IgG was used to control for nonspecific interactions.
To confirm binding between Ku70 and BRG1 in mammalian cells, both proteins were C-terminally tagged with either FLAG or V5, respectively, and transiently coexpressed in SW-13 cells. Immunoprecipitation using FLAG- or V5-specific antibodies showed that both proteins associate in vivo (Fig. 1C). To determine if the BRG1-Ku70 association occurs within different human cell types, we performed immunoprecipitation analysis using SW-13, HeLa, and U2OS osteosarcoma cells transiently expressing BRG1-FLAG or GFP-FLAG. Total cell lysate from each cell line was immunoprecipitated using anti-FLAG antibody, and the immunopurified complexes were analyzed by Western blotting using anti-Ku70 antibody.
Our coimmunoprecipitation data show that Ku70 preferentially associates with BRG1 compared to the nonspecific binding control GFP (Fig. 1D, compare lanes 3 and 4). These data suggest that the Ku interaction with BRG1 is not specific to SW-13 cells and may play an important role in a variety of cell lines. To further examine if endogenous Ku also interacts with core BAF subunits of the SWI/SNF chromatin-remodeling complex, we performed a series of coimmunoprecipitation assays using antibodies specific for various BAF and Ku proteins, including BRG1, BAF250a, BAF170, BAF155, Ku70, or Ku86. Interestingly, endogenous Ku strongly interacts with BRG1 and is also found to associate with all members of the SWI/SNF complex assayed (Fig. 1E). The preferential binding of Ku to BRG1 may result from the direct interaction between these proteins, while the weaker association with the core BAF proteins may result from indirect, BRG1-mediated interactions between Ku and member subunits of the remodeling complex, as suggested by the Y2H and recombinant binding assays.
To determine whether the HSA or BRK domain of BRG1 was required for association with Ku protein, we performed a series of coimmunoprecipitation assays in SW-13 cells expressing BRG1 or BRG1 deletion mutants, ΔHSA, ΔBRK, or Δ475-565 (ΔHSA and ΔBRK). As reported in a previous study, each of these mutants retains intact ATPase activity (40). Immunoprecipitation buffer was supplemented with benzonase nuclease (5 U/ml) to eliminate both DNA- and RNA-dependent interactions. SWI/SNF complexes were immunopurified using antibodies specific for Ku70 or BRG1 and detected by Western blotting with anti-BRG1 antibody. As shown by immunoblot analyses, Ku70 associates with wild-type BRG1 and the ΔBRK mutant protein; however, when the HSA region is removed (as in ΔHSA or Δ475-656), the Ku70 interaction with BRG1 is significantly diminished, suggesting that the HSA motif may serve as the critical binding module for the association with Ku (Fig. 1F).
The HSA domain of BRG1 is required for recruitment of Ku to chromatin MMTV and to endogenous promoters.
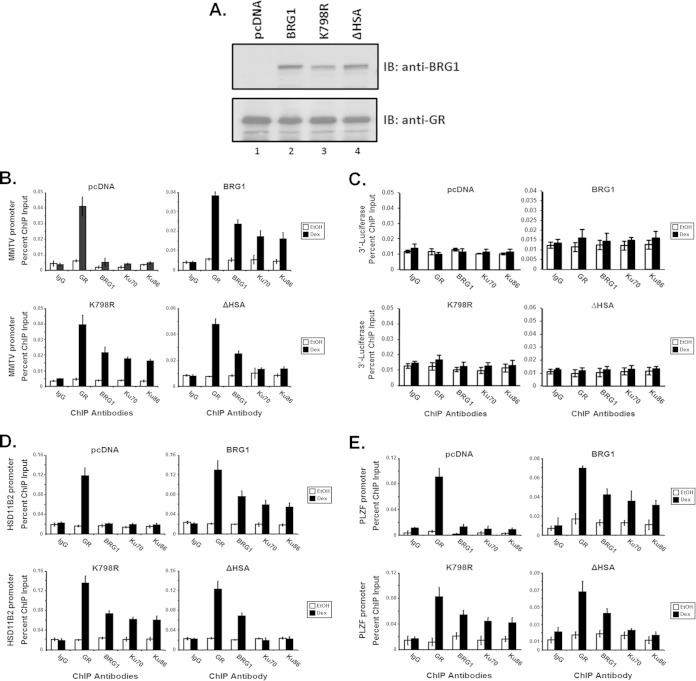
Previous studies have shown that BRG1 is recruited to chromatin MMTV in a hormone-dependent manner, suggesting that ligand-induced binding of GR may lead to recruitment of the remodeling complex to target promoters (38). To determine whether Ku proteins are recruited to GR-responsive promoters, we performed ChIP analysis in treated SW-13/MMTV cells (Fig. 2). Cells expressing GR and either empty vector (pcDNA) or BRG1-, K798R-, or ΔHSA-3×Flag protein (Fig. 2A) were treated with Dex or vehicle, subjected to ChIP analysis using anti-GR, -FLAG, -Ku70, or -Ku86 antibodies, and analyzed by real-time PCR with primers specific for MMTV-NucB, HSD11B2, or PLZF promoters. Our data suggest that Ku proteins are recruited to the GR-responsive promoters in a hormone-dependent fashion (Fig. 2B, D, and E). Interestingly, Ku promoter binding appears to be dependent on the presence of BRG1 protein (compare pcDNA to BRG1) but is not dependent on BRG1-ATPase activity (compare BRG1 to K798R). However, in cells expressing the BRG1-ΔHSA mutant protein, Ku promoter recruitment is significantly decreased (compare BRG1 and K798R to ΔHSA) (Fig. 2B, D, and E). When purified ChIP DNA was analyzed using primers specific for the downstream luciferase reporter, recruitment of GR, BRG1, Ku70, or Ku86 was not detected, indicating that the binding of these proteins is specific for the promoter region (Fig. 2C). Taken together, these data suggest that Ku is recruited to GR-responsive promoters, including the chromatin-integrated MMTV promoter and endogenous promoters HSD11B2 and PLZF, in a hormone-dependent manner and that this recruitment requires the HSA domain of BRG1.
FIG 2.
Hormone-dependent recruitment of Ku to chromatin MMTV and endogenous promoters requires the HSA domain of BRG1. Recruitment of Ku70/86 to stably integrated MMTV or endogenous PLZF or HSD11B2 promoters requires the N-terminal HSA domain of BRG1, as demonstrated by chromatin immunoprecipitation analysis. (A) Western blot analysis using anti-BRG1 or -GR antibodies in order to monitor protein expression of transfected plasmids, with stably expressed GR serving as protein loading control. (B to E) Purified immunoprecipitated DNA fragments from treated SW-13/MMTV cells expressing BRG1, K798R, or ΔHSA proteins were analyzed by real-time PCR using primer sets covering the MMTV promoter nucleosome B region (B) and the 3′ coding region of the integrated luciferase reporter (C) or gene-specific primers targeting the endogenous promoters of HSD11B2 (D) and PLZF (E). Cross-linked DNA-protein complexes were isolated using antibodies specific for GR, BRG1, Ku70, or Ku86 protein. Normal IgG was used as a control for nonspecific interactions. Quantitative analysis was performed, with results displayed as percentage of ChIP input. Values are shown as mean ± standard deviation from 3 biological replicates.
Ku70 enhances nuclear receptor-mediated BRG1-dependent transcriptional activation of chromatin MMTV and endogenous genes.
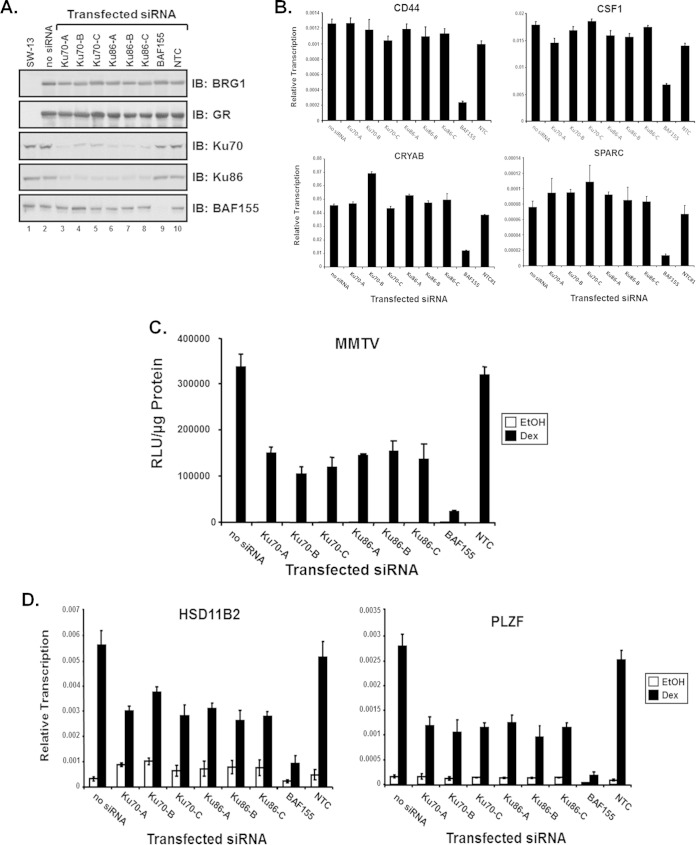
To determine whether Ku70 is required for SWI/SNF-dependent transcriptional activation, we performed a series of siRNA studies in conjunction with transcriptional activation assays. Parental SW-13 cells were cotransfected with BRG1 expression plasmid and siRNA duplexes specific for Ku70, Ku86, BAF155, or a nontargeting control (NTC). For these transcriptional activation assays, three independent siRNA duplexes for both Ku70 and Ku86 were used to evaluate the effects of Ku on BRG1-dependent transcription. Immunoblot analysis was performed using antibodies specific for BRG1, Ku70, Ku86, or BAF155 to determine the extent of protein expression and to evaluate the level of siRNA protein knockdown within the transfected cells. Transfecting SW-13 cells with Ku70 or Ku86 siRNA resulted in a significant reduction in Ku protein within 48 h (Fig. 3A). Expression of endogenous BRG1-dependent genes was evaluated in the presence of siRNA-mediated knockdown of Ku protein. At 48 h after siRNA transfection, total RNA from SW-13 cells was used for cDNA synthesis and evaluated to determine relative transcription levels of BRG1-dependent genes. Notably, expression of CD44 (59), CRYAB, SPARC (38, 40), and CSF1 (60) was not altered upon protein knockdown of Ku, yielding similar relative expression, indicating that Ku protein is not required for general BRG1-mediated transcription (Fig. 3B).
