Abstract
Giardia intestinalis parasites contain mitosomes, one of the simplest mitochondrion-related organelles. Strategies to identify the functions of mitosomes have been limited mainly to homology detection, which is not suitable for identifying species-specific proteins and their functions. An in vivo enzymatic tagging technique based on the Escherichia coli biotin ligase (BirA) has been introduced to G. intestinalis; this method allows for the compartment-specific biotinylation of a protein of interest. Known proteins involved in the mitosomal protein import were in vivo tagged, cross-linked, and used to copurify complexes from the outer and inner mitosomal membranes in a single step. New proteins were then identified by mass spectrometry. This approach enabled the identification of highly diverged mitosomal Tim44 (GiTim44), the first known component of the mitosomal inner membrane translocase (TIM). In addition, our subsequent bioinformatics searches returned novel diverged Tim44 paralogs, which mediate the translation and mitosomal insertion of mitochondrially encoded proteins in other eukaryotes. However, most of the identified proteins are specific to G. intestinalis and even absent from the related diplomonad parasite Spironucleus salmonicida, thus reflecting the unique character of the mitosomal metabolism. The in vivo enzymatic tagging also showed that proteins enter the mitosome posttranslationally in an unfolded state and without vesicular transport.
INTRODUCTION
Giardia intestinalis causes intestinal infection in diverse vertebrate species, including humans, where it causes the disease giardiasis (1). In addition to its medical and veterinary importance, Giardia is an interesting unicellular eukaryote (protist) from cell biology and evolutionary perspectives (2).
The binucleated Giardia trophozoite is equipped with eight flagella and an adhesive disc, which mediates attachment to its host's intestine. The interior of the cell is dominated by an endoplasmic reticulum (ER) network (3) and lysosome-like peripheral vacuoles that mediate the uptake and digestion of nutrients (4). There are also Golgi body-like encystation vesicles that distribute the cyst wall material to the cell surface during encystation of the parasite (5).
The mitosomes of Giardia are highly adapted forms of mitochondria and are approximately 100 nm in size. These organelles are surrounded by two membranes, but unlike mitochondria, they do not contain DNA. The mitosomal proteome is currently limited to 21 proteins, which primarily participate in iron-sulfur cluster biosynthesis and protein import and folding (6–8). The identification of mitosomal proteins has been accomplished mostly using bioinformatics techniques, such as phylogenetics (9, 10) and hidden Markov model (HMM)-based searches (6, 11), that detect homologous proteins. Thus, in contrast to hydrogenosomes and mitochondria, in which 20 to 50% of proteins have no assigned function (12–14), the vast majority of mitosomal proteins have known functions and orthologs in the mitochondria of other eukaryotes. Attempts to identify the mitosomal proteome using cell fractionation techniques have been largely stymied by the abundance of the ER and cytoskeletal structures in the cell (7). Analogous studies of the proteomes of encystation vesicles and peripheral vacuoles of Giardia using sophisticated organelle purification procedures have demonstrated the limits of direct organelle isolation approaches (15). As a result, several essential aspects of the mitosome, such as the nature of the translocase of the inner membrane (TIM) complex or the protein composition of the outer mitosomal membrane, remain unknown.
Here, we addressed the difficulty of the biochemical characterization of giardial mitosomes by employing an in vivo enzymatic tagging approach. The highly specific purification of biotinylated mitosomal proteins led to the identification of divergent GiTim44, the first component of the mitosomal TIM complex. In addition, over 10 novel mitosomal proteins from the mitosomal matrix and the outer mitosomal membrane were also identified, increasing the known mitosomal proteome by one-half. Most of these proteins reflect the unique and unpredictable character of giardial mitosome biology. Moreover, the compartment-specific protein tagging allowed us to identify the mode of mitosomal protein transport.
MATERIALS AND METHODS
Cell culture and fractionation.
Trophozoites of G. intestinalis strain WB (ATCC 30957) were grown in TY-S-33 medium (16) supplemented with 10% heat-inactivated bovine serum (PAA Laboratories), 0.1% bovine bile, and antibiotics. Cells expressing the dihydrofolate reductase (DHFR) fusion protein were grown in medium supplemented with 100 μM pyrimethamine (PM).
Preparation of cell fractions.
The cells were harvested by centrifugation at 1,000 × g at 4°C for 10 min in ice-cold phosphate-buffered saline (PBS), washed once in SM buffer (250 mM sucrose, 20 mM MOPS [morpholinepropanesulfonic acid], pH 7.4), and resuspended in SM buffer with protease inhibitors (cOmplete, EDTA-free; Roche). Cells were disrupted on ice by sonication with 1-s pulses and an amplitude of 40 for 1 min (Bioblock Scientific Vibra-Cell 72405). The lysate was then centrifuged at 2,750 × g for 10 min. The centrifugation step was repeated until the pellet containing unbroken cells, nuclei, and the cytoskeleton was no longer visible. The clear supernatant was centrifuged at 180,000 × g at 4°C for 30 min. The resulting high-speed supernatant represented the cytosolic fraction; the high-speed pellet (HSP) containing the mitosomes was resuspended in SM buffer containing protease inhibitors.
Fluorescence microscopy.
G. intestinalis trophozoites were fixed and immunolabeled as previously described (17). Mitosomal GiTom40 was detected with a specific polyclonal antibody raised in rabbits (18), and the hemagglutinin epitope (HA tag) was recognized by a rat monoclonal antibody (Roche). The primary antibodies were detected by a donkey Alexa Fluor 594 (red)-conjugated anti-rabbit antibody and Alexa Fluor 594 (red)- or Alexa Fluor 488 (green)-conjugated anti-rat antibodies (Life Technologies), respectively. Alexa Fluor 488 (green)-conjugated streptavidin (Life Technologies) was used to detect biotinylation. Slides were mounted with Vectashield containing DAPI (4′,6-diamidino-2-phenylindole; Vector Laboratories). The slides were imaged with an Olympus Cell-R, IX81 microscope system, and the images were processed using ImageJ 1.41e software (NIH).
Cloning and transfection.
The pTG vector was used for Escherichia coli biotin ligase (BirA) cloning and protein expression (17). The gene encoding BirA (WP_023308552) was amplified from pET21a-BirA (19). Table S1 in the supplemental material lists all the primers used in this study. To coexpress proteins with BirA, biotin acceptor peptide (BAP) was introduced into the pONDRA vector (6) using a reverse primer for GiPam18-BAP. This vector carrying the C-terminal BAP was used for the subsequent cloning of the other genes. All Giardia genes were amplified from genomic DNA. Mouse DHFR was amplified from pARL2-GDG (20) (kindly provided by Jude Przyborski, Philipps University Marburg). G. intestinalis transfection was performed as previously described (6). Briefly, 300 μl of G. intestinalis trophozoites (3.3 × 107 cells/ml) was electroporated with a Bio-Rad Gene Pulser using an exponential protocol (U = 350 V; C = 1,000 μF; R = 750 Ω). The transfected cells were grown in medium supplemented with antibiotics (57 μg/ml puromycin and 600 μg/ml G418).
Cross-linking, protein isolation, and mass spectrometry (MS).