FIG 3.
Ku is required for GR-mediated BRG1-dependent transcriptional activation. Three siRNA duplexes specific for human Ku70 or Ku86 were used to evaluate the requirement of Ku in GR-mediated BRG1-dependent transcriptional activation of chromatin MMTV and endogenous genes. (A) Western blot analysis using total protein lysates from transfected SW-13/MMTV cells and antibodies specific for GR, BRG1, Ku70, Ku86, or BAF155 to monitor protein expression levels from transfected plasmids and to evaluate the extent of siRNA-induced protein knockdown. (B) Reverse transcription–real-time PCR analysis was used to determine the expression levels of endogenous BRG1-dependent genes in parental SW-13 cells cotransfected with BRG1 expression plasmid and the indicated siRNA duplexes specifically targeting Ku70, Ku86, or the core SWI/SNF subunit BAF155. Equal amounts of cDNA and gene-specific primers for CRYAB or SPARC were used for real-time PCR analysis. A nontargeting control (NTC) duplex siRNA used as a negative control. Quantitative analysis was performed with data normalized to GAPDH, and results are displayed as relative expression. Values are shown as mean ± standard deviation from 3 biological replicates. (C) SW-13 cells, containing a stably integrated MMTV reporter, were cotransfected with GR and BRG1 expression plasmids, and the indicated siRNAs were treated with 100 nM Dex or ethanol (EtOH) and assayed for relative luciferase activity. Relative MMTV luciferase activity was normalized to total protein measured and represented as relative light units (RLU). Values are shown as mean ± standard deviation (n = 3). (D) Reverse transcription-PCR analysis was used to determine the expression levels of endogenous GR/BRG1-dependent genes in SW-13 cells upon Ku siRNA protein knockdown. Total RNA extracted from treated SW-13 cells cotransfected with GR and BRG1 expression vectors and Ku70 or Ku86 specific siRNA duplexes was used as the template for cDNA synthesis. A nontargeting control (NTC) siRNA used as a negative control for protein knockdown. Values are shown as mean ± standard deviation from 3 biological replicates.
To determine if Ku is required for nuclear receptor-mediated BRG1-dependent transcriptional activation, we performed similar siRNA protein knockdown studies in SW-13 cells containing an integrated MMTV promoter reporter driving expression of luciferase. SW-13/MMTV cells were cotransfected with expression plasmids for GR and BRG1 along with siRNA duplexes specific for Ku70, Ku86, BAF155, or a NTC, treated with Dex or vehicle, and assayed for reporter activity (Fig. 3C). Cells expressing GR and BRG1 (no siRNA) showed an increase in reporter activity from the chromatin promoter upon hormone treatment, as previously described (38). Interestingly, this hormone-stimulated GR/BRG1 activity from chromatin MMTV was significantly attenuated upon Ku protein knockdown, suggesting that Ku may be necessary for GR-mediated BRG1-dependent transcriptional activation (Fig. 3C). To test whether Ku is required for transcriptional initiation of endogenous GR-mediated, BRG1-dependent genes, we evaluated expression of HSD11B2 (38) and PLZF/ZBTB16 (61) upon siRNA protein knockdown of Ku70 and Ku86. Expression of both genes was reduced in the presence of Ku70 or Ku86 siRNA but not upon transfection with the NTC (Fig. 3D). Taken together, these results suggest that Ku may play an important role that is specific to hormone-stimulated nuclear receptor-mediated transcriptional regulation by the SWI/SNF chromatin-remodeling complex.
GR-dependent BRG1-mediated recruitment of TOP2β/PARP1 complex components is critical for transcriptional regulation.
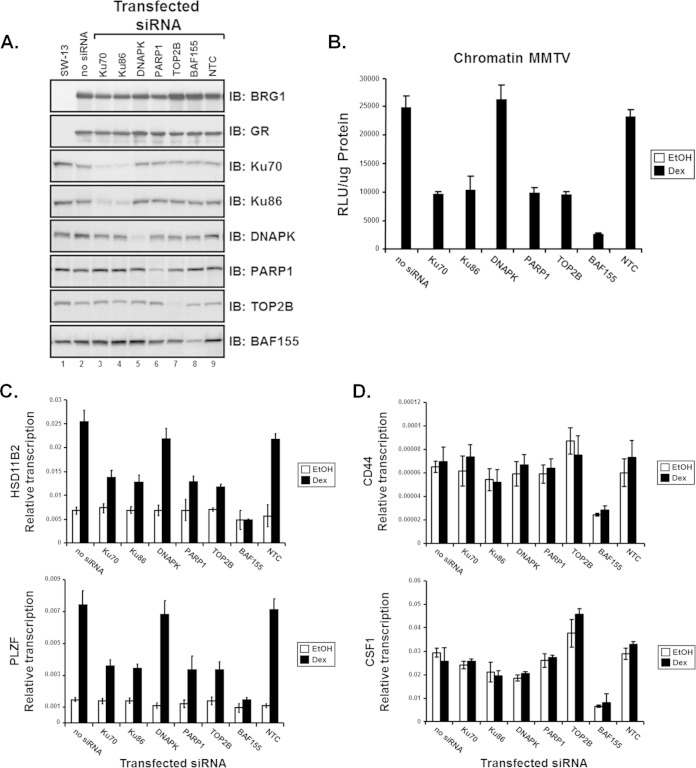
Previous reports suggest that components of the DNA damage/repair machinery, including the DNA-dependent protein kinase (DNAPK), Ku70, and Ku86, along with TOP2β and poly(ADP-ribose) polymerase 1 (PARP1), are all recruited to the estrogen receptor-responsive pS2 promoter (62). These observations may suggest a role for the DNA damage and repair apparatus in hormone-dependent transcriptional activation by nuclear receptors. To determine if components of the TOP2β/DNAPK/Ku complex are necessary for GR-mediated BRG1-dependent transcriptional activation, we performed siRNA protein knockdown reporter assays using duplexed siRNAs that target each member protein of the TOP2β/PARP1 complex. SW-13/MMTV reporter cells were cotransfected with GR and BRG1 expression plasmids along with siRNA duplexes specific for Ku70, Ku86, DNAPK, PARP1, or TOP2β (Fig. 4A), treated with hormone (Dex), and assayed for luciferase reporter activity.
FIG 4.
Components of the TOP2β/PARP-1 complex are required for the transcriptional activation of chromatin MMTV and endogenous GR-mediated BRG1-dependent genes. siRNA duplexes specific for members of the TOP2β/PARP-1 complex were used to evaluate the requirement of Ku70, Ku86, DNAPK, TOP2β, and PARP-1 proteins in GR-mediated BRG1-dependent transcriptional activation. (A) Western blot analysis was used to monitor levels of protein expression of transfected plasmids and to evaluate the extent of protein knockdown upon introduction of siRNA. Total protein lysates from transfected SW-13 cells were resolved by SDS-PAGE and Western analysis performed using antibodies specific for BRG1, GR, Ku70, Ku86, DNAPK, PARP1, TOB2β, or BAF155. (B) SW-13/MMTV cells cotransfected with GR and BRG1 expression plasmids and the indicated siRNA were treated with 100 nM Dex or vehicle (EtOH) and assayed for luciferase activity. Relative MMTV luciferase activity was normalized to total protein measured and represented as relative light units (RLU). (C and D) Reverse transcription–real-time PCR analysis was used to determine expression levels of endogenous BRG1-mediated and GR-dependent (C) or -independent (D) genes in SW-13 cells upon siRNA protein knockdown. Total RNA, extracted from Dex- or vehicle-treated cells cotransfected with GR and BRG1 expression vectors and siRNA duplexes, was used as the template for cDNA synthesis. Equal amounts of cDNA were used for real-time PCR analysis with primers specific for GR-dependent genes HSD11B2 or PLZF (C) or GR-independent genes CD44 and CSF1 (D). Quantitative analysis was performed with data normalized to GAPDH, and results are displayed as relative transcription. A nontargeting control (NTC) siRNA used as a negative control for protein knockdown. Values are shown as mean ± standard deviation from 3 biological replicates.
As previously shown, knockdown of Ku protein resulted in a decrease in GR-mediated BRG1-dependent reporter levels; however, transcriptional activity in cells with reduced DNAPK protein appeared to be unchanged. Notably, transcription from the MMTV promoter in cells transfected with siRNAs targeting PARP-1 and TOP2β was reduced to levels similar to that observed in cells with reduced Ku protein (Fig. 4B). To determine whether members of the TOP2β/DNAPK/Ku complex are required for GR-mediated, BRG1-dependent transcriptional activation of endogenous genes, we evaluated expression of HSD11B2 or PLZF. Transcription from each endogenous promoter was attenuated in the presence of reduced Ku, PARP-1, and TOP2β siRNA but not upon transfection with the NTC (Fig. 4C). In contrast, expression of CD44 and CSF1, two BRG1-dependent genes not regulated by glucocorticoids, was not significantly reduced. Thus, components of the TOP2β/DNAPK/Ku complex may be limited to hormone-induced transcription or other pathways involving nuclear receptor signaling (Fig. 4D). GR-mediated BRG1-dependent transcriptional activation of chromatinized MMTV and endogenous promoters HSD11B2 or PLZF was unaffected by NTC control siRNA but significantly reduced with knockdown of the SWI/SNF core subunit, BAF155 (Fig. 4B to D). Together, these data suggest that members of the TOP2β/PARP1 complex, including Ku70 and Ku86, play an important role in the rapid response to hormone that occurs during NR-mediated transcriptional regulation by the SWI/SNF chromatin-remodeling complex.
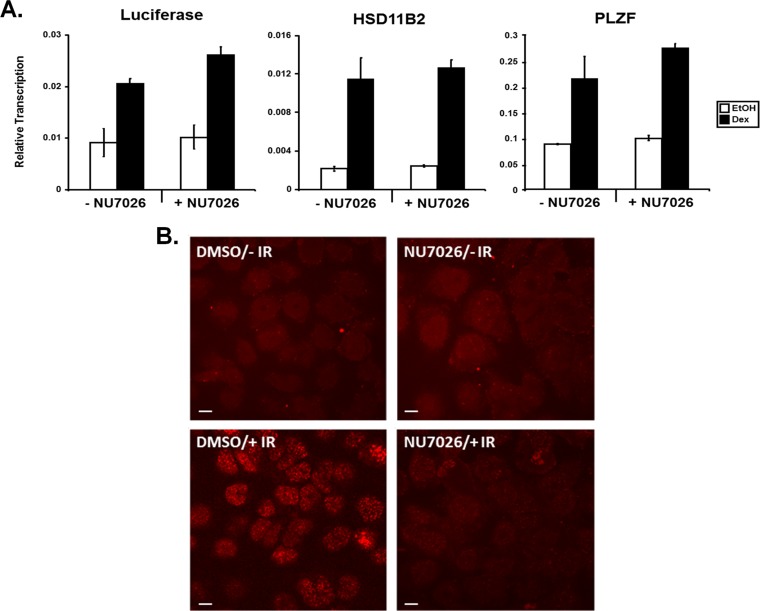
Our data suggest that DNAPK is recruited to GR-responsive promoters in a hormone-dependent fashion; however, inhibition of DNAPK by siRNA did not seem to alter gene expression levels in SW-13 cells. To address whether the enzymatic activity of DNAPK is required for hormone-mediated transcription, we treated SW-13/MMTV cells expressing GR and BRG1 with the specific DNAPK inhibitor NU7026 prior to Dex treatment. NU7026 did not affect expression from endogenous glucocorticoid-regulated genes (Fig. 5A). Although previous studies indicate a functional link between DNAPK and both ER and AR transcriptional activity (63–66), our results suggest that the DNAPK catalytic activity is not involved in the initial steps required for regulated gene expression in response to glucocorticoid signaling. To test the efficacy of the NU7026 used in this assay, cells pretreated with inhibitor or vehicle were irradiated with 5 Gy infrared (IR). At 15 min post-IR, cells were immune stained with anti-gamma-H2A.X (phosphor S139) antibody. As predicted, cells treated with the inhibitor show reduced γ-H2A.X staining, indicating that NU7026 efficiently inhibits phosphorylation of S139 of histone H2A.X by DNAPK (Fig. 5B).
FIG 5.
DNAPK activity is not required for GR-mediated BRG1-dependent transcriptional activation from chromatinized MMTV or endogenous promoters. (A) SW-13/MMTV cells expressing GR and BRG1 were treated with 25 μM DNAPK-specific inhibitor NU7026 (+NU7026) or DMSO vehicle (−NU7026) for 24 h. After exposure, cells were treated with Dex or vehicle (EtOH) for 6 h, followed by total RNA extraction and cDNA synthesis. Equal amounts of cDNA and gene-specific primers for luciferase, HSD11B2, or PLZF were used for real-time PCR analysis. Quantitative analysis was performed with data normalized to GAPDH and displayed as relative expression. Values are shown as mean ± standard deviation (n = 3). (B) To evaluate the effectiveness of NU7026, cells were pretreated with NU7026 or vehicle (DMSO), followed by irradiation with 5 Gy IR. At 15 min post-IR, cells were fixed, permeabilized, and stained with anti-gamma-H2A.X (phosphor S139) antibody. Representative images, collected using an LSM 5 Pa confocal microscope at ×63 magnification, are shown for each treatment group. Scale bars, 10 μm.
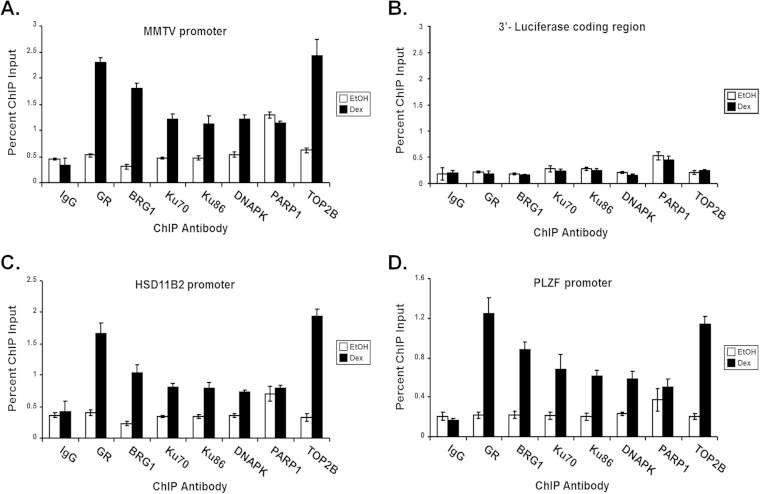
To determine whether components of the TOP2β/DNAPK/Ku complex are recruited to GR-responsive SWI/SNF-dependent promoters, we performed chromatin immunoprecipitation (ChIP) analysis using treated SW-13/MMTV cells expressing GR and BRG1. Cells expressing GR and BRG1 were treated with Dex or vehicle and subjected to ChIP analysis using antibodies specific for GR, BRG1 and members of the TOP2β/DNAPK/Ku complex. As previously shown, GR, BRG1, and Ku proteins are recruited to GR-responsive promoters in a hormone-dependent manner (Fig. 2). Interestingly, DNAPK, PARP1, and TOP2β also target GR-responsive promoters, with PARP1-binding promoters independent of hormone (Fig. 6A to D). DNAPK, PARP1, and TOP2β did not bind to the luciferase 3′ untranslated region (UTR) negative control (Fig. 6B). These data suggest that members of the TOP2β/DNAPK/Ku complex are recruited to GR-responsive promoters in a hormone-dependent manner and that this recruitment is required for nuclear receptor-mediated SWI/SNF-dependent transcriptional regulation (Fig. 3).
FIG 6.
Components of the TOP2β/PARP-1 complex are recruited to chromatinized MMTV and endogenous GR/BRG1-responsive promoters in a hormone-dependent manner. DNAPK, TOP2β, and PARP-1 are recruited to stably integrated MMTV and endogenous promoters in a GR/BRG1-dependent manner, as demonstrated by chromatin immunoprecipitation analysis (ChIP). Purified immunoprecipitated DNA fragments from treated SW-13/MMTV cells expressing GR and BRG1 were analyzed by real-time PCR using primer sets covering the nucleosome B promoter region of MMTV (A) and to the 3′ coding region of the integrated luciferase reporter (B) or gene-specific primers targeting the GRs of HSD11B2 (C) and PLZF (D). Normal IgG was used as a control for nonspecific interactions. Quantitative analysis was performed and results displayed as percentages of ChIP input. Values are shown as mean ± standard deviation from biological replicates.