G. intestinalis cells were grown in standard medium supplemented with 50 μM biotin for 24 h prior to harvesting. The cells were harvested and fractionated as described above. The HSP (40 mg) was used for the cross-linking and protein isolation. The pellet was resuspended in PBS (pH 7.4) supplemented with protease inhibitors (Roche) at a final protein concentration of 1.5 mg/ml. Then, a 25 μM concentration of the cross-linker DSP (dithiobis [succinimidyl propionate]; Thermo Scientific) was added, followed by 1 h of incubation on ice. After the incubation, Tris (pH 8) was added at a final concentration of 50 mM, and the sample was incubated at room temperature for 15 min. The sample was centrifuged at 30,000 × g for 10 min at 4°C, and the resulting pellet was resuspended in boiling buffer (50 mM Tris, 1 mM EDTA, 1% SDS, pH 7.4) supplemented with protease inhibitors at a final protein concentration of 1.5 mg/ml. The sample was incubated at 80°C for 10 min and was centrifuged at 30,000 × g for 10 min at room temperature. The resulting supernatant was diluted 1:10 in incubation buffer (50 mM Tris, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, pH 7.4) supplemented with protease inhibitors. Then, 200 μl of streptavidin-coupled magnetic beads (Dynabeads MyOne Streptavidin C1; Invitrogen) was washed 3 times with incubation buffer, mixed with the sample, and incubated overnight at 4°C with gentle rotation. The beads were then subjected to the following washes: 3 times for 5 min each in incubation buffer supplemented with 0.1% SDS, once for 5 min in boiling buffer, once for 5 min in washing buffer (60 mM Tris, 2% SDS, 10% glycerol), and twice for 5 min each in incubation buffer supplemented with 0.1% SDS. Finally, proteins were eluted from the beads in SDS-PAGE sample buffer supplemented with 20 mM biotin for 5 min at 95°C.
The samples were analyzed by Western blotting using streptavidin-conjugated Alexa Fluor 488 and were visualized using a Molecular Imager FX imager (Bio-Rad). The eluate was resolved by SDS-PAGE and stained with Coomassie brilliant blue. The gel was cut, destained, trypsin digested, and analyzed on a mass spectrometer.
Mass spectrometry and MS/MS analyses.
Spectra were acquired using a (4800 Plus MALDI-TOF/TOF) analyzer (Applied Biosystems/MDS Sciex) equipped with an Nd:YAG laser (355 nm) with a firing rate of 200 Hz. The tandem mass spectrometry (MS/MS) analyses were performed as previously described (21).
Protease protection assay.
To determine whether proteins were embedded in the outer mitosomal membrane, 150 μg of the HSP fraction in SM buffer supplemented with protease inhibitors was incubated with 200 μg/ml trypsin for 10 min at 37°C. The control sample also contained 0.1% Triton X-100 to completely digest the proteins of the solubilized organelles. The samples were separated by SDS-PAGE and blotted onto a nitrocellulose membrane, and proteins were detected with antibodies.
To determine whether proteins were in the mitosomal matrix, 1 mg of the HSP fraction was resuspended in 400 μl of either hypotonic buffer (1 mM EDTA, 10 mM MOPS, pH 7.2), isotonic buffer (hypotonic buffer supplemented with 250 mM sucrose), or NaCl buffer (500 mM NaCl, 10 mM Tris, pH 7.4). Pellets were resuspended by gentle pipetting or by sonication with 1-s pulses and amplitude of 60 for 2 times 1 min (Bioblock Scientific Vibra Cell 72405). Subsequently, 100 μl of each sample was treated with a different concentration of proteinase K (PK) and incubated on ice for 20 min. The reaction was stopped by the addition of 2 μl of 1 mM phenylmethanesulfonyl fluoride (PMSF), and the mixture was incubated on ice for 10 min. For the protein precipitation, 20 μl of 100% trichloroacetic acid (TCA) was added, and the samples were incubated on ice for 30 min. The samples were then centrifuged at 30,000 × g at 4°C for 30 min. The resulting pellets were washed in acetone and centrifuged at 30,000 × g at 4°C for 30 min, air dried, and resuspended in SDS-PAGE sample buffer.
Electron microscopy.
For transmission electron microscopy (TEM) studies, G. intestinalis cell pellets were fixed for 24 h in 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2) and were postfixed in 2% OsO4 in the same buffer. The fixed samples were dehydrated by passage through an ascending ethanol and acetone series and were embedded in an Araldite-Poly/Bed 812 mixture. Thin sections were cut on a Reichert-Jung Ultracut E ultramicrotome and were stained using uranyl acetate and lead citrate. The sections were examined and photographed with a JEOL JEM-1011 electron microscope. Fine-structure measurements were performed with a Veleta camera and iTEM 5.1 software (Olympus Soft Imaging Solution GmbH).
Bioinformatic analyses.
To identify the copurified proteins, their amino acid sequences were analyzed by BLASTP against the NCBI nr database using the following algorithms: HHpred at http://toolkit.tuebingen.mpg.de/hhpred# (22); HMMER3 at http://hmmer.janelia.org/ (23); and I-TASSER at http://zhanglab.ccmb.med.umich.edu/I-TASSER/ (24). The subcellular localization and topology of the proteins were predicted using TargetP at http://www.cbs.dtu.dk/services/ (25) and Phobius at http://phobius.sbc.su.se (26), respectively.
RESULTS
In vivo enzymatic tagging in Giardia intestinalis.
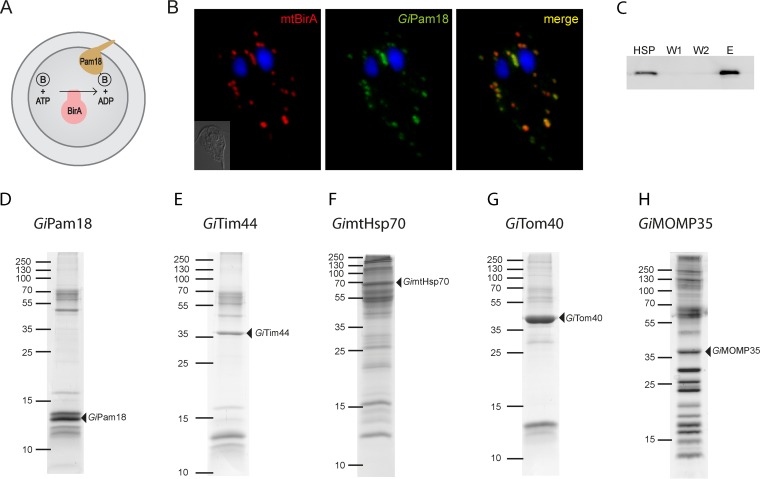
To gain insights into the composition of protein import pathways and other processes in giardial mitosomes, we took a direct biochemical approach involving highly specific protein pulldown assays followed by mass spectrometry analyses. To this end, an in vivo enzymatic tagging technique based on the biotin-avidin interaction was introduced into Giardia. This tagging relies on the highly specific E. coli biotin ligase (BirA), which uses one ATP molecule to catalyze the covalent attachment of biotin to the side chain of lysine within a biotin acceptor peptide (BAP) (27). A chimeric construct composed of E. coli BirA preceded by the N-terminal region of mitosomal GiMge1 and followed by a double HA tag was expressed in Giardia. The resulting strain contained mitosomally localized BirA (mtBirA). This construct was cotransformed with a second plasmid carrying a gene encoding mitosomal GiPam18 with the C-terminal BAP (Fig. 1A). Detection using a fluorescent streptavidin conjugate revealed the specific biotinylation of BAP (Fig. 1B). GiPam18-BAP-specific biotinylation was confirmed by Western blotting of a Giardia trophozoite lysate, which produced a single band of approximately 13 kDa, which corresponded to GiPam18-BAP. These results demonstrated that BirA remained active when delivered to Giardia mitosomes and that no nonspecific biotinylation was detected. Moreover, the use of mitosomal ATP during the biotinylation of the BAP had no apparent effects on mitosomal morphology, mitosomal distribution, or parasite growth.
FIG 1.
GiPam18 is biotinylated within mitosomes. (A) Schematic representation of mitosome-specific in vivo enzymatic tagging. E. coli biotin ligase (BirA) specifically biotinylates the biotin acceptor peptide when present in the same compartment. B, biotin. (B) BAP-tagged GiPam18 was successfully biotinylated by mtBirA. Cells were stained with an anti-HA tag antibody (red) and streptavidin-conjugated Alexa Fluor 488 (green) to detect mtBirA. Nuclei were stained with DAPI (blue). (C) Example of the purification steps (GiPam18) as analyzed on the Western blot by Alexa Fluor Fluor 488-streptavidin conjugate. (D to H) Protein profiles of the particular eluates from the streptavidin-coupled magnetic beads resolved by SDS-PAGE. The triangles indicate the proteins carrying the BAP tag.
Search for the TIM components.
GiPam18-BAP was further used to identify putative components of the mitosomal TIM complex. As a part of the PAM complex at the inner mitochondrial membrane, Pam18 interacts with the translocation channel via Tim44 (28). HSPs, which were enriched for mitosomes, were obtained from Giardia trophozoites expressing mtBirA and GiPam18-BAP. The purification of the biotinylated proteins was initially performed under native conditions; however, the resulting eluates contained numerous contaminating proteins (data not shown). Thus, chemical cross-linking and denaturation conditions were used instead. The HSP was treated with a low concentration of the membrane-permeable reversible cross-linker DSP, which is commonly used to identify interacting proteins in various cellular compartments, including mitochondria (29, 30).
Upon cross-linking, the detergent-solubilized samples were passed over streptavidin-coupled magnetic beads, and the resulting protein fractions were analyzed via SDS-PAGE and Western blotting (Fig. 1C and D). The samples were then trypsin cleaved and analyzed by mass spectrometry. Analogous purification experiments were performed in parallel using HSPs isolated from wild-type Giardia cells and from Giardia cells expressing mtBirA only. These two samples were used as negative controls for the mass spectrometry protein identification. After the results of the negative controls were subtracted, the identified proteins were ordered according to their Mascot score. Although none of the known mitosomal proteins were present in the negative controls, these proteins were abundant among the hits derived from the GiPam18-BAP samples. The high specificity of the purification procedures suggests that GiPam18-interacting partners were present among the top identified proteins. The remainder of the refined data set largely represented proteins of unknown function, and their amino acid sequences were analyzed using homology and topology detection software.
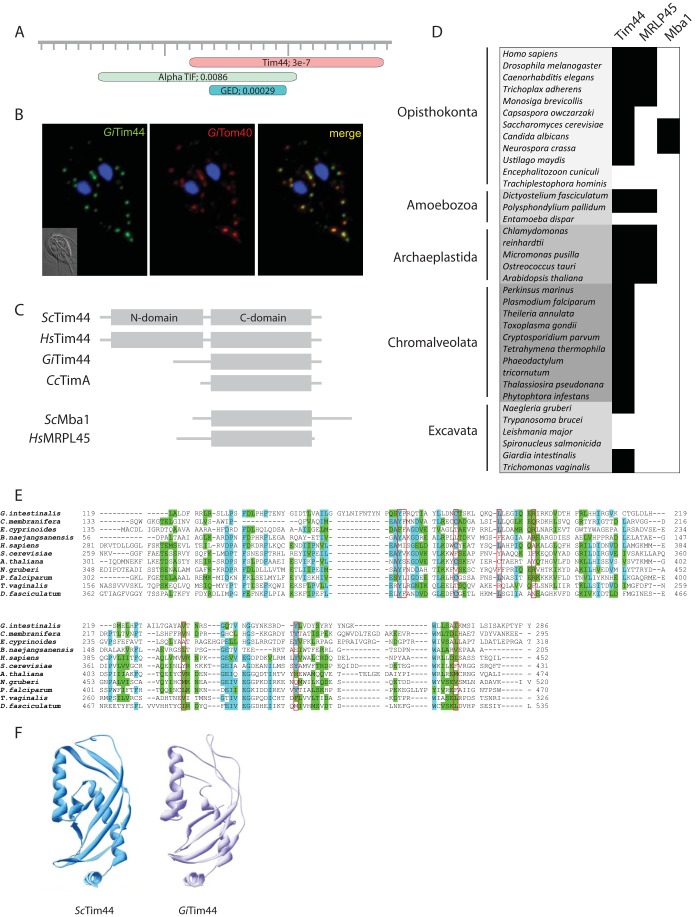
Mitosomes contain highly diverged Tim44.
Of the proteins that copurified with GiPam18-BAP, GL50803_14845 had the highest Mascot score of the unknown proteins (see Fig. S3 in the supplemental material). Although pairwise sequence analyses of GL50803_14845 showed no obvious homology to known protein families, profile-sequence comparisons conducted with HHpred showed clear homology to Tim44, a key component of the TIM complex (Fig. 2A). The mitosomal localization of GL50803_14845, here referred to as GiTim44, was confirmed by its episomal expression in Giardia (Fig. 2B). Further comparisons with mitochondrial and hydrogenosomal Tim44 proteins revealed that GiTim44 is one of the most divergent Tim44 orthologs identified and that it consists of the C-terminal domain of Tim44, which has been suggested to interact with mitochondrial lipids. However, GiTim44 lacks recognizable N-terminal domain of Tim44 (Fig. 2C), which binds the import motor molecule mtHsp70 and the core subunit of the protein-conducting channel, Tim23 (31, 32).
FIG 2.
A Tim44 homolog is present in giardial mitosomes. (A) HHpred analysis of GL50803_14845 shows the presence of a Tim44 domain. (B) HA-tagged Giardia Tim44 (GiTim44) localizes to mitosomes. Green, anti-HA antibody; red, anti-GiTom40 antibody; blue, nuclei stained with DAPI. (C) Domain structure of Tim44 orthologs in eukaryotes and bacteria. Sc, Saccharomyces cerevisiae; Hs, Homo sapiens; Gi, Giardia intestinalis; Cc, Caulobacter crescentus. (D) Distribution of Tim44 paralogs in eukaryotes. (E) Protein sequence alignment of GiTim44 with the C-terminal domains of Tim44 orthologs from Carpediemonas membranifera, Ergobibamus cyprinoides, Brevundimonas naejangsanensis, Homo sapiens, Saccharomyces cerevisiae, Arabidopsis thaliana, Naegleria gruberi, Plasmodium falciparum, Trichomonas vaginalis, and Dictyostelium discoideum. The sequences were aligned using MAFFT at http://mafft.cbrc.jp/alignment/server/. The residues involved in forming a hydrophobic pocket are framed in red (57). (F) Model of the C-terminal domain of GiTim44 obtained by Swiss-Model (58) using human Tim44 as a template (48).
The homology model of the C-terminal domain of GiTim44 indicated that this protein was capable of forming a conserved Tim44 structure containing a hydrophobic cavity, indicating its possible attachment to the mitosomal membrane (Fig. 2F). To test whether mitosomal GiTim44 interacts with its putative mitochondrial partner, GimtHsp70, Giardia trophozoites coexpressing mtBirA and GiTim44-BAP were generated. Following chemical cross-linking and purification, the proteins that copurified with GiTim44-BAP were analyzed by mass spectrometry. Similar to what was seen in the initial experiment, the purified sample was highly enriched for known mitosomal proteins (see Fig. S3 in the supplemental material). The five most highly enriched identified proteins included mitosomal GimtHsp70 and its nucleotide exchange factor, GiMge1, which strongly supports the hypothesis that Tim44 and Hsp70 interact within mitosomes.
Distant Tim44 orthologs in eukaryotes.
The discovery of a divergent Tim44 in Giardia led us to search for other Tim44 orthologs in eukaryotes. Using Tim44-specific HMMs, we identified Tim44 orthologs in two free-living metamonads, Carpediemonas membranifera and Ergobibamus cyprinoides. However, no Tim44 orthologs were identified in the fish parasite Spironucleus salmonicida or in the group Euglenozoa, which includes medically important trypanosomes and leishmania.
Surprisingly, the HMMs identified two mitochondrial proteins, MRLP45 and Mba1, as belonging to the Tim44 protein family (Fig. 2C and D). Whereas MRLP45 is a subunit of the mitochondrial ribosome (33), Mba1 serves as a mitochondrial ribosome receptor during the membrane insertion of mitochondrially translated proteins (34). Their distribution in eukaryotes suggests that both proteins represent independent paralogs of Tim44 that are specialized for mitochondrial protein translation (Fig. 2D).
Search for the translocase of the TOM components.