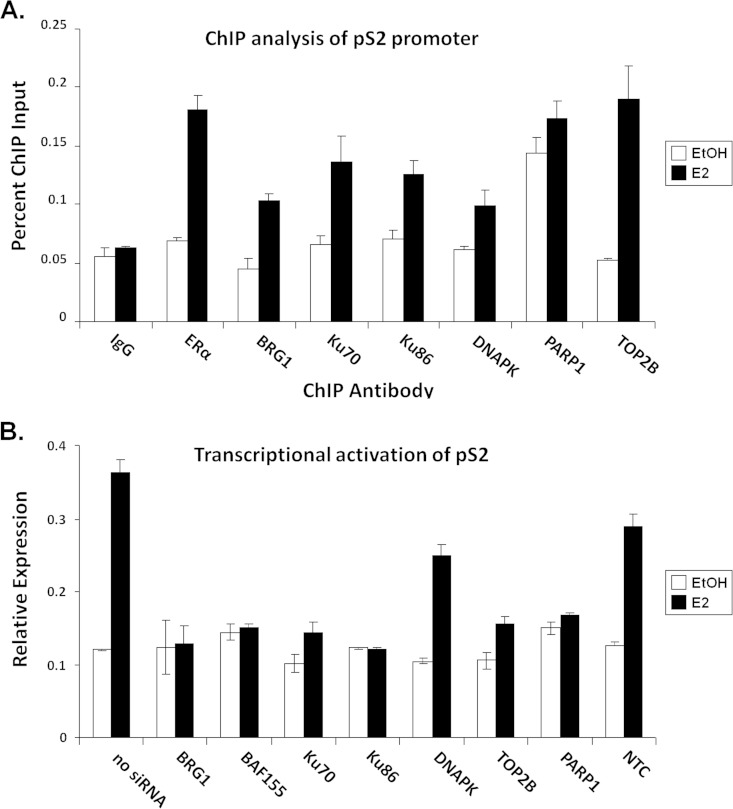
To determine if components of the SWI/SNF and the Ku/TOP2β/DNAPK complexes are recruited to and required for transcriptional activation of the 17β-estradiol (E2)-dependent pS2 promoter, we performed RT-PCR and ChIP analysis. MCF7 cells, transfected with siRNA duplexes specific for BRG1, BAF170, Ku70, Ku86, DNAPK, PARP1, or TOP2β were treated with E2 or vehicle and subjected to transcriptional activation assay using the estrogen-responsive region within the pS2 promoter. As previously reported, components of the TOP2β/DNAPK/Ku complex are required for hormone-dependent transcriptional activation of pS2 (65). Notably, members of the SWI/SNF complex, including the central ATPase, BRG1, and the core subunit, BAF155, are also needed for ER-mediated transcription of pS2 (Fig. 7A). Chromatin immunoprecipitation analysis was performed to determine if BRG1 is recruited to the pS2 promoter in a hormone-dependent manner. After E2 treatment, ER and all members of the TOP2β/DNAPK/Ku complex as well as BRG1 were recruited to the pS2 promoter (Fig. 7B), suggesting that the SWI/SNF complex may be involved in ER-mediated transcriptional regulation.
FIG 7.
Components of the SWI/SNF and the TOP2β/PARP-1 complexes are recruited to and required for the transcriptional activation of endogenous pS2 in MCF-7 cells. (A) siRNA-mediated protein knockdown analysis to evaluate the requirement of the SWI/SNF chromatin remodeling complex and members of the TOP2β/PARP-1 complex in ER-mediated transcriptional activation. Reverse transcription–real-time PCR analysis was used to determine the level of transcriptional activation from the endogenous ER-responsive gene pS2 in MCF-7 cells upon siRNA protein knockdown. Total RNA, extracted from estradiol (E2)- or vehicle (EtOH)-treated MCF-7 cells transfected with indicated siRNA duplexes, was used as the template for cDNA synthesis. (B) BRG1 and members of the TOP2β/PARP-1 complex are recruited to the endogenous pS2 promoters in MCF7 cells, as demonstrated by ChIP analysis using antibodies specific for ERα, BRG1, Ku70, Ku86, DNAPK, PARP1, and TOP2β.
Taken together, these data suggest that the enzymatic activities of TOP2β and its complex members Ku and PARP1 may be the critical components involved in initiating the multistep process required for SWI/SNF-regulated nuclear receptor-mediated transcription. Chromatin immunoprecipitation analysis suggests that components of the Ku/TOP2β/PARP1 complex are recruited to NR target promoters in a ligand-dependent manner. Inhibition of Ku70, Ku86, TOP2β, and PARP1 by siRNA led to reduced GR- and ER-mediated transcriptional activation in SW-13 and MCF7 cells, respectively. Interestingly, our data further establish the functional link between DNA repair and transcriptional regulation by nuclear receptors and the SWI/SNF chromatin-remodeling complex (18, 67–69).
Ku70 and SWI/SNF subunits, BRG1 and BAF250a, cooccupy chromatinized MMTV after stimulation with hormone.
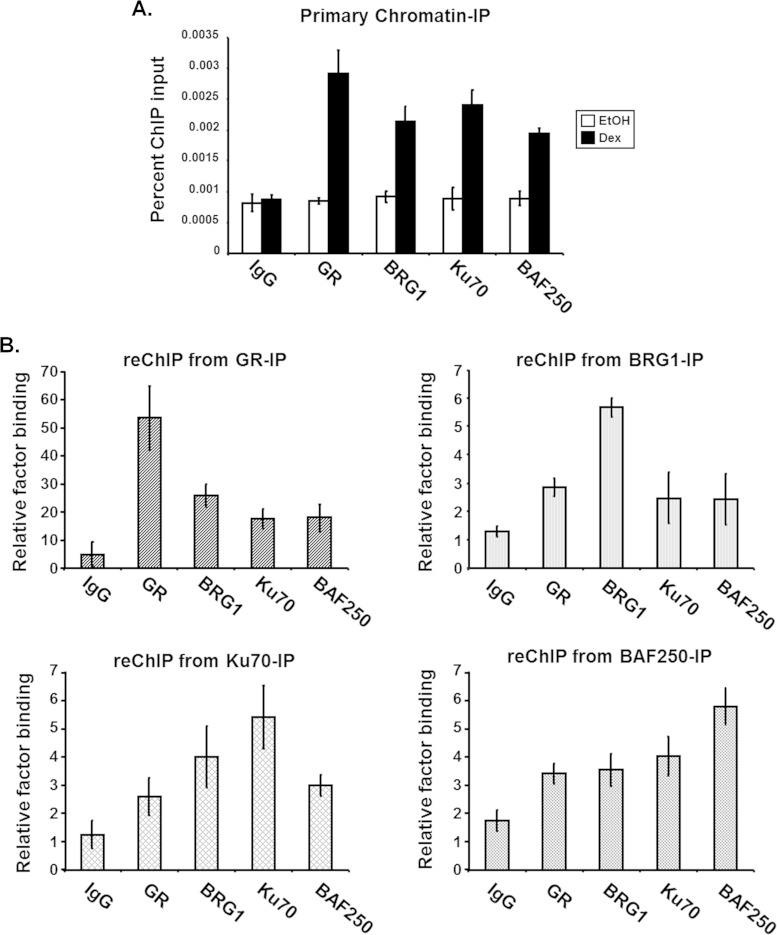
Helicase-SANT-associated (HSA) domains have been identified in several chromatin-remodeling proteins, usually present adjacent to the central ATPase domain, and serve as a primary binding module for regulatory or structural proteins, including actin-related proteins (ARP) and actin (40), (18). The N-terminal region of BRG1 contains an HSA domain that is similar to those found within BRG1 homologs as well as the related INO80 chromatin remodeler (41). Studies using BRG1 deletion mutants suggest that the HSA domain is required for the association with ARID1A/BAF250a, which is able to bind GR through its C-terminal domain, and is necessary for SWI/SNF-mediated transcriptional activation (40). At present, our studies, including Y2H-cDNA library screens, GST-protein pulldowns, and immunoprecipitation assays, suggest that the HSA domain of BRG1 is required for the association with Ku70 (Table 1; Fig. 1 and 2). To determine if BAF250a and Ku70 cooccupy the BRG1-HSA domain and bind cooperatively to the MMTV promoter, we performed a ChIP-reChIP assay. The first ChIP was performed using extracts from ligand-treated SW-13 cells expressing GR and BRG1 and antibodies specific for GR, BRG1, Ku70, or BAF250a. Consistent with previous ChIP data, hormone-dependent binding to chromatin MMTV was observed for GR, BRG1, Ku70, and BAF250a (Fig. 8A). Using the samples immunoprecipitated with GR, BRG1, Ku70, or BAF250a antibody, we reChIPed each sample with each antibody used for the initial immunoprecipitation. Intriguingly, in each case, GR, Brg1, Ku70, and Baf250a reChIPs showed enrichment over IgG, indicating that the four proteins cooccupy sites on chromatin only in the presence of hormone (Fig. 8B). This corecruitment may involve their association with the HSA domain of BRG1, which is required for stimulating both nuclear receptor-dependent and -independent transcriptional regulation by SWI/SNF. Taken together, these results indicate that the HSA domain is an important binding surface for the association of regulatory proteins with BRG1 and may confer specificity regarding the targeting, function, and/or regulation of SWI/SNF activity during hormone-activated transcription.