To identify outer mitosomal membrane components, GiTom40-BAP was coexpressed with the cytosolic version of BirA (cytBirA). As a result, mitosome-specific biotinylation was observed (see Fig. S1 in the supplemental material). Employing the same strategy as the one used for GiPam18 and GiTim44, the proteins obtained by GiTom40-BAP purification were identified using mass spectrometry. The proteins obtained from wild-type Giardia cells and Giardia cells expressing cytBirA only were subtracted from the data set.
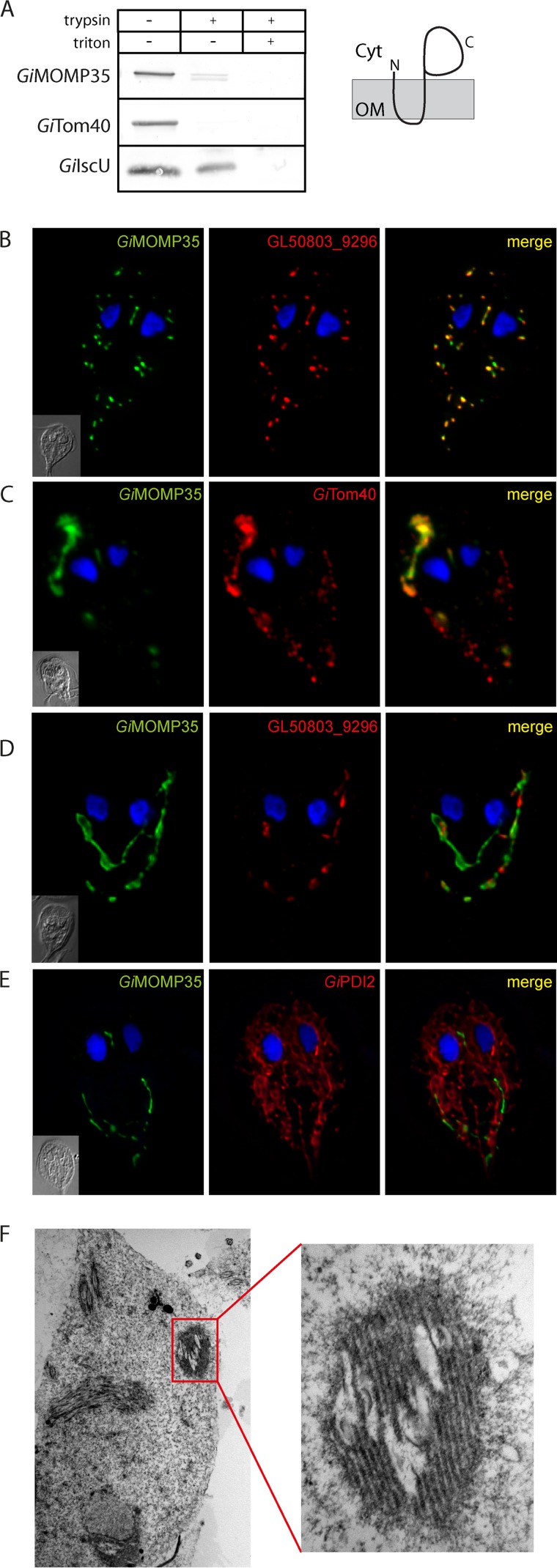
Because GiTom40 is the only known outer mitosomal membrane protein, the specificity of the purification procedure could not be determined. Nevertheless, the absence of mitosomal matrix proteins among the most significant hits (see Fig. S3 in the supplemental material) indicated that a distinct subset of mitosomal proteins was obtained. However, the previously identified mitosomal protein GL50803_14939 was found among the hits (7) (see Fig. S3 in the supplemental material). According to transmembrane topology predictors, GL50803_14939 contains two transmembrane domains in its N-terminal region. To determine whether the protein is embedded in the outer or inner mitosomal membrane, an HSP isolated from Giardia expressing C-terminally HA-tagged GL50803_14939 was subjected to a protease protection assay. Similar to GiTom40, GL50803_14939 was sensitive to protease activity even without the addition of detergent, which suggests that GL50803_14939 is inserted into the outer membrane (Fig. 3A). Taken together, these data suggest that GL50803_14939, here referred to as mitosomal outer membrane protein 35 (GiMOMP35), is anchored by two N-terminal transmembrane domains in the outer mitosomal membrane and that its C-terminal domain is in the cytosol. Whether the transmembrane domains of GiMOMP35 are also responsible for its mitosomal targeting was tested by analyzing the expression of an N-terminally truncated version of the protein. Indeed, the removal of the transmembrane domains resulted in the cytosolic localization of the truncated GiMOMP35 (see Fig. S2A in the supplemental material).
FIG 3.
GiMOMP35 is an outer mitosomal membrane protein. (A) Protease protection assay of high-speed pellets isolated from Giardia expressing HA-tagged GiMOMP35. After incubation with trypsin, the samples were immunolabeled with antibodies against the HA tag, the outer membrane protein GiTom40, and the matrix protein GiIscU. The sensitivity of GiMOMP35 to the protease indicates its outer membrane localization. The drawing shows the suggested topology of GiMOMP35. Cyt, cytosol; OM, outer mitosomal membrane. (B) Cells expressing HA-tagged GiMOMP35 were stained with an anti-HA tag antibody (green) and an anti-GL50803_9296 antibody (red). Nuclei were stained with DAPI (blue). In addition to exhibiting typical mitosomal morphology (B), approximately 50% of the observed trophozoites contained elongated tubular structures (C to E). These structures colocalized with GiTom40 (red) (C); however, only a small fraction exhibited costaining for GL50803_9296 (red) (D). The structures were devoid of the ER marker GiPDI2 (red) (E). These data indicate that the elongated tubular structures represent an enlarged outer mitosomal membrane. Under transmission electron microscopy, the structures appeared as organized membrane layers (F).
The function of the exposed soluble domain could not be predicted using bioinformatic analyses, which revealed no significant similarity of the domain to any known protein families. To examine the function of GiMOMP35, we attempted to overexpress the full-length protein using a strong promoter (ornithine carbamoyltransferase) (35). However, a stable Giardia line could not be established after numerous attempts, indicating that the overexpression of GiMOMP35 was lethal. Milder GiMOMP35 overexpression (using the 5′ untranslated region [5′UTR] of glutamate dehydrogenase as a promoter) allowed a stable line of Giardia transformants to be established and inspected for mitosome-related defects. Approximately one half of the cells retained typical mitosomal distribution and morphology (Fig. 3B), whereas the other half exhibited dramatic membrane protrusions and aggregation (Fig. 3C to E).
Further analyses indicated that GiTom40 colocalized with these elongated structures (Fig. 3C). However, these structures were largely devoid of the mitosomal protein GL50803_9296, which localized to the mitosomal matrix (see Fig. S2B in the supplemental material). When examined with a transmission electron microscope, the structures were observed as tightly packed multimembrane complexes (Fig. 3F). These data suggest that the membrane protrusions corresponded to the enlarged and aggregated outer mitosomal membrane. In contrast, the overexpression of GiMOMP35 did not result in an ER-related phenotype, as illustrated by the lack of colocalization between the ER and the enlarged mitosomes (Fig. 3E).
As an alternate means of investigating the function of GiMOMP35, the cross-linked BAP-tagged protein was purified and subjected to mass spectrometry. As expected, GiTom40 was included in the significant hits; however, the obtained data set contained no clear indication of the function of GiMOMP35 (see Fig. S3 in the supplemental material).
Newly identified mitosomal proteins.
In addition to GiTim44 and GiMOMP35, a number of other proteins of unknown function were identified from the pulldown experiments. The proteins that coprecipitated with BAP-tagged mitosomal Hsp70 (GimtHsp70) were added to the data sets derived from the GiPam18, GiTim44, GiTom40, and GiMOP35 coimmunoprecipitations, and the data were analyzed together (Fig. 1D to H). GimtHsp70 is thought to be a central component of mitosomal metabolism and to participate in protein import and iron-sulfur cluster assembly.