FIG 8.
Ku and SWI/SNF subunits, BRG1 and BAF250a, cooccupy chromatin MMTV after stimulation with hormone. ChIP-reChIP analysis of integrated MMTV promoter in treated SW-13 cells expressing GR and BRG1 was performed. Cells were treated with hormone, formaldehyde cross-linked, and subjected to ChIP analysis. (A) For the primary ChIP, DNA immunocomplexes were isolated using antibodies specific for GR, BRG1, Ku70, or BAF250a. (B) ReChIP analysis was performed using Dex-treated samples from primary ChIP immunoprecipitated with GR, BRG1, Ku70, or BAF250a antibodies. Immunopurified DNA complexes from both ChIPs were reverse cross-linked and analyzed by real-time PCR using MMTV nucleosome B-specific primers. Quantitative analysis of data from primary ChIP (A) and from reChIP (B) was performed, with results displayed as percentages of ChIP input. The data are from a representative experiment with standard deviations of technical replicates.
Hormone-dependent SWI/SNF-mediated transcriptional activation induces topoisomerase IIβ DSBs within chromatin MMTV.
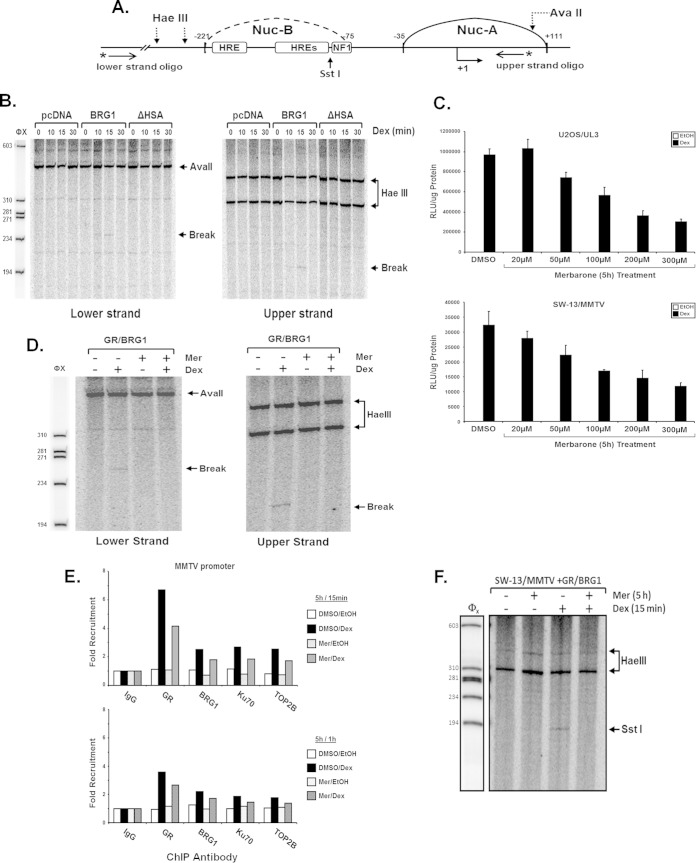
Growing evidence suggests that the enzymatic activity of TOP2β may be a critical component for hormone-stimulated and often rapidly upregulated gene expression by nuclear receptors such as ER and AR. In these signaling pathways, TOP2β is recruited to hormone-responsive promoters, where its catalytic activity produces a transient double-strand DNA break (DSB) which may help resolve the topological constraints resulting from rapid changes in chromatin architecture that must occur for nuclear receptor binding and/or transcriptional activation (65, 70). To investigate whether the enzymatic activity of TOP2β produces DSBs within chromatin MMTV upon glucocorticoid signaling, we developed a protocol that detects DNA break formation in the promoter region using a modified iterative primer extension technique (50). SW-13/MMTV cells expressing GR were transfected with empty vector (pcDNA), BRG1, or BRG1-ΔHSA mutant plasmids and stimulated with hormone for various time periods (0, 10, 15, or 30 min). Immediately after treatment, genomic DNA was isolated and digested to completion with AvaII or HaeIII for analysis of the upper strand or lower strand of DNA, respectively (Fig. 9A). We were intrigued to find that genomic DNA isolated from cells expressing BRG1 yielded a band indicative of a break within the promoter sequence and that this band appears to be specific at 15 min of hormone treatment. These DNA breaks were observed on both the upper and lower strands in hormone-treated cells expressing wild-type BRG1, indicating that this double-strand DNA (dsDNA) break may be required for hormone-mediated transcriptional regulation by the SWI/SNF complex. These breaks were not observed in the genomic DNA purified from treated SW-13 cells expressing the BRG1-ΔHSA mutant, suggesting that the HSA domain within the N-terminal region of BRG1 is important for DSB formation with chromatin MMTV in response to hormonal signaling (Fig. 9B). The Dex-induced cleavage sites within the MMTV promoter were mapped to the nucleosome B region. Using this strategy, the position of the ligand-dependent DNA breaks within chromatin MMTV appears to border an active hormone response element (HRE).
FIG 9.
Identification of hormone-dependent BRG1-mediated DNA breaks within chromatin MMTV. Transient DNA breaks were detected using a modified reiterative primer extension technique. (A) Schematic of the proximal portion of the MMTV promoter indicating nucleosomal location, restriction endonuclease cleavage sites, transcriptional factor binding regions, and positions of oligonucleotides used for primer extension. (B) SW-13/MMTV cells expressing GR and BRG1 or ΔHSA mutant protein were treated with 100 nM Dex for 0, 10, 15, or 30 min prior to isolation of nuclei and DNA. Purified genomic DNA was digested to completion with AvaII or HaeIII and analyzed for DNA breaks using iterative primer extension with 32P-labeled primers specific for the upper or lower strand of MMTV. (C) Merbarone (Mer) treatment attenuates hormone-induced transcriptional activation of chromatin MMTV. U2OS/UL3 cells containing integrated copies of MMTV reporter or SW-13/MMTV cells expressing GR and BRG1 were incubated for 5 h with 100 μM Mer at the indicated concentrations, followed by treatment with Dex. Posttreatment, cells were lysed and assayed for luciferase activity. Relative luciferase units (RLU) values were normalized to total protein measured and reported as the average from three independent experiments. Error bars are standard deviations (n = 3). (D) Inhibition of TOP2β activity blocks hormone-dependent BRG1-mediated DNA break formation. SW-13/MMTV cells expressing GR and BRG1 were treated with Mer and/or Dex. Following treatment, purified genomic DNA was digested with AvaII or HaeIII and analyzed for DNA break formation as described above. Extension products were resolved on 7% denaturing polyacrylamide gels and exposed to a phosphorimager screen. DNA breaks were identified using ImageQuant peak-finder analysis software. (E) Hormone-stimulated recruitment of GR, BRG1, Ku70, or TOP2β to chromatin MMTV is not affected by treatment with Mer. SW-13/MMTV cells expressing GR and BRG1 were treated with Mer or vehicle (DMSO) for 5 h prior to addition of Dex or ethanol (EtOH) for 15 min or 1 h. Data are reported as the average from three independent experiments. (F) In vivo restriction enzyme accessibility assay to determine chromatin architecture at MMTV-NucB upon treatment with Mer and hormone (Dex). Nuclei purified from cells expressing GR and BRG1 were pretreated with Mer (5 h) and then subjected to treatment with Dex (15 min) followed by digestion with SstI (in vivo) restriction endonuclease. Purified products were further digested to completion with HaeIII (in vitro) and used as the template for iterative primer extension with an MMTV-specific primer. Extension products were resolved by 6% denaturing PAGE.
To determine whether TOP2β mediates the formation of ligand-dependent DNA breaks within the integrated MMTV promoter, we treated SW-13 cells expressing GR and BRG1 with the topoisomerase II-specific inhibitor merbarone (Mer) [5-(N-phenyl carboxamido)-2-thiobarbituric acid], which inhibits the catalytic activity of DNA TOP2β and TOP2α without damaging DNA or stabilizing the DNA-TOP2 cleavable complexes (71, 72). Transcriptional activation in Mer-treated Dex-stimulated U2OS/UL3 or SW-13 cells expressing GR and BRG1 was significantly diminished compared to that in cells treated with vehicle (Fig. 9C). Treatment with Mer also inhibited the formation of BRG1-dependent DNA breaks within chromatin MMTV when stimulated with hormone for 15 min (Fig. 9D). To determine if Mer-induced transcriptional attenuation and inhibition of promoter-specific dsDNA breaks upon hormone stimulation resulted from impaired recruitment of SWI/SNF or TOP2β/PARP1 complexes, we performed ChIP analysis of chromatin MMTV. Merbarone treatment did not appear to alter the recruitment patterns of components of SWI/SNF or TOP2β/PARP1 complexes at either 15 min of 1 h of Dex treatment (Fig. 9E). However, we did reproducibly observe a slight diminution in the binding of GR, BRG1 Ku70, and TOP2β with the prior treatment with Mer. Finally, the ability of BRG1 to remodel the MMTV nucleosome B seems to be lost after Mer treatment, indicating that topoisomerase II is required not only for transcriptional activation but also to alter the nucleosomal architecture within MMTV upon hormonal induction (Fig. 9F).