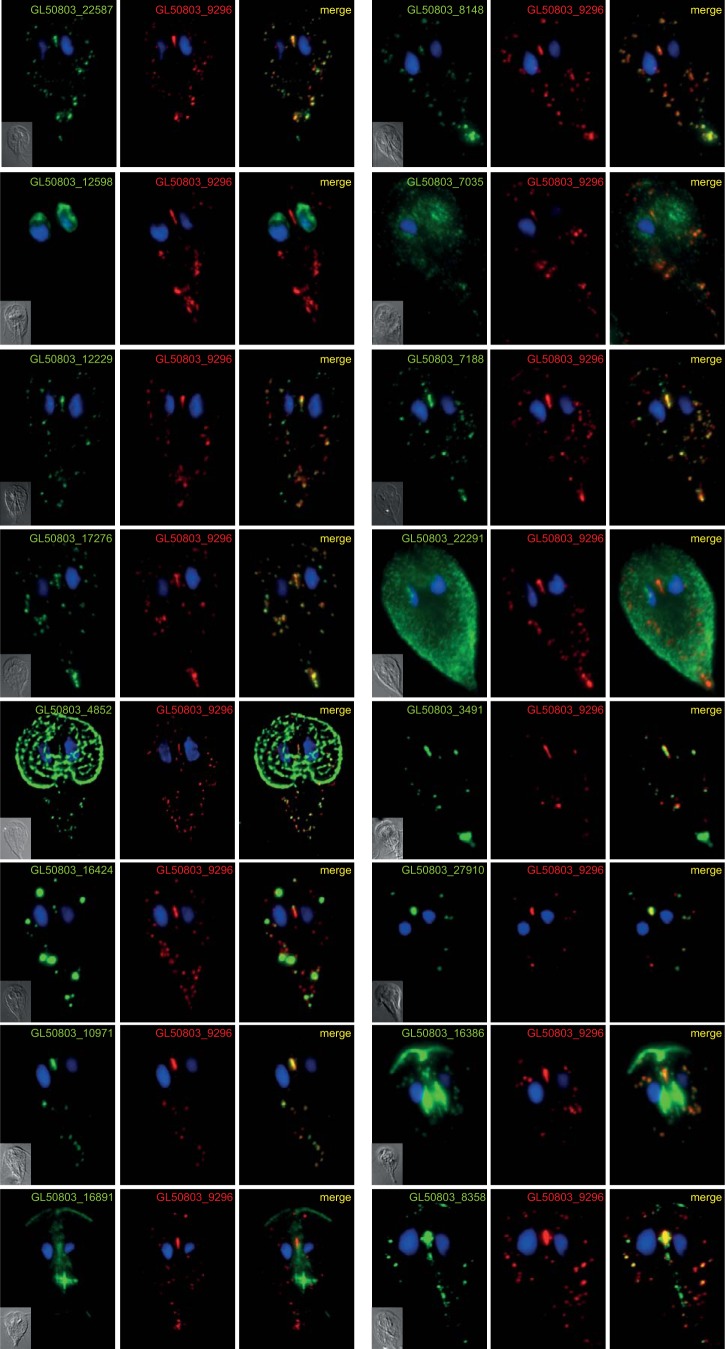
Seventeen proteins (see Table S2 in the supplemental material) were subcloned into Giardia expression vectors to verify their mitosomal localization. These proteins were selected according to three criteria: (i) the protein copurified with more than one target molecule, (ii) the identification of the protein was highly significant, or (iii) homology predictors showed an affiliation with a particular protein family. Using this approach, mitosomal localization was confirmed for 13 of the proteins, including 3 with dual localization (Fig. 4). The localization of one protein (GL50803_92741) could not be confirmed because no viable transformants were obtained after three independent transfections. Particular attention was paid to GL50803_27910 and GL50803_16424. The first represents an ortholog of rhodanese, a protein involved in various aspects of sulfur metabolism (36), including the repair of iron-sulfur clusters (37). The latter was the only protein identified in all the pulldown experiments performed in this study (see Fig. S3 in the supplemental material); i.e., GL50803_16424 coprecipitated with the outer membrane, the inner membrane, and the matrix proteins, which might indicate its complicated topology. Moreover, the episomal expression of GL50803_16424 often but not always resulted in the formation of enlarged structures at the mitosomal sites (Fig. 4). Strikingly, this protein appears to be a member of the myelodysplasia-myeloid leukemia factor 1-interacting protein (Mlf1IP) family, which has been considered exclusive to metazoans (38). For the remainder of the confirmed mitosomal proteins, no recognizable homology could be identified. Moreover, with the exception of GL50803_27910, GL50803_3491, and GL50803_16424, these proteins appear to lack orthologs, even in other metamonad species; thus, they currently represent Giardia-specific molecules (see Table S3 in the supplemental material).
FIG 4.
Localization of putative mitosomal proteins. Selected HA-tagged proteins were expressed in Giardia, and their cellular localization was determined using immunofluorescence microscopy. The cells were stained with anti-HA tag (green) and anti-GL50803_9296 (red) antibodies. Nuclei were stained with DAPI (blue).
Mode of mitosomal protein import.
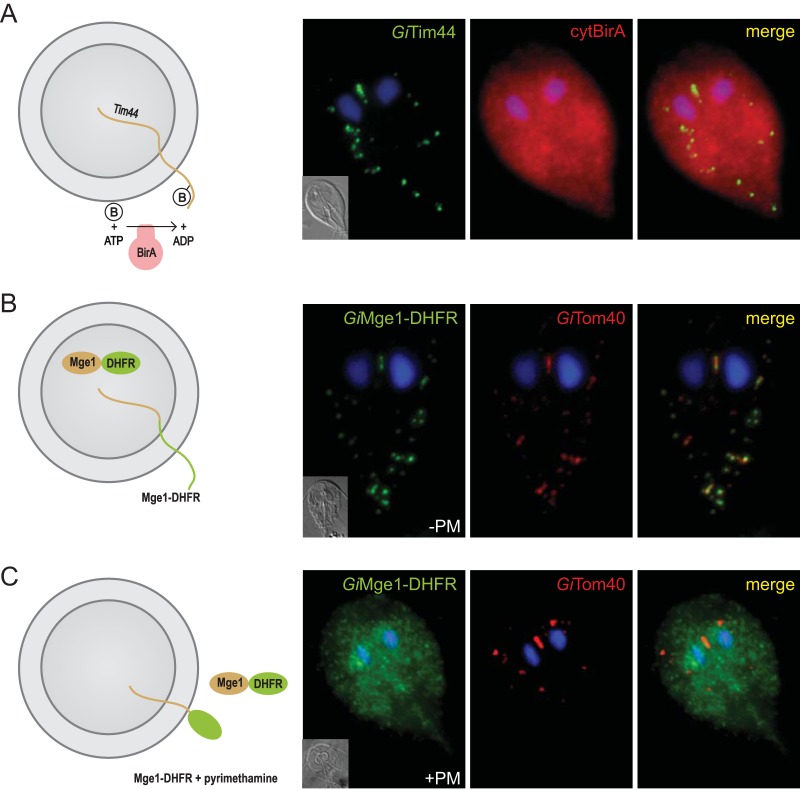
Compartment-specific biotinylation allows one to determine whether a given protein is transported into an organelle co- or posttranslationally. To this end, we generated a Giardia strain expressing a cytosolic version of BirA (cytBirA) (Fig. 5). The ability of cytBirA to biotinylate the BAP on a mitosomal protein indicates that the protein is transported posttranslationally. Indeed, the biotinylation of GiTim44-BAP was observed upon coexpression with cytBirA (Fig. 5A). Similarly, the use of compartment-specific biotinylation allowed us to assess whether the reported presence of a SNARE protein, Sec20, in the mitosomes (39) indicated the fusion of secretory vesicles with the mitosomal surface. However, because no biotinylation of the mitosomal proteins was observed when BirA was targeted to the ER (data not shown), the integration of mitosomes into the secretory pathway could not be confirmed. The posttranslational transport of mitosomal proteins raised the question of whether these proteins are required to retain their unfolded state during import. To address that question, a chimeric construct encoding mitosomal GiMge1, mouse dihydrofolate reductase (DHFR), and a C-terminal HA tag was expressed in Giardia. The DHFR domain is a classical experimental substrate used in protein translocation studies due to its ability to fold upon the addition of a folate analog (40). Usually, the use of folate analogs requires the experiment to be performed in vitro on isolated organelles due to the effect of these analogs to the endogenous enzyme (40). However, Giardia lacks DHFR and instead relies on the purine salvage pathway (41), which allows for the in vivo use of DHFR-containing constructs. The localization of the chimeric protein was examined in cells incubated with or without the folate analog pyrimethamine (PM). As expected, in the absence of the folate analog, the targeting information on GiMge1 mediated the efficient delivery of this protein to the mitosomes (Fig. 5B). In contrast, the addition of PM resulted in an entirely cytosolic localization (Fig. 5C). These results demonstrate that the protein must remain unfolded before and during its import into mitosomes.