Collectively, these results suggest that the SWI/SNF chromatin-remodeling complex is required for the formation of site-specific DNA breaks within the MMTV promoter and that these dsDNA breaks are mediated through the enzymatic activity of TOP2β and are required for ligand-dependent transcriptional activation. Our investigation has also revealed the important role of the HSA domain of BRG1 in establishing short-lived, transient DNA breaks during GR-mediated transcriptional activation.
DISCUSSION
The central catalytic subunit of the SWI/SNF complex, BRG1, provides ATP-dependent enzymatic activity and is required for direct or indirect associations with various proteins involved in cell growth and differentiation, cell cycle regulation, and immune response, such as β-catenin, p130, p107, BRCA1, p53, Rb, and cyclin E (18).
The SWI/SNF chromatin-remodeling complex also participates in ligand-mediated signaling to regulate hormone-induced transcription through associations with type I nuclear receptors, including glucocorticoid, progesterone, estrogen, and androgen receptors. These interactions lead to the hormone-dependent recruitment of the BRG1 complex to target promoters for chromatin-remodeling-dependent changes in gene expression (28). Transcriptional regulation studies using BRG1 chimeras and/or N-terminal mutant proteins indicate that the BRG1 ATPase domain is necessary but not sufficient for ligand-dependent transcriptional activation by GR. Collectively, these observations suggest that the N-terminal HSA domain of BRG1 is required in conjunction with the ATPase activity for GR-mediated BRG1-dependent transcriptional activation of endogenous hormone-responsive promoters (40). The HSA region has also been implicated as an important binding domain responsible for the association of BRG1 with members of the SWI/SNF complex, BAF250a, BAF53, and β-actin (18, 28, 40, 41).
Our yeast 2-hybrid screen identified a novel interaction of BRG1 with the double-strand break repair protein Ku70/XRCC6/G22P1, and subsequent biochemical analysis suggested that the Ku heterodimer (Ku70/86) association with the SWI/SNF chromatin-remodeling complex is mediated through the N-terminal HSA domain of BRG1. This was subsequently confirmed via recombinant binding assays using protein fragments containing the BRG1 HSA-BRK domain fragment and the Ku surface fragment that were identified in the original yeast 2-hybrid assay.
The segment of Ku70 involved in the BRG1 interaction includes the central DNA-binding beta-barrels, polypeptide rings, and C-terminal arm of the protein. This segment includes many surface residues that would be exposed in the Ku70/86 dimer structure that has been determined crystallographically (58, 73). To determine if the enzymatic function of Ku70, a single-stranded DNA-dependent ATP-dependent helicase, is involved in SWI/SNF-mediated transcriptional activation, we performed a series of siRNA protein knockdown studies in SW-13 cells expressing BRG1 or in ligand-treated cells expressing GR and BRG1. Data we have shown from these studies (Fig. 2) suggest that Ku is not required for transcriptional activation of all BRG1-dependent genes; however, transcription from hormone-responsive promoters is significantly attenuated with reduced Ku protein.
Ligand-dependent recruitment of Ku to GR-responsive promoters was observed by chromatin immunoprecipitation, providing further evidence that Ku may play a vital role in hormone response pathways. Ku protein has also been implicated in nuclear receptor signaling events involving the estrogen receptor (28, 65, 66). In these experiments, treatment of MCF7 cells with 17β-estradiol induced the recruitment of a complex containing Ku, DNAPK, TOP2β, and PARP1 to the pS2 promoter, resulting in transcriptional activation (65). Our data suggest that SWI/SNF may work in conjunction with the TOP2β/Ku70/86/DNAPK/PARP1 complex during nuclear receptor-dependent transcriptional regulation. Through a series of siRNA protein knockdown studies, we were able to show that Ku70/86, TOP2β, and PARP1 are required for GR-mediated transcription activation of chromatinized MMTV and of endogenous genes HSD11B2 and PLZF. Chromatin immunoprecipitation assays indicate that these proteins, along with BRG1 and DNAPK, are recruited to GR-responsive promoters in a ligand-dependent manner.
In contrast to the reduction in transcriptional output observed with the reduction in protein levels for Ku70/86, TOP2β, and PARP1, we did not see as strong an effect when DNAPK was inhibited. This is intriguing because while the role and activity of DNAPK in NHEJ is well established, previous studies have also indicated a role for DNAPK in ER-dependent transcription (65, 66). In these cases, the kinase activity of DNAPK was thought to activate a variety of coregulators know to be required for ER activation (74). We examined this possibility directly with NU7026, a cell-permeative, potent, specific, and ATP-competitive inhibitor of DNAPK, and did not discern a significant impact (Fig. 5) (75). Thus, the results from our studies suggest that in the case of GR, this requirement may be somewhat relaxed; loss of DNAPK is less deleterious than loss of Ku70/86 or TOP2β. Collectively, our data reveal that members of the Ku/TOP2β/PARP1 and SWI/SNF complexes are recruited together to target promoters to promote NR-mediated transcriptional regulation.
During DNA replication and transcription, nuclear factors bind DNA and alter the local chromatin structure, producing significant steric tension among nucleosomes that surround the target region. This tension is resolved by TOP2β, a protein with enzymatic activity that catalyzes transient DNA double-strand breaks to resolve the topological constraints produced by genomic events (62). To test whether TOP2β alters the topology of the MMTV nucleosome B region, we developed a protocol that enables the detection of double-strand DNA break formation using a modified iterative primer extension technique. Using this assay, we have detected BRG1-dependent DNA breaks within the MMTV promoter upon treatment with hormone. To determine if these hormone-responsive, BRG1-dependent double-strand DNA breaks are mediated by TOP2β, we made use of a specific inhibitor of TOP2, Mer, which inhibits the cleavage activity of TOP2 without damaging the DNA or stabilizing the DNA-TOP2 cleavable complexes. We then evaluated the presence of DNA breaks within the chromatin promoter and found that Mer treatment not only prevents DNA breaks upon hormone stimulation but also attenuates transcriptional activation. These observations are consistent with the inhibitor preventing DNA breaks on the stimulated pS2 promoter and blocking pS2 transcription in E2-treated MCF7 cells, suggesting that the enzymatic activity of TOP2A plays a role at that promoter as well (65). Consistent with the inhibitor studies, reduction of TOP2β protein levels diminished the transcriptional response from ligand-treated cells, suggesting that the enzymatic activity of TOP2β is required for NR-induced transcription.
Our studies have defined a specific role for Ku/BRG1/TOP2β in glucocorticoid-dependent transcription, and, notably, we do not find a role in the BRG1-dependent induction of a number of non-steroid-inducible genes, such as CD44 and CRYAB (38). These results, however, do not preclude additional functions for the BRG1/TOP2A/B complexes. Indeed, recent experiments demonstrate that in mouse embryonic stem cells and in mouse embryonic fibroblasts, BRG1 and TOP2A function to resolve anaphase bridges, and in cancer cells mutations that impair BRG1 contribute to genomic instability and cancer etiology (76). The elegant demonstration of TOP2A/BRG1 activities in decatenation reveals a clear role for regions of BRG1 where mutations have been shown to be important for tumorigenesis (77). Interestingly, two of these BRG1 mutants, G1232D and T910M, that are shown to be important for decatenation are also distinct from those HSA/BRK deletion (475-656) mutants critical for the interaction with the GR/Ku70/86 and TOP2β that we describe in our analysis.
Collectively, our data reveal that members of the TOP2β-containing complexes function within and are required components of NR-specific SWI/SNF-dependent regulated transcription. Hormone stimulation leads to corecruitment of ligand-bound GR and the SWI/SNF chromatin-remodeling complex in association with Ku to the enhancer/promoter. Among the coactivators binding with the SWI/SNF chromatin-remodeling complex, TOP2β provides double-strand DNA break activity that may function to alleviate the topological and torsional stress expected in the local chromatin architecture of the promoter. The DNA breaks occur quickly and appear short-lived, observed within 10 min of the addition of hormone and potentially repaired by mechanisms that are at present unclear but may involve enzymatic activities critical for processes including DNA replication and repair. This model provides insights into nuclear receptor-mediated transcriptional regulation and suggests that it is possible to recruit previously well-described molecular entities to accomplish a new function.
ACKNOWLEDGMENTS
We thank C. Burd for intellectual input and J. Hoffman and E. Milliman for technical support and critical review of the manuscript.
REFERENCES
- 1.Chen J, Kinyamu HK, Archer TK. 2006. Changes in attitude, changes in latitude: nuclear receptors remodeling chromatin to regulate transcription. Mol Endocrinol 20:1–13. doi: 10.1210/me.2005-0192. [DOI] [PubMed] [Google Scholar]
- 2.Hansen JC. 2002. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu Rev Biophys Biomol Struct 31:361–392. doi: 10.1146/annurev.biophys.31.101101.140858. [DOI] [PubMed] [Google Scholar]
- 3.Knezetic JA, Luse DS. 1986. The presence of nucleosomes on a DNA template prevents initiation by RNA polymerase II in vitro. Cell 45:95–104. doi: 10.1016/0092-8674(86)90541-6. [DOI] [PubMed] [Google Scholar]
- 4.Wan Y, Nordeen SK. 2002. Overlapping but distinct gene regulation profiles by glucocorticoids and progestins in human breast cancer cells. Mol Endocrinol 16:1204–1214. doi: 10.1210/mend.16.6.0848. [DOI] [PubMed] [Google Scholar]
- 5.Workman JL, Kingston RE. 1998. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem 67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 6.Aoyagi S, Trotter KW, Archer TK. 2005. ATP-dependent chromatin remodeling complexes and their role in nuclear receptor-dependent transcription in vivo. Vitam Horm 70:281–307. [DOI] [PubMed] [Google Scholar]
- 7.Edmondson DG, Roth SY. 1996. Chromatin and transcription. FASEB J 10:1173–1182. [DOI] [PubMed] [Google Scholar]
- 8.Kouzarides T. 2007. Chromatin modifications and their function. Cell 128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Trotter KW, Archer TK. 2007. Nuclear receptors and chromatin remodeling machinery. Mol Cell Endocrinol 265-266:162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aoyagi S, Archer TK. 2008. Dynamics of coactivator recruitment and chromatin modifications during nuclear receptor mediated transcription. Mol Cell Endocrinol 280:1–5. doi: 10.1016/j.mce.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ausio J, Dong F, van Holde KE. 1989. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone “tails” in the stabilization of the nucleosome. J Mol Biol 206:451–463. doi: 10.1016/0022-2836(89)90493-2. [DOI] [PubMed] [Google Scholar]
- 12.Hayes JJ, Clark DJ, Wolffe AP. 1991. Histone contributions to the structure of DNA in the nucleosome. Proc Natl Acad Sci U S A 88:6829–6833. doi: 10.1073/pnas.88.15.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson CN, Adkins NL, Georgel P. 2005. Chromatin remodeling complexes: ATP-dependent machines in action. Biochem Cell Biol 83:405–417. doi: 10.1139/o05-115. [DOI] [PubMed] [Google Scholar]
- 14.Eisen JA, Sweder KS, Hanawalt PC. 1995. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res 23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fairman-Williams ME, Guenther UP, Jankowsky E. 2010. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol 20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hauk G, Bowman GD. 2011. Structural insights into regulation and action of SWI2/SNF2 ATPases. Curr Opin Struct Biol 21:719–727. doi: 10.1016/j.sbi.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phelan ML, Sif S, Narlikar GJ, Kingston RE. 1999. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell 3:247–253. doi: 10.1016/S1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 18.Trotter KW, Archer TK. 2008. The BRG1 transcriptional coregulator. Nucl Recept Signal 6:e004. doi: 10.1621/nrs.06004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W, Cote J, Xue Y, Zhou S, Khavari PA, Biggar SR, Muchardt C, Kalpana GV, Goff SP, Yaniv M, Workman JL, Crabtree GR. 1996. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J 15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 20.Hargreaves DC, Crabtree GR. 2011. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res 21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. 2009. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci U S A 106:5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohrmann L, Verrijzer CP. 2005. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim Biophys Acta 1681:59–73. doi: 10.1016/j.bbaexp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 23.Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG. 2004. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature 432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 24.Wu JI, Lessard J, Olave IA, Qiu Z, Ghosh A, Graef IA, Crabtree GR. 2007. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron 56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Cheng MB, Zhang YJ, Zhong X, Dai H, Yan L, Wu NH, Shen YF. 2010. A switch from hBrm to Brg1 at IFNgamma-activated sequences mediates the activation of human genes. Cell Res 20:1345–1360. doi: 10.1038/cr.2010.155. [DOI] [PubMed] [Google Scholar]
- 26.Fryer CJ, Archer TK. 1998. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature 393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- 27.Hebbar PB, Archer TK. 2003. Chromatin remodeling by nuclear receptors. Chromosoma 111:495–504. doi: 10.1007/s00412-003-0232-x. [DOI] [PubMed] [Google Scholar]
- 28.King HA, Trotter KW, Archer TK. 2012. Chromatin remodeling during glucocorticoid receptor regulated transactivation. Biochim Biophys Acta 1819:716–726. doi: 10.1016/j.bbagrm.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nie Z, Xue Y, Yang D, Zhou S, Deroo BJ, Archer TK, Wang W. 2000. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol Cell Biol 20:8879–8888. doi: 10.1128/MCB.20.23.8879-8888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belandia B, Orford RL, Hurst HC, Parker MG. 2002. Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. EMBO J 21:4094–4103. doi: 10.1093/emboj/cdf412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia-Pedrero JM, Kiskinis E, Parker MG, Belandia B. 2006. The SWI/SNF chromatin remodeling subunit BAF57 is a critical regulator of estrogen receptor function in breast cancer cells. J Biol Chem 281:22656–22664. doi: 10.1074/jbc.M602561200. [DOI] [PubMed] [Google Scholar]
- 32.Ichinose H, Garnier JM, Chambon P, Losson R. 1997. Ligand-dependent interaction between the estrogen receptor and the human homologues of SWI2/SNF2. Gene 188:95–100. doi: 10.1016/S0378-1119(96)00785-8. [DOI] [PubMed] [Google Scholar]
- 33.Inoue H, Furukawa T, Giannakopoulos S, Zhou S, King DS, Tanese N. 2002. Largest subunits of the human SWI/SNF chromatin-remodeling complex promote transcriptional activation by steroid hormone receptors. J Biol Chem 277:41674–41685. doi: 10.1074/jbc.M205961200. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Fu J, Toumazou C, Yoon HG, Wong J. 2006. A role of the amino-terminal (N) and carboxyl-terminal (C) interaction in binding of androgen receptor to chromatin. Mol Endocrinol 20:776–785. doi: 10.1210/me.2005-0298. [DOI] [PubMed] [Google Scholar]
- 35.Link KA, Burd CJ, Williams E, Marshall T, Rosson G, Henry E, Weissman B, Knudsen KE. 2005. BAF57 governs androgen receptor action and androgen-dependent proliferation through SWI/SNF. Mol Cell Biol 25:2200–2215. doi: 10.1128/MCB.25.6.2200-2215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marshall TW, Link KA, Petre-Draviam CE, Knudsen KE. 2003. Differential requirement of SWI/SNF for androgen receptor activity. J Biol Chem 278:30605–30613. doi: 10.1074/jbc.M304582200. [DOI] [PubMed] [Google Scholar]
- 37.Archer TK, Cordingley MG, Wolford RG, Hager GL. 1991. Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Mol Cell Biol 11:688–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trotter KW, Archer TK. 2004. Reconstitution of glucocorticoid receptor-dependent transcription in vivo. Mol Cell Biol 24:3347–3358. doi: 10.1128/MCB.24.8.3347-3358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan HY, Trotter KW, Archer TK, Kingston RE. 2005. Swapping function of two chromatin remodeling complexes. Mol Cell 17:805–815. doi: 10.1016/j.molcel.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 40.Trotter KW, Fan HY, Ivey ML, Kingston RE, Archer TK. 2008. The HSA domain of BRG1 mediates critical interactions required for glucocorticoid receptor-dependent transcriptional activation in vivo. Mol Cell Biol 28:1413–1426. doi: 10.1128/MCB.01301-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szerlong H, Hinata K, Viswanathan R, Erdjument-Bromage H, Tempst P, Cairns BR. 2008. The HSA domain binds nuclear actin-related proteins to regulate chromatin-remodeling ATPases. Nat Struct Mol Biol 15:469–476. doi: 10.1038/nsmb.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Featherstone C, Jackson SP. 1999. Ku, a DNA repair protein with multiple cellular functions? Mutat Res 434:3–15. doi: 10.1016/S0921-8777(99)00006-3. [DOI] [PubMed] [Google Scholar]