FIG 5.
Mitosomal proteins are transported posttranslationally and in an unfolded state. BAP-tagged GiTim44 was coexpressed with cytosolic BirA (cytBirA), and the biotinylation of the tag was observed using fluorescence microscopy. Cells were stained with an anti-HA tag antibody (red) to detect cytBirA and with streptavidin-conjugated Alexa Fluor 488 (green) to detect the biotinylation of the BAP tag. Nuclei were stained with DAPI (blue). (A) A fusion protein of mouse DHFR, mitosomal GiMge1, and a C-terminal HA tag was expressed in Giardia. (B and C) The localization of the chimeric construct was assessed in the absence (B) or presence (C) of 100 μM pyrimethamine (+PM), which induces the folding of the DHFR domain. The cells were stained with anti-HA (green) and anti-GiTom40 antibodies (red). Nuclei were stained with DAPI (blue).
DISCUSSION
The Giardia mitosome remains one of the least well characterized forms of mitochondria. This is especially true for its biogenesis pathways, which ensure that the organelle maintains its integrity and functions. The aim of this study was to identify new mitosomal proteins, which might have diverged from known proteins beyond the sensitivity of homology detection algorithms or have been replaced by lineage-specific components. Because mitosomes represent one of the smallest membrane-bound cellular compartments of eukaryotes (42), biochemical approaches using cell fractionation techniques are highly challenging (7). The in vivo enzymatic tagging approach utilizing E. coli BirA introduced in this study allows proteins of interest to be purified and their transport through cellular organelles and their subcompartments to be monitored.
First, two key proteins involved in mitosomal protein import, which reside in different mitosomal membranes, were used to search for new mitosomal components. GiPam18 was the best available candidate to identify putative TIM components in mitosomes. The protein identified with this approach, GiTim44, represents one of the most diverged eukaryotic Tim44 proteins. The homology of GiTim44 is limited to the C-terminal membrane interaction domain, an arrangement reminiscent of the distant Tim44 ortholog found exclusively in alphaproteobacteria (43). However, despite the absence of the N-terminal domain, which has been shown to mediate an interaction with mtHsp70 (31), GimtHsp70 was found among the most significant proteins that copurified with GiTim44. This finding may indicate that the interaction between these proteins is conserved in mitosomes, although it is mediated by different amino acid residues. Unfortunately, no protein translocase candidate was found among the obtained data set, which lacked polytopic membrane proteins. This absence was likely due to the experimental conditions used, particularly the cross-linking chemistry and the preparation of samples for mass spectrometry (44). A customized procedure will be necessary to identify such proteins.
Using GiTim44 as a query, related sequences were found in metamonads such as C. membranifera and E. cyprinoides. Surprisingly, no Tim44 ortholog was identified in the recently published genome of the hydrogenosome-bearing fish parasite S. salmonicida (45). According to further HMM-based searches, Tim44 has been lost several times in the evolution of eukaryotes. Previous reports have shown that this protein is absent from Entamoeba (46) and microsporidian species (47), which also carry highly adapted mitosomes. Strikingly, the Tim44 protein is also missing from the entire group of kinetoplastida, which contain complex aerobic mitochondria. However, our Tim44-specific HMM identified additional new Tim44 paralogs in the mitochondria of opisthokonts, amoebozoa, and plants. Specifically, the mitochondrial proteins MRLP45 and Mba1 participate in mitochondrial translation and membrane protein insertion, respectively (33, 34). MRLP45 is a component of the large subunit of the mitoribosome, the structure of which was recently been resolved (33). The structure of MRLP45 clearly demonstrates its homology to the C-terminal domain of Tim44 (48). Although Mba1 is not a mitoribosome component, it binds the large subunit of the mitoribosome and cooperates with OxaI in the membrane insertion of mitochondrially translated proteins (34). Despite the differences in the molecular architecture of the complexes containing MRLP45 and Mba1, it is likely that these proteins perform analogous functions.
The only known outer mitosomal membrane protein has been GiTom40. This eukaryotic porin is a hallmark of all mitochondria (49), in which it constitutes the general import pore (18, 50) and interacts with the components of the SAM and ERMES complexes (51). The purification of GiTom40 led to the identification of GiMOMP35. The protein had already been identified in giardial mitosomes, but neither its localization within the mitosome nor its topology had been determined (7). According to our results, the GiMOMP35 protein is anchored in the outer mitosomal membrane with its C-terminal domain exposed to the cytosol. The phenotype of mitochondrial aggregation triggered by the overexpression of GiMOMP35 is reminiscent of the overexpression of some of the outer membrane proteins involved in protein import (52) or mitochondrial dynamics (53). However, without further characterization of its C-terminal domain, the exact function of GiMOMP35 in mitosome biology will remain unknown. This protein is exclusive to Giardia; no related sequences have been found in S. salmonicida or in other parasitic or free-living metamonads.
Due to the presence of Tom40 in the outer mitosomal membrane, the occurrence of Sam50 was expected (54). Sam50 is an essential component of the SAM complex, the β-barrel protein folding machine (55) that is considered an important evolutionary feature linking mitochondria to Gram-negative bacteria (56). Despite the omnipresence of Sam50 in eukaryotes, no ortholog has been identified in the Giardia genome (18) or among the proteins that copurified with GiTom40 or GiMOMP35 in the present work. Surprisingly, while missing in the G. intestinalis and S. salmonicida genomes, Sam50 orthologs are present in the expressed sequence tag (EST) data of C. membranifera and E. cyprinoides (M. Kolísko and A. J. Roger, unpublished results). This strongly suggests that the unique loss of Sam50 in the evolution of eukaryotes occurred in the common ancestor of diplomonads.
However, 5 of the 13 novel mitosomal proteins seem to be specific to the outer mitosomal membrane, as these were exclusive to GiTom40- and GiMOMP35-derived data sets. Further investigation of these proteins may bring more information on the biogenesis of the outer mitosomal membrane as well as on the interaction between the mitosomes and other cellular organelles.
In general, the identification of novel mitosomal proteins, the vast majority of which are specific to Giardia, demonstrates that metabolic processes other than the formation of iron-sulfur clusters occur in mitosomes. The presence of a rhodanese ortholog indicates the existence of additional sulfur metabolism at a minimum. In addition, the striking presence of an Mlf1IP ortholog in mitosomes may shed light on the exact function of the protein, for which a precise role has not been assigned in the Metazoa.
In addition to the identification of new proteins, the techniques used in this study enabled us to demonstrate that proteins maintain an unfolded state while traveling to mitosomes posttranslationally. However, no sign of mixing of the ER and mitosome lumina was detected. The reported mitosomal localization of Giardia Sec20 ortholog indicated that a vesicular transport may play a role in mitosomal protein import (39). Our data on the localization of the endogenous Sec20 (not shown in this work) using specific polyclonal antibody indicate that its mitosomal localization is a result of experimental artifact, a phenomenon often observed for the overexpression of tail-anchored proteins. These results provide new evidence that mitosomal biogenesis follows the same rules as mitochondrial biogenesis despite the absence of some of the core components.