- 43.Tuteja R, Tuteja N. 2000. Ku autoantigen: a multifunctional DNA-binding protein. Crit Rev Biochem Mol Biol 35:1–33. doi: 10.1080/10409230091169177. [DOI] [PubMed] [Google Scholar]
- 44.Smider V, Chu G. 1997. The end-joining reaction in V(D)J recombination. Semin Immunol 9:189–197. doi: 10.1006/smim.1997.0070. [DOI] [PubMed] [Google Scholar]
- 45.Jones JM, Gellert M, Yang W. 2001. A Ku bridge over broken DNA. Structure 9:881–884. doi: 10.1016/S0969-2126(01)00658-X. [DOI] [PubMed] [Google Scholar]
- 46.Muller C, Paupert J, Monferran S, Salles B. 2005. The double life of the Ku protein: facing the DNA breaks and the extracellular environment. Cell Cycle 4:438–441. doi: 10.4161/cc.4.3.1565. [DOI] [PubMed] [Google Scholar]
- 47.DeGrado-Warren J, Dufford M, Chen J, Bartel PL, Shattuck D, Frech GC. 2008. Construction and characterization of a normalized yeast two-hybrid library derived from a human protein-coding clone collection. Biotechniques 44:265–273. doi: 10.2144/000112674. [DOI] [PubMed] [Google Scholar]
- 48.Veuger SJ, Curtin NJ, Richardson CJ, Smith GC, Durkacz BW. 2003. Radiosensitization and DNA repair inhibition by the combined use of novel inhibitors of DNA-dependent protein kinase and poly(ADP-ribose) polymerase-1. Cancer Res 63:6008–6015. [PubMed] [Google Scholar]
- 49.Hebbar PB, Archer TK. 2007. Chromatin-dependent cooperativity between site-specific transcription factors in vivo. J Biol Chem 282:8284–8291. doi: 10.1074/jbc.M610554200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trotter KW, Archer TK. 2012. Assaying chromatin structure and remodeling by restriction enzyme accessibility. Methods Mol Biol 833:89–102. doi: 10.1007/978-1-61779-477-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaeser MD, Aslanian A, Dong MQ, Yates JR III, Emerson BM. 2008. BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J Biol Chem 283:32254–32263. doi: 10.1074/jbc.M806061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nielsen AL, Sanchez C, Ichinose H, Cervino M, Lerouge T, Chambon P, Losson R. 2002. Selective interaction between the chromatin-remodeling factor BRG1 and the heterochromatin-associated protein HP1alpha. EMBO J 21:5797–5806. doi: 10.1093/emboj/cdf560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. 1996. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev 10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 54.de Bruijn DR, Peters WJ, Chuva de Sousa Lopes SM, van Dijk AH, Willemse MP, Pfundt R, de Boer P, Geurts van Kessel A. 2006. Targeted disruption of the synovial sarcoma-associated SS18 gene causes early embryonic lethality and affects PPARBP expression. Hum Mol Genet 15:2936–2944. doi: 10.1093/hmg/ddl235. [DOI] [PubMed] [Google Scholar]
- 55.Hu G, Schones DE, Cui K, Ybarra R, Northrup D, Tang Q, Gattinoni L, Restifo NP, Huang S, Zhao K. 2011. Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome Res 21:1650–1658. doi: 10.1101/gr.121145.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu Z, Meng X, Cai Y, Koury MJ, Brandt SJ. 2006. Recruitment of the SWI/SNF protein Brg1 by a multiprotein complex effects transcriptional repression in murine erythroid progenitors. Biochem J 399:297–304. doi: 10.1042/BJ20060873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mori K, Kato H. 2002. A putative nuclear receptor coactivator (TMF/ARA160) associates with hbrm/hSNF2 alpha and BRG-1/hSNF2 beta and localizes in the Golgi apparatus. FEBS Lett 520:127–132. doi: 10.1016/S0014-5793(02)02803-X. [DOI] [PubMed] [Google Scholar]
- 58.Rivera-Calzada A, Spagnolo L, Pearl LH, Llorca O. 2007. Structural model of full-length human Ku70-Ku80 heterodimer and its recognition of DNA and DNA-PKcs. EMBO Rep 8:56–62. doi: 10.1038/sj.embor.7400847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reisman DN, Strobeck MW, Betz BL, Sciariotta J, Funkhouser W Jr, Murchardt C, Yaniv M, Sherman LS, Knudsen ES, Weissman BE. 2002. Concomitant down-regulation of BRM and BRG1 in human tumor cell lines: differential effects on RB-mediated growth arrest vs CD44 expression. Oncogene 21:1196–1207. doi: 10.1038/sj.onc.1205188. [DOI] [PubMed] [Google Scholar]
- 60.Liu H, Mulholland N, Fu H, Zhao K. 2006. Cooperative activity of BRG1 and Z-DNA formation in chromatin remodeling. Mol Cell Biol 26:2550–2559. doi: 10.1128/MCB.26.7.2550-2559.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Otsuki T, Furukawa Y, Ikeda K, Endo H, Yamashita T, Shinohara A, Iwamatsu A, Ozawa K, Liu JM. 2001. Fanconi anemia protein, FANCA, associates with BRG1, a component of the human SWI/SNF complex. Hum Mol Genet 10:2651–2660. doi: 10.1093/hmg/10.23.2651. [DOI] [PubMed] [Google Scholar]
- 62.Ju BG, Rosenfeld MG. 2006. A breaking strategy for topoisomerase IIbeta/PARP-1-dependent regulated transcription. Cell Cycle 5:2557–2560. doi: 10.4161/cc.5.22.3497. [DOI] [PubMed] [Google Scholar]
- 63.Mayeur GL, Kung WJ, Martinez A, Izumiya C, Chen DJ, Kung HJ. 2005. Ku is a novel transcriptional recycling coactivator of the androgen receptor in prostate cancer cells. J Biol Chem 280:10827–10833. doi: 10.1074/jbc.M413336200. [DOI] [PubMed] [Google Scholar]
- 64.Shank LC, Kelley JB, Gioeli D, Yang CS, Spencer A, Allison LA, Paschal BM. 2008. Activation of the DNA-dependent protein kinase stimulates nuclear export of the androgen receptor in vitro. J Biol Chem 283:10568–10580. doi: 10.1074/jbc.M800810200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. 2006. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science 312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 66.Medunjanin S, Weinert S, Schmeisser A, Mayer D, Braun-Dullaeus RC. 2010. Interaction of the double-strand break repair kinase DNA-PK and estrogen receptor-alpha. Mol Biol Cell 21:1620–1628. doi: 10.1091/mbc.E09-08-0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lans H, Marteijn JA, Vermeulen W. 2012. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenet Chromatin 5:4. doi: 10.1186/1756-8935-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang L, Zhang Q, Jones K, Patel M, Gong F. 2009. The chromatin remodeling factor BRG1 stimulates nucleotide excision repair by facilitating recruitment of XPC to sites of DNA damage. Cell Cycle 8:3953–3959. doi: 10.4161/cc.8.23.10115. [DOI] [PubMed] [Google Scholar]
- 69.Lee HS, Park JH, Kim SJ, Kwon SJ, Kwon J. 2010. A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J 29:1434–1445. doi: 10.1038/emboj.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haffner MC, Aryee MJ, Toubaji A, Esopi DM, Albadine R, Gurel B, Isaacs WB, Bova GS, Liu W, Xu J, Meeker AK, Netto G, De Marzo AM, Nelson WG, Yegnasubramanian S. 2010. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat Genet 42:668–675. doi: 10.1038/ng.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fortune JM, Osheroff N. 1998. Merbarone inhibits the catalytic activity of human topoisomerase IIalpha by blocking DNA cleavage. J Biol Chem 273:17643–17650. doi: 10.1074/jbc.273.28.17643. [DOI] [PubMed] [Google Scholar]
- 72.Khelifa T, Beck WT. 1999. Merbarone, a catalytic inhibitor of DNA topoisomerase II, induces apoptosis in CEM cells through activation of ICE/CED-3-like protease. Mol Pharmacol 55:548–556. [PubMed] [Google Scholar]
- 73.Spagnolo L, Rivera-Calzada A, Pearl LH, Llorca O. 2006. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol Cell 22:511–519. doi: 10.1016/j.molcel.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 74.Foulds CE, Feng Q, Ding C, Bailey S, Hunsaker TL, Malovannaya A, Hamilton RA, Gates LA, Zhang Z, Li C, Chan D, Bajaj A, Callaway CG, Edwards DP, Lonard DM, Tsai SY, Tsai MJ, Qin J, O'Malley BW. 2013. Proteomic analysis of coregulators bound to ERalpha on DNA and nucleosomes reveals coregulator dynamics. Mol Cell 51:185–199. doi: 10.1016/j.molcel.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nutley BP, Smith NF, Hayes A, Kelland LR, Brunton L, Golding BT, Smith GC, Martin NM, Workman P, Raynaud FI. 2005. Preclinical pharmacokinetics and metabolism of a novel prototype DNA-PK inhibitor NU7026. Br J Cancer 93:1011–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lada AG, Waisertreiger IS, Grabow CE, Prakash A, Borgstahl GE, Rogozin IB, Pavlov YI. 2011. Replication protein A (RPA) hampers the processive action of APOBEC3G cytosine deaminase on single-stranded DNA. PLoS One 6:e24848. doi: 10.1371/journal.pone.0024848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, Crabtree GR. 2013. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet 45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]