Taken together, the data presented here demonstrate that techniques such as in vivo enzymatic tagging are extremely valuable tools to investigate the biology of organelles as small as Giardia mitosomes. The identification of Giardia-specific proteins also demonstrates that our current concept of mitosomes as highly simplified mitochondria may not entirely reflect the true biology of these organelles. Future studies will likely reveal yet-unknown mitosomal functions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Veronika Klápštová, Vladimíra Najdrová, and Zuzana Drašnarová for valuable technical assistance.
This work was funded by a grant from the Czech Science Foundation (P305-10-0651), by the European Regional Development Fund to the Biomedicine Center of the Academy of Sciences and Charles University (CZ.1.05/1.1.00/02.0109), and by a grant from Charles University Grant Agency (98214).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00448-15.
REFERENCES
- 1.Adam RD. 2001. Biology of Giardia lamblia. Clin Microbiol Rev 14:447–475. doi: 10.1128/CMR.14.3.447-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ankarklev J, Jerlström-Hultqvist J, Ringqvist E, Troell K, Svärd SG. 2010. Behind the smile: cell biology and disease mechanisms of Giardia species. Nat Rev Microbiol 8:413–422. doi: 10.1038/nrmicro2317. [DOI] [PubMed] [Google Scholar]
- 3.Hehl AB, Marti M. 2004. Secretory protein trafficking in Giardia intestinalis. Mol Microbiol 53:19–28. doi: 10.1111/j.1365-2958.2004.04115.x. [DOI] [PubMed] [Google Scholar]
- 4.Lanfredi-Rangel A, Attias M, de Carvalho TM, Kattenbach WM, De Souza W. 1998. The peripheral vesicles of trophozoites of the primitive protozoan Giardia lamblia may correspond to early and late endosomes and to lysosomes. J Struct Biol 123:225–235. doi: 10.1006/jsbi.1998.4035. [DOI] [PubMed] [Google Scholar]
- 5.Konrad C, Spycher C, Hehl AB. 2010. Selective condensation drives partitioning and sequential secretion of cyst wall proteins in differentiating Giardia lamblia. PLoS Pathog 6:e1000835. doi: 10.1371/journal.ppat.1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dolezal P, Smíd O, Rada P, Zubácová Z, Bursać D, Suták R, Nebesárová J, Lithgow T, Tachezy J. 2005. Giardia mitosomes and trichomonad hydrogenosomes share a common mode of protein targeting. Proc Natl Acad Sci U S A 102:10924–10929. doi: 10.1073/pnas.0500349102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jedelský PL, Doležal P, Rada P, Pyrih J, Smíd O, Hrdý I, Sedinová M, Marcinčiková M, Voleman L, Perry AJ, Beltrán NC, Lithgow T, Tachezy J. 2011. The minimal proteome in the reduced mitochondrion of the parasitic protist Giardia intestinalis. PLoS One 6:e17285. doi: 10.1371/journal.pone.0017285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Regoes A, Zourmpanou D, León-Avila G, van der Giezen M, Tovar J, Hehl AB. 2005. Protein import, replication, and inheritance of a vestigial mitochondrion. J Biol Chem 280:30557–30563. doi: 10.1074/jbc.M500787200. [DOI] [PubMed] [Google Scholar]
- 9.Roger AJ, Svärd SG, Tovar J, Clark CG, Smith MW, Gillin FD, Sogin ML. 1998. A mitochondrial-like chaperonin 60 gene in Giardia lamblia: evidence that diplomonads once harbored an endosymbiont related to the progenitor of mitochondria. Proc Natl Acad Sci U S A 95:229–234. doi: 10.1073/pnas.95.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tachezy J, Sánchez LB, Müller M. 2001. Mitochondrial type iron-sulfur cluster assembly in the amitochondriate eukaryotes Trichomonas vaginalis and Giardia intestinalis, as indicated by the phylogeny of IscS. Mol Biol Evol 18:1919–1928. doi: 10.1093/oxfordjournals.molbev.a003732. [DOI] [PubMed] [Google Scholar]
- 11.Likic VA, Dolezal P, Celik N, Dagley M, Lithgow T. 2010. Using hidden markov models to discover new protein transport machines. Methods Mol Biol 619:271–284. doi: 10.1007/978-1-60327-412-8_16. [DOI] [PubMed] [Google Scholar]
- 12.Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schönfisch B, Perschil I, Chacinska A, Guiard B, Rehling P, Pfanner N, Meisinger C. 2003. The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci U S A 100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider RE, Brown MT, Shiflett AM, Dyall SD, Hayes RD, Xie Y, Loo JA, Johnson PJ. 2011. The Trichomonas vaginalis hydrogenosome proteome is highly reduced relative to mitochondria, yet complex compared with mitosomes. Int J Parasitol 41:1421–1434. doi: 10.1016/j.ijpara.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Panigrahi AK, Ogata Y, Zíková A, Anupama A, Dalley RA, Acestor N, Myler PJ, Stuart KD. 2009. A comprehensive analysis of Trypanosoma brucei mitochondrial proteome. Proteomics 9:434–450. doi: 10.1002/pmic.200800477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wampfler PB, Tosevski V, Nanni P, Spycher C, Hehl AB. 2014. Proteomics of secretory and endocytic organelles in Giardia lamblia. PLoS One 9:e94089. doi: 10.1371/journal.pone.0094089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keister DB. 1983. Axenic culture of Giardia lamblia in TYI-S-33 medium supplemented with bile. Trans R Soc Trop Med Hyg 77:487–488. doi: 10.1016/0035-9203(83)90120-7. [DOI] [PubMed] [Google Scholar]
- 17.Martincová E, Voleman L, Najdrová V, De Napoli M, Eshar S, Gualdron M, Hopp CS, Sanin DE, Tembo DL, Van Tyne D, Walker D, Marcinčiková M, Tachezy J, Doležal P. 2012. Live imaging of mitosomes and hydrogenosomes by HaloTag technology. PLoS One 7:e36314. doi: 10.1371/journal.pone.0036314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dagley MJ, Dolezal P, Likic VA, Smid O, Purcell AW, Buchanan SK, Tachezy J, Lithgow T. 2009. The protein import channel in the outer mitosomal membrane of Giardia intestinalis. Mol Biol Evol 26:1941–1947. doi: 10.1093/molbev/msp117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howarth M, Takao K, Hayashi Y, Ting AY. 2005. Targeting quantum dots to surface proteins in living cells with biotin ligase. Proc Natl Acad Sci U S A 102:7583–7588. doi: 10.1073/pnas.0503125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gehde N, Hinrichs C, Montilla I, Charpian S, Lingelbach K, Przyborski JM. 2009. Protein unfolding is an essential requirement for transport across the parasitophorous vacuolar membrane of Plasmodium falciparum. Mol Microbiol 71:613–628. doi: 10.1111/j.1365-2958.2008.06552.x. [DOI] [PubMed] [Google Scholar]
- 21.Rada P, Doležal P, Jedelský PL, Bursac D, Perry AJ, Šedinová M, Smíšková K, Novotný M, Beltrán NC, Hrdý I, Lithgow T, Tachezy J. 2011. The core components of organelle biogenesis and membrane transport in the hydrogenosomes of Trichomonas vaginalis. PLoS One 6:e24428. doi: 10.1371/journal.pone.0024428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Söding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol 7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 26.Käll L, Krogh A, Sonnhammer ELL. 2007. Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res 35:W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howarth M, Ting AY. 2008. Imaging proteins in live mammalian cells with biotin ligase and monovalent streptavidin. Nat Protoc 3:534–545. doi: 10.1038/nprot.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chacinska A, van der Laan M, Mehnert CS, Guiard B, Mick DU, Hutu DP, Truscott KN, Wiedemann N, Meisinger C, Pfanner N, Rehling P. 2010. Distinct forms of mitochondrial TOM-TIM supercomplexes define signal-dependent states of preprotein sorting. Mol Cell Biol 30:307–318. doi: 10.1128/MCB.00749-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tieu Q, Okreglak V, Naylor K, Nunnari J. 2002. The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J Cell Biol 158:445–452. doi: 10.1083/jcb.200205031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoon Y, Krueger EW, Oswald BJ, McNiven MA. 2003. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol Cell Biol 23:5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merlin A, Voos W, Maarse AC, Meijer M, Pfanner N, Rassow J. 1999. The J-related segment of tim44 is essential for cell viability: a mutant Tim44 remains in the mitochondrial import site, but inefficiently recruits mtHsp70 and impairs protein translocation. J Cell Biol 145:961–972. doi: 10.1083/jcb.145.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ting S-Y, Schilke BA, Hayashi M, Craig EA. 2014. Architecture of the TIM23 inner mitochondrial translocon and interactions with the matrix import motor. J Biol Chem 289:28689–28696. doi: 10.1074/jbc.M114.588152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown A, Amunts A, Bai X-C, Sugimoto Y, Edwards PC, Murshudov G, Scheres SHW, Ramakrishnan V. 2014. Structure of the large ribosomal subunit from human mitochondria. Science 346:718–722. doi: 10.1126/science.1258026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ott M, Prestele M, Bauerschmitt H, Funes S, Bonnefoy N, Herrmann JM. 2006. Mba1, a membrane-associated ribosome receptor in mitochondria. EMBO J 25:1603–1610. doi: 10.1038/sj.emboj.7601070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lauwaet T, Davids BJ, Torres-Escobar A, Birkeland SR, Cipriano MJ, Preheim SP, Palm D, Svärd SG, McArthur AG, Gillin FD. 2007. Protein phosphatase 2A plays a crucial role in Giardia lamblia differentiation. Mol Biochem Parasitol 152:80–89. doi: 10.1016/j.molbiopara.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cipollone R, Ascenzi P, Visca P. 2007. Common themes and variations in the rhodanese superfamily. IUBMB Life 59:51–59. doi: 10.1080/15216540701206859. [DOI] [PubMed] [Google Scholar]
- 37.Bonomi F, Pagani S, Cerletti P, Cannella C. 1977. Rhodanese-mediated sulfur transfer to succinate dehydrogenase. Eur J Biochem 72:17–24. doi: 10.1111/j.1432-1033.1977.tb11219.x. [DOI] [PubMed] [Google Scholar]
- 38.Ohno K, Takahashi Y, Hirose F, Inoue YH, Taguchi O, Nishida Y, Matsukage A, Yamaguchi M. 2000. Characterization of a Drosophila homologue of the human myelodysplasia/myeloid leukemia factor (MLF). Gene 260:133–143. doi: 10.1016/S0378-1119(00)00447-9. [DOI] [PubMed] [Google Scholar]
- 39.Elias EV, Quiroga R, Gottig N, Nakanishi H, Nash TE, Neiman A, Lujan HD. 2008. Characterization of SNAREs determines the absence of a typical Golgi apparatus in the ancient eukaryote Giardia lamblia. J Biol Chem 283:35996–36010. doi: 10.1074/jbc.M806545200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eilers M, Schatz G. 1986. Binding of a specific ligand inhibits import of a purified precursor protein into mitochondria. Nature 322:228–232. [DOI] [PubMed] [Google Scholar]
- 41.Wang CC, Aldritt S. 1983. Purine salvage networks in Giardia lamblia. J Exp Med 158:1703–1712. doi: 10.1084/jem.158.5.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Giezen M, Tovar J. 2005. Degenerate mitochondria. EMBO Rep 6:525–530. doi: 10.1038/sj.embor.7400440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clements A, Bursac D, Gatsos X, Perry AJ, Civciristov S, Celik N, Likic VA, Poggio S, Jacobs-Wagner C, Strugnell RA, Lithgow T. 2009. The reducible complexity of a mitochondrial molecular machine. Proc Natl Acad Sci U S A 106:15791–15795. doi: 10.1073/pnas.0908264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schey KL, Grey AC, Nicklay JJ. 2013. Mass spectrometry of membrane proteins: a focus on aquaporins. Biochemistry 52:3807–3817. doi: 10.1021/bi301604j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu F, Jerlström-Hultqvist J, Einarsson E, Astvaldsson A, Svärd SG, Andersson JO. 2014. The genome of Spironucleus salmonicida highlights a fish pathogen adapted to fluctuating environments. PLoS Genet 10:e1004053. doi: 10.1371/journal.pgen.1004053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dolezal P, Dagley MJ, Kono M, Wolynec P, Likić VA, Foo JH, Sedinová M, Tachezy J, Bachmann A, Bruchhaus I, Lithgow T. 2010. The essentials of protein import in the degenerate mitochondrion of Entamoeba histolytica. PLoS Pathog 6:e1000812. doi: 10.1371/journal.ppat.1000812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waller RF, Jabbour C, Chan NC, Celik N, Likic VA, Mulhern TD, Lithgow T. 2009. Evidence of a reduced and modified mitochondrial protein import apparatus in microsporidian mitosomes. Eukaryot Cell 8:19–26. doi: 10.1128/EC.00313-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Handa N, Kishishita S, Morita S, Akasaka R, Jin Z, Chrzas J, Chen L, Liu Z-J, Wang B-C, Sugano S, Tanaka A, Terada T, Shirouzu M, Yokoyama S. 2007. Structure of the human Tim44 C-terminal domain in complex with pentaethylene glycol: ligand-bound form. Acta Crystallogr D Biol Crystallogr 63:1225–1234. doi: 10.1107/S0907444907051463. [DOI] [PubMed] [Google Scholar]
- 49.Zarsky V, Tachezy J, Dolezal P. 2012. Tom40 is likely common to all mitochondria. Curr Biol 22:R479–R481; author reply, R481–R482. doi: 10.1016/j.cub.2012.03.057. [DOI] [PubMed] [Google Scholar]
- 50.Baker KP, Schaniel A, Vestweber D, Schatz G. 1990. A yeast mitochondrial outer membrane protein essential for protein import and cell viability. Nature 348:605–609. doi: 10.1038/348605a0. [DOI] [PubMed] [Google Scholar]
- 51.Yamano K, Tanaka-Yamano S, Endo T. 2010. Mdm10 as a dynamic constituent of the TOB/SAM complex directs coordinated assembly of Tom40. EMBO Rep 11:187–193. doi: 10.1038/embor.2009.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yano M, Kanazawa M, Terada K, Namchai C, Yamaizumi M, Hanson B, Hoogenraad N, Mori M. 1997. Visualization of mitochondrial protein import in cultured mammalian cells with green fluorescent protein and effects of overexpression of the human import receptor Tom20. J Biol Chem 272:8459–8465. doi: 10.1074/jbc.272.13.8459. [DOI] [PubMed] [Google Scholar]
- 53.Rojo M, Legros F, Chateau D, Lombès A. 2002. Membrane topology and mitochondrial targeting of mitofusins, ubiquitous mammalian homologs of the transmembrane GTPase Fzo. J Cell Sci 115:1663–1674. [DOI] [PubMed] [Google Scholar]
- 54.Dolezal P, Likic V, Tachezy J, Lithgow T. 2006. Evolution of the molecular machines for protein import into mitochondria. Science 313:314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- 55.Kozjak V, Wiedemann N, Milenkovic D, Lohaus C, Meyer HE, Guiard B, Meisinger C, Pfanner N. 2003. An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J Biol Chem 278:48520–48523. doi: 10.1074/jbc.C300442200. [DOI] [PubMed] [Google Scholar]
- 56.Gentle I, Gabriel K, Beech P, Waller R, Lithgow T. 2004. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J Cell Biol 164:19–24. doi: 10.1083/jcb.200310092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Josyula R, Jin Z, Fu Z, Sha B. 2006. Crystal structure of yeast mitochondrial peripheral membrane protein Tim44p C-terminal domain. J Mol Biol 359:798–804. doi: 10.1016/j.jmb.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 58.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, Schwede T. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